Conspectus
The development of various chemical methods has enabled scientists to decipher the distribution features and biological functions of RNA modifications in the past decade. In addition to modifying noncoding RNAs such as tRNAs and rRNAs, N6-methyladenosine (m6A) has been proven to be the most abundant internal chemical modification on mRNAs in eukaryotic cells and is also the most widely studied mRNA modification to date. Extensive studies have repeatedly demonstrated the important functions of m6A in various biological conditions, ranging from embryonic organ development to adult organ function and pathogenesis. Unlike DNA methylation which is relatively stable, the reversible m6A modification on mRNA is highly dynamic and easily influenced by various internal or external factors, such as cell type, developmental stage, nutrient supply, circadian rhythm, and environmental stresses.
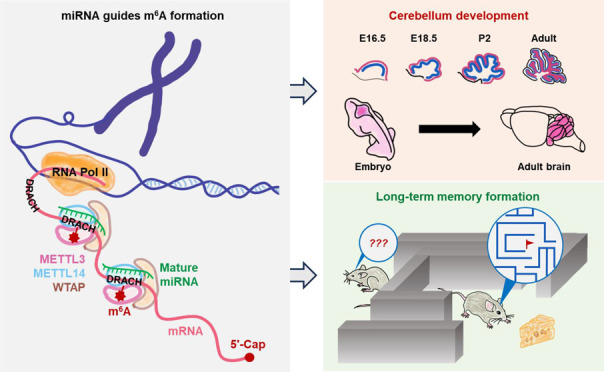
In this Account, we review our previous findings on the site selectivity mechanisms regulating m6A formation, as well as the physiological roles of m6A modification in cerebellum development and long-term memory consolidation. In our initial efforts to profile m6A in various types of mouse and human cells, we surprisingly found that the sequence motifs surrounding m6A sites were often complementary with the seed sequences of miRNAs. By manipulating the abundance of the miRNA biogenesis enzyme Dicer or individual miRNAs or mutating miRNA sequences, we were able to reveal a new role of nucleus localized miRNAs, which is to guide the m6A methyltransferase METTL3 to bind to mRNAs and to promote m6A formation. As a result, we partially answered the question of why only a small proportion of m6A motifs within an mRNA could have m6A modification at a certain time point. We further explored the functions of m6A modification in regulating brain development and brain functions. We found that cerebellum had the most severe defects when Mettl3 was knocked out in developing mouse embryonic brain and revealed that the underlying mechanisms could be attributed to aberrant mRNA splicing and enhanced cell apoptosis under m6A deficit conditions. On the other hand, knocking out Mettl3 in postnatal hippocampus did not cause morphological defects in the mouse brain but impaired the efficacy of long-term memory consolidation. Under learning stimuli, formation of m6A modifications could be detected on transcripts encoding proteins related to dendrite growth, synapse formation, and other memory related functions. Loss of m6A modifications on these transcripts would result in translation deficiency and reduced protein production, particularly in the translation of early response genes, and therefore would compromise the efficacy of long-term memory consolidation. Interestingly, excessive training sessions or increased training intensity could overcome such m6A deficiency related memory defects, which is likely due to the longer turnover cycle and the cumulative abundance of proteins throughout the training process. In addition to revealing the roles of m6A modification in regulating long-term memory formation, our work also demonstrated an effective method for studying memory formation efficacy. As the lack of an appropriate model for studying memory formation efficacy has been a long-lasting problem in the field of neural science, our hippocampus-specific postnatal m6A knockout model could also be utilized to study other questions related to memory formation efficacy.
Key References
Chen T.; Hao Y.-J.; Zhang Y.; Li M.-M.; Wang M.; Han W.; Wu Y.; Lv Y.; Hao J.; Wang L.; Li A.; Yang Y.; Jin K.-X.; Zhao X.; Li Y.; Ping X.-L.; Lai W.-Y.; Wu L.-G.; Jiang G.; Wang H.-L.; Sang L.; Wang X.-J.; Yang Y.-G.; Zhou Q.. m6A RNA Methylation Is Regulated by microRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 2015, 16, 289–301.1This study discovered the miRNA-mediated site selectivity mechanism underlying m6A modification formation. Manipulating miRNA abundance or sequences could alter the amount of m6A modification or create new m6A modification sites.
Wang C.-X.; Cui G.-S.; Liu X.; Xu K.; Wang M.; Zhang X.-X.; Jiang L.-Y.; Li A.; Yang Y.; Lai W.-Y.; Sun B.-F.; Jiang G.-B.; Wang H.-L.; Tong W.-M.; Li W.; Wang X.-J.; Yang Y.-G.; Zhou Q.. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biology 2018, 16, e2004880.2This study revealed the functions and underlying mechanisms of m6A modification in regulating cerebellar development in mouse embryonic brain. Reduced sizes and enhanced cell apoptosis were observed in m6A deficient mouse cerebellum.
Zhang Z.; Wang M.; Xie D.; Huang Z.; Zhang L.; Yang Y.; Ma D.; Li W.; Zhou Q.; Yang Y.-G.; Wang X.-J.. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Research 2018, 28, 1050–1061.3This study identified the role of m6A in modulating the efficacy of long-term memory formation. Mice without m6A in their hippocampus exhibited reduced memory ability; however, such defect could be compensated by excessive training sessions or increased training intensity.
