Abstract
A competitive PCR technique was used to enumerate the proteolytic bacterium Clostridium proteoclasticum from the rumen. A PCR primer, which circumscribes this organism and several closely related strains, was designed for a variable region within their 16S rRNA genes and was used in conjunction with a universal forward primer. This primer pair was tested for specificity against 85 ruminal bacterial strains. An internal control DNA was constructed for use in competitive PCRs and was shown to amplify under the same reaction conditions and with the same amplification efficiency as the target DNA. DNA from a known number of C. proteoclasticum cells was coamplified with the internal control to construct a standard curve. Rumen samples were collected from eight dairy cows fed four diets in rotation: high nitrogen, high nitrogen supplemented with carbohydrate, low nitrogen, and low nitrogen supplemented with carbohydrate. DNA extracted from these and spiked with internal control DNA was amplified with the C. proteoclasticum primer pair. The relative intensities of the PCR products were used to quantitate the numbers of C. proteoclasticum cell equivalents from the rumen samples. The numbers ranged from 2.01 × 106 ml−1 to 3.12 × 107 ml−1. There was no significant effect on the numbers of C. proteoclasticum detected in rumen samples among cows fed the four diets. The utility of the competitive PCR approach for quantifying ruminal bacterial populations in vivo and the occurrence of C. proteoclasticum in forage-fed dairy cows are discussed.
New Zealand ruminants graze a fresh forage diet which is high in protein and low in soluble carbohydrates (10). Up to 50% of the protein available from this diet can be lost due to the rapid microbial breakdown of plant protein (16). Bacteria are thought to be responsible for the majority of this protein degradation in the rumen (8). The proteolytic bacteria in forage-fed New Zealand cattle are dominated by species of the genera Streptococcus, Eubacterium, and Butyrivibrio (2), while a novel, highly proteolytic strain, Clostridium proteoclasticum, has also been isolated. To determine the significance of these particular proteolytic bacteria within the New Zealand ruminant, we have set out to develop a sensitive and specific method of quantitation of microbial populations directly from rumen samples.
rRNA probes have been successfully used for the detection and enumeration of bacteria within the rumen (7, 11, 17–20, 27). This technique expresses the abundance of a particular rRNA sequence in relation to total RNA extracted from a sample. However, it is relatively insensitive, only detecting down to 106 bacteria per ml of rumen fluid (27). PCR has been used to detect bacteria in many environments (5, 6), and its sensitivity has allowed the detection of a single bacterium (28). Conventional PCR amplifies target DNA exponentially, making it difficult to use the technique in a quantitative manner. Competitive PCR (cPCR), however, has been used to determine bacterial numbers in a range of environments (12, 14, 15). The addition of an internal DNA standard controls for the variation among reactions, allowing reliable PCR quantitation. The internal DNA standard contains the same primer binding sites as the target, and the two DNAs compete for reaction reagents to produce PCR products of different sizes, which can be separated in an agarose gel. The log ratio of intensities of amplified target DNA to internal control DNA is determined by the equation log (Nn1/Nn2) = log (N01/N02) + n log (eff1/eff2) (29). If the efficiencies of amplification (eff1 and eff2) are equal, the ratio of amplified products (Nn1/Nn2) is dependent on the log ratio of starting products (N01/N02) (29). Even if the efficiencies of the two reactions are not equal, the values still hold assuming that eff1/eff2 is constant and amplification is in the exponential phase. With this technique, amounts of target DNA can be determined by amplification with known amounts of internal control DNA. The resulting log ratio of intensities of PCR products is compared to standard curves derived from serial dilutions of known target DNA amplified with known amounts of internal control DNA.
C. proteoclasticum is a gram-positive, straight to slightly curved rod which was first isolated from a pasture-grazed cow in New Zealand (4). Its most distinguishing feature is its extremely high proteinase activity, and because of this feature we were interested in quantifying this organism to estimate its contribution to rumen proteolysis. Since its description as a new species, it has become apparent that the 16S rRNA gene (rDNA) sequence of C. proteoclasticum is very similar to those of Butyrivibrio fibrisolvens NCDO 2435, 2434, 2222, and 2398. However, the description of C. proteoclasticum as a new species is justified since C. proteoclasticum and these Butyrivibrio fibrisolvens strains are phylogenetically closely related to each another (96.9 to 99.5% 16S rRNA sequence similarity) but are distantly related to the Butyrivibrio fibrisolvens type strain NCDO 2221T (93.1 to 94.1% similarity). We have developed a cPCR technique which detects C. proteoclasticum and these closely related B. fibrisolvens strains directly from rumen samples. PCR primers were designed against a common region of the C. proteoclasticum and B. fibrisolvens 16S rRNA genes. These primers have been tested for specificity against DNA from 85 rumen bacterial strains, and the sensitivity of detection in a mixed rumen bacterial background has been investigated. We have used this cPCR approach to enumerate C. proteoclasticum cell equivalents in rumen samples from dairy cows fed four different diets: high nitrogen, high nitrogen supplemented with carbohydrate, low nitrogen, and low nitrogen supplemented with carbohydrate.
MATERIALS AND METHODS
Bacterial strains and growth.
