Abstract
Large-scale whole-genome sequencing (WGS) studies have improved our understanding of the contributions of coding and noncoding rare variants to complex human traits. Leveraging association effect sizes across multiple traits in WGS rare variant association analysis can improve statistical power over single-trait analysis, and also detect pleiotropic genes and regions. Existing multi-trait methods have limited ability to perform rare variant analysis of large-scale WGS data. We propose MultiSTAAR, a statistical framework and computationally-scalable analytical pipeline for functionally-informed multi-trait rare variant analysis in large-scale WGS studies. MultiSTAAR accounts for relatedness, population structure and correlation among phenotypes by jointly analyzing multiple traits, and further empowers rare variant association analysis by incorporating multiple functional annotations. We applied MultiSTAAR to jointly analyze three lipid traits (low-density lipoprotein cholesterol, high-density lipoprotein cholesterol and triglycerides) in 61,861 multi-ethnic samples from the Trans-Omics for Precision Medicine (TOPMed) Program. We discovered new associations with lipid traits missed by single-trait analysis, including rare variants within an enhancer of NIPSNAP3A and an intergenic region on chromosome 1.
Advances in next generation sequencing technologies and the decreasing cost of whole-exome/whole-genome sequencing (WES/WGS) have made it possible to study the genetic underpinnings of rare variants (i.e. minor allele frequency (MAF) < 1%) in complex human traits. Large nationwide consortia and biobanks, such as the National Heart, Lung and Blood Institute (NHLBI)’s Trans-Omics for Precision Medicine (TOPMed) Program1, the National Human Genome Research Institute’s Genome Sequencing Program (GSP) , the National Institute of Health’s All of Us Research Program2, and the UK’s Biobank WGS Program3, are expected to sequence more than a million of individuals in total, at more than 1 billion genetic variants in both coding and noncoding regions of the human genome, while also recording thousands of phenotypes. To mitigate the lack of power of single-variant analyses to identify rare variant associations4, variant set tests have been proposed to analyze the joint effects of multiple rare variants 5-9, where most of the work has focused single trait analysis.
Pleiotropy occurs when genetic variants influence multiple traits10. There is growing empirical evidence from genome-wide association studies (GWASs) that many variants have pleiotropic effects11,12. Identifying these effects can provide valuable insights into the genetic architecture of complex traits13. As such, it is of increasing interest to identify pleiotropic rare variants by jointly analyzing multiple traits in WGS rare variant association studies (RVASs).
Several existing methods for multi-trait rare variant association analysis, such as MSKAT14, Multi-SKAT15 and MTAR16, have shown that leveraging the cross-phenotype correlation structure can improve the power of multi-trait analyses compared to single-trait analyses when analyzing pleiotropic genes14-17. However, existing methods do not scale well, and are not feasible when analyzing large-scale WGS studies with hundreds of millions of rare variants in samples exhibiting relatedness and population structure. Furthermore, none of the existing multi-trait rare variant analysis methods leverages functional annotations that predict the biological functionality of variants, resulting in limited interpretability and power loss. While the STAAR method18 dynamically incorporates multiple variant functional annotations to maximize the power of rare variant association tests, it is designed for single-trait analysis and cannot be directly applied to multiple traits.
To overcome these limitations, we propose the Multi-trait variant-Set Test for Association using Annotation infoRmation (MultiSTAAR), a statistical framework for multi-trait rare variant analyses of large-scale WGS studies and biobanks. It has several features. First, by fitting a null Multivariate Linear Mixed Model (MLMM)19 for multiple quantitative traits simultaneously, adjusting for ancestry principal components (PCs)20 and using a sparse genetic relatedness matrix (GRM)21,22, MultiSTAAR scales well but also accounts for relatedness and population structure, as well as correlations among the multiple traits. Second, MultiSTAAR enables the incorporation of multiple variant functional annotations as weights to improve the power of RVASs. Furthermore, we provide MultiSTAAR via a comprehensive pipeline for large-scale WGS studies, that facilitates functionally-informed multi-trait analysis of both coding and noncoding rare variants. Third, MultiSTAAR enables conditional multi-trait analysis to assess rare variant association signals beyond known common and low frequency variants.
In the current study, we conducted extensive simulation studies to demonstrate the validity of MultiSTAAR and to assess the power gain of MultiSTAAR by incorporating multiple relevant variant functional annotations, and its ability in preserving Type I error rates. We then applied MultiSTAAR to perform WGS RVAS of 61,838 ancestrally diverse participants from 20 studies from NHLBI’s TOPMed consortium by jointly analyzing three circulating lipid traits: low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C) and triglycerides (TG). We show that MultiSTAAR is computationally feasible for large-scale WGS multi-trait rare variant analysis, and in conditional analysis of LDL-C, HDL-C and TG, MultiSTAAR identifies signals that were missed either by the existing multi-trait rare variant analysis methods that overlook variant functional annotations, or by single-trait functionally-informed analysis that ignore correlations between phenotypes.
Results
Overview of the methods
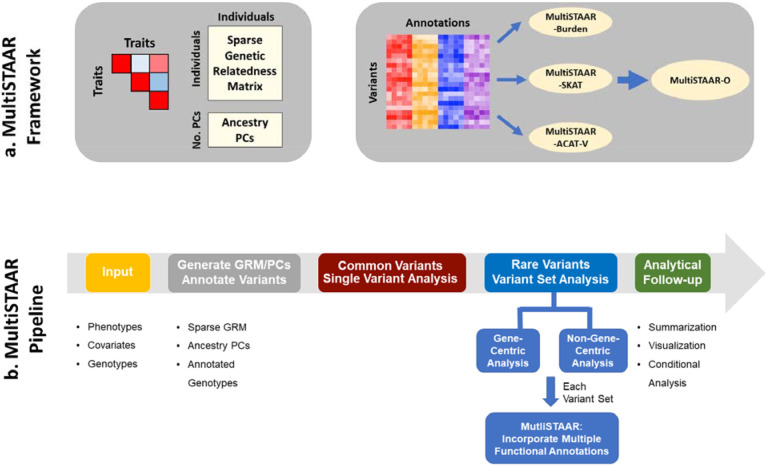
MultiSTAAR is a statistical framework and an analytic pipeline for jointly analyzing multiple traits in large-scale WGS rare variant association studies. There are two main components in the MultiSTAAR framework: (i) fitting null MLMMs using ancestry PCs and sparse GRMs to account for population structure, relatedness and the correlation between phenotypes, and (ii) testing for associations between each aggregated variant set and multiple traits by dynamically incorporating multiple variant functional annotations18 (Fig. 1a).
Fig. 1 ∣. MultiSTAAR framework and pipeline.
a, MultiSTAAR framework. (i) Fit null Multivariate Linear Mixed Models (MLMMs) using sparse GRM and ancestry PCs to account for population structure, relatedness and the correlation between phenotypes. (ii) Test for associations between each variant set and multiple traits by dynamically incorporating multiple variant functional annotations. b, MultiSTAAR pipeline. (i) Prepare the input data of MultiSTAAR, including genotypes, multiple phenotypes and covariates. (ii) Calculate sparse GRM, ancestry PCs and annotate all variants in the genome. (iii) Perform single variant analysis for common variants. (iv) Define the rare variant analysis units, including gene-centric analysis of five coding functional categories and eight noncoding functional categories and non-gene-centric analysis of sliding windows. (v) Provide result summarization and perform analytical follow-up via conditional analysis.
In WGS RVASs, an important but often underemphasized challenge is selecting biologically-meaningful and functionally-interpretable analysis units, especially for the noncoding genome23,24. In gene-centric analyses of multiple traits, MultiSTAAR provides five functional categories (masks) to aggregate coding rare variants of each protein-coding gene, as well as an additional eight masks of regulatory regions to aggregate noncoding rare variants. In non-gene-centric analyses of multiple traits, MultiSTAAR performs agnostic genetic region analyses using sliding windows18,25 (Fig. 1b).
For each rare variant set analyzed, MultiSTAAR first constructs the multi-trait burden, SKAT and ACAT-V test statistics (Methods). For each type of rare variant test, MultiSTAAR calculates multiple candidate P values using different variant functional annotations as weights, following the STAAR framework18. MultiSTAAR then aggregates the association strength by combining the P values from all annotations using the ACAT method, that provides robustness to correlation between tests9, and proposes an omnibus test, MultiSTAAR-O, that leverages the advantages of different type of tests (Methods). Furthermore, MultiSTAAR can test multi-trait rare variants’ associations conditional on a set of known associations (Fig. 1b).
Simulation studies
To evaluate the type I error rates and the power of MultiSTAAR, we performed simulation studies under several configurations. Following the steps described in Data Simulation (Methods), we generated three quantitative traits with a correlation matrix similar to the empirical correlation in the three lipid traits26-28. We then generated genotypes by simulating 20,000 sequences for 100 different 1 megabase (Mb) regions, each of them were generated to mimic the linkage disequilibrium structure of an African American population by using the calibration coalescent model29. Throughout the simulation studies, we randomly and uniformly selected 5-kilobase (kb) regions from these 1-Mb regions and considered sample sizes of 10,000 for each replicate. The simulation studies focused on aggregating uncommon variants with an MAF < 5%.
Type I error rate evaluations
We performed 108 simulations to evaluate the type I error rates of the multi-trait burden, SKAT, ACAT-V and MultiSTAAR-O tests at α = 10−4, 10−5 and 10−6 (Supplementary Table 1). The results show that, for multi-trait rare variant analysis, all four MultiSTAAR tests controlled the type I error rates at very close to the nominal α levels.
Empirical power simulations
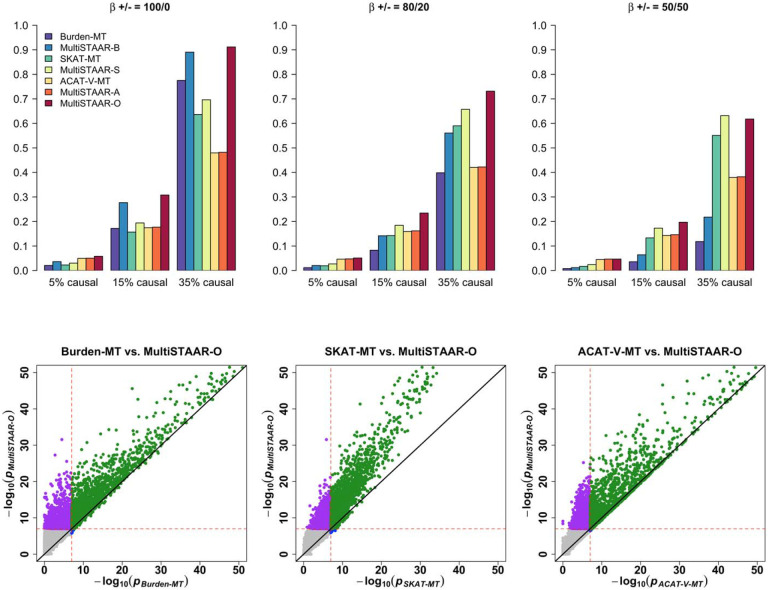
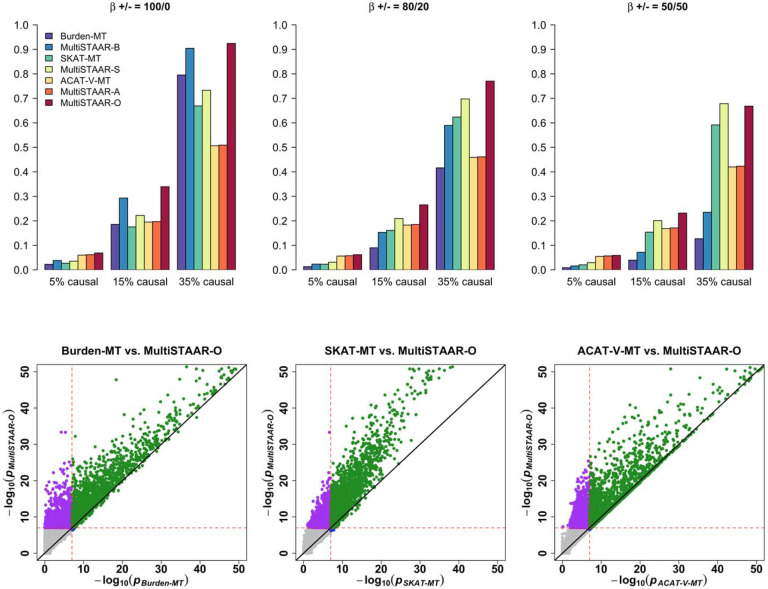
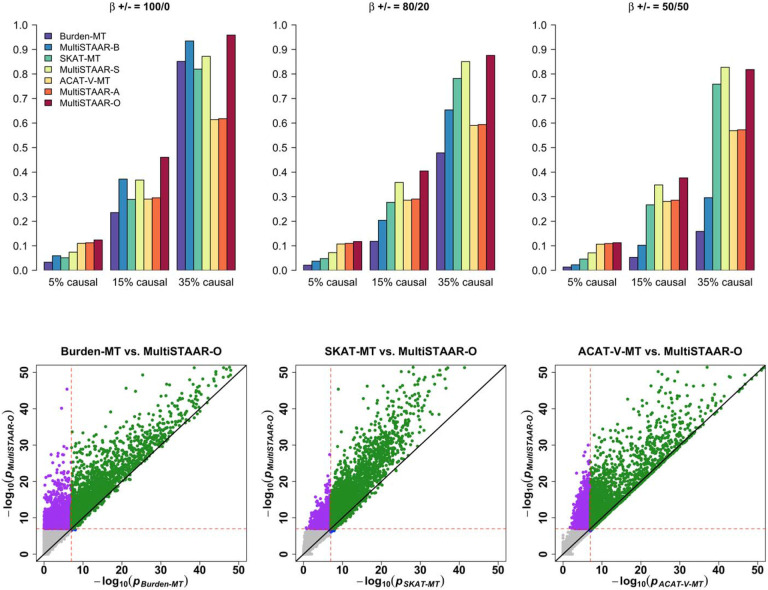
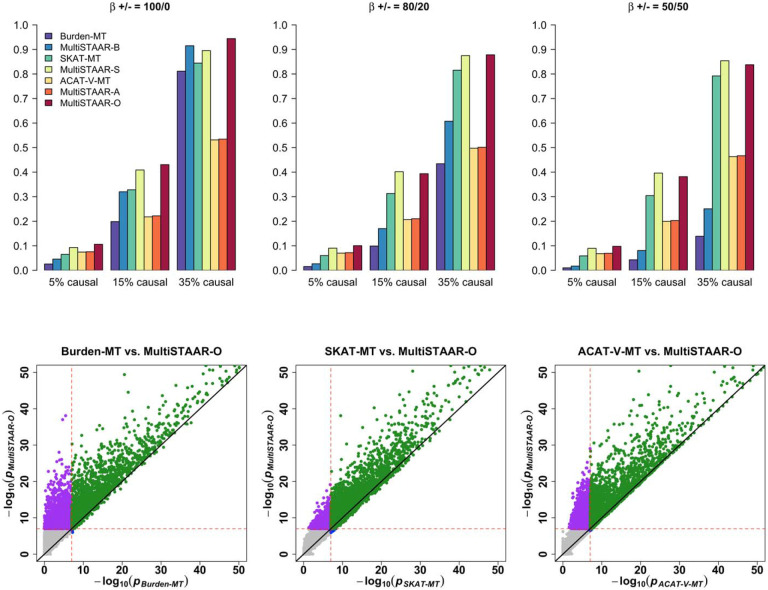
We next assessed the power of MultiSTAAR-O for the analysis of multiple phenotypes under different genetic architectures, while also comparing its power with existing methods. Specifically, we considered four models, in which variants in the signal region (variant-phenotype association regions) were associated with (1) one phenotype only, (2) two positively correlated phenotypes, (3) two negatively correlated phenotypes and (4) all three phenotypes. In addition, we considered different proportions (5%, 15% and 35% on average) of causal variants in the signal region, where causality of variants depended on different sets of annotations, and the effect size directions of causal variants were allowed to vary (Methods). Power was evaluated as the proportions of P values less than α = 10−7 based on 104 simulations. Overall, MultiSTAAR-O consistently delivered higher power to detect signal regions compared to multi-trait burden, SKAT and ACAT-V tests, through its incorporation of multiple annotations (Extended Data Figs. 2-5, Supplementary Figs. 1-4). This power advantage was also robust to the existence of noninformative annotations.
