Summary
Cilia-derived extracellular vesicles (EVs) contain signaling proteins and act in intercellular communication. Polycystin-2 (PKD-2), a transient receptor potential channel, is a conserved ciliary EVs cargo. Caenorhabditis elegans serves as a model for studying ciliary EV biogenesis and function. C. elegans males release EVs in a mechanically-induced manner and deposit PKD-2-labeled EVs onto the hermaphrodite vulva during mating, suggesting an active release process. Here, we study the dynamics of ciliary EV release using time-lapse imaging and find that cilia can sustain the release of PKD-2-labeled EVs for a two-hour duration. Intriguingly, this extended release doesn’t require neuronal synaptic transmission. Instead, ciliary intrinsic mechanisms regulate PKD-2 ciliary membrane replenishment and dynamic EV release. The ciliary kinesin-3 motor KLP-6 is necessary for both initial and extended ciliary EV release, while the transition zone protein NPHP-4 is required only for sustained EV release. The dihydroceramide desaturase DEGS1/2 ortholog TTM-5 is highly expressed in the EV-releasing sensory neurons, localizes to cilia, and is required for sustained but not initial ciliary EV release, implicating ceramide in ciliary ectocytosis. The study offers a comprehensive portrait of real-time ciliary EV release, and mechanisms supporting cilia as proficient EV release platforms.
Keywords: C. elegans, cilia, extracellular vesicles, polycystin, kinesin-3, KLP-6, NPHP-4, PKD-2, DEGS1/2, ceramide
Results and Discussion
Extracellular vesicles (EVs) are submicron membranous structures and key mediators of intercellular communication, shuttling bioactive cargo and signaling molecules to recipient cells1,2. Recent research has highlighted roles for cilia-derived EVs in signal transduction, underscoring their importance as bioactive extracellular organelles containing conserved ciliary signaling proteins3,4 . Members of the TRP channel Polycystin-2 (PKD-2) family are found in ciliary EVs of the green algae Chlamydomonas and the nematode Caenorhabditis elegans5,6 and in EVs in the mouse embryonic node and isolated from human urine7,8.
In C. elegans, PKD-2 is exclusively expressed in male-specific EV releasing sensory neurons, which extend their ciliary tips to ciliary pore and directly release EVs into the environment6,9. Males release EVs in a mechanically-stimulated manner, regulate EV cargo content in response to mating partners, and deposit PKD-2::GFP-labeled EVs on the vulval cuticle of hermaphrodites during mating9,10. Combined, our findings suggest that ciliary EV release is a dynamic process. Herein, we identify mechanisms controlling dynamic EV shedding.
We discovered that cilia are capable of releasing PKD-2::GFP-labeled EVs for a duration of up to two hours. Surprisingly, synaptic transmission is not required for the extended PKD-2 ciliary EV release from sensory neurons. Rather, the ciliary transport machinery involving the kinesin-3 motor KLP-6 and the transition zone protein NPHP-4 are necessary for the replenishment of PKD-2 in the ciliary membrane and this prolonged release of EVs. These findings emphasize the critical role of intrinsic ciliary mechanisms in governing dynamic EV release. We also show that the dihydroceramide desaturase DEGS1/2 ortholog TTM-5 is required for sustained release of PKD-2 ciliary EVs, highlighting the role of ceramide in ciliary ectocytosis. In summary, we visualize real-time ciliary EV release and identify basic principles that govern its dynamic regulation. This work presents a glimpse into how ciliated sensory neurons release EVs in real time in a living animal.
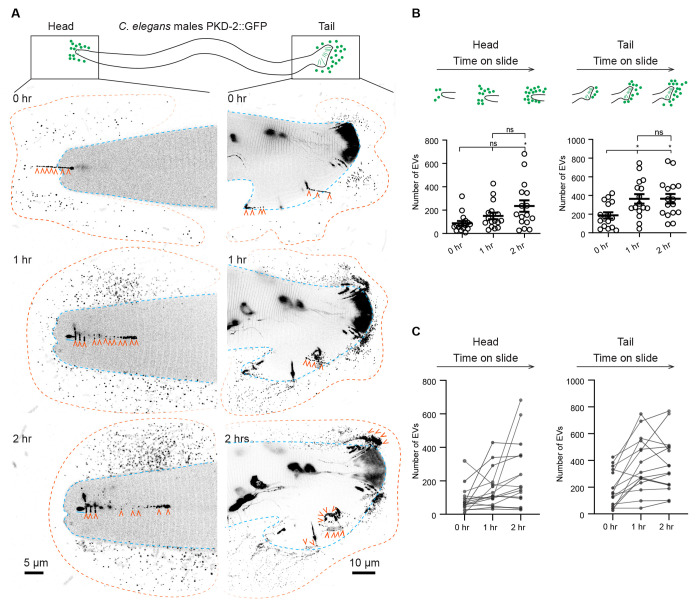
Extended release of PKD-2::GFP labeled ciliary EVs for two hours
PKD-2-laden ciliary EVs are generated from both the ciliary base and tip, with the latter being directly released into the surrounding environment and mediating inter-animal communication6,9,11. C. elegans males release PKD-2::GFP-labeled ciliary EVs in response to contact with a coverslip while the animals are mounted on glass slides for microscopic imaging10. To gain insight into the dynamics of ciliary EV release, we conducted a series of time-lapse imaging experiments. We found that C. elegans males consistently released PKD-2::GFP-labeled ciliary EVs over a span of two hours (Figure 1). We observed environmental EV accumulation at release sites at the head and tail, attributed to the immobilization of animals through anesthesia (Figure 1A). In the head region, EV counts gradually increased, with statistical significance achieved at the two-hour time point (Figure 1B). In the tail region, we observed a more consistent increase in EV numbers, culminating in statistically significant maximum at the one-hour time point (Figure 1B). Further analysis of EV number trajectories of individual animals revealed that, although both the head and tail displayed extended EV release patterns, the tail exhibited a more consistent EV number increase (Figure 1C). We therefore used the more consistent male tail assay for subsequent analysis of EV dynamic release.
