Abstract
Neuroscience studies are often carried out in animal models for the purpose of understanding specific aspects of the human condition. However, the translation of findings across species remains a significant challenge. Network science approaches can enhance the translational impact of cross-species studies by providing a means of mapping small-scale cellular processes identified in animal model studies to larger-scale interregional circuits observed in humans. In this Review, we highlight the contributions of network science approaches to the development of cross-species translational research in neuroscience. We lay the foundation for our discussion by exploring the objectives of cross-species translational models. We then discuss how the development of new tools that enable the acquisition of whole-brain data in animal models with cellular resolution provides unprecedented opportunity for cross-species applications of network science approaches for understanding large-scale brain networks. We describe how these tools may support the translation of findings across species and imaging modalities and highlight future opportunities. Our overarching goal is to illustrate how the application of network science tools across human and animal model studies could deepen insight into the neurobiology that underlies phenomena observed with noninvasive neuroimaging methods and could simultaneously further our ability to translate findings across species.
Introduction
Through translational studies in animal models, we endeavour to understand aspects of disease that remain inaccessible through noninvasive studies of the human brain. The behavioural, genetic, molecular and neuroanatomical similarities between humans and many other species1–4, including non-human primates, rodents, zebrafish, the nematode Caenorhabditis elegans, and others, make these animals suitable for modelling aspects of the human condition. Like humans, each of these organisms engages in observable forms of navigation5, information-seeking6,7 and social interaction8,9. Nonetheless, establishing environmental conditions and approaches to studying the brain that are analogous across species remains a significant challenge for effective cross-species translation. To address this latter point, we propose that network science approaches are of great benefit.
In its most basic form, a network science approach to studying the brain entails representing the brain as a system or graph composed of nodes and edges10. Nodes may represent interacting entities ranging in scale from molecules to whole brain regions11,12. Edges that connect nodes are often structural or functional. Structural edges represent anatomical connectivity between nodes, such as white matter tracts, and functional edges represent statistical dependencies in neural activity between nodes13. Network models of the brain are often characterized using metrics from the subfield of mathematics known as graph theory (BOX 1). These metrics can define local, mesoscale or global properties of the network14,15, which are particularly useful for descriptive (not causal) characterizations. More recently, network control theory (NCT) has been used to identify causal processes underlying changes in whole-brain state16–18 and graph neural network models constructed from whole-brain datasets have been used to demonstrate how network connectivity supports neural computations19. Detailed explanations of available tools for understanding network models of the brain are provided in other recent reviews12,20,21.
Box 1. A brief overview of network models.
Graph theory
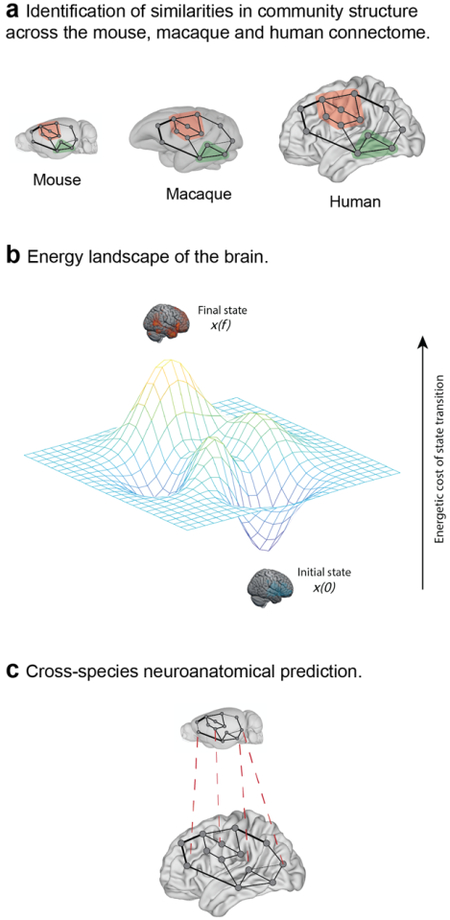
Graph theory is a branch of mathematics that provides descriptive tools for understanding properties of brain networks. Graph theory metrics are broadly categorized as local, mesoscale or global14,15. Local metrics provide insight into node-level properties of a network; for example, degree reflects the sum of connections to a given node. Mesoscale metrics describe clustering characteristics; for example, community structure refers to the presence of subgroups of nodes that are more strongly connected to one another than to the rest of the network. Finally, global metrics describe network-wide features; for example, global efficiency is a measure of the efficiency of long-range communication within a network. In cross-species studies, graph theory metrics have been used to identify similarities and differences in the brain’s topology across species29 (see the figure, part a).
Network control theory
Network control theory (NCT) is a systems engineering approach that can be used to model the relationship between structure and function in the brain16,18. Within an NCT framework, brain states are defined as neural activity across brain regions. These activity states are constrained by the brain’s structural network16. Brain states may be represented as peaks and valleys in an energy landscape (see the figure, part b), in which x(0) and x(f) represent the initial and final brain states, respectively, and the distance between states represents the cost of transitioning between them. NCT has been used to identify ‘control points’ in the brain that are particularly influential in driving brain state transitions185,186. In cross-species studies, NCT may be useful for the translation of therapeutic targets from one species to another because it enables predictions about the brain’s response to perturbation after accounting for its anatomical connectivity.
Graph neural networks
Graph neural networks are a type of deep learning model that can derive inferences from graph structures187. These models are thus well suited to modelling brain networks, including interactions between brain structure and function188,189. Graph neural networks enable predictions about the behaviour of the whole network, its constituent nodes or edges. Graph neural networks have previously shown utility for predicting cell types190 and transcription factor binding sites162 across species, suggesting that they might also prove useful in the cross-species translation of neural data (see the figure, part c).
Brain images in parts a and c based on data in the Scalable Brain Atlas182. Mouse brain images in parts a and c based on data from ref.184. Macaque brain image in part a based on data from ref.183. Human brain images in parts a and c based on data from refs.191–194.
As the data acquired through studies of the nervous system becomes increasingly complex, network models have gained significant traction owing to their capacity to distill meaningful structure from large, intricate datasets. Network measures have been applied extensively to data from functional MRI (fMRI) and structural MRI studies in humans, and connectivity-based measures derived via network analysis of human neuroimaging data have informed our understanding of cognition22,23 and the relationship between structure and function24. They also show promise as biomarkers for disease risk, diagnosis and the prediction of treatment outcomes25,26. The application of network approaches to studies in preclinical animal models, in which environmental and genetic variables are tractable, can facilitate the identification and translation of such biomarkers.
The use of network models for cross-species translation is supported by work in the field of cross-species connectomics, which has identified common elements of brain network architecture across species27–30. Work in this area has primarily used graph theory metrics to identify topological features common to the structural connectomes of species ranging from C. elegans to humans, including community structure and small-world properties29. Important differences have also been investigated28. For instance, comparative studies across primate species have identified scaling principles that account for differences in the proportion of white matter connectivity31. These studies support the notion that cross-species network models of the brain could be used to translate findings from animal models of disease to humans.
Network models are particularly useful in the context of cross-species studies because they balance abstraction and specificity. Cross-species studies must acknowledge species differences while also excavating species similarities. Simple models that engage a level of abstraction can remain agnostic to species differences, whereas simple models that engage specificity can embrace species similarities. Network models are simple models that provide a balance between these two objectives and hence offer promise in cross-species analysis.
In this Review, we begin by discussing the goals of cross-species translational research in neuroscience. We illustrate the kinds of questions that animal models have enabled us to address about the networked architecture of the brain. We then review studies in the areas of neurodevelopment, neuromodulation, neuropsychiatric disorders and neurodegeneration that exemplify the translational potential of cross-species network models. We discuss opportunities for future work that would further our ability to translate neuroscientific findings across species and scales. Finally, we conclude by summarizing the contributions of network science tools in cross-species investigations and the role of network science tools in further developing translational models. Given that a summary of this nature has not been put together previously, our overarching goal is to clarify the state of the field and stimulate effective use of network models to support the future goals of translational neuroscience.
Translational models in neuroscience
The long-term goals of translational research in neuroscience are to identify causal processes that underlie cognitive and behavioural phenomena observed in humans, and to transform basic science findings into approaches that may be used to treat disorders of the nervous system (FIG. 1). Many high-level cognitive processes can only be studied in humans, while in vivo cellular and molecular characterizations may only be carried out in non-human animals. To fill gaps in our understanding of how cellular processes give rise to cognition in health and disease states, cross-species translational research aims to relate insights about the structure and function of the non-human animal brain to the human brain.
Figure 1. Cross-species translational research workflow.
