Abstract
Highly enriched methanotrophic communities (>25 serial transfers) were obtained from acidic ombrotrophic peat bogs from four boreal forest sites. The enrichment strategy involved using media conditions that were associated with the highest rates of methane uptake by the original peat samples, namely, the use of diluted mineral medium of low buffering capacity, moderate incubation temperature (20°C), and pH values of 3 to 6. Enriched communities contained a mixture of rod-shaped bacteria arranged in aggregates with a minor contribution of Hyphomicrobium-like cells. The growth stoichiometry of isolates was characteristic of methanotrophic bacteria (CH4/O2/CO2=1:1.1:0.59), with an average apparent yield of 0.41 ± 0.03 g of biomass C/g of CH4-C. DNA from each enrichment yielded a PCR product of the expected size with primers for both mmoX and mmoY genes of soluble methane monooxygenase. Two types of sequences were obtained for PCR-amplified fragments of mmoX. One of them exhibited high identity to the mmoX protein of the Methylocystis-Methylosinus group, whereas the other showed an equal level of divergence from both the Methylosinus-Methylocystis group and Methylococcus capsulatus (Bath) and formed a distinct branch. The pH optimum for growth and for CH4 uptake was 4.5 to 5.5, which is very similar to that for the optimum CH4 uptake observed in the original peat samples. These methanotrophs are moderate acidophiles rather than acidotolerant organisms, since their growth rate and methane uptake were much lower at neutral pH. The growth of the methanotrophic community was enhanced by using media with a very low salt content (20 to 200 mg/liter), more typical of their natural environment. All four enriched communities grew on N-free medium.
Northern wetlands have attracted considerable attention in the last decade as a possible significant source of atmospheric methane (12). Acidic ombrotrophic peat bogs are the most extensive type of wetland, occupying about 3% of total land area and being one of the dominant terrestrial ecosystems in the boreal forest zone of North America and Eurasia. There is a great body of evidence that peat bogs are inhabited by active methanotrophic bacteria that reduce the emission of methane to the atmosphere to 10 to 90% of that generated in the anaerobic layers of the bog profile (9, 17, 19, 21, 22). Although intensive methanotrophic activity in this habitat was recognized many years ago, the microorganisms responsible for this process have eluded isolation. All described methanotrophs are incapable of growth at pH values below 5.0 (7) and thus apparently are unable to oxidize methane in these Sphagnum peat bogs, which have a pH of 3.5 to 5. Other characteristic features of the Sphagnum bog potentially important to the microbial community are the low content of mineral elements in peat water (5 to 50 mg/liter), the presence of inhibitory products from mosses, and a broad annual temperature range from −30 to +30°C. Methane consumption, in particular, is very sensitive to temperature variation, especially during cold seasons. Routine enrichment techniques have failed to yield isolates of methanotrophs from this hostile environment. The only exception is a report on the isolation of a bacterium ascribed to the genus Methylosinus from an acidic peat lake (9), but no experimental confirmation of its activity at low pH was provided. Furthermore, no evidence was provided that the isolated bacterium exhibited any activity in situ, an especially important point since Methylosinus has the ability to form exospores and survive for a long time under unfavorable conditions (6).
The ecological application of molecular techniques has opened up a new opportunity for direct detection of methane-oxidizing bacteria in environmental samples. Indeed, primers designed for amplification of the soluble methane monooxygenase (sMMO) gene cluster have shown the predicted PCR products from DNA from acidic peat, suggesting that these habitats contain numerous methanotrophs (13). Other evidence for the existence of acidophilic methanotrophs was obtained by screening 16S rDNA libraries from several peat samples by means of hybridization with specific probes (14). A few of these clones were found to be representatives of a potentially novel group of methanotrophs related to the Methylosinus-Methylocystis cluster.
Our recent studies (4, 5) dealt with measurements of methanotrophic activity in samples of native peat from four different bogs under various environmental conditions. We found that indigenous methanotrophic populations, as reflected by their activity in peat, have temperature optima of 15 to 20°C and pH optima of 4.5 to 5.5 and are extremely sensitive to salt stress. The aim of the present study was to undertake the next step in the characterization of acidophilic methanotrophs and obtain a highly enriched methanotrophic population able to grow in acidic peat. We also report on the kinetic and physiological features of the organisms adapted to this unique habitat.
MATERIALS AND METHODS
Sampling sites.
The peat samples were collected from different layers of ombrotrophic and minerotrophic bogs of West Siberia and the European plain of Russia. The locations and descriptions of the bogs are as follows.
(i) Sosvyatskoe ombrotrophic bog.
This bog is located in the Tver region, West Dvinskiy district, field station of the Institute of Forestry, Russian Academy of Sciences (56°10′N, 32°12′E). The vegetation consists of Pinus sylvestris and Sphagnum fuscum on the periphery of the bog and Pinus sylvestris with the Andromeda-Eriophorum-Sphagnum plant community in the raised center. The pH of the peat water is 3.5 to 4.2, and the water table varies seasonally from 10 to 30 cm. The measurements of the methane uptake rate within bog profile have revealed the maximum activity to be located in the 15- to 20-cm layer (4, 17).
(ii) Kyrgyznoye ombrotrophic bog.
This bog is located in West Siberia, Tomsk region, field station of the Institute of Forestry, Siberian Branch of Russian Academy of Sciences (56°N, 85°E). The vegetation consists of Pinus sylvestris with Ledum palustre, as well as Sphagnum fuscum and Eriophorum vaginatum. Other properties are a pH of 3.5 to 4.0, a water table of 10 to 20 cm, and a maximum methane uptake activity located in the 15- to 25-cm layer.
(iii) Krugloye minerotrophic bog.
