Abstract
We have developed a PCR procedure to amplify DNA for quick identification of the economically important species from each of the six taxonomic groups in the plant pathogen genus Phytophthora. This procedure involves amplification of the 5.8S ribosomal DNA gene and internal transcribed spacers (ITS) with the ITS primers ITS 5 and ITS 4. Restriction digests of the amplified DNA products were conducted with the restriction enzymes RsaI, MspI, and HaeIII. Restriction fragment patterns were similar after digestions with RsaI for the following species: P. capsici and P. citricola; P. infestans, P. cactorum, and P. mirabilis; P. fragariae, P. cinnamomi, and P. megasperma from peach; P. palmivora, P. citrophthora, P. erythroseptica, and P. cryptogea; and P. megasperma from raspberry and P. sojae. Restriction digests with MspI separated P. capsici from P. citricola and separated P. cactorum from P. infestans and P. mirabilis. Restriction digests with HaeIII separated P. citrophthora from P. cryptogea, P. cinnamomi from P. fragariae and P. megasperma on peach, P. palmivora from P. citrophthora, and P. megasperma on raspberry from P. sojae. P. infestans and P. mirabilis digests were identical and P. cryptogea and P. erythroseptica digests were identical with all restriction enzymes tested. A unique DNA sequence from the ITS region I in P. capsici was used to develop a primer called PCAP. The PCAP primer was used in PCRs with ITS 1 and amplified only isolates of P. capsici, P. citricola, and P. citrophthora and not 13 other species in the genus. Restriction digests with MspI separated P. capsici from the other two species. PCR was superior to traditional isolation methods for detection of P. capsici in infected bell pepper tissue in field samples. The techniques described will provide a powerful tool for identification of the major species in the genus Phytophthora.
Phytophthora species are responsible for economically important diseases of a wide range of agronomic and ornamental crops. Species identification for Phytophthora has traditionally been based upon microscopic examination of morphological characters and growth characteristics of the pathogen on specific media (27, 35). Variations in the morphological characters of both the sexual and asexual stages of this group of pathogens exist, leading to difficulties in accurate identification by traditional methods. In addition, identification based on pathogenicity assays or growth characteristics are time-consuming. Accurate and rapid identification of Phytophthora species in plant material is important for several reasons. First, in many hosts, such as citrus, walnut, strawberry, raspberry, potato, and tomato, multiple species of Phytophthora can infect the plant, and the relative severity of disease and the plant part infected can vary among pathogen species (6, 25, 37, 43). Preplant identification of Phytophthora species can be important for quarantine purposes and is important for restricting the spread of pathogens in plant material (23). In addition, accurate diagnosis of the species of Phytophthora is important in disease management and control.
Molecular tools including isozyme analysis, restriction fragment length polymorphisms in nuclear and mitochondrial DNA, randomly amplified polymorphic DNA PCRs, serological assays, DNA probes, and PCR of internal transcribed spacer (ITS) regions and nuclear small- and large-subunit ribosomal DNA (rDNA) have been used to evaluate intraspecific and interspecific variation in Phytophthora species (1, 5, 8, 11, 13, 14, 22, 28). Molecular techniques have also been used to study genetic diversity and evolutionary origins in populations of many different fungal genera (2). Nucleotide sequences of rRNA genes have been used in studies of phylogenetic relationships over a wide range of taxonomic levels with many organisms (2, 9, 31, 41). The nuclear small-subunit rDNA sequences evolve relatively slowly and are useful for studying distantly related organisms, whereas the ITS regions and intergenic region of the nuclear rRNA repeat units evolve the fastest and may vary among species and populations (41). Mitochondrial rRNA genes also evolve rapidly and can be useful at the ordinal or family level (41). The evolutionary lineage of the oomycetes has been elucidated by sequencing studies with small-subunit rRNA sequences (9).
We have adopted a quick extraction procedure for DNA and a reliable PCR technique for amplification of DNA from Phytophthora species. This method is based on procedures developed by Lee and Taylor (21) and Lee et al. (22) for Phytophthora species and involves amplification of the ITS and 5.8S rDNA. We used ITS primers 5 and 4 and PCR to amplify the entire 5.8S rDNA gene, both ITS regions I and II, and a portion of the 18S nuclear small-subunit rDNA gene. The amplified DNA was then cut with a series of restriction enzymes to develop species-specific restriction fragment patterns for rapid identification of the important plant-pathogenic Phytophthora species from all the different taxonomic groups in the genus that infect economically important hosts (35). In addition, we devised a PCR primer to specifically amplify P. capsici, an important pathogen of pepper, and used this primer (PCAP) to compare PCR to traditional isolation methods for identification of the pathogen in infected pepper tissue from the field.
MATERIALS AND METHODS
Culture preparation and PCR methods.
Mycelium of each Phytophthora species was grown in pea broth. Pea broth was prepared by autoclaving 120 g of frozen peas in 500 ml of distilled water for 5 min. The filtrate was brought to 1 liter with distilled water and autoclaved for 25 min. Multiple cultures of authenticated isolates from each of the six taxonomic groups in the genus, including group I, P. cactorum (Lebert & Cohn) Schroter; group II, P. capsici Leonian, P. citrophthora (R. E. Sm & E. H. Smith), P. nicotianae Breda de Haan (15), P. palmivora (E. Butler); group III, P. citricola (Saw.); group IV, P. infestans (Mont.) de Bary and P. mirabilis; group V, P. fragariae (C. J. Hickman), P. megasperma (Drechs.), and P. sojae (Hildebr.); and group VI, P. cinnamomi (Rands), P. cryptogea (Pethybr. & Laff.), and P. erythroseptica (Pethybr.), were collected from researchers (Table 1). These taxonomic groups are based on growth characteristics of the pathogen on media, morphological characters of the sexual and asexual propagules, and cardinal temperatures for growth (35). Isolates of P. infestans and P. fragariae were grown in pea broth for 1 week at 18 and 20°C, respectively, while the other species were grown in pea broth for 1 week at 25°C. Mycelium was filtered from the pea broth and frozen in cryogenic vials at −20°C for subsequent work.
TABLE 1.