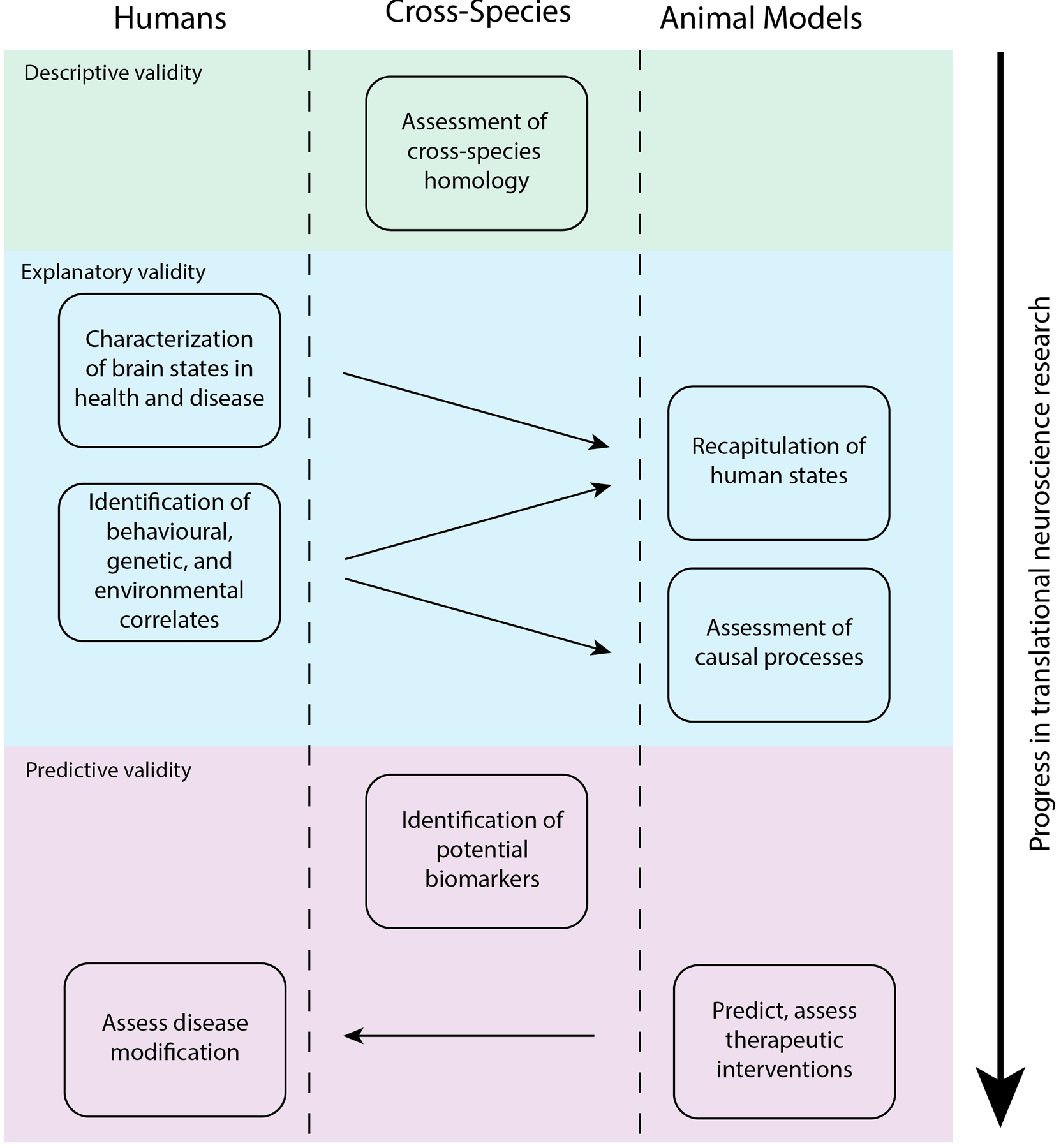
Schematic illustrating the interplay between studies in humans and animal models in translational research. Each box represents an area of research focus. Movement from top to bottom represents progress in the field of translational neuroscience, from identifying appropriate model species to predicting treatment outcomes. Arrows between boxes represent the flow of information between studies in different species. Shading indicates the categories of model assessment that each of these processes rely on. Descriptive validity refers to the extent to which an animal model mirrors the processes it is intended to model in humans and may be evaluated in terms of cross-species homology of the brain and behaviour. The development of animal models that recapitulate human states and enable the assessment of their underlying causes rely on explanatory validity. The predictive validity of an animal model is demonstrated by its utility for identifying biomarkers and therapeutic interventions that show efficacy in humans.
Early neuroscience studies carried out in animals provided fundamental insights into properties of neurons, synapses and neural activity, thereby critically expanding our understanding of the brain’s structure and function. These observations were made possible by the capacity to directly record neuronal activity, measure neurotransmitter release and study the structure of the nervous system at high resolution32. These approaches also contributed to the development of a variety of animal models of CNS disorders.
Whereas animal models have enabled fundamental discoveries about the nature of the brain’s architecture and provided a testbed for examining cellular mechanisms of CNS disorders, the development of noninvasive imaging techniques has enabled the study of whole-brain network dynamics in humans. The first application of positron emission tomography (PET) for mapping brain activity measured changes in cerebral blood flow while participants were presented with visual stimuli33. Subsequently, early fMRI studies mapped cerebral blood volume in participants viewing visual stimuli34 and measured changes in blood oxygenation in the primary visual and motor cortices in response to both motor commands and images35. These studies built on earlier work that linked alterations in neuronal activity, metabolism and cerebral blood flow36 and provided initial demonstrations of the utility of MRI for studying brain activity during a task. Ultimately, these noninvasive neuroimaging tools have given rise to the field of human brain mapping, which has shaped and significantly deepened our understanding of the neural bases of human cognition37 and has further contributed to the identification of circuit-level biomarkers for CNS disorders38.
Historically, differences in the tools used to study the brains of humans and other animals have supported two distinct conceptions of the brain. In the first, the brain is treated as a complex, networked system composed of interacting elements, whereas in the second, individual brain regions, projections or small circuits are treated as distinct entities. Though early studies of correlations in interregional or inter-neuronal activity were carried out in both humans39 and non-human primates40,41, the application of connectivity-based approaches in non-human species has remained considerably less widespread. This may be attributable to the fact that whole-brain, noninvasive imaging approaches that are typically employed in human studies naturally lend themselves to network-level approaches to understanding the brain, whereas studies in animal models often focus on microscale neural dynamics within a particular brain region. Network science approaches to analyzing human neuroimaging data have provided insight into the contributions of individual brain regions to whole-brain dynamics, but the relationship between cellular and regional dynamics is not always clear. This lack of clarity presents a challenge in the translation of findings from animals to humans: how does one place single-region processes in the context of a networked system?
The development of new tools that enable the collection of whole-brain data across species offers an unprecedented opportunity to understand the network organization of the brain across scales ranging from single-cell firing activity to regional changes in metabolic activity. Network models have the potential to facilitate cross-species translation in two major ways: first, by enabling the identification of similarities and differences in connectome organization across species29,42, and second, by contributing to more readily translatable models of human disease. In the following sections, we discuss studies that illustrate the utility of network models across species, first by reviewing studies in animal models that have contributed to the development of network science tools, and then by providing examples of translational studies in animal models that have effectively leveraged network approaches.
Insights into the brain’s networked architecture
Studies carried out in C. elegans, macaques and cats have identified fundamental network properties of neural systems, including their short average path lengths and clustering characteristics27,43,44. Network science has since revolutionized the field of human neuroimaging by providing a mathematical language and associated statistical approaches to characterize large-scale brain networks and to probe neural processes that underlie cognition in health and disease. The application of these tools in animal models has lagged behind their widespread adoption in human studies, arguably owing to differences in how investigators approach the study of the brain in these different species. As the repertoire of available tools for measuring whole-brain structural connectivity, gene expression and neural activity expands, so do opportunities to align theoretical models with large-scale biological processes. In this section, we begin by discussing approaches that enable whole-brain assessments of brain structure and function across scales. We then explore how network analyses of whole-brain datasets acquired using different imaging modalities across species have furthered our understanding of the brain’s architecture, including the causal relations among gene expression, structure, and function. Finally, we discuss how studies in animal models can be used to assess the validity of theoretical principles underlying network models of the CNS.
Tools for studying connectivity across species and scales
Advances in imaging techniques have begun to enable analysis of whole-brain structure and function throughout diverse species and across many levels of spatial and temporal resolution (FIG. 2). fMRI in animal models enables whole-brain imaging at the mesoscale and has produced results that are comparable to those obtained from human fMRI studies. For example, a conserved functional network, the default mode network (DMN), has been identified. Among the functional subnetworks of the brain that have been widely studied in humans, the DMN is a system of brain regions that exhibit coordinated activity during rest. In addition to humans, the DMN has now been identified in mice45, rats46,47 and multiple non-human primate species, including marmosets48 and macaques49. The consistency of the DMN’s anatomy and function suggests that it is an evolutionarily conserved system, although small variations in DMN topology also point to modest differences in regional contributions to cognition across species50. Direct imaging of neuronal activity (DIANA), the nascent approach for acquiring fMRI with high temporal precision in rodents, holds the potential to afford further insights into the relations between neuronal and metabolic activity in the brain51.
Figure 2. Measuring large-scale neural activity across species and scales.
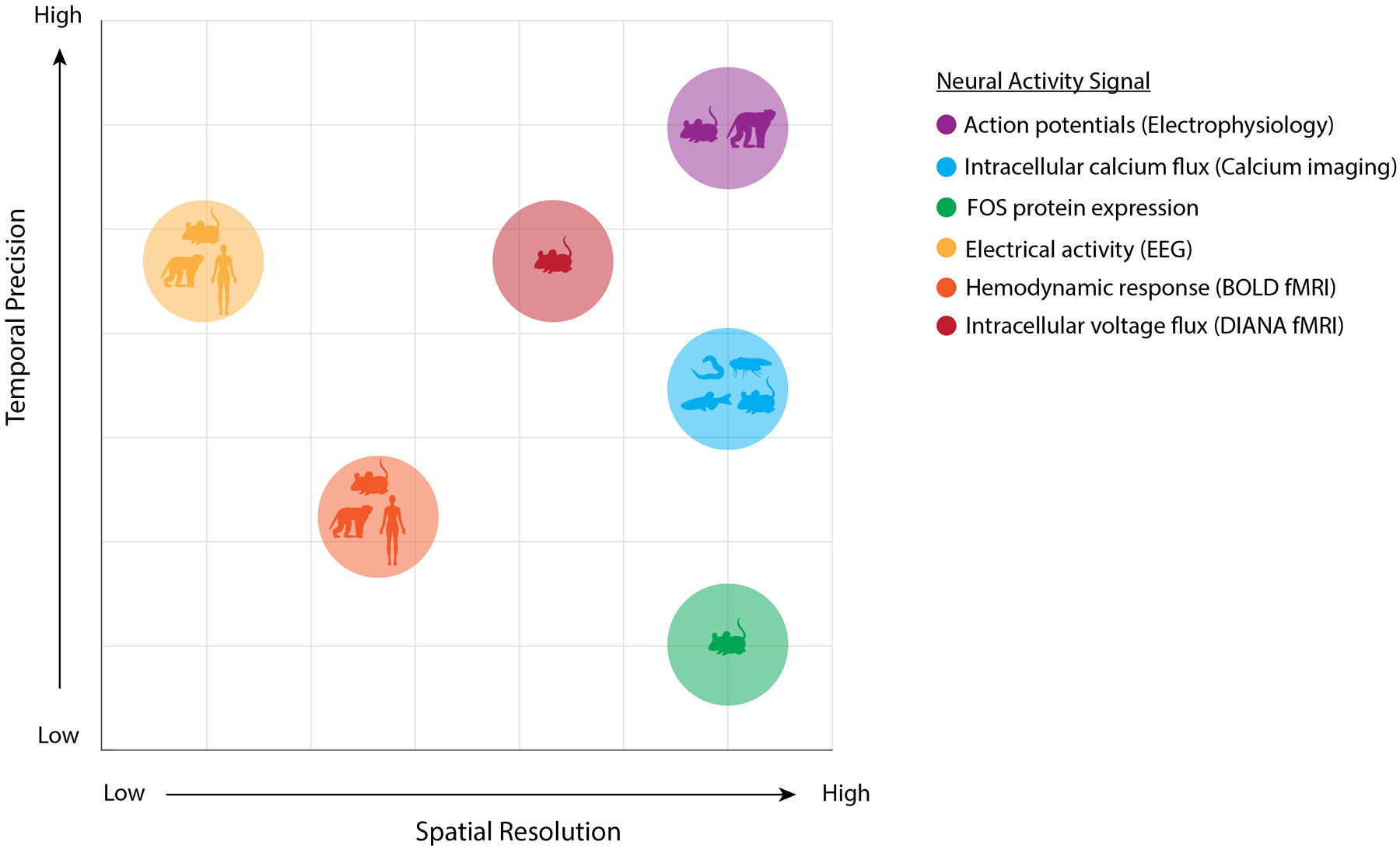
The application of network science tools to datasets acquired using different neural activity markers enables network-level inferences across scales ranging from cells to brain regions. This figure represents the types of signals that may be used to infer large-scale neural dynamics, including action potentials (measurable at large scale via Neuropixels181), intracellular calcium flux (measurable via calcium imaging), expression of FOS (the protein product of the immediate early gene Fos), electrical activity measured via electroencephalography (EEG), hemodynamic response measured via blood-oxygen-level-dependent (BOLD) functional MRI (fMRI), and intracellular voltage flux measured via direct imaging of neuronal activity (DIANA) fMRI. Temporal precision refers to the relative temporal proximity of the signal to neuronal activity. Spatial resolution ranges from individual cells to coarse brain regions. Species are ordered according to the number of neurons present in their nervous systems. Each type of neural signal is represented as a point on the graph, where the x coordinate represents spatial resolution and the y coordinate represents temporal precision. The species from which a given signal may be acquired are represented as symbols within each point and comprise C. elegans, Drosophila, zebrafish, rodents, non-human primates and humans.