Introduction
The development and survival of humans and other organisms are the results of coordinated functions of billions of molecules, including large biomolecules and small chemical molecules. With the advancements in chemical biology and high-throughput sequencing technologies, multiple layers of new regulatory types have been identified in cells over the past few decades. This has complicated our understanding of the regulatory mechanisms and networks that govern cellular behaviors.4,5 For example, in addition to the well-recognized regulations controlled by DNA sequences, RNA transcription, RNA splicing, and protein abundance, many new types of regulations have been discovered in the past few decades, including DNA higher structures, DNA methylation, histone codes, noncoding RNAs, and RNA modifications, and have been proven to play essential roles in all types of biological events.5−12
RNA modification refers to the addition or changes of chemical compounds to RNA molecules. For example, over 38 types of modifications on adenosine have been recorded (Figure 1).13 For only the N6 position of adenosine, there could be at least 13 types of modifications, including N6-methyladenosine (m6A), N6-hydroxymethyladenosine (hm6A), N6-acetyladenosine (ac6A), N6-formyladenosine (f6A), etc. In the recently updated version of the MODOMICS database, the number of modified RNA residue entries increased from less than 170 to 334, including 180 modified nucleotide residues, 152 modified nucleoside residues, and 3 modified bases.13,14 Among these, m6A is the most abundant internal modification found on mRNAs in eukaryotic cells.15
Figure 1.
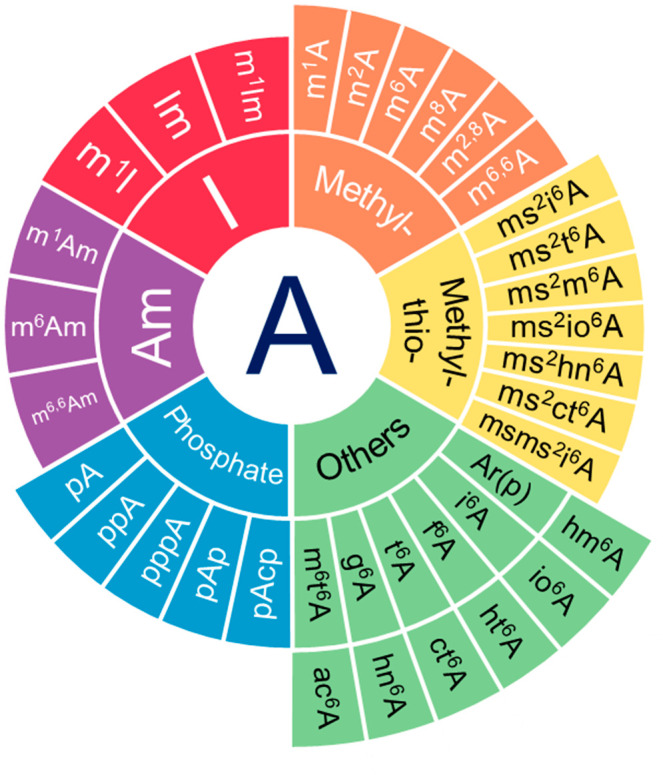
Pie chart summary of chemical modifications on adenosine documented in the MODOMICS database. Adenosine modifications are classified into 6 groups: 1. Deamination (I), 2. Methylation on adenosine (Methyl-), 3. Methylthiolation on adenosine (Methyl-thio-), 4. Methylation on 2′-O-methyladenosine (Am), 5. Phosphorylation (Phosphate), 6. Other types of chemical moieties added on adenosine (Others).
The discovery of m6A modification on RNAs can be traced back to the late 1960s;16,17 however, due to technical limitations, the distribution and functions of m6A modification were not elucidated until 2010s.18−20 With the development of antibodies against m6A modification and new biochemical methods to detect m6A modification,5 scientists nowadays are able to profile m6A modification sites genome-wide at single nucleotide resolution21 or at single cell level22,23 and also discovered the diverse functions of m6A modification in regulating organism development as well as various physiological or pathological responses. Although scientists were excited about these new discoveries, an important question remains to be addressed: how are the selectivity and dynamics of m6A modification regulated? Our previous studies have revealed some clues to these questions. In this Account, we give a review of the mechanisms underlying the site selectivity of m6A modification and the functions of m6A modification in regulating cerebellum development and long-term memory formation.
Site Selectivity of m6A Modifications
Unlike DNA methylation, modifications on RNAs are more dynamic and emerge or diminish in a more rapid manner.24,25 The formation of m6A modification is mainly catalyzed by a methyltransferase complex (known as the m6A writer complex) with METTL3, METTL14, and WTAP as the core components, of which METTL3 serves as the primary catalytic methyltransferase.26 Reversely, the demethylation of m6A is mediated by FTO or ALKBH5 (termed as m6A erasers).25 Multiple proteins within cells can recognize and bind to m6A modifications (termed as m6A readers), including the well-known group of proteins with a YTH (YT521-B homology) domain (namely YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2) and some newly identified m6A readers without a YTH domain, such as HNRNPC, HNRNPG, and IGF2BPs.27
m6A modification tends to occur on adenosine within the DRACH motif in human and mouse cells, where D represents G/A/U, R represents G/A, and H represents A/U/C. However, although the DRACH motif could theoretically occur in every 57 randomly ordered nucleotides (3/4 × 1/2 × 1/4 × 1/4 × 3/4), most RNA transcripts contain fewer than 3 m6A modification sites.28 Moreover, which DRACH motif is selected for adding m6A modification is dynamically regulated, even for transcripts from the same gene. In different cell types or the same cell type under different physiological conditions, the profiles of m6A modification could be quite different. When we started to work on m6A modification about ten years ago, how cells know which DRACH motif on mRNAs to modify with m6A was an important yet unaddressed question.
