Abstract
Tumor-associated macrophages (TAMs) are frequently categorized as being ‘M1’ or ‘M2’ polarized, even as substantial data challenges this binary modeling of macrophage cell state. One molecule consistently referenced as a delineator of a putative immunosuppressive ‘M2’ state is the surface protein CD206. We thus made a novel conditional CD206 (Mrc1) knock-in mouse to specifically visualize and/or deplete CD206+ ‘M2-like’ TAMs and assess their correspondence with pro-tumoral immunity. Early, but not late depletion of CD206+ macrophages and monocytes (here, ‘Mono/Macs’) led to an indirect loss of a key anti-tumor network of NK cells, conventional type I dendritic cells (cDC1) and CD8 T cells. Among myeloid cells, we found that the CD206+ TAMs are the primary producers of CXCL9, and able to differentially attract activated CD8 T cells. In contrast, a population of stress-responsive TAMs (“Hypoxic” or Spp1+) and immature monocytes, which lack CD206 expression and become prominent following early depletion, expressed markedly diminished levels of CXCL9. Those NK and CD8 T cells which enter CD206-depleted tumors express vastly reduced levels of the corresponding receptor Cxcr3, the cDC1-attracting chemokine Xcl1 and cDC1 growth factor Flt3l transcripts. Consistent with the loss of this critical network, early CD206+ TAM depletion decreased tumor control by antigen specific CD8 T cells in mice. Likewise, in humans, the CD206Replete, but not the CD206Depleted Mono/Mac gene signature correlated robustly with CD8 T cell, NK cell and stimulatory cDC1 gene signatures and transcriptomic signatures skewed towards CD206Replete Mono/Macs associated with better survival. Together, these findings negate the unqualified classification of CD206+ ‘M2-like’ macrophages as immunosuppressive by illuminating contexts for their role in organizing a critical tumor-reactive archetype of immunity.
Introduction:
Macrophages have diverse roles in homeostasis and disease and a refined understanding of the direct and indirect effects of targeting them in tumors is an imperative, given the current impetus in developing myeloid targeting therapies for cancer(1, 2). A widely used shortcut for describing macrophage function in tumors involves an ‘M1’ versus ‘M2’ nomenclature, derived from in vitro skewing with Th1 versus Th2 cytokines, and often equated with pro and anti-inflammatory functions respectively. However, this binary M1/M2 delineation of macrophage phenotype does not capture the heterogeneity at the single cell level (3–5). In addition, there is scant evidence of these markers being part of coordinated gene programs in vivo. In wound healing, Arg1 and Mrc1(gene corresponding to the mannose-binding C-type lectin CD206), both purportedly key markers of an M2 state, have distinct expression patterns (6). In both mouse and human tumors, there is also a complete lack of correlation among genes characterizing M1 or M2 phenotypes within Mono/Macs (3). In fact, M1 and M2 signatures in Mono/Macs often show correlated instead of opposing expression patterns in tumors (4, 5). Nonetheless, myeloid cells expressing CD206, sometimes therefore designated as ‘M2-like’, continue to be used as a marker of an immunosuppressive state. The detrimental effects of tumor-associated macrophages (TAMs) on anti-tumor immunity have indeed been highlighted by a number of critical studies (7–11). However, a holistic dissection of the role of CD206-expressing Mono/Macs and the precise effects of targeting them in tumors in vivo is lacking. We therefore developed a conditional knock-in reporter mouse using the Mrc1 (CD206) allele that allows specific visualization and depletion of those cells to define their true impact on anti-tumor immunity.
Results:
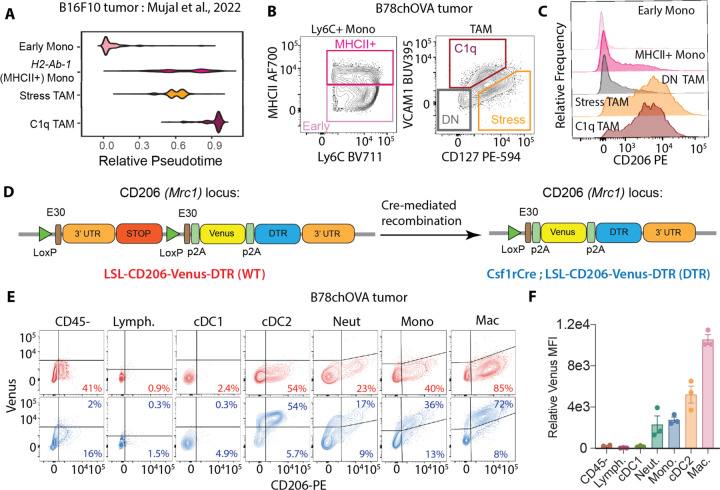
To highlight CD206 surface expression variation across Mono/Mac differentiation in tumors, we identified relevant subsets from previously published single cell transcriptomics in B16F10 tumors (Fig. 1A, (3)), and applied flow cytometry to gate on those populations in a related B78chOVA ((11); B78: an amelanotic clone of B16 to allow imaging of tumors, chOVA: mCherry and Ovalbumin) tumor model. CD206 was most prominently expressed by terminal VCAM-1hiIL-7Rαlo C1q TAMs (3) followed by the VCAM-1loIL-7Rαhi stress-responsive population (associated with enriched glycolysis, increased Arg1 and Il7r expression(3, 12) and possibly hypoxic(13)), where TAMs were defined as CD45+Lin(CD90.2, Ly6G, B220, NK1.1, Siglec-F)−Ly6C−F480+CD24− (Supplementary Fig. S1A). In contrast, CD206 was expressed at low levels by other less differentiated VCAM−IL-7Ra− (DN) TAMs (Fig. 1B, C). Among monocytes, defined as CD45+Lin(CD90.2, Ly6G, B220, NK1.1, Siglec-F)−Ly6C+, the MHCII+ subset was the prominent CD206 expressor as opposed to early (immature, MHCII-) monocytes, albeit at lower levels than Stress and C1q TAMs (Fig. 1B, C). Overall, this analysis showed that CD206 is variably expressed across multiple monocyte and macrophage subsets, and generally increases with differentiation. However, when considering the use of this protein and the gene encoding it as a means of eliminating these macrophages and thereby studying their function, we noted that this scavenger receptor is frequently also expressed on other cells including endothelial cells and keratinocytes (14, 15) and may further be ectopically produced by other cells in the TME.