Species, plant host, source and designation of isolates of Phytophthora species used in PCR experiments
| Species and taxo- nomic groupa | Plant host (no. of isolates) | Isolate designation and sourceb |
|---|---|---|
| Group I | ||
| P. cactorum | Unknown (3) | 127 77, 234 81 (L. Cooke); 1298 (G. Weidemann) |
| Group II | ||
| P. capsici | Pepper (24) | 1, 17–33, 82–88 (J. Ristaino) |
| Tomato (5) | 34–38 (J. Ristaino) | |
| Pumpkin (3) | 39–41 (J. Ristaino) | |
| Squash (3) | 52, 55, 57 (J. Ristaino) | |
| Cucumber (1) | 61 (J. Ristaino) | |
| P. citrophthora | Citrus (5) | M86, M139, M140, M189, M259 (J. Menge) |
| Walnut (1) | 34-4-7 (J. Mircetich) | |
| P. nicotianae | Tobacco (5) | Rmt 6, 332, 340, 335, 435 (D. Shew) |
| Tomato (5) | 1-3A, 6-1A, 5-3A, 2HB, 6-H (J. Ristaino) | |
| Walnut (1) | 35-1-5 (J. Mircetich) | |
| Boxwood (1) | 2107 (M. Benson) | |
| Vinca (1) | 2127 (M. Benson) | |
| Rhododendron (1) | 2109, 116 (M. Benson) | |
| Azalea (1) | 2121 (M. Benson) | |
| Citrus (5) | D-1, R-1, H-2, BHG-1, B-1 (J. Graham) | |
| P. palmivora | Milkweed (1) | P66 (J. Graham) |
| Citrus (6) | P8, P29, P40, P44, P48, Shaw (J. Graham) | |
| Group III | ||
| P. citricola | Avocado (5) | M213, M215, M220, M265, M266 (J. Menge) |
| Group IV | ||
| P. infestans | Tomato (5) | US-7 NY (W. Fry); PINC 93-2, PINC 93-1, PINC 93-4, PINC 93-5 (P. Shoemaker) |
| Potato (8) | US-6 NY (W. Fry); PINC 94-8-1, PINC 94-19, PINC 94-1, PINC 94-7, PINC 94-37 (P. Shoemaker); US-1, US-8 (B. Christ) | |
| P. mirabilis | Mirabilis jalapa (1) | 0S0016 (W. Fry) |
| Group V | ||
| P. fragariae | Strawberry (5) | A-8, R-4, NC-1, R-1, R-6 (B. Milholland) |
| P. megasperma | Raspberry (2) | NY 318, NY 321 (W. Wilcox) |
| Apricot (1) | NY 222 (W. Wilcox) | |
| Cherry (2) | NY 344, NY 346 (W. Wilcox) | |
| Peach (1) | NY 412 (W. Wilcox) | |
| Walnut (1) | 33-2-9 (J. Mircetich) | |
| P. sojae | Soybean (6) | R1, R3, R4, R8, R13, R25 (X. Yang) |
| Group VI | ||
| P. cinnamomi | Rhododendron (1) | 2301 (M. Benson) |
| Fraser fir (1) | 2302 (M. Benson) | |
| Camellia (1) | 2322 (M. Benson) | |
| Shore juniper (1) | 2325 (M. Benson) | |
| Leucothe (1) | 2349 (M. Benson) | |
| Walnut (1) | 34-2-8 (J. Mircetich) | |
| P. cryptogea | Safflower (2) | PCR-1, 34-1-7 (J. Duniway) |
| P. erythroseptica | Potato (3) | 4, 10, 1 (J. Duniway) |
Groups I to VI are taxonomic groups devised by Waterhouse (40) and Stamps et al. (35) to separate the species within the genus. Taxonomic groups are based on morphological and physiological characters of the pathogen.
Isolate designation and source indicates the isolate number and name of the investigator from whom the culture was obtained.
DNA was extracted from frozen mycelium by a CTAB (hexadecyltrimethylammonium bromide) procedure (19). Frozen mycelium was placed in 1.5-ml microtubes, 150 μl of extraction buffer (0.35 M sorbitol, 0.1 M Tris, 0.005 M EDTA [pH 7.5], 0.02 M sodium bisulfite) was added, and the tubes were vortexed. Nuclear lysis buffer (150 μl) containing 0.2 M Tris, 0.05 M EDTA (pH 7.5), 2.0 M NaCl, and 2% CTAB (pH 7.5) was added, followed by 60 μl of 5% Sarkosyl (5 g N-lauroylsarcosine per 100 ml of H2O), and the tubes were vortexed and then incubated at 65°C for 15 to 30 min. Chloroform-isoamyl alcohol (24:1 mixture of chloroform and isoamyl alcohol) (1 volume) was added to each tube, and the tubes were mixed and centrifuged for 15 min at 13,000 × g. The aqueous phase was transferred to a new tube, and the chloroform extraction was repeated. DNA was precipitated overnight at −20°C after the addition of 0.1 volume of 3 M sodium acetate (pH 8.0) and 2 volumes of cold 100% ethanol. The supernatant was discarded, and the pellets were washed with 70% ethanol and then dried by vacuum centrifugation. DNA was resuspended in 100 μl of TE (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]) and then diluted 1:100 for use in PCRs in TE. Extracted DNA was electrophoresed in 1% agarose gels at 25 mA for 3 h. The gels were stained for 15 min in ethidium bromide (0.5 μg/ml) and destained for 15 min in distilled water; alternatively, ethidium bromide was incorporated directly into the gels at a rate of 0.5 μg/ml. The gels were photographed under UV light, and digital images were scanned onto diskettes with a gel scanner (UVP Imagestore 7500).
PCRs were conducted in 50-μl reaction volumes. Each reaction tube contained approximately 1 μl of a 1-ng/μl DNA template, 5 μl of 10× PCR buffer (Boehringer Mannheim, Indianapolis, Ind.), 36.6 μl of sterile distilled water, 2 μl (each) of 1.25 mM deoxynucleoside triphosphates (Pharmacia Biotech, Piscataway, N.J.), 2 μl of 10 mM MgCl2 (Sigma, St. Louis, Mo.), 2 μl each of 10 μM forward and reverse primers (41), and 0.4 μl of Taq (5 U/μl; Boehringer Mannheim). Two drops of mineral oil was placed on the top of each reaction mixture before thermal cycling. The thermal cycling parameters were initial denaturation at 96°C for 2 min followed by 35 cycles consisting of denaturation at 96°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. A final extension at 72°C for 10 min was done at the end of the amplification. Negative controls (no DNA template) were used in every experiment to test for the presence of contamination in reagents. Separate pipettes fitted with filter pipette tips were used in a UV-irradiated hood to prepare master mix reagents for PCR. DNA was pipetted in a separate location with different pipettes.