Application to the TOPMed lipids WGS data
We applied MultiSTAAR to identify rare variant associations with three quantitative lipid traits (LDL-C, HDL-C and TG) through a multi-trait analysis using TOPMed Freeze 8 WGS data, comprising 61,838 individuals from 20 multi-ethnic studies (Supplementary Note). LDL-C values were adjusted for the usage of lipid-lowering medication26,30 (Methods), and DNA samples were sequenced at >30x target coverage. Sample- and variant-level quality control were performed for each participating study1,26,30.
Race/ethnicity was measured using a combination of self-reported race/ethnicity and study recruitment information31 (Supplementary Note). Of the 61,838 samples, 15,636 (25.3%) were Black or African American, 27,439 (44.4%) were White, 4,461 (7.2%) were Asian or Asian American, 13,138 (21.2%) were Hispanic/Latino American and 1,164 (1.9%) were Samoans. There were 414 million single-nucleotide variants (SNVs) observed overall, with 6.5 million (1.6%) common variants (MAF > 5%), 5.2 million (1.2%) low-frequency variants (1% ≤ MAF ≤ 5%) and 402 million (97.2%) rare variants (MAF < 1%). The study-specific demographics and baseline characteristics are given in Supplementary Table 2.
Gene-centric multi-trait analysis of coding and noncoding rare variants
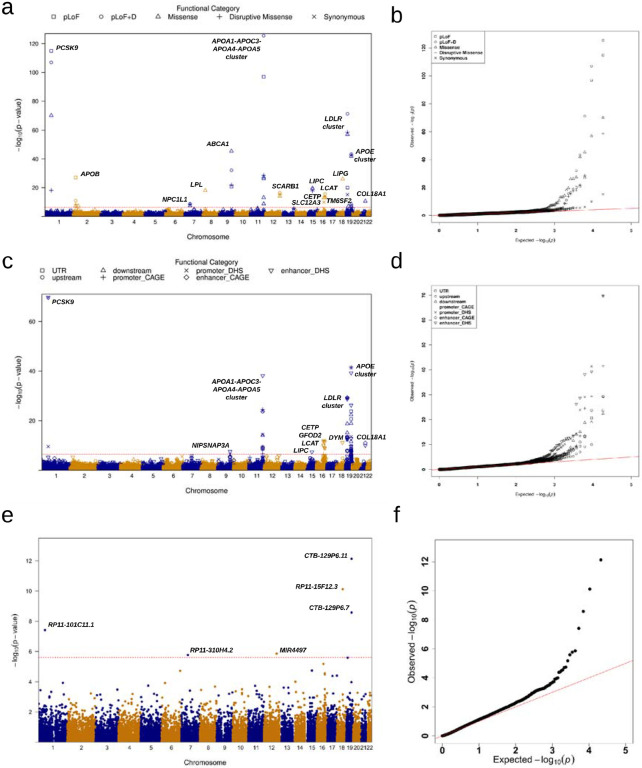
We applied MultiSTAAR-O on gene-centric multi-trait analysis of coding and noncoding rare variants of genes with lipid traits in TOPMed. For coding variants, rare variants (MAF < 1%) from five coding functional categories (masks) were aggregated, separately, and analyzed using a joint model for LDL-C, HDL-C and TG, including (1) putative loss-of-function (stop gain, stop loss and splice) rare variants, (2) missense rare variants, (3) disruptive missense rare variants, (4) putative loss-of-function and disruptive missense rare variants and (5) synonymous rare variants of each protein-coding gene. The putative loss-of-function, missense and synonymous RVs were defined by GENCODE Variant Effect Predictor (VEP) categories32. The disruptive variants were further defined by MetaSVM33, which measures the deleteriousness of missense mutations. We incorporated 9 annotation principal components (aPCs)18,26,34, CADD35, LINSIGHT36, FATHMM-XF37 and MetaSVM33 (for missense rare variants only) along with the two MAF-based weights4 in MultiSTAAR-O (Supplementary Table 3). The overall distribution of MultiSTAAR-O P values was well-calibrated for the multi-trait analysis of coding rare variants (Extended Data Fig. 1b). At a Bonferroni-corrected significance threshold of α = 0.05/(20,000 × 5) = 5.00 × 10−7 accounting for five different coding masks across protein-coding genes, MultiSTAAR-O identified 51 genome-wide significant associations using unconditional multi-trait analysis (Extended Data Fig. 1a, Supplementary Table 4). After conditioning on previously reported variants associated with LDL-C, HDL-C or TG located within a 1 Mb broader region of each coding mask in the GWAS Catalog and Million Veteran Program (MVP)26,38,39, 34 out of the 51 associations remained significant at the Bonferroni-corrected threshold of α = 0.05/51 = 9.80 × 10−4 (Table 1).
Table 1 ∣. TOPMed Gene-centric coding multi-trait analysis results of both unconditional analysis and analysis conditional on known lipids-associated variants.
A total of 61,838 samples from the TOPMed Program were considered in the analysis. Results for the conditionally significant genes (unconditional MultiSTAAR-O P < 5.00 × 10−7; conditional MultiSTAAR-O P < 9.80 × 10−4) are presented in the table. MultiSTAAR-O is a two-sided test. Chr. no., chromosome number; Category, functional category; No. of SNVs, number of rare variants (MAF < 1%) of the particular coding functional category in the gene; MultiSTAAR-O, MultiSTAAR-O P value; Variants (adjusted), adjusted variants in the conditional analysis.
| Gene | Chr. no. |
Category | No. of SNVs |
MultiSTAAR-O (Unconditional) |
MultiSTAAR-O (Conditional) |
Variants (adjusted) |
|---|---|---|---|---|---|---|
| PCSK9 | 1 | Putative loss-of-function | 14 | 1.14E-115 | 2.66E-08 | rs12117661, rs2495491, rs11591147, rs67608943, rs72646508, rs693668, rs28362261, rs28362263, rs141502002, rs505151, rs28362286 |
| APOB | 2 | Putative loss-of-function | 29 | 8.04E-28 | 5.76E-27 | rs12478327, rs72654432, rs1042034, rs676210, rs533617, rs17240441, rs34722314, rs563290, rs10692845 |
| ABCA1 | 9 | Putative loss-of-function | 28 | 2.04E-21 | 5.41E-21 | rs2150867, rs33918808, rs112853430, rs4149307, rs9282541, rs1883025, rs1800978 |
| LDLR | 19 | Putative loss-of-function | 19 | 8.81E-21 | 7.16E-21 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
| PCSK9 | 1 | Missense | 271 | 8.94E-71 | 1.29E-10 | rs12117661, rs2495491, rs11591147, rs67608943, rs72646508, rs693668, rs28362261, rs28362263, rs141502002, rs505151, rs28362286 |
| APOB | 2 | Missense | 1407 | 5.57E-08 | 4.31E-08 | rs12478327, rs72654432, rs1042034, rs676210, rs533617, rs17240441, rs34722314, rs563290, rs10692845 |
| ABCG5 | 2 | Missense | 242 | 5.75E-08 | 9.81E-08 | rs114780578, rs11887534, rs4245791 |
| NPC1L1 | 7 | Missense | 477 | 3.10E-08 | 1.60E-07 | rs217381 |
| LPL | 8 | Missense | 149 | 9.57E-19 | 7.14E-04 | rs6996383, rs268, rs328, rs3289, rs13702, rs15285, rs78810414, rs28550053, rs12676079, rs55682243 |
| ABCA1 | 9 | Missense | 597 | 3.63E-46 | 1.75E-33 | rs2150867, rs33918808, rs112853430, rs4149307, rs9282541, rs1883025, rs1800978 |
| SCARB1 | 12 | Missense | 192 | 6.77E-15 | 3.55E-15 | rs6488913, rs4765127, rs1716407, rs825456, rs1672875, rs10846744, rs10773112, rs187471874, rs10773119 |
| LIPC | 15 | Missense | 246 | 2.54E-20 | 6.66E-15 | rs1973688, rs1601935, rs2043082, rs10468017, rs1532085, rs436965, rs35980001, rs1800588, rs2070895, rs113298164 |
| CETP | 16 | Missense | 168 | 8.84E-14 | 2.09E-04 | rs35571500, rs247617, rs17231506, rs34498052, rs34119551, rs34065661, rs1597000001*, rs7499892, rs5883, rs289719, rs11860407, rs189866004, rs5880 |
| LCAT | 16 | Missense | 107 | 9.18E-14 | 3.06E-17 | rs111315946, rs150660813, rs4986970, rs35673026, rs1109166, rs548291389 |
| LDLR | 19 | Missense | 342 | 7.92E-58 | 2.12E-57 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
| TM6SF2 | 19 | Missense | 120 | 7.06E-08 | 6.16E-07 | rs3761077, rs150641967, rs187429064, rs2074304 |
| PCSK9 | 1 | Putative loss-of-function and disruptive missense | 71 | 1.14E-107 | 8.22E-17 | rs12117661, rs2495491, rs11591147, rs67608943, rs72646508, rs693668, rs28362261, rs28362263, rs141502002, rs505151, rs28362286 |
| APOB | 2 | Putative loss-of-function and disruptive missense | 75 | 9.96E-12 | 9.86E-12 | rs12478327, rs72654432, rs1042034, rs676210, rs533617, rs17240441, rs34722314, rs563290, rs10692845 |
| NPC1L1 | 7 | Putative loss-of-function and disruptive missense | 303 | 1.79E-09 | 8.29E-09 | rs217381 |
| ABCA1 | 9 | Putative loss-of-function and disruptive missense | 357 | 7.85E-33 | 2.66E-33 | rs2150867, rs33918808, rs112853430, rs4149307, rs9282541, rs1883025, rs1800978 |
| APOC3 | 11 | Putative loss-of-function and disruptive missense | 15 | 2.86E-126 | 3.01E-06 | rs509728, rs61905072, rs66505542, rs7102314, rs964184, rs75198898, rs142958146, rs2075291, rs3135506, rs651821, rs45611741, rs662799, rs10750097, rs9804646, rs978880643, rs2070669, rs76353203, rs138326449, rs147210663, rs140621530, rs525028, rs141469619, rs188287950, rs202207736 |
| SCARB1 | 12 | Putative loss-of-function and disruptive missense | 60 | 3.49E-17 | 2.14E-17 | rs6488913, rs4765127, rs1716407, rs825456, rs1672875, rs10846744, rs10773112, rs187471874, rs10773119 |
| LIPC | 15 | Putative loss-of-function and disruptive missense | 130 | 1.01E-19 | 1.49E-17 | rs1973688, rs1601935, rs2043082, rs10468017, rs1532085, rs436965, rs35980001, rs1800588, rs2070895, rs113298164 |
| LCAT | 16 | Putative loss-of-function and disruptive missense | 88 | 2.38E-16 | 5.07E-17 | rs111315946, rs150660813, rs4986970, rs35673026, rs1109166, rs548291389 |
| LDLR | 19 | Putative loss-of-function and disruptive missense | 221 | 6.97E-72 | 1.57E-71 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
| PCSK9 | 1 | Disruptive missense | 57 | 7.03E-19 | 1.33E-12 | rs12117661, rs2495491, rs11591147, rs67608943, rs72646508, rs693668, rs28362261, rs28362263, rs141502002, rs505151, rs28362286 |
| APOB | 2 | Disruptive missense | 46 | 5.78E-09 | 4.48E-09 | rs12478327, rs72654432, rs1042034, rs676210, rs533617, rs17240441, rs34722314, rs563290, rs10692845 |
| NPC1L1 | 7 | Disruptive missense | 276 | 3.34E-09 | 1.57E-08 | rs217381 |
| ABCA1 | 9 | Disruptive missense | 329 | 1.17E-22 | 1.59E-23 | rs2150867, rs33918808, rs112853430, rs4149307, rs9282541, rs1883025, rs1800978 |
| APOC3 | 11 | Disruptive missense | 6 | 2.38E-29 | 3.93E-04 | rs509728, rs61905072, rs66505542, rs7102314, rs964184, rs75198898, rs142958146, rs2075291, rs3135506, rs651821, rs45611741, rs662799, rs10750097, rs9804646, rs978880643, rs2070669, rs76353203, rs138326449, rs147210663, rs140621530, rs525028, rs141469619, rs188287950, rs202207736 |
| SCARB1 | 12 | Disruptive missense | 51 | 4.44E-16 | 2.86E-16 | rs6488913, rs4765127, rs1716407, rs825456, rs1672875, rs10846744, rs10773112, rs187471874, rs10773119 |
| LIPC | 15 | Disruptive missense | 112 | 2.19E-18 | 2.65E-16 | rs1973688, rs1601935, rs2043082, rs10468017, rs1532085, rs436965, rs35980001, rs1800588, rs2070895, rs113298164 |
| LCAT | 16 | Disruptive missense | 84 | 2.85E-14 | 6.44E-15 | rs111315946, rs150660813, rs4986970, rs35673026, rs1109166, rs548291389 |
| LDLR | 19 | Disruptive missense | 203 | 2.22E-59 | 5.13E-59 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
Samoan-specific missense variant.