Advances in microscopy and tissue clearing have enabled measurements with cellular resolution at the whole-brain scale. Examples include measurement of immediate early genes52, amyloid β (Aβ) plaques53 or vasculature54 across the entire rodent brain and automated registration to a whole-brain atlas. As an alternative to tissue clearing, block-face serial microscopy tomography (FAST) can be used to image sectioned tissue and obtain high-resolution, three-dimensional renderings of whole-brain immunostaining for neural markers55,56. Multiplex immunofluorescence can be used in combination with deep learning to perform automated cell phenotyping across the entire rodent brain57. Whole-brain imaging of neuronal activity in an awake, behaving state can also be performed in some species, including C. elegans58 and larval zebrafish59, and in mice, recent advances have enabled imaging of the entire cortical mantle60. Collectively, these approaches afford profiling of cell type-specific activity and simultaneous measurements of whole-brain cellular activity and behaviour.
Linking network structure, function and gene expression
The ability to perform whole-brain network analysis in animal models enables the identification and assessment of biological principles that support connectivity and synchronization in human studies. Animal models are amenable to genetic manipulation and stimulation or inhibition of neuronal activity in brain regions that have been identified as essential for function at the network level. For example, a series of recent studies used chemogenetic approaches to demonstrate the impact of silencing or stimulating activity in a particular region on whole-brain functional network topology61–63 or system-wide functional connectivity64,65. Notably, chemogenetically activating the locus coeruleus, the brain’s primary source of norepinephrine, shifted whole-brain connectivity patterns61 and strengthened functional connections in the DMN65. By contrast, suppressing activity in the anterior cingulate cortex decreased DMN connectivity64. These studies established causal roles for specific regions in modulating network connectivity.
In addition to assessments of causality in functional networks, animal models have afforded key insights into the properties of the brain’s structural architecture. Tract-tracing of the structural connectome has been performed in mice66, rats67, cats68 and several non-human primate species30,69. Importantly, anatomical connectivity has been mapped in animal models at a resolution that is not possible in humans. Diffusion MRI (dMRI) in humans has been used to reveal the density of interregional structural connections and the degree of myelination, but dMRI cannot provide information regarding the directionality of axonal projections. Since major white matter connections are conserved across many species, directionality of connectivity in animal models can guide inferences about directionality in studies of the human brain, in which such information is lacking. Knowledge of directionality is critical for understanding and predicting the propagation of information through the brain’s structural network.
Axonal connections can be visualized in animal models by virally expressing fluorophores that illuminate axons in an anterograde (from cell body to synapse) direction or cross synapses in a retrograde (from synaptic terminal to cell body) manner, thus providing information about the direction of interregional connections66,70,71. Connectivity assessment can also be confined to a subclass of neurons by conditional expression under a specific promoter72,73, or single neurons can be traced74,75, providing high-resolution information about regional connectivity. Using these strategies, the whole mouse brain anatomical connectome has been mapped at the mesoscale66. A recent study developed a connectivity blueprint for translating structural connectivity maps across primate species76. This framework could allow for structural changes identified in non-human primate studies to be mapped on to the human brain, enabling translational predictions.
To understand how structural and functional networks are biologically related, several studies in animal models have investigated the relationship between gene expression and whole-brain structural and functional connectivity. These studies have yielded insights into the molecular underpinnings of the brain’s structural topology and coordinated neural activity77–79. One study integrated structural, functional and gene expression data to demonstrate that both axonal connectivity between regions and similarities in gene expression give rise to functional connectivity measured with fMRI. In mice, both structural and functional connectivity are primarily associated with a subset of genes involved in synaptic activity, including genes that encode ion channels and synaptic membrane proteins77,79.
Tools for genetic manipulation in rodents and other species allow for causally linking changes in gene expression with changes in connectivity. In one recent investigation, deletion of the mu opioid receptor-encoding gene was found to alter functional connectivity between brain regions implicated in reward and aversion, while minimally impacting structural connectivity78. This finding suggests that genetic alterations can change the relationship between structure and function in the brain. Though a number of studies have linked gene expression and connectivity in the human brain80, these studies are correlative in nature. Recent work that has performed cross-species analysis of gene expression in the mouse and human brain81 has the potential to facilitate the translation of findings from causal studies in animal models to humans.
The application of network science approaches to data from animal models can also be used to directly test theoretical principles that underlie network science approaches to conceptualizing the brain. For example, NCT offers a powerful set of approaches for causally relating brain structure and function and for predicting the impact of perturbing activity in a particular region on whole-brain activity states. In human studies, NCT has been used to identify brain regions or systems that drive transitions in brain states associated with health and disease21,82–85. NCT applications depend on the assumption that a system can be driven toward a desired state by administering control signals to a defined set of input nodes86. In C. elegans, NCT-derived predictions about the role of specific neurons in behavioural responses to touch were validated by ablating these neurons and observing the impact on locomotor activity87. This endeavour was made possible by the tractability of mapping the C. elegans nervous system in its entirety at the cellular level, and by the animal’s well-defined repertoire of behavioural responses to stimuli.
In summary, studies of the brain’s networked architecture across species have identified genetic and regional drivers of network connectivity and enabled validation of predictions made by theoretical models. The use of these approaches across species has also identified important differences in the organization of the brain in humans and other animals, potentially enabling us to account for these differences when translating findings across species.
Enhancing the translational potential of animal model research
In this section, we discuss how cross-species applications of network science tools have contributed to the development of translational models for understanding the impact of causal manipulations on large-scale brain networks in the context of neurodevelopment, neuromodulation, neuropsychiatric disorders and neurodegeneration (FIG. 3).
Figure 3. Translational models in the areas of neurodevelopment, neuromodulation and neurodegeneration.
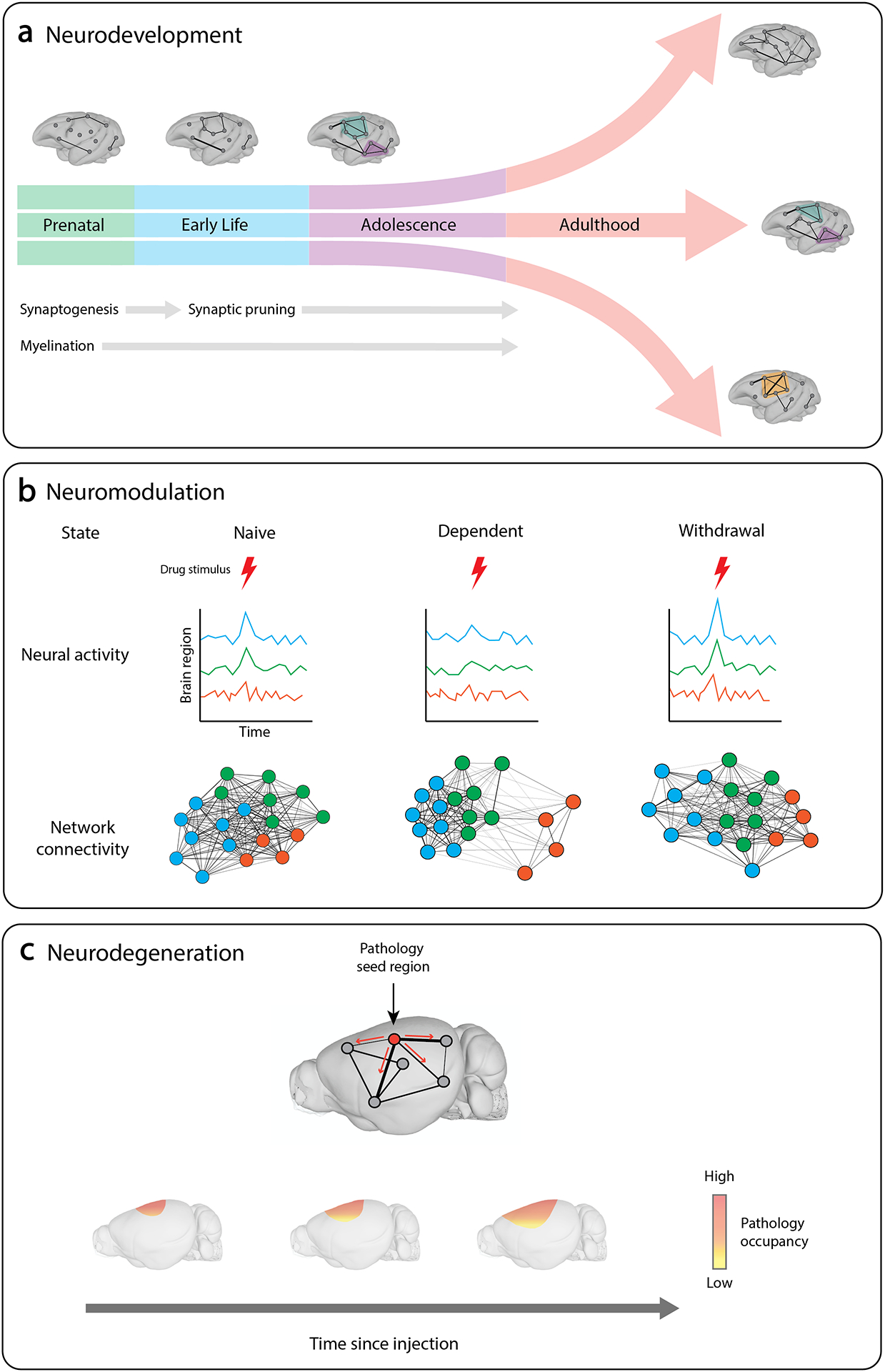
In translational studies of neurodevelopment, network models can be used to chart changes in the brain’s architecture over the course of development and to determine how genetic and environmental factors influence developmental outcomes. a | Illustration showing how developmental divergence during adolescence can result in differences in the network architecture of the adult brain. Studies of neuromodulation assess the brain’s response to stimuli under different conditions. b | Network models of drug dependence can identify differences in coordinated activity in response to drug stimuli. c | In seeding studies of neurodegeneration, pathological proteins are injected into specific regions of the brain. Network models can then be used to model the spread of pathology along the brain’s structural tracts and to predict disease progression. Brain images based on data in the Scalable Brain Atlas182. Brain images in part a based on data from ref.183. Brain images in part c based on data from ref.184.