Fig. 1: Genetic Myeloid-specific Labeling of CD206+ Macrophages in Tumors:
(A) Pseudotime plots of select Mono/Mac subsets in B16F10 tumors from Mujal et al(3); (B) Gating on the equivalent subsets in B78chOVA tumors by flow cytometry and (C) CD206 expression in each of these subsets; (D) Schematic representation of the Mrc1LSL-Venus-DTR knock-in construct before (WT) and after (DTR) Cre-mediated recombination by crossing to the Csf1rCre allele; (E) Flow cytometry plots showing reporter (Venus) and CD206 expression in different immune cells in d18 B78chOVA tumors in WT (red) and DTR (blue) mice with (F) quantification of relative reporter expression (DTR – WT) in the different subsets. data are mean +/− SEM, from 3 biological replicates, WT levels averaged from 2 biological replicates.
We thus generate a conditional system where a lineage-specific Cre could drive the recombination of a 3’ knock-in Mrc1LSL-Venus-DTR allele (Venus : Yellow Fluorescent Protein variant for visualization; DTR: Diphtheria toxin receptor for depletion) (Fig. 1D). Then, using a Csf1rCre; Mrc1LSL-Venus-DTR cross (DTR), compared to a Mrc1LSL-Venus-DTR (WT) control, we assessed the reporter expression in various immune and CD45- non-immune compartments in the subcutaneous melanoma model B78chOVA (Fig. 1D, Supplementary Fig. S1A). As predicted from CD206 expression, nearly 75% of TAMs showed robust Venus expression tightly correlated with surface expression of CD206 protein (Fig. 1F). We also found that nearly half of cDC2s, consistent with their monocytic origin as previously described(16), expressed Venus in this system. A small subset of CD206+ monocytes expressed the reporter, again consistent with Csf1r driven expression. Weak expression was also found in a yet smaller population of neutrophils. When viewed by the level of CD206-driven expression of the Venus marker, macrophages were 2–3x brighter than the other populations (Fig. 1F). Importantly, no reporter expression was detected in non-immune cells (which include endothelial cells and keratinocytes and where a small fraction is CD206+), lymphocytes and cDC1s (Fig. 1E, F). Likewise, in the proximal tumor-draining lymph nodes (tdLN), the same hierarchy of expression patterns was observed, albeit at much lower levels (Supplementary Fig. S1B). In addition, some tissue-resident macrophages, such as alveolar macrophages in the lung express high levels of CD206 and therefore the reporter, while interstitial macrophages, monocytes, neutrophils, and non-immune cells showed very modest to no expression (Supplementary Fig. S1C, D). Therefore, in Csf1rCre; Mrc1LSL-Venus-DTR mice, a robust marking of mature CD206+ macrophages in the subcutaneous tumor was observed, along with faithful marking of CD206+ subsets of monocytes, neutrophils and cDC2s, but not cDC1s, lymphocytes and non-immune cells.
CD206+ TAM depletion leads to indirect loss of a reactive immune archetype:
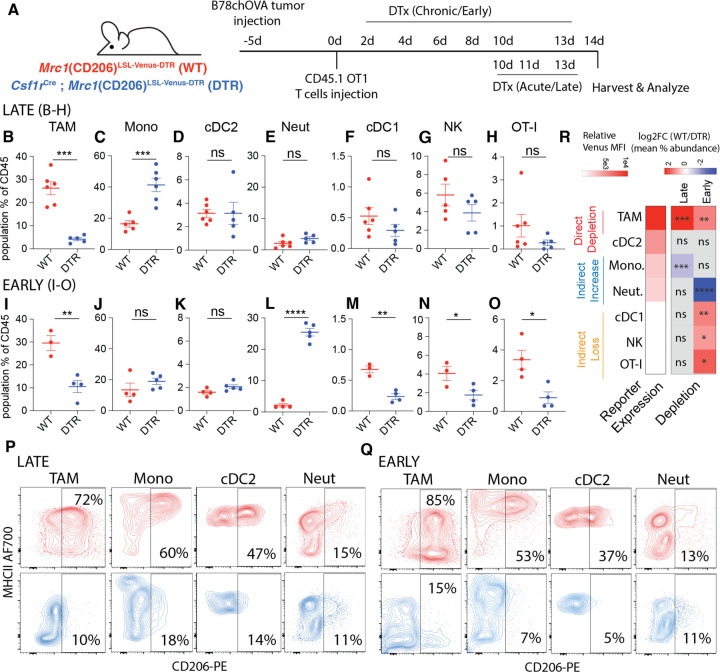
To test the impact of CD206+ macrophage targeting on the overall tumor immune microenvironment, we took advantage of the linkage of Venus and DTR expression in this background to deplete those cells. In our setup using subcutaneous B78chOVA tumors as described previously, adoptively transferred ovalbumin-specific OT-I cells allow the tracking of antigen-specific CD8 T cell responses, which nevertheless do not mediate tumor control(11, 17). We first confirmed that Cre-mediated induction of reporter expression without diphtheria toxin (DTx) administration did not alter the immune composition of these OVA-expressing tumors with OT-I transfer in the WT (Mrc1LSL-Venus-DTR) vs. DTR mice (Csf1rCre; Mrc1LSL-Venus-DTR) (Supplementary Fig. S2A). With this baseline, we administered DTx either ‘late/acute’, namely in the last 4 days prior to the tumor harvest or ‘early/chronic’, i.e., every day 2–3 days, starting 2 days post T cell injection until harvest to parse out the role of CD206+ Mono/Macs in the TME (Figure 2A). These two modes of depletion represent perturbations at different phases of the establishment of the tumor immune microenvironment.