For non-coding variants, rare variants from eight noncoding masks were analyzed in a similar fashion, including (1) promoter rare variants overlaid with CAGE sites40, (2) promoter rare variants overlaid with DHS sites41, (3) enhancer rare variants overlaid with CAGE sites42,43, (4) enhancer rare variants overlaid with DHS sites41,43, (5) untranslated region (UTR) rare variants, (6) upstream region rare variants, (7) downstream region rare variants of each protein-coding gene and (8) rare variants in ncRNA genes24. The promoter rare variants were defined as rare variants in the ±3-kilobase (kb) window of transcription start sites with the overlap of CAGE sites or DHS sites. The enhancer rare variants were defined as RVs in GeneHancer-predicted regions with the overlap of CAGE sites or DHS sites. The UTR, upstream, downstream and ncRNA rare variants were defined by GENCODE VEP categories32. With a well-calibrated overall distribution of MultiSTAAR-O P values (Extended Data Fig. 1d) and at a Bonferroni-corrected significance threshold of α = 0.05/(20,000 × 7) = 3.57 × 10−7, accounting for seven different noncoding masks across protein-coding genes, MultiSTAAR-O identified 76 genome-wide significant associations using unconditional multi-trait analysis (Extended Data Fig. 1c, Supplementary Table 5). After conditioning on known lipids-associated variants26,38,39, 6 out of the 76 associations remained significant at the Bonferroni-corrected threshold of α = 0.05/76 = 6.58 × 10−4 (Table 2). These included promoter CAGE and enhancer CAGE rare variants in APOA1, promoter DHS rare variants in CETP, enhancer CAGE rare variants in SPC24, and enhancer DHS rare variants in NIPSNAP3A and LIPC.
Table 2 ∣. TOPMed Gene-centric noncoding multi-trait analysis results of both unconditional analysis and analysis conditional on known lipids-associated variants.
A total of 61,838 samples from the TOPMed Program were considered in the analysis. Results for the conditionally significant genes (unconditional MultiSTAAR-O P < 3.57 × 10−7 and conditional MultiSTAAR-O P < 6.58 × 10−4 for 7 different noncoding masks across protein-coding genes; unconditional MultiSTAAR-O P < 2.50 × 10−6 and conditional MultiSTAAR-O P < 8.33 × 10−3 for ncRNA genes) are presented in the table. MultiSTAAR-O is a two-sided test. Chr. no., chromosome number; Category, functional category; No. of SNVs, number of rare variants (MAF < 1%) of the particular noncoding functional category in the gene; MultiSTAAR-O, MultiSTAAR-O P value; Variants (adjusted), adjusted variants in the conditional analysis; n/a, no variant adjusted in the conditional analysis.
| Gene | Chr. no. |
Category | No. of SNVs |
MultiSTAAR-O (Unconditional) |
MultiSTAAR-O (Conditional) |
Variants (adjusted) |
|---|---|---|---|---|---|---|
| APOA1 | 11 | Promoter (CAGE) | 230 | 2.33E-07 | 9.45E-07 | rs509728, rs61905072, rs66505542, rs7102314, rs964184, rs75198898, rs142958146, rs2075291, rs3135506, rs651821, rs45611741, rs662799, rs10750097, rs9804646, rs978880643, rs2070669, rs76353203, rs138326449, rs147210663, rs140621530, rs525028, rs141469619, rs188287950, rs202207736 |
| CETP | 16 | Promoter (DHS) | 411 | 1.21E-12 | 5.75E-04 | rs35571500, rs247617, rs17231506, rs34498052, rs34119551, rs34065661, rs1597000001*, rs7499892, rs5883, rs289719, rs11860407, rs189866004, rs5880 |
| APOA1 | 11 | Enhancer (CAGE) | 642 | 1.88E-24 | 6.23E-04 | rs509728, rs61905072, rs66505542, rs7102314, rs964184, rs75198898, rs142958146, rs2075291, rs3135506, rs651821, rs45611741, rs662799, rs10750097, rs9804646, rs978880643, rs2070669, rs76353203, rs138326449, rs147210663, rs140621530, rs525028, rs141469619, rs188287950, rs202207736 |
| SPC24 | 19 | Enhancer (CAGE) | 366 | 1.33E-08 | 4.88E-04 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
| NIPSNAP3A | 9 | Enhancer (DHS) | 767 | 2.63E-08 | 8.46E-06 | rs2150867, rs33918808, rs112853430, rs4149307, rs9282541, rs1883025, rs1800978 |
| LIPC | 15 | Enhancer (DHS) | 3714 | 4.26E-08 | 1.25E-04 | rs1973688, rs1601935, rs2043082, rs10468017, rs1532085, rs436965, rs35980001, rs1800588, rs2070895, rs113298164 |
| RP11-310H4.2 | 7 | ncRNA | 154 | 1.69E-06 | 1.69E-06 | n/a |
| MIR4497 | 12 | ncRNA | 23 | 1.37E-06 | 1.42E-06 | rs5800864 |
| RP11-15F12.3 | 18 | ncRNA | 64 | 7.53E-11 | 7.50E-03 | rs77960347, rs117623631, rs9958734, rs7229562, rs8086351, rs10048323, rs8084172 |
Samoan-specific missense variant.
MultiSTAAR-O further identified 6 genome-wide significant associations using unconditional multi-trait analysis at α = 0.05/20,000 = 2.50 × 10−6 accounting for ncRNA genes (Extended Data Fig. 1e, Supplementary Table 5), with 3 rare variant associations in RP11-15F12.3, RP11-310H4.2 and MIR4497 remained significant at α = 0.05/6 = 8.33 × 10−3 after conditioning on known lipids-associated variants26,38,39 (Table 2).
Notably, among the 9 conditionally significant noncoding rare variants associations with lipid traits, 4 of them were not detected by any of the three single-trait analysis (LDL-C, HDL-C or TG) using unconditional analysis of STAAR-O, including the associations of enhancer DHS rare variants in NIPSNAP3A and LIPC as well as ncRNA rare variants in RP11-310H4.2 and MIR4497 (Supplementary Table 5). These results demonstrate that MultiSTAAR-O can increase power over existing methods, and identify additional trait-associated signals by leveraging cross-phenotype correlations between multiple traits.
Genetic region multi-trait analysis of rare variants
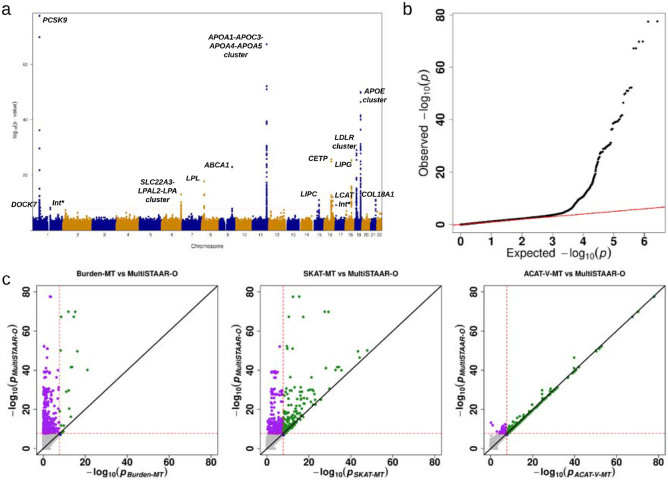
We next applied MultiSTAAR-O to perform genetic region multi-trait analysis to identify rare variants associated with lipid traits in TOPMed. Rare variants residing in 2-kilobase (kb) sliding windows with a 1-kb skip length were aggregated and analyzed using a joint model for LDL-C, HDL-C and TG. We incorporated 12 quantitative annotations, including 9 aPCs, CADD, LINSIGHT, FATHMM-XF along with the two MAF weights in MultiSTAAR-O (Methods). The overall distribution of MultiSTAAR-O P values was well-calibrated for the multi-trait analysis (Fig. 2b). At a Bonferroni-corrected significance threshold of α = 0.05/(2.65 × 106) = 1.89 × 10−8 accounting for 2.65 million 2-kb sliding windows across the genome, MultiSTAAR-O identified 502 genome-wide significant associations using unconditional multi-trait analysis (Fig. 2a, Supplementary Table 6). By dynamically incorporating multiple functional annotations capturing different aspects of variant function, MultiSTAAR-O detected more significant sliding windows and showed consistently smaller P values for top sliding windows compared with multi-trait analysis using only MAFs as the weight (Fig. 2c). After conditioning on known lipids-associated variants26,38,39, 7 out of the 502 associations remained significant at the Bonferroni-corrected threshold of α 0.05/502 = 9.96 × 10−5 (Table 3), including two sliding windows in DOCK7 (chromosome 1: 62,651,447 - 62,653,446 bp; chromosome 1: 62,652,447 - 62,654,446 bp) and an intergenic sliding window (chromosome 1: 145,530,447 - 145,532,446 bp) that were not detected by any of the three single-trait analysis (LDL-C, HDL-C or TG) using STAAR-O (Supplementary Table 6). Notably, all known lipids-associated variants indexed in the previous literature were at least 1-Mb away from the intergenic sliding window.
Fig. 2 ∣. TOPMed Genetic region (2-kb sliding window) unconditional multi-trait analysis results of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C) and triglycerides (TG) using TOPMed data.
a, Manhattan plot showing the associations of 2.65 million 2-kb sliding windows versus −log10(P) of MultiSTAAR-O. The horizontal line indicates a genome-wide P value threshold of 1.89 × 10−8 (n = 61,838). b, Quantile-quantile plot of 2-kb sliding window MultiSTAAR-O P values (n = 61,838). c, Scatterplot of P values for the 2-kb sliding windows comparing MultiSTAAR-O with Burden-MT, SKAT-MT and ACAT-V-MT tests (MT is short for Multi-Trait). Each dot represents a sliding window with x-axis label being the −log10(P) of the conventional multi-trait test and y-axis label being the −log10(P) of MultiSTAAR-O (n = 61,838). Burden-MT, SKAT-MT, ACAT-V-MT and MultiSTAAR-O are two-sided tests. Int*, intergenic sliding window.
