Abstract
Background:
Huntington’s disease (HD) is a hereditary, neurodegenerative disorder characterized by motor, cognitive, and psychiatric symptoms. Currently, HD can only be managed symptomatically, including a large variety of prescribed drugs. Many HD patients experience negative medication effects (e.g. side effects or non-response). Pharmacogenetic (PGx) studies show how genetic variation affects both medication efficacy and toxicity and holds the potential to improve these outcomes of drug treatment.
Primary objective:
To classify the effect of the PGx profile of CYP2C19 and CYP2D6 in HD gene expansion carriers on negative medication effects of HD-related medication.
Design:
Multicenter, observational study with 1-year follow-up. Adult HD gene expansion carriers who use one or more HD-related medications are eligible to participate.
Methods and analysis:
A detailed overview of medication use, medication efficacy, and side effects is retrospectively and prospectively collected via medication diaries, questionnaires, phone calls, and pharmacy medication verification schemes. PGx analysis on whole blood-extracted DNA is performed with Agena Bioscience VeriDose® Core Panel and long-range polymerase chain reaction copy number variation analysis. Per the study protocol-defined negative medication effects in HD gene expansion carriers with a genotype predicted poor or ultrarapid metabolizer phenotype will be compared with HD gene expansion carriers with a predicted intermediate and normal metabolizer phenotype. Frequencies will be analyzed via χ2 and logistic multivariate regression analysis. In addition, we summarize in this manuscript HD-relevant PGx prescription recommendations to improve drug therapy.
Ethics:
The original study protocol was approved by the medical research ethics committee Leiden Den Haag Delft on 26 November 2019.
Discussion:
HD-MED is a low-risk study that will generate personalized PGx results that can immediately be implemented in clinical practice, thus potentially improving pharmacovigilance and patients’ quality of life.
Registration:
This study is registered in the International Clinical Trial Registry Platform under registration number NL8251, URL https://trialsearch.who.int/Trial2.aspx?TrialID=NL8251.
Keywords: biobank, drug type, Huntington’s disease, medication efficacy, medication use, pharmacogenetics, pharmacogenomics, pharmacovigilance, protocol, rare disorder
Introduction
Huntington’s disease (HD) is a rare, autosomal dominantly inherited, neurodegenerative disease that is caused by a cytosine-adenine-guanine (CAG) trinucleotide repeat expansion on chromosome 4.1,2 HD symptoms and severity vary widely between patients and include three main domains: motor dysfunction, behavioral symptoms, and cognitive impairment. 3 Chorea is the most prominent motor symptom of HD but dystonia, abnormal eye movements, and postural instability are often present as well. Furthermore, a variety of neuropsychiatric symptoms may arise, including mood instability, anxiety, and aggression. 4
Up until today, no disease-modifying treatment exists. Although several new therapeutic options are being explored, most of these are still in the developmental phase and not likely to become available for the larger HD population on short notice. 5 Hence, HD can only be managed symptomatically with non-pharmacological and pharmacological treatment, focused on improving quality of daily life by alleviating the impact of symptoms. Although some drugs for the treatment of HD symptoms, such as tetrabenazine against chorea, have been systematically examined in randomized controlled trials, 6 the large majority is not properly assessed according to the present standards. Combined with each physician’s own specific drug preferences, this results in a large variety of prescribed medications in clinical practice. 7
Currently, there is no accurate overview of medications used by HD patients. A preliminary attempt was made by the TRACK-HD investigators, who classified medication use by retrospectively using cross-sectional, annual data from an observational biomarker study. 8 However, these data were incomplete and often lacked information on the duration of medication use, medication efficacy, side effects, and details on the indication of use. Especially the latter is of utmost importance in HD, since the same drug can be prescribed for different symptomatology. For instance, olanzapine, an often prescribed neuroleptic, is frequently prescribed to treat both chorea and/or irritability. 8
The large variability of prescribed medications in clinical care is accompanied by a similar unpredictable variability in medication efficacy and side effect profile, both for motor and non-motor symptoms. 7 Therefore, many HD patients experience side effects, varying from minor inconveniences to disruptions of daily life. A part of this variability could be explained by pharmacogenetics (PGx). Pharmacogenetics is the study of individual genetic differences affecting medication response, in terms of efficacy and side effects. 9 Individuals are categorized into distinct genotype-predicted phenotype groups for a given actionable drug–gene combination; poor metabolizer (PM), intermediate metabolizer (IM), normal metabolizer (NM), or ultra-rapid metabolizer (UM). The Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG), part of the Royal Dutch Pharmacist’s Association (KNMP) are world leading in developing PGx guidelines.10,11 Currently, more than 50 drugs have been linked with multiple pharmacogenes and clinical guidelines have been formulated. 12 The Cytochrome P450 (CYP)2C19 and 2D6 enzymes, encoded for by the CYP2C19 and CYP2D6 pharmacogenes, play an important role in the metabolism of often used HD drugs. Approximately 7–14% of the general Dutch population is a PM or UM for CYP2C19 (3.0% and 4.0%, respectively) and/or CYP2D6 (5.0% and 1.5%, respectively). 13 Because of their phenotype, alternative dosing or switching to a different type of medication is recommended. 14 However, PGx guidelines are poorly incorporated in daily clinical HD practice, mainly because the relevance of these gene–drug interactions has never been assessed in the HD population. Besides, clinicians are often unaware of which drugs could benefit from implementing PGx in prescribing methods. Clinicians are therefore prone to focus on the classical parameters, such as age and comorbidities, when pharmacologically treating symptoms rather than implementing PGx recommendations.
Eventually, almost all HD patients will use the medication in their life course and most HD-related drugs (60–80%) are metabolized by CYP2C19 and/or CYP2D6.14,15 The current study will be the first PGx study in HD. Using a longitudinal, observational design medication efficacy and toxicity under the current standard clinical care will be summarized. Therewith, this study aims to explore PGx interactions in HD to better guide the long-term pharmacological management of HD in a more personalized manner by reducing negative medication effects.
Primary objective
To classify the effect of the pharmacogenetic profile of CYP2C19 and CYP2D6 in HD gene expansion carriers on negative medication effects of HD-related medication with an actionable drug–gene interaction in the corresponding CYP genes.
Secondary objectives
○ To assess and categorize current detailed medication use in HD gene expansion carriers.
○ To determine the eligibility of a 1-year medication diary compared to pharmacy medication history surveys in HD patients.