Figure 1. Extended release of PKD-2::GFP-labeled extracellular vesicles (EVs) by C. elegans male sensory neurons.
(A) Representative images capturing the head and tail regions of C. elegans males at 0, 1, and 2 hours following mounting on a glass slide.
Blue lines indicate the outline of the male head or tail. Orange lines indicate the outline of the EV clouds released by the head and the tail. Orange arrowheads point to EVs that are released in a string-like pattern.
(B) Quantification of EV counts from the head and tail regions at 0, 1, and 2 hours. The scatter plot with lines indicates the mean ± SEM. Each data point represents the total EV count released by an individual C. elegans male, either from its head (left) or tail (right). n = 16 and the data were scored over 4 days. Statistical analysis was performed using one-way ANOVA with Bonferroni correction. ns denotes not significant (p ≥ 0.05), and * denotes p < 0.05.
(C) Individual trajectories depicting the EV release pattern from the head and tail regions of C. elegans males. Each trajectory represents EV counts from a single animal. The EV count at new time points includes newly released EVs in addition to previously released EVs that remain visible post-photobleaching. Photobleaching explains the decline in EV counts among certain animals at the 2-hour mark, where newly released EVs are fewer than the photobleached older EVs (photobleached twice).
To further characterize the dynamics of EV release during the first hour, we imaged EV release every 10 minutes (Figure S1, Video S1). The number of EVs consistently increased over the course of an hour and reached statistical significance at the 30-minute mark (Figure S1A). Analysis of individual EV trajectories in animals revealed that 5 out of 8 males continuously released EVs over the course of an hour, whereas in 3 out of 8 animals, EV numbers plateaued at the 30-minute mark (Figure S1B). This observation may explain the variability in EV release during the two-hour assay. To visualize real-time EV release, we performed time-lapse imaging for one minute (Video S2, 39 seconds per frame). As the cilium was deflected and moved along the coverslip, the ciliary tip released EVs that appeared as strings and that formed clouds (Figure 1A, Video S2). Our time-lapse imaging confirms that these continuously released EVs originate from the cilia tips. Collectively, these findings demonstrate the capacity of cilia to release EVs for up to two hours from sensory neurons within living animals.
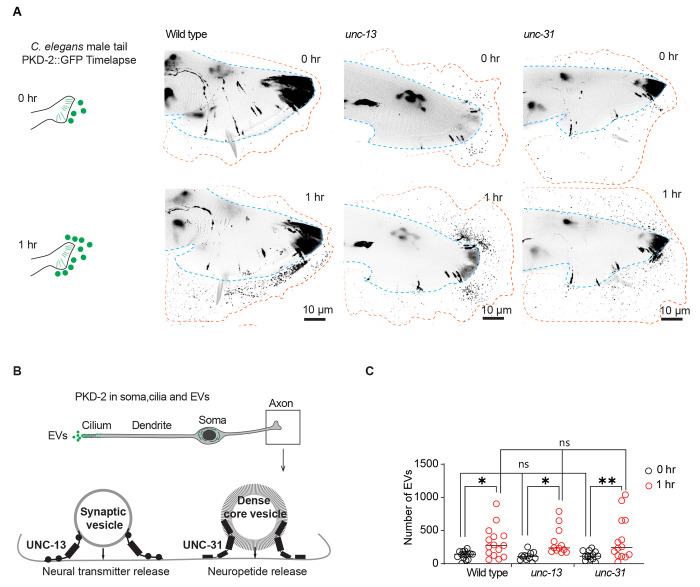
Synaptic transmission is not required for dynamic PKD-2 ciliary EV release from sensory neurons
Sensory neurons release EVs from cilia when mechanically stimulated and during mating10. Therefore, we asked whether synaptic transmission and communication from other neurons was necessary for the dynamic PKD-2 ciliary EV release. We examined mutants defective in docking synaptic vesicles (unc-13) and in dense core vesicle exocytosis (unc-31)12–14 (Figure 2). Neither unc-13 nor unc-31 mutant males displayed deficiencies in PKD-2 ciliary EV release (see Figure 2A–C). At the initial time point (0 hours), both unc-13 and unc-31 mutants exhibited PKD-2 EV release at wild-type levels. The cumulative EV release over one hour was comparable between unc-13, unc-31, and wild-type males (Figure 2). This data indicates that synaptic vesicle and dense core vesicle exocytosis are not required for PKD-2 initial or extended ciliary EV release.
Figure 2. Neuronal transmission-independent release of PKD-2::GFP-labeled ciliary EVs.
(A) Representative images showing EV release from the tail at the initial imaging (0 hr) and the second imaging (1 hr) while the animals were mounted on slides, in wild-type, unc-13, and unc-31 males.
(B) Schematic diagram depicts the functional roles of UNC-13 and UNC-31 in synaptic vesicle- and dense core vesicle-mediated neuronal transmission in the axon. (C) Quantification of EV release from the tail at the initial imaging (0 hr) and second imaging (1 hr) in wild-type, unc-13, and unc-31 males. The scatter plot with lines indicates the mean ± SEM. Each data point represents the total EV count released by an individual C. elegans male. 12-15 animals were imaged for each genotype. Statistical analysis was performed by two-way ANOVA with Bonferroni correction. ns denotes not significant, * denotes p < 0.05, and ** denotes p < 0.01.
