Abstract
The incorporation of histone variants, distinct paralogs of core histones, into chromatin affects all DNA-templated processes in the cell, including the regulation of transcription. In recent years, much research has been focused on H2A.Z, an evolutionarily conserved H2A variant found in all eukaryotes. In order to investigate the functional conservation of H2A.Z histones during eukaryotic evolution we transformed h2a.z deficient plants with three human H2A.Z proteins to assess their ability to rescue the mutant defects. We discovered that human H2A.Z.1 and H2A.Z.2.1 fully complement the phenotypic abnormalities of h2a.z plants despite the fact that Arabidopsis and human H2A.Z N-terminal tail sequences are quite divergent. In contrast, the brain-specific splice variant H2A.Z.2.2 has a dominant-negative effect in wild-type plants. Furthermore, H2A.Z.1 almost completely re-establishes normal H2A.Z chromatin occupancy in h2a.z plants and restores the transcript levels of more than 84 % of misexpressed genes. Finally, our hypothesis that the N-terminal tail of Arabidopsis H2A.Z is not crucial for its developmental functions was supported by the ability of N-terminal end truncations of Arabidopsis HTA11 to largely rescue the defects of h2a.z mutants.
INTRODUCTION
Nucleosomes, the basic units of chromatin organization, are comprised of approximately 147 bp of DNA wrapped around a histone octamer, which contains the two copies of each of the four canonical histones H2A, H2B, H3, and H4. While the core histones are synthesized during the S-phase of the cell cycle for deposition onto a newly replicated DNA molecule, their counterparts, histone variants, are expressed throughout the cell cycle and replace evicted canonical histones in a replication-independent manner [1–3]. The incorporation of histone variants can have a profound effect on nucleosome dynamics and function, thereby affecting virtually all DNA-templated cellular processes, including the regulation of transcription [2–4].
H2A variants represent the largest and most diverse family of histones [2, 4, 5]. One member of this family is H2A.Z, a highly conserved variant of canonical H2A histone found in all eukaryotic organisms. Even though H2A.Z shares about 60% amino acid identity with canonical H2A [6, 7], the primary sequence differences between these two histones, particularly in the C-terminus of the proteins involved in intra- and internucleosomal interactions, led researchers to hypothesize that incorporation of H2A.Z into chromatin could alter the stability of nucleosomes and affect DNA folding [7, 8]. H2A.Z has been extensively studied, especially in recent years, to elucidate its essential role in regulating diverse cellular processes, such as DNA repair and maintenance of genome stability, prevention of heterochromatin spreading, telomere silencing, cellular differentiation, as well as positive and negative regulation of transcription in both animals and plants [9–16].
In the human genome, two genes encode H2A.Z proteins: Homo Sapiens H2A.Z.1 and H2A.Z.2.1 (HsH2A.Z.1 and HsH2A.Z.2.1). Mammalian H2A.Zs are necessary for proper development as null mutations are embryonic-lethal [17]. HsH2A.Z.1 and HsH2A.Z.2.1 differ in only three amino acids, indicating a high level of functional redundancy. Interestingly, experimental evidence also suggests distinct functional roles for each HsH2A.Z protein [4, 16, 18, 19]. Recently, an alternatively spliced form of HsH2A.Z.2.1, named HsH2A.Z.2.2, was discovered and found to be predominantly expressed in brain tissues [20, 21]. Thus, humans possess three functional H2A.Z proteins with both redundant and specific roles during development.
In Arabidopsis thaliana (At), three genes encode H2A.Z proteins: AtHTA8, AtHTA9, and AtHTA11. Arabidopsis H2A.Z proteins act redundantly since mutations in two of the most highly expressed H2A.Z genes (AtHTA9 and AtHTA11) are necessary to detect pleiotropic morphological abnormalities. H2A.Z mutant plants experience a variety of phenotypic defects, including early flowering, lack of shoot apical dominance, altered flower development and reduced fertility, serrated leaves, and inability to respond to various biotic and abiotic stresses [22–25]. However, unlike animals, Arabidopsis h2a.z mutants are viable.
Even though H2A.Zs are variants of canonical H2A histones, Arabidopsis H2A.Z proteins share more amino acid identity with human H2A.Zs than with other Arabidopsis H2A histones, indicating a high degree of H2A.Z evolutionary conservation [14]. The majority of sequence differences between Arabidopsis and human H2A.Zs are found at the N-terminal end of the proteins. In humans, many amino acids at the N-terminus, including multiple lysine residues, are known substrates for post-translational modifications (PTMs), which are shown to play an important role in H2A.Z-mediated regulation of transcription [15, 26, 27]. For instance, H2A.Z acetylation of lysine residues has been positively correlated with gene activation in many studies [26]. An intriguing question is how much functional conservation exists between human and Arabidopsis H2A.Zs, considering that the most diverse parts of the proteins are at the N-terminal tails, which, in humans, harbor functionally significant amino acid residues.