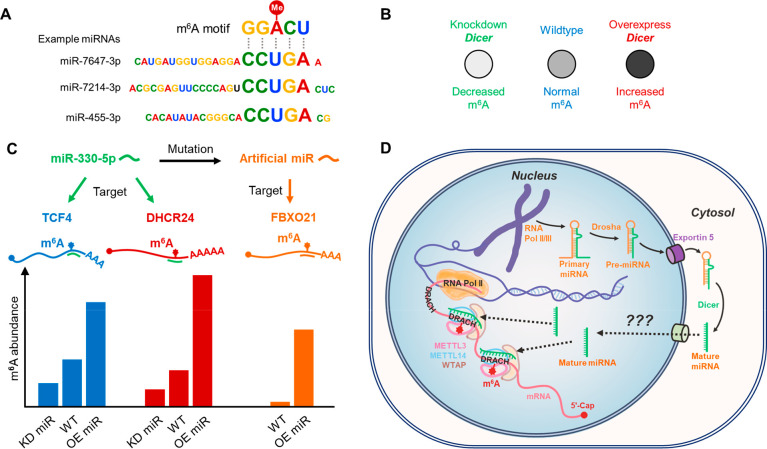
Based on our years of research experience on microRNAs (miRNAs), we tried to align the enriched m6A motifs with miRNA sequences when analyzing the m6A modification profiles in mouse embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), neural stem cells (NSCs) and testicular sertoli cells (SCs).1 We surprisingly found that most 8-nt long sequence motifs surrounding m6A modification sites exhibited reverse complementary pairing relationships with the seed sequences of miRNAs (Figure 2A). In contrast, such preference could not be found among randomly generated control sequences with similar nucleotide composition to miRNAs. Thus, we hypothesized that the selectivity of m6A modification sites might in part be mediated by miRNA binding. Inspired by this hypothesis, we first tested the effects of Dicer (the major miRNA processing enzyme29) on the overall m6A abundance in NSCs and HeLa cells. In line with our hypothesis, down-regulating Dicer expression resulted in a decrease in m6A abundance, while upregulating Dicer led to an increase in m6A level (Figure 2B).1 We further used individual miRNAs to demonstrate that manipulating miRNA levels could indeed impact the abundance of m6A on adenosines within the miRNA binding sites (Figure 2C).1 Moreover, by introducing mutations into the miRNA sequences to target sites that were previously unmodified by m6A, we successfully generated new m6A modifications on these artificial miRNA target sites (Figure 2C).1 These findings provide evidence that miRNAs could serve as guide sequences to direct site-specific m6A formation (Figure 2D).
Figure 2.
Illustration of miRNA-guided regulation on site-specific m6A formation. (A) Pairing relationships between m6A motif and example miRNAs. (B) Knockdown or overexpression of Dicer leads to decreased or increased m6A abundance in cells, respectively. (C) Alteration of m6A abundance on specific genes by miRNA. Knockdown or overexpression of miR-330-5p can specifically decrease or increase m6A abundance on its native targets TCF4 and DHCR24; mutating miR-330-5p to make it artificially target FBXO21 can create a new m6A modification site on FBXO21. (D) Proposed model for miRNA-guided regulation on site-specific m6A formation. m6A is installed co-transcriptionally inside the cell nucleus by the m6A writer complex (METTL3–METTL14–WTAP). Primary miRNAs are mostly transcribed by RNA polymerase II or III and are first processed into pre-miRNAs (stem–loop structure) in the nucleus, then transported into the cytosol and further processed into mature miRNAs. Mediated by unknown mechanisms, some mature miRNAs could be shuttled back into cell nucleus, where they bind to complementary sequences around DRACH motifs on nascent mRNAs. Such miRNA pairing can enhance the binding of m6A writer complex to mRNAs and facilitate m6A formation.
Although the above lines of evidence are quite strong, still something appears to be contradictory to the traditional understanding of miRNAs. This contradiction lies in the inconsistency between the locations of miRNAs and m6A formation. The m6A methyltransferase METTL3 is localized in the nucleus,1 whereas previous studies have shown that in mammalian cells, miRNAs are processed from their precursors to the ∼22 nt mature functional forms in the cytosol.30 Thus, it seems that the cytosolic miRNAs are unable to influence nucleic m6A formation. However, nuclear localization of some miRNAs in mouse and human cells has been detected by many research groups31 and ourselves (Figure 3), demonstrating that some miRNAs could be transported back to the nucleus after cytosolic processing, thereby having the opportunity to physically interact with the writer complex and their target DRACH sites. Indeed, we have also found that miRNAs could affect the ability of METTL3 to bind to their target mRNAs,1 further supporting the function of miRNAs in mediating m6A formation.