Neurodevelopment
During development, the brain undergoes periods of marked growth in which neural pathways are shaped and reshaped. Though the precise timing of developmental processes varies across species, major developmental events are largely conserved88,89. In humans, the formation of synapses (synaptogenesis) and myelination of axonal connections commences during the prenatal period. Synaptic pruning and refinement begin during early childhood and continue into adulthood88,90. Together, these processes underlie the growth and plasticity of whole-brain network connectivity91,92 (FIG. 3). Studies of neurodevelopment seek to define changes in the brain over the course of development, and often do so by identifying factors that may contribute to an individual’s risk for developing neuropsychiatric or neurological disorders93. Network approaches are well-suited to two key efforts: (i) characterizing large-scale changes associated with brain maturation and (ii) investigating the genesis of circuit-level dysfunction associated with neurodevelopmental disorders94,95. We discuss both in turn below.
Studies in animal models have charted adaptations in the topology of the brain over the course of development. Functional and structural MRI studies in non-human primates have revealed how connectivity evolves during infancy and how it is impacted by environmental influences96–98. In macaques, changes in functional connectivity in visual cortical pathways have been found to proceed in a region-specific manner, such that anterior regions develop before posterior regions97. Other studies have identified specific nutrients that are critical for the development of mature patterns of connectivity in the brain’s functional subnetworks98 and modularity in the cortex96. At the cellular level, network science approaches have been used to identify changes in the modular structure of the nervous system over the course of development in C. elegans99. This work identified both consistent and dynamic connections in the network, the latter of which contribute to an increase in modularity across maturation. Importantly, this study identified features of early neurons, such as the extent of their physical contact with other cells and their number of connections, that correlate with the likelihood that they will form new connections during development99 and provides a template that can be used to assess whether this principle is maintained across other species.
Network approaches have also been applied to models of neurodevelopmental disorders. In a rabbit model of intrauterine growth restriction (IUGR) — a prevalent risk factor for neurodevelopmental disorders — characterization with graph theory metrics identified lower average degree and global efficiency in the brain’s structural network100. Importantly, this work built on a prior study of IUGR in human infants that had also identified altered structural network topology and lower global efficiency, suggesting impaired communication between brain regions101. The similarity of findings across species highlights the potential utility of the rabbit IUGR model for identifying biomarkers of neurodevelopmental disorders. Taken together, the studies discussed in this section illustrate the utility of network approaches for modelling the impact of environmental conditions on developmental trajectories.
Neuromodulation
In the context of this Review, we broadly define neuromodulation as the manipulation of neural activity via interventions such as targeted stimulation or drug exposure. The resulting change in whole-brain activity state may be accompanied by altered cognition or behaviour102. In this section, we focus on pharmacological forms of neuromodulation because of their relevance to modelling neuropsychiatric disorders such as addiction and to treating CNS disorders via pharmaceutical approaches. We provide examples of studies in this area that have been informed by network approaches to modeling data acquired from multiple modalities, including (i) fMRI and (ii) single-cell immediate early gene activity markers.
Human neuroimaging studies suggest that substance use disorders may be effectively characterized by large-scale changes in relations between brain regions, rather than a pathology of any one region or system103,104. In animal models of drug dependence, network approaches have been used to examine the modulatory effects of substances of abuse on network connectivity. Longitudinal studies using fMRI have probed circuit-level biomarkers in a rat model of nicotine dependence105,106. In this work, whole-brain modularity analysis was used to partition the naïve rat brain into five functional subnetworks (modules). Connectivity of an insular–frontal cortex module with other modules was found to predict the severity of nicotine dependence following chronic exposure105. Subsequently, innate differences in insular-frontal and insular-striatal connectivity observed prior to nicotine exposure were found to moderate the strength of an inverse relationship between cingulate–striatal connectivity and nicotine dependence observed following chronic exposure106. Functional connectivity strength between the cingulate and striatum was previously found to correlate with addiction severity in human cigarette smokers, such that weaker connectivity was associated with greater nicotine dependence107,108. In addition to highlighting the consistent role of cingulate–striatal connectivity in nicotine dependence across species, the rodent studies determined that dysfunction of this circuit is a result of nicotine dependence rather than a predispositional risk factor106. More broadly, this work underscores how the application of network approaches to analogous datasets from different species can facilitate the translation of findings.
Network models have also been used to characterize cellular-level changes in functional responses to drug stimuli. Specifically, a number of recent studies have used protein expression levels of the immediate early gene Fos as a surrogate for brain-wide activity mapping with cellular resolution in mouse models of drug exposure109–112. In these studies, neural activity is inferred from quantification of FOS-expressing cells following administration of a stimulus. Within network representations of these data, edges represent correlations in neural activity across animals at a given time point. In one such study, pharmacologically increasing or decreasing dopamine was found to reduce whole-brain functional connectivity, suggesting that modulating dopaminergic function in either direction disrupts interregional communication109. Network science tools, including graph theory and network control theory metrics, have also been used to characterize network-level changes in the brain following chronic exposure to alcohol, morphine, nicotine, cocaine and methamphetamine110–112. These studies have identified altered modularity during withdrawal from psychostimulants and alcohol110,111 and have pinpointed regional differences in the control energy required to drive the brain between opioid-naive, dependent, and protracted withdrawal states112. Collectively, this work has begun to bridge the gap between characterizations of state-specific neural activity patterns that are commonly assessed using FOS and systems-level conceptions of brain states in response to drug stimuli.
Neuropsychiatric disorders
It is increasingly appreciated that many neuropsychiatric conditions are associated with altered whole-brain network dynamics rather than localized dysfunction113. In this section we focus on network studies of major depressive disorder (MDD), which has been characterized and modelled in a range of species.
Studies in animal models are uniquely suited to identifying factors that promote vulnerability to MDD and other neuropsychiatric conditions. In a mouse model, network analysis of low-frequency potentials recorded from multiple cortical and subcortical regions identified distinct connectivity profiles that predict resilience or susceptibility to depression following social defeat stress114. Subsequently, work in zebrafish has identified a role for the habenula in coping behaviour during a stress-inducing behavioural challenge115.Although network-level biomarkers of depression vulnerability have not yet been identified in humans116, several studies have identified changes in connectivity that are associated with MDD. For instance, functional connectivity networks that characterize distinct depression subtypes have been identified117. Work in animal models could potentially assess whether these connectivity profiles are associated with susceptibility to or result from depressive phenotypes.
In general, the field has begun to recognize neuropsychiatric disorders as occurring along a spectrum rather than falling neatly in distinct diagnostic categories. Recent work has identified connectivity–symptom relationships that cross diagnostic boundaries118 and has used machine learning to predict treatment outcomes among patients who have been diagnosed with different conditions but show overlap in their symptomatology119. As human studies in this area continue to progress, work in animal models has the capacity to advance our understanding of the neurobiology underlying transdiagnostic connectivity profiles.
Neurodegeneration
Network studies of neurodegeneration seek to capture large-scale changes associated with disease-induced neuronal atrophy and identify mechanisms by which neuronal loss may be slowed or halted. Neurodegenerative disorders including Parkinson disease and Alzheimer disease are characterized pathologically by the accumulation of misfolded proteins in specific brain circuits120. Neurodegenerative disease symptoms are progressive, and postmortem staging studies have found that symptom severity is associated with progressive accumulation of pathological proteins in an increasing number of brain regions. This progressive accumulation of pathology has led to the hypothesis that pathology spreads through interconnected regions of the brain120–125. This hypothesis has been difficult to test, however, given the heterogeneous nature of disease presentation in patients, and the limited availability to examine pathological processes during life. PET imaging has shown promise for the assessment of Aβ pathology, which is a hallmark of Alzheimer disease126, and tau pathology, which is associated with both Alzheimer disease and Parkinson disease127,128. However, PET ligands for Aβ and tau have only recently become available. Further, the ability to longitudinally image using these ligands is limited and the resolution of signal is low compared with histological methods. Finally, imaging ligands for ɑ-synuclein pathology, which is implicated in Parkinson disease and other Lewy body diseases129, and TDP-43 pathology, which is associated with amyotrophic lateral sclerosis and frontotemporal dementia130, are not currently available. It is therefore generally not possible to pinpoint sites of early pathology development or to examine the causal processes underlying their progression using available tools in human studies.
Animal models allow for precise spatial and temporal control of pathology progression. Misfolded tau protein seeding models are particularly appropriate as they do not rely on transgenic overexpression of protein, but instead use a small amount of misfolded protein to seed pathology in a defined injection site131 (FIG. 3). The formation of pathological tau aggregates in these models is progressive and relies on the endogenous factors present in the animal model brain. Initial studies suggested that the progression of pathology was related to anatomical projection to the injection site, and later to secondary connections132. However, it was not possible to attribute progression quantitatively to connectivity or gene expression. Recent work using network models has begun to overcome this limitation.