Here, we address this question by first generating a complete h2a.z knockout in Arabidopsis using CRISPR methodology, followed by the transformation of h2a.z plants with each of the three human H2A.Z genes. We show that human H2A.Z.1 and H2A.Z.2.1 phenotypically fully rescue the severe pleiotropic defects of Arabidopsis h2a.z plants, while the brain-specific splice variant H2A.Z.2.2 fails to complement the defects and in fact has a dominant negative effect in wild-type plants. At the molecular level, human H2A.Z.1 occupied nearly 100% of normal Arabidopsis H2A.Z deposition sites. Out of 8,399 mis-expressed genes in h2a.z plants, human H2A.Z.1 restored the expression of more than 84% of those genes back to wild-type (WT) levels. Taken together, these results indicate a high degree of functional conservation between Arabidopsis and human H2A.Zs. Importantly, these results also led us to hypothesize that the N-terminal tails of Arabidopsis H2A.Zs are not essential for their major developmental functions. Surprisingly, this hypothesis was supported by the ability of various N-terminal end truncations of Arabidopsis HTA11 to rescue the h2a.z phenotypic defects. Future experimental evaluation, including the full identification of post-translational modifications of H2A.Z amino acids, is necessary to further dissect the role of the N-terminal tail in H2A.Z-mediated control of gene expression.
RESULTS
Human H2A.Z.1 and H2A.Z.2.1, but not H2A.Z.2.2, completely rescue the severe phenotypic defects of h2a.z plants
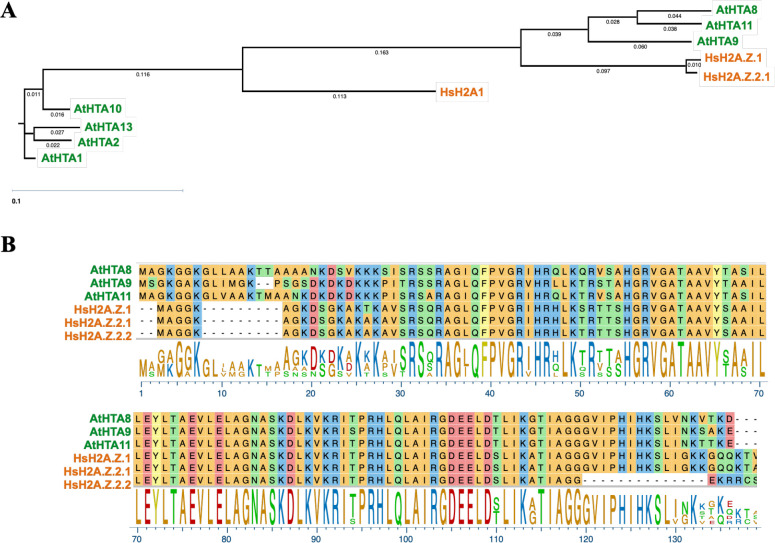
Other studies, together with our phylogenetic analysis of the H2A family of proteins from Arabidopsis and humans revealed an interesting phenomenon: amino acid sequences of Arabidopsis H2A.Z proteins, AtHTA8, AtHTA9, and AtHTA11, are more similar to human H2A.Zs than to other Arabidopsis HTA (core H2A) histones (Figure 1A, [7, 14]). In fact, Arabidopsis and human H2A.Zs share about 80% amino acid identity, with the most diverse amino acid sequences among the proteins residing in their N-terminal ends (Figure 1B). This observation suggested a high degree of evolutionary conservation between human and Arabidopsis H2A.Zs and raised the intriguing question whether human H2A.Zs can function in plants and rescue the Arabidopsis h2a.z mutant defects.
Figure 1. Arabidopsis H2A.Zs share about 80% amino acid identity with human H2A.Zs and are more similar to human H2A.Zs than to the other Arabidopsis H2A histones.
(A) Neighbor-joining phylogenetic tree of human and Arabidopsis H2A and H2A.Z proteins. (B) ClustalW alignment of three human H2A.Zs and three Arabidopsis H2A.Zs. Phylogenetic tree and sequence alignments were generated using DNASTAR.