Table 3 ∣. TOPMed Genetic region (2-kb sliding window) multi-trait analysis results of both unconditional analysis and analysis conditional on known lipid-associated variants.
A total of 61,838 samples from the TOPMed Program were considered in the analysis. Results for the conditionally significant sliding windows (unconditional MultiSTAAR-O P < 1.89 × 10−8 and conditional MultiSTAAR-O P < 9.96 × 10−5) are presented in the table. MultiSTAAR-O is a two-sided test. Chr. no., chromosome number; Start location, start location of the 2-kb sliding window; End location, end location of the 2-kb sliding window; No. of SNVs, number of rare variants (MAF < 1%) in the 2-kb sliding window; MultiSTAAR-O, MultiSTAAR-O P value; Variants (adjusted), adjusted variants in the conditional analysis; n/a, no variant adjusted in the conditional analysis. Physical positions of each window are on build hg38.
| Chr. no. |
Start location |
End location |
Gene | No. of SNVs |
MultiSTAAR-O (Unconditional) |
MultiSTAAR-O (Conditional) |
Variants (adjusted) |
|---|---|---|---|---|---|---|---|
| 1 | 55,051,447 | 55,053,446 | PCSK9 | 327 | 7.11E-11 | 6.60E-08 | rs12117661, rs2495491, rs11591147, rs67608943, rs72646508, rs693668, rs28362261, rs28362263, rs141502002, rs505151, rs28362286 |
| 1 | 55,052,447 | 55,054,446 | PCSK9 | 320 | 9.37E-09 | 9.07E-06 | rs12117661, rs2495491, rs11591147, rs67608943, rs72646508, rs693668, rs28362261, rs28362263, rs141502002, rs505151, rs28362286 |
| 1 | 62,651,447 | 62,653,446 | DOCK7 | 277 | 5.08E-09 | 7.56E-10 | rs67461605 |
| 1 | 62,652,447 | 62,654,446 | DOCK7 | 257 | 4.87E-09 | 7.24E-10 | rs67461605 |
| 1 | 145,530,447 | 145,532,446 | intergenic | 233 | 5.12E-09 | 5.12E-09 | n/a |
| 19 | 11,104,367 | 11,106,366 | LDLR | 336 | 1.15E-12 | 8.33E-13 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
| 19 | 11,105,367 | 11,107,366 | LDLR | 338 | 5.97E-14 | 5.55E-15 | rs140753491, rs138294113, rs17242353, rs17242843, rs10422256, rs72658860, rs11669576, rs2738447, rs72658867, rs2738464, rs6511728, rs3760782, rs59168178, rs2278426, rs112942459 |
Samoan-specific missense variant.
Comparison of MultiSTAAR-O with existing multi-trait rare variant tests
Using TOPMed Freeze 8 WGS data, our gene-centric multi-trait analysis of coding rare variants identified 34 conditionally significant associations with lipid traits (Table 1), including NPC1L1 and SCARB1 missense rare variants that were missed by multi-trait burden, SKAT and ACAT-V tests (Supplementary Table 4). Among the 9 and 7 conditionally significant associations detected in gene-centric multi-trait analysis of noncoding rare variants and genetic region multi-trait analysis, MultiSTAAR-O identified 1 and 2 associations, respectively, that were missed by multi-trait burden, SKAT and ACAT-V tests (Supplementary Tables 5-6). These associations included enhancer CAGE rare variants in SPC24 and two sliding windows in LDLR (chromosome 19: 11,104,367 - 11,106,366 bp; chromosome 19: 11,105,367 - 11,107,366 bp).
Computation cost
The computational cost for MultiSTAAR-O to perform WGS multi-trait rare variant analysis of n = 61,838 related TOPMed lipids samples was 2 hours using 250 2.10-GHz computing cores with 12-GB memory for gene-centric coding analysis; or 20 hours using 250 2.10-GHz computing cores with 24-GB memory for gene-centric noncoding analysis; 2 hours using 250 2.10-GHz computing cores with 12-GB memory of ncRNA analysis; and 20 hours using 500 2.10-GHz computing cores with 24-GB memory for sliding window analysis. Runtime for all analyses scales linearly with the sample size24.
Discussion
In this study, we have introduced MultiSTAAR as a general statistical framework and a flexible analytical pipeline for performing functionally-informed multi-trait RVAS in large-scale WGS studies. MultiSTAAR improves power by analyzing multiple traits simultaneously and dynamically incorporating multiple functional annotations, while accounting for relatedness and population structure among study samples.
By jointly analyzing multiple quantitative traits using a multivariate linear mixed model, MultiSTAAR explicitly leverages the correlation among multiple phenotypes to enhance power for detecting additional association signals, outperforming single-trait analyses of the individual phenotypes. MultiSTAAR also enables conditional multi-trait analysis to identify putatively novel rare variant associations independent of a set of known variants. Using TOPMed Freeze 8 WGS data, our gene-centric multi-trait analysis of noncoding rare variants identified 9 conditionally significant associations with lipid traits (Table 2), including 4 noncoding associations that were missed by single-trait analysis using STAAR (Supplementary Table 5). Our genetic region multi-trait analysis of rare variants identified 7 conditionally significant 2-kb sliding windows associated with lipid traits (Table 3), including 3 associations that were missed by single-trait analysis using STAAR (Supplementary Table 6).
By dynamically incorporating multiple annotations capturing diverse aspects of variant biological function in the second step, MultiSTAAR further improves power over existing multi-trait rare variant analysis methods. Our simulation studies demonstrated that MultiSTAAR-O maintained accurate type I error rates while achieving considerable power gains over multi-trait burden, SKAT and ACAT-V tests that do not incorporate functional annotation information (Extended Data Figs. 2-5, Supplementary Figs. 1-4). Notably, the existing ACAT-V method9 does not support multi-trait analysis. We extended it to accommodate multi-trait settings and incorporated the multi-trait ACAT-V test into the MultiSTAAR framework (Methods).
Implemented as a flexible analytical pipeline, MultiSTAAR allows for customized input phenotype selection, variant set definition and user-specified annotation weights to facilitate functionally-informed multi-trait analyses. In addition to rare variant association analysis of coding and noncoding regions, MultiSTAAR also provides single-variant multi-trait analysis for common and low-frequency variants under a given MAF or minor allele count (MAC) cutoff (e.g. MAC ≥ 20). Using 61,838 TOPMed lipids samples, it took 8 hours using 250 2.10-GHz computing cores with 12-GB memory for single-variant multi-trait analysis, which is scalable for large WGS/WES datasets. On the other hand, MultiSTAAR could be further extended to allow for dynamic windows with data-adaptive sizes in genetic region analysis24,44, to properly leverage synthetic surrogates in the presence of partially missing phenotypes45, and to incorporate summary statistics for meta-analysis of multiple WGS/WES studies46.
In summary, MultiSTAAR provides a powerful statistical framework and a computationally scalable analytical pipeline for large-scale WGS multi-trait analysis with complex study samples. Compared to single-trait analysis, MultiSTAAR offers a notable increase in statistical power when analyzing multiple moderately to highly correlated traits, all while maintaining control over type I error rates across various genetic architectures. As the sample sizes and number of available phenotypes increase in biobank-scale sequencing studies, our proposed method may contribute to a better understanding of the genetic architecture of complex traits by elucidating the role of rare variants with pleiotropic effects.
Methods
Ethics statement
This study relied on analyses of genetic data from TOPMed cohorts. The study has been approved by the TOPMed Publications Committee, TOPMed Lipids Working Group and all the participating cohorts, including Old Order Amish (phs000956.v1.p1), Atherosclerosis Risk in Communities Study (phs001211), Mt Sinai BioMe Biobank (phs001644), Coronary Artery Risk Development in Young Adults (phs001612), Cleveland Family Study (phs000954), Cardiovascular Health Study (phs001368), Diabetes Heart Study (phs001412), Framingham Heart Study (phs000974), Genetic Study of Atherosclerosis Risk (phs001218), Genetic Epidemiology Network of Arteriopathy (phs001345), Genetic Epidemiology Network of Salt Sensitivity (phs001217), Genetics of Lipid Lowering Drugs and Diet Network (phs001359), Hispanic Community Health Study - Study of Latinos (phs001395), Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (phs001293), Jackson Heart Study (phs000964), Multi-Ethnic Study of Atherosclerosis (phs001416), San Antonio Family Heart Study (phs001215), Genome-wide Association Study of Adiposity in Samoans (phs000972), Taiwan Study of Hypertension using Rare Variants (phs001387), and Women’s Health Initiative (phs001237), where the accession numbers are provided in parenthesis. The use of human genetics data from TOPMed cohorts was approved by the Harvard T.H. Chan School of Public Health IRB (IRB13-0353).
Notation and model
Suppose there are subjects with a total of variants sequenced across the whole genome. For the i-th subject, let denote a vector of K quantitative phenotypes; denotes covariates, such as age, gender and ancestral principal components; denotes the genotype matrix of the genetic variants in a variant set. Since these K phenotypes may be defined on different measurement scales, we assume that each phenotype has been rescaled to have zero mean and unit variance.
When the data consist of unrelated samples, we consider the following Multivariate Linear Model (MLM)
| (1) |
where is an intercept, and are column vectors of regression coefficients for covariates and genotype in phenotype , respectively. The error terms are independent and identically distributed and follow a multivariate normal distribution with mean a vector of zeros and variance-covariance matrix , assumed identical for all subjects. For all subjects, using matrix notation we can write model (1) as
| (2) |
where is a column vector of 1’s with length , is a column vector of regression intercepts, the -th columns of and are and , respectively, and follows a matrix normal distribution. We calculate the scaled residual for each subject on each phenotype, defined as , where (a matrix of fitted values) and are estimated under the null MLM , where no variant has any effect on any outcome.
When the data consist of related samples, we consider the following Multivariate Linear Mixed Model (MLMM)19,47,48
| (3) |
where the random effects account for relatedness and remaining population structure unaccounted by ancestral PCs20. We assume that with a variance component matrix and a sparse genetic relatedness matrix 21,22. For all subjects, using matrix notation we can rewrite equation (3) as
| (4) |
We calculate the scaled residual for each subject on each phenotype, defined as , where and are estimated under the null MLMM . Under both MLM and MLMM, our goal is to test for an association between a set of genetic variants and quantitative phenotypes, adjusting for covariates and relatedness. This corresponds to testing .
Multi-trait rare variant association tests using MultiSTAAR
Single-trait score-based aggregation methods5-9 can be extended to allow for jointly testing the association between rare variants in a variant set and multiple quantitative phenotypes. For a given variant set, let denote the matrix of score statistics where is the score statistic for the -th variant on the -th phenotype. For multi-trait burden test using MultiSTAAR (Burden-MT), we consider test statistic
where is the weight defined as a function of the MAF for the -th variant4,18, is the -th row of and is the estimated variance-covariance matrix of . asymptotically follows a standard chi-square distribution with degrees of freedom under the null hypothesis, and its P value can be obtained analytically while accounting for LD between variants and correlation between phenotypes.
For multi-trait SKAT using MultiSTAAR (SKAT-MT), we consider the statistic
asymptotically follows a mixture of chi-square distributions under the null hypothesis, and its P value can be obtained analytically while accounting for LD between variants and correlation between phenotypes14,15.
For multi-trait ACAT-V using MultiSTAAR (ACAT-V-MT), we propose test statistic
where is the number of variants with a minor allele count (MAC) greater than 10 and is the multi-trait association P value of individual variant for those variants with a MAC > 10, whose test statistic is given by the K degrees of freedom multivariate score test
where . is the estimated variance-covariance matrix of ; is the multi-trait burden test P value of extremely rare variants with an MAC ≤ 10 as described above and is the average of the weights among the extremely rare variants with an MAC ≤ 10. is approximated well by a scaled Cauchy distribution under the null hypothesis, and its P value can be obtained analytically while accounting for LD between variants and correlation between phenotypes9,49. Note that when K = 1, the multi-trait burden, SKAT, and ACAT-V tests reduce to the original single-trait burden, SKAT and ACAT-V tests.
Suppose we have a collection of annotations, let denote the -th annotation for the jth variant in the variant set. We define the functionally-informed multi-trait burden, SKAT and ACAT-V test statistics weighted by the -th annotation as follows
where , with , is the estimated variance-covariance matrix of . and is the average of the weights among the extremely rare variants with MAC ≤ 10. Finally, we define the omnibus MultiSTAAR-O test statistic as
and the P value of can be calculated by
Data simulation
Type I error rate simulations
We performed simulation studies to evaluate how accurately MultiSTAAR controls the type I error rate. We generated three quantitative traits from a multivariate linear model, conditional on two covariates
Where Bernoulli(0.5) and
The correlation matrix of error terms was chosen to mimic the correlations between three lipid traits LDL-C, HDL-C and TG, estimated from the TOPMed data26. We considered a sample size of 10,000 and generated genotypes by simulating 20,000 sequences for 100 different regions each spanning 1 Mb. The data generation used the calibration coalescent model (COSI)29 with parameters set to mimic the LD structure of African Americans. In each simulation replicate, 10 annotations were generated as A1, …, A10 all independently and identically distributed as N(0,1) for each variant, and we randomly selected 5-kb regions from these 1-Mb regions for type I error rate simulations. We applied MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A and MultiSTAAR-O by incorporating MAFs and the 10 annotations together with Burden-MT, SKAT-MT and ACAT-V-MT tests. We repeated the procedure with 108 replicates to examine the type I error rate at levels α = 10−4, 10−5. and 10−6.