The bacterial strains used are listed in Table 1. Bacteria were grown on CC medium (13), except that the rumen fluid was not preincubated to remove soluble carbohydrates and carbon sources were replaced by either 1.0% (wt/vol) glucose or cellobiose. Clostridium aminophilum, C. sticklandii, and Peptostreptococcus anaerobius were grown in CC medium in which no carbon source was added and tryptone (1.5% [wt/vol]; Difco, Detroit, Mich.) replaced trypticase.
TABLE 1.
Bacterial strains used in this study
| Species | Strain | Source or reference | Reaction with:
|
|
|---|---|---|---|---|
| Universal primersa | Specific primersb | |||
| Butyrivibrio fibrisolvens | C130a, C211, C219 | 2 | + | − |
| Butyrivibrio fibrisolvens | CF3, H17c | M. P. Bryant, University of Illinois | + | − |
| Butyrivibrio fibrisolvens | WV1 | Laboratory strain | + | − |
| Butyrivibrio fibrisolvens | OR509 | R. Teather, Centre for Food and Animal Research, Agriculture and Agri-food, Canada | + | − |
| Butyrivibrio fibrisolvens like | C122a, C21b | 2 | + | − |
| Clostridium aminophilum | F | J. B. Russell, Cornell University | + | − |
| Clostridium clostridioforme | ATCC 25537, ATCC 29084 | ATCCc | + | − |
| Clostridium proteoclasticum | ATCC 51982 | 2 | + | + |
| Clostridium sticklandii | SR | J. B. Russell, Cornell University | + | − |
| Enterococcus faecalis | NCTC 775 | S. Flint, New Zealand Dairy Research Institute | + | − |
| Enterococcus faecalis | 68a | Laboratory strain | + | − |
| Eubacterium cellulosolvens | 5494 | M. P. Bryant | + | − |
| Eubacterium limosum | ATCC 8486 | M. P. Bryant | + | − |
| Eubacterium ruminantium | M. P. Bryant | + | − | |
| Eubacterium sp. | C11, C12b, C13b, C14b#2, C118b, C119b, C120a, C124b, C125b, C260 | 2 | + | − |
| Fibrobacter succinogenes | AC3 | M. P. Bryant | + | − |
| Lachnospira multiparus | ATCC 19307 | M. P. Bryant | + | − |
| Megasphaera elsdenii | T81 | M. P. Bryant | + | − |
| Peptostreptococcus anaerobius | C | J. B. Russell | + | − |
| Peptostreptococcus productus | SF-50 | M. P. Bryant | + | − |
| Prevotella ruminicola | C21a | 2 | + | − |
| Prevotella ruminicola subsp. brevis | 118b, B14 | M. P. Bryant | + | − |
| Prevotella ruminicola subsp. ruminicola | 23 | M. P. Bryant | + | − |
| Ruminococcus albus | 7, 8 | M. P. Bryant | + | − |
| Ruminococcus flavefaciens | ATCC 19208, FD1 | M. P. Bryant | + | − |
| Selenomonas ruminantium subsp. lactilytica | GA31 | M. P. Bryant | + | − |
| Selenomonas ruminantium subsp. ruminantium | ATCC 12561, ATCC 27209 | M. P. Bryant | + | − |
| Streptococcus bovis | JB1 | M. P. Bryant | + | − |
| Streptococcus bovis | NCFB 2476 | A. G. Williams, Hannah Research Institute | + | − |
| Streptococcus bovis | 3-31, 3-36, 3-39, 5-21, 7-2, 7-25, 7-26 | J. B. Russell | + | − |
| Streptococcus bovis | A12, A14, A120, A166, A191b, B11, B32a, B314, B315, B327, B337, B342, B346, B348, B350, B360, B372, B382, B385, B395, B396, B398, C14b#1, C17, C123b, C271 | 2 | + | − |
| Streptococcus bovis | RF-1, RF-2, UDY-KF | Laboratory strains | + | − |
| Succinomonas amylolytica | ATCC 19206 | M. P. Bryant | + | − |
| Succinovibrio dextrinosolvens | ATCC 27209 | M. P. Bryant | + | − |
PCR amplification with universal forward and universal reverse primers.
PCR amplification with universal forward primer and S-S-Cprot-0832-a-A-21.
ATCC, American Type Culture Collection, Rockville, Md.
Rumen samples.
Eight fistulated, lactating, Friesian dairy cows were fed four different diets in rotation: high nitrogen, high nitrogen with carbohydrate, low nitrogen, and low nitrogen with carbohydrate. Ryegrass pasture, containing less than 5% clover, was top dressed with 20 kg of nitrogen (as urea) per ha for low-nitrogen diets and 90 kg of nitrogen per ha for high-nitrogen diets. Urea was applied 21 to 28 days before cutting, and the nitrogen was 2.11 and 2.82% of dry matter for the low- and high-nitrogen diets, respectively. Carbohydrate was supplied as a 50:50 mixture of dextrose and corn flour on an energy basis and was drenched at 9:00 a.m., 11:00 a.m., 4:00 p.m., and 6:00 p.m., supplying 10% of the minimum energy intake. The cows were fed at 9:00 a.m. and 4:00 p.m. and were maintained on each diet for 12 days before the samples were taken. Whole rumen samples of approximately 250 ml were collected before the last 4:00 p.m. feed, frozen immediately in liquid nitrogen, and stored at −80°C until DNA extraction. Immediately before DNA extraction, the samples were thawed in a 37°C water bath and diluted with an equal weight of mineral salts (MS) buffer (3) before homogenization in a Sorvall tissue homogenizer (Du Pont Co., Wilmington, Del.).