This bog is in the same location as above. The raised bog center is covered by the Sphagnum-Andromeda community under pine and birch forest; the peripheral plant community consists of Equisetum, Carex, Phragmites, Comarum, and Menyanthes. Other properties are a pH of 4.0 to 4.5, a water table of 0 to 10 cm, and a maximally active methanotrophic layer at 2 to 15 cm.
(iv) Bakchar ombrotrophic bog.
This bog is located in settlement Plotnikovo, Tomsk region, West Siberia, field station of the Institute of Soil Science and Agrochemistry, Siberian Branch of Russian Academy of Sciences (56°53′N, 82°50′E). The main unforested part of the bog is covered with continuous Sphagnum (S. angustifolium and S. magellanicum) and patches of vascular plants (Carex rostrata, Menyanthes trifoliata, and Equisetum fluviatile). The forest on the periphery of the bog consists of Pinus sylvestris, Pinus sibirica, and Betula pubescens; the surface is covered with tussocks of Eriophorum vaginatum, bushes (Andromeda polifolia, Ledum palustre, Rubus chamaemorus, and Eriophorum vaginatum), and Sphagnum fuscum. It has a pH of 3.6 to 4.5, a water table of 0 to 5 cm, and maximal methane consumption in the 5- to 15-cm layer.
Enrichment procedures.
Methanotrophic bacteria were enriched in a liquid mineral medium, M1, containing (in grams per liter of distilled water) KNO3, 1; KH2PO4, 0.2; MgSO4 · 7H2O, 0.1; and CaCl2 · 2H2O, 0.02, with the addition of 0.1% (by volume) of a trace elements stock solution containing (in grams per liter) EDTA, 5; FeSO4 · 7H2O, 2; ZnSO4 · 7H2O, 0.1; MnCl2 · 4H2O, 0.03; CoCl2 · 6H2O, 0.2; CuCl2 · 5H2O, 0.1; NiCl2 · 6H2O, 0.02; and Na2MoO4, 0.03. For enrichment of nitrogen-fixing methanotrophic bacteria, the same medium without KNO3 was used. A series of media with different acidities (the pH varied from 6 to 3) were prepared by adjusting the initial pH of the medium with concentrated phosphoric acid. The inoculation was done with peat samples exhibiting the highest methanotrophic activity (4). Incubations were carried out for 6 to 8 weeks at 20°C in desiccators under a 50:50 methane-air mixture. Control incubations were run in parallel under the same conditions but without methane. As soon as visual turbidity developed, an aliquot was transferred to fresh medium of the same composition.
Maintenance of methanotrophic communities.
Screw-cap serum bottles with a capacity of 500 ml were used for methanotroph cultivation. In addition to the basic M1 medium, we used this medium diluted 5- and 10-fold (M2 and M3, respectively) where indicated. The initial pH of the medium used for maintenance of the communities was 4.5 to 5.0. The total amount of liquid was 20% of the bottle volume. After inoculation, the bottles were closed with silicone rubber septa, and methane was added aseptically via a syringe equipped with a disposable filter (pore size, 0.22 μm) to achieve a 15 to 20% concentration in the headspace. The bottles were incubated on a rotary shaker at 20°C. Methanotrophic bacteria were subcultured at 2- to 3-week intervals.
Test strains of methanotrophic bacteria.
Methylococcus capsulatus (Bath) ATCC 33009 was obtained from the American Type Culture Collection. Methylocystis pyreformis 44 and Methylomonas albus 85 were kindly supplied by Y. A. Trotsenko (IBPM, Pushchino, Russia), and Methylosinus trichosporium 44 was kindly supplied by V. F. Gal’chenko (Institute of Microbiology, Moscow, Russia). All the strains were cultivated on NMS medium (20).
Assay of the effects of pH on growth.
The cells of exponentially growing methanotrophic enrichments at pH 5.0 were collected by centrifugation, washed twice with fresh M2 N-free medium, and transferred to bottles containing the same medium at pH values ranging from 3.5 to 8.5. Variations in the acidity level were achieved by mixing 0.1 M solutions of H3PO4, KH2PO4, K2HPO4, and K3PO4 to create media with the same ionic strength. Two replicates of each pH value were incubated. Control growth experiments with two cultures of neutrophilic methanotrophs—Methylocystis pyreformis 44 and Methylosinus trichosporium 44—were run in parallel under the same conditions. After inoculation and injection of methane into the headspace, the bottles were incubated for 2 weeks on a rotary shaker at 20°C. Growth was monitored by nephelometry, and the pH of culture liquid was determined at the same time. All measurements were performed in triplicate.
Assay of the effects of pH on methanotrophic activity.
The methanotrophic enrichments were grown on M2 medium at an initial pH of 5.0 up to the midpoint of the exponential phase. The cells were pelleted, washed with the same fresh medium, and transferred to bottles containing M2 medium at pH values ranging from 2.5 to 8.5 as described above. Methane was injected into the headspace, and the bottles were incubated for 2 h on a rotary shaker at 20°C. The specific rate of CH4 consumption was determined by linear regression of the residual substrate in the gas phase determined every 0.5 h relative to the cell concentration. The pH values were determined at the beginning and end of the experiment. All values are means of three analyses of each of two replicates.
Cultivation on N-free medium.
To account for any residual N in the inoculum, methanotrophic communities were grown on both nitrate-containing (N+) and nitrate-free (N−) M2 medium to the midpoint of the exponential phase. Then the cells were pelleted, washed twice with fresh nitrogen-free or nitrogen-sufficient medium, and used to inoculate both N− and N+ M2 medium (pH 5.0). Growth was monitored by measuring CO2 evolution and cell biomass accumulation.
DNA extraction.