Fig. 2: Early CD206+ TAM depletion leads to a coordinated and indirect loss of NK, cDC1 and CD8 T cells in the TME:
(A) Schematic representation of the experimental setup for early and late CD206+ TAM depletion in B78chOVA tumors using Mrc1(CD206)LSL-Venus-DTR (WT) and Csf1rCre; CD206LSL-Venus-DTR (DTR) mice; Relative abundance of different immune populations as a percentage of CD45+ cells with (B-H) late and (I-O) early depletion regimens; Representative flow cytometry plots showing CD206 vs. MHCII expression in different myeloid subsets in WT (red) and DTR (blue) mice in the (P) late and (Q) early depletion regimens. (R) heatmap representation of the log fold change of the ratio of mean abundances in WT and DTR mice (data from B-O), alongside the extent of reporter expression (mean relative Venus MFI from Fig. 1F) to indicate direct depletion and indirect loss or enrichment. (statistical significance is indicated on the respective squares; ***p <0.001, **p<0.01, *p <0.05, ns = no significance by Student’s t-tests).
In the context of late DTx administration, we found that this regimen specifically depleted the cells of interest and otherwise had little overall effect on other non-targeted cells. Thus, there was a strong reduction in the TAMs (Figure 2B) which corresponded to a specific loss of the CD206+ population (Fig. 2P). We found a compensatory rise in monocytes overall, accompanied by a specific loss of its CD206+ subset (Figure 2C, 2P). No significant loss was observed in cDC2s, cDC1s, NK cells, the adoptively transferred OTI, or neutrophils (2D-H), although the regimen did select against the CD206+ populations in the case of cDC2s and neutrophils (Figure 2P). Thus, direct depletion was reliably restricted to the highest expressors of the construct, namely the CD206+ TAM and monocyte populations.
When we depleted CD206+ populations with DTx early, i.e., starting 2d after OTI adoption, again we found robust depletion of TAMs (Fig. 2I) expectedly with a strong selection against those expressing CD206 (Fig. 2Q). As before, the CD206+ subsets of other populations were still depleted—robustly in monocytes and cDC2s and mildly in neutrophils (Fig. 2K, Q). A compensatory increase in neutrophils was observed, while overall abundance of monocytes, albeit biased towards CD206-, remained similar, in contrast to acute depletion. (Fig. 2J, L). Strikingly, under this early elimination regime, we also observed a decrease in intratumoral cDC1 abundance (Fig. 2M). Further, NK cells and transferred OT-I abundance in the tumor was also significantly compromised (Fig. 2N, O).
These trends in abundances were similar when expressed as percentage of live cells, indicating numerical changes, in all cases except the increase in monocytes following acute depletion (Supplementary Fig. S2B–O), which only trended higher. When we similarly treated non-tumor bearing DTR mice with DTx with six doses akin to the early depletion regimen in tumors, and analyzed the immune compositions in the skin (site of the ectopic tumor injections)(Supplementary Fig. S2P), no robust indirect loss of populations were observed, but an increase of neutrophils in an otherwise scarcely immune-populated skin was recorded (Supplementary Fig. S2Q). Given the associated increase in neutrophils, we repeated the same early depletion experiment in tumors, now with the addition of anti-Ly6G neutrophil depleting antibody or isotype control (Supplementary Fig. S2R) to assess whether the gained neutrophils played a role in the indirect loss of lymphocytes and cDC1s. As expected, both in terms of the total number of cells per gram of tumor (Supplementary Fig. S2S) and the percentage of CD45+ (immune) cells (Supplementary Fig. S2T), the abundance of immune cell types in WT and DTR mice treated with isotype control mirrored those of the early depletion regimen. With anti-Ly6G treatment in DTR mice, neutrophils were reduced to levels below those of untreated controls, without any concomitant effect on the indirect depletion of cDC1s, NK cells and OT-I T cells (Supplementary Fig. S2S, T). Noting that none of these CD8, NK and cDC1 populations express the reporter, we concluded that a direct, targeted ablation of ‘M2-like’ CD206+ Mono/Macs by early DTx treatment in tumors led to the indirect loss of this key anti-tumor reactive archetype comprising of NK cells, cDC1s and antigen specific CD8 T cells(18), (Fig. 2R). This suggested that CD206+ Mono/Macs were involved in the recruitment and early establishment of this module in the TME.
Early depletion skews Mono/Macs towards immature and hypoxic subsets:
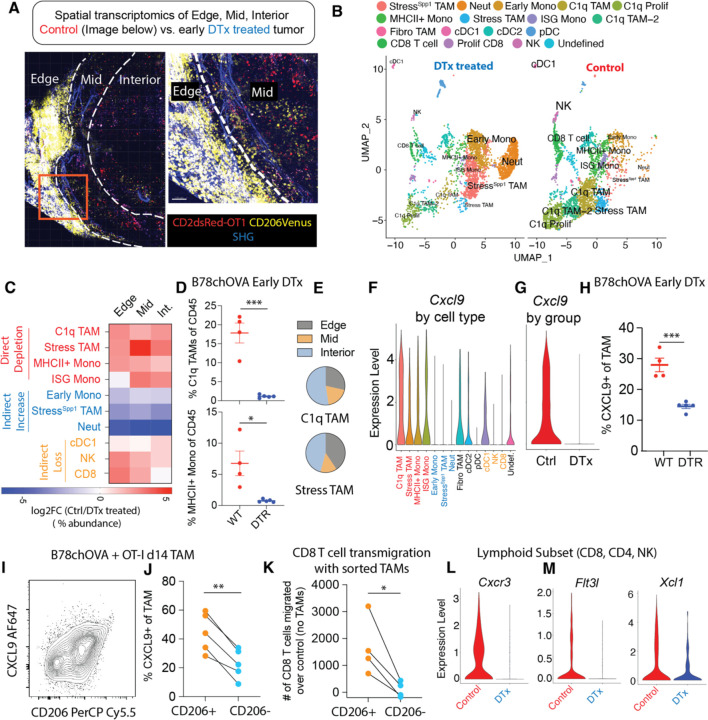
To define the macrophage subtypes associated with reactive immunity and their potential spatially segregated modes of action, we performed spatial transcriptomics of B78chOVA tumors, guided by Venus (CD206 reporter) expression. For this, we first spatially mapped the CD206+ macrophage population by Venus expression, using two-photon microscopy of B78chOVA tumor slices with transferred OT-I T cells marked by the CD2dsRed allele (Fig. 3A). Doing this revealed three distinct niches of CD206+ macrophage and T cell localization. The ‘edge’, which is macrophage and collagen-rich with modest T cell presence, ‘mid’, the interfacial layer with abundant T cell: macrophage interaction zones and ‘interior’. The interior is sparser in both immune cell types but represents the bulk of the tumor by volume (Fig. 3A). We then performed post-imaging spatial transcriptomics by ZipSeq (19) on CD45+ cells in these three zones (11) of B78chOVA tumors, with or without early DTx treatment harvested at d12 post T cell injection. UMAP projection of non-linear dimensional reduction and louvain clustering clearly showed the remarkable shift in tumor immune composition among control and DTx treated groups (Fig. 3B, C, Supplementary Fig. S3A). Notably, previously defined C1q and Stress-responsive (Stress) TAMs, which most robustly express CD206 at the protein level, along with MHCII+ and Interferon-stimulatory gene (ISG) -expressing monocytes were expectedly depleted by direct DTx action (Fig. 3C), with the robust depletion of C1q TAMs (VCAM-1hiIL-7Rαlo) and MHCII+ Monocytes verified by flow cytometry (Fig. 3D). On the other hand, early monocytes, neutrophils and a Spp1, Hif1α-expressing subset related to the Stress TAMs by shared expression of Arg1, Il7r (i.e., StressSpp1 TAM), became prominent in the DTx treated condition (Fig. 3C, Supplementary Fig. S3A). The loss of cDC1:NK:CD8 populations was again evident in the analysis of relative abundance from the scRNASeq data (Fig. 3C). Even though the small area of the tumor edge was much denser in the CD206+ Mono/Macs as shown by imaging, the transcriptomic data suggests high CD206-expressing C1q and Stress TAMs were more or equally as abundant in the interior than the edge (Fig. 3E). consistent with the trajectory of increasing Mono/Mac differentiation towards the interior of the tumor(11). Overall, in this subcutaneous tumor model, the changes in immune subpopulations were not limited to a specific region of the tumor (Fig. 3C), but permeated throughout as a holistic overhaul of the tumor immune microenvironment.
Fig. 3: Loss of CXCL9-positive TAMs and CXCR3-expressing, cDC1 supportive lymphocytes with CD206+ TAM depletion.
(A) Two-photon imaging of representative (control) B78chOVA tumors d12 post adoptive transfer of CD2dsRed; OT-I CD8 T cells showing three zones of Venus-expressing macrophage and associated CD8 T cell localization – edge, mid and interior (Int.) mapped by spatial transcriptomic barcoding ZipSeq; Boxed region is magnified (right) to show corresponding edge-mid interface SHG: Second Harmonic Generation; (B) UMAP representation of major immune cell populations obtained from Control and early DTx treated B78chOVA tumors d12 post OT-I injection aggregated across all three regions; (C) Summary heatmap showing relative log fold change of the abundance (calculated as the % of the total number of cells recovered within that region) of each major cluster in Ctrl/DTx treated conditions, split by region of tumor; Cxcl9 expression; (D) Flow cytometry data showing abundance of C1q TAMs and MHCII+ Monocytes in Ctrl and DTx treated conditions; (E) Distribution of C1q and Stress-responsive TAMs in the three spatial regions in control B78chOVA tumors; Cxcl9 expression (F) aggregated across treatment conditions by cluster and (G) aggregated across clusters by condition; (I) Representative flow cytometry plot showing intracellular CXCL9 vs. surface CD206 expression in TAMs in B78chOVA tumors at d14 post OT-I adoptive transfer and (J) the same CXCL9 expression split by CD206 positivity; (K) in vitro activated CD8 T cell migration at 3h through a 5μm transwell insert in the presence of sorted CD206+ vs. CD206- TAMs from B78chOVA tumors, normalized to migration with no TAMs; (L) Cxcr3, (M) Flt3l and Xcl1 expression in the lymphocyte subset (CD8 T cell, NK cell and CD4 T cell) by treatment group. ***p <0.001, **p<0.01, *p <0.05, ns = no significance by Mann-Whitney test (D), unpaired t-test (H) and ratio paired t-test (J, K).
CD206+ TAMs attract CXCR3-expressing, cDC1-supportive lymphocytes to the tumor:
A well-established positive functional role of TAMs is the production of CXCL9 and CXCL10, inducing CXCR3-dependent lymphocyte recruitment in tumors(20). Given the indirect loss of lymphocytes upon early removal of CD206+ Mono/Macs, we hypothesized that this axis is prominent in CD206 positive myeloid populations. Analyzing the scSeq data in detail, we found that expression of Cxcl10 (Supplementary Fig. S3C) and Cxcl9 (Fig. 3G) in particular were markedly reduced in the DTx treated tumors and this corresponded to substantial expression by the directly depleted subsets (CD206+MHCII+ Mono/Macs) and none of the indirectly increased ones (Early Mono, StressSpp1 TAM and Neutrophils) (Supplementary Fig. S3B, Fig. 3F). We confirmed this finding by flow cytometry for intracellular CXCL9 expression in TAMs from WT and DTR mice with early depletion (~50% decline, Fig. 3H, Supplementary Fig. S3D). This analysis further revealed a positive association between this chemokine and CD206 expression in B78chOVA TAMs (Fig. 3I), resulting in a significant difference (again ~50%) in CXCL9 expression in CD206+ vs. CD206- TAMs (Fig. 3J) in the WT mice. This finding was substantiated in another subcutaneously injected tumor model MC38chOVA and the spontaneous breast tumor model PyMTchOVA, both with lower overall CXCL9 positivity in the absence of OT-I adoptive transfer, but a consistent ~50% or more difference between the CD206+ and CD206- groups (Supplementary Fig. S3E). CD206+ monocytes also showed higher CXCL9 expression, compared to CD206- counterparts (Supplementary Fig. S3F), but CXCL9+ monocytes were only 1/4th as abundant as CXCL9+ TAMs in the B78chOVA TME, thus limiting their relative role in myeloid CXCL9 production (Supplementary Fig. S3G). We therefore sorted CD206+ vs. CD206- TAMs from B78chOVA tumors (d14 post tumor injection without OT-I treatment) and interrogated their relative effects on in vitro activated CD8 T cell transmigration in a 3h window. Consistent with their CXCL9 expression, the CD206+ but not the CD206- TAMs induced enhanced transmigration over no TAM-added controls. (Fig. 3K). Since CD206 TAM-depleted tumors still had small numbers of lymphocytes, we compared their levels of CXCR3 at the transcript level, which reflects receptor-ligand engagement avoiding the confounding effect of receptor internalization(21), and found that Cxcr3 expression was markedly lower in the DTx treated condition in all the lymphocyte subsets (Fig. 3L, Supplementary Fig. S3H). Taken together, these data point to the role of CD206+ TAMs in the recruitment of CXCR3-expressing lymphocytes to the TME.