In recent studies, network science approaches have been used to identify factors that contribute to the spread of α-synuclein and tau pathology in animal models133–139. These studies have found that pathology progression can be well explained by spread through neuroanatomic connections, mostly in the retrograde direction. Further, unexplained variance in the network model predictions appears related to regional gene expression patterns137. These animal model studies suggest that anatomical connectivity is a major constraint on pathology progression in neurodegenerative diseases. However, are these findings translatable to human disease? Human brain imaging studies of Parkinson disease, Alzheimer disease, behavioural variant frontotemporal dementia and semantic variant primary progressive aphasia have found that similar network models are able to predict brain atrophy140–144 or PET-imaged brain pathology145, suggesting that disease progression may be best understood, and treated, on a network level.
As our molecular understanding of disease grows, so does our ability to interpret molecular findings in a network context using mathematical models146. For example, previous studies have found that microglia, especially reactive microglia, contribute to the spread of tau pathology147–150. A recent study used an in silico model to assess the modulatory effects of the microglia-associated gene Trem2 on pathology spread, determining that Trem2 gene expression inhibits local accumulation of tau protein but increases its transmission to other brain regions139. This work thereby links a molecular finding (reactive microglia facilitate the propagation of tau pathology) with a network level explanation and demonstrates that the predictive power of connectivity-based spread models can be enhanced by accounting for molecular factors139.
Future challenges and opportunities
In the previous section, we provided examples to illustrate how network science approaches can contribute to translationally relevant animal models in the areas of neurodevelopment, neuromodulation, neuropsychiatric disorders and neurodegeneration. While not the focus of this Review, animal models of nervous system disorders are also challenged by the fact that the full extent of a human condition is unlikely to be reproducible in another animal, and that there is not often a one-to-one mapping of symptoms between humans and other species151. In addition to benefiting data translation, network approaches can highlight differences in the organization of the brain across species50,152, contributing to judicious use of animal models in neuroscience research. In this final section of our Review, we discuss some of the challenges we could overcome and questions we could address in the future with cross-species network models.
Despite their promise, network models of the brain are not without limitations. Translating mathematical concepts across datasets presents distinct challenges. For example, correlations in protein expression levels across individuals and correlations in the blood-oxygen-level-dependent (BOLD) signal across time can both be used to construct edges in a functional network, but each represent coordinated neural activity in ways that are not clearly related. Further, network models provide simplified representations of brain connectivity that may not reflect underlying biology in its entirety. Lack of comparable datasets across species can also limit the utility of cross-species network models. We devote the remainder of this section to discussing opportunities for further development that could help overcome these limitations.
Although network science tools have the capacity to provide analogous models of the brain across species, the translation of findings across different imaging modalities remains a significant challenge. Given the unique and important insights offered by each type of imaging, the long-term goal of relating findings from one species to another may be best served by developing tools that would enable translation of findings across neural signals derived from distinct imaging modalities. The application of network science tools across species could further our understanding of how cellular dynamics contribute to activity patterns across brain regions, which would enable predictions about how perturbations at the cellular level might lead to changes in regional or brain-wide dynamics. This capacity could aid the development of targeted therapeutic interventions by enabling investigators to extrapolate from animal studies knowledge of where to apply cell type specific neuromodulation, for example, or predict the effect of a pharmacotherapy on cognition. It could also contribute to the development of biomarkers for disease in model species and further our ability to effectively translate findings to humans.
Several studies in animal models have made progress toward translating neural signals across different modalities. In rats implanted with an electrode in the striatum, low-frequency potentials were recorded during fMRI scans to identify specific frequency bands underlying the BOLD signal153. Other studies in rats and mice have performed simultaneous recordings of calcium signals and fMRI154,155. One study determined that neuronal coupling between the DMN and salience network regions measured with fiber photometry is analogous to that previously identified with fMRI154, and another demonstrated that the calcium signal predicts the BOLD signal155. This work represents an important step toward translating between cell-type specific and nonspecific signals. Finally, simultaneous recordings of neuronal spiking activity and calcium imaging data from the same population of neurons have led to the development of tools that can be used to generate synthetic calcium imaging data from electrophysiological data156 and to infer neuronal spiking rate from calcium imaging data157,158. Collectively, these studies have begun to lay the groundwork for translating whole-brain neural signals ranging in scale from direct measurements of cells firing to metabolic signals representing regional neuron population dynamics.
Network science approaches could be used to develop tools for translating whole-brain neural data across different modalities. Using data from simultaneous recordings like those described in the previous paragraph, we could construct multiscale, multilayer network models to represent neuronal activity across different levels of spatial resolution and imaging modalities. In such a representation, intralayer edges would represent correlations in neural activity recorded by a given modality, and interlayer edges would represent the relationship between each pair of signals for every region (FIG. 4). The development of network frameworks for inferring one type of signal from another could enable us to relate electrophysiological recordings to calcium imaging to metabolic signals such as BOLD, while accounting for interregional differences in the relationship between these signals. By integrating cell-type specificity with coarser signals, these approaches could also enable us to draw inferences about the properties of cell types or connections observed via noninvasive imaging methods. Given knowledge of cell phenotypes or the valence of projections (that is, whether they are excitatory or inhibitory), one could gain a better understanding of how these features of the brain relate to the BOLD signal.
Figure 4. A multilayer network conception of neural signals across modalities in the mouse brain.
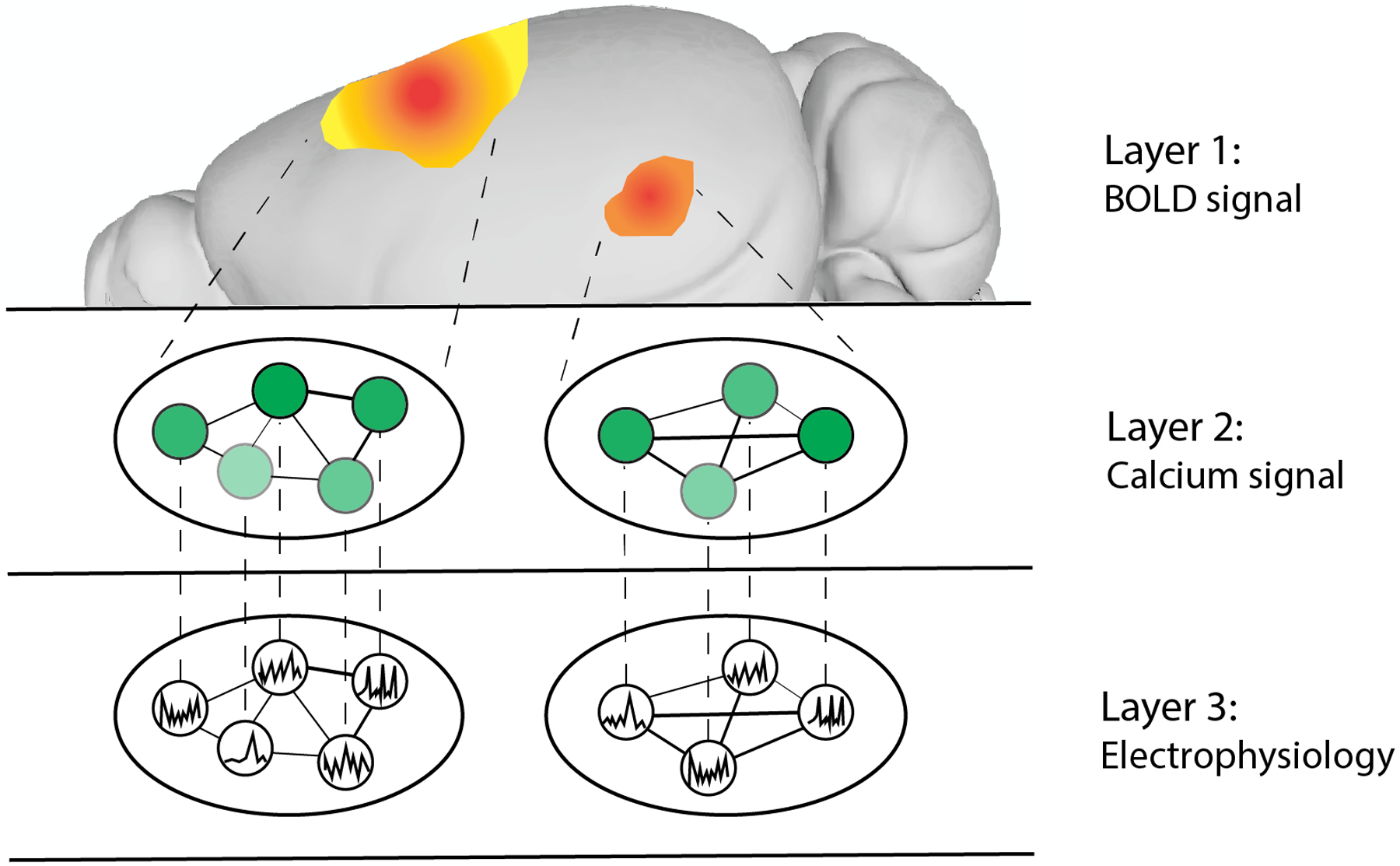
In this multilayer network, each layer represents neural activity inferred from one of three modalities: functional MRI (fMRI) blood-oxygen-level-dependent (BOLD) signal, calcium imaging and electrophysiology (using Neuropixels probes). In layer 1, the fMRI BOLD signal represents the coarse-grained dynamics of the two neuron populations illustrated in layers 2 and 3. In layer 2, intralayer edges between cells represent correlations in calcium signal, which are derived from fluorescence intensity. In layer 3, intralayer edges represent correlations in electrophysiological signal recorded from these population of cells. Interlayer edges represent the relations between each pair of signals. Brain image based on data in the Scalable Brain Atlas182. Brain image based on data from ref.184.
Graph neural network (GNN) models represent another promising approach for translating signals across species and modalities. GNNs are a class of deep learning model that preserve their graph topology while learning to perform tasks159. In recent years, these models have been used to predict large-scale neuronal dynamics across time and across individuals160,161. In the field of genomics, neural networks have been used to accurately predict transcription factor binding sites across species (mouse and human)162. Specifically, this strategy involves implementing a neural network comprised of two subnetworks, one of which is trained using data from one species, and the second of which aims to predict the species from which randomly selected datasets are derived. These two subnetworks share a convolutional layer that learns to effectively translate findings from one species to another by minimizing discriminative features across species162. Such an approach could be usefully applied to translating neural data across species by overcoming differences in neuroanatomy or signal resolution that serve as barriers to cross-species interpretability.