Both total-community and test strain DNA extractions were performed by a sodium dodecyl sulfate-based lysis method (23), with slight modifications. Cells from 15-ml samples of late-exponential batch cultures were collected by centrifugation. The pellet was ground with sterile sand for disruption of aggregates and suspended in 4 ml of extraction buffer containing 100 mM Tris-HCl (pH 8.0), 100 mM EDTA (pH 8.0), 1.5 M NaCl, and 1% (wt/vol) hexadecyltrimethylammonium bromide (CTAB). To achieve complete lysis, the cells were subjected to three repeated cycles of rapid freezing (−70°C) and thawing (65°C). Proteinase K was added to a final concentration of 50 μg/ml and mixed; then 400 μl of 20% (wt/vol) sodium dodecyl sulfate was added, and the mixture was incubated at 65°C for 2 h with gentle inversion every 15 to 20 min. The purification was performed once with 2 ml of phenol plus 2 ml of chloroform-isoamyl alcohol (24:1) and once with 4 ml of chloroform and was followed by centrifugation. The DNA was precipitated from the aqueous phase with isopropanol, washed with cold 70% ethanol, and dissolved in 400 μl of sterile distilled water.
PCR amplification.
The mmoX and mmoY structural genes of the sMMO were amplified with the primers of McDonald et al. (13). All the oligonucleotides were synthesized at the Macromolecular Structure, Sequencing and Synthesis Facility, Michigan State University. PCR amplification was performed with 20-μl (total volume) reaction mixtures in 0.2-ml Eppendorf tubes by using a programmable temperature cycler (GeneAmp PCR system 9600; Perkin-Elmer Corp., Norwalk, Conn.). The reaction mixture was composed of 1× PCR amplification buffer II (10× PCR amplification buffer contains 500 mM KCl and 100 mM Tris-Cl [pH 8.3]), 1.5 mM MgCl2, each deoxynucleoside triphosphate at 200 μM, each primer at 0.1 mM, 0.5 μl of template DNA (corresponding to approximately 20 to 30 ng of genomic DNA), and 0.5 U of Taq polymerase (Perkin-Elmer Corp.). The reaction conditions were as follows: denaturation at 94°C for 3 min; 35 cycles consisting of 94°C for 30 s, 55°C for 1 min, and 72°C for 2 min 10 s; and one additional 6-min cycle for chain elongation. The reaction products were checked for size and purity on 1.5% agarose gel stained with ethidium bromide.
Cloning and sequencing of mmoX gene PCR products.
The PCR-amplified mmoX gene products were quantified and ligated to the pCR II vector provided with a TA cloning kit from Invitrogen (San Diego, Calif.). The positive recombinant clones were screened by direct amplification of the cloned inserts from transformant cells with vector-specific primers (24). The constructed mmoX clone library was analyzed to reveal the groups of clones with similar inserts. For these purposes, the mmoX inserts were reamplified by the above approach and restricted with different tetrameric endonucleases: MspI, RsaI, HhaI, and HaeIII (Gibco BRL Life Technologies, Gaithersburg, Md.). The resulting restriction products were separated by gel electrophoresis in 3.5% MetaPhor agarose (FMS Bioproducts, Rockland, Maine) in 1× Tris-borate-EDTA (TBE) at 4°C for 5 h and stained with ethidium bromide. Representative clones were sequenced with the plasmid DNA as the template after purification with the Wizard 373 DNA purification system (Promega, Madison, Wis.). Nucleotide sequences were determined by automated fluorescent Taq cycle sequencing with the ABI 373A sequencer (Applied Biosystems, Foster City, Calif.). Approximately 1.5 to 2 μg of the purified plasmid DNA was used for the sequencing reaction performed with forward and reverse M13/pUC sequencing primers (Boehringer GmbH, Mannheim, Germany). Sequence data were assembled and edited with the Sequencher package (version 3.0; Gene Codes Corp., Inc., Ann Arbor, Mich.). Nucleotide and derived amino acid sequences were aligned and compared to similar database sequences with the BCM Search Launcher project (Human Genome Center, Houston, Tex.) and LASERGENE NAVIGATOR program of the DNASTAR package (DNASTAR, Inc., Madison, Wis.).
The sequences of the sMMO gene clusters of Methylococcus capsulatus (Bath) and Methylocystis sp. strain M were obtained from GenBank (accession no. M58499 and U81594, respectively). The Methylosinus trichosporium OB3b sMMO sequence data were obtained from EMBL (accession no. X55394).
Analytical techniques.
CH4 was measured with a gas chromatograph equipped with flame ionization detector. CO2 and O2 were analyzed by gas chromatography with a thermoconductivity detector. All measurements were performed with five replications. The biomass carbon was measured periodically by dichromate oxidation of particulate material and systematically by nephelometry at 410 nm followed by conversion to cell carbon from the respective calibration curve. The acidity of peat extracts was determined with glass electrodes (pH meter I-130). pH dynamics in culture liquids were measured with a Φ12pH/ISE meter (Beckman Instruments, Fullerton, Calif.).
Scanning electron microscopy.
Microscopic examination was done on batch cultures in the late exponential growth phase. Cells were collected by centrifugation and fixed at 4°C for 1 to 2 h in 4% glutaraldehyde buffered with 0.1 M sodium phosphate (pH 7.4). The samples were prepared by a poly-l-lysine procedure to adhere the bacteria to the coverslip. After a brief rinse in the buffer, the samples were dehydrated in an ethanol series (25, 50, 75, and 95%) for 15 min in each solution and three times for 15 min in 100% ethanol. After dehydration, the samples were dried in a Balzers critical-point dryer to prevent any shape alterations and were then coated with gold in an Emscope sputter coater (model SC 500). The cells were examined with a JEOL JSM-6400V scanning electron microscope at the Center for Electron Optics, Michigan State University.