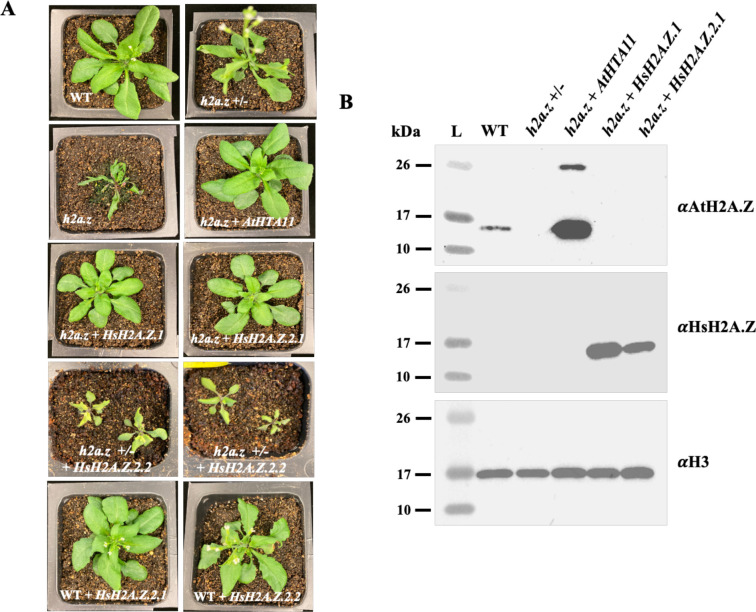
To objectively assess the ability of human H2A.Z proteins to complement the phenotypic and molecular defects caused by the lack of endogenous H2A.Zs in Arabidopsis we needed to use complete loss-of-function h2a.z plants. However, except for one example [28], all h2a.z mutant plants described in the literature are T-DNA-based double and triple insertion mutants, which are not complete h2a.z knockouts. Therefore, we first produced h2a.z null plants by employing the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) methodology. Arabidopsis plants identified as the CRISPR-generated h2a.z homozygous triple knockouts (or simply h2a.z plants) have much more severe phenotypic defects than h2a.z T-DNA mutants, including drastically delayed germination, extremely stunted growth, pointy and twisted leaves, and complete sterility (Figure 2A, Supplemental Figure 1). In fact, very few of the h2a.z plants transitioned to flowering and instead died prematurely. The few that do successfully transition to the reproductive stage of development have severely impaired flower development and are sterile (Supplemental Figure 1). We then identified the plants that are homozygous for CRISPR-generated mutant alleles of hta9 and hta11 and heterozygous for the hta8 CRISPR allele (hereafter named h2a.z +/− plants). These plants display phenotypic defects typical of h2a.z T-DNA mutants: serrated leaves, loss of apical dominance, aberrant petal number, partial fertility, and early flowering (Figure 2A, Supplemental Figures 1 and 2). Since h2a.z +/− plants are fertile, they were used for transformation with three plant codon-optimized human H2A.Z transgenes (HsH2A.Z.1, HsH2A.Z.2.1, and the HsH2A.Z.2.1 splice variant known as H2A.Z.2.2). Importantly, we also included the endogenous genomic HTA11 construct (AtHTA11) as our positive control for complementation, and a canonical H2A-encoding gene HTA2, as our negative control in rescue experiments. All constructs were driven by the native HTA11 promoter, except for HTA2, which was driven by the constitutively active 35S promoter.
Figure 2. Human H2A.Z.1 and H2A.Z.2.1 but not H2A.Z.2.2 rescue phenotypic defects of crispr-generated h2a.z null plants.
(A) 2.5 week old WT and mutant plants were grown under long-day conditions and individually photographed. (B) Western-blot analysis using 10ul of leaf chromatin extracts from wild type (WT), h2a.z +/−, and transgenic plants, were probed with either plant specific (top panel), or human-specific (middle panel) H2A.Z antibodies. H3 antibody was used as a loading control (bottom panel).