Explorative objectives
○ To explore the effect of the pharmacogenetic profile of CYP2C19 and CYP2D6 in HD gene expansion carriers who use HD-related medication and who either use a relatively low or a relatively high drug dose with regard to the symptom the drug is prescribed for.
○ To explore associations between genetic variants and extraordinary side effect profiles or lack of medication response in HD gene expansion carriers.
The SPIRIT (Standard Protocol Items: Recommendations for Interventional Trials) statement and checklist were followed when preparing this study protocol.
METHODS: Participants and outcomes
HD-MED is a longitudinal, observational cohort study in adult HD gene expansion carriers who are either just starting with or are already using one or more HD-related drugs. The study will be conducted in three academic Dutch hospitals: Leiden University Medical Center, Maastricht University Medical Center +, and University Medical Center Groningen, and is also open to HD gene expansion carriers who do not regularly visit these neurology outpatient clinics. The total follow-up period per participant will be 1 year (± 6 weeks) in which medication use is recorded at designated intervals.
Inclusion and exclusion criteria
To be eligible to participate in this study, a subject must meet all of the following criteria:
○ A capacitated individual, aged ⩾ 18 years.
○ Genetically confirmed CAG-repeat expansion of ⩾ 36 in the Huntingtin (HTT) gene.
○ Either about to start with medication related to HD or already using one or more HD-related drugs. HD-related drugs are drugs considered to treat symptoms that either are related to manifest HD or are prescribed in the pre-motor manifest HD stage for symptoms that may be attributed to HD.
○ Sufficient knowledge of the Dutch language to understand the subject information letter and to give written informed consent.
Subjects with any medical condition, in the view of the investigator, which might endanger the subject’s safety and/or satisfactory participation in the study, will be excluded from the HD-MED study. No restrictions to concomitant care, concomitant therapy, or interventions apply.
Main outcome
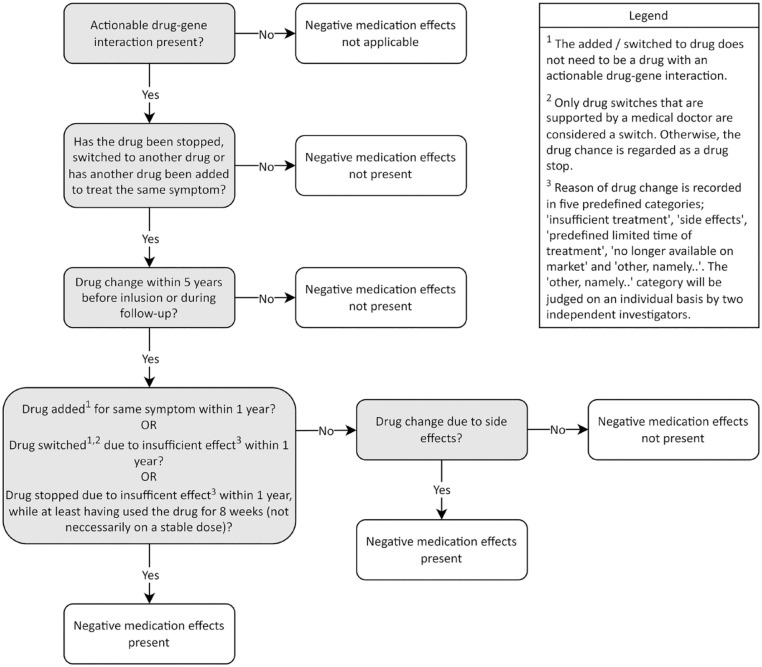
Percentage per study protocol-defined negative medication effects in HD gene expansion carriers with a CYP2C19 or CYP2D6 predicted PM and UM phenotype versus those with a predicted IM and NM phenotype. Negative medication effects are classified as non-response or side effects of the prescribed drug, which lead to definite prescription changes in that drug. Definite prescription changes are classified as follows: discontinuation of the drug, switching to another type of drug, or adding another drug to treat the same symptom. Each subject can contribute once in CYP2C19 and once in CYP2D6 to a negative medication effect.
Secondary outcomes
○ Overview of medication use by HD gene expansion carriers in 1 year. Both prescription drugs and over-the-counter medications, including supplements, are recorded. Information on drug name, dosage, start date, end date (if applicable), reasons for prescription, reasons to stop treatment (if applicable), drug efficacy, and side effects will be collected. For more details, see Section ‘Methods: Data collection, management, and analysis’.
○ Number of prescription drug discrepancies between the official pharmacy medication verification scheme and the patient-reported medication diary for the HD-MED follow-up period. Discrepancies in the amount of medications taken, naming, dosage, start date, and end date (if applicable) of the medications are reported.
Exploratory outcomes
○ Genetic variants associated with extraordinary side effect profiles or lack of medication response.
Participant timeline
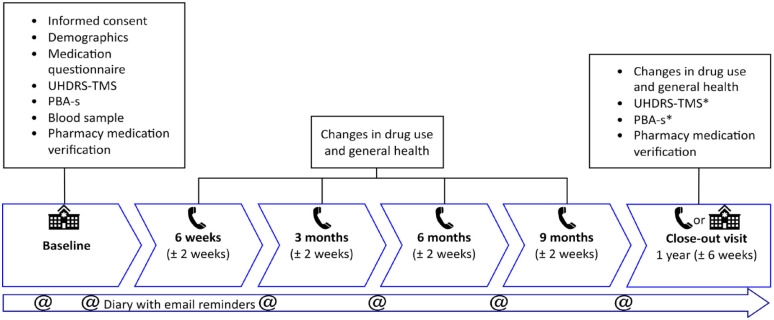
The HD-MED study has a follow-up period of 1 year, in which six visits are scheduled. The baseline visit will always be in the neurology outpatient clinic. Successive visits are phone contacts and the last visit may either be in the clinic or can be a phone contact when this is more convenient for the subject or necessary due to COVID-19 restrictions. Figure 1 provides a timeline and overview of study procedures. Details on all assessments and study instruments are shown in the Section ‘Methods: Data collection, management, and analysis’.
Figure 1.
Study timeline and overview of study procedures.
*Only performed during in-clinic visits.
PBA-s, Problem Behaviors Assessment-short; UHDRS-TMS, Unified Huntington’s Disease Rating Scale – Total Motor Score.