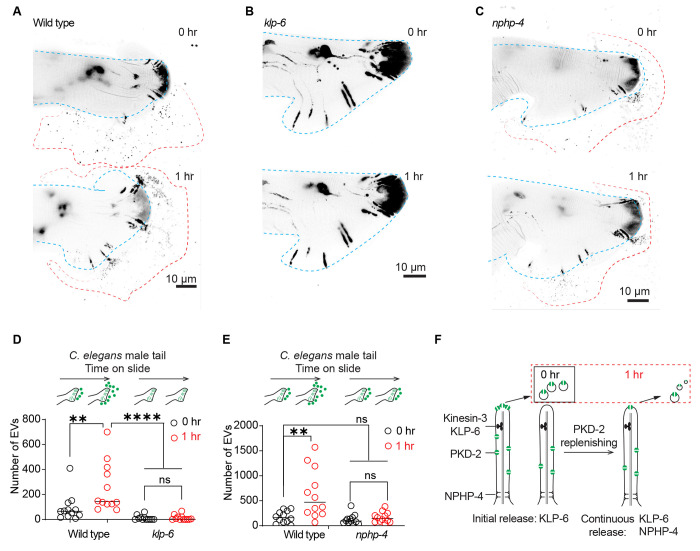
Continuous EV release relies on the membrane replenishment of PKD-2 at the ciliary tip
The ciliary kinesin-3 motor KLP-6 (kinesin like protein 6) is essential for PKD-2 environmental EV release at the ciliary tip. In klp-6 mutant animals, PKD-2 is not released in environmental EVs, leading to excessive shedding of EVs at the ciliary base, into the glial lumen surrounding the ciliary base6 (Figure S2A–D). To explore the potential contribution of the ciliary base EV reservoir to the sustained release of PKD-2 EVs, we conducted a time-lapse analysis of PKD-2 EV release in klp-6 mutant males (Figure 3). klp-6 mutant animals exhibited a defect in environmental EV release at both the initial and the one-hour time point (Figure 3A, B, D). These data show that ciliary base EVs are not released into the environment during the prolonged one-hour time-lapse assay, and that KLP-6 kinesin motor-facilitated enrichment of EV cargos at the ciliary tip is necessary for the dynamic release of PKD-2 ciliary EVs.
Figure 3. Continued PKD-2 ciliary EV release relies on effective replenishment of PKD-2 at the ciliary tip.
(A-C) Representative images of PKD-2::GFP ciliary EV release in male tail of wild-type, klp-6, and nphp-4 males.
(D) Quantification of EV counts from wild-type and klp-6 mutant males in the tail regions at 0 and 1 hour. The scatter plot with lines indicates the mean ± SEM. Each data point represents the total EV count released by an individual C. elegans male.
(E) Quantification of EV counts from wild-type and nphp-4 mutant males in the tail regions at 0 and 1 hour. The scatter plot with lines indicates the mean ± SEM. Each data point represents the total EV count released by an individual C. elegans male.
For D-E, statistical analysis was performed by two-way ANOVA with Bonferroni correction. n = 12-15 animals per genotype. ns denotes not significant, * denotes p < 0.05, and ** denotes p < 0.01., **** p<0.0001.
(F) Model of PKD-2 ciliary EV dynamics. NPHP-4 regulates the release of PKD-2::GFP ciliary EVs by replenishing PKD-2::GFP in the ciliary membrane. Meanwhile, KLP-6 controls ciliary PKD-2 EV release by concentrating PKD-2 at the ciliary tip. The sustained release of PKD-2 ciliary EVs over an hour necessitates the replenishment of PKD-2 in both the ciliary membrane and tip.
See also Figure S2.
To test the hypothesis that continuous EV release originates from the ciliary tip, we reasoned that a mutant defective in PKD-2 ciliary membrane replenishment would also be defective in sustained EV release (model shown in Figure 3F). In cultured mammalian cells, the KLP-6 homolog KIF13B physically interacts with the transition zone protein NPHP4 (nephronophthisis gene 4) to regulate ciliary membrane composition and Sonic hedgehog signaling15. Therefore, we determined whether NPHP-4 was important for PKD-2 ciliary EV release. We previously found that the nphp-4 mutant does not grossly affect PKD-2 ciliary localization16. At the initial time point, the nphp-4 mutant did not show defects in PKD-2::GFP ciliary EV release. However, at the one-hour time point, nphp-4 mutant males failed to release additional EVs (Figure 3 C, F). Using transmission electron microscopy (TEM), we found that nphp-4 mutants displayed excessive EV accumulation surrounding the ciliary base (Figure S2E–F). The nphp-4 ciliary base EV accumulation phenotype resembles the klp-6 mutant phenotype (Figure S2), consistent with the two genes acting in a common process. The finding that the nphp-4 mutant is required for sustained PKD-2 ciliary EV release, but not for initial EV release, suggests that the prolonged PKD-2 EV release requires efficient ciliary replenishment of PKD-2. Like their mammalian homologs, NPHP-4 might interact with KLP-6 to facilitate the efficient import of ciliary membrane proteins. The transition zone plays important roles in regulating ciliary protein and lipid content17, the latter raising the question of the potential function of lipids in ciliary EV biogenesis.