As expected, Arabidopsis HTA11 was able to fully rescue all phenotypic defects of h2a.z plants (Figure 2A). Interestingly, h2a.z plants carrying either human H2A.Z.1 or H2A.Z.2.1 transgenes were also phenotypically rescued and indistinguishable from the WT and h2a.z + AtHTA11 transgenic plants (Figure 2A, Supplemental Figure 2). On the other hand, T1 transgenic h2a.z +/− plants carrying a brain-specific HsH2A.Z.2.2 variant had more severe defects than h2a.z +/− plants alone and more closely resembled h2a.z null phenotypes (Figure 2A). Moreover, 26 out of 29 T1 HsH2A.Z.2.2 transgenic h2a.z +/− plants died before transitioning to the reproductive stage and, therefore, we were unable to analyze these plants further. Based on these results we hypothesized that the presence of human H2A.Z.2.2 in plants can somehow interfere with the normal function of endogenous Arabidopsis H2A.Zs. To test this possibility, we expressed HsH2A.Z.2.2 in WT plants and, as a control, we also expressed HsH2A.Z.2.1 in WT plants. We found that T1 plants carrying HsH2A.Z.2.2 phenotypically resembled h2a.z +/− plants, including the early flowering phenotype, while the primary transgenic plants expressing HsH2A.Z.2.1 were morphologically indistinguishable from WT plants (Figure 2A, Supplemental Figure 2). These results support the notion that, when expressed in plants, human H2A.Z.2.2 can hinder the function of endogenous Arabidopsis H2A.Zs.
On the other hand, T1 h2a.z +/− plants that expressed the canonical Arabidopsis H2A transgene, HTA2, at high levels could not rescue h2a.z +/− phenotypes (Supplemental Figure 3), suggesting that only H2A.Z proteins, and not canonical H2A proteins, can complement the h2a.z +/− and h2a.z plant morphological defects.
Overall, the results from our rescue experiments with human H2A.Zs suggest a high degree of functional conservation between Arabidopsis and human H2A.Z histones.
Human H2A.Z.1 restores WT H2A.Z occupancy in h2a.z plants
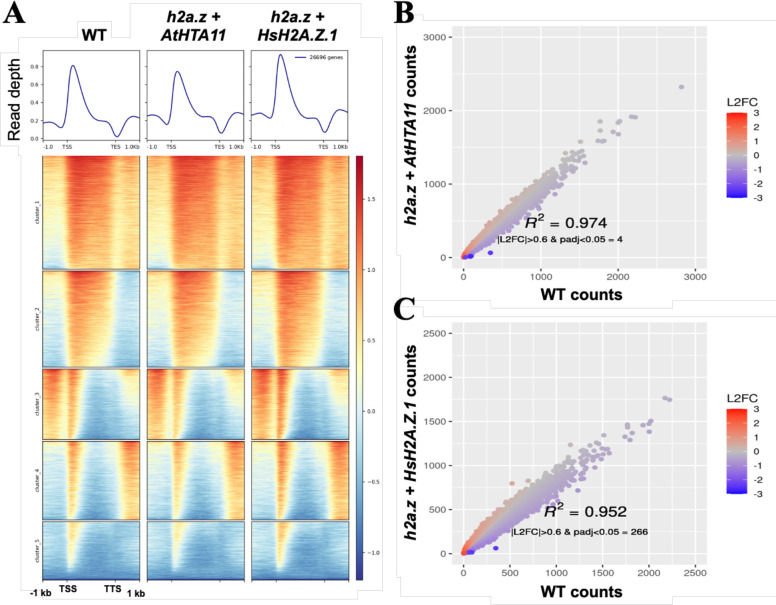
Since both HsH2A.Z.1 and HsH2A.Z.2.1 equally complemented the phenotypic defects of h2a.z plants, we decided to use h2a.z + HsH2A.Z.1 transgenic plants to examine the degree of molecular rescue in h2a.z plants mediated by human H2A.Z.1. To measure the ability of HsH2A.Z.1 to be deposited into chromatin, we performed two biological replicates of chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) using a human-specific H2A.Z antibody on h2a.z + HsH2A.Z.1 transgenic plants, and on WT and h2a.z + AtHTA11 transgenic plants using an Arabidopsis-specific H2A.Z antibody (Figure 2B). When we compared the average profiles and heat maps of H2A.Z enrichment across genes in WT plants, h2a.z + AtHTA11, and in h2a.z + HsH2A.Z.1 transgenic plants we found almost identical profiles, indicating that both AtHTA11 and human H2A.Z.1 are able to properly re-establish H2A.Z deposition sites in h2a.z plants (Figure 3A). To quantify these results, we examined a correlation of H2A.Z ChIP-seq read counts between WT and h2a.z + AtHTA11 plants, and between WT and h2a.z + HsH2A.Z.1 plants, and discovered that only four genes were differentially enriched for H2A.Z between WT and h2a.z + AtHTA11, and only 266 genes between WT and h2a.z + HsH2A.Z.1 transgenic plants (Figure 3B–C).