Nucleotide sequence accession numbers.
The nucleotide sequences of the mmoX clones amplified from community DNAs have been deposited into GenBank and assigned accession no. AF004554 and AF004555.
RESULTS
Enrichment and morphological diversity.
Methanotrophic communities were successfully enriched from all peat samples, as judged by surface film formation and increase of medium turbidity up to an optical density of 0.6 to 0.8. Growth was obvious after 6 to 8 weeks of incubation on media of pHs from 4 to 6. Preliminary examinations under phase-contrast microscopy revealed no differences among cultures enriched at different pH values from the same inoculum. Thus, we selected for future studies the methanotrophic communities enriched at pH 4, since this condition was most typical of the natural condition. All but one of the enriched cultures were obtained on nitrogen-containing medium. In one case, (Bakchar bog), we obtained an enrichment culture on both nitrogen-free and nitrogen-sufficient media. All the enriched communities were maintained under laboratory conditions for 6 years (since 1991) and underwent at least 25 to 30 serial transfers with 5% inoculum and medium with an initial pH of 4.5 to 5. Both regular phase-contrast microscopic examinations and growth parameter measurements indicated that communities were stable over the entire study period; i.e., the maximum specific growth rates were 0.0191, 0.0247, and 0.0244 h−1 for 1991, 1992, and 1995, respectively.
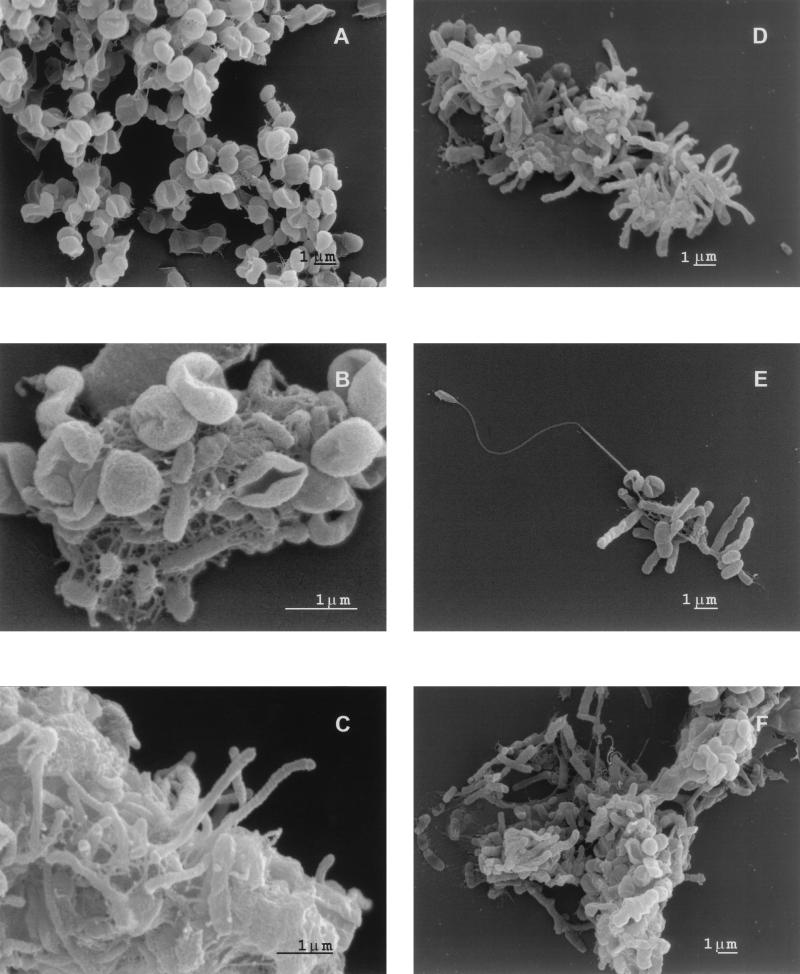
Examination of the methanotrophic communities by electron microscopy revealed wide morphological diversity (Fig. 1). The main component of the microbial consortia was rod-shaped bacteria of different sizes. Another common component in all communities was flattened cells with a concave center (Fig. 1a, b, e, and f). The minor components were pleomorphic, e.g., Hyphomicrobium-like cells in the Sosviatskoe community (Fig. 1e) and long, thin cells in Bakchar community (Fig. 1c). Cell aggregation was a characteristic feature of all cultures, with the number of cells per aggregate varying from 10 to 50 cells in the Sosviatskoe and Krugloye communities to approximately 102 to 103 cells in one macroaggregate in the Bakchar community.
FIG. 1.
Morphological diversity of cells in methanotrophic communities. (A and B) Flattened cells with concave center, Sosvyatskoe. (C) Long thin cells, Bakchar, nitrogen-containing medium. (D) Whimsical aggregates, Bakchar, nitrogen-containing medium. (E) Hyphomicrobium-like cell as a center of aggregation, Sosvyatskoe. (F) Macroaggregates of cells, Bakchar, nitrogen-free medium.
Growth dynamics and stoichiometry.
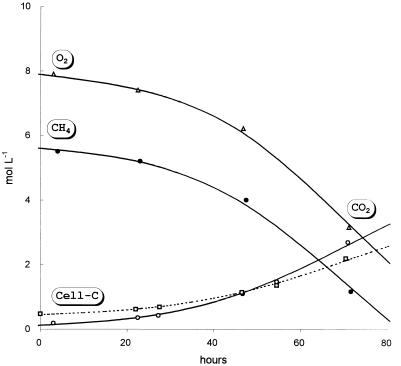
All communities were capable of growth on minimal mineral media without any vitamins and growth factors and with methane as the sole source of carbon and energy. We did not observe growth in control experiments on the same mineral medium without methane. Methane and oxygen were consumed at approximately the same rate, and the two main products were carbon dioxide and biomass (Fig. 2). The maximum specific growth rate of different communities was in the range from 0.02 to 0.04 h−1.