DNA extraction.
General techniques of DNA precipitation and phenol-chloroform extractions were performed as described by Sambrook et al. (26). For determination of primer specificity, bacterial cultures were grown overnight and DNA was extracted by the enzymatic lysis procedure described by Saito and Miura (25). To eliminate possible bias introduced by enzymatic cell wall lysis and to maximize DNA extraction efficiencies, physical disruption was used to extract DNA from rumen samples and from C. proteoclasticum for sensitivity experiments and standard curve construction. Unless otherwise stated, DNA was extracted from triplicate samples. A 1-ml volume of homogenized rumen contents or 1010 C. proteoclasticum cells was added to 1.2 g of sterile zirconia-silica beads (Biospec Products) followed by centrifugation at 12,000 × g for 10 min at room temperature. The pellet and beads were rinsed twice in saline-EDTA solution (0.15 M NaCl, 0.1 M EDTA) before final resuspension in 750 μl of saline-EDTA. Physical disruption was then performed with a Mini-beadbeater (Biospec Products) at maximum speed for two intervals of 2 min each, with a 1-min incubation on ice between each treatment. Phenol-chloroform-isoamyl alcohol (25:24:1) was added and mixed, and the mixture was centrifuged at 12,000 × g for 5 min at room temperature. The aqueous phase was removed, and the interface was reextracted with TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The combined aqueous phases were repeatedly extracted with the phenol-chloroform-isoamyl alcohol until no protein remained at the interface. A final chloroform-isoamyl alcohol extraction was performed before the nucleic acids were precipitated with ethanol and centrifuged at 10,000 × g for 20 min at 4°C. The air-dried pellet was resuspended in TE buffer, RNase A (1 mg ml−1 [final concentration]) was added, and the mixture was incubated at 37°C for 60 min. RNase A was removed by phenol-chloroform-isoamyl alcohol extraction followed by ethanol precipitation and centrifugation. The air-dried DNA pellet was resuspended in TE buffer and was stored at −20°C, until required. DNAzol reagent (Gibco BRL, Life Technologies, Auckland, New Zealand) was used for chemical extraction of DNA. Cells were suspended in 1 ml of DNAzol reagent before being subjected to bead beater treatment as described above. The DNA was then precipitated as specified by the manufacturer. The concentration and purity of the DNA were determined spectrophotometrically by measuring the absorbances at 260 and 280 nm (A260/280) with a Spectramax microplate spectrophotometer (Molecular Dynamics).
To check that there was no amplification of plant material with the primer pair, DNA was extracted from 20 g of pasture plant tissue. This plant tissue was diluted with 4 volumes of MS buffer to ensure complete homogenization. The DNA was extracted by the mechanical disruption method described above.
PCR primers and amplification.
The primers used for the amplification of the 16S rRNA genes were as follows: universal forward primer (S-*-Univ-0008-a-S-19; 5′ GAG TTT GAT CCT GGC TCA G 3′), universal reverse primer (S-*-Univ-1528-a-A-17; 5′ AAG GAG GTG ATC CAG CC 3′), and the C. proteoclasticum primer (S-S-Cprot-0832-a-A-21; 5′ CTG AAT GCC TAT GGC ACC CAA 3′). The S-S-Cprot-0832-a-A-21 primer was screened for specificity with the PROBE CHECK program of the Ribosomal Database Project (21) and the BLAST (Basic Local Alignment Search Tool) facility at the National Center for Biotechnology. PCR amplification of C. proteoclasticum DNA produces an 830-bp product when amplified with the universal forward primer and S-S-Cprot-0832-a-A-21. Because the S-S-Cprot-0832-a-A-21 primer also detects some closely related B. fibrisolvens strains in rumen samples, in these instances cell numbers are expressed as C. proteoclasticum cell equivalents.
The PCR mixtures contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM each dATP, dCTP, dGTP and dTTP, 1 μM each primer, and 0.5 U of Taq DNA polymerase (Gibco BRL). The PCRs were performed in a final volume of 20 μl sealed in a capillary tip, and thermocycling was carried out in a model FTS-1 capillary thermal sequencer (Corbett Research, Sydney, Australia). The PCR amplification conditions for S-S-Cprot-0832-a-A-21 and the universal forward primers were as follows: denaturation at 95°C for 3 min followed by 6 cycles of 95°C for 30 s, 62°C for 15 s, and 72°C for 30 s and 25 cycles of 95°C for 15 s, 62°C for 5 s, and 72°C for 30 s, with a final cycle of 72°C for 3 min. Amplification with the universal forward and reverse primers differed only in the annealing temperature, which was 55°C. PCR products were separated by electrophoresis in agarose gels, stained with ethidium bromide, and visualized by UV transillumination.
Construction of the internal control.