Sample size
CYP2C19 and CYP2D6 are currently the most important pharmacogenes in HD. Frequently prescribed drugs in HD, including antidepressants and neuroleptics, show an interaction with either of these CYP genes (see also Table 1). 14 Approximately 60% of HD patients are using one or more drugs with an actionable variant in these two pharmacogenes. 15 Records from our own HD outpatient clinic with patients in a more advanced stage of the disease show that these numbers are even up to 80%. In the general Dutch population, approximately 7% have a CYP2C19 and/or CYP2D6 extreme phenotype, defined as, respectively, 4% and 1.5% UMs, 3% and 5% PMs. 13 Currently, at our own outpatient clinic, approximately 20% of the HD population do not show a beneficial drug effect and discontinue their medication use or need to switch to or add a different type of medication (unpublished data). It is expected, with a conservative estimate, that 50% of patients with a poor or ultra-rapid predicted CYP2C19 and CYP2D6 phenotype need medication adjustments.
Table 1.
Actionable drug–gene interactions for CYP2C19 and CYP2D6, for drugs commonly used by the HD population, including suggested dosing recommendations.
| CYP2C19 | PM | IM | UM |
|---|---|---|---|
| Citalopram | Do not exceed the following daily doses (50% of the standard maximum dose): adults <65 years: 20 mg as tablets or 16 mg as drops, adults 65 years or older: 10 mg as tablets or 8 mg as drops. | Do not exceed the following daily doses: adults <65 years: 30 mg as tablets or 22 mg as drops, adults 65 years or older: 15 mg as tablets or 10 mg as drops. | No action is needed for this gene–drug interaction. The gene variation increases the conversion of citalopram to a weakly active metabolite. However, there is no significant effect on the plasma concentration of citalopram, the tolerance, or the response. |
| Clopidogrel a | Determine the level of inhibition of platelet aggregation by clopidogrel. Consider an alternative for poor responders. Prasugrel and ticagrelor are not metabolized by CYP2C19 (or to a lesser extent). | No action is required. | The genetic variation results in increased conversion of clopidogrel to the active metabolite. However, this can result in both positive effects (reduction in the risk of serious cardiovascular and cerebrovascular events) and negative effects (increase in the risk of bleeding). |
| Escitalopram | Do not exceed the following doses (50% of the standard maximum dose): adults <65 years: 10 mg/day, adults 65 years or older: 5 mg/day. | Do not exceed the following doses (75% of the standard maximum dose): adults <65 years: 15 mg/day, adults 65 years or older: 7.5 mg/day. | Avoid escitalopram. Antidepressants that are not metabolized or that are metabolized to a lesser extent by CYP2C19 are, for example, paroxetine or fluvoxamine. |
| Imipramine b | Use 70% of the standard dose and monitor the effect and side effects of the imipramine and active metabolite desipramine plasma concentrations to determine the maintenance dose or avoid imipramine. Antidepressants that are not or to a lesser extent metabolized by CYP2C19 include, for example, nortriptyline, fluvoxamine, and mirtazapine. | No action is required. | No action is required. |
| Sertraline | Do not give doses exceeding 75 mg/day. Guide the dose by the response and side effects and/or sertraline plasma concentration. | No action is required. The gene variation has a minor effect on the sertraline plasma concentration. No effect on side effects was found. | No action is required. The gene variation has a negligible effect on the plasma concentration of sertraline. Moreover, no significant effect on response and side effects has been found. |
| CYP2D6 | PM | IM | UM |
| Amitriptyline | Use 70% of the standard dose and monitor the effect and side effects or the plasma concentrations of amitriptyline and nortriptyline to adjust the maintenance dose. | Use 75% of the standard dose and monitor the efficacy and side effects or the plasma concentrations of amitriptyline and nortriptyline to adjust the maintenance dose. | Increase the dose to 1.4 times the standard dose, monitor the effect and side effects of the plasma concentrations, and be alert to increased plasma concentrations of the cardiotoxic Z-10-hydroxy metabolites. If a dose increase is not desirable due to the cardiotoxic hydroxy metabolite, avoid amitriptyline. Consider anti-depressants that are not metabolized by CYP2D6, or to a lesser extent, such as citalopram and sertraline. |
| Aripiprazole | Administer no more than 10 mg/day or 300 mg/month (68–75% of the standard maximum dose of aripiprazole). | No action is needed for this gene–drug interaction. The genetic variation increases the plasma concentration of the sum of aripiprazole and the active metabolite dehydroaripiprazole to a limited degree. There is insufficient evidence that this increases the risk of side effects. | No action is needed for this gene-drug interaction. The genetic variation decreases the plasma concentration of the sum of aripiprazole and the active metabolite dehydroaripiprazole to a limited degree. There is no evidence that this increases the risk of reduced effectiveness. |
| CYP2D6 | PM | IM | UM |
| Clomipramine | For depression, use 50% of the standard dose. For anxiety disorders and obsessive-compulsive disorder, use 50% of the standard dose if side effects occur. Monitor the effect and side effects or the plasma concentrations of clomipramine and desmethylclomipramine to set a maintenance dose. If dose reduction does not have the desired effect, avoid clomipramine. | Use 70% of the standard dose and monitor the effect and side effects or the plasma concentrations of clomipramine and desmethylclomipramine. | Use 1.5 times the standard dose and monitor the effect and side effects of the plasma concentrations of clomipramine and desmethylclomipramine to set the maintenance dose. If a dose increase is not wanted due to potential cardiotoxic hydroxy metabolites, avoid clomipramine. |
| Codeine | For cough, no action is required. For pain, choose an alternative. Do not select tramadol, as this is also metabolized by CYP2D6. Morphine is not metabolized by CYP2D6. Oxycodone is metabolized by CYP2D6 to a limited extent, but this does not result in differences in analgesia in patients. | For cough, no action is required. For pain, try a dose increase in the case of inadequate effectiveness. Choose an alternative if this does not work. Do not select tramadol, as this is also metabolized by CYP2D6. Morphine is not metabolized by CYP2D6. Oxycodone is metabolized by CYP2D6 to a limited extent, but this does not result in differences in analgesia in patients. | Doses >20 mg every 6 h for adults and 10 mg every 6 h for children aged 12 years or older and/or additional risk factors, such as co-medication with CYP3A4 inhibitors and/or reduced kidney function: Codeine is contra-indicated, if possible select an alternative: For cough, noscapine is not metabolized by CYP2D6. For pain, see alternatives under IM. |