FIG. 2.
Growth dynamics of the methanotrophic community at pH 5.2 in a batch culture in M2 medium. Primary experimental data are smoothed within experimental error by splines.
The ratios between methane and oxygen consumed, as well as CO2 and cell biomass produced, were used to analyze growth stoichiometry and assess the contribution of true methanotrophs to the activity of the mixed enrichment culture. The following empirical stoichiometric equation for the microbial growth of this culture was derived to satisfy the mass balance of three elements (C, O, and H): CH4 + 1.1O2 = 0.41 CH5.9O0.5 + 0.59 CO2 + 0.8 H2O.
The empirically derived formula for the biomass, CH5.9O0.5, is similar to one characterizing the elemental composition of other bacteria (16) except for an unusually high content of hydrogen, which can be a consequence of lipid or poly-β-hydroxybutyrate accumulation. More than half (59%) of the consumed methane carbon was oxidized to CO2, while the rest (41%) was incorporated into the biomass of growing cells, with an average yield of 0.41 ± 0.03 g of biomass carbon per g of consumed CH4-C. During the growth cycle, the yield consistently decreased from 0.45 at zero time to 0.35 g of cell C/g of CH4-C after 100 h, indicating diversion of energy flux to maintenance:
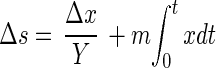 |
1 |
where x is total microbial biomass, Δx is the increase in microbial biomass resulting from the use of substrate Δs (methane), t is time, and m is the maintenance coefficient.
The apparent maintenance coefficient, m, as calculated from equation 1, increased during the course of cultivation from 0.5 to 8.0 mg of CH4-C/h · g of biomass C. It may be explained by inhibitory effects of metabolic products accumulating in batch culture.
The yield, maintenance coefficient, and stoichiometric ratios of these methanotrophic communities are similar to the respective values obtained for pure cultures of known methanotrophs. This suggests that the enrichment has typical methanotrophic physiology and that the nonmethanotrophic members did not contribute significantly to the total mass balance of the community.
MMO amplification and sequencing.
The primers for mmoX and mmoY amplified products of 524 and 602 bp, respectively, from sMMO+ control DNA [Methylococcus capsulatus (Bath) (sMMO+)] and from the Sosvyatskoe, Kyrgyznoye, and Krugloye bog community DNAs. No product was obtained with template DNA from the Bakchar community or from the negative controls, Methylomonas albus 85 (sMMO−) and Methylocystis pyreformis 44 (sMMO−). The mmoX gene PCR-amplified products were cloned, and the library of 43 clones was screened by restriction digestion with four different endonucleases. The Sosvyatskoe and Kyrgyznoye bog communities yielded just one type of clone, whereas the clones obtained from Krugloye bog community form two distinct groups: one of them possessed the same restriction pattern as that found in the other two communities and accounted for 30% of the clones, and the other showed a different pattern and accounted for 70% of the clones.
The nucleotide sequences of clones with similar restriction patterns (clones 2 [Sosvyatskoe], 22 [Kyrgyznoye], and 25 [Krugloye]) were identical and showed 76.7% identity to the clone with the unique restriction pattern (clone 24 [Krugloye]). The PCR products exhibited high nucleotide sequence identity to the mmoX genes from Methylococcus capsulatus (Bath), Methylosinus trichosporium OB3b, and Methylocystis sp. strain M (1, 15, 18). Translated amino acid sequences of mmoX clones correspond to amino acid residues 293 to 466 of the MmoX amino acid sequences of the above cultures at a high level of identity (Table 1). The mmoX gene in the group of clones 2, 22, and 25 diverged from these gene segments in known methanotrophs and showed the same level of identity (79.9%) to the same region in both Methylococcus capsulatus (Bath) and the Methylocystis-Methylosinus group. The amino acid sequence of clone 24 was more closely related to the Methylosinus-Methylocystis group (93.7%) (Table 1).
TABLE 1.
Comparison of the derived amino acid sequences of mmoX clones of PCR-amplified community DNAs with the amino acid sequences of the mmoX proteins from the indicated reference strains
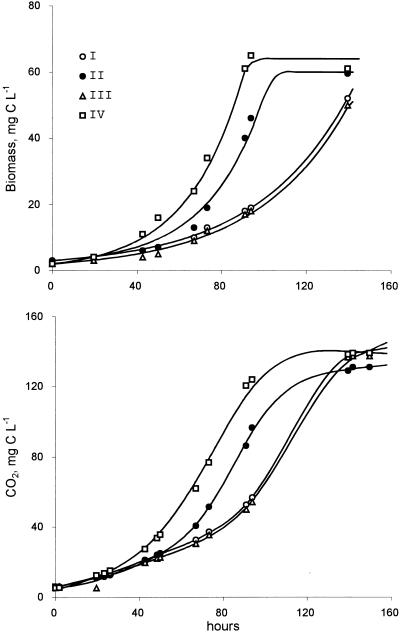
Growth on nitrogen-free medium.
All methanotrophic communities, including those which were enriched on full-strength nitrogen-containing medium, grew on nitrogen-free medium (Fig. 3). The experimental data were fit to the following equation to account for a lag phase (16):
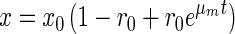 |
2 |
where x is the biomass concentration, x0 is the starting biomass, μm is the maximum specific growth rate, and r0 is a variable reflecting the physiological state at time t = 0 (r is interpreted as the relative content of cell components responsible for intensive growth, e.g., the amount of ribosomes; the smaller r0, the longer the lag phase).