The 830-bp PCR product from C. proteoclasticum DNA amplified with the universal forward and S-S-Cprot-0832-a-A-21 primers contains two AluI sites (Fig. 1). To produce an AluI deletion, the 830-bp fragment was digested with AluI as specified by the manufacturer (Boehringer, Mannheim, Germany) and the restriction endonuclease was removed with phenol treatment followed by ethanol precipitation. The AluI fragments were ligated with T4 DNA ligase (Gibco BRL), 2 μl of the ligation reaction was used in a PCR with the universal forward and S-S-Cprot-0832-a-A-21 primers, and the products were separated by agarose gel electrophoresis. One of the PCR products was a 480-bp fragment expected from the deletion of the 350-bp internal AluI fragment. The 480-bp fragment was purified from the gel and used as an internal control for the cPCRs. The concentration of the internal control was estimated from photographs of the gel to be approximately 100 ng ml−1. However, the DNA concentration was too low to obtain an accurate A260/280 reading, and so the internal control was expressed as a dilution of the concentrated mixture.
FIG. 1.
Construction of the internal control. The 830-bp PCR product was digested with AluI to give three fragments of 90, 350, and 390 bp. The fragments were ligated and PCR amplified. The 480-bp product was purified and used as the internal control.
Preparation of C. proteoclasticum cells for sensitivity testing.
C. proteoclasticum was grown in 100-ml overnight cultures, and a sample was counted by phase-contrast microscopy with a WSI counting chamber (Weber Scientific International Ltd., Middlesex, England). The cells were pelleted by centrifugation and resuspended to a final concentration of 1010 cells ml−1. DNA extractions were performed on 1010 cells; 10-fold serial dilutions of this DNA were amplified to determine the detection limit.
Quantitation of PCR products.
PCR products were quantitated by photographing agarose gels with Polaroid 665 film (Polaroid, St. Albans, England), which produces a negative image of the photograph. The negative was scanned with a GS-670 imaging densitometer (Bio-Rad, Hercules, Calif.) and analyzed with Molecular Analyst software version 1.4 (Bio-Rad). To correct for differences in the fluorescence of ethidium bromide-stained PCR fragments of different sizes (23), the intensity of the internal control was multiplied by the ratio 830/480.
RESULTS
Primer specificity and sensitivity.
Alignment of the C. proteoclasticum 16S rDNA sequence with other sequences from the Ribosomal Database Project and closely related B. fibrisolvens sequences retrieved from BLAST searches identified a region at bp 832 to 851 (E. coli numbering) that was common to C. proteoclasticum and B. fibrisolvens NCDO 2435, 2222, 2398, and 2434. A primer, S-S-Cprot-0832-a-A-21, complementary to the sense strand of this region facing the 5′ end of the 16S rRNA gene was designed. This enabled it to be used with the universal forward primer in PCRs to generate an 830-bp product. The primer pair was tested for specificity against DNA from 85 bacterial strains, mostly of rumen origin (Table 1). Only C. proteoclasticum genomic DNA amplified with these two primers at an annealing temperature of 62°C (Table 1); closely related strains (B. fibrisolvens-like C21b and C122b) failed to amplify under these conditions. At an annealing temperature of 60°C, some amplification occurred from unrelated Enterococcus faecalis NCTC 775 and Streptococcus bovis RF-2. This nonspecific amplification was eliminated once the annealing temperature was raised to 62°C. The DNA from each of the rumen strains was also tested with the universal forward and reverse primers at an annealing temperature of 55°C. All the strains produced a PCR fragment of approximately 1,500 bp, corresponding to the approximate size of the 16S rRNA gene, demonstrating that the DNA from each strain was amplifiable.
The sensitivity of the S-S-Cprot-0832-a-A-21 primer in PCRs was investigated by amplification of a serial dilution of DNA from known numbers of C. proteoclasticum cells. The results show that 50 fg of DNA (the equivalent of DNA from 25 C. proteoclasticum cells) was the lower limit of detection when coamplified with a 2 × 106 dilution of the internal control DNA (Fig. 2).
FIG. 2.
Detection limit of cPCR. DNA extracted from 1010 C. proteoclasticum cells ml−1 was serially diluted and coamplified with a 2 × 106 dilution of the internal control to determine the detection limit of the cPCR assay. The results are expressed as C. proteoclasticum cell equivalents based on the amount of DNA extracted per cell.
DNA extraction.
To determine the most efficient method of recovering DNA from bacterial samples, 1010 C. proteoclasticum cells were subjected to three methods of DNA extraction: enzymatic cell lysis, chemical extraction, and physical disruption. A260/280 readings demonstrated that physical disruption was the most efficient method of extraction, recovering 67 μg of DNA from 1010 cells. Enzymatic lysis and chemical extraction recovered 43 and 1.01 μg of DNA per 1010 cells, respectively.
Internal-control amplification efficiency.