| Doxepin | Use 40% of the standard dose and monitor the effect and side effects or the plasma concentrations of doxepin and nordoxepin to set the maintenance dose. | Use 80% of the standard dose and monitor the effect and side effects or the plasma concentrations of doxepin and nordoxepin to set the maintenance dose. | Double the standard dose and monitor the effect and side effects or the plasma concentrations of doxepin and nordoxepin to set the maintenance dose. If a dose increase is not wanted due to the potentially cardiotoxic hydroxy metabolites, avoid doxepin. |
| Haloperidol | Use 60% of the standard. This gene variation increases the plasma concentration 1.7 times, indicating an increased risk for side effects. | No action is required. | Use 1.5 times the standard dose or choose an alternative. Antipsychotics that are not metabolized by CYP2D6 – or to a much lesser extent – include, for example, flupentixol, penfluridol, quetiapine, olanzapine, or clozapine. |
| Imipramine b | Use 30% of the standard dose and monitor the effect and side effects or the plasma concentrations of imipramine and desipramine to set the maintenance dose. | Use 70% of the standard dose and monitor the effect and side effects or the plasma concentrations of imipramine and desipramine to set the maintenance dose. | Use 1.7 times the standard dose and monitor the effect and side effects or the plasma concentrations of imipramine and desipramine to set the maintenance dose. If a dose increase is not wanted due to the potentially cardiotoxic hydroxy metabolites, avoid imipramine. |
| Metoprolol | If a gradual reduction in heart rate is desired, or in the event of symptomatic bradycardia, use smaller steps in dose titration and/or prescribe no more than 25% of the standard dose. | If a gradual reduction in heart rate is desired, or in the event of symptomatic bradycardia, use smaller steps in dose titration and/or prescribe no more than 50% of the standard dose. | Use the maximum dose for the relevant indication as a target dose. Increase the dose based on effectiveness and side effects to 2.5 times the standard dose or select an alternative. |
| Nortriptyline | Use 40% of the standard dose and monitor the effect and side effects or the plasma concentration of nortriptyline to set the maintenance dose. | Use 60% of the standard dose and monitor the effect and side effects or the plasma concentration of nortriptyline to set the maintenance dose. | Use 1.7 times the standard dose and monitor the effect and side effects or the plasma concentration. Be alert to an increase in the cardiotoxic metabolite Z-10-hydroxynortriptyline. Alternatively, avoid nortriptyline. |
| CYP2D6 | PM | IM | UM |
| Paroxetine | No action is required. | No action is required. | It is not possible to offer substantiated advice for dose adjustment based on the literature. Efficacy is likely to be absent. Avoid paroxetine. |
| Pimozide | Use no more than 10 mg/day in adults (50% of the standard maximum dose). | Use no more than 16 mg/day in adults (80% of the standard maximum dose). | No action is required. |
| Risperidone | Use 67% of the standard dose. If problematic side effects originating in the central nervous system occur despite this reduced dose, then reduce the dose further to 50% of the standard dose. | No action is required. | Choose an alternative or titrate the dose according to the maximum dose for the active metabolite paliperidone. |
| Tramadol | It is not possible to provide a recommendation for dose adjustment, because the total analgesic effect changes when the ratio between the mother compound and the active metabolite changes. Try a dose increase or choose an alternative (see codeine) in the case of inadequate effectiveness. | Try a dose increase or choose an alternative (see codeine) in the case of inadequate effectiveness. | As the total analgesic effect changes when the ratio between the mother compound and the active metabolite changes, the effect of a dose reduction cannot be predicted with certainty. Select an alternative (see codeine). If an alternative is not possible, use 40% of the standard dose. |
| Venlafaxine | Avoid venlafaxine or reduce the dose. Monitor the effect and side effects or check the plasma concentrations of venlafaxine and O-desmethylvenlafaxine. Antidepressants that are not metabolized by CYP2D6 – or to a lesser extent – include, for example, duloxetine, mirtazapine, citalopram, and sertraline. | Avoid venlafaxine or reduce the dose. Monitor the effect and side effects or check the plasma concentrations of venlafaxine and O-desmethylvenlafaxine. Antidepressants that are not metabolized by CYP2D6 – or to a lesser extent – include, for example, duloxetine, mirtazapine, citalopram, and sertraline. | No action is required. |
| Zuclopenthixol | Use 50% of the standard dose. | Use 75% of the standard dose. | Try a dose increase if the effectiveness is insufficient. Do not exceed 1.5 times the standard dose. |
Other recommendations apply in case of percutaneous coronary intervention, stroke, and transient ischemic attack.
Consult a pharmacist when both CYP2C19 and CYP2D6 polymorphic variants are present.Normal metabolizers are not included since they are considered the reference category that does not require specific clinical recommendations. Guidelines and content are subject to updates and modifications, and users should confirm they are accessing the most current content.
IM, intermediate metabolizer; PM, poor metabolizer; UM, ultra-rapid metabolizer.
By performing a sample size calculation on two proportions with a two-sided test, at least 216 participants are needed (power 0.80, alpha 0.05, proportion group A 0.2, proportion group B 0.5, ratio between sample sizes of the two groups 7):
24 participants with a poor or ultra-rapid predicted CYP2C19 phenotype;
24 participants with a poor or ultra-rapid predicted CYP2D6 phenotype and;
168 participants with an intermediate or normal predicted CYP2C19/CYP2D6 phenotype.
The number of participants needed with a poor or ultra-rapid predicted CYP2C19 and/or CYP2D6 phenotype is dependent on the sampling ratio. However, group size B will only differ slightly (N = 23) when adjusting for the real-world sampling ratio of 100/7 ≈ 14.3. To retain the option to not include too many patients in this study and to adhere to the unequal distribution of PGx phenotypes in the general population, we have chosen to set the sampling ratio to 7 and to include a set number of participants as mentioned above. This study will continue to include participants until the 24-24-168 distribution is achieved. However, the investigators are aware that, since there is no method to assess beforehand who has a poor or ultra-rapid predicted CYP2C19 or CYP2D6 phenotype, more subjects with an intermediate or normal predicted CYP2C19 and/or CYP2D6 phenotype than n = 168 will be sampled. These additional subjects will be of high value by better assessing the prevalence of different drugs used in the HD population, which is one of the secondary objectives.
Overall, it is expected that approximately 100/7 × 24 ≈ 343 controls, 24 participants with a poor or ultra-rapid predicted CYP2C19 phenotype, and 24 participants with a poor or ultra-rapid predicted CYP2D6 phenotype are required. In total, 391 subjects will be included in the HD-MED study.