FIG. 3.
Growth dynamics of methanotrophic community (Sosvyatskoe) as influenced by the nitrogen source. The curves are as follows: I, nitrogen-free medium inoculated by bacteria grown on nitrogen-free medium; II, nitrogen-containing medium inoculated with bacteria grown on nitrogen-free medium; III, nitrogen-free medium inoculated with bacteria grown on full medium; IV, nitrogen-containing medium inoculated with bacteria grown on full medium.
The identified parameters (Table 2) indicate that the nitrogen-free medium yielded a slightly lower growth rate than the nitrogen-containing medium, but the difference is small. Transition from a nitrogen-free to a nitrogen-containing medium caused a considerable lag phase. The reverse transition (from rich to poor medium) did not result in a lag phase, indicating the constitutive nature of the basic enzyme system responsible for survival under oligotrophic conditions.
TABLE 2.
Growth parameters of methanotrophic bacteria relative to the nitrogen availability
| Medium for growth of inoculum | Medium for cultivation | r0 | Lag phase (h)a | μ (h−1) |
|---|---|---|---|---|
| N free | Without N | 0.71 | 9.70 | 0.023 |
| N free | N added | 0.40 | 40.24 | 0.037 |
| N sufficient | Without N | 1.00 | 0.00 | 0.023 |
| N sufficient | N added | 1.00 | 0.00 | 0.037 |
The duration of the lag phase was calculated as the time required to raise the instant r variable from r0 to the submaximal value r = 0.75.
Effects of pH on methanotrophic activity and growth.
The methane consumption by pregrown communities was linear throughout a 2-h experiment at all pH values. Both nitrogen-fixing and nitrogen-dependent communities displayed similar patterns. Methane consumption peaked at pH 4.5 to 5.5 and was detectable over the pH range from 2.5 to 8 (Fig. 4A). The specific rate of methane oxidation was rather high (up to 0.5 mmol of CH4/h · g of biomass) at all pH values characteristic of natural conditions (pH 3.5 to 4.5) but exhibited a dramatic decrease in activity at neutral pH (Fig. 4B). The effect of pH on methane uptake kinetics was similar for the original peat samples (5), except that the peat samples had a sharper peak (Fig. 4C), indicating a slightly wider pH tolerance of bacteria selected during enrichment.
FIG. 4.
Dependence of instant methanotrophic activity on extracellular pH. The curves and pK values are calculated from equation 3. (A) Methanotrophic community from Sosvyatskoe; (B) methanotrophic community from Bakchar; (C) data for peat samples from Sosvyatskoe taken from reference 5.
The pH curves can be approximated by equations used in enzymatic kinetics (2):
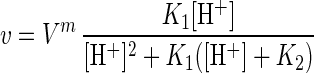 |
3 |
where v is observed reaction rate, Vm is the upper limit of v, and [H+] is the proton concentration. The derivation of equation 3 is based on the assumption that the active center of the enzyme is a dibasic acid, H2E, which undergoes two-step dissociation (H2E ↔ HE− ↔ E2−) with equilibrium constants K1 and K2, and only the singly ionized complex, HE−, is active. Although the real mechanism is much more complex, this equation is useful since it provides a good fit to the experimental data and allows us to make unbiased comparisons among different organisms and experimental conditions. The shape of the pH profile is determined by two pK values, the optimal pH is found as (pK1 + pK2)/2, while the difference pK2 − pK1 determines the width of the peak at 50% of maximal activity. It follows from a least-squares fitting procedure that enriched cultures and the intact peat community have almost the same pH optimum (4.7 to 5.1 and 5.14 respectively), while pH tolerance is higher for isolated bacteria: pK range of 2.5 to 2 units as compared with 1.34 units in the original peat community.
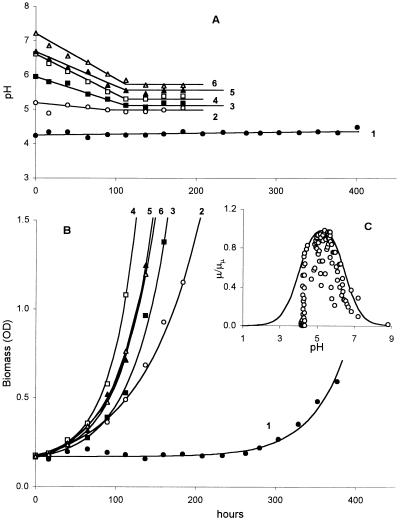
We also evaluated the growth rate of these communities as a function of pH. This experiment was complicated by the fact that cultures inoculated in all media of pH > 5.0 acidified the medium before achieving their maximum growth rate (Fig. 5A). However, we can estimate the maximum growth rate as a function of pH by calculating the instantaneous growth rate (Fig. 5C), at each measured pH, during the growth response for all cultures (Fig. 5B). The highest specific growth rates were established when the acidity of the extracellular liquid reached the minimal steady-state values, which were in the pH range 5.0 to 5.7 depending on the initial pH. The higher the initial pH, the higher the final steady-state pH values (Fig. 5A).
FIG. 5.
Growth of methanotrophic bacteria in media of different initial acidities. (A) Measured pH of media during growth. (B) Growth of cultures in media of different initial pH values; the curves were calculated from equation 2 with parameters listed in Table 3. The numbers with the curves and symbols correspond to the pH conditions in panel A. (C) Plot of the instantaneous specific growth rate versus measured pH values for that time point. The growth rates were calculated from the fitted experimental curves for growth.