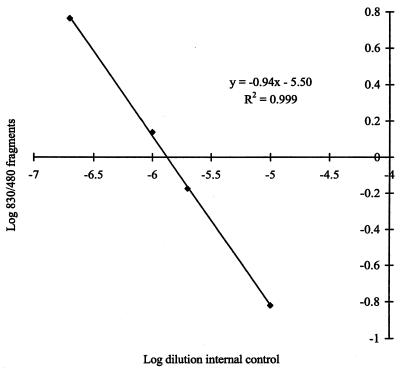
The relative amplification efficiencies of target and internal-control DNAs can be determined from a plot of the ratio of log target intensity to internal control intensity against the log concentration of internal control DNA. Coamplification of DNA from 103 C. proteoclasticum cells with dilutions of the internal control (Fig. 3) results in a line with a slope of 0.94 and a regression of 0.999. This indicates equivalent amplification efficiencies of the target and control DNAs. The line intersects the x axis at −5.9, indicating that the optimal dilution of internal control for detection of 103 C. proteoclasticum cells is 1.26 × 106.
FIG. 3.
Internal-control amplification efficiency. Dilutions of the internal control were coamplified with DNA from 103 C. proteoclasticum cells, and the ratio of the intensities of internal control to the target DNA was plotted against the dilution of the internal control on a log scale.
Standard curve.
Since cPCR is most accurate when the target and internal control are coamplified in equimolar proportions, it was necessary to determine the optimal internal-control concentrations to use with DNA extracted from rumen samples. Dilutions of internal control were coamplified with DNA from selected samples, and the optimal dilution of the internal control was found to be 10−6. This concentration of internal control was used to construct a standard curve by coamplification with C. proteoclasticum DNA extracted from a known number of cells (Fig. 4). The results show that DNA from 1 × 104 to 5 × 101 cells gave a linear response and could be used for quantitation of samples within this range.
FIG. 4.
Standard curve construction. (a) DNA extracted from 1010 C. proteoclasticum cells ml−1 was serially diluted and coamplified with a 106 dilution of the internal control. The results are expressed as C. proteoclasticum cell equivalents based on the amount of DNA extracted per cell. (b) Ratios of the intensities of internal control to target DNA were quantitated by scanning densitometry of negative images of Polaroid photographs of ethidium bromide-stained gels.
Detection of C. proteoclasticum added to rumen fluid.
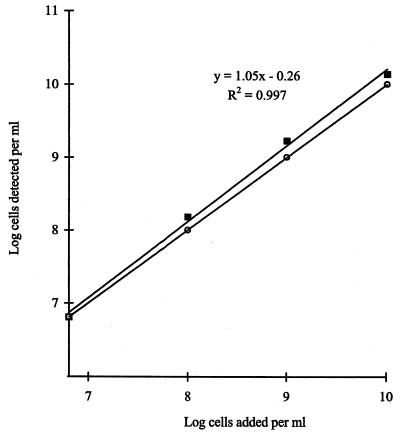
To test whether C. proteoclasticum could be detected by cPCR within a background of nonspecific DNA from rumen fluid, samples of rumen contents were spiked with known numbers of C. proteoclasticum cells. DNA from each of the samples was coamplified under optimal cPCR conditions, and the results show a linear relationship between the number of cells added and the number of cells detected. The assay slightly overestimated the number of C. proteoclasticum cells added compared to the ideal (y = x), and this was particularly evident at the higher concentrations of cells (Fig. 5). A population of 6.25 × 106 C. proteoclasticum cell equivalents ml−1 was detected in the unspiked rumen samples.
FIG. 5.
In vivo detection of C. proteoclasticum. Rumen samples were spiked with known numbers of C. proteoclasticum cells. DNA was extracted from these mixtures and assayed for the presence of C. proteoclasticum. Solid squares denote numbers of C. proteoclasticum cell equivalents detected in each sample. Open circles denote y = x. The y intercept of the line denotes no C. proteoclasticum cells added. This represents a background population of C. proteoclasticum and closely related B. fibrisolvens strains of 6.46 × 106 ml−1.
Detection of C. proteoclasticum and closely related strains in vivo.
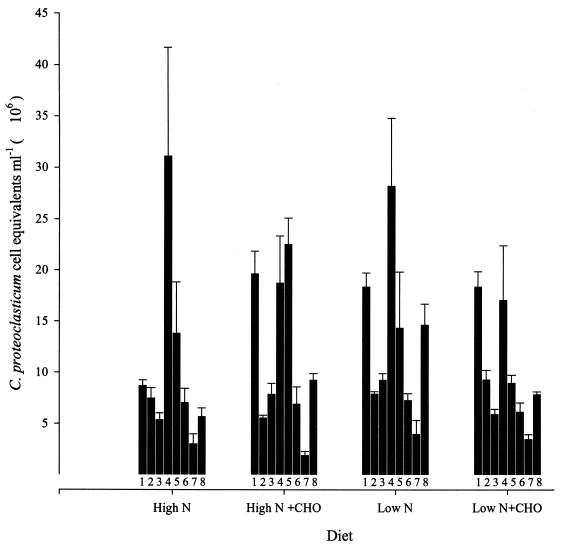
To examine the application of the cPCR approach to determining bacterial numbers directly from animals, rumen samples were collected from eight lactating dairy cows fed four different diets in rotation. The number of C. proteoclasticum cell equivalents detected ranged from 2.01 × 106 to 3.12 × 107 ml−1 of rumen contents (Fig. 6). Within individual animals there were significant responses to diet, but overall there was no significant difference between the diets.