Tools for translating neural data across signals could be integrated with existing biophysical models to generate cross-species simulations. The development of platforms that enable brain simulations in primates163 and mice164 are useful for predicting changes in the brain in response to perturbation. Such models enable individual-specific predictions by linking brain structure and function with variability in behavior165. By further developing these models for cross-species applications, conditions simulated in one species could readily be translated to another.
Using network approaches for translating signals across species, we could more readily develop novel therapeutics. For example, neuromodulation via targeted stimulation of the brain represents an emerging and promising approach to treating severe neurological and neuropsychiatric diseases. Optogenetics, which has been successfully used to treat human retinal degeneration166, could potentially be usefully applied to treat other conditions via cell type-specific neuromodulation167. Optogenetics has recently been applied to human neocortical and hippocampal tissue, providing proof of principle for its potential utility as a therapeutic tool168. Translating network-level data from animal models could enable us to effectively predict where this stimulation should occur for therapeutic benefit. By contrast, the ‘reverse translation’ of findings from humans to animal models could further our understanding of causal processes that contribute to the efficacy of emerging therapeutic approaches, such as transcranial magnetic stimulation. This would also open the door to developing novel treatments by uncovering processes that could be targeted by other types of therapeutic interventions.
Conclusions
In this Review, we illustrated the utility of network science approaches for translationally relevant, cross-species investigations in neuroscience. The advent of tools that enable whole-brain recording with cellular resolution presents new opportunities to apply network science approaches to understanding cellular and molecular dynamics and relating them to interregional dynamics. We reviewed literature that has integrated molecular, structural, and functional connectivity in the brain, leading to a deeper understanding of the neurophysiological bases of phenomena observed with noninvasive neuroimaging methods. By providing an overview of studies in animal models of neurodevelopment, neuromodulation, neuropsychiatric disorders, and neurodegeneration, we highlighted the contributions of network approaches to the identification of biomarkers and treatments for human nervous system disorders. Finally, we discussed how network science approaches can be used to further efforts toward translating neural signals across modalities and across species, leading to the development of novel therapeutic approaches. Our key take-home message is that the field is now well-positioned to leverage network models in studies that bridge species and scales, enabling more effective translational research. Ultimately, we hope that this work will stimulate interactions between investigators conducting studies in humans and other animals, setting the stage for a future in which making connections across species and scales towards a more cohesive understanding of the brain is possible.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grant F32AA030475-01A1 to J.K.B.
Glossary
- Average path lengths
The average number of edges that connect each pair of nodes in a network.
- Modularity
A measure of how readily a network can be partitioned into subgroups of nodes that are more strongly connected to one another than to the rest of the network.
- Degree
The sum of connections to a given node.
- Global efficiency
A measure of the efficiency of long-range communication in a network.
- Control energy
The magnitude of input required to drive the brain from one activity state to another while accounting for its structural topology, time and the number of nodes into which input is given.
- Multilayer network
A graph structure in which nodes are organized into multiple layers; intralayer edges represent relations between nodes within a layer, and interlayer edges represent relations between nodes in different layers.
Citation diversity statement
Recent work in several fields of science has identified a bias in citation practices such that papers from women and other minority scholars are under-cited relative to the number of such papers in the field169–177. Here we sought to proactively consider choosing references that reflect the diversity of the intellectual contributions made in the areas covered in this review in thought, form of contribution, gender, race, ethnicity and other factors. First, we obtained the predicted gender of the first and last author of each reference by using databases that store the probability of a first name being carried by a woman173,178. By this measure and excluding self-citations to the first and last authors of our current paper), our references contain 5.84% woman(first)/woman(last), 8.94% man/woman, 19.15% woman/man, and 66.08% man/man. This method is limited in that a) names, pronouns, and social media profiles used to construct the databases may not, in every case, be indicative of gender identity and b) it cannot account for intersex, non-binary, or transgender people. Second, we obtained predicted racial/ethnic category of the first and last author of each reference by databases that store the probability of a first and last name being carried by an author of colour179,180. By this measure (and excluding self-citations), our references contain 18.98% author of color (first)/author of color(last), 18.90% white author/author of color, 20.98% author of color/white author, and 41.14% white author/white author. This method is limited in that a) names and Florida Voter Data to make the predictions may not be indicative of racial/ethnic identity, and b) it cannot account for Indigenous and mixed-race authors, or those who may face differential biases due to the ambiguous racialization or ethnicization of their names. We look forward to future work that could help us to better understand how to support equitable practices in science.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.elegans Sequencing Consortium C. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Howe K et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakken TE et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson BJ, Fatima GL, Oh S & Gire DH Many Paths to the Same Goal: Balancing Exploration and Exploitation during Probabilistic Route Planning. eNeuro 7, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calhoun AJ, Chalasani SH & Sharpee TO Maximally informative foraging by Caenorhabditis elegans. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidd C & Hayden BY The Psychology and Neuroscience of Curiosity. Neuron 88, 449–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bono M & Maricq AV Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci 28, 451–501 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Kalueff AV, Stewart AM & Gerlai R Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol Sci 35, 63–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornito A, Zalesky A & Bullmore E Fundamentals of Brain Network Analysis. (2016).
- 11.Betzel RF & Bassett DS Multi-scale brain networks. Neuroimage 160, 73–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassett DS, Zurn P & Gold JI On the nature and use of models in network neuroscience. Nat Rev Neurosci 19, 566–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friston KJ Functional and effective connectivity in neuroimaging: A synthesis. Human Brain Mapping 2, 56–78 (1994). [Google Scholar]
- 14.Rubinov M & Sporns O Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Sporns O Graph theory methods: applications in brain networks. Dialogues Clin Neurosci 20, 111–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu S et al. Controllability of structural brain networks. Nat Commun 6, 8414–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muldoon SF et al. Stimulation-Based Control of Dynamic Brain Networks. PLoS Comput Biol 12, e1005076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karrer TM et al. A practical guide to methodological considerations in the controllability of structural brain networks. J Neural Eng 17, 026031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, Yang GR, Laurent P, Schultz DH & Cole MW Constructing neural network models from brain data reveals representational transformations linked to adaptive behavior. Nat Commun 13, 673–16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perich MG & Rajan K Rethinking brain-wide interactions through multi-region ‘network of networks’ models. Curr Opin Neurobiol 65, 146–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava P, Fotiadis P, Parkes L & Bassett DS The expanding horizons of network neuroscience: From description to prediction and control. Neuroimage 258, 119250 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sporns O Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci 17, 652–660 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Medaglia JD, Lynall M-E & Bassett DS Cognitive network neuroscience. J Cogn Neurosci 27, 1471–1491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Dai Z, Gong G, Zhou C & He Y Understanding structural-functional relationships in the human brain: a large-scale network perspective. Neuroscientist 21, 290–305 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Deco G & Kringelbach ML Great expectations: using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron 84, 892–905 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Woo C-W, Chang LJ, Lindquist MA & Wager TD Building better biomarkers: brain models in translational neuroimaging. Nat Neurosci 20, 365–377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scannell JW & Young MP The connectional organization of neural systems in the cat cerebral cortex. Curr Biol 3, 191–200 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Mars RB et al. Comparing brains by matching connectivity profiles. Neurosci Biobehav Rev 60, 90–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Heuvel MP, Bullmore ET & Sporns O Comparative Connectomics. Trends Cogn Sci 20, 345–361 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Liu Z-Q, Zheng Y-Q & Misic B Network topology of the marmoset connectome. Netw Neurosci 4, 1181–1196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardesch DJ et al. Scaling Principles of White Matter Connectivity in the Human and Nonhuman Primate Brain. Cereb Cortex 32, 2831–2842 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eccles JC The synapse: from electrical to chemical transmission. Annu Rev Neurosci 5, 325–339 (1982). [DOI] [PubMed] [Google Scholar]
- 33.Fox PT et al. Mapping human visual cortex with positron emission tomography. Nature 323, 806–809 (1986). [DOI] [PubMed] [Google Scholar]
- 34.Belliveau JW et al. Functional mapping of the human visual cortex by magnetic resonance imaging. Science 254, 716–719 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Kwong KK et al. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A 89, 5675–5679 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fox PT & Raichle ME Focal physiological uncoupling of cerebral blood flow and oxidative metabolism during somatosensory stimulation in human subjects. Proc Natl Acad Sci U S A 83, 1140–1144 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raichle ME A brief history of human brain mapping. Trends Neurosci 32, 118–126 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Calhoun VD, Pearlson GD & Sui J Data-driven approaches to neuroimaging biomarkers for neurological and psychiatric disorders: emerging approaches and examples. Curr Opin Neurol 34, 469–479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Büchel C, Coull JT & Friston KJ The predictive value of changes in effective connectivity for human learning. Science 283, 1538–1541 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Averbeck BB & Lee D Neural noise and movement-related codes in the macaque supplementary motor area. J Neurosci 23, 7630–7641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romo R, Hernández A, Zainos A & Salinas E Correlated neuronal discharges that increase coding efficiency during perceptual discrimination. Neuron 38, 649–657 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Rilling JK & van den Heuvel MP Comparative Primate Connectomics. Brain Behav Evol 91, 170–179 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Watts DJ & Strogatz SH Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Hilgetag CC, Burns GA, O’Neill MA, Scannell JW & Young MP Anatomical connectivity defines the organization of clusters of cortical areas in the macaque monkey and the cat. Philos Trans R Soc Lond B Biol Sci 355, 91–110 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitesell JD et al. Regional, Layer, and Cell-Type-Specific Connectivity of the Mouse Default Mode Network. Neuron 109, 545–559.