Recruitment
Since HD is a rare disease that requires expert knowledge for treatment, subjects will be primarily recruited from the specialized HD outpatient clinics in the Netherlands. These clinics are located at the participating study sites; Leiden University Medical Center, Maastricht University Medical Center + and University Medical Center Groningen. To increase nationwide coverage, an update on the start of inclusion will be published on HD-specific websites and by advertisement in the magazine of the Dutch HD patient organization. Those eligible to participate will be informed about the study by the coordinating investigator or research assistant. Individuals who show an interest in the study will receive the subject information letter and have the opportunity to ask questions, either to one of the investigating researchers and/or the independent expert before obtaining informed consent. The total inclusion period of the study is dependent on the rate of inclusion.
Methods: Data collection, management, and analysis
Demographic and clinical variables
The following demographic variables will be recorded: sex, age, CAG-repeat length, height, weight, self-reported ethnicity, medical history, smoking status, recreational drug use and alcohol, and caffeine and citrus fruit consumption. If applicable, comorbidities and previous surgeries are recorded as well. HTT CAG-repeat length will be verified by the official laboratory report from the electronic patient file. To assess motor symptoms of HD, the established and widely used Unified Huntington’s Disease Rating Scale – Total Motor Score (UHDRS-TMS) will be used. 16 Each rater is trained and certified to assess this scale. Training and examination have to be performed every year. Scores range from 0 (no dysfunction) to 124 (severe dysfunction).
Neuropsychological assessments
The short version of the Problem Behaviors Assessment (PBA-s) is a semi-structured interview that evaluates the neuropsychologic and behavioral state of participants. 17 It focuses on 11 symptoms of HD: depression, suicidal ideation, anxiety, irritability, aggressive behavior, apathy, perseveration, obsessions and compulsions, paranoid thinking, hallucinations, and disorientation. PBA-s will be administered to objectively assess and pursue neuropsychologic problems. Raters are certified neuropsychologists. Currently, no effective pharmacological treatment against cognitive decline in HD exists. 18 Therefore, no cognitive measurements will be performed.
Medication questionnaires and diary
At baseline, a medication questionnaire will be filled in. This will be done together with the subject to improve data quality and completeness. To promote recruitment, the subject may complete the questionnaire at home when he/she is unable to fill in the questionnaire with an investigator. Return envelopes will be provided. If necessary, an additional phone call for clarification may be performed. All participants will be queried about their complete current medication use and previous HD-specific medication use. Both prescription drugs and over-the-counter medications, including supplements and occasional drugs, are recorded. Information on drug name, dosage, start date, end date (if applicable), reasons for prescription, reasons to stop treatment (if applicable), drug efficacy, and side effects will be collected. Reasons to stop treatment are recorded in predefined categories; ‘insufficient effect’, ‘side effects’, ‘predefined limited time of treatment’, ‘no longer available on market’, and ‘other, namely. . .’. Drug efficacy is rated on a six-item scale (0 = no effect; 1 = some effect; 2 = moderate effect; 3 = sufficient effect; 4 = good effect; and 5 = perfect effect). Information on side effects includes a description of the side effect, its frequency of occurrence, start date, end date (if applicable), severity of the side effect, patient’s level of certainty that the reported side effect is a true side effect of the medication taken and whether the reported side effect is a documented side effect according to the drug information leaflet. Frequency of occurrence is reported in six categories: ‘never’, ‘seldom (less than once a week)’, ‘sometimes (1–4 times a week)’, ‘regularly (most days/4–6 days in a week)’, ‘every day’, and ‘always (present during the whole day)’. The severity of the side effect is reported in four categories: ‘not severe’, ‘mild’, ‘moderate’, and ‘extremely severe’. Patients report their certainty on whether the experienced side effect is a true side effect on a five item Likert scale: ‘very unsure’, ‘slightly unsure’, ‘don’t know’, ‘slightly sure’, and ‘very sure’. In this study protocol, patient-reported data are leading. However, the causality of the adverse drug reactions will be assessed by a medical doctor. When in doubt about whether or not a causal relation exists between a reported side effect and medication, two medical experts will be consulted until a consensus is reached.
Subjects are asked to record any changes in medication use in their personal, paper medication diary. A translated copy of the medication diary (Dutch to English) is included as Supplementary Material A. The diary does not need to be filled in daily but provides an easy-to-use handout and acts as a mnemonic for detailed information regarding the medication changes. Investigators will query subjects on their diaries when follow-up questionnaires are performed by phone. In case of lost or filled diaries, additional ones can be sent via post. A semi-structured follow-up questionnaire will take place after 6 weeks and after 3, 6, 9, and 12 months via telephone. This questionnaire will address changes in general health and medication use compared to the previous visit. Up to five phone call attempts will be performed, and voicemail messages will be left if the subject cannot be reached at the first attempt. A letter will be sent to the subject’s home address as a final attempt to prevent loss of follow-up.
At baseline and after 1 year, a medication verification scheme will be requested at the subjects’ local pharmacy, to verify drug use at the beginning of the study and during follow-up. To further improve subject involvement and minimize recall bias a total of six reminders via email will be sent in-between visits, at 2, 4, 10, 19, 32, and 45 weeks. All tests will be performed by qualified site staff. Study procedures are summarized in Figure 1.
Pharmacogenetic laboratory procedures
A blood sample (10 mL EDTA tube) for PGx analysis will be drawn at baseline. DNA will be isolated from whole blood according to automated PerkinElmer’s Chemagen magnetic particles based on polyvinyl alcohol technology. Based on current evidence, the frequency of prescribed HD-related medication and in line with the CPIC and DPWG guidelines on pharmacogenetics, CYP2C19, and CYP2D6 polymorphisms will be tested using Agena Bioscience’s VeriDose® Core Panel combined with copy number variation analysis via long-range polymerase chain reaction to detect CYP2D6 duplications and deletions.19,20 An overview of the analyzed polymorphisms is included as Supplementary Material B. Routine diagnostic workflow procedures from the clinical pharmacogenomics laboratory in the Leiden University Medical Center will be followed. All blood samples will be analyzed at the end of the study, to ensure comparability of data and guarantee cost- and time-efficient results. The subject’s pharmacogenetic profile will not be used to inform or alter treatment decisions during the study. Subjects will be classified according to their CYP2C19 and CYP2D6 predicted phenotype in four categories: PM, IM, NM, and UM. Details on actionable drug-gene interactions for CYP2C19 and CYP2D6 in HD patients that should require dose adjustments based on the Clinical Pharmacogenetics Implementation Consortium (CPIC) and DPWG (Royal Dutch Pharmacists Association – Pharmacogenetics Working Group) guidelines are summarized in Table 1. With future identifications of novel variations between variant alleles and clinically relevant drug responses, this pharmacogenetic panel can be expanded accordingly during the study for all subjects. Drug–drug interactions and corresponding co-medication-induced phenoconversion will be taken into account if present, according to the predictions23,24 displayed in Table 2. According to the exploratory objective, subjects who either have a remarkable side effect profile or do not have any drug response will be more extensively genetically characterized. Additional consent for this analysis will be required. In case of incidental significant findings, the investigator will inform the participant and treating physician to plan appropriate follow-up care. After study participation, the participant’s pharmacogenetic profile will be recorded in their electronic patient file and reported to their local pharmacy and general practitioner to tailor future drug prescriptions and improve clinical care.