Lymphocytes are well-established as key producers of cDC1-formative chemokines FLT3L (18) and XCL1(22). Therefore, given the loss of cDC1s in concert with lymphocytes with this depletion regime, we probed for these chemokines in the intratumoral lymphocytes in control vs. CD206-depleted dataset. This demonstrated that both Flt3l and Xcl1 (Fig. 3M, Supplementary Fig. S3I) transcripts were markedly reduced in the NK cells and CD8 T cells in the DTx-treated condition. These changes on a per cell basis, in addition to the overall decrease in CD8 T cells and NK cells are consistent with the loss of cDC1s in the TME, as a result of the disruption of the CD8:NK:cDC1 module (18).
Depletion of CD206+ TAMs thwarts CD8 T cell mediated anti-tumor immunity in mice:
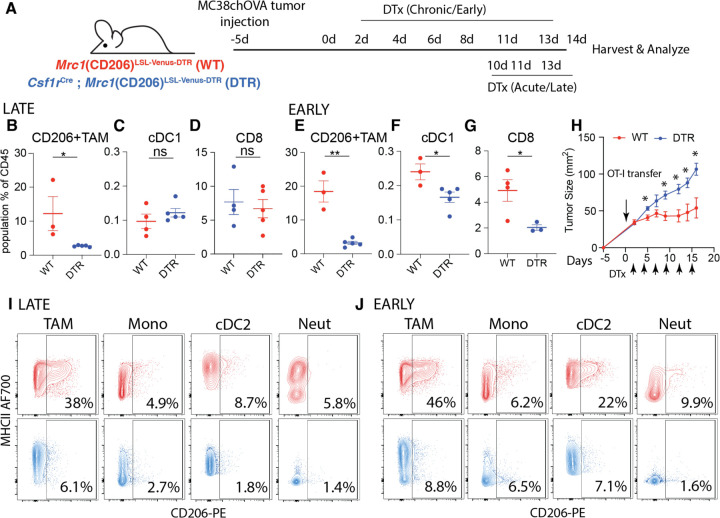
Given our finding that CD206+ ‘M2-like’ macrophages are critical to the organization a key node of antitumor immunity, we asked whether they were necessary for successful CD8 T cell mediated tumor regression. To test this, we used a MC38chOVA model where an adoptive transfer of OT-I T cells that results in efficient tumor control(17) (Supplementary Fig. S3J). We confirmed first that reporter expression in these tumors followed largely the same pattern as the B78chOVA tumors, with substantial expression only in TAMs and cDC2s, albeit at lower levels due to the overall lower CD206+ fraction, and little to no expression in neutrophils and monocytes in this model. (Supplementary Fig. S3K, L). Importantly, lymphocytes and cDC1s again showed no reporter expression (Supplementary Fig. S3K, L). As with the B78chOVA model, we applied early and late depletion regimens to the MC38chOVA tumors, first without the addition of OT-I T cells, to assess differential effects on early establishment and maintenance of immune cells without the confounding variable of tumor regression (Fig. 4A). Some differences were observed compared to the B78chOVA model, including an overall maintenance of TAM abundance in late and only a modest (~25%) decline in early depletion (Supplementary Fig. S3N, R) despite robust depletion of the CD206+ populations (Fig. 4B, E). Indeed, following reporter expression patterns, the CD206+ subsets were depleted robustly in TAMs and cDC2s and modestly in monocytes and neutrophils in both the late (Fig. 4I) and early (Fig. 4J) depletion regimens. We also noted other variations namely increased monocyte abundance in early but not late depletion (Supplementary Fig. S3O, S) and neutrophil enrichment in both regimens (albeit with much lower effect size in late depletion; Supplementary Fig. S3Q, U). As in the case of B78chOVA tumors, CD206+ cDC2s were depleted (Fig. 4I, J) but no change was detected in overall cDC2 abundance (Supplementary Fig. S3P, T) with both early and late DTx treatment. Importantly, there was once again a robust indirect loss of cDC1s and CD8 T cells specifically under the early but not the late depletion regimen (Fig. 4C, D, F, G). When evaluated further by directly measuring total number of cells per g of tumor, we observed once again the direct depletion of CD206+ TAMs, indirect increase in neutrophils and the indirect loss of cDC1s and CD8 T cells, but not NK cells (Supplementary Fig. S3U) in MC38chOVA tumors with early DTx treatment. With confirmation of this key indirect effect of CD206+ TAM depletion in the MC38chOVA model, we treated subcutaneous MC38chOVA tumors in WT and DTR mice with OT-Is and concomitant early DTx administration. With the prediction that depletion of CD206+ TAMs would thwart the tumor control ability of OT-Is, we tracked changes in tumor size and indeed observed significantly reduced OT-I-mediated tumor control of MC38chOVA tumors (Fig. 4H) in the DTR group.