FIG. 6.
Populations of C. proteoclasticum and closely related B. fibrisolvens strains in dairy cows under four different feeding regimens. Numbers 1 through 8 represent cows numbered 709, 710, 727, 788, 1758, 8702, 9754, and 9775, respectively. The results are the means of triplicate determinations and error bars represent standard error of the mean. The diets were fed in rotation, and rumen samples were taken during each feeding regime. CHO, carbohydrate.
DISCUSSION
C. proteoclasticum is a proteolytic bacterium that was isolated from the rumen contents of a forage-fed cow (2). It has a predominately serine-type proteinase activity (3) but also some cysteine- and metallo-type proteinase activity. It seems most likely to be involved with primary hydrolysis of feed protein, but the significance of C. proteoclasticum to rumen microbial ecology has not yet been determined. Since no suitable selective media are available for enumeration of C. proteoclasticum, we set out to develop a method to quantify this organism directly from rumen samples.
PCR is an extremely sensitive and specific method for the amplification of DNA, and when it is used in conjunction with an internal DNA control, the products of the amplification reactions can be quantified. This technique, cPCR, was originally developed for quantitation of human immunodeficiency virus type 1 3B long terminal repeat DNA (29). Its use has since been extended to bacterial quantitation in the environment (12, 14, 15). In the present study, a cPCR method was developed to quantify C. proteoclasticum and closely related strains from rumen samples. PCR primers, based on both conserved and hypervariable regions of the 16S rRNA gene, were used to amplify DNA in the presence of an internal control which allowed quantitation of the PCR products. The primer pair used circumscribes C. proteoclasticum and four closely related B. fibrisolvens strains and did not amplify DNA from any other bacterium tested. The specificity of the assay stems from the selection of 16S rDNA as the target for amplification and the primer design. The semiconserved nature of rRNA genes allows the use of regions of hypervariable sequence as targets for group-, genus-, species-, and strain-specific amplification (22). The primer was designed to match, as closely as possible, the length, G+C content, and Tm of the universal forward primer so that the forward primer “anchored” the PCR at the 5′ end of the 16S rRNA gene. This approach limits the range of suitable specific primers which can be designed within a particular region but avoids the need to design two specific primers for a given organism. Using the 16S rRNA gene as the amplification target also allows primers to be checked against the existing entries in DNA sequence databases, thereby reducing the effort needed to test primer specificity.
A critical factor in ensuring the accuracy of cPCR is to demonstrate that coamplifications of the target and internal control are equivalent (24). This can be done by plotting the log ratio of target to internal-control DNA intensities against the log dilution of internal-control DNA. Coamplification of C. proteoclasticum DNA with dilutions of internal control gave a straight line with a slope of −0.94, indicating that the amplification efficiencies of the two DNAs are essentially equal. The small deviation from equivalence may be due to a slightly more efficient amplification of the smaller internal control (480 bp) than of the target (830 bp). This effect was taken into account by using standard curves to relate the log ratio of target to internal control to the log of cell numbers. These standard curves also account for the rRNA gene copy number, which can vary anywhere from 2 to 10 copies per genome (1, 9). Each standard curve, generated by coamplification of a single dilution of internal control with DNA from different numbers of C. proteoclasticum cells, gives a substantial working range of cell numbers of approximately 103- to 104-fold. Indeed, in the application of the assay to rumen samples, only a single standard curve was required to encompass the entire range of C. proteoclasticum numbers encountered in differently fed dairy cows.
In a similar manner, dilutions of internal control can be used to determine the absolute sensitivity of the assay. Theoretically, the detection of one copy of the target sequence is possible (28). However, in cPCR, the target and internal control compete for amplification reagents, reducing the sensitivity of detection. In our assay, DNA from the equivalent of 25 C. proteoclasticum cells could be detected when coamplified with a 2 × 106 dilution of the internal control. However, it was found that a 100-fold dilution was necessary to overcome an inhibitory effect in DNA extracted from rumen samples. Thus, in practice, the sensitivity of the assay is limited to 2.5 × 103 cells. The exact nature of the inhibitory factor is not known, but high A260/280 readings from these DNA samples indicate that it is not proteinaceous and that it may be a water-soluble polysaccharide or polyphenolic compound similar in nature to the humic acids described by Leser (14) as interfering with the PCRs. The level of detection compares favorably with that in similar studies of bacterial populations from other environments. Leser (14) detected DNA from 40 Pseudomonas cells in cPCR assays of samples from a marine environment. Radiolabelled oligonucleotides used to probe rRNA from rumen bacteria (7, 11, 17–20, 27) are less sensitive, being able to detect 0.01% of the total rumen population, or approximately 106 cells ml−1 (27). It should be noted that the cPCR technique does not discriminate between living and dead cells and therefore is likely to overestimate viable populations. This is in contrast with oligonucleotide probing, where microbial abundance is expressed in terms of a proportion of total rRNA. Since the rRNA content in cells changes according to the growth phase, this technique provides an approximation of relative cell numbers (20), which reflects their contribution to total metabolic activity (27). However, the estimation of total rRNA abundance depends on universal probes, and recent evidence suggests that domain-specific variations in dissociation temperatures of universal probes could lead to significant biases in quantifying microbial populations from environmental samples (30). Estimates of both absolute cell numbers and the percent contribution to total rRNA may be the best approach to gain an accurate assessment of the importance of microbial populations in environmental samples.