The ITS primers ITS 5 (5′-GGAAGTAAAAGTCGTAACAAGG) and ITS 4 (5′-TCCTCCGCTTATTGATATGC) (41) amplify the ITS region I between the 18S and 5.8S rDNAs, the 5.8S rDNA, the ITS region II, and a portion of the 28S rDNA. In the first experiments with P. infestans, we used the three primer pairs ITS 5 and ITS 4, ITS 5 and ITS 2 (5′ GCTGCGTTCTTCATCGATGC), and ITS 3 (5′-GCATCGATGAAGAACGCAGC) and ITS 4. All the primer sequences are written 5′ to 3′. Odd-numbered primers are 5′-to-3′ primers, and even-numbered primers are 3′-to-5′ primers. ITS region I between the 18S rDNA and the 5.8S rDNA is flanked by ITS 5 and ITS 2 (41). ITS region II between the 5.8S rDNA and the 28S rDNA is flanked by ITS 3 and ITS 4 (41). In subsequent experiments, primers ITS 4 and ITS 5 were used with all the Phytophthora species.
Amplified fragments were digested with the restriction enzymes RsaI, HaeIII, and/or MspI. Restriction digests consisted of 3 μl of enzyme mixture (1 μl of REact buffer [Gibco BRL, Gaithersburg, Md.], 1 μl of restriction enzyme, and 8 μl of sterile distilled water) and 30 μl of amplified PCR product. DNA was digested at 37°C for 1.5 h and then at 65°C for 10 min. Digested DNA was electrophoresed on a 2% agarose gel at 25 mA for 3 h. The gels were stained in ethidium bromide (0.5 μg/ml) to visualize polymorphisms in amplified DNA fragments. The sizes of the restriction fragments of all the species were measured directly from the same gels and compared to standards ladders. Fragment sizes in base pairs were calculated with the shareware program SEQAID II (32). Representative restriction fragment patterns of individual isolates are shown in the figures; however, all the isolates in Table 1 were tested in individual experiments.
Development of a P. capsici-specific primer.
DNA from two isolates of P. capsici (B1HB14 and B2HH4) was amplified with PCR primers ITS 1 (5′-TCCGTAGGTGAACCTGCGG) and ITS 4. The amplified DNA was cleaned with a Gene Clean kit (Bio 101, Vista, Calif.) by standard procedures. DNA from the two isolates was subjected to automated DNA sequencing on a Perkin-Elmer DNA sequencer at the Iowa State University DNA Sequencing Facility (Ames, Iowa). The DNA sequences were aligned with published sequences from five other Phytophthora species (21) by using the sequence alignment program CLUSTAL (18). Regions of dissimilarity in the ITS region I were used to design and construct a primer specific for P. capsici, called the PCAP primer. The best sequence for the PCAP primer was 5′-TAATCAGTTTTGTGAAATGG. This sequence was published by Lee and Taylor (22) and was developed as an oligonucleotide probe for P. capsici. The PCAP primer was paired with primer ITS 1 and tested with 38 isolates of P. capsici obtained from a variety of vegetable hosts including pepper, tomato, pumpkin, squash, and cucumber (Table 1). The PCAP primer was also tested on isolates comprising 13 different species of Phytophthora (Table 1) in PCRs as described above.
PCR detection in plant tissue.
Field samples of bell pepper that either were asymptomatic, contained visible lesions, or were dead from infections caused by P. capsici were sampled from field plots in 1995. The lesions were cut in half to compare recovery after culture on isolation media to the PCR method. The tissue was surface disinfested in 0.05% sodium hypochlorite and plated on a semiselective medium for isolation of the pathogen (20). For PCR, a portion of the remaining lesion (10 mg) was lysed with 0.5 N NaOH (10 μl/mg), and then 5 μl was diluted immediately in 495 μl of 100 mM Tris buffer (pH 8.0) (39). A 1-μl volume of this extract was used as the DNA template for PCR with the PCAP and ITS 1 primers. Twenty-five plants from each symptom category were sampled, and the PCR experiments were repeated twice.
Nucleotide sequence accession numbers.
The complete ITS sequences of the two pepper isolates of P. capsici have been submitted to the GenBank at the National Center for Biotechnology Information (accession no. AF007021 and AF007022).
RESULTS
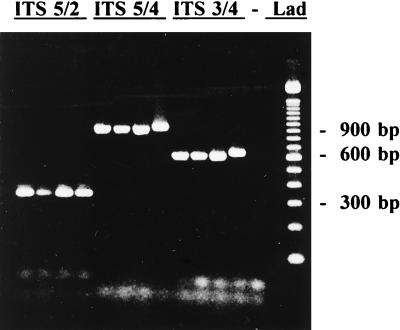
DNA extracted from P. infestans was amplified with ITS primer pairs ITS 5/2, ITS 5/4, and ITS 3/4 (Fig. 1). Pythium ultimum, a related oomycete in a different genus, was amplified for comparison (Fig. 1, lanes 4, 8, and 12). PCR amplification of P. infestans with ITS primers 5/2, 5/4, and 3/4 yielded an estimated 363-bp product, a 946-bp product, and a 612-bp product, respectively. PCR amplification of P. ultimum with ITS primers 5/4 and 3/4 yielded slightly larger products than did amplification of P. infestans, whereas amplification of P. ultimum with ITS 5/2 yielded a similar-size product (Fig. 1).
FIG. 1.
Extracted DNA of P. infestans was amplified with primer pairs ITS 5 and 2, ITS 5 and 4, and ITS 3 and 4. Pythium ultimum DNA amplified with the same primer pairs is shown in the intervening lanes. The no-template control (−) and 100-bp DNA ladder (Lad) are also shown.
P. infestans ITS DNA was digested with a panel of restriction enzymes. Restriction analysis of ITS DNA and 5.8S rDNA from P. infestans amplified with primer pairs ITS 5/2, ITS 5/4, and ITS 3/4 was conducted. Restriction digests with BstNI, HhaI, HinfI, RsaI, PstI, and HaeIII were done. None of the enzymes tested digested the 363-bp product from P. infestans, but RsaI cut the larger 946-bp product into smaller fragments approximately 433, 286, 100, and 79 bp in length. Restriction sites for enzymes BstNI, HhaI, and HinfI were also found in the amplified 946-bp fragment and the 612-bp fragment.