Empirical power simulations
Next, we carried out simulation studies under a variety of configurations to assess the the power of MultiSTAAR-O, and how its incorporation of multiple functional annotations affects power compared to the multi-trait burden, SKAT, and ACAT-V tests implemented in MultiSTAAR. In each simulation replicate, we randomly selected 5-kb regions from a 1-Mb region for power evaluations. For each selected 5-kb region, we generated three quantitative traits from a multivariate linear model
where were defined as in the type I error rate simulations, and were the genotypes and effect sizes of the genetic variants in the signal region.
The genetic effect of variant on phenotype was defined as to allow for heterogeneous effect sizes among variants and phenotypes. Specifically, we generated the causal variant indicator according to a logistic model
where were randomly sampled for each region. For different regions, causality of variants depended on different sets of annotations. We set for all annotations and varied the proportions of causal variants in the signal region by setting δ0 = logit(0.0015), logit(0.015) and logit(0.18) which corresponds to averaging 5%, 15% and 35% causal variants in the signal region, respectively. We considered four scenarios of phenotypic indicator that reflect different underlying genetic architectures across phenotypes: and . These correspond to causal variants in the signal region being associated with (1) one phenotype only, (2) two positively correlated phenotypes, (3) two negatively correlated phenotypes and (4) all three phenotypes. We modeled the absolute effect sizes of causal variants using , such that it was a decreasing function of MAF. was set to be 0.13, 0.1, 0.1 and 0.07, respectively, to ensure a decent power of tests under each scenario. We additionally varied the proportions of causal variant effect size directions (signs of ) by randomly generating 100%, 80%, and 50% variants on average to have positive effects. We applied MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A, and MultiSTAAR-O using MAFs and all 10 annotations together with Burden-MT, SKAT-MT and ACAT-V-MT tests. We repeated the procedure with 104 replicates to examine the power at level α = 10−7. The sample size was 10,000 across all scenarios.
Lipid Traits
Conventionally measured plasma lipids, including LDL-C, HDL-C, and triglycerides, were included for analysis. LDL-C was either calculated by the Friedewald equation when triglycerides were <400 mg/dl or directly measured. Given the average effect of statins, when statins were present, LDL-C was adjusted by dividing by 0.7. Triglycerides were natural log transformed for analysis. Phenotypes were harmonized by each cohort and deposited into the dbGaP TOPMed Exchange Area.
Multi-trait analysis of lipid levels in the TOPMed WGS data
The TOPMed WGS data consist of multi-ethnic related samples1. Race/ethnicity was defined using a combination of self-reported race/ethnicity from participant questionnaires and study recruitment information (Supplementary Note)31. In this study, we applied MultiSTAAR to perform multi-trait rare variant analysis of three quantitative lipid traits (LDL-C, HDL-C and TG) using 20 study cohorts from the TOPMed Freeze 8 WGS data. LDL-C was adjusted for the presence of medications as before30. For each study, we first fit a linear regression model adjusting for age, age2, sex for each race/ethnicity-specific group. In addition, for Old Order Amish (OOA), we also adjusted for APOB p.R3527Q in LDL-C and TC analyses and adjusted for APOC3 p.R19Ter in TG and HDL-C analyses30.
We performed rank-based inverse normal transformation of the residuals of LDL-C, HDL-C and TG within each race/ethnicity-specific group. We then fit a multivariate linear mixed model for the rank normalized residuals, adjusting for 11 ancestral principal components, ethnicity group indicators, and a variance component for empirically derived sparse kinship matrix to account for population structure, relatedness and correlation between phenotypes.
We next applied MultiSTAAR-O to perform multi-trait variant set analyses for rare variants (MAF < 1%) by scanning the genome, including gene-centric analysis of each protein-coding gene using five coding variant functional categories (putative loss-of-function rare variants, missense rare variants, disruptive missense rare variants, putative loss-of-function and disruptive missense rare variants and synonymous rare variants); seven noncoding variant functional categories (promoter rare variants overlaid with CAGE sites, promoter rare variants overlaid with DHS sites, enhancer rare variants overlaid with CAGE sites, enhancer rare variants overlaid with DHS sites, UTR rare variants, upstream region rare variants, downstream region rare variants) and rare variants in ncRNA genes; and genetic region analysis using 2-kb sliding windows across the genome with a 1-kb skip length. The WGS multi-trait rare variant analysis was performed using the R packages MultiSTAAR (version 0.9.7, https://github.com/xihaoli/MultiSTAAR) and STAARpipeline (version 0.9.7, https://github.com/xihaoli/STAARpipeline). The WGS rare variant single-trait analysis of LDL-C, HDL-C and TG was performed using the R package STAARpipeline (version 0.9.7, https://github.com/xihaoli/STAARpipeline). Both multi-trait and single-trait analyses results were summarized and visualized using the R package STAARpipelineSummary (version 0.9.7, https://github.com/xihaoli/STAARpipelineSummary).
Genome build
All genome coordinates are given in NCBI GRCh38/UCSC hg38.
Statistics and reproducibility
Sample size was not predetermined. The multi-trait analysis consists of 20 study cohorts of TOPMed Freeze 8 and had 61,838 samples with lipid traits. We did not use any study design that required randomization or blinding.
Extended Data
Extended Data Fig. 1 ∣. Manhattan plots and Q-Q plots for unconditional gene-centric coding, noncoding and ncRNA analysis of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C) and triglycerids (TG) using TOPMed data (n = 61,838).
a, Manhattan plots for unconditional gene-centric coding analysis of protein-coding gene. The horizontal line indicates a genome-wide MultiSTAAR-O P value threshold of 5.00 × 10−7. The significant threshold is defined by multiple comparisons using the Bonferroni correction (0.05/(20,000 × 5) = (5.00 × 10−7). Different symbols represent the MultiSTAAR-O P value of the protein-coding gene using different functional categories (putative loss-of-function, putative loss-of-function and disruptive missense, missense, disruptive missense, synonymous). b, Quantile-quantile plots for unconditional gene-centric coding analysis of protein-coding gene. Different symbols represent the MultiSTAAR-O P-value of the gene using different functional categories. c, Manhattan plots for unconditional gene-centric noncoding analysis of protein-coding gene. The horizontal line indicates a genome-wide MultiSTAAR-O P value threshold of 3.57 × 10−7. The significant threshold is defined by multiple comparisons using the Bonferroni correction (0.05/(20,000 × 7) = 3.57 × 10−7). Different symbols represent the MultiSTAAR-O P value of the protein-coding gene using different functional categories (upstream, downstream, UTR, promoter_CAGE, promoter_DHS, enhancer_CAGE, enhancer_DHS). Promoter_CAGE and promoter_DHS are the promoters with overlap of Cap Analysis of Gene Expression (CAGE) sites and DNase hypersensitivity (DHS) sites for a given gene, respectively. Enhancer_CAGE and enhancer_DHS are the enhancers in GeneHancer predicted regions with the overlap of CAGE sites and DHS sites for a given gene, respectively. d, Quantile-quantile plots for unconditional gene-centric noncoding analysis of protein-coding gene. Different symbols represent the MultiSTAAR-O P-value of the gene using different functional categories. e, Manhattan plots for unconditional gene-centric noncoding analysis of ncRNA gene. The horizontal line indicates a genome-wide MultiSTAAR-O P value threshold of 2.50 × 10−6. The significant threshold is defined by multiple comparisons using the Bonferroni correction (0.05/20,000 = 2.50 × 10−6). f, Quantile-quantile plots for unconditional gene-centric noncoding analysis of ncRNA gene. In panels, a, c and e, the chromosome number are indicated by the colors of dots. In all panels, MultiSTAAR-O is a two-sided test.
Extended Data Fig. 2 ∣. Power comparisons of Burden-MT, SKAT-MT, ACAT-V-MT (MT is short for Multi-Trait) and MultiSTAAR methods when variants in the signal region are associated with one phenotype.
Multi-trait Burden, SKAT and ACAT-V tests implemented in MultiSTAAR are denoted by Burden-MT, SKAT-MT and ACAT-V-MT. MultiSTAAR methods incorporating ten functional annotations are denoted by MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A and MultiSTAAR-O. In each simulation replicate, a 5-kb region was randomly selected as the signal region. Within each signal region, variants were randomly generated to be causal based on the multivariate logistic model and on average there were 5%, 15% or 35% causal variants in the signal region. The effect sizes of causal variants were , where was set to be 0.13. The barplot of power in the top panel consider settings in which the effect sizes for the causal variants are 100% positive (0% negative), 80% positive (20% negative), and 50% positive (50% negative). The scatterplot of P values in the bottom panel compare MultiSTAAR-O to Burden-MT, SKAT-MT and ACAT-V-MT when 15% of variants in the signal region are causal variants with all positive effect sizes. Power was estimated as the proportion of the P values less than α = 10−7 based on 104 replicates. Burden-MT, SKAT-MT, ACAT-V-MT, MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A and MultiSTAAR-O are two-sided tests. Total sample size considered was 10,000.
Extended Data Fig. 3 ∣. Power comparisons of Burden-MT, SKAT-MT, ACAT-V-MT (MT is short for Multi-Trait) and MultiSTAAR methods when variants in the signal region are associated with two positively correlated phenotypes.
In each simulation replicate, a 5-kb region was randomly selected as the signal region. Within each signal region, variants were randomly generated to be causal based on the multivariate logistic model and on average there were 5%, 15% or 35% causal variants in the signal region. The effect sizes of causal variants were , where was set to be 0.1. The barplot of power in the top panel consider settings in which the effect sizes for the causal variants are 100% positive (0% negative), 80% positive (20% negative), and 50% positive (50% negative). The scatterplot of P values in the bottom panel compare MultiSTAAR-O to Burden-MT, SKAT-MT and ACAT-V-MT when 15% of variants in the signal region are causal variants with all positive effect sizes. Power was estimated as the proportion of the P values less than α = 10−7 based on 104 replicates. Burden-MT, SKAT-MT, ACAT-V-MT, MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A and MultiSTAAR-O are two-sided tests. Total sample size considered was 10,000.
Extended Data Fig. 4 ∣. Power comparisons of Burden-MT, SKAT-MT, ACAT-V-MT (MT is short for Multi-Trait) and MultiSTAAR methods when variants in the signal region are associated with two negatively correlated phenotypes.
In each simulation replicate, a 5-kb region was randomly selected as the signal region. Within each signal region, variants were randomly generated to be causal based on the multivariate logistic model and on average there were 5%, 15% or 35% causal variants in the signal region. The effect sizes of causal variants were , where was set to be 0.1. The barplot of power in the top panel consider settings in which the effect sizes for the causal variants are 100% positive (0% negative), 80% positive (20% negative), and 50% positive (50% negative). The scatterplot of P values in the bottom panel compare MultiSTAAR-O to Burden-MT, SKAT-MT and ACAT-V-MT when 15% of variants in the signal region are causal variants with all positive effect sizes. Power was estimated as the proportion of the P values less than α = 10−7 based on 104 replicates. Burden-MT, SKAT-MT, ACAT-V-MT, MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A and MultiSTAAR-O are two-sided tests. Total sample size considered was 10,000.
Extended Data Fig. 5 ∣. Power comparisons of Burden-MT, SKAT-MT, ACAT-V-MT (MT is short for Multi-Trait) and MultiSTAAR methods when variants in the signal region are associated with three phenotypes.
In each simulation replicate, a 5-kb region was randomly selected as the signal region. Within each signal region, variants were randomly generated to be causal based on the multivariate logistic model and on average there were 5%, 15% or 35% causal variants in the signal region. The effect sizes of causal variants were , where was set to be 0.07. The barplot of power in the top panel consider settings in which the effect sizes for the causal variants are 100% positive (0% negative), 80% positive (20% negative), and 50% positive (50% negative). The scatterplot of P values in the bottom panel compare MultiSTAAR-O to Burden-MT, SKAT-MT and ACAT-V-MT when 15% of variants in the signal region are causal variants with all positive effect sizes. Power was estimated as the proportion of the P values less than α = 10−7 based on 104 replicates. Burden-MT, SKAT-MT, ACAT-V-MT, MultiSTAAR-B, MultiSTAAR-S, MultiSTAAR-A and MultiSTAAR-O are two-sided tests. Total sample size considered was 10,000.