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H et al. Rat brains also have a default mode network. Proc Natl Acad Sci U S A 109, 3979–3984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu L-M et al. Constituents and functional implications of the rat default mode network. Proc Natl Acad Sci U S A 113, E4541–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belcher AM et al. Large-scale brain networks in the awake, truly resting marmoset monkey. J Neurosci 33, 16796–16804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantini D et al. Default mode of brain function in monkeys. J Neurosci 31, 12954–12962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garin CM et al. An evolutionary gap in primate default mode network organization. Cell Rep 39, 110669 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toi PT et al. In vivo direct imaging of neuronal activity at high temporospatial resolution. Science 378, 160–168 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Renier N et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liebmann T et al. Three-Dimensional Study of Alzheimer’s Disease Hallmarks Using the iDISCO Clearing Method. Cell Rep 16, 1138–1152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirst C et al. Mapping the Fine-Scale Organization and Plasticity of the Brain Vasculature. Cell 180, 780–795.e25 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Seiriki K et al. High-Speed and Scalable Whole-Brain Imaging in Rodents and Primates. Neuron 94, 1085–1100.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Seiriki K et al. Whole-brain block-face serial microscopy tomography at subcellular resolution using FAST. Nat Protoc 14, 1509–1529 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Maric D et al. Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat Commun 12, 1550–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen JP et al. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc Natl Acad Sci U S A 113, E1074–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cong L et al. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barson D et al. Simultaneous mesoscopic and two-photon imaging of neuronal activity in cortical circuits. Nat Methods 17, 107–113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zerbi V et al. Rapid Reconfiguration of the Functional Connectome after Chemogenetic Locus Coeruleus Activation. Neuron 103, 702–718.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Tu W, Ma Z & Zhang N Brain network reorganization after targeted attack at a hub region. Neuroimage 237, 118219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rocchi F et al. Increased fMRI connectivity upon chemogenetic inhibition of the mouse prefrontal cortex. Nat Commun 13, 1056–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tu W, Ma Z, Ma Y, Dopfel D & Zhang N Suppressing Anterior Cingulate Cortex Modulates Default Mode Network and Behavior in Awake Rats. Cereb Cortex 31, 312–323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oyarzabal EA et al. Chemogenetic stimulation of tonic locus coeruleus activity strengthens the default mode network. Sci Adv 8, eabm9898 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oh SW et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitt O & Eipert P neuroVIISAS: approaching multiscale simulation of the rat connectome. Neuroinformatics 10, 243–267 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Scannell JW, Blakemore C & Young MP Analysis of connectivity in the cat cerebral cortex. J Neurosci 15, 1463–1483 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Markov NT et al. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb Cortex 24, 17–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu F et al. High-throughput mapping of a whole rhesus monkey brain at micrometer resolution. Nat Biotechnol 39, 1521–1528 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Saleeba C, Dempsey B, Le S, Goodchild A & McMullan S A Student’s Guide to Neural Circuit Tracing. Front Neurosci 13, 897 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu H, Williams J & Nathans J Complete morphologies of basal forebrain cholinergic neurons in the mouse. Elife 3, e02444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winnubst J et al. Reconstruction of 1,000 Projection Neurons Reveals New Cell Types and Organization of Long-Range Connectivity in the Mouse Brain. Cell 179, 268–281.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao L et al. Single-neuron projectome of mouse prefrontal cortex. Nat Neurosci 25, 515–529 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Mars RB et al. Whole brain comparative anatomy using connectivity blueprints. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richiardi J et al. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 348, 1241–1244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mechling AE et al. Deletion of the mu opioid receptor gene in mice reshapes the reward-aversion connectome. Proc Natl Acad Sci U S A 113, 11603–11608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mills BD et al. Correlated Gene Expression and Anatomical Communication Support Synchronized Brain Activity in the Mouse Functional Connectome. J Neurosci 38, 5774–5787 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arnatkeviciute A, Fulcher BD, Bellgrove MA & Fornito A Where the genome meets the connectome: Understanding how genes shape human brain connectivity. Neuroimage 244, 118570 (2021). [DOI] [PubMed] [Google Scholar]
- 81.Beauchamp A et al. Whole-brain comparison of rodent and human brains using spatial transcriptomics. Elife 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cui Z et al. Optimization of energy state transition trajectory supports the development of executive function during youth. Elife 9, 17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scheid BH et al. Time-evolving controllability of effective connectivity networks during seizure progression. Proc Natl Acad Sci U S A 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Braun U et al. Brain network dynamics during working memory are modulated by dopamine and diminished in schizophrenia. Nat Commun 12, 3478–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parkes L et al. Network Controllability in Transmodal Cortex Predicts Positive Psychosis Spectrum Symptoms. Biol Psychiatry 90, 409–418 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y-Y & Barabási A-L Control principles of complex systems. Rev. Mod. Phys (2016). [Google Scholar]
- 87.Yan G et al. Network control principles predict neuron function in the Caenorhabditis elegans connectome. Nature 550, 519–523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Semple BD, Blomgren K, Gimlin K, Ferriero DM & Noble-Haeusslein LJ Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol 106–107, 1–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iwata R Temporal differences of neurodevelopment processes between species. Neurosci Res 177, 8–15 (2022). [DOI] [PubMed] [Google Scholar]
- 90.Huttenlocher PR Synaptic density in human frontal cortex - developmental changes and effects of aging. Brain Res 163, 195–205 (1979). [DOI] [PubMed] [Google Scholar]
- 91.Power JD, Fair DA, Schlaggar BL & Petersen SE The development of human functional brain networks. Neuron 67, 735–748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uddin LQ, Supekar K & Menon V Typical and atypical development of functional human brain networks: insights from resting-state FMRI. Front Syst Neurosci 4, 21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levitt P & Veenstra-VanderWeele J Neurodevelopment and the origins of brain disorders. Neuropsychopharmacology 40, 1–3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grayson DS & Fair DA Development of large-scale functional networks from birth to adulthood: A guide to the neuroimaging literature. Neuroimage 160, 15–31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Graham AM, Marr M, Buss C, Sullivan EL & Fair DA Understanding Vulnerability and Adaptation in Early Brain Development using Network Neuroscience. Trends Neurosci 44, 276–288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grayson DS, Kroenke CD, Neuringer M & Fair DA Dietary omega-3 fatty acids modulate large-scale systems organization in the rhesus macaque brain. J Neurosci 34, 2065–2074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kovacs-Balint Z et al. Early Developmental Trajectories of Functional Connectivity Along the Visual Pathways in Rhesus Monkeys. Cereb Cortex 29, 3514–3526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miranda-Dominguez O et al. Carotenoids improve the development of cerebral cortical networks in formula-fed infant macaques. Sci Rep 12, 15220–13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Witvliet D et al. Connectomes across development reveal principles of brain maturation. Nature 596, 257–261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Batalle D et al. Long-term reorganization of structural brain networks in a rabbit model of intrauterine growth restriction. Neuroimage 100, 24–38 (2014). [DOI] [PubMed] [Google Scholar]
- 101.Batalle D et al. Altered small-world topology of structural brain networks in infants with intrauterine growth restriction and its association with later neurodevelopmental outcome. Neuroimage 60, 1352–1366 (2012). [DOI] [PubMed] [Google Scholar]
- 102.Lee S-H & Dan Y Neuromodulation of brain states. Neuron 76, 209–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tolomeo S & Yu R Brain network dysfunctions in addiction: a meta-analysis of resting-state functional connectivity. Transl Psychiatry 12, 41–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joutsa J et al. Brain lesions disrupting addiction map to a common human brain circuit. Nat Med 28, 1249–1255 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsu L-M et al. Intrinsic Insular-Frontal Networks Predict Future Nicotine Dependence Severity. J Neurosci 39, 5028–5037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Keeley RJ et al. Intrinsic differences in insular circuits moderate the negative association between nicotine dependence and cingulate-striatal connectivity strength. Neuropsychopharmacology 45, 1042–1049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hong LE et al. Association of nicotine addiction and nicotine’s actions with separate cingulate cortex functional circuits. Arch Gen Psychiatry 66, 431–441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hong LE et al. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc Natl Acad Sci U S A 107, 13509–13514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cruces-Solis H, Nissen W, Ferger B & Arban R Whole-brain signatures of functional connectivity after bidirectional modulation of the dopaminergic system in mice. Neuropharmacology 178, 108246 (2020). [DOI] [PubMed] [Google Scholar]
- 110.Kimbrough A et al. Brain-wide functional architecture remodeling by alcohol dependence and abstinence. Proc Natl Acad Sci U S A 117, 2149–2159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kimbrough A et al. Characterization of the Brain Functional Architecture of Psychostimulant Withdrawal Using Single-Cell Whole-Brain Imaging. eNeuro 8, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brynildsen JK et al. Gene coexpression patterns predict opiate-induced brain-state transitions. Proc Natl Acad Sci U S A 117, 19556–19565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li B-J et al. A brain network model for depression: From symptom understanding to disease intervention. CNS Neurosci Ther 24, 1004–1019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hultman R et al. Brain-wide Electrical Spatiotemporal Dynamics Encode Depression Vulnerability. Cell 173, 166–180.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Andalman AS et al. Neuronal Dynamics Regulating Brain and Behavioral State Transitions. Cell 177, 970–985.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grossman Y & Dzirasa K Is depression a disorder of electrical brain networks? Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 45, 230–231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Drysdale AT et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 23, 28–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xia CH et al. Linked dimensions of psychopathology and connectivity in functional brain networks. Nat Commun 9, 3003–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y et al. Identification of psychiatric disorder subtypes from functional connectivity patterns in resting-state electroencephalography. Nat Biomed Eng 5, 309–323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brettschneider J, Del Tredici K, Lee VMY & Trojanowski JQ Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat Rev Neurosci 16, 109–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Braak H & Braak E Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 122.Del Tredici K, Rüb U, de Vos RAI, Bohl JRE & Braak H Where does parkinson disease pathology begin in the brain? J Neuropathol Exp Neurol 61, 413–426 (2002). [DOI] [PubMed] [Google Scholar]