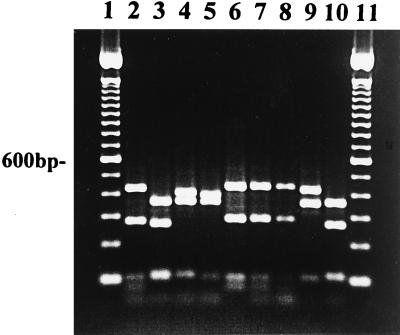
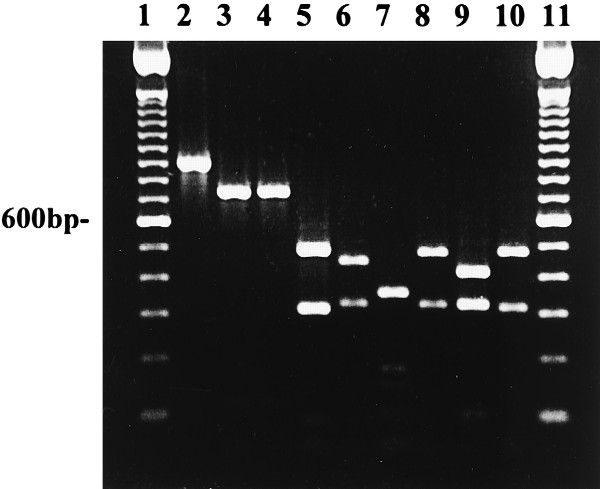
Amplified rDNA from isolates of P. cactorum from taxonomic group I was digested with RsaI, MspI, and HaeIII. Four restriction fragments were observed in P. cactorum after digestion with RsaI (Fig. 2, lane 2; Table 2). The restriction fragment pattern for P. cactorum was identical to the patterns observed for P. infestans and P. mirabilis (Fig. 3, lanes 2 to 4). MspI digests of amplified DNA distinguished P. cactorum (Fig. 4, lane 2; Table 3) from P. infestans and P. mirabilis (Fig. 4, lanes 3 and 4; Table 3). P. infestans and P. mirabilis had identical restriction fragment patterns when digested with MspI (Fig. 4, lanes 3 and 4). Restriction sites for HaeIII were not found in amplified DNA from P. infestans and P. mirabilis, but two fragments of approximately 717 and 189 bp were observed in digested rDNA of P. cactorum (Table 3).
FIG. 2.
Restriction analysis with RsaI of DNA amplified with primer pair ITS 5 and ITS 4 from P. cactorum 1298 (lane 2), P. capsici B1HB14 (lane 3), P. citrophthora M86 (lane 4), P. citrophthora 34-4-7 (lane 5), P. nicotianae D-1 (lane 6), P. nicotianae Rmt 6 (lane 7), P. nicotianae 1-3A (lane 8), P. palmivora P8 (lane 9), and P. citricola M213 (lane 10). Lanes 1 and 11 contain 100-bp ladders.
TABLE 2.
Restriction fragment sizes from ITS and 5.8S rDNA of Phytophthora species amplified with ITS primers 5 and 4a
| Species and taxonomic groupb | Fragment sizes (bp) | Nondigestedproduct size (bp) |
|---|---|---|
| Group I | ||
| P. cactorum** | 436, 286, 111, 79 | 925 |
| Group II | ||
| P. capsici* | 369, 278, 116 | 899 |
| P. citrophthora from citrus**** | 426, 378, 122 | 925 |
| P. citrophthora from walnut | 416, 378, 122 | 925 |
| P. nicotianae | 456, 310, 116, 92 | 925–980 |
| P. palmivora**** | 436, 369, 116 | 952 |
| Group III | ||
| P. citricola* | 378, 286, 122 | 925 |
| Group IV | ||
| P. infestans** | 433, 286, 100, 79 | 925 |
| P. mirabilis** | 433, 286, 100, 79 | 925 |
| Group V | ||
| P. fragariae*** | 454, 225, 178, 111 | 980 |
| P. megasperma from raspberry***** | 454, 393, 111 | 980 |
| P. megasperma from peach*** | 454, 218, 184, 111 | 980 |
| P. sojae***** | 454, 393, 111 | 980 |
| Group VI | ||
| P. cinnamomi*** | 454, 218, 184, 111 | 980 |
| P. cryptogea**** | 423, 383, 111 | 925 |
| P. erythroseptica**** | 423, 383, 111 | 925 |
Amplified DNA was digested with RsaI.
Species with the same number of asterisks have similar restriction fragment patterns after digestions with RsaI.
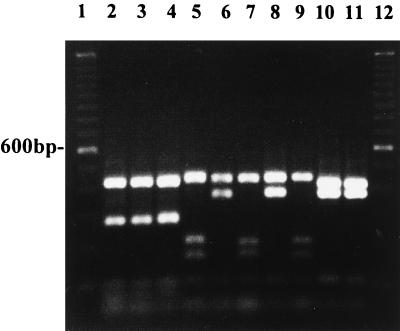
FIG. 3.
Restriction analysis with RsaI of DNA amplified with primer pair ITS 5 and ITS 4 from P. infestans US-6 NY (lane 2), P. infestans 93-1 (lane 3), P. mirabilis 0S0016 (lane 4), P. fragariae A-8 (lane 5), P. megasperma NY 318 (lane 6), P. megasperma NY 412 (lane 7), P. sojae R1 (lane 8), P. cinnamomi 2302 (lane 9), P. cryptogea PCR-1 (lane 10), and P. erythroseptica 4 (lane 11). Lanes 1 and 12 contain 100-bp ladders.
FIG. 4.
Restriction analysis with MspI of DNA amplified with ITS 5 and ITS 4 from P. cactorum 1298 (lane 2), P. infestans US-6 NY (lane 3), P. mirabilis 0S0016 (lane 4), P. capsici B1HB14 (lane 5), and P. citricola M213 (lane 6). Lanes 1 and 7 contain 100-bp ladders.
TABLE 3.
Restriction fragment sizes from ITS and 5.8S rDNA of Phytophthora species amplified with ITS primers 5 and 4a
| Species | Fragment sizes (bp) |
|---|---|
| Digestion with MspI | |
| P. capsici | 290, 219, 194, 140 |
| P. citricola | 340, 274, 111 |
| P. cactorum | 396, 225, 189, 109 |
| P. infestans | 397, 293, 225 |
| P. mirabilis | 397, 293, 225 |
| Digestion with HaeIII | |
| P. cactorum | 717, 189 |
| P. palmivora | 920 |
| P. erythroseptica | 749, 122 |
| P. cryptogea | 749, 122 |
| P. citrophthora | 481, 306 |
| P. cinnamomi | 446, 319 |
| P. fragariae | 347, 179, 145, 106 |
| P. megasperma from peach | 463, 315, 128 |
| P. sojae | 402, 315, 101 |
| P. megasperma from raspberry | 475, 302, 108 |
Amplified DNA was digested with either MspI or HaeIII.