Figure 3.
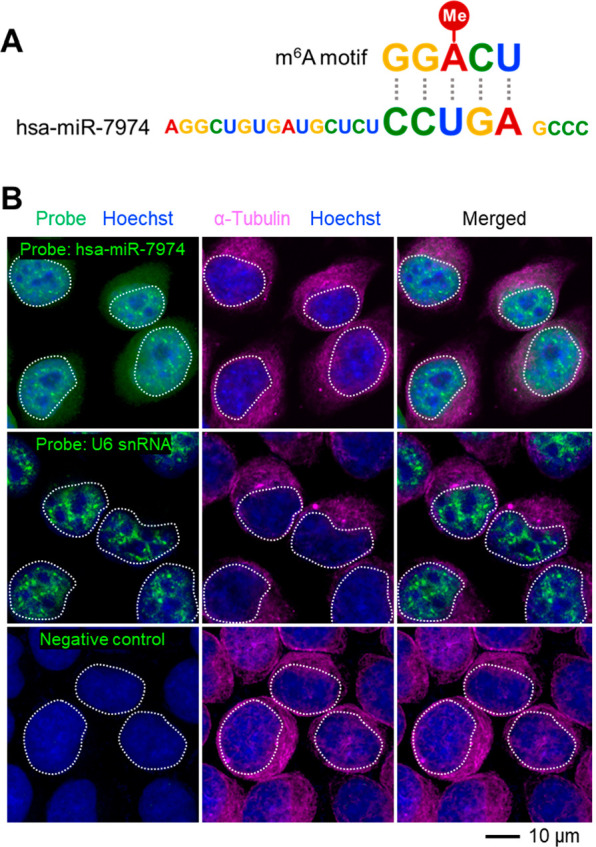
Subcellular localization of miRNA hsa-miR-7974. (A) Complementary relationship between m6A motif and hsa-miR-7974 mature sequence. (B) Fluorescent in situ hybridizations show the nuclear localization of hsa-miR-7974 in HeLa cells. Nuclei are highlighted in white dashed circles. Probes targeting nuclear localized U6 snRNA are used as positive controls; probes with no predictable targets are used as negative controls.
Later on, Prof. Jianjun Chen, Prof. Jianhua Yang, Prof. Chuan He and their colleagues reported that H3K36me3 could guide m6A formation co-transcriptionally by interacting with METTL14,32 which explains another possible mechanism that regulates the site selectivity of m6A formation. However, as H3K36me3 is a transcription elongation-associated histone modification which can be found throughout entire mRNAs,33 the H3K36me3 guidance theory still cannot explain why most mRNAs have only 1–3 m6A modification sites, although there are many other positions that could be methylated. It is possible that the sequence guidance function of miRNAs is still required in the theory of H3K36me3-directed m6A formation, further experiments are needed to explore the relationships among miRNAs, H3K36me3, and the site selectivity of m6A modification.
Requirement for m6A Modification in Cerebellum Development
The primary function of m6A modification is to regulate embryogenesis and organ formation. Mouse epiblasts or embryonic stem cells without the m6A methyltransferase METTL3 experienced early embryonic lethality;34 accordingly, our previous collaborative research also showed that knocking down Mettl3 reduced the expression of pluripotent genes (Oct4, Sox2, and Nanog) and impaired cell reprogramming efficacy.1 A large number of publications have demonstrated that m6A modification is necessary for the development of nearly every organ in mouse.24 Therefore, we directed our attention to studying the role of m6A modification in brain development.
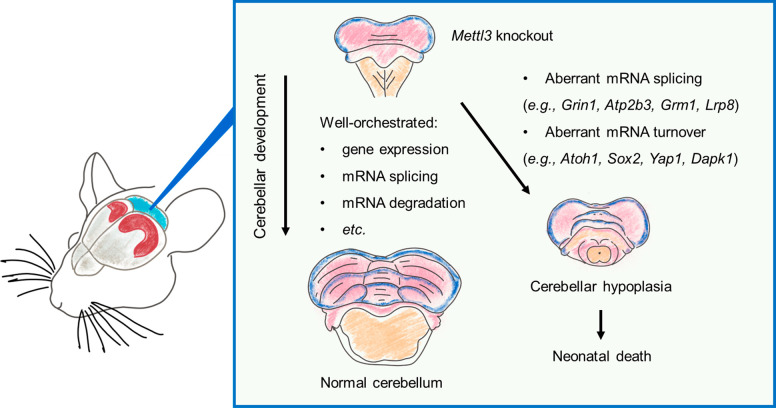
By utilizing Nestin-Cre transgenic animals,35 we were able to specifically knockout Mettl3 in the neural system (Mettl3-Nestin-cKO) of mouse embryos. Although the Mettl3-Nestin-cKO embryos were able to produce live pups, the newborns were significantly smaller in body size, had impaired movement ability, and died within 3 weeks after birth.2 Similar to previously published Mettl14-Nestin-Cre cKO results,36 the Mettl3-Nestin-cKO mice also had enlarged brain ventricles but reduced overall brain sizes, with the most significant shrinkage observed in the cerebellum compared to other brain regions.2 Such more severe developmental defects in cerebellum may be attributed to the fact that cerebellum has higher intrinsic m6A abundance than other brain regions,37 which makes it more vulnerable to Mettl3 knockout. Through various histological and molecular experimental approaches, we demonstrated that such Mettl3-Nestin-cKO related defects in the cerebellum were caused by abnormal expression and aberrant splicing of m6A modified genes involved in neural development (e.g., Atoh1, Sox2, Yap1, and Dapk1) and apoptotic signaling pathways (e.g., Grin1, Atp2b3, Grm1, and Lrp8) (Figure 4).2
Figure 4.