After the development of the cPCR assay in vitro, it was necessary to test its ability to detect C. proteoclasticum cells in the complex mixture of plant and microbial DNA present in rumen fluid. Mechanical disruption was chosen for DNA extraction from rumen contents since the efficiency of extraction was greatest with bead beating followed by phenol-chloroform extractions. Also, this method allowed entire rumen contents to be sampled, thereby enabling the quantitation of populations adherent to plant tissue. Previous methods have sampled only filtered rumen contents (7, 17, 27). The addition of C. proteoclasticum cells to rumen fluid followed by cPCR quantitation demonstrated the usefulness of the technique in vivo. There was good correlation between the number of cells added and those detected by the assay. A slight overestimation of absolute numbers of cells compared to the ideal (y = x) is probably due to the resident population of C. proteoclasticum and closely related B. fibrisolvens strains in the rumen fluid used.
When the assay was applied to rumen samples collected from animals fed diets differing in nitrogen and carbohydrate content, the C. proteoclasticum cell equivalents detected ranged from 2.01 × 106 cells per ml in the carbohydrate-supplemented, high-nitrogen diet to 3.12 × 107 cells per ml in the high-nitrogen diet. These numbers represent the population of C. proteoclasticum and closely related B. fibrisolvens strains present in the samples and are consistent with the original isolation of C. proteoclasticum from a 108 dilution of rumen contents (4). These results indicate that this group of organisms is common among forage-fed ruminants in New Zealand. Within individual animals, the diet had some effects on C. proteoclasticum cell equivalents, but overall, between the animals there were no significant differences (Fig. 6). The relatively small range of cell numbers detected indicates that the population is stable and unresponsive to changes in dietary content. This is a little surprising since one might expect the proteolytic population of C. proteoclasticum to be influenced by nitrogen supply. However, the population density of C. proteoclasticum does not necessarily reflect its overall contribution to proteolytic activity, which may well vary significantly under the conditions tested. Previously, it was shown that C. proteoclasticum has a high chymotrypsin-like proteinase activity (3), and, based on its specific activity for the artificial chymotrypsin substrate N-succinyl alanine-alanine-proline-phenylalanine-p-nitroanilide and the populations detected in this study, it may be responsible for up to 20% of this type of activity in forage-fed dairy cows. More detailed investigations of specific C. proteoclasticum proteinase activities are required before the contribution of this organism to ruminal protein breakdown can be estimated more precisely.
We have found cPCR to be an easy, accurate, and reliable method of bacterial quantitation. Collection of rumen samples, DNA extraction, preliminary PCR amplification with internal control dilutions, and cPCR followed by scanning densitometry can be accomplished within 10 h. Testing primers for specificity and constructing internal DNA controls are the most time-consuming steps in developing the method, but once these have been carried out, they need not be repeated. The precision of the technique is good, with the coefficient of variation between replicate PCRs of the same sample averaging 2.5% and that between samples from the same animal averaging 7.5%. The technique combines the specificity of 16S rDNA-targeted oligonucleotides with the sensitivity of PCR in a format which allows quantitation of bacterial populations from a complex microbial ecosystem. The technique is currently being extended to other protein-degrading genera and will eventually allow us to examine how proteolytic bacterial populations are influenced by changes in the rumen ecosystem.
ACKNOWLEDGMENTS
This work was funded by Public Good Science Funding from the Foundation for Research in Science and Technology and a Lottery Science Research Grant from the New Zealand Lotteries Grant Board.
We thank Vicky Carruthers of the Dairying Research Corporation for the rumen samples and Raymond Bennett and Doug Hopcroft of HortResearch for the development of photographs. Constructive criticisms of the manuscript by Barry Scott and K. N. Joblin are also gratefully acknowledged.