Restriction fragment patterns were similar between all taxonomic group II isolates of P. capsici tested (Fig. 2, lane 3; Table 1) and taxonomic group III isolates of P. citricola (Fig. 2, lane 10). Since P. capsici and P. citricola had similar restriction fragment patterns in this amplified region of DNA, further digests with other restriction enzymes were done. Isolates of P. capsici were differentiated from isolates of P. citricola after digestion with MspI (Fig. 4, lanes 5 and 6; Table 3). In contrast, taxonomic group II isolates of P. citrophthora from citrus (Fig. 2, lane 4; Table 2) had different restriction fragment length patterns from P. capsici and P. citricola after digestion with RsaI (Fig. 2, lanes 3 and 10). Only one isolate of P. citrophthora from walnut was tested in our study (lane 5), and it had a slightly different restriction fragment pattern from the citrus isolates of P. citrophthora (lane 4). Other isolates from walnut need to be tested to confirm or refute this restriction fragment pattern for the walnut P. citrophthora.
Isolates of P. nicotianae (formerly P. parasitica) from tobacco, tomato, walnut, boxwood, vinca, rhododendron, azalea, and citrus are classified into taxonomic group II (Table 1). All isolates of P. nicotianae tested had the same restriction fragment patterns in this amplified region of ITS and 5.8S rDNA, and four fragments were visible after restriction digestion with RsaI (Fig. 2, lanes 6 to 8; Tables 1 and 2).
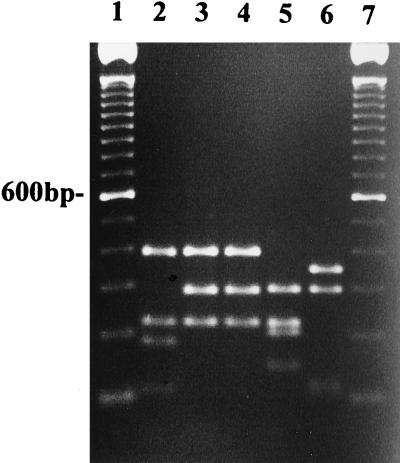
P. palmivora from citrus (Fig. 2, lane 9) had a similar restriction fragment pattern to P. citrophthora from citrus (lane 4) when digested with RsaI but had a different pattern from isolates of P. nicotianae from citrus (lane 6). P. palmivora could be distinguished from P. citrophthora after digestion with HaeIII (Fig. 5, lanes 2 and 5; Table 3). P. palmivora was not digested by HaeIII, but P. citrophthora was digested (Fig. 5, lanes 2 and 5; Table 3).
FIG. 5.
Restriction analysis with HaeIII of DNA amplified with ITS 5 and ITS 4 from P. palmivora P8 (lane 2), P. erythroseptica 4 (lane 3), P. cryptogea PCR-1 (lane 4), P. citrophthora M86 (lane 5), P. cinnamomi 2302 (lane 6), P. fragariae A-8 (lane 7), P. megasperma NY 412 (lane 8), P. sojae R1 (lane 9), and P. megasperma NY 318 (lane 10). Lanes 1 and 11 contain 100-bp ladders.
All taxonomic group IV isolates of P. infestans from potato and tomato showed the same restriction fragment pattern after digestion with RsaI (Fig. 3, lanes 2 and 3). Four fragments were observed, and the restriction patterns were identical to those of P. mirabilis (Fig. 3, lane 4; Table 2). In contrast, P. erythroseptica, which causes pink rot of potato, gave a restriction fragment pattern different from that of P. infestans in this amplified region when digested with RsaI (Fig. 3, lane 11; Table 2).
All isolates of P. fragariae (taxonomic group V) from strawberry had identical restriction fragment patterns when amplified DNA was digested with RsaI and yielded four fragments (Fig. 3, lane 5). Variation in the restriction fragment patterns was observed within the group of isolates identified as P. megasperma. Isolates of P. megasperma from raspberry (Fig. 3, lane 6), apricot, and cherry had the same restriction fragment patterns when digested with RsaI (Tables 1 and 2). However, the putative isolates of P. megasperma from peach (Fig. 3, lane 7) and walnut (not shown) had restriction fragment patterns similar to P. fragariae (lane 5) and P. cinnamomi (lane 9) after digestion with RsaI. P. sojae isolates from soybean (lane 8) had identical restriction fragment patterns to P. megasperma from raspberry, apricot, and cherry when digested with RsaI (Fig. 3, lane 6; Table 2). However, restriction digestion with HaeIII separated these two species (Fig. 5, lanes 8 and 9; Table 3).
All the isolates of P. cinnamomi (taxonomic group VI) from a variety of hosts including rhododendron, fraser fir, camellia, shore juniper, and leucothe (Table 1) had similar restriction fragment patterns when digested with RsaI and yielded four fragments (Fig. 3, lane 9; Table 2). These bands were similar in size to the restriction fragments observed when DNA from P. fragariae and P. megasperma from peach were digested with RsaI (Fig. 3, lanes 5 and 7; Table 2). Digestion of amplified DNA with HaeIII differentiated P. cinnamomi from P. fragariae (Fig. 5, lanes 6 and 7). Both P. cryptogea isolates from safflower had the same restriction fragment patterns when digested with RsaI and yielded three fragments (Fig. 3, lanes 10). These restriction fragments were similar to those of RsaI-digested P. citrophthora (Fig. 2, lane 4) and P. erythroseptica (Fig. 3, lane 11). However, digestion of amplified DNA with HaeIII differentiated isolates of P. cryptogea and P. erythroseptica (Fig. 5, lanes 3 and 4; Table 3) from P. citrophthora (Fig. 5, lane 5). P. erythroseptica and P. cryptogea had the same restriction fragment patterns after digestion with HaeIII, and we were unable to distinguish between these two species with the range of restriction enzymes tested.
Development of the PCAP primer.