Fig. 4: CD206+ TAM depletion disrupts the cDC1:CD8 module and attenuates T cell-mediated tumor control in an immune-responsive tumor model:
(A) Schematic representation of the experimental setup for early and late CD206+ TAM depletion in MC38chOVA tumors using Csf1rCre; Mrc1LSL-Venus-DTR mice; Relative abundance of (B, E) CD206+ TAMs, (C, F) cDC1s and (D, G) CD8 T cells as a percentage of CD45+ cells with late and early depletion regimens respectively; (H) tumor growth kinetics of MC38chOVA tumors in WT and DTR mice with DTx treatment beginning 2d post OT-I adoptive transfer at Day 0; Representative flow cytometry plots showing CD206 vs. MHCII expression in different myeloid subsets in WT (red) and DTR (blue) mice in the (I) late and (J) early depletion regimens; **p<0.01, *p <0.05, ns = no significance by Mann-Whitney U test t-tests.
CD206+ Mono/Mac signatures associate with anti-tumor immunity in human cancers:
The data presented thus far provided substantial evidence of a context in which CD206+ populations of Mono/Macs were in fact positive contributors to reactive anti-tumor immunity in mice, rather than being simply immunosuppressive. Consistent with this understanding, we found that higher levels of MRC1 RNA alone correlated with slightly better survival from patient data rather than worse in a large cohort curated from The Cancer Genome Atlas (TCGA) (23) (Fig. 5A). We also sought to determine whether the revealed relationships between CD206+ TAMs, CXCL9 and the cDC1:CD8:NK module in our study might similarly extended to human disease. To do so, we first applied differential gene expression (DGE) analysis of the Ctrl vs. DTx treated Mono/Mac populations (Fig. 5B, C) (excluding neutrophils, cDC1, cDC2 and lymphocyte subsets) from our scRNASeq dataset (Fig. 3B). We then used the top 10 DEGs (by average log fold change and having an adjusted p-value <0.01) to create CD206 ‘Replete’ and ‘Depleted’ gene signatures (Fig. 5D). The former are DEGs associated with the presence of CD206+ populations and not only included C1qa, Cxcl9, Apoe, but also several MHC-II related genes (Fig. 5D, E), consistent with flow cytometry data on C1q TAM, CD206, CXCL9 and MHC-II expression described above. The Depleted signature contains genes differentially expressed in macrophages that remain post CD206+ Mono/Mac depletion and included Il1b, S100a8, along with Spp1 (Fig. 5D, E). Even though we obtained these gene signatures based upon depletion of Mono/Macs using the prominent “M2” marker CD206, both M1 and M2-associated genes were differentially upregulated in Replete signature (Fig. 5D), reiterating the lack of coordination among such markers when studied in vivo.
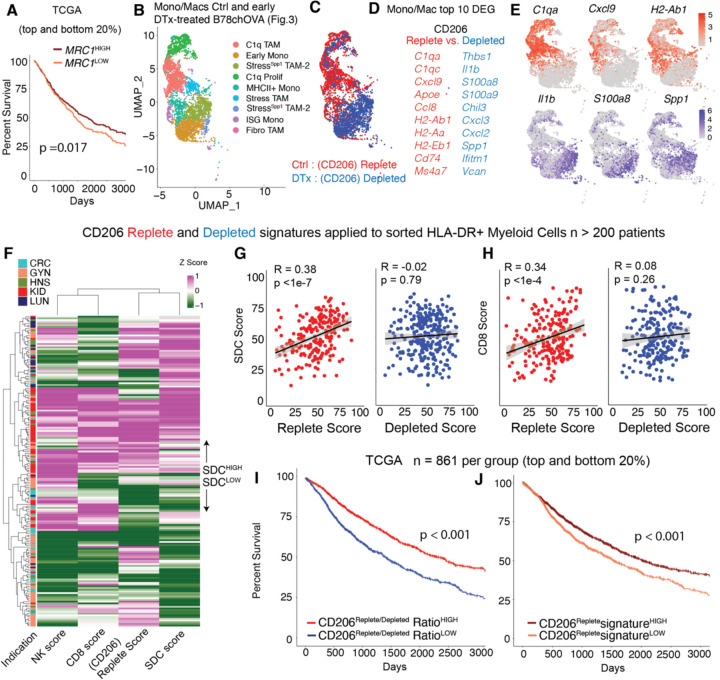
Fig. 5: CD206Replete Mono/Mac signature associates with anti-tumor immunity in human cancers:
(A) Kaplan-Meier survival curves of patients in TCGA grouped by the expression of MRC1 gene; (B) UMAP representation of the Mono/Mac subsets and (C) overlay of the CD206 Replete (Ctrl) and Depleted (DTx) groups on the UMAP from spatial scSeq described in Fig. 3; (D) Top 10 genes from Differential Gene Expression (DGE) of Mono/Macs in the Ctrl vs. DTx treated conditions, which were used to generate CD206 Replete and CD206 Depleted Mono/Mac signature scores (F) Heatmap of z-scored CD206Replete, CD8, NK and Stimulatory dendritic cell (SDC) score, calculated from sorted immune compartments, as previously described (23); Scatter plots of the Myeloid-specific CD206 Replete and Depleted score per patient with the (G) stimulatory dendritic cell (SDC) score and (H) CD8 T cell score (Pearson R and p value for the null hypothesis that there is not a correlation are noted); Kaplan-Meier survival curves of patients grouped by the value of the (I) CD206Replete: CD206Depleted signature ratio and (J) CD206Replete signature in TCGA, p values for the log-rank test are noted in (A, I, J).