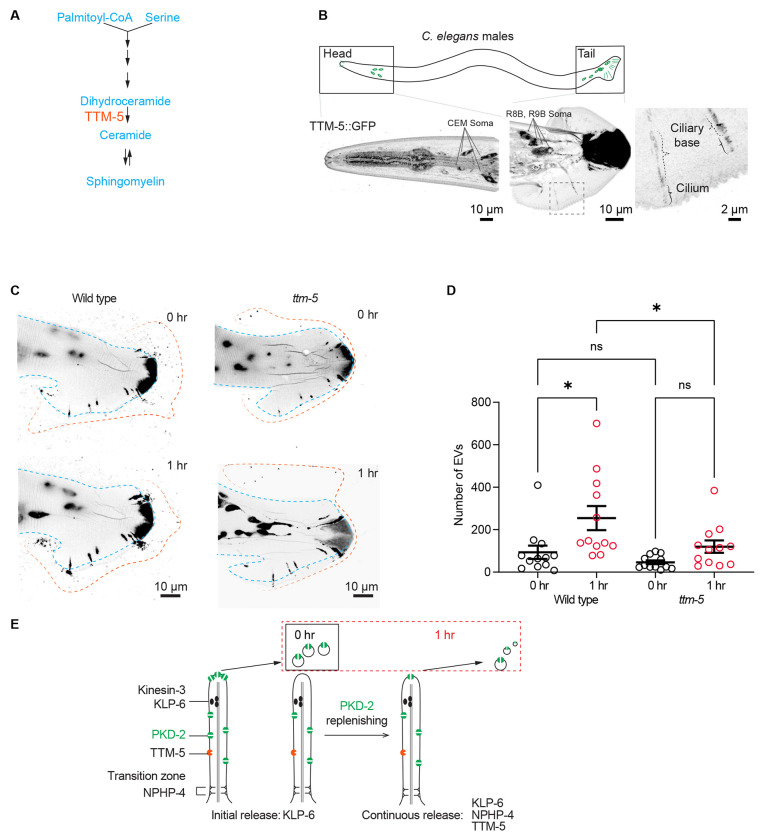
Dihydroceramide desaturase TTM-5 is required for the extended PKD-2 ciliary EV release
The sustained release of ciliary EVs requires an abundant supply of membrane lipids. As per our estimation, the membrane of approximately 60 EVs equates to the ciliary membrane (Figure S3A). Ceramide is a pivotal membrane lipid essential for EV formation2,18. Ceramide is also an important lipid for cilia formation, ciliary length and ciliary signaling19. TTM-5 (Toxin-regulated Targets of MAPK 5) is an ortholog of human DEGS1/DEGS2 (delta 4-desaturase, sphingolipid) that plays a role in de novo ceramide synthesis20–22 (Figures 4A). ttm-5 shows high expression in IL2 ciliated sensory neurons23, which are the sex-shared EV-releasing neurons (Figure S3B) and express ciliary EV biogenesis genes such as klp-6 and cil-7 24–26.
Figure 4. TTM-5 facilitates the dynamic release of PKD-2 ciliary EVs.
(A) Schematic cartoon of TTM-5 action site. TTM-5 is the ortholog of DEGS1/2 (delta-4-desaturase, sphingolipid 1/2), an enzyme that converts dihydroceramides to ceramides. The unsaturated fatty acid chain creates a kink in the chain, making the cone shape of mature ceramides to support the high curvature membrane of cilia and EV.
(B) A CRISPR reporter of TTM-5::GFP is expressed in male-specific EV-releasing neurons, including four cephalic male (CEM) neurons and 17 neurons in the tail (soma of R8B and R9B are shown, for the full tail images, see Figure S3C). In the tail EV-releasing neurons, TTM-5 primarily localizes to the soma, ciliary base, and cilium and is not detectable in EVs.
(C) Representative images of PKD-2::GFP ciliary EV release in male tail sensory neurons at 0 and 1 hour in wild-type and ttm-5 mutant males. Blue lines indicate the outline of the male tail. Orange lines indicate the outline of the EV clouds released by the tail.
(D) Quantification of EV counts at 0 and 1 hour of wild-type and ttm-5 mutant males. The scatter plot with lines indicates the mean ± SEM. Each data point represents the total EV count released by an individual C. elegans male. n = 12. Statistical analysis was performed using one-way ANOVA with Bonferroni correction. ns denotes not significant (p ≥ 0.05), and * denotes p < 0.05.
To test whether TTM-5 might function in the PKD-2 EV releasing neurons, we edited the ttm-5 genomic location using a CRISPR/Cas9 based methodology to generate an endogenous reporter by fusing ttm-5 to the GFP encoding sequence. TTM-5::GFP is primarily expressed in the EV-releasing neurons as demonstrated by co-expression of TTM-5::GFP with LOV-1::mSc27, as well as the single reporter TTM-5::GFP. In the male head, TTM-5 is expressed in IL2 and CEM neurons, along with several amphid neurons. In the male tail, TTM-5 is coexpressed with LOV-1 in HOB and RnB neurons (Figures 4B and Figure S3C). TTM-5::GFP localizes to the cilia; however, TTM-5 is not discernibly present in ciliary EVs (Figures 4B and Figure S3C). This is consistent with our prior EVome profiling data from which TTM-5 is absent28.
TTM-5 expression in the EV-releasing neurons and localization in cilia suggest a role in ciliary EV biogenesis. In the ttm-5(tm6585) loss-of-function mutant (Figure S4), the localization of PKD-2::GFP appears largely normal (Figure 4C). We next examined EV release. In the ttm-5 mutant, initial EV release was comparable to wild type. However, the ttm-5 mutant displayed an inability to release additional EVs in the one-hour assay (see Figures 4C–D). In summary, our findings indicate that the enzyme TTM-5, associated with the ceramide metabolism pathway, is enriched in the EV-releasing neurons, localizes to the ciliary membrane, and is vital for the sustained release of PKD-2 ciliary EVs.
C. elegans males release ciliary EVs in response to mechanical stimuli and in response to mating partners9,10. Here we comprehensively characterize real-time ciliary EV release and identify components that govern its dynamic regulation. Our study offers novel insights into the dynamic regulation of EV release from cilia of sensory neurons in C. elegans. The prolonged release of PKD-2::GFP-labeled ciliary EVs for up to two hours underscores the robust capability of cilia to produce EVs. This work opens new avenues for exploring the regulators of ciliary EV biogenesis, cargo sorting, and signaling.
Synaptic transmission is not required for dynamic PKD-2 ciliary EV release. This finding is consistent with a previous report showing that touch-induced calcium responses in these ray RnB neurons do not rely on synaptic transmission29. This ciliary EV shedding, independent of synaptic transmission, aligns with the finding that ciliary EVs carrying polycystin-2 are released from single-celled Chlamydomonas5. Our results are consistent with a ciliary intrinsic mechanism mediating the dynamic release of PKD-2 ciliary EVs.
Both UNC-13 and UNC-31 are conserved bridge molecules between fusing vesicles and target membranes. Neither unc-13 nor unc-31 is required for PKD-2 ciliary EV release. However, we do not rule out that unc-13 and unc-31 regulate MVB-mediated exosome biogenesis in C. elegans. The Drosophila UNC-13 homolog, stac, localizes to the multivesicular body (MVB) and is required for MVB-mediated EV release and tracheal cell fusion in embryos30. The human ortholog of UNC-13, Munc13-4, is required for MVB maturation and exosome release in cancer cells31. The UNC-31 ortholog, CAPS1, overexpression promotes exosome-regulated cancer cell migration32.