Cartoon summary for the functions of m6A in cerebellar development. Embryonic brain development is a complex process that must be tightly controlled by well-orchestrated gene expression, mRNA splicing, and mRNA degradation. Depletion of m6A writer Mettl3 in the embryonic mouse brain leads to aberrant mRNA splicing, turnover of essential genes, and cerebellar hypoplasia and neonatal death.
Around the same time as our work, a paper published in Genome Biology also investigated the role of m6A modification in regulating the postnatal development of mouse cerebellum.38 They observed significant changes of m6A modifications in mouse cerebellum from postnatal day 7 to day 60. Specifically, they found an increase of m6A peaks around the start codon regions of mRNAs and a decrease of m6A peaks around the stop codon regions. As expected, genes with dynamic m6A modifications during cerebellum development were enriched in functions related to cell cycle, DNA damage response, neural development, and synaptic plasticity. In line with our findings, they also observed abnormal cerebellar development when knocking down Mettl3 or knocking out the m6A eraser enzyme Alkbh5.38
m6A Mediates the Efficacy of Long-Term Memory Formation
After discovering the developmental regulatory function of m6A modification in the cerebellum, we proceeded to explore its roles in adult brains. As a dynamic RNA modification type, m6A modification has been proven to function in multiple physiological and pathological processes, such as circadian rhythm, immune responses, metabolism, and various cancers.24 We believe that the necessity of RNA modifications for cells lies in their responses to various internal or external stimuli. For adults, one of the major stimuli to the neural system is learning and memory. Thus, we hypothesized that m6A modification could play a role in memory regulation.
To test this hypothesis, we generated Mettl3 conditional knockout mice using CaMKIIα-Cre (Mettl3-CaMKIIα-cKO), which depleted Mettl3 in the excitatory neurons in hippocampus and cerebral cortex from postnatal day 1.39 The Mettl3-CaMKIIα-cKO mice exhibited normal brain morphology and motor ability, without any detectable developmental or psychological defects.3 The Mettl3-CaMKIIα-cKO mice behaved normally in short-term memory tests but showed reduced long-term memory formation efficacy in both the Morris water maize test and fear conditioning test.3 We further proved that such Mettl3 depletion associated long-term memory defects were indeed caused by the lack of m6A modification using mutagenesis of the key enzymatic site in the METTL3 methyltransferase domain.3 Interestingly, after repeated training sessions or increased training intensity (10 training times for Morris water maize test or 3 consecutive electronic shocks for fear conditioning test), the Mettl3-CaMKIIα-cKO mice performed similarly to the wild-type control mice,3 indicating that extended training sessions or increased training intensity can compensate for the memory defects caused by m6A modification deficiency.
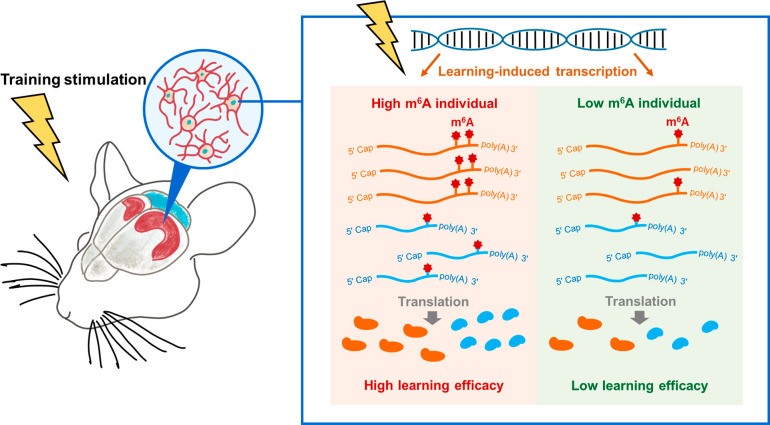
Molecularly, the m6A modification related memory consolidation is achieved by modulating the translation efficacy of genes that regulate dendrite development, synapse organization, cellular protein localization, and other processes related to memory formation, especially the translation of immediate early genes (e.g., c-Fos, Egr1, Arc, Npas4, and Nr4a1) that are essential for neuronal quick responses to learning training (Figure 5).3 By manipulating the abundance of m6A reader protein YTHDF1, another collaborative team also demonstrated that m6A facilitates hippocampus-dependent learning and memory by promoting protein translation, which aligns with our findings.40 Yet they did not observe the compensatory effects of extensive training for the lack of m6A modification, probably due to differences in m6A perturbation approaches (knockout of m6A methyltransferase Mettl3 vs knockout of m6A reader Ythdf1) or training procedures.
Figure 5.