- 123.Braak H et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 24, 197–211 (2003). [DOI] [PubMed] [Google Scholar]
- 124.Braak H, Ghebremedhin E, Rüb U, Bratzke H & Del Tredici K Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res 318, 121–134 (2004). [DOI] [PubMed] [Google Scholar]
- 125.Braak H, Alafuzoff I, Arzberger T, Kretzschmar H & Del Tredici K Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta neuropathologica 112, 389–404 (Springer-Verlag, 2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bloom GS Amyloid-β and Tau: The Trigger and Bullet in Alzheimer Disease Pathogenesis. JAMA Neurol 71, 505–508 (2014). [DOI] [PubMed] [Google Scholar]
- 127.Irwin DJ, Lee VMY & Trojanowski JQ Parkinson’s disease dementia: convergence of α-synuclein, tau and amyloid-β pathologies. Nat Rev Neurosci 14, 626–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van der Kant R, Goldstein LSB & Ossenkoppele R Amyloid-β-independent regulators of tau pathology in Alzheimer disease. Nat Rev Neurosci 21, 21–35 (2020). [DOI] [PubMed] [Google Scholar]
- 129.Calabresi P et al. Alpha-synuclein in Parkinson’s disease and other synucleinopathies: from overt neurodegeneration back to early synaptic dysfunction. Cell Death Dis 14, 176–16 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Keating SS, San Gil R, Swanson MEV, Scotter EL & Walker AK TDP-43 pathology: From noxious assembly to therapeutic removal. Prog Neurobiol 211, 102229 (2022). [DOI] [PubMed] [Google Scholar]
- 131.Robert A, Schöll M & Vogels T Tau Seeding Mouse Models with Patient Brain-Derived Aggregates. Int J Mol Sci 22, 6132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clavaguera F et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol 11, 909–913 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mezias C, LoCastro E, Xia C & Raj A Connectivity, not region-intrinsic properties, predicts regional vulnerability to progressive tau pathology in mouse models of disease. Acta Neuropathol Commun 5, 61–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Henderson MX et al. Spread of α-synuclein pathology through the brain connectome is modulated by selective vulnerability and predicted by network analysis. Nat Neurosci 22, 1248–1257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Henderson MX et al. Glucocerebrosidase Activity Modulates Neuronal Susceptibility to Pathological α-Synuclein Insult. Neuron 105, 822–836.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mezias C, Rey N, Brundin P & Raj A Neural connectivity predicts spreading of alpha-synuclein pathology in fibril-injected mouse models: Involvement of retrograde and anterograde axonal propagation. Neurobiol Dis 134, 104623 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cornblath EJ et al. Computational modeling of tau pathology spread reveals patterns of regional vulnerability and the impact of a genetic risk factor. Sci Adv 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rahayel S et al. Differentially targeted seeding reveals unique pathological alpha-synuclein propagation patterns. Brain 145, 1743–1756 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Anand C, Maia PD, Torok J, Mezias C & Raj A The effects of microglia on tauopathy progression can be quantified using Nexopathy in silico (Nexis) models. Sci Rep 12, 21170–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raj A, Kuceyeski A & Weiner M A network diffusion model of disease progression in dementia. Neuron 73, 1204–1215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dagher A & Zeighami Y Testing the Protein Propagation Hypothesis of Parkinson Disease. J Exp Neurosci 12, 1179069518786715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brown JA et al. Patient-Tailored, Connectivity-Based Forecasts of Spreading Brain Atrophy. Neuron 104, 856–868.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pandya S et al. Predictive model of spread of Parkinson’s pathology using network diffusion. Neuroimage 192, 178–194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zheng Y-Q et al. Local vulnerability and global connectivity jointly shape neurodegenerative disease propagation. PLoS Biol 17, e3000495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ossenkoppele R et al. Tau covariance patterns in Alzheimer’s disease patients match intrinsic connectivity networks in the healthy brain. Neuroimage Clin 23, 101848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Warren JD et al. Molecular nexopathies: a new paradigm of neurodegenerative disease. Trends Neurosci 36, 561–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Asai H et al. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci 18, 1584–1593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Maphis N et al. Reactive microglia drive tau pathology and contribute to the spreading of pathological tau in the brain. Brain 138, 1738–1755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leyns CEG et al. TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc Natl Acad Sci U S A 114, 11524–11529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gratuze M et al. TREM2-independent microgliosis promotes tau-mediated neurodegeneration in the presence of ApoE4. Neuron 111, 202–219.e7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nestler EJ & Hyman SE Animal models of neuropsychiatric disorders. Nat Neurosci 13, 1161–1169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu T et al. Cross-species functional alignment reveals evolutionary hierarchy within the connectome. Neuroimage 223, 117346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jaime S et al. Delta Rhythm Orchestrates the Neural Activity Underlying the Resting State BOLD Signal via Phase-amplitude Coupling. Cereb Cortex 29, 119–133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liang Z, Ma Y, Watson GDR & Zhang N Simultaneous GCaMP6-based fiber photometry and fMRI in rats. J Neurosci Methods 289, 31–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lake EMR et al. Simultaneous cortex-wide fluorescence Ca2+ imaging and whole-brain fMRI. Nat Methods 17, 1262–1271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei Z et al. A comparison of neuronal population dynamics measured with calcium imaging and electrophysiology. PLoS Comput Biol 16, e1008198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jewell SW, Hocking TD, Fearnhead P & Witten DM Fast nonconvex deconvolution of calcium imaging data. Biostatistics 21, 709–726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fleming W, Jewell S, Engelhard B, Witten DM & Witten IB Inferring spikes from calcium imaging in dopamine neurons. PLoS One 16, e0252345 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bessadok A, Mahjoub MA & Rekik I Graph Neural Networks in Network Neuroscience. IEEE Trans Pattern Anal Mach Intell 45, 5833–5848 (2023). [DOI] [PubMed] [Google Scholar]
- 160.Wang PY, Sapra S, George VK & Silva GA Generalizable Machine Learning in Neuroscience Using Graph Neural Networks. Front Artif Intell 4, 618372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wein S et al. Forecasting brain activity based on models of spatiotemporal brain dynamics: A comparison of graph neural network architectures. Netw Neurosci 6, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cochran K et al. Domain-adaptive neural networks improve cross-species prediction of transcription factor binding. Genome Res 32, 512–523 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sanz Leon P et al. The Virtual Brain: a simulator of primate brain network dynamics. Front Neuroinform 7, 10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Melozzi F, Woodman MM, Jirsa VK & Bernard C The Virtual Mouse Brain: A Computational Neuroinformatics Platform to Study Whole Mouse Brain Dynamics. eNeuro 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kumar VG, Dutta S, Talwar S, Roy D & Banerjee A Biophysical mechanisms governing large-scale brain network dynamics underlying individual-specific variability of perception. Eur J Neurosci 52, 3746–3762 (2020). [DOI] [PubMed] [Google Scholar]
- 166.McClements ME, Staurenghi F, MacLaren RE & Cehajic-Kapetanovic J Optogenetic Gene Therapy for the Degenerate Retina: Recent Advances. Front Neurosci 14, 570909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shen Y, Campbell RE, Côté DC & Paquet M-E Challenges for Therapeutic Applications of Opsin-Based Optogenetic Tools in Humans. Front Neural Circuits 14, 41 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Andersson M et al. Optogenetic control of human neurons in organotypic brain cultures. Sci Rep 6, 24818–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mitchell SM, Lange S & Brus H Gendered citation patterns in international relations journals. International Studies Perspectives 14, 485–492 (2013). [Google Scholar]
- 170.Dion ML, Sumner JL & Mitchell SM Gendered citation patterns across political science and social science methodology fields. Political Analysis 26, 312–327 (2018). [Google Scholar]
- 171.Caplar N, Tacchella S & Birrer S Quantitative evaluation of gender bias in astronomical publications from citation counts. Nature Astronomy 1, 0141 (2017). [Google Scholar]
- 172.Maliniak D, Powers R & Walter BF The gender citation gap in international relations. International Organization 67, 889–922 (2013). [Google Scholar]
- 173.Dworkin JD et al. The extent and drivers of gender imbalance in neuroscience reference lists. Nat Neurosci 23, 918–926 (2020). [DOI] [PubMed] [Google Scholar]
- 174.Bertolero MA et al. Racial and ethnic imbalance in neuroscience reference lists and intersections with gender. bioRxiv 2020.10.12.336230 (2020). doi: 10.1101/2020.10.12.336230 [DOI] [Google Scholar]
- 175.Wang X et al. Gendered Citation Practices in the Field of Communication. Ann Int Commun Assoc 45, 134–153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chatterjee P & Werner RM Gender Disparity in Citations in High-Impact Journal Articles. JAMA Netw Open 4, e2114509–e2114509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fulvio JM, Akinnola I & Postle BR Gender (Im)balance in Citation Practices in Cognitive Neuroscience. J Cogn Neurosci 33, 3–7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou D et al. dalejn/cleanBib: v1.1.2. (2022). doi: 10.5281/zenodo.4104748 [DOI] [Google Scholar]
- 179.Ambekar A, Ward C, Mohammed J, Male S & Skiena S Name-ethnicity classification from open sources. in (eds. Elder JF IV, Fogelman-Soulie F, Flach P & Zaki M) (2009). [Google Scholar]
- 180.Sood G & Laohaprapanon S Predicting Race and Ethnicity From the Sequence of Characters in a Name. (2018).
- 181.Jun JJ et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bakker R, Tiesinga P & Kötter R The Scalable Brain Atlas: Instant Web-Based Access to Public Brain Atlases and Related Content. Neuroinformatics 13, 353–366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Calabrese E et al. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage 117, 408–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 185.Betzel RF, Gu S, Medaglia JD, Pasqualetti F & Bassett DS Optimally controlling the human connectome: the role of network topology. Sci Rep 6, 30770–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bassett DS & Sporns O Network neuroscience. Nat Neurosci 20, 353–364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wu Z et al. A Comprehensive Survey on Graph Neural Networks. IEEE Trans Neural Netw Learn Syst 32, 4–24 (2021). [DOI] [PubMed] [Google Scholar]
- 188.Wein S et al. A graph neural network framework for causal inference in brain networks. Sci Rep 11, 8061–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li X et al. BrainGNN: Interpretable Brain Graph Neural Network for fMRI Analysis. Med Image Anal 74, 102233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu X, Shen Q & Zhang S Cross-species cell-type assignment from single-cell RNA-seq data by a heterogeneous graph neural network. Genome Res 33, 96–111 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Makris N et al. Decreased volume of left and total anterior insular lobule in schizophrenia. Schizophr Res 83, 155–171 (2006). [DOI] [PubMed] [Google Scholar]
- 192.Frazier JA et al. Structural brain magnetic resonance imaging of limbic and thalamic volumes in pediatric bipolar disorder. Am J Psychiatry 162, 1256–1265 (2005). [DOI] [PubMed] [Google Scholar]
- 193.Desikan RS et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980 (2006). [DOI] [PubMed] [Google Scholar]
- 194.Goldstein JM et al. Hypothalamic abnormalities in schizophrenia: sex effects and genetic vulnerability. Biol Psychiatry 61, 935–945 (2007). [DOI] [PubMed] [Google Scholar]