Table 2.
Phenoconversion for CYP2C19 and CYP2D6 phenotypes.
| Enzyme | Genotype-predicted phenotype | Phenotype predicted following drug-induced phenoconversion |
|---|---|---|
| In the presence of a strong inhibitor | ||
| CYP2C19 | PM | PM a |
| CYP2C19 | IM, NM | PM |
| CYP2C19 | UM | IM |
| CYP2D6 | PM | PM a |
| CYP2D6 | IM, NM | PM |
| CYP2D6 | UM | PM |
| In the presence of a moderate inhibitor | ||
| CYP2C19 | PM | PM a |
| CYP2C19 | IM, NM | PM |
| CYP2C19 | UM | IM |
| CYP2D6 | PM | PM a |
| CYP2D6 | IM, NM | IM |
| CYP2D6 | UM | NM or UM b |
| In the presence of an inducer c | ||
| CYP2C19 | PM | PM a |
| CYP2C19 | IM | NM |
| CYP2C19 | NM | UM |
| CYP2C19 | UM | UM d |
Genotype-predicted CYP2C19 and CYP2D6 PMs are not affected by inhibitors or inducers and the phenotype remains as PM.
Dependent on the adjusted genotype activity score.
There are no known moderate or strong inducers for CYP2D6.
Genotype-predicted UMs are likely to be further induced; however, the phenotype remains as UM.
IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultra-rapid metabolizer.
Data management
Data will be recorded on paper forms and combined in an electronic data capture system provided by Castor Ciwit Private Limited company. The Castor database is compliant with ISO 27001, ISO 9001, FDA 21 CFR Part 11, EU Annex 11, ICH E6 Good Clinical Practice, European General Data Protection Regulation, and the U.S. Health Insurance Portability and Accountability Act. Data verification steps, such as range control for age, weight, and CAG-repeat length are built in to promote data quality. Medical information (e.g. CAG-repeat length and medical history) that is important for this study will be retrieved from and/or cross-checked in the hospital’s electronic patient files. If a subject does not have a patient file at the including site, details on their medical history can be retrieved from their healthcare provider, if the subject has given consent. All data will be encrypted with a unique participant identification code and therewith pseudonymized. The de-anonymization code is only available for a limited number of authorized investigating personnel. These include the coordinating investigator, principal investigator, and research nurse. Names of personnel who have access to the password-protected, de-anonymization coding file, will be listed in a separate file. To perform quality assessments and in line with current legislation, deanonymized data is accessible to the monitor and regulatory national bodies such as the Dutch Health and Youth Care Inspectorate. Blood samples will be likewise anonymized with the corresponding participant code and stored in freezers inside the Leiden University Medical Center. Data will be stored for 15 years in accordance with the privacy regulations of the Leiden University Medical Center, the Dutch Act on Implementation of the General Data Protection Regulation, and the European Union General Data Protection Regulation.
Statistical methods
Demographic data will be analyzed using parametric and nonparametric tests when applicable. For each pharmacogene (CYP2C19 or CYP2D6), subjects are divided into four groups based on a divergent phenotype (yes/no) and the presence of per study protocol-defined negative medication effects (yes/no). In this study protocol, a divergent phenotype is classified as either PM or UM. Negative medication effects are classified as non-response or severe side effects of the prescribed drug, leading to definite prescription changes in that drug. Definite prescription changes are classified as follows: discontinuation of the drug, switching to another type of drug, or adding another drug to treat the same symptom. Using these definitions, actionable drugs are clustered in pharmacogene groups, either CYP2C19 or CYP2D6. Each subject can contribute only once to a negative medication effect in either pharmacogene to prevent effect measure inflation. When a subject is receiving more than one actionable drug, the drug that leads to a negative medication effect will be used in the analysis. Subjects with multiple negative medication effects and/or an extraordinary side effect profile will be looked at in closer detail as part of the exploratory objectives. A standardized operating procedure to determine whether negative medication effects have occurred is available in Figure 2. To test the assumption that subjects with a poor or ultra-rapid predicted phenotype experience more negative side effects, a two-sided parametric χ2 test, or if necessary its nonparametric counterpart will be performed. In addition, a logistic multivariate regression analysis with covariates for age, gender, and concomitant medication will be performed for both CYP2C19 and CYP2D6. To examine the completeness of the paper home diary versus standardized pharmacy medication history surveys a χ2 test will be performed. To investigate the exploratory objective regarding the effect of the PGx profile in those who are outliers regarding drug dose versus severity of the symptom prescribed for, a similar logistic multivariate regression will be performed. Defining when a subject is regarded as an outlier is based on clinical consensus between two HD specialists, who are unaware of the PGx profile of the subjects involved. In case of sufficient subjects, the exploratory objective ‘to explore associations between genetic variants and extra-ordinary side effect profiles or lack of medication response in HD gene expansion carriers’ will be statistically analyzed using unpaired t-tests with Bonferroni correction for multiple testing. In case of missing data, the reason for missingness will be recovered (if possible). Complete case analysis and an analysis with imputation of conservative estimates (missingness imputed with no effect variables) will be performed. The final appropriate statistical models will be selected based on the characteristics and distribution of the variables.
Figure 2.
Standardized operating procedures to classify negative medication effects. To complete separately for actionable drug–gene interactions in CYP2C19 and CYP2D6.