Cartoon illustrating the positive regulatory functions of m6A on the efficacy of long-term memory formation. Behavior training will induce gene expression in mouse hippocampus. Such learning-based gene activation is essential for the brain to reshape its neural network to encode new information as memory. After learning stimulation, m6A modifications would be added to transcripts of learning-induced genes and further enhance mRNA translation, neural network long-term potentiation, and the efficacy of mouse long-term memory formation.
We also demonstrated that overexpressing Mettl3 in hippocampus could significantly improve memory ability to create “super smart” mice; however, such superiority only exists in the early training stages; after repeated training, wild-type mice could also reach the same memory level as the “super smart” mice.3 Such phenomenon is in accordance with one Chinese proverb which is “diligence can make up dullness”, and may be explained by the accumulative effects of memory-related proteins and their regulations on the neural circuits.41 Due to the longer turnover rate of proteins, the abundance of memory regulatory proteins would accumulate after each round of training. If there is a functional saturation level for proteins in regulating neural connections and long-term memory formation, under excessive training conditions, the depletion or overexpression of Mettl3 would only affect the time needed to reach the saturated protein level of m6A modified transcripts, but not the final learning outcomes. In a comment on our work written by Prof. Pico Caroni, he pointed out that the underlying mechanisms of memory strength were poorly understood; our work identified “an endogenous learning-related molecular process with a role in modulating memory strength” and thus “makes an important contribution to molecular studies of learning and memory”.42
Conclusions and Outlook
In summary, the past years of effort from our team and our collaborators have identified a class of guiders used by cells to select specific adenosine sites for adding m6A modifications when necessary and also revealed the functions of m6A in regulating cerebellum development and long-term memory formation. Based on these, there are a few questions worth exploring in the future.
First, how do miRNAs and m6A-writer/reader/eraser proteins function together to respond to different biological signals? As both m6A modification and miRNA expression play essential roles in daily physiological activities such as metabolism, circadian rhythm, learning, and immune responses,24,43 they could be highly dynamic in cells. One potential working model for the cause of such dynamic m6A changes could be that certain internal or external signals trigger the conditional generation and nuclear shuttling of miRNAs to guide m6A modifications on mRNAs. However, the mechanisms by which these signals are converted into miRNA expression and nucleus transportation remain to be investigated. Similar questions also arise regarding pathological related m6A changes. In addition, as miRNAs would affect the binding of METTL3 to mRNAs, how METTL3 can efficiently interact with specific miRNAs under different conditions also awaits attention.
Second, regarding the enhancement role of m6A in long-term memory consolidation, most of the reported works trained mice using context-dependent behavior protocols, which primarily examine the spatial memory (e.g., spatial cues in the Morris water maze test and the conditioned box in the fear conditioning test). But for human beings, learning is a complicated process that involves the combination of multiple types of memories, such as emotional memory, social memory, and implicit memory.44 Therefore, it is important to examine the extent to which m6A modification participates in other types of memory, and whether other types of RNA modifications also play roles in learning and memory formation. Knocking-out Mettl3 in mouse brain regions other than the hippocampus and designing new behavioral test protocols are also desired. In addition, due to the lack of m6A profiling information in human samples, it is unclear whether aging associated memory decline is related to reduced m6A abundance or METTL3 activity. On the other hand, designing or screening for chemical compounds that could enhance m6A modifications would be beneficial for improving long-term memory formation efficacy. However, due to the diverse functions of m6A, such a strategy should be applied with caution regarding the target specificity of m6A modifications.
Finally, most reported m6A studies either focus on the global profiling of m6A modifications or the dynamic changes of m6A on particular genes;5 little attention has been paid to the coordinated changes of m6A modifications on different genes as well as the coordinated functions of m6A and other types of modifications on the same transcripts. The latter is to a large extent constrained by technical limitations. Hopefully, with the development of more sensitive RNA modification detection methods, we will be able to uncover the relationship between m6A and other RNA modifications in the near future.
Acknowledgments
The authors would like to thank former and present members in Xiu-Jie Wang’s lab and all collaborators who participated in the research described in this Account, especially Prof. Yun-Gui Yang for his long-term collaboration.
Biographies
Zeyu Zhang received his Ph.D. in Bioinformatics from Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, in 2022 under the guidance of Prof. Xiu-Jie Wang. Then he continued his research work as a postdoctoral fellow in Prof. Wang’s lab. His major research interests include stem cell biology, neuroscience, and RNA modifications.
Xiu-Jie Wang received her Ph.D. in Bioinformatics from Rockefeller University in 2004 and joined the Institute of Genetics and Developmental Biology of Chinese Academy of Sciences as a principal investigator in the same year. She is now also a professor at the University of Chinese Academy of Sciences. A major research focus of her lab is to utilize bioinformatics and experimental approaches to decipher the functions and underlying mechanisms of noncoding RNAs and RNA modifications in mouse and human.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. CRediT: Zeyu Zhang conceptualization, formal analysis, investigation, methodology, project administration, resources, validation, visualization, writing-review & editing; Xiujie Wang conceptualization, funding acquisition, methodology, project administration, resources, supervision, writing-original draft, writing-review & editing.
We acknowledge funding from the National Key Research and Development Program of China (2019YFA0802203), Beijing Natural Science Foundation (Z200020), and Chinese Academy of Sciences for Young Scientists in Basic Research Project (YSBR-073) to X.-J.W.
The authors declare the following competing financial interest(s): The authors have filed a patent pertaining to mechanisms described in this Account.