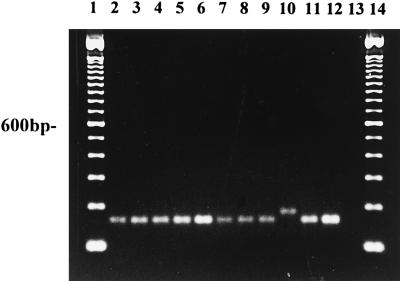
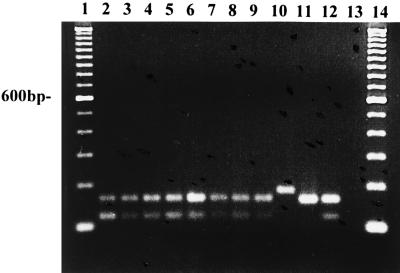
The PCAP primer amplified an approximately 172-bp fragment of DNA in all isolates of P. capsici tested from a range of hosts (Fig. 6, lanes 2 to 9 and 12; Table 1). The primer also amplified a similar-size fragment in isolates of P. citricola (Fig. 6, lane 11). Isolates of P. citrophthora were also amplified by the PCAP primer, but the amplified product was larger than that of P. capsici or P. citricola (lane 10). Digestions of the 172-bp fragment with MspI differentiated P. capsici from P. citrophthora and P. citricola, which were not digested by this enzyme (Fig. 7). Apparently two different PCR products, both approximately 172 bp in size, were amplified by the PCAP and ITS 1 primer pair. Restriction digestion with MspI yielded a 172-bp product and several smaller products in isolates of P. capsici (Fig. 7, lanes 2 to 9 and 12). P. capsici and P. citricola can also be differentiated by restriction digestion of ITS DNA with MspI (Fig. 4, lanes 5 and 6; Table 3). None of the other species of Phytophthora tested, including P. cactorum, P. palmivora, P. nicotianae, P. infestans, P. mirabilis, P. fragariae, P. sojae, P. megasperma, P. cinnamomi, P. cryptogea, and P. erythroseptica, were amplified with the PCAP primer.
FIG. 6.
DNA amplified with the PCAP primer and ITS 1 from P. capsici 17 (lane 2), P. capsici 18 (lane 3), P. capsici 19 (lane 4), P. capsici 20 (lane 5), P. capsici 21 (lane 6), P. capsici 22 (lane 7), P. capsici 23 (lane 8), P. capsici 25 (lane 9), P. citrophthora M86 (lane 10), P. citricola M213 (lane 11), and P. capsici 87 (lane 12). Lanes 1 and 14 contain 100-bp ladders; lane 13 contains a no-template control.
FIG. 7.
Restriction digest with MspI of DNA amplified with the PCAP primer and ITS 1 from P. capsici 17 (lane 2), P. capsici 18 (lane 3), P. capsici 19 (lane 4), P. capsici 20 (lane 5), P. capsici 21 (lane 6), P. capsici 22 (lane 7), P. capsici 23 (lane 8), P. capsici 25 (lane 9), P. citrophthora M86 (lane 10), P. citricola M213 (lane 11), and P. capsici 87 (lane 12). Lanes 1 and 14 contain 100-bp ladders; lane 13 contains a no-template control.
PCR was more rapid and efficient than traditional isolation methods in identification of P. capsici in field-infected plant samples. Of infected pepper plants with visible lesions that were positive by traditional isolation on media, 92% were also positive by PCR. The PCR method also detected 32% of the infections in samples where the pathogen was not identified previously by traditional isolation on agar media. Neither method was successful in detection of the pathogen in severely decayed tissue.
DISCUSSION
Species identification in the genus Phytophthora is difficult and requires the use of taxonomic keys and knowledge of the host range of the pathogen. The PCR procedures we describe in this work will provide a powerful tool for plant disease diagnosticians and researchers who are interested in the identification of many of the major species in the genus. Currently, the taxonomic key of Stamps et al. (35), which is based on earlier work by Waterhouse (40), is the standard reference for identification of pathogens in the genus Phytophthora by classical methods. The tabular key divides the genus into six morphological groups based on characteristics of the sporangia, gametangia, growth at specific temperatures, and culture characters. We analyzed restriction fragment patterns of amplified ITS DNA from a sample of many isolates from each of six morphological groups described previously (35).
Based on morphological characteristics, isolates of P. capsici and P. citricola are placed in taxonomic groups II and III; however, these two species had common restriction fragment patterns when digested with RsaI. The PCAP primer also amplified P. capsici, P. citricola, and P. citrophthora, indicating that there is sequence homology in the spacer I region between the 18S and 5.8S rDNA among these three species from taxonomic groups II and III. Forster et al. (11) and Cooke and Duncan (3) sequenced the ITS region I of DNA in a large number of Phytophthora species and identified a cluster among isolates of P. capsici, P. citricola, and P. citrophthora isolates. In addition, isozyme, mitochondrial DNA restriction fragment length polymorphism, and ITS DNA sequence studies have demonstrated close relationships among these three species (8, 21, 29). Lee and Taylor (21) analyzed ITS variability in several Phytophthora species, and their data support a close relationship between cacao isolates of P. capsici and P. citrophthora (21). The PCR primer developed by Ersek et al. (5) for P. citrophthora also amplified DNA of P. capsici. These three species have papillate or semipapillate sporangia. Our data and the data of others support the phylogenetic grouping of P. capsici, P. citricola, and P. citrophthora into a distinct cluster (3, 11).
Isolates of P. nicotianae tested from citrus, tomato, tobacco, and many ornamental plants were genetically similar after restriction digests with RsaI in this amplified region of ITS DNA. Restriction fragment length polymorphism analysis of genomic DNA of this species indicated little variation among tobacco and citrus isolates (30). Hall (15) redescribed this species as a single species and suggested elimination of the forma specialis designations after extensive testing of 31 morphological, physiological, and biochemical characters (15). We did not test the host pathogenicity or physiological and biochemical traits of isolates in our work, but our data indicate little genetic variation among the isolates we tested.
P. fragariae, P. cactorum, P. nicotianae, and P. citricola are pathogens of strawberry plants in the United States (6, 24). These pathogens can be transported in infected propagation material and introduced into fields. P. fragariae, P. cactorum, P. nicotianae, and P. citricola are from four different taxonomic groups (V, I, II, and III, respectively) but can be easily distinguished after digestion of amplified ITS DNA with RsaI. In addition, the PINF primer we developed in related work for the potato and tomato late blight pathogen P. infestans also amplifies P. cactorum (38), and the PCAP primer described in our present work for P. capsici amplifies P. citricola. Primer sequences which amplify P. fragariae isolates from strawberry, P. fragariae var. rubi from raspberry, and P. nicotianae have been reported (4, 5, 33, 34). Both specific and universal PCR primers could be used to screen plant material and improve the detection of all the major Phytophthora pathogens of strawberry. Strawberry plants are vegetatively propagated, and Phytophthora species can spread readily in infected plant material.
A number of Phytophthora species, including P. nicotianae, P. citrophthora, P. palmivora, P. citricola, P. syringae, and P. hibernalis, infect citrus (14, 42). Four of these six species were examined in this study. P. nicotianae and P. citrophthora are the two most common species on citrus, and they were easily distinguished after restriction digestion with RsaI. P. citrophthora and P. nicotianae both cause gummosis and root rot of citrus, but P. citrophthora is more active in the fruit and aerial plant parts than P. nicotianae. The incidence of brown rot on fruit caused by P. palmivora has increased in recent years in Florida (14a).