All growth data were in excellent agreement with equation 2 (Table 3). The kinetic analysis allows us to clarify the following growth features. First, in contrast to the pH profile of methanotrophic activity, there is no single relationship between pH and microbial growth rate. Instead, there is a cloud of experimental points (Fig. 5C) over a broader pH range, although the calculated maximum is pH 5.25. The likely reason for the greater scatter is the effect of the pH history of the batch culture. Second, the lag phase is longer for cultures started at pHs more acidic and basic than the optimum. The lag is particularly long for the two cultures started at pH 4.2, but it is noteworthy that they achieved a high maximum growth rate (Table 3). After a more or less transient phase, all the cultures attained maximum growth rate in the range 0.013 to 0.030 h−1 independent of the initial conditions. Third, the most optimum growth shown by kinetic analysis (Table 3) corresponds to a moderately acidic medium at pH values around 5.
TABLE 3.
Growth parameters of methanotrophic bacteria relative to the initial acidity of the medium
| pH | r0 | Lag phase (h) | μm (h−1) |
|---|---|---|---|
| 4.21 | 0.0005 | 393 | 0.022 |
| 4.23 | 0.0005 | 380 | 0.023 |
| 5.19 | 0.5275 | 74 | 0.013 |
| 5.23 | 0.6908 | 23 | 0.013 |
| 6.44 | 0.3139 | 84 | 0.022 |
| 6.61 | 0.1789 | 87 | 0.030 |
| 6.68 | 0.2071 | 98 | 0.025 |
| 7.21 | 0.1908 | 102 | 0.025 |
By contrast, both Methylocystis pyreformis 44 and Methylosinus trichosporium 44 were not able to grow at pH values below 5.6 to 5.8 whereas their highest growth rates were observed at pH values around 6.5 to 7.5 (data not shown). Thus, we conclude that the enriched cultures do differ from neutrophilic methanotrophs and belong to the category of moderately acidophilic organisms.
Effect of salt concentration.
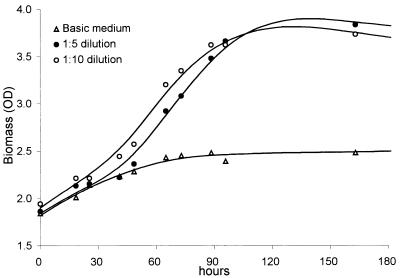
To test the effect of salts, we diluted the basic M1 medium to obtain media M2 (5-fold diluted) and M3 (10-fold diluted). The initial acidity of all the tested media was the same, pH 5.0. Maximum growth was achieved on the two diluted media (Fig. 6).
FIG. 6.
Effect of more dilute media on the growth of the methanotrophic community. OD, optical density.
DISCUSSION
The problem of microbial isolation from natural sources remains the most crucial step in studies of microbial diversity. It is commonplace to state that collections of environmental isolates represent only a minor part of microbial communities functioning in natural habitats. Evidence from both direct microscopy and molecular methods supports this notion. Unculturable microorganisms are usually offered as the explanation. However, most cases of unsuccessful isolation are probably due to unknown growth requirements; i.e., they are due to organisms which are difficult to culture rather than impossible to culture. Hence, more successful isolation probably requires more accurate simulation of the natural environment in the laboratory.
The methanotrophs are a ubiquitous group of microorganisms that are widespread in freshwater, marine, and terrestrial environments (6). As reviewed by King (11), numerous isolates have been obtained from diverse habitats (8, 20), but the approaches used for isolation have been limited. The media routinely employed were based on nitrate or ammonia mineral salts (20). In the case of isolation of marine methanotrophs, adaptation of standard media by means of dilution and supplementation with NaCl have proven useful (10). Generally, however, the media used are not consistent with the conditions of the natural habitat. The Sphagnum peat bogs, characterized by low pH, low mineral content, and weak buffering capacity, are an example. Hence, it is hardly surprising that the usual neutral media with a salt content of about 1.5 to 3 g/liter (in contrast to 5 to 50 mg/liter in peat water) are not appropriate for the isolation of indigenous methanotrophic populations.
We used a two-step habitat-specific approach for the enrichment of these elusive methanotrophs. In the first step, a kinetic study of the target microbial process is performed on in intact soil samples incubated with added substrate (CH4) under various temperatures, pH values, and mineral salt contents. This provides data on the growth kinetics of the dominant microbial components in situ and on the optimum conditions for the process under study. In our case, we have found a low specific growth rate, inhibition of metabolic activity by inorganic salts at concentrations above 100 mg liter−1, and optimal growth at 15 to 20°C and pH 5 (4, 5). These features become an “identity card” for the microbial population in question. The second step is the development of laboratory culture conditions which imitate the natural habitat in its essential features: low salt content, decreased buffering capacity, temperature, and pH. The selected enriched methanotrophic communities turned out to be unique to acid peat bogs and responsible for methane oxidation in this ecosystem. All essential growth parameters were the same or very similar to those found previously in intact peat samples. Furthermore, the pH and temperature range and susceptibility to salt stress were similar for the peat and the enrichments.
This study provides evidence that there are some methanotrophs which are moderate acidophiles. The basis for this acidophily is not known. Perhaps higher salt contents provide higher buffer capacity, which prevents metabolic pH tuning (shift to more neutral values) in the microenvironment surrounding the cells. Also, heterogeneity within the aggregates typical of this community (Fig. 1) could enhance tolerance to the conditions of this site. A similar cell arrangement was revealed for acidotolerant chemolithotrophic nitrifying bacteria enriched from acid soils and nitrifying at pH 4 (3).
Another essential property of the methanotrophic community from ombrotrophic bogs is its ability to develop on nitrogen-free media. This feature is particularly important since the concentration of available nitrogen (both nitrate and ammonium) in wetlands of this type varies in the range 0.1 to 1.0 mg of nitrogen per liter. Thus, methane consumers in the bog are independent of a bound nitrogen source. On the other hand, if nitrogen becomes available, methanotrophic communities are able to switch to another type of nitrogen acquisition.