The requirement for KLP-6 in extended EV shedding is in line with previous findings indicating KLP-6’s essential role in EV shedding from the ciliary tip6,9. This observation supports the hypothesis that prolonged PKD-2 ciliary EV release originates from the ciliary tip rather than the base EVs. Within primary cilia, NPHP4 interacts with KIF13B, a KLP-6 homolog, to regulate ciliary membrane composition and Hedgehog signaling15. Intriguingly, NPHP4 also plays a role in whole cilia shedding, where cilia break off at the transition zone in Paramecium33. In Chlamydomonas, NPHP4 and other transition zone components influence the proteome of ciliary ectosomes34. We propose a cilia-intrinsic regulatory mechanism whereby NPHP-4 operates through KLP-6 to facilitate PKD-2 ciliary membrane replenishment, thus supporting effective PKD-2 ciliary EV release (Figure 4E). Furthermore, our findings highlight a role for NPHP-4 as a conserved regulator of ciliary EV biogenesis. Mutations in transition zone components like NPHP4 cause human ciliopathies35, suggesting that EV defects may contribute to ciliopathies such as nephronophthisis and polycystic kidney disease4.
TTM-5 is a dihydroceramide desaturase (DEGS1/2) ortholog, which acts in a ceramide biosynthesis pathway to convert dihydroceramide to ceramide22. Ceramide is a bioactive signaling lipid that plays roles in ciliogenesis, ciliary TGF-β receptor/sonic hedgehog signaling, and exosome biogenesis2,19. With respect to the latter, the cone-shaped structure of ceramide induces negative membrane curvature that leads to invagination into the endosome membrane and formation of inner luminal vesicles of the MVB – a prerequisite for exosome biogenesis. Here, we show that TTM-5 is required for the sustained, but not initial, release of ciliary-derived EVs. We propose that, similar to exosome biogenesis, the cone shape of ceramide induces membrane curvature and ectocytosis from the ciliary tip.
The high expression of ttm-5 specifically in EV releasing neurons and the ciliary localization of TTM-5 suggests that EV releasing neurons optimize a membrane lipid synthesis pathway for sustained ciliary EV release and constant renewal of the ciliary membrane. Vertebrate photoreceptors generate an enormous amount of light sensitive membrane daily and are capable of producing vast quantities of ectosomes36,37. The ability to form ectosome is blocked by peripherin-2 to allow outer segment formation38. Like photoreceptors, the C. elegans EV releasing neurons produce copious amounts of ciliary EVs. The DEGS1/2 TTM-5-mediated lipid synthesis pathway may be important for generating the ciliary membrane required to support sustained release of EVs. Sphingolipid metabolism enzymes play a conserved role in cilium formation in Chlamydomonas, C. elegans, and vertebrate cells, linking compromised sphingolipid production with ciliopathies39.
Our previous super-resolution imaging studies revealed that cilia release EVs from two sites – the cilia base and tip9. Here we developed a powerful one-hour imaging approach that provides spatiotemporal information on ciliary EV shedding in real-time. This extended imaging strategy revealed that different mechanisms control initial versus prolonged EV release. Not surprisingly, kinesin-3 KLP-6 is required for both initial and prolonged EV release. In contrast, the transition zone component NPHP-4 and the dihydroceramide desaturase TTM-5 are important for prolonged but not initial EV release. These results suggest that the transition zone – the ciliary gate keeper – and the sphingolipid ceramide play important roles in maintaining steady-state ciliary ectocytosis. Our C. elegans system enables discovery of conserved mechanisms regulating ciliary membrane renewal and ciliary ectocytosis, which provides much needed in vivo insight to EV biogenesis and signaling.
STAR★METHODS
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts Juan Wang (juwang07@rutgers.edu) and Maureen Barr (mmbarr@rutgers.edu).
Materials availability
Plasmids and transgenic C. elegans strains are available upon request.
Data and code availability
Microscopy data reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
C. elegans culture and genetics were performed as described42. C. elegans strains were maintained at 20°C on nematode growth medium (NGM) plates seeded with Escherichia coli (OP50 strain) as a food source.