Using these signatures, we queried a previously described immune compartment-specific bulk RNA-Seq data derived from sorted HLA-DR+ myeloid (to capture Mono/Macs and DCs), T and total live cells from >200 human tumor biopsies(23) (Fig. 5F, G) belonging to five common solid tumor indications (CRC: Colorectal Cancer, GYN: Gynecological Cancer, HNSC: Head and Neck Squamous Cell Carcinoma, KID: Kidney Cancer; LUNG: Lung Carcinoma). Given our finding that the CD206+ CXCL9-expressing TAMs recruit CXCR3-expressing cDC1-supportive lymphocytes, we predicted that the CD206 “Replete” but not the “Depleted” signature in the myeloid compartment would associate with previously established CD8, NK and stimulatory cDC1s (stimulatory dendritic cell or SDC) gene signatures (10) (23). Indeed, the Replete signature, but not the Depleted signature correlated significantly with those of each component of the tumor-reactive immune module (Fig. 5F) of SDCs (Fig. 5G), CD8 T cells (Fig. 5H) and NK cells (Supplementary Fig. 3W). Given that it is by now well-established that SDCs are associated with survival (10), we also queried whether the relative abundance of CD206 Replete Mono/Macs (i.e., CD206Replete/CD206Depleted ratio) was correlated with better survival in patients. In the TCGA dataset, we observed a large (~20% in 5-year survival) and significant shift in survival for patients biased towards the CD206Replete Mono/Mac signature (Fig. 5I). Indeed, patients scoring high on the CD206Replete Mono/Mac signature alone were also found to have significantly better survival but with a reduced effect size (Fig. 5J) as compared to the ratio. Among specific indications, the Replete/Depleted signature ratio was associated with overall survival in Lung, Liver, Pancreatic, Bladder, Kidney Cancer and Melanoma (Supplementary Fig. S3X). Thus, contrary to the simplistic labeling of CD206 expressing macrophages as immunosuppressive, this data establishes contexts in which these Mono/Macs are a critical organizing fulcrum for the reactive archetype of NK cells, cDC1s and CD8 T cells. Taken together, these data contribute to a nuanced understanding of the context-dependent role of TAMs in the TME, necessary to rationally design next-generation myeloid-targeting immunotherapies in cancer.
Discussion:
cDC1s have been previously linked to FLT3L and XCL1 producing NK cells and activated CD8 T cells(18, 22), and this network represents one module of immunity that predisposes to immune checkpoint blockade response (reviewed in(24)). The same CD8 T cells in turn may be recruited and expanded by chemokines and antigen-presentation by cDC1s, creating a virtuous feedback loop for anti-tumor immunity. It was however, previously unexplored how specific macrophage subsets support or thwart this anti-tumor archetype. Here, we demonstrate that CD206 expression in macrophages is robustly correlated with their expression of CXCL9 and these macrophages play a critical role in initiating the assembly of the cDC1:NK-CD8 anti-tumor reactive immune archetype in tumors.
This work is the latest in a series of publications (3, 5, 12). that force re-evaluation of the prevalent but insufficient M1/M2 classification of macrophages in tumors. Notably, CD206 expression is still often used to categorize macrophages as immunosuppressive and ‘M2-like’, even though strong in vivo data supporting this assertion is lacking. Here, we show that CD206 should not be used as an unqualified indicator of immunosuppressive function. Indeed in the context of ongoing anti-tumor responses studied here in the early depletion setting, these TAMs are crucial for effective recruitment of critical immune cells.
When thinking about these revealed functions of TAMs expressing CD206, we also note that this study specifically found them critical in early T cell recruitment. However, our work and others have shown that some mature TAMs which include those that express CD206 may also be involved in coupling with CD8 T cells and promote T cell exhaustion (11, 25). Thus, TAMs may have distinct phenotypes and functions depending on the immunological state of the tumor – perhaps reflected in the early and late depletion conditions shown here. Future studies to understand this balance of pro and anti-tumor effects of TAMs is critical. At present, one should not simplistically take our study to indicate that CD206+ macrophages are universally favorable for anti-tumor immunity. However, the M1/M2 dichotomy—and particularly a version that equates CD206 with pro-tumoral functions—appears to be a misleading lens through which to view macrophage functional heterogeneity.
Other recent data using CXCL9 versus SPP1 gene expression to functionally classify macrophages in human tumors (12) as anti- or pro-tumor respectively, are aligned with our findings. In our earlier studies of SPP1 in macrophages (3), these were observed in human tumors, likely embedded within an Arg1 (Stress) TAM subset in mice, and here they only emerged as a distinct cluster due to their disproportionate enrichment post depletion. As we have previously noted(3), these non-CD206 expressing ‘Stress’ macrophages are distinctly glycolytic, express Hif1α, and are likely the cells that have previously been defined as hypoxic macrophages (26, 27) and now associated with poor patient outcomes.
Studies prior to ours and using more universal Mono/Mac depletion have also variably reported compensatory neutrophil influx when depleting cells of monocytic origin in tumors (28–30). Our data also shows an increase in neutrophils in the early CD206-gated depletion condition, but a lack of similar influx in the late depletion regimen. One interpretation of our data is that a microenvironment-dependent opportunistic filling of the early myeloid niche by neutrophils takes place in the absence of sufficient Mono/Macs and the reactive immune components. While further studies may uncover key nodes of this balance of myeloid populations, our results show that the compensatory neutrophils do not contribute to the reduction of CXCR3-dependent lymphocyte recruitment (20), which is the primary driver leading to the loss of the key tumor-reactive archetype.
One key success of our study is the ability to differentially target subsets of TAMs within the TME. Prior to our work, many questions regarding the specific role of TAMs have remained obscured or unanswered partly owing to the lack of sufficiently specific and penetrant tools to manipulate them in vivo. Commonly used methods, while useful lack sufficient specificity, including the depletion of all monocytes and monocyte-derived dendritic cells (CSF1R blocking antibody;(31–33)) and the depletion of all phagocytic cells and arrest of neutrophils (Clodronate; (34)). In this context, the novel conditional CD206 reporter introduced here—paired with Csf1r-Cre to avoid depleting other non-myeloid cells that express CD206—provides a more selective marking and depletion tool for CD206+ TAMs, with a further potential to target various subpopulations by altering the Cre driver alleles.
Overall, our results indicate that even this subset-dependent depletion of Mono/Mac populations may not be prudent in all contexts. To this extent, while anti-CSF1R antibodies have failed to show benefits in clinical trials (35), other strategies using for example, drugs that modulate specific subsets such as those expressing TREM2 or trigger others by engaging TREM1 may prove more surgical(36). Systematically dissecting the role of individual TAM subtypes will continue to be crucial to deciphering their context-dependent and complex roles in the TME, with a view towards harnessing them for better immunotherapy outcomes.