Methods: Monitoring
Based on the nature and procedures of this study, a monitoring plan has been drafted before study initiation. The HD-MED study has been recognized by its judging medical research ethics committee (MREC) as a low-risk profile study. Blood draws are the single invasive procedure in this study. Therefore, the accredited MREC has given dispensation for the need to conclude an additional research subject insurance covering damage to participants through injury or death caused by the study. Furthermore, no Data Safety Monitoring Board will be installed since injury to subjects is negligible. Instead, independent monitoring will be performed by the Leiden University Medical Center monitoring group when 10% of participants are included. Monitoring criteria are described in the aforementioned monitoring plan and include but are not limited to the informed consent procedure, inclusion and exclusion criteria, source data verification, accuracy and completeness of the trial master file, and general research practice. Results will be described in a formal written report and sent to the accredited MREC. No interim analyses will be performed. Yearly progress reports will detail recruitment progress, premature study termination by patients, and the occurrence of (serious) adverse events ((S)AE). If required, additional monitoring can be compelled by the MREC.
All drugs in this study are officially registered for human use. Since drug use in routine care will be observed without any study-related drug interventions, no additional safety issues are expected. Side effects of routine-care prescribed drugs will be investigated in this study and therefore not considered (S)AEs. However, to support safety monitoring, SAEs not directly related to routine-care prescribed drugs and spontaneously reported to the investigator or her staff will be recorded. Only adverse events related to the invasive study procedure (blood draw) will be regarded as SAE. All (S)AEs will be followed until they have abated, or until a stable situation has been reached. In accordance with section 10, subsection 4, of the Medical Research Involving Human Subjects Act, the study will be suspended if there is sufficient ground that continuation of the study will jeopardize the subject’s health or safety. The accredited MREC will be notified without undue delay of a temporary halt including the reason for such an action. The study will be suspended pending a further positive decision by the accredited MREC. The investigator will take care that all subjects are kept informed.
Collaboration Leiden Huntington’s Disease Biobank
The Leiden Huntington’s Disease Biobank is initiated together with the current HD-MED protocol to build a structured and durable registry for medical data and biological specimens of Dutch HD patients and healthy controls. All patients who will be included in the HD-MED study are asked to participate in the Leiden Huntington’s Disease Biobank. If consent is given, medication details and PGx profiles are incorporated into this biobank. Various data, including but not limited to HD disease characteristics and biological specimens (blood and cerebrospinal fluid when collected for other HD studies) will be stored for an unlimited amount of time. Every year, data will be updated and biological specimens collected. From the samples, proteins, lipids, metabolites, inflammation markers, peripheral blood mononuclear cells, DNA and microRNA can be analyzed. Signs of hemolysis and/or any deviation from the standardized operating procedures will be recorded. The MREC Leiden Den Haag Delft approved the Leiden Huntington’s Disease Biobank under number B19.055. Written informed consent will be obtained from all patients. Optional consent includes information on the date and cause of death (if applicable). For additional information and/or collaboration, author Susanne T. de Bot may be contacted.
Supplemental Material
Supplemental material, sj-pdf-1-trd-10.1177_26330040231204643 for Study protocol of the HD-MED study aiming to personalize drug treatment in Huntington’s disease: a longitudinal, observational study to assess medication use and efficacy in relation to pharmacogenetics by Stephanie Feleus, Maaike van der Lee, Jesse J. Swen, Raymund A. C. Roos and Susanne T. de Bot in Therapeutic Advances in Rare Disease
Supplemental material, sj-pdf-2-trd-10.1177_26330040231204643 for Study protocol of the HD-MED study aiming to personalize drug treatment in Huntington’s disease: a longitudinal, observational study to assess medication use and efficacy in relation to pharmacogenetics by Stephanie Feleus, Maaike van der Lee, Jesse J. Swen, Raymund A. C. Roos and Susanne T. de Bot in Therapeutic Advances in Rare Disease
Acknowledgments
We thank all patients who have contributed and will contribute to this study. Without their efforts and time spent, this study would not have been possible. We also thank Wessel van der Zande for his assistance in laboratory analyses. Furthermore, we would like to thank all staff involved. In specific we would like to thank Maraike A. Coenen and professor H.P.H. Kremer from the University Medical Center Groningen and Daisy M.J. Ramakers and Mayke Oosterloo from the Maastricht University Medical Center + for their collaboration. Authors Stephanie Feleus and Susanne T. de Bot are members of the European Reference Network for Rare Neurological Diseases – Project ID No 739510.
Footnotes
ORCID iDs: Stephanie Feleus  https://orcid.org/0000-0002-1160-0920
https://orcid.org/0000-0002-1160-0920
Jesse J. Swen  https://orcid.org/0000-0002-3965-5552
https://orcid.org/0000-0002-3965-5552
Susanne T. de Bot  https://orcid.org/0000-0002-3512-2468
https://orcid.org/0000-0002-3512-2468
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Stephanie Feleus, Department of Neurology, Leiden University Medical Center, Leiden, The Netherlands; Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands.
Maaike van der Lee, Department of Clinical Pharmacy and Toxicology, Leiden University Medical Center, Leiden, The Netherlands.
Jesse J. Swen, Department of Clinical Pharmacy and Toxicology, Leiden University Medical Center, Leiden, The Netherlands
Raymund A. C. Roos, Department of Neurology, Leiden University Medical Center, Leiden, The Netherlands
Susanne T. de Bot, Department of Neurology, Leiden University Medical Center, Leiden, The Netherlands
Declarations
Ethics approval and consent to participate: This study will be conducted according to the World Medical Association of Helsinki and has been approved by the MREC Leiden Den Haag Delft based in the Leiden University Medical Center on 26 November 2019 under number NL70391.058.19. This study has been amended once from a single center to a multicenter protocol. The most recent protocol (version 2.0) has been approved by the accredited MREC on 5 October 2021. The principal investigator is responsible for communicating all important protocol modifications to relevant parties. Research staff will obtain written informed consent from all study participants. Additional optional written consent may be obtained to request medical records from other healthcare providers, for medical data and biological specimens storage and evaluation related to other HD research, and for extensive genetic analysis to identify potential new genetic polymorphisms with pharmacogenetic action. Participation is always voluntary. Subjects can leave the study at any time for any reason if they wish to do so without any consequences. Study withdrawal will not affect the clinical treatment given. Subjects do not need to inform the investigator of the reason for withdrawal. The investigator can decide to withdraw a subject from the study for urgent medical reasons.
Consent for publication: Not applicable, no identifying data from participants are used in this manuscript.
Author contributions: Stephanie Feleus: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Software; Validation; Visualization; Writing – original draft; Writing – review & editing.
Maaike van der Lee: Data curation; Formal analysis; Writing – review & editing.
Jesse J. Swen: Conceptualization; Methodology; Supervision; Writing – review & editing.