There was variation in restriction fragment patterns of ITS DNA among the group V isolates of P. megasperma. Variation within this species has also been noted by others (7, 10, 11, 16, 17, 30, 43). The host-specialized form species of isolates that infect legumes within P. megasperma have been given species designations and include P. sojae, P. medicaginis, and P. trifolii (17). However, the isolates of P. megasperma from woody hosts were placed into the broad-host-range (BHR) lineage and separated into electrophoretic types BHR, AC, and DF karyotypes (16, 17). Sequence differences in the ITS spacer I region indicate that the pathogens in this BHR group do not represent a single biological species (11). In our work, two isolates of P. megasperma from peach and walnut had restriction patterns that differed from the other isolates from raspberry, apricot, and cherry. One of these isolates from peach (NY 412) was studied previously by others and is the A/C electrophoretic type sensu Hansen et al. (17, 42a). This isolate is in a morphologically, culturally, and electrophoretically distinct group that has been referred to as the small-oospore, high-temperature type group of P. megasperma (44). Further work must be done with a larger number of isolates of P. megasperma from the woody-host group to further delineate fruit tree isolates. Separate species designations for the fruit tree isolates are probably warranted, as has been done with the legume isolates (7), to clarify the taxonomy of Phytophthora megasperma.
P. infestans, P. mirabilis, and P. cactorum had identical restriction fragment patterns when their ITS DNA was digested with RsaI. These three species also yielded an identical product when amplified with the PINF primer (38). Others also developed a PCR primer that amplifies both P. infestans and P. mirabilis and found similarities in the ITS region II between these species (36, 37). P. cactorum can be easily differentiated from P. infestans after restriction digests with MspI. The ITS region I DNA sequences of P. cactorum and P. infestans were similar, and these species also formed a cluster in phylogenetic analysis (3, 11). In our work, P. mirabilis and P. infestans were not distinguishable after restriction digestion with a number of enzymes including RsaI, MspI, EcoRI, and HaeIII. These data suggest that the two species have considerable sequence homology in this region of ITS DNA. P. mirabilis was first described as a new species on Mirabilis jalapa in Mexico in 1985 (12). Other authors have suggested that P. mirabilis should be called a forma specialis of P. infestans (26). P. infestans and P. mirabilis have similar mitochondrial DNA restriction patterns (26). An oligonucleotide probe, pL121-3, was developed to differentiate P. mirabilis from P. infestans, but the reaction of the probe with other potato pathogens was not examined (26). We have not yet sequenced the ITS DNA of P. mirabilis, but our data also suggest that two species designations may be unwarranted.
It is evident from our work and that of others (3, 11, 28, 29) that the morphological differentiations of the major species of Phytophthora do not necessarily represent genetic differences among the species. Isolates with divergent sporangial characters, temperature requirements, and hosts have sequence homology in their ITS DNA. The isolates used in our present study were well characterized by morphological criteria before the advent of PCR. Identification by classical methods requires expertise and is time-consuming. The molecular methods we describe will provide useful and rapid tools for identification of the economically important species within this plant-pathogenic genus.
ACKNOWLEDGMENTS
This work was funded in part by a research experience for undergraduates (REU) supplemental grant from the National Science Foundation (M. Madritch) and in part by a USDA National Research Initiatives Competitive Grant.
J.B.R. is grateful to Dina St. Clair, University of California, Davis, and David Francis, Ohio State University, Wooster, for sharing techniques, intellectual expertise, and enthusiasm during a visit in November 1994 of the senior author to UC Davis. Appreciation is also expressed to Michael Benson, Barbara Christ, John Duniway, William Fry, Jim Graham, John Menge, Robert Milholland, John Mircetich, David Shew, Paul Shoemaker, Greg Weidemann, Wayne Wilcox, and X. B. Yang and former graduate student Dawn Fraser for generously providing cultures of the Phytophthora species used in this work. The assistance of former graduate student Ronald French with PCR work with P. capsici and discussions with Lee Campbell and Marcia Gumpertz on sampling strategies are also acknowledged.
REFERENCES
- 1.Benson D M. Detection of Phytophthora cinnamomi in azalea with commercial serological assay kits. Plant Dis. 1991;75:478–482. [Google Scholar]
- 2.Bruns T D, White T J, Taylor J W. Fungal molecular systematics. Annu Rev Ecol Syst. 1991;22:525–564. [Google Scholar]
- 3.Cooke D E L, Duncan J M. Phylogenetic analysis of Phytophthora species based on ITS 1 and 2 sequences of the rDNA gene repeat. Mycol Res. 1997;101:667–677. [Google Scholar]
- 4.Duncan J. Fungal and bacterial diseases. Dundee, Scotland: Report of the Scottish Crops research Institute; 1995. [Google Scholar]
- 5.Ersek T, Schoeltz J E, English J T. PCR amplification of species-specific DNA sequences can distinguish among Phytophthora species. Appl Environ Microbiol. 1994;60:2616–2621. doi: 10.1128/aem.60.7.2616-2621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farr D F, Bill G F, Chamuris G P, Rossman A Y. Fungi on plants and plant products in the United States. St. Paul, Minn: American Phytopathological Society Press; 1989. [Google Scholar]
- 7.Forster H, Kinscherf T G, Leong S A, Maxwell D P. Restriction fragment length polymorphism of the mitochondrial DNA of Phytophthora megasperma isolated from soybean, alfalfa, and fruit trees. Can J Bot. 1989;67:529–537. [Google Scholar]
- 8.Forster H, Oudemans P, Coffey M D. Mitochondrial and nuclear DNA diversity within six species of Phytophthora. Exp Mycol. 1990;14:18–31. [Google Scholar]
- 9.Forster H, Coffey M, Elwood H, Sogin M L. Sequence analysis of the small subunit ribosomal RNA’s of three zoosporic fungi and implications for fungal evolution. Mycologia. 1990;82:306–312. [Google Scholar]
- 10.Forster H, Coffey M D. Molecular taxonomy of Phytophthora megasperma based on mitochondrial and nuclear DNA polymorphisms. Mycol Res. 1993;97:1101–1112. [Google Scholar]
- 11.Forster H, Learn G, Coffey M D. Towards a better understanding of the evolutionary history of the species of the genus Phytophthora using isozymes, DNA RFLP’s, and rDNA spacer sequences. In: Dowley L J, Bannon E, Cooke L R, Keane T, O’Sullivan E, editors. Phytophthora 150. Dublin, Ireland: European Association for Potato Research. Boole, Ltd.; 1995. pp. 42–54. [Google Scholar]
- 12.Galindo A J, Hohl H R. Phytophthora mirabilis a new species of Phytophthora. Sydowia. 1985;38:87–96. [Google Scholar]
- 13.Goodwin P H, Kirkpatrick B C, Duniway J M. Cloned DNA probes for identification of P. parasitica. Phytopathology. 1989;79:716–721. [Google Scholar]
- 14.Goodwin P H, Kirkpatrick B C, Duniway J M. Identification of Phytophthora citrophthora with cloned DNA probes. Appl Environ Microbiol. 1990;56:669–674. doi: 10.1128/aem.56.3.669-674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Graham, J. Personal communication.