Supplementary Material
Acknowledgments:
Funding:
National Institutes of Health Grants: NIH R01CA197363 and NIH R37AI052116
AR was supported by a Cancer Research Institute Postdoctoral Fellowship (CRI2940)
KHH was supported by an American Cancer Society and Jean Perkins Foundation Postdoctoral Fellowship
NFK was supported by the Cancer Research Institute / Merck Postdoctoral Fellowship (CRI4546)
We thank members of the Krummel lab for their inputs to the manuscript.
Funding Statement
National Institutes of Health Grants: NIH R01CA197363 and NIH R37AI052116
AR was supported by a Cancer Research Institute Postdoctoral Fellowship (CRI2940)
KHH was supported by an American Cancer Society and Jean Perkins Foundation Postdoctoral Fellowship
NFK was supported by the Cancer Research Institute / Merck Postdoctoral Fellowship (CRI4546)
We thank members of the Krummel lab for their inputs to the manuscript.
Footnotes
Declaration of Interests:
The authors declare no competing interests
Data and materials availability:
Relevant data will be made publicly available before publication in its final form. Meanwhile, data will be available upon reasonable request, please contact the authors directly.
References:
- 1.Binnewies M. et al. , Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goswami S., Anandhan S., Raychaudhuri D., Sharma P., Myeloid cell-targeted therapies for solid tumours. Nat Rev Immunol 23, 106–120 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Mujal A. M. et al. , Holistic Characterization of Tumor Monocyte-to-Macrophage Differentiation Integrates Distinct Immune Phenotypes in Kidney Cancer. Cancer immunology research 10, 403–419 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng S. et al. , A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809 e723 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Azizi E. et al. , Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu K. H. et al. , Transcriptional space-time mapping identifies concerted immune and stromal cell patterns and gene programs in wound healing and cancer. Cell Stem Cell 30, 885–903 e810 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doedens A. L. et al. , Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res 70, 7465–7475 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruffell B. et al. , Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 26, 623–637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peranzoni E. et al. , Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Natl Acad Sci U S A 115, E4041–E4050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broz M. L. et al. , Dissecting the Tumor Myeloid Compartment Reveals Rare Activating Antigen-Presenting Cells Critical for T Cell Immunity. Cancer Cell 26, 938 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Kersten K. et al. , Spatiotemporal co-dependency between macrophages and exhausted CD8+ T cells in cancer. Cancer Cell 40, 624–638.e629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bill R. et al. , CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science 381, 515–524 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei J. et al. , Characterizing Intercellular Communication of Pan-Cancer Reveals SPP1+ Tumor-Associated Macrophage Expanded in Hypoxia and Promoting Cancer Malignancy Through Single-Cell RNA-Seq Data. Front Cell Dev Biol 9, 749210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi J. T. et al. , Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A 100, 10623–10628 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szolnoky G. et al. , A mannose-binding receptor is expressed on human keratinocytes and mediates killing of Candida albicans. J Invest Dermatol 117, 205–213 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Liu Z. et al. , Dendritic cell type 3 arises from Ly6C(+) monocyte-dendritic cell progenitors. Immunity 56, 1761–1777 e1766 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Ray A. et al. , Multimodal identification of rare potent effector CD8 T cells in solid tumors. Biorxiv, (2023). [Google Scholar]
- 18.Barry K. C. et al. , A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med 24, 1178–1191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu K. H. et al. , ZipSeq: barcoding for real-time mapping of single cell transcriptomes. Nat Methods 17, 833–843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.House I. G. et al. , Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin Cancer Res 26, 487–504 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Meiser A. et al. , The chemokine receptor CXCR3 is degraded following internalization and is replenished at the cell surface by de novo synthesis of receptor. J Immunol 180, 6713–6724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bottcher J. P. et al. , NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037 e1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Combes A. J. et al. , Discovering dominant tumor immune archetypes in a pan-cancer census. Cell 185, 184–203.e119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Im K., Combes A. J., Spitzer M. H., Satpathy A. T., Krummel M. F., Archetypes of checkpoint-responsive immunity. Trends Immunol 42, 960–974 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nixon B. G. et al. , Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity 55, 2044–2058 e2045 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White J. R. et al. , Genetic amplification of the transcriptional response to hypoxia as a novel means of identifying regulators of angiogenesis. Genomics 83, 1–8 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Bosco M. C. et al. , Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol 177, 1941–1955 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kumar V. et al. , Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell 32, 654–668 e655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien S. A. et al. , Activity of tumor-associated macrophage depletion by CSF1R blockade is highly dependent on the tumor model and timing of treatment. Cancer Immunol Immunother 70, 2401–2410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ries C. H. et al. , Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25, 846–859 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Naik S. et al. , Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greter M. et al. , GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity 36, 1031–1046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swierczak A. et al. , The promotion of breast cancer metastasis caused by inhibition of CSF-1R/CSF-1 signaling is blocked by targeting the G-CSF receptor. Cancer Immunol Res 2, 765–776 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Culemann S. et al. , Stunning of neutrophils accounts for the anti-inflammatory effects of clodronate liposomes. J Exp Med 220, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Roca C. A. et al. , Phase I study of emactuzumab single agent or in combination with paclitaxel in patients with advanced/metastatic solid tumors reveals depletion of immunosuppressive M2-like macrophages. Ann Oncol 30, 1381–1392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juric V. et al. , TREM1 activation of myeloid cells promotes antitumor immunity. Sci Transl Med 15, eadd9990 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Ruhland M. K. et al. , Visualizing Synaptic Transfer of Tumor Antigens among Dendritic Cells. Cancer Cell 37, 786–799.e785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engelhardt J. J. et al. , Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell 21, 402–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korsunsky I. et al. , Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A. et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B., Dewey C. N., RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant data will be made publicly available before publication in its final form. Meanwhile, data will be available upon reasonable request, please contact the authors directly.