Special Issue
Published as part of the Accounts of Chemical Research special issue “RNA Modifications”.
References
- Chen T.; Hao Y.-J.; Zhang Y.; Li M.-M.; Wang M.; Han W.; Wu Y.; Lv Y.; Hao J.; Wang L.; Li A.; Yang Y.; Jin K.-X.; Zhao X.; Li Y.; Ping X.-L.; Lai W.-Y.; Wu L.-G.; Jiang G.; Wang H.-L.; Sang L.; Wang X.-J.; Yang Y.-G.; Zhou Q. m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 2015, 16, 289–301. 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Wang C.-X.; Cui G.-S.; Liu X.; Xu K.; Wang M.; Zhang X.-X.; Jiang L.-Y.; Li A.; Yang Y.; Lai W.-Y.; Sun B.-F.; Jiang G.-B.; Wang H.-L.; Tong W.-M.; Li W.; Wang X.-J.; Yang Y.-G.; Zhou Q. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biology 2018, 16, e2004880 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Wang M.; Xie D.; Huang Z.; Zhang L.; Yang Y.; Ma D.; Li W.; Zhou Q.; Yang Y.-G.; Wang X.-J. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Research 2018, 28, 1050–1061. 10.1038/s41422-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X.; Shen L. Advances and Trends in Omics Technology Development. Frontiers in Medicine 2022, 9, 911861. 10.3389/fmed.2022.911861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.; Ma J.; Li P.; Wu Y.; Yu H. Recent advances in the plant epitranscriptome. Genome Biology 2023, 24, 43. 10.1186/s13059-023-02872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Moral-Morales A.; Salgado-Albarrán M.; Sánchez-Pérez Y.; Wenke N. K.; Baumbach J.; Soto-Reyes E. CTCF and Its Multi-Partner Network for Chromatin Regulation. Cells 2023, 12, 1357. 10.3390/cells12101357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney E. R.; Nolan C. M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- Sun H.; Li K.; Liu C.; Yi C. Regulation and functions of non-m(6)A mRNA modifications. Nat. Rev. Mol. Cell Biol. 2023, 24, 714. 10.1038/s41580-023-00622-x. [DOI] [PubMed] [Google Scholar]
- Statello L.; Guo C.-J.; Chen L.-L.; Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. J.; Stoilov P.; Xing Y. Chromatin and epigenetic regulation of pre-mRNA processing. Hum. Mol. Genet. 2012, 21, R90–R96. 10.1093/hmg/dds353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.; Jiang Y.; Hao L.; Hui J.; Xing Y. Genetic variation and microRNA targeting of A-to-I RNA editing fine tune human tissue transcriptomes. Genome Biol. 2021, 22, 77. 10.1186/s13059-021-02287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E.; Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014, 6, a019133. 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P.; Stefaniak F.; Ray A.; Cappannini A.; Mukherjee S.; Purta E.; Kurkowska M.; Shirvanizadeh N.; Destefanis E.; Groza P.; Avşar G.; Romitelli A.; Pir P.; Dassi E.; Conticello S. G.; Aguilo F.; Bujnicki J. M. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–d235. 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P.; Machnicka M. A.; Purta E.; Piatkowski P.; Baginski B.; Wirecki T. K.; de Crécy-Lagard V.; Ross R.; Limbach P. A.; Kotter A.; Helm M.; Bujnicki J. M. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–d307. 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S.; Ries R. J.; Jaffrey S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Saneyoshi M.; Harada F.; Nishimura S. Isolation and characterization of N6-methyladenosine from Escherichia coli valine transfer RNA. Biochim. Biophys. Acta 1969, 190, 264–273. 10.1016/0005-2787(69)90078-1. [DOI] [PubMed] [Google Scholar]
- Tuck M. T. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int. J. Biochem 1992, 24, 379–386. 10.1016/0020-711X(92)90028-Y. [DOI] [PubMed] [Google Scholar]
- Bodi Z.; Button J. D.; Grierson D.; Fray R. G. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010, 38, 5327–5335. 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D.; Saletore Y.; Zumbo P.; Elemento O.; Mason C. E.; Jaffrey S. R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D.; Moshitch-Moshkovitz S.; Schwartz S.; Salmon-Divon M.; Ungar L.; Osenberg S.; Cesarkas K.; Jacob-Hirsch J.; Amariglio N.; Kupiec M.; Sorek R.; Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Linder B.; Grozhik A. V.; Olarerin-George A. O.; Meydan C.; Mason C. E.; Jaffrey S. R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegowski M.; Flamand M. N.; Meyer K. D. scDART-seq reveals distinct m(6)A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 2022, 82, 868–878. 10.1016/j.molcel.2021.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Gao C. C.; Zhang D.; Xu J.; Song G.; Fan X.; Liang D. B.; Chen Y. S.; Li Q.; Guo Y.; Cai Y. T.; Hu L.; Zhao Y. L.; Sun Y. P.; Yang Y.; Han J.; Yang Y. G. scm(6)A-seq reveals single-cell landscapes of the dynamic m(6)A during oocyte maturation and early embryonic development. Nat. Commun. 