Strain identifiers and genotypes are cataloged in the Key Resources Table.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| E. coli, OP50 strain | http://www.cgc.umn.edu | RRID: WB-STRAIN:OP50 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Levamisole | http://www.acros.com | Cat # 187870100 |
| Agarose | https://www.sigmaaldrich.com/ | Cat # A9539 |
| Experimental Models: Organisms/Strains | ||
| Strain name | Genotype | Reference |
| C. elegans Strain PT621: him-5(e1490) myIs4 [PKD-2::GFP+Punc-122::GFP]V | Bae et al (2006)40 | RRID: WB-STRAIN: PT621 |
| PT3716: unc-13(e51)I; him-5(e1490), myIs4V | this paper | RRID: WB-STRAIN: PT3716 |
| PT3038: unc-31(e169)IV; him-5(e1490), myIs4V | This paper | RRID: WB-STRAIN: PT3038 |
| PT443: myIs1[PKD2::GFP] pkd-2(sy606) IV; him-5(e1490) | Bae et al (200640) | RRID: WB-STRAIN: PT443 |
| PT1195: klp-6(my8) III; him-5 (e1490), myIs4 V | Morsci and Barr (2011)41 | RRID: WB-STRAIN: PT1195 |
| PT835: pkd-2(sy606) myIs1 IV; nphp-4(tm925) him-5(e1490) V | Jauregui et al (2008)16 | RRID: WB-STRAIN: PT835 |
| PT3502: ttm-5(6585) I; him-5 (e1490), myIs4 V | This paper | RRID: WB-STRAIN: PT3502 |
| PT3555: ttm-5(my92[TTM-5::GFP]) I; him-5(e1490)V | This paper | RRID: WB-STRAIN:PT3555 |
| PT4108: ttm-5(my92[TTM-5::GFP]) I; lov-1(my78[lov-1::mSC])II; him-5(e1490)V | This paper | RRID: WB-STRAIN: PT4108 |
| PT3629: lov-1(my78[lov-1::mSC])II; him-5(e1490)V | Walsh et al (2022)27 | RRID: WB-STRAIN: PT3629 |
| Software and Algorithms | ||
| Zen Blue 2.0 | https://www.zeiss.com | N/A |
| Zen Black 2.0 | https://www.zeiss.com | N/A |
| Imaris 9.5 | https://imaris.oxinst.com | N/A |
| Prism 9 | http://www.graphpad.com | N/A |
| Oligonucleotides | ||
| 5′- GTTTGATCTCAACAATAACAGCAAGAAGCTCAAC GGAGGTGGCGGATCTGGAGGTGGAGGCTCTGGA GGAGGTGGATCTAGTAAAGGAGAAGAACTTTTC - 3′ |
This paper | TTM-5::GFP forward homology donor Primer |
| 5′- gtttaaaagagaacatctgtatttaatccttcccaaaCTATTTG TATAGTTCATCCATG – 3′ |
This paper | TTM-5::GFP reverse homology donor Primer |
| 5′ - AGCAAGAAGCUCAACUAGUU –3′ | This paper | crRNA target sequence |
Method details
Airyscan super-resolution microscopy
Super-resolution imaging was performed on the Zeiss LSM880 confocal system equipped with Airyscan Super-Resolution Detector with 7 Single Photon Lasers (405, 458.488, 514, 561, 594, 633nm), Axio Observer 7 Motorized Inverted Microscope, Motorized X-Y Stage with Z-Piezo, T-PMT.
Time-lapse imaging and PKD-2::GFP EVs quantification
The day before imaging, 20-30 L4 larval males were picked to NGM plates freshly seeded with E. coli OP50 to attain a synchronized population. On the day of imaging, now-adult male C. elegans were picked from the NGM plates to a coverslip containing 1 μL drops of 10 mM levamisole in water and mounted on a 10% agarose pad on a slide, and imaged in 1-minute, 10-minute or one-hour intervals. All images were taken with Airyscan super-resolution on an LSM 880 confocal microscope (Zeiss). Images were saved and processed using Zen Black software (Zeiss) to generate maximum intensity projections and Zen Blue software (Zeiss) image analysis wizard to quantify the number of EVs. Images were processed and prepared for publication using Imaris, FIJI software, Adobe Illustrator and Adobe Photoshop. Maximum intensity projects of Z-stack images were inverted and adjusted to grayscale in Adobe Photoshop.
Transmission Electron Microscopy (TEM)
For method details, refer to Jauregui et al (2008)16. Briefly, worms were fixed in 3% glutaraldehyde in cacodylate buffer on ice. Then, heads were cut off, moved to fresh fixative, and held in fix overnight at 4°C. Animals were rinsed in buffer and stained with 1% osmium tetroxide in cacodylate buffer for 1 hr at 4°C. After embedding in small groups in agarose, the specimens were en bloc stained with 1% uranyl acetate in sodium acetate buffer, dehydrated, and embedded in Embed812 resin according to the general procedures described by Hall (1995)43. Thin sections were collected on a diamond knife and post-stained before being viewed on a CM10 electron microscope.
Quantification and statistical analysis
The Prism software package (GraphPad Software 8) was used to carry out statistical analyses. Information about statistical tests, p values and n numbers are provided in the respective figures and figure legends.
CRISPR/Cas9-mediatad editing
To generate the TTM-5::GFP CRISPR endogenous CRISPR tags, we followed the Mello Lab protocol and used partially single-stranded dsDNA donors44 with 35-bp of flanking homology45 that included a short flexible linker sequence between the 3′ end of ttm-5 and the start of GFP. The guide sequence was designed using CRISPOR46 [http://crispor.tefor.net/].
Supplementary Material
Video S1, Timelapse capture of dynamic EV release at 10-minute intervals per frame, related to Figure 1.
The video presents time-lapse imaging captured at 10-minute intervals over a one-hour period, demonstrating the continuous release of EVs throughout that duration. The first frame displays a cartoon of a C. elegans male; the rectangle indicates the imaging area for the micrograph shown below. The micrograph is an overlap of bright field and a fluorescence image of the male tail, demonstrating that PKD-2::GFP is enriched at the ciliary tip and is released from the ray pore structure into the environment outside the worm. The image is adapted from Figure 1 in Wang et al (2020)10.
Video S2, related to Figure 1. Timelapse capture of dynamic EV release at 39 second intervals per frame.
The video presents time-lapse imaging captured at 39-second intervals over five minutes demonstrating the continuous release of EVs from the ciliary tip. The left panel displays a cartoon of a C. elegans male; the rectangle indicates the imaging area for the micrograph shown on the right. The micrograph is an overlap of bright field and a fluorescence image of the male tail, demonstrating that PKD-2::GFP is enriched at the ciliary tip and is released from the ray pore structure into the environment outside the worm. The image is adapted from Figure 1 in Wang et al (2020)10.
Acknowledgments
This work was supported by National Institutes of Health (NIH) DK059418, DK116606 and NS120745 (M.M.B). We thank Gloria Androwski for excellent technical assistance, Barr lab mates and the Rutgers C. elegans community for feedback and constructive criticism throughout this project. We also thank WormBase, Japan National Bioresource Project for the nematode and Caenorhabditis Genetics Center (CGC) for resources information and strains. The CGC is supported by the National Institutes of Health - Office of Research Infrastructure Programs (P40OD010440).
Footnotes
Declaration of interests. The authors declare no competing interests.