Supplementary Material
Acknowledgments
This work was supported by grants R35-CA197449, U19-CA203654, U01-HG012064, and U01-HG009088 (X. Lin), NHLBI TOPMed Fellowship (X. Li), R01-HL142711 and R01-HL127564 (P.N. and G.M.P.), 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR001881, DK063491, R01-HL071051, R01-HL071205, R01-HL071250, R01-HL071251, R01-HL071258, R01-HL071259, and UL1-RR033176 (J.I.R.), HHSN268201800001I and U01-HL137162 (K.M.R.), 1R35-HL135818, R01-HL113338, and HL046389 (S.R.), HL105756 (B.M.P.), HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C (C.K.), R01-MD012765 and R01-DK117445 (N.F.), 18CDA34110116 from American Heart Association (P.S.d.V.), R01-HL153805, R03-HL154284 (B.E.C.), HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, and HHSN268201700004I (E.B.), U01-HL072524, R01-HL104135-04S1, U01-HL054472, U01-HL054473, U01-HL054495, U01-HL054509, and R01-HL055673-18S1 (D.K.A.), U01-HL72518, HL087698, HL49762, HL59684, HL58625, HL071025, HL112064, NR0224103, and M01-RR000052 (to the Johns Hopkins General Clinical Research Center). This work was supported by R01 HL92301, R01 HL67348, R01 NS058700, R01 AR48797, R01 DK071891, R01 AG058921, the General Clinical Research Center of the Wake Forest University School of Medicine (M01 RR07122, F32 HL085989), the American Diabetes Association, and a pilot grant from the Claude Pepper Older Americans Independence Center of Wake Forest University Health Sciences (P60 AG10484). The Framingham Heart Study (FHS) acknowledges the support of contracts NO1-HC-25195, HHSN268201500001I and 75N92019D00031 from the National Heart, Lung and Blood Institute and grant supplement R01 HL092577-06S1 for this research. We also acknowledge the dedication of the FHS study participants without whom this research would not be possible. R.S.V. is supported in part by the Evans Medical Foundation and the Jay and Louis Coffman Endowment from the Department of Medicine, Boston University School of Medicine. The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. Support for GENOA was provided by the National Heart, Lung and Blood Institute (U01HL054457, U01HL054464, U01HL054481, R01HL119443, and R01HL087660) of the National Institutes of Health. Collection of the San Antonio Family Study data was supported in part by National Institutes of Health (NIH) grants P01 HL045522, MH078143, MH078111 and MH083824; and whole genome sequencing of SAFS subjects was supported by U01 DK085524 and R01 HL113323. Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The full study specific acknowledgements are detailed in Supplementary Note.
NHLBI Trans-Omics for Precisition Medicine (TOPMed) Consortium
Namiko Abe50, Gonçalo Abecasis51, Francois Aguet52, Christine Albert53, Laura Almasy54, Alvaro Alonso55, Seth Ament56, Peter Anderson57, Pramod Anugu58, Deborah Applebaum-Bowden59, Kristin Ardlie52, Dan Arking60, Allison Ashley-Koch61, Stella Aslibekyan62, Tim Assimes63, Paul Auer64, Dimitrios Avramopoulos60, Najib Ayas65, Adithya Balasubramanian66, John Barnard67, Kathleen Barnes68, R. Graham Barr69, Emily Barron-Casella60, Lucas Barwick70, Terri Beaty60, Gerald Beck71, Diane Becker72, Lewis Becker60, Rebecca Beer73, Amber Beitelshees56, Emelia Benjamin74, Takis Benos75, Marcos Bezerra76, Larry Bielak51, Thomas Blackwell51, Nathan Blue77, Russell Bowler78, Ulrich Broeckel79, Jai Broome57, Deborah Brown80, Karen Bunting50, Esteban Burchard81, Carlos Bustamante82, Erin Buth83, Jonathan Cardwell84, Vincent Carey85, Julie Carrier86, Cara Carty87, Richard Casaburi88, Juan P Casas Romero89, James Casella60, Peter Castaldi90, Mark Chaffin52, Christy Chang56, Yi-Cheng Chang91, Daniel Chasman92, Sameer Chavan84, Bo-Juen Chen50, Wei-Min Chen93, Michael Cho85, Seung Hoan Choi52, Lee-Ming Chuang94, Mina Chung95, Ren-Hua Chung96, Clary Clish97, Suzy Comhair98, Matthew Conomos83, Elaine Cornell99, Adolfo Correa100, Carolyn Crandall88, James Crapo101, L. Adrienne Cupples102, Jeffrey Curtis103, Brian Custer104, Coleen Damcott56, Dawood Darbar105, Sean David106, Colleen Davis57, Michelle Daya84, Michael DeBaun107, Dawn DeMeo85, Ranjan Deka108, Scott Devine56, Huyen Dinh66, Harsha Doddapaneni109, Qing Duan110, Shannon Dugan-Perez111, Ravi Duggirala112, Jon Peter Durda113, Susan K. Dutcher114, Charles Eaton115, Lynette Ekunwe58, Adel El Boueiz116, Patrick Ellinor117, Leslie Emery57, Serpil Erzurum118, Charles Farber93, Jesse Farek66, Tasha Fingerlin119, Matthew Flickinger51, Chris Frazar57, Mao Fu56, Stephanie M. Fullerton57, Lucinda Fulton120, Stacey Gabriel52, Weiniu Gan73, Shanshan Gao84, Yan Gao58, Margery Gass121, Heather Geiger122, Bruce Gelb123, Mark Geraci124, Soren Germer50, Robert Gerszten125, Auyon Ghosh85, Richard Gibbs66, Chris Gignoux63, Mark Gladwin126, David Glahn127, Stephanie Gogarten57, Da-Wei Gong56, Harald Goring128, Sharon Graw129, Kathryn J. Gray130, Daniel Grine84, Colin Gross51, Yue Guan56, Xiuqing Guo131, Namrata Gupta132, Jeff Haessler121, Michael Hall133, Yi Han66, Patrick Hanly134, Daniel Harris135, Nicola L. Hawley136, Ben Heavner83, Susan Heckbert137, Ryan Hernandez81, David Herrington138, Craig Hersh139, Bertha Hidalgo62, James Hixson140, Brian Hobbs141, John Hokanson84, Elliott Hong56, Karin Hoth142, Chao (Agnes) Hsiung143, Jianhong Hu66, Haley Huston144, Chii Min Hwu145, Rebecca Jackson146, Deepti Jain57, Cashell Jaquish147, Jill Johnsen148, Andrew Johnson73, Craig Johnson57, Rich Johnston55, Kimberly Jones60, Hyun Min Kang149, Shannon Kelly150, Eimear Kenny123, Michael Kessler56, Alyna Khan57, Ziad Khan66, Wonji Kim151, John Kimoff152, Greg Kinney153, Barbara Konkle154, Holly Kramer155, Christoph Lange156, Ethan Lange84, Leslie Lange157, Cathy Laurie57, Cecelia Laurie57, Meryl LeBoff85, Jonathon LeFaive51, Jiwon Lee85, Sandra Lee66, Wen-Jane Lee145, David Levine57, Daniel Levy73, Joshua Lewis56, Xiaohui Li131, Yun Li110, Henry Lin131, Honghuang Lin158, Simin Liu159, Yongmei Liu160, Yu Liu161, Steven Lubitz117, Kathryn Lunetta162, James Luo73, Ulysses Magalang163, Barry Make60, Ani Manichaikul93, Alisa Manning164, JoAnn Manson85, Melissa Marton122, Susan Mathai84, Susanne May83, Patrick McArdle56, Merry-Lynn McDonald165, Sean McFarland151, Stephen McGarvey166, Daniel McGoldrick167, Caitlin McHugh83, Becky McNeil168, Hao Mei58, James Meigs169, Vipin Menon66, Luisa Mestroni129, Ginger Metcalf66, Deborah A Meyers170, Emmanuel Mignot171, Julie Mikulla73, Nancy Min58, Mollie Minear172, Matt Moll90, Zeineen Momin66, Courtney Montgomery173, Donna Muzny66, Josyf C Mychaleckyj93, Girish Nadkarni123, Rakhi Naik60, Take Naseri174, Sergei Nekhai175, Sarah C. Nelson83, Bonnie Neltner84, Caitlin Nessner66, Deborah Nickerson176, Osuji Nkechinyere66, Kari North110, Jeff O'Connell177, Tim O'Connor56, Heather Ochs-Balcom178, Geoffrey Okwuonu66, Allan Pack179, David T. Paik180, James Pankow181, George Papanicolaou73, Cora Parker182, Juan Manuel Peralta112, Marco Perez63, James Perry56, Ulrike Peters183, Lawrence S Phillips55, Jacob Pleiness51, Toni Pollin56, Wendy Post184, Julia Powers Becker185, Meher Preethi Boorgula84, Michael Preuss123, Pankaj Qasba73, Dandi Qiao85, Zhaohui Qin55, Nicholas Rafaels186, Mahitha Rajendran66, D.C. Rao120, Laura Rasmussen-Torvik187, Aakrosh Ratan93, Robert Reed56, Catherine Reeves188, Elizabeth Regan189, Muagututi‘a Sefuiva Reupena190, Rebecca Robillard191, Nicolas Robine122, Dan Roden192, Carolina Roselli52, Ingo Ruczinski60, Alexi Runnels122, Pamela Russell84, Sarah Ruuska193, Kathleen Ryan56, Ester Cerdeira Sabino194, Danish Saleheen195, Shabnam Salimi196, Sejal Salvi66, Steven Salzberg60, Kevin Sandow197, Vjay G. Sankaran198, Jireh Santibanez66, Karen Schwander120, David Schwartz84, Frank Sciurba126, Christine Seidman199, Jonathan Seidman200, Vivien Sheehan201, Stephanie L. Sherman202, Amol Shetty56, Aniket Shetty84, Wayne Hui-Heng Sheu145, M. Benjamin Shoemaker203, Brian Silver204, Edwin Silverman85, Robert Skomro205, Albert Vernon Smith206, Josh Smith57, Nicholas Smith207, Tanja Smith50, Sylvia Smoller208, Beverly Snively209, Michael Snyder210, Tamar Sofer125, Nona Sotoodehnia57, Adrienne M. Stilp57, Garrett Storm211, Elizabeth Streeten56, Jessica Lasky Su212, Yun Ju Sung120, Jody Sylvia85, Adam Szpiro57, Frédéric Sériès213, Daniel Taliun51, Hua Tang210, Margaret Taub60, Matthew Taylor129, Simeon Taylor56, Marilyn Telen61, Timothy A. Thornton57, Machiko Threlkeld214, Lesley Tinker215, David Tirschwell57, Sarah Tishkoff216, Catherine Tong217, Russell Tracy218, Michael Tsai181, Dhananjay Vaidya60, David Van Den Berg219, Peter VandeHaar51, Scott Vrieze181, Tarik Walker84, Robert Wallace142, Avram Walts84, Fei Fei Wang57, Heming Wang220, Jiongming Wang221, Karol Watson88, Jennifer Watt66, Daniel E. Weeks222, Joshua Weinstock149, Bruce Weir57, Scott T Weiss223, Lu-Chen Weng117, Jennifer Wessel224, Cristen Willer103, Kayleen Williams83, L. Keoki Williams225, Scott Williams226, Carla Wilson85, James Wilson227, Lara Winterkorn122, Quenna Wong57, Baojun Wu228, Joseph Wu180, Huichun Xu56, Ivana Yang84, Ketian Yu51, Seyedeh Maryam Zekavat52, Yingze Zhang229, Snow Xueyan Zhao101, Wei Zhao230, Xiaofeng Zhu231, Elad Ziv232, Michael Zody50, Sebastian Zoellner51, Mariza de Andrade233, Lisa de las Fuentes234
50 - New York Genome Center, New York, New York, 10013, US; 51 - University of Michigan, Ann Arbor, Michigan, 48109, US; 52 - Broad Institute, Cambridge, Massachusetts, 2142, US; 53 - Cedars Sinai, Boston, Massachusetts, 2114, US; 54 - Children's Hospital of Philadelphia, University of Pennsylvania, Philadelphia, Pennsylvania, 19104, US; 55 - Emory University, Atlanta, Georgia, 30322, US; 56 - University of Maryland, Baltimore, Maryland, 21201, US; 57 - University of Washington, Seattle, Washington, 98195, US; 58 - University of Mississippi, Jackson, Mississippi, 38677, US; 59 - National Institutes of Health, Bethesda, Maryland, 20892, US; 60 - Johns Hopkins University, Baltimore, Maryland, 21218, US; 61 - Duke University, Durham, North Carolina, 27708, US; 62 - University of Alabama, Birmingham, Alabama, 35487, US; 63 - Stanford University, Stanford, California, 94305, US; 64 - Medical College of Wisconsin, Milwaukee, Wisconsin, 53211, US; 65 - Providence Health Care, Medicine, Vancouver, CA; 66 - Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas, 77030, US; 67 - Cleveland Clinic, Cleveland, Ohio, 44195, US; 68 - Tempus, University of Colorado Anschutz Medical Campus, Aurora, Colorado, 80045, US; 69 - Columbia University, New York, New York, 10032, US; 70 - The Emmes Corporation, LTRC, Rockville, Maryland, 20850, US; 71 - Cleveland Clinic, Quantitative Health Sciences, Cleveland, Ohio, 44195, US; 72 - Johns Hopkins University, Medicine, Baltimore, Maryland, 21218, US; 73 - National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland, 20892, US; 74 - Boston University, Massachusetts General Hospital, Boston University School of Medicine, Boston, Massachusetts, 2114, US; 75 - University of Florida, Epidemiology, Gainesville, Florida, 32610, US; 76 - Fundação de Hematologia e Hemoterapia de Pernambuco - Hemope, Recife, 52011-000, BR; 77 - University of Utah, Obstetrics and Gynecology, Salt Lake City, Utah, 84132, US; 78 - National Jewish Health, National Jewish Health, Denver, Colorado, 80206, US; 79 - Medical College of Wisconsin, Pediatrics, Milwaukee, Wisconsin, 53226, US; 80 - University of Texas Health at Houston, Pediatrics, Houston, Texas, 77030, US; 81 - University of California, San Francisco, San Francisco, California, 94143, US; 82 - Stanford University, Biomedical Data Science, Stanford, California, 94305, US; 83 - University of Washington, Biostatistics, Seattle, Washington, 98195, US; 84 - University of Colorado at Denver, Denver, Colorado, 80204, US; 85 - Brigham & Women's Hospital, Boston, Massachusetts, 2115, US; 86 - University of Montreal, US; 87 - Washington State University, Pullman, Washington, 99164, US; 88 - University of California, Los Angeles, Los Angeles, California, 90095, US; 89 - Brigham & Women's Hospital, US; 90 - Brigham & Women's Hospital, Medicine, Boston, Massachusetts, 