The methanotrophic potential of these communities was confirmed by growth stoichiometry and finding PCR-amplifiable products of the predicted size for both mmoX and mmoY genes of sMMO and a high degree of sequence identity to 524 bp of the mmoX gene. One site, Krugloye, yielded a mmoX clone, whose sequence was very similar (identity of 93.7% and similarity of 96.6%) to the previously known mmoX sequence from Methylocystis sp. strain M. However, the most commonly found clones were much more divergent in sequence from the two previously known mmoX sequence clusters, those of the Methylosinus-Methylocystis group and of Methylococcus capsulatus (Bath). These sequence differences are 77 to 80% and 80%, respectively. This result suggests that the sMMO of these acidophilic methanotrophs is also novel. Furthermore, the clones of this group were obtained from three of the four bog communities. The last fact supports the hypothesis that such organisms could be typical of acid peat environments.
ACKNOWLEDGMENTS
This research was supported in part by the Russian Fund of Fundamental Research, grant 96-04-49321, and by NSF grants INT9315089 for Russian collaborative work and BIR9120006.
We thank Carol Flegler, MSU Center for Electron Optics, for her professional assistance in electron microscopy, and we thank Tamara Tsoi, Center for Microbial Ecology, for sharing her experience of molecular techniques. S.D. and N.P. appreciate very much the great technical advantages and friendly atmosphere at the Center of Microbial Ecology during their experimental work at MSU.
REFERENCES
- 1.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 2.Cornish-Bowden A. Principles of enzyme kinetics. London, United Kingdom: Butterworths; 1976. pp. 142–159. [Google Scholar]
- 3.De Boer W, Klein Gunnewiek P J A, Veenhuis M, Bock E, Laanbroek H J. Nitrification at low pH by aggregated chemolithotrophic bacteria. Appl Environ Microbiol. 1991;57:3600–3604. doi: 10.1128/aem.57.12.3600-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dedysh S N. Abstracts of the XIth International Symposium on Environmental Biogeochemistry. 1993. Methane consumption in forested acidic peat bogs; pp. 27.IX–1.X. [Google Scholar]
- 5.Dedysh S N, Panikov N S. Effect of pH, temperature, and concentration of salts on methane oxidation kinetics in Sphagnum peat. Microbiology (transl form Mikrobiologiya) 1997;66:569–573. [Google Scholar]
- 6.Hanson R S, Netrusov A I, Truji K. The obligate methanotrophic bacteria: Methylococcus, Methylomonas and Methylosinus. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The procaryotes. New York, N.Y: Springer-Verlag; 1991. pp. 661–684. [Google Scholar]
- 7.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyer J, Malashenko Y, Berger U, Budkova E. Vertreitung methanotropher Bakterien. Z Allg Mikrobiol. 1984;24:725–744. [Google Scholar]
- 9.Heyer J, Suckow R. Ecological studies of methane oxidation in an acid bog lake. Limnologica. 1985;16:247–266. [Google Scholar]
- 10.Holmes A J, Owens N J P, Murrell J C. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiology. 1995;141:1947–1955. doi: 10.1099/13500872-141-8-1947. [DOI] [PubMed] [Google Scholar]
- 11.King G M. Ecological aspects of methane oxidation, a key determinant of global methane dynamics. Adv Microb Ecol. 1992;12:431–461. [Google Scholar]
- 12.Matthews E, Fung I. Methane emission from natural wetlands: global distribution, area, and environmental characteristics of sources. Global Biochem Cycles. 1987;1:61–86. [Google Scholar]
- 13.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology technigues. FEMS Microbiol Ecol. 1996;21:197–211. [Google Scholar]
- 15.McDonald I R, Uchiyama H, Kambe S, Yagi O, Murrell J C. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl Environ Microbiol. 1997;63:1898–1904. doi: 10.1128/aem.63.5.1898-1904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panikov N S. Microbial growth kinetics. London, United Kingdom: Chapman & Hall, Ltd.; 1995. pp. 187–236. [Google Scholar]
- 17.Panikov N S, Semenov A M, Tarasov A L, Belyaev A S, Kravchenko I K, Smagina M V, Palejeva M V, Zelenev V V, Skupchenko K V. Methane production and uptake in soils of the European part of the USSR. J Ecol Chem. 1993;1:7–18. [Google Scholar]
- 18.Stainthorpe A C, Lees V, Salmond G P C, Dalton H, Murrell J C. The methane monooxygenase cluster of Methylococcus capsulatus (Bath) Gene. 1990;91:27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- 19.Topp E, Hanson R S. Metabolism of radiatively important trace gases by methane-oxidizing bacteria. In: Rogers J E, Whitman W B, editors. Microbial production and consumption of greenhouse gases: methane, nitrogen oxides and halomethanes. Washington, D.C: American Society for Microbiology; 1991. pp. 71–90. [Google Scholar]
- 20.Whittenbury R, Phillips K C, Wilkinson T F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 21.Yavitt J B, Downey D M, Lancaster E, Lang G E. Methane consumption in decomposing Sphagnum-derived peat. Soil Biol Biochem. 1990;22:441–447. [Google Scholar]
- 22.Yavitt J B, Lang G E, Downey D M. Potential methane production and methane oxidation rates in peatland ecosystems of the Appalachian Mountains, United States. Global Biogeochem Cycles. 1988;2:253–268. [Google Scholar]
- 23.Zhou J-Z, Fries M R, Chee-Sanford J C, Tiedje J M. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol. 1995;45:500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]