References
- 1.Buzas E.I. (2023). The roles of extracellular vesicles in the immune system. Nat Rev Immunol 23, 236–250. 10.1038/s41577-022-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixson A.C., Dawson T.R., Di Vizio D., and Weaver A.M. (2023). Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat Rev Mol Cell Biol 24, 454–476. 10.1038/s41580-023-00576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ojeda Naharros I., and Nachury M.V. (2022). Shedding of ciliary vesicles at a glance. J Cell Sci 135. 10.1242/jcs.246553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., and Barr M.M. (2018). Cell-cell communication via ciliary extracellular vesicles: clues from model systems. Essays Biochem 62, 205–213. 10.1042/EBC20170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood C.R., Huang K., Diener D.R., and Rosenbaum J.L. (2013). The cilium secretes bioactive ectosomes. Curr Biol 23, 906–911. S0960-9822(13)00427-2 [pii] 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., Silva M., Haas L.A., Morsci N.S., Nguyen K.C., Hall D.H., and Barr M.M. (2014). C. elegans Ciliated Sensory Neurons Release Extracellular Vesicles that Function in Animal Communication. Curr Biol 24, 519–525. S0960-9822(14)00003-7 [pii] 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka Y., Morozumi A., and Hirokawa N. (2023). Nodal flow transfers polycystin to determine mouse left-right asymmetry. Dev Cell 58, 1447–1461 e1446. 10.1016/j.devcel.2023.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Hogan M.C., Manganelli L., Woollard J.R., Masyuk A.I., Masyuk T.V., Tammachote R., Huang B.Q., Leontovich A.A., Beito T.G., Madden B.J., et al. (2009). Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol 20, 278–288. ASN.2008060564 [pii] 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Nikonorova I.A., Silva M., Walsh J.D., Tilton P.E., Gu A., Akella J.S., and Barr M.M. (2021). Sensory cilia act as a specialized venue for regulated extracellular vesicle biogenesis and signaling. Curr Biol 31, 3943–3951 e3943. 10.1016/j.cub.2021.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Nikonorova I.A., Gu A., Sternberg P.W., and Barr M.M. (2020). Release and targeting of polycystin-2-carrying ciliary extracellular vesicles. Current Biology 30(13), R755–R756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva M., Morsci N., Nguyen K.C., Rizvi A., Rongo C., Hall D.H., and Barr M.M. (2017). Cell-Specific alpha-Tubulin Isotype Regulates Ciliary Microtubule Ultrastructure, Intraflagellar Transport, and Extracellular Vesicle Biology. Curr Biol 27, 968–980. 10.1016/j.cub.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palfreyman M.T., and Jorgensen E.M. (2017). Unc13 Aligns SNAREs and Superprimes Synaptic Vesicles. Neuron 95, 473–475. 10.1016/j.neuron.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Richmond J.E., Davis W.S., and Jorgensen E.M. (1999). UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci 2, 959–964. 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speese S., Petrie M., Schuske K., Ailion M., Ann K., Iwasaki K., Jorgensen E.M., and Martin T.F. (2007). UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27, 6150–6162. 10.1523/JNEUROSCI.1466-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schou K.B., Mogensen J.B., Morthorst S.K., Nielsen B.S., Aleliunaite A., Serra-Marques A., Fürstenberg N., Saunier S., Bizet A.A., Veland I.R., et al. (2017). KIF13B establishes a CAV1-enriched microdomain at the ciliary transition zone to promote Sonic hedgehog signalling. Nat. Commun. 8, 14177. 10.1038/ncomms14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jauregui A.R., Nguyen K.C., Hall D.H., and Barr M.M. (2008). The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J Cell Biol 180, 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derderian C., Canales G.I., and Reiter J.F. (2023). Seriously cilia: A tiny organelle illuminates evolution, disease, and intercellular communication. Dev Cell 58, 1333–1349. 10.1016/j.devcel.2023.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horbay R., Hamraghani A., Ermini L., Holcik S., Beug S.T., and Yeganeh B. (2022). Role of Ceramides and Lysosomes in Extracellular Vesicle Biogenesis, Cargo Sorting and Release. Int J Mol Sci 23. 10.3390/ijms232315317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser F., Huebecker M., and Wachten D. (2020). Sphingolipids controlling ciliary and microvillar function. FEBS Lett 594, 3652–3667. 10.1002/1873-3468.13816. [DOI] [PubMed] [Google Scholar]
- 20.Kamal M., Moshiri H., Magomedova L., Han D., Nguyen K.C.Q., Yeo M., Knox J., Bagg R., Won A.M., Szlapa K., et al. (2019). The marginal cells of the Caenorhabditis elegans pharynx scavenge cholesterol and other hydrophobic small molecules. Nat Commun 10, 3938. 10.1038/s41467-019-11908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao C.Y., Los F.C., Huffman D.L., Wachi S., Kloft N., Husmann M., Karabrahimi V., Schwartz J.L., Bellier A., Ha C., et al. (2011). Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog 7, e1001314. 10.1371/journal.ppat.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watts J.L., and Ristow M. (2017). Lipid and Carbohydrate Metabolism in Caenorhabditis elegans. Genetics 207, 413–446. 10.1534/genetics.117.300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammarlund M., Hobert O., Miller D.M. 3rd, and Sestan N. (2018). The CeNGEN Project: The Complete Gene Expression Map of an Entire Nervous System. Neuron 99, 430–433. 10.1016/j.neuron.2018.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Schwartz H.T., and Barr M.M. (2010). Functional specialization of sensory cilia by an RFX transcription factor isoform. Genetics 186, 1295–1307. genetics. 110.122879 [pii] 10.1534/genetics.110.122879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Kaletsky R., Silva M., Williams A., Haas L.A., Androwski R.J., Landis J.N., Patrick C., Rashid A., Santiago-Martinez D., et al. (2015). Cell-Specific Transcriptional Profiling of Ciliated Sensory Neurons Reveals Regulators of Behavior and Extracellular Vesicle Biogenesis. Curr Biol 25, 3232–3238. 10.1016/j.cub.2015.