2023, 14, 315. 10.1038/s41467-023-35958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.; Hu Y.; Zhou B.; Bao Y.; Li Z.; Gong C.; Yang H.; Wang S.; Xiao Y. The role of m(6)A modification in physiology and disease. Cell Death Dis 2020, 11, 960. 10.1038/s41419-020-03143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D.; Jaffrey S. R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.; Li S.; Deng T.; Gao M.; Yin Y.; Wu B.; Peng C.; Liu J.; Ma J.; Zhang K. Cryo-EM structures of human m(6)A writer complexes. Cell Res. 2022, 32, 982–994. 10.1038/s41422-022-00725-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. I.; Pan T. An additional class of m(6)A readers. Nat. Cell Biol. 2018, 20, 230–232. 10.1038/s41556-018-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S.; Ries R. J.; Jaffrey S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Lee Y. Y.; Lee H.; Kim H.; Kim V. N.; Roh S. H. Structure of the human DICER-pre-miRNA complex in a dicing state. Nature 2023, 615, 331–338. 10.1038/s41586-023-05723-3. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell 2001, 107, 823–826. 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- Jie M.; Feng T.; Huang W.; Zhang M.; Feng Y.; Jiang H.; Wen Z. Subcellular Localization of miRNAs and Implications in Cellular Homeostasis. Genes (Basel) 2021, 12, 856. 10.3390/genes12060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Weng H.; Zhou K.; Wu T.; Zhao B. S.; Sun M.; Chen Z.; Deng X.; Xiao G.; Auer F.; Klemm L.; Wu H.; Zuo Z.; Qin X.; Dong Y.; Zhou Y.; Qin H.; Tao S.; Du J.; Liu J.; Lu Z.; Yin H.; Mesquita A.; Yuan C. L.; Hu Y.-C.; Sun W.; Su R.; Dong L.; Shen C.; Li C.; Qing Y.; Jiang X.; Wu X.; Sun M.; Guan J.-L.; Qu L.; Wei M.; Müschen M.; Huang G.; He C.; Yang J.; Chen J. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R. J. 3rd; Reinberg D. Processing the H3K36me3 signature. Nat. Genet. 2009, 41, 270–271. 10.1038/ng0309-270. [DOI] [PubMed] [Google Scholar]
- Geula S.; Moshitch-Moshkovitz S.; Dominissini D.; Mansour A. A.; Kol N.; Salmon-Divon M.; Hershkovitz V.; Peer E.; Mor N.; Manor Y. S.; Ben-Haim M. S.; Eyal E.; Yunger S.; Pinto Y.; Jaitin D. A.; Viukov S.; Rais Y.; Krupalnik V.; Chomsky E.; Zerbib M.; Maza I.; Rechavi Y.; Massarwa R.; Hanna S.; Amit I.; Levanon E. Y.; Amariglio N.; Stern-Ginossar N.; Novershtern N.; Rechavi G.; Hanna J. H. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Betz U. A.; Vosshenrich C. A.; Rajewsky K.; Müller W. Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr. Biol. 1996, 6, 1307–1316. 10.1016/S0960-9822(02)70717-3. [DOI] [PubMed] [Google Scholar]
- Yoon K. J.; Ringeling F. R.; Vissers C.; Jacob F.; Pokrass M.; Jimenez-Cyrus D.; Su Y.; Kim N. S.; Zhu Y.; Zheng L.; Kim S.; Wang X.; Doré L. C.; Jin P.; Regot S.; Zhuang X.; Canzar S.; He C.; Ming G. L.; Song H. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 2017, 171, 877–889. 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.; Lv H.; Zhang W.; Ma C.; He X.; Zhao S.; Zhang Z. W.; Zeng Y. X.; Song S.; Niu Y.; Tong W. M. Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017, 7, 170166. 10.1098/rsob.170166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Chang M.; Lv H.; Zhang Z. W.; Zhang W.; He X.; Wu G.; Zhao S.; Zhang Y.; Wang D.; Teng X.; Liu C.; Li Q.; Klungland A.; Niu Y.; Song S.; Tong W. M. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68. 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navabpour S.; Kwapis J. L.; Jarome T. J. A neuroscientist’s guide to transgenic mice and other genetic tools. Neurosci Biobehav Rev. 2020, 108, 732–748. 10.1016/j.neubiorev.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Zhang X.; Weng Y. L.; Lu Z.; Liu Y.; Lu Z.; Li J.; Hao P.; Zhang Y.; Zhang F.; Wu Y.; Delgado J. Y.; Su Y.; Patel M. J.; Cao X.; Shen B.; Huang X.; Ming G. L.; Zhuang X.; Song H.; He C.; Zhou T. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A.; Caroni P. Functional and structural underpinnings of neuronal assembly formation in learning. Nat. Neurosci 2016, 19, 1553–1562. 10.1038/nn.4418. [DOI] [PubMed] [Google Scholar]
- Krüttner S.; Caroni P. m(6)A-epitranscriptome modulates memory strength. Cell Res. 2019, 29, 4–5. 10.1038/s41422-018-0121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr A. M.; Mott J. L. Overview of microRNA biology. Semin Liver Dis 2015, 35, 3–11. 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L. T. Memory and the brain. J. Dent Educ 2002, 66, 30–42. 10.1002/j.0022-0337.2002.66.1.tb03506.x. [DOI] [PubMed] [Google Scholar]