- 15.Hall G. An integrated approach to the analysis of variation in Phytophthora nicotianae and a redescription of the species. Mycol Res. 1993;97:559–574. [Google Scholar]
- 16.Hansen E M, Braiser C M, Shaw D S, Hamm P B. The taxonomic structure of Phytophthora megasperma: evidence for emerging biological species groups. Trans Br Mycol Soc. 1986;87:557–573. [Google Scholar]
- 17.Hansen E M, Maxwell D P. Species of the Phytophthora megasperma complex. Mycologia. 1991;83:376–381. [Google Scholar]
- 18.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 19.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. [Google Scholar]
- 20.Kannwischer M E, Mitchell D J. The influence of a fungicide on the epidemiology of black shank of tobacco. Phytopathology. 1978;68:1760–1765. [Google Scholar]
- 21.Lee S B, Taylor J W. Phylogeny of five fungus-like protoctistan Phytophthora species, inferred from internal transcribed spacers of ribosomal DNA. Mol Biol Evol. 1992;9:636–653. doi: 10.1093/oxfordjournals.molbev.a040750. [DOI] [PubMed] [Google Scholar]
- 22.Lee S B, White T J, Taylor J W. Detection of Phytophthora species by oligonucleotide hybridization to amplified ribosomal DNA spacers. Phytopathology. 1993;83:177–181. [Google Scholar]
- 23.MacDonald J D, Stites J, Kabashima J. Comparison of serological and culture plate methods for detection of Phytophthora species. Plant Dis. 1990;74:655–659. [Google Scholar]
- 24.Milholland R D. A monograph of Phytophthora fragariae and the red stele disease of strawberry. North Carolina Agricultural Research Service technical bulletin 306. Raleigh: North Carolina Agricultural Research Service; 1994. [Google Scholar]
- 25.Mircetich S M, Browne G T, Krueger W, Schreader W. Phytophthora species isolated from surface water irrigation sources in California. Phytopathology. 1985;75:1346–1347. [Google Scholar]
- 26.Moller E M, De Cock A W A M, Prell H H. Mitochondrial and nuclear DNA restriction enzyme analysis of the closely related Phytophthora species, P. infestans, P. mirabilis, and P. phaseoli. J Phytopathol. 1993;139:309–321. [Google Scholar]
- 27.Newhook F J, Waterhouse G M, Stamps O J. Tabular key to the species of Phytophthora. Mycotaxon. 1976;32:199–217. [Google Scholar]
- 28.Oudemans P, Coffey M D. A revised systematics of twelve papillate Phytophthora species based on isozyme analysis. Mycol Res. 1991;95:1025–1046. [Google Scholar]
- 29.Oudemans P, Forster H, Coffey M D. Evidence for distinct isozyme subgroups within Phytophthora citricola and close relationships with P. capsici and P. citrophthora. Mycol Res. 1994;98:189–199. [Google Scholar]
- 30.Panabieres F, Marais A, Trentin F, Bonnet P, Ricci P. Repetitive DNA polymorphism analysis as a tool for identifying Phytophthora species. Phytopathology. 1989;79:1105–1109. [Google Scholar]
- 31.Redecker D, Thierfelder H, Walker C, Werner D. Restriction analysis of PCR-amplified internal transcribed spacers of ribosomal DNA as a tool for species identification in different genera of the order Glomales. Appl Environ Microbiol. 1997;63:1756–1761. doi: 10.1128/aem.63.5.1756-1761.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes D D, Roufa D J. SEQAID II. Manhattan: University of Kansas; 1989. [Google Scholar]
- 33.Stammler G, Seemuller E. Specific and sensitive detection of Phytophthora fragariae var. rubi in raspberry roots by PCR amplification. J Plant Dis Prot. 1993;100:394–400. [Google Scholar]
- 34.Stammler G, Seemuller E, Duncan J M. Taxonomic characterization of Phytophthora fragariae by DNA hybridization and restriction fragment length polymorphism analysis. Mycol Res. 1993;97:150–156. [Google Scholar]
- 35.Stamps D J, Waterhouse G M, Newhook F J, Hall G S. Revised tabular key to the species of Phytophthora. Mycology papers no. 162. C.A.B. Egham, England: International Mycological Institute; 1990. [Google Scholar]
- 36.Tooley P W, Carras M M, Falkenstein K F. Relationships among group IV Phytophthora species inferred by restriction analysis of ITS 2 region. J Phytopathol. 1996;144:363–369. [Google Scholar]
- 37.Tooley P W, Bunyard B A, Carras M M, Hatziloukas E. Development of PCR primers from internal transcribed spacer region 2 for detection of Phytophthora species infecting potatoes. Appl Environ Microbiol. 1997;63:1467–1475. doi: 10.1128/aem.63.4.1467-1475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trout C L, Ristaino J B, Madritch M, Wangsoomboondee T. Rapid detection of Phytophthora infestans in late blight infected potatoes and tomatoes using PCR. Plant Dis. 1997;81:1042–1048. doi: 10.1094/PDIS.1997.81.9.1042. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Qi M, Cutler A J. A simple method of preparing plant samples for PCR. Nucleic Acids Res. 1993;21:4153–4154. doi: 10.1093/nar/21.17.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse G M. Key to the species of Phytophthora de Bary. Mycological Papers 92. Kew, Surrey, England: Commonwealth Mycological Institute; 1963. [Google Scholar]
- 41.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 42.Whiteside J O, Garnsey S M, Timmer L W. Compendium of citrus diseases. St. Paul, Minn: American Phytopathological Society Press; 1988. [Google Scholar]
- 42a.Wilcox, W. F. Personal communication.
- 43.Wilcox W F. Identity, virulence and isolation frequency of seven Phytophthora species causing root rot of raspberry in New York. Phytopathology. 1989;79:93–101. [Google Scholar]
- 44.Wilcox W F, Mircetich J. Lack of host specificity among isolates of P. megasperma. Phytopathology. 1987;77:1132–1137. [Google Scholar]