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire J.E., Silva M., Nguyen K.C., Hellen E., Kern A.D., Hall D.H., and Barr M.M. (2015). Myristoylated CIL-7 regulates ciliary extracellular vesicle biogenesis. Mol Biol Cell 26, 2823–2832. 10.1091/mbc.E15-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh J.D., Wang J., DeHart M., Nikonorova I.A., Srinivasan J., and Barr M.M. (2022). Tracking N- and C-termini of C. elegans polycystin-1 reveals their distinct targeting requirements and functions in cilia and extracellular vesicles. PLoS Genet 18, e1010560. 10.1371/journal.pgen.1010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikonorova I.A., Wang J., Cope A.L., Tilton P.E., Power K.M., Walsh J.D., Akella J.S., Krauchunas A.R., Shah P., and Barr M.M. (2022). Isolation, profiling, and tracking of extracellular vesicle cargo in Caenorhabditis elegans. Curr Biol. 10.1016/j.cub.2022.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Yue X., Cheng H., Zhang X., Cai Y., Zou W., Huang G., Cheng L., Ye F., and Kang L. (2018). OSM-9 and an amiloride-sensitive channel, but not PKD-2, are involved in mechanosensation in C. elegans male ray neurons. Scientific reports 8, 7192. 10.1038/s41598-018-25542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camelo C., Korte A., Jacobs T., and Luschnig S. (2022). Tracheal tube fusion in Drosophila involves release of extracellular vesicles from multivesicular bodies. J Cell Sci 135. 10.1242/jcs.259590. [DOI] [PubMed] [Google Scholar]
- 31.Messenger S.W., Woo S.S., Sun Z., and Martin T.F.J. (2018). A Ca(2+)-stimulated exosome release pathway in cancer cells is regulated by Munc13-4. J Cell Biol 217, 2877–2890. 10.1083/jcb.201710132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu B., Sun D., Ma L., Deng Y., Zhang S., Dong L., and Chen S. (2019). Exosomes isolated from CAPS1-overexpressing colorectal cancer cells promote cell migration. Oncol Rep 42, 2528–2536. 10.3892/or.2019.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gogendeau D., Lemullois M., Le Borgne P., Castelli M., Aubusson-Fleury A., Arnaiz O., Cohen J., Vesque C., Schneider-Maunoury S., Bouhouche K., et al. (2020). MKSNPHP module proteins control ciliary shedding at the transition zone. PLoS Biol 18, e3000640. 10.1371/journal.pbio.3000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Wen X., Wang Z., Lin Z., Li C., Zhou H., Yu H., Li Y., Cheng Y., Chen Y., et al. (2022). Ciliary transition zone proteins coordinate ciliary protein composition and ectosome shedding. Nat Commun 13, 3997. 10.1038/s41467-022-31751-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiter J.F., and Leroux M.R. (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer W.J., Lewis T.R., Pearring J.N., and Arshavsky V.Y. (2020). Photoreceptor Discs: Built Like Ectosomes. Trends Cell Biol 30, 904–915. 10.1016/j.tcb.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer W.J. (2023). Extracellular vesicles highlight many cases of photoreceptor degeneration. Front Mol Neurosci 16, 1182573. 10.3389/fnmol.2023.1182573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salinas R.Y., Pearring J.N., Ding J.D., Spencer W.J., Hao Y., and Arshavsky V.Y. (2017). Photoreceptor discs form through peripherin-dependent suppression of ciliary ectosome release. J Cell Biol 216, 1489–1499. 10.1083/jcb.201608081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu D., Huang J., Zhu H., Chen Z., Chai Y., Ke J., Lei K., Peng Z., Zhang R., Li X., et al. (2022). Ciliogenesis requires sphingolipid-dependent membrane and axoneme interaction. Proc Natl Acad Sci U S A 119, e2201096119. 10.1073/pnas.2201096119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae Y.-K., Qin H., Knobel K.M., Hu J., Rosenbaum J.L., and Barr M.M. (2006). General and cell-type specific mechanisms target TRPP2/PKD-2 to cilia. Development 133, 3859–3870. 10.1242/dev.02555. [DOI] [PubMed] [Google Scholar]
- 41.Morsci N.S., and Barr M.M. (2011). Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of Caenorhabditis elegans. Curr Biol 21, 1239–1244. S0960-9822(11)00700-7 [pii] 10.1016/j.cub.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall D.H. (1995). Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol 48, 395–436. [DOI] [PubMed] [Google Scholar]
- 44.Dokshin G.A., Ghanta K.S., Piscopo K.M., and Mello C.C. (2018). Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics 210, 781–787. 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paix A., Folkmann A., Goldman D.H., Kulaga H., Grzelak M.J., Rasoloson D., Paidemarry S., Green R., Reed R.R., and Seydoux G. (2017). Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. U. S. A. 114, E10745–E10754. 10.1073/pnas.1711979114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1, Timelapse capture of dynamic EV release at 10-minute intervals per frame, related to Figure 1.
The video presents time-lapse imaging captured at 10-minute intervals over a one-hour period, demonstrating the continuous release of EVs throughout that duration. The first frame displays a cartoon of a C. elegans male; the rectangle indicates the imaging area for the micrograph shown below. The micrograph is an overlap of bright field and a fluorescence image of the male tail, demonstrating that PKD-2::GFP is enriched at the ciliary tip and is released from the ray pore structure into the environment outside the worm. The image is adapted from Figure 1 in Wang et al (2020)10.
Video S2, related to Figure 1. Timelapse capture of dynamic EV release at 39 second intervals per frame.
The video presents time-lapse imaging captured at 39-second intervals over five minutes demonstrating the continuous release of EVs from the ciliary tip. The left panel displays a cartoon of a C. elegans male; the rectangle indicates the imaging area for the micrograph shown on the right. The micrograph is an overlap of bright field and a fluorescence image of the male tail, demonstrating that PKD-2::GFP is enriched at the ciliary tip and is released from the ray pore structure into the environment outside the worm. The image is adapted from Figure 1 in Wang et al (2020)10.
Data Availability Statement
Microscopy data reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.