2115, US; 91 - National Taiwan University, Taipei, 10617, TW; 92 - Brigham & Women's Hospital, Division of Preventive Medicine, Boston, Massachusetts, 2215, US; 93 - University of Virginia, Charlottesville, Virginia, 22903, US; 94 - National Taiwan University, National Taiwan University Hospital, Taipei, 10617, TW; 95 - Cleveland Clinic, Cleveland Clinic, Cleveland, Ohio, 44195, US; 96 - National Health Research Institute Taiwan, Miaoli County, 350, TW; 97 - Broad Institute, Metabolomics Platform, Cambridge, Massachusetts, 2142, US; 98 - Cleveland Clinic, Immunity and Immunology, Cleveland, Ohio, 44195, US; 99 - University of Vermont, Burlington, Vermont, 5405, US; 100 - University of Mississippi, Population Health Science, Jackson, Mississippi, 39216, US; 101 - National Jewish Health, Denver, Colorado, 80206, US; 102 - Boston University, Biostatistics, Boston, Massachusetts, 2115, US; 103 - University of Michigan, Internal Medicine, Ann Arbor, Michigan, 48109, US; 104 - Vitalant Research Institute, San Francisco, California, 94118, US; 105 - University of Illinois at Chicago, Chicago, Illinois, 60607, US; 106 - University of Chicago, Chicago, Illinois, 60637, US; 107 - Vanderbilt University, Nashville, Tennessee, 37235, US; 108 - University of Cincinnati, Cincinnati, Ohio, 45220, US; 109 - Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas, 77030; 110 - University of North Carolina, Chapel Hill, North Carolina, 27599, US; 111 - Baylor College of Medicine Human Genome Sequencing Center, BCM, Houston, Texas, 77030, US; 112 - University of Texas Rio Grande Valley School of Medicine, Edinburg, Texas, 78539, US; 113 - University of Vermont, Pathology and Laboratory Medicine, Burlington, Vermont, 5405, US; 114 - Washington University in St Louis, Genetics, St Louis, Missouri, 63110, US; 115 - Brown University, Providence, Rhode Island, 2912, US; 116 - Harvard University, Channing Division of Network Medicine, Cambridge, Massachusetts, 2138, US; 117 - Massachusetts General Hospital, Boston, Massachusetts, 2114, US; 118 - Cleveland Clinic, Lerner Research Institute, Cleveland, Ohio, 44195, US; 119 - National Jewish Health, Center for Genes, Environment and Health, Denver, Colorado, 80206, US; 120 - Washington University in St Louis, St Louis, Missouri, 63130, US; 121 - Fred Hutchinson Cancer Research Center, Seattle, Washington, 98109, US; 122 - New York Genome Center, New York City, New York, 10013, US; 123 - Icahn School of Medicine at Mount Sinai, New York, New York, 10029, US; 124 - University of Pittsburgh, Pittsburgh, Pennsylvania, US; 125 - Beth Israel Deaconess Medical Center, Boston, Massachusetts, 2215, US; 126 - University of Pittsburgh, Pittsburgh, Pennsylvania, 15260, US; 127 - Boston Children's Hospital, Harvard Medical School, Department of Psychiatry, Boston, Massachusetts, 2115, US; 128 - University of Texas Rio Grande Valley School of Medicine, San Antonio, Texas, 78229, US; 129 - University of Colorado Anschutz Medical Campus, Aurora, Colorado, 80045, US; 130 - Mass General Brigham, Obstetrics and Gynecology, Boston, Massachusetts, 2115, US; 131 - Lundquist Institute, Torrance, California, 90502, US; 132 - Broad Institute, Broad Institute, Cambridge, Massachusetts, 2142, US; 133 - University of Mississippi, Cardiology, Jackson, Mississippi, 39216, US; 134 - University of Calgary, Medicine, Calgary, CA; 135 - University of Maryland, Genetics, Philadelphia, Pennsylvania, 19104, US; 136 - Yale University, Department of Chronic Disease Epidemiology, New Haven, Connecticut, 6520, US; 137 - University of Washington, Epidemiology, Seattle, Washington, 98195-9458, US; 138 - Wake Forest Baptist Health, Winston-Salem, North Carolina, 27157, US; 139 - Brigham & Women's Hospital, Channing Division of Network Medicine, Boston, Massachusetts, 2115, US; 140 - University of Texas Health at Houston, Houston, Texas, 77225, US; 141 - Regeneron Genetics Center, Boston, Massachusetts, 2115, US; 142 - University of Iowa, Iowa City, Iowa, 52242, US; 143 - National Health Research Institute Taiwan, Institute of Population Health Sciences, NHRI, Miaoli County, 350, TW; 144 - Blood Works Northwest, Seattle, Washington, 98104, US; 145 - Taichung Veterans General Hospital Taiwan, Taichung City, 407, TW; 146 - Oklahoma State University Medical Center, Internal Medicine, DIvision of Endocrinology, Diabetes and Metabolism, Columbus, Ohio, 43210, US; 147 - National Heart, Lung, and Blood Institute, National Institutes of Health, NHLBI, Bethesda, Maryland, 20892, US; 148 - University of Washington, Medicine, Seattle, Washington, 98109, US; 149 - University of Michigan, Biostatistics, Ann Arbor, Michigan, 48109, US; 150 - University of California, San Francisco, San Francisco, California, 94118, US; 151 - Harvard University, Cambridge, Massachusetts, 2138, US; 152 - McGill University, Montréal, QC H3A 0G4, CA; 153 - University of Colorado at Denver, Epidemiology, Aurora, Colorado, 80045, US; 154 - Blood Works Northwest, Medicine, Seattle, Washington, 98104, US; 155 - Loyola University, Public Health Sciences, Maywood, Illinois, 60153, US; 156 - Harvard School of Public Health, Biostats, Boston, Massachusetts, 2115, US; 157 - University of Colorado at Denver, Medicine, Aurora, Colorado, 80048, US; 158 - Boston University, University of Massachusetts Chan Medical School, Worcester, Massachusetts, 1655, US; 159 - Brown University, Epidemiology and Medicine, Providence, Rhode Island, 2912, US; 160 - Duke University, Cardiology, Durham, North Carolina, 27708, US; 161 - Stanford University, Cardiovascular Institute, Stanford, California, 94305, US; 162 - Boston University, Boston, Massachusetts, 2215, US; 163 - The Ohio State University, Division of Pulmonary, Critical Care and Sleep Medicine, Columbus, Ohio, 43210, US; 164 - Broad Institute, Harvard University, Massachusetts General Hospital; 165 - University of Alabama, University of Alabama at Birmingham, Birmingham, Alabama, 35487, US; 166 - Brown University, Epidemiology, Providence, Rhode Island, 2912, US; 167 - University of Washington, Genome Sciences, Seattle, Washington, 98195, US; 168 - RTI International, US; 169 - Massachusetts General Hospital, Medicine, Boston, Massachusetts, 2114, US; 170 - University of Arizona, Tucson, Arizona, 85721, US; 171 - Stanford University, Center For Sleep Sciences and Medicine, Palo Alto, California, 94304, US; 172 - National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland, 20892, US; 173 - Oklahoma Medical Research Foundation, Genes and Human Disease, Oklahoma City, Oklahoma, 73104, US; 174 - Ministry of Health, Government of Samoa, Apia, WS; 175 - Howard University, Washington, District of Columbia, 20059, US; 176 - University of Washington, Department of Genome Sciences, Seattle, Washington, 98195, US; 177 - University of Maryland, Balitmore, Maryland, 21201, US; 178 - University at Buffalo, Buffalo, New York, 14260, US; 179 - University of Pennsylvania, Division of Sleep Medicine/Department of Medicine, Philadelphia, Pennsylvania, 19104-3403, US; 180 - Stanford University, Stanford Cardiovascular Institute, Stanford, California, 94305, US; 181 - University of Minnesota, Minneapolis, Minnesota, 55455, US; 182 - RTI International, Biostatistics and Epidemiology Division, Research Triangle Park, North Carolina, 27709-2194, US; 183 - Fred Hutchinson Cancer Research Center, Fred Hutch and UW, Seattle, Washington, 98109, US; 184 - Johns Hopkins University, Cardiology/Medicine, Baltimore, Maryland, 21218, US; 185 - University of Colorado at Denver, Medicine, Denver, Colorado, 80204, US; 186 - University of Colorado at Denver, CCPM, Denver, Colorado, 80045, US; 187 - Northwestern University, Chicago, Illinois, 60208, US; 188 - New York Genome Center, New York Genome Center, New York City, New York, 10013, US; 189 - National Jewish Health, Medicine, Denver, Colorado, 80206, US; 190 - Lutia I Puava Ae Mapu I Fagalele, Apia, WS; 191 - University of Ottawa, Sleep Research Unit, University of Ottawa Institute for Mental Health Research, Ottawa, ON K1Z 7K4, CA; 192 - Vanderbilt University, Medicine, Pharmacology, Biomedicla Informatics, Nashville, Tennessee, 37235, US; 193 - University of Washington, Seattle, Washington, 98104, US; 194 - Universidade de Sao Paulo, Faculdade de Medicina, Sao Paulo, 1310000, BR; 195 - Columbia University, New York, New York, 10027, US; 196 - University of Maryland, Pathology, Seattle, Washington, 98195, US; 197 - Lundquist Institute, TGPS, Torrance, California, 90502, US; 198 - Harvard University, Division of Hematology/Oncology, Boston, Massachusetts, 2115, US; 199 - Harvard Medical School, Genetics, Boston, Massachusetts, 2115, US; 200 - Harvard Medical School, Boston, Massachusetts, 2115, US; 201 - Emory University, Pediatrics, Atlanta, Georgia, 30307, US; 202 - Emory University, Human Genetics, Atlanta, Georgia, 30322, US; 203 - Vanderbilt University, Medicine/Cardiology, Nashville, Tennessee, 37235, US; 204 - UMass Memorial Medical Center, Worcester, Massachusetts, 1655, US; 205 - University of Saskatchewan, Saskatoon, SK S7N 5C9, CA; 206 - University of Michigan; 207 - University of Washington, Epidemiology, Seattle, Washington, 98195, US; 208 - Albert Einstein College of Medicine, New York, New York, 10461, US; 209 - Wake Forest Baptist Health, Biostatistical Sciences, Winston-Salem, North Carolina, 27157, US; 210 - Stanford University, Genetics, Stanford, California, 94305, US; 211 - University of Colorado at Denver, Genomic Cardiology, Aurora, Colorado, 80045, US; 212 - Brigham & Women's Hospital, Channing Department of Medicine, Boston, Massachusetts, 2115, US; 213 - Université Laval, Quebec City, G1V 0A6, CA; 214 - University of Washington, University of Washington, Department of Genome Sciences, Seattle, Washington, 98195, US; 215 - Fred Hutchinson Cancer Research Center, Cancer Prevention Division of Public Health Sciences, Seattle, Washington, 98109, US; 216 - University of Pennsylvania, Genetics, Philadelphia, Pennsylvania, 19104, US; 217 - University of Washington, Department of Biostatistics, Seattle, Washington, 98195, US; 218 - University of Vermont, Pathology & Laboratory Medicine, Burlington, Vermont, 5405, US; 219 - University of Southern California, USC Methylation Characterization Center, University of Southern California, California, 90033, US; 220 - Brigham & Women's Hospital, Mass General Brigham, Boston, Massachusetts, 2115, US; 221 - University of Michigan, US; 222 - University of Pittsburgh, Department of Human Genetics, Pittsburgh, Pennsylvania, 15260, US; 223 - Brigham & Women's Hospital, Channing Division of Network Medicine, Department of Medicine, Boston, Massachusetts, 2115, US; 224 - Indiana University, Epidemiology, Indianapolis, Indiana, 46202, US; 225 - Henry Ford Health System, Detroit, Michigan, 48202, US; 226 - Case Western Reserve University; 227 - Beth Israel Deaconess Medical Center, Cardiology, Cambridge, Massachusetts, 2139, US; 228 - Henry Ford Health System, Department of Medicine, Detroit, Michigan, 48202, US; 229 - University of Pittsburgh, Medicine, Pittsburgh, Pennsylvania, 15260, US; 230 - University of Michigan, Department of Epidemiology, Ann Arbor, Michigan, 48109, US; 231 - Case Western Reserve University, Department of Population and Quantitative Health Sciences, Cleveland, Ohio, 44106, US; 232 - University of California, San Francisco, Medicine, San Francisco, California, 94143, US; 233 - Mayo Clinic, Health Quantitative Sciences Research, Rochester, Minnesota, 55905, US; 234 - Washington University in St Louis, Department of Medicine, Cardiovascular Division, St. Louis, Missouri, 63110, US
Footnotes
Competing interests
Z.R.M. is an employee of Insitro. M.E.M. receives research funding from Regeneron Pharmaceutical Inc., unrelated to this project. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. L.M.R. is a consultant for the TOPMed Administrative Coordinating Center (via Westat). X. Lin is a consultant of AbbVie Pharmaceuticals and Verily Life Sciences. The remaining authors declare no competing interests.