Raymund A.C. Roos: Conceptualization; Funding acquisition; Methodology; Supervision; Writing – review & editing.
Susanne T. de Bot: Conceptualization; Funding acquisition; Methodology; Project administration; Resources; Supervision; Visualization; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research is supported by private HD community donations. We received no funding from commercial or not-for-profit parties. HD-MED is an investigator-initiated study set up by the Department of Neurology and Department of Clinical Pharmacy and Toxicology of the Leiden University Medical Center.
Leiden University Medical Center receives grants from the European Huntington’s Disease Network (EHDN) and Cure HD Initiative (CHDI), participates in an EU Horizon 2020 project: Innovative Medicines Initiative (IMI) 2 (IDEA_FAST), and participates in clinical trials sponsored by PRILENIA, PTC Therapeutics, WAVE and VICO Therapeutics. The aforementioned sponsors had no role in the design, execution, interpretation, or writing of this current study protocol. Authors MvdL, JJS, and RACR declare no conflict of interest.
Availability of data and materials: The final results of the HD-MED study, irrespective of findings, will be submitted for publication in scientific journals and therewith publicly disclosed according to current international and local academic standards. Study data from the HD-MED protocol will not be publicly released since this study is still ongoing. Parties that wish to collaborate on data generated by this study protocol are encouraged to contact the corresponding author.
References
- 1. MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell 1993; 72: 971–983. [DOI] [PubMed] [Google Scholar]
- 2. Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat rev Dis Primers 2015; 1: 15005. [DOI] [PubMed] [Google Scholar]
- 3. McColgan P, Tabrizi SJ. Huntington’s disease: a clinical review. Eur J Neurol 2018; 25: 24–34. [DOI] [PubMed] [Google Scholar]
- 4. Roos RAC. Clinical neurology. In: Bates GP, Tabrizi SJ, Jones L. (eds) Huntington’s disease. 4th ed. New York: Oxford University Press, 2014, pp.25–35. [Google Scholar]
- 5. Tabrizi SJ, Estevez-Fraga C, van Roon-Mom WMC, et al. Potential disease-modifying therapies for Huntington’s disease: lessons learned and future opportunities. Lancet Neurol 2022; 21: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huntington Study Group. Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurology 2006; 66: 366–372. [DOI] [PubMed] [Google Scholar]
- 7. Coppen EM, Roos RA. Current pharmacological approaches to reduce chorea in huntington’s disease. Drugs 2017; 77: 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keogh R, Frost C, Owen G, et al. Medication use in Early-HD participants in track-HD: an investigation of its effects on clinical performance. PLoS Curr 2016; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corvol JC, Devos D, Hulot JS, et al. Clinical implications of neuropharmacogenetics. Rev Neurol 2015; 171: 482–497. [DOI] [PubMed] [Google Scholar]
- 10. Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte – an update of guidelines. Clin Pharmacol Ther 2011; 89: 662–673. [DOI] [PubMed] [Google Scholar]
- 11. Relling MV, Klein TE. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther 2011; 89: 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. PharmGKB. Guideline Annotations, https://www.pharmgkb.org/guidelineAnnotations (2022, accessed 28 February 2023).
- 13. Bank PCD, Swen JJ, Schaap RD, et al. A pilot study of the implementation of pharmacogenomic pharmacist initiated pre-emptive testing in primary care. Eur J Hum Genet 2019; 27: 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bank PCD, Caudle KE, Swen JJ, et al. Comparison of the guidelines of the clinical pharmacogenetics implementation consortium and the dutch pharmacogenetics working group. Clin Pharmacol Ther 2018; 103: 599–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enroll-HD Consortium. Enroll-HD database; A worldwide observational study for Huntington’s disease families. CHDI Foundation Inc. [Google Scholar]
- 16. Huntington Study Group. Unified Huntington’s disease rating scale: reliability and consistency Mov Disord 1996; 11: 136–142. [DOI] [PubMed] [Google Scholar]
- 17. Kingma EM, van Duijn E, Timman R, et al. Behavioural problems in Huntington’s disease using the problem behaviours assessment. General Hosp Psychiatry 2008; 30: 155–161. [DOI] [PubMed] [Google Scholar]
- 18. Wyant KJ, Ridder AJ, Dayalu P. Huntington’s disease-update on treatments. Curr Neurol Neurosci Rep 2017; 17: 33. [DOI] [PubMed] [Google Scholar]
- 19. Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the clinical pharmacogenetics implementation consortium and dutch pharmacogenetics working group. Clin Transl Sci 2020; 13: 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brouwer J, Nijenhuis M, Soree B, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2C19 and CYP2D6 and SSRIs. Eur J Hum Genet 2022; 30: 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clinical Pharmacogenetics Implementation Consortium (CPIC). Guidelines, https://cpicpgx.org/guidelines/ (2023, accessed 20 July 2023).
- 22. Royal Dutch Pharmacists Association – Pharmacogenetics Working Group. Kennisbank Farmacogenetica [Knowledge bank pharmacogenetics], https://kennisbank.knmp.nl/article/farmacogenetica/intro.html (accessed 22 March 2023).
- 23. Cicali EJ, Elchynski AL, Cook KJ, et al. How to integrate CYP2D6 phenoconversion into clinical pharmacogenetics: a tutorial. Clin Pharmacol Ther 2021; 110: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mostafa S, Kirkpatrick CMJ, Byron K, et al. An analysis of allele, genotype and phenotype frequencies, actionable pharmacogenomic (PGx) variants and phenoconversion in 5408 Australian patients genotyped for CYP2D6, CYP2C19, CYP2C9 and VKORC1 genes. J Neural Transm (Vienna) 2019; 126: 5–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-trd-10.1177_26330040231204643 for Study protocol of the HD-MED study aiming to personalize drug treatment in Huntington’s disease: a longitudinal, observational study to assess medication use and efficacy in relation to pharmacogenetics by Stephanie Feleus, Maaike van der Lee, Jesse J. Swen, Raymund A. C. Roos and Susanne T. de Bot in Therapeutic Advances in Rare Disease
Supplemental material, sj-pdf-2-trd-10.1177_26330040231204643 for Study protocol of the HD-MED study aiming to personalize drug treatment in Huntington’s disease: a longitudinal, observational study to assess medication use and efficacy in relation to pharmacogenetics by Stephanie Feleus, Maaike van der Lee, Jesse J. Swen, Raymund A. C. Roos and Susanne T. de Bot in Therapeutic Advances in Rare Disease