REFERENCES
- 1.Amikam D, Glaser G, Razin S. Mycoplasmas (Mollicutes) have a low number of rRNA genes. J Bacteriol. 1984;158:376–378. doi: 10.1128/jb.158.1.376-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attwood G T, Reilly K. Identification of proteolytic rumen bacteria isolated from New Zealand cattle. J Appl Bacteriol. 1995;79:22–29. doi: 10.1111/j.1365-2672.1995.tb03119.x. [DOI] [PubMed] [Google Scholar]
- 3.Attwood G T, Reilly K. Characterization of proteolytic activities of rumen bacterial isolates from forage-fed cattle. J Appl Bacteriol. 1996;81:545–552. doi: 10.1111/j.1365-2672.1996.tb03545.x. [DOI] [PubMed] [Google Scholar]
- 4.Attwood G T, Reilly K, Patel B K C. Clostridium proteoclasticum sp. nov., a novel proteolytic bacterium from the bovine rumen. Int J Syst Bacteriol. 1996;46:753–758. doi: 10.1099/00207713-46-3-753. [DOI] [PubMed] [Google Scholar]
- 5.Bej A K, Mahbubani M H, Atlas R M. Amplification of nucleic acids by polymerase chain reaction (PCR) and other methods and their applications. Crit Rev Biochem Mol Biol. 1991;26:301–334. doi: 10.3109/10409239109114071. [DOI] [PubMed] [Google Scholar]
- 6.Bej A K, Mahbubani M H. Application of the polymerase chain reaction in environmental microbiology. PCR Methods Applic. 1992;1:151–159. doi: 10.1101/gr.1.3.151. [DOI] [PubMed] [Google Scholar]
- 7.Briesacher S L, May T, Grigsby K N, Kerley M S, Anthony R V, Paterson J A. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J Anim Sci. 1992;70:289–295. doi: 10.2527/1992.701289x. [DOI] [PubMed] [Google Scholar]
- 8.Brock F M, Forsberg C W, Buchanan-Smith J G. Proteolytic activity of rumen microorganisms and the effects of proteinase inhibitors. Appl Environ Microbiol. 1982;44:561–569. doi: 10.1128/aem.44.3.561-569.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvis E D, Widom R L, LaFauci G, Setoguchi Y, Richter I R, Rudner R. Chromosomal organization of rRNA operons in Bacillus subtilis. Genetics. 1988;120:625–635. doi: 10.1093/genetics/120.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johns A T. Pasture quality and ruminant digestion. 1. Seasonal change in botanical and chemical composition of pasture. N Z J Sci Technol Sect A. 1955;37:301–311. [Google Scholar]
- 11.Krause D O, Russell J B. An rRNA approach for assessing the role of obligate amino acid-fermenting bacteria in ruminal amino acid deamination. Appl Environ Microbiol. 1996;62:815–821. doi: 10.1128/aem.62.3.815-821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of abundance of an uncultured soil bacterium strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leedle J A Z, Hespell R B. Differential carbohydrate media and anaerobic replica plating techniques in delineating carbohydrate-utilizing subgroups in rumen bacterial populations. Appl Environ Microbiol. 1980;39:709–719. doi: 10.1128/aem.39.4.709-719.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leser T D. Quantitation of Pseudomonas sp. strain B13(FR1) in the marine environment by competitive polymerase chain reaction. J Microbiol Methods. 1995;22:249–262. [Google Scholar]
- 15.Leser T D, Boye M, Hendricksen N B. Survival and activity of Pseudomonas sp. strain B13(FR1) in a marine microcosm determined by quantitative PCR and an rRNA-targeting probe and its effect on the indigenous bacterioplankton. Appl Environ Microbiol. 1995;61:1201–1207. doi: 10.1128/aem.61.4.1201-1207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacRae J C, Ulyatt M J. Quantitative digestion of fresh herbage by sheep. II. The sites of digestion of some nitrogenous constituents. J Agric Sci. 1974;82:309–319. [Google Scholar]
- 17.May T, Kerley M S, Williams J E. Supplemental protein influences on carbohydrate degradation and bacterial 16S ribosomal ribonucleic acid. J Dairy Sci. 1993;76:3479–3489. doi: 10.3168/jds.S0022-0302(93)77687-0. [DOI] [PubMed] [Google Scholar]
- 18.McSweeney C S, Mackie R I, Odenyo A A, Stahl D A. Development of an oligonucleotide probe targeting 16S rRNA and its application for detection and quantitation of the ruminal bacterium Synergistes jonesii in a mixed population chemostat. Appl Environ Microbiol. 1993;59:1607–1612. doi: 10.1128/aem.59.5.1607-1612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odenyo A A, Mackie R I, Stahl D A, White B A. The use of 16S rRNA-targeted oligonucleotide probes to study the competition between ruminal fibrolytic bacteria: development of probes for Ruminococcus species and evidence for bacteriocin production. Appl Environ Microbiol. 1994;60:3688–3696. doi: 10.1128/aem.60.10.3688-3696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odenyo A A, Mackie R I, Stahl D A, White B A. The use of rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: pure-culture studies with cellulose and alkaline peroxide-treated wheat straw. Appl Environ Microbiol. 1994;60:3697–3703. doi: 10.1128/aem.60.10.3697-3703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen G J, Overbeek R, Larsen N, Marsh T L, McCaughey M J, Maciukenas M A, Kuan W M, Macke T J, Xing Y Q, Woese C R. The ribosomal database project. Nucleic Acids Res. 1992;20:2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace N R, Stahl D A, Lane D J, Olsen G L. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 23.Piatak M, Luk K C, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–80. [PubMed] [Google Scholar]
- 24.Raeymaekers L. Quantitative PCR: theoretical considerations with practical implications. Anal Biochem. 1993;214:582–585. doi: 10.1006/abio.1993.1542. [DOI] [PubMed] [Google Scholar]
- 25.Saito H, Miura K-I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based probes for studies of ruminal microbial populations. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kuppeveld F J M, van der Logt J T M, Angulo A F, van Zoest M J, Quint W G V, Niesters H G M, Galama J M D, Melchers W J G. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl Environ Microbiol. 1992;58:2606–2615. doi: 10.1128/aem.58.8.2606-2615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zachar V, Thomas R A, Goustin A S. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;8:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]