Data availability
This paper used the TOPMed Freeze 8 WGS data and lipids phenotype data. Genotype and phenotype data are both available in database of Genotypes and Phenotypes. The TOPMed WGS data were from the following twenty study cohorts (accession numbers provided in parentheses): Old Order Amish (phs000956.v1.p1), Atherosclerosis Risk in Communities Study (phs001211), Mt Sinai BioMe Biobank (phs001644), Coronary Artery Risk Development in Young Adults (phs001612), Cleveland Family Study (phs000954), Cardiovascular Health Study (phs001368), Diabetes Heart Study (phs001412), Framingham Heart Study (phs000974), Genetic Study of Atherosclerosis Risk (phs001218), Genetic Epidemiology Network of Arteriopathy (phs001345), Genetic Epidemiology Network of Salt Sensitivity (phs001217), Genetics of Lipid Lowering Drugs and Diet Network (phs001359), Hispanic Community Health Study - Study of Latinos (phs001395), Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (phs001293), Jackson Heart Study (phs000964), Multi-Ethnic Study of Atherosclerosis (phs001416), San Antonio Family Heart Study (phs001215), Genome-wide Association Study of Adiposity in Samoans (phs000972), Taiwan Study of Hypertension using Rare Variants (phs001387), and Women’s Health Initiative (phs001237). The sample sizes, ancestry and phenotype summary statistics of these cohorts are given in Supplementary Table 2.
The functional annotation data are publicly available and were downloaded from the following links: GRCh38 CADD v1.4 (https://cadd.gs.washington.edu/download); ANNOVAR dbNSFP v3.3a (https://annovar.openbioinformatics.org/en/latest/user-guide/download); LINSIGHT (https://github.com/CshlSiepelLab/LINSIGHT); FATHMM-XF (http://fathmm.biocompute.org.uk/fathmm-xf); FANTOM5 CAGE (https://fantom.gsc.riken.jp/5/data); GeneCards (https://www.genecards.org; v4.7 for hg38); and Umap/Bismap (https://bismap.hoffmanlab.org; ‘before March 2020’ version). In addition, recombination rate and nucleotide diversity were obtained from Gazal et al50. The whole-genome individual functional annotation data was assembled from a variety of sources and the computed annotation principal components are available at the Functional Annotation of Variant-Online Resource (FAVOR) site (https://favor.genohub.org)51 and the FAVOR database (https://doi.org/10.7910/DVN/1VGTJI)52.
Code availability
MultiSTAAR is implemented as an open source R package available at https://github.com/xihaoli/MultiSTAAR and https://content.sph.harvard.edu/xlin/software.html. Data analysis was performed in R (4.1.0). STAAR v0.9.7 and MultiSTAAR v0.9.7 were used in simulation and real data analysis and implemented as open-source R packages available at https://github.com/xihaoli/STAAR and https://github.com/xihaoli/MultiSTAAR. The assembled functional annotation data were downloaded from FAVOR using Wget (https://www.gnu.org/software/wget/wget.html).
References
- 1.Taliun D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The “All of Us” Research Program. New England Journal of Medicine 381, 668–676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halldorsson B.V. et al. The sequences of 150,119 genomes in the UK Biobank. Nature 607, 732–740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S., Abecasis Gonçalo R., Boehnke M. & Lin X. Rare-Variant Association Analysis: Study Designs and Statistical Tests. The American Journal of Human Genetics 95, 5–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B. & Leal S.M. Methods for Detecting Associations with Rare Variants for Common Diseases: Application to Analysis of Sequence Data. The American Journal of Human Genetics 83, 311–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madsen B.E. & Browning S.R. A Groupwise Association Test for Rare Mutations Using a Weighted Sum Statistic. PLOS Genetics 5, e1000384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris A.P. & Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genetic Epidemiology 34, 188–193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Michael C. et al. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. The American Journal of Human Genetics 89, 82–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y. et al. ACAT: A Fast and Powerful p Value Combination Method for Rare-Variant Analysis in Sequencing Studies. The American Journal of Human Genetics 104, 410–421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solovieff N., Cotsapas C., Lee P.H., Purcell S.M. & Smoller J.W. Pleiotropy in complex traits: challenges and strategies. Nature Reviews Genetics 14, 483–495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivakumaran S. et al. Abundant Pleiotropy in Human Complex Diseases and Traits. The American Journal of Human Genetics 89, 607–618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdellaoui A., Yengo L., Verweij K.J.H. & Visscher P.M. 15 years of GWAS discovery: Realizing the promise. The American Journal of Human Genetics (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nature Genetics 51, 1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Wu B. & Pankow J.S. Sequence Kernel Association Test of Multiple Continuous Phenotypes. Genetic Epidemiology 40, 91–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta D., Scott L., Boehnke M. & Lee S. Multi-SKAT: General framework to test for rare-variant association with multiple phenotypes. Genetic Epidemiology 43, 4–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo L. et al. Multi-trait analysis of rare-variant association summary statistics using MTAR. Nature Communications 11, 2850 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broadaway K.A. et al. A Statistical Approach for Testing Cross-Phenotype Effects of Rare Variants. The American Journal of Human Genetics 98, 525–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X. et al. Dynamic incorporation of multiple in silico functional annotations empowers rare variant association analysis of large whole-genome sequencing studies at scale. Nature Genetics 52, 969–983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sammel M., Lin X. & Ryan L. Multivariate linear mixed models for multiple outcomes. Statistics in Medicine 18, 2479–2492 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Conomos M.P., Miller M.B. & Thornton T.A. Robust Inference of Population Structure for Ancestry Prediction and Correction of Stratification in the Presence of Relatedness. Genetic Epidemiology 39, 276–293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conomos Matthew P., Reiner Alexander P., Weir Bruce S. & Thornton Timothy A. Model-free Estimation of Recent Genetic Relatedness. The American Journal of Human Genetics 98, 127–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gogarten S.M. et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics 35, 5346–5348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee P.H. et al. Principles and methods of in-silico prioritization of non-coding regulatory variants. Human Genetics 137, 15–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z. et al. A framework for detecting noncoding rare-variant associations of large-scale whole-genome sequencing studies. Nature Methods 19, 1599–1611 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison A.C. et al. Practical approaches for whole-genome sequence analysis of heart-and blood-related traits. The American Journal of Human Genetics 100, 205–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selvaraj M.S. et al. Whole genome sequence analysis of blood lipid levels in >66,000 individuals. Nature Communications 13, 5995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z. & Lin X. Multiple phenotype association tests using summary statistics in genome-wide association studies. Biometrics 74, 165–175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teslovich T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaffner S.F. et al. Calibrating a coalescent simulation of human genome sequence variation. Genome research 15, 1576–1583 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Natarajan P. et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nature Communications 9, 3391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stilp A.M. et al. A System for Phenotype Harmonization in the National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) Program. American Journal of Epidemiology (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frankish A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic acids research 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong C. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Human Molecular Genetics 24, 2125–2137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z. et al. A framework for detecting noncoding rare variant associations of large-scale whole-genome sequencing studies. bioRxiv, 2021.11.05.467531 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kircher M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y.-F., Gulko B. & Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nature Genetics 49, 618–624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers M.F. et al. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34, 511–513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buniello A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Research 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klarin D. et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature Genetics 50, 1514–1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forrest A.R. et al. A promoter-level mammalian expression atlas. Nature 507, 462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abascal F. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fishilevich S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z. et al. Dynamic Scan Procedure for Detecting Rare-Variant Association Regions in Whole-Genome Sequencing Studies. The American Journal of Human Genetics 104, 802–814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaw Z.R., Gao J., Lin X. & Gronsbell J. Leveraging a machine learning derived surrogate phenotype to improve power for genome-wide association studies of partially missing phenotypes in population biobanks. bioRxiv, 2022.12.12.520180 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Li X. et al. Powerful, scalable and resource-efficient meta-analysis of rare variant associations in large whole genome sequencing studies. Nature Genetics 55, 154–164 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H. et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. The American Journal of Human Genetics 98, 653–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H. et al. Efficient Variant Set Mixed Model Association Tests for Continuous and Binary Traits in Large-Scale Whole-Genome Sequencing Studies. The American Journal of Human Genetics 104, 260–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y. & Xie J. Cauchy combination test: a powerful test with analytic p-value calculation under arbitrary dependency structures. Journal of the American Statistical Association 115, 393–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gazal S. et al. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nature Genetics 49, 1421–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H. et al. FAVOR: functional annotation of variants online resource and annotator for variation across the human genome. Nucleic Acids Research 51, D1300–D1311 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H., Arapoglou T., Li X., Li Z. & Lin X. FAVOR Essential Database. V1 Edition (Harvard Dataverse, 2022). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper used the TOPMed Freeze 8 WGS data and lipids phenotype data. Genotype and phenotype data are both available in database of Genotypes and Phenotypes. The TOPMed WGS data were from the following twenty study cohorts (accession numbers provided in parentheses): Old Order Amish (phs000956.v1.p1), Atherosclerosis Risk in Communities Study (phs001211), Mt Sinai BioMe Biobank (phs001644), Coronary Artery Risk Development in Young Adults (phs001612), Cleveland Family Study (phs000954), Cardiovascular Health Study (phs001368), Diabetes Heart Study (phs001412), Framingham Heart Study (phs000974), Genetic Study of Atherosclerosis Risk (phs001218), Genetic Epidemiology Network of Arteriopathy (phs001345), Genetic Epidemiology Network of Salt Sensitivity (phs001217), Genetics of Lipid Lowering Drugs and Diet Network (phs001359), Hispanic Community Health Study - Study of Latinos (phs001395), Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (phs001293), Jackson Heart Study (phs000964), Multi-Ethnic Study of Atherosclerosis (phs001416), San Antonio Family Heart Study (phs001215), Genome-wide Association Study of Adiposity in Samoans (phs000972), Taiwan Study of Hypertension using Rare Variants (phs001387), and Women’s Health Initiative (phs001237). The sample sizes, ancestry and phenotype summary statistics of these cohorts are given in Supplementary Table 2.
The functional annotation data are publicly available and were downloaded from the following links: GRCh38 CADD v1.4 (https://cadd.gs.washington.edu/download); ANNOVAR dbNSFP v3.3a (https://annovar.openbioinformatics.org/en/latest/user-guide/download); LINSIGHT (https://github.com/CshlSiepelLab/LINSIGHT); FATHMM-XF (http://fathmm.biocompute.org.uk/fathmm-xf); FANTOM5 CAGE (https://fantom.gsc.riken.jp/5/data); GeneCards (https://www.genecards.org; v4.7 for hg38); and Umap/Bismap (https://bismap.hoffmanlab.org; ‘before March 2020’ version). In addition, recombination rate and nucleotide diversity were obtained from Gazal et al50. The whole-genome individual functional annotation data was assembled from a variety of sources and the computed annotation principal components are available at the Functional Annotation of Variant-Online Resource (FAVOR) site (https://favor.genohub.org)51 and the FAVOR database (https://doi.org/10.7910/DVN/1VGTJI)52.
MultiSTAAR is implemented as an open source R package available at https://github.com/xihaoli/MultiSTAAR and https://content.sph.harvard.edu/xlin/software.html. Data analysis was performed in R (4.1.0). STAAR v0.9.7 and MultiSTAAR v0.9.7 were used in simulation and real data analysis and implemented as open-source R packages available at https://github.com/xihaoli/STAAR and https://github.com/xihaoli/MultiSTAAR. The assembled functional annotation data were downloaded from FAVOR using Wget (https://www.gnu.org/software/wget/wget.html).