Figure 3. Chromatin immunoprecipitation with sequencing (ChIP-seq) analysis reveals that human H2A.Z.1 restores H2A.Z deposition sites in h2a.z plants to nearly identical WT levels.
(A) Heatmap of normalized ChIP-seq signal (H2A.Z/input) for 26,669 genes. Average ChIP-seq H2A.Z profiles of WT, h2a.z + AtHTA11, and h2a.z + HsH2A.Z.1 plants were plotted over gene body coordinates of 26,669 genes, from the transcript start site (TSS) to the transcript end site (TES). 5 k-means clusters were sorted by mean signal value. One replicate is used as a representative of each genotype. (B and C) Correlation of H2A.Z ChIP-seq read counts at genes between WT and h2a.z + AtHTA11 plants (B), and between WT and h2a.z + HsH2A.Z.1 plants (C). Read counts at genes were defined by mapped reads from TSS to TES. Counts were normalized via DESeq2. Color represents log2 fold change of normalized counts in sample vs WT.
Overall, these results suggest that human H2A.Z.1 is able to almost completely restore WT H2A.Z deposition sites in h2a.z plants.
The expression of more than 84% of mis-expressed genes in h2a.z plants is restored to the WT levels by human H2A.Z.1
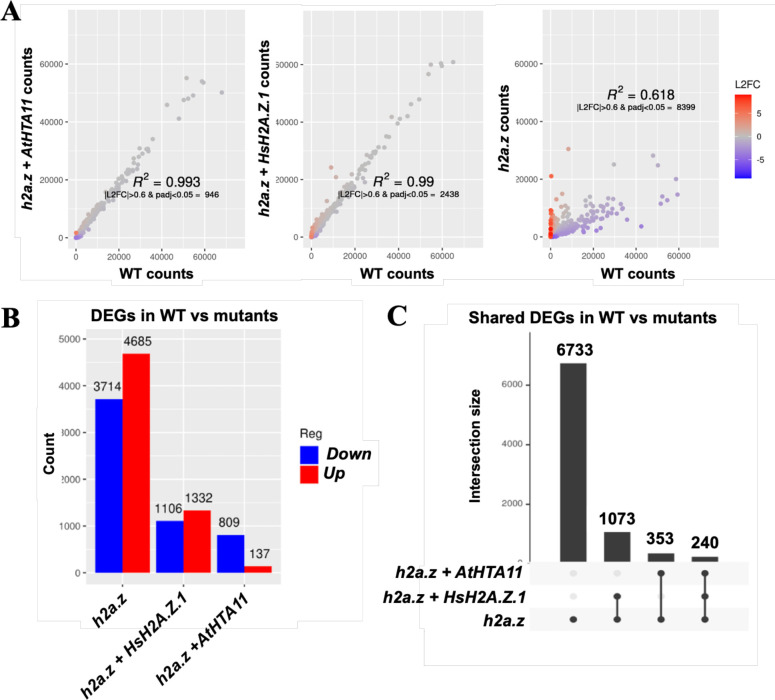
To investigate the ability of human H2A.Z.1 to restore the expression of misregulated genes in h2a.z plants we performed RNA-seq experiments using three biological replicates of WT, h2a.z plants, h2a.z + AtHTA11, and h2a.z + HsH2A.Z.1 transgenic plants. We identified 8,399 differentially expressed genes (DEGs) in h2a.z plants when compared to the WT, with 3714 downregulated genes and 4685 upregulated genes (with p adj ≤ 0.05, and absolute log2 fold change ³ 0.6), Figure 4A–B). Significantly lower number of genes were misexpressed in h2a.z + AtHTA11 plants (809 downregulated genes and 137 upregulated genes, total of 946 DEGs), and in h2a.z + HsH2A.Z.1 plants (1106 downregulated genes and 1332 upregulated genes, total of 2438 DEGs) when compared to the WT (Figure 4A–B). We then examined the overlap of all misexpressed genes in h2a.z plants with all misexpressed genes in h2a.z + AtHTA11 plants and discovered that only 593 genes, out of 8399, were still misregulated in h2a.z + AtHTA11 plants (Figure 4C). These results demonstrated that the AtHTA11 transgene restores the expression of 93% of misexpressed genes in h2az plants. Similarly, we analyzed the overlap of all misexpressed genes in h2a.z plants with all misexpressed genes in h2a.z + HsH2A.Z.1 plants and found that 1,313 genes were still misregulated in h2a.z + HsH2A.Z.1 plants (Figure 4C). Based on these results we calculated that human H2A.Z.1 is able to restore the expression of more than 84% of h2a.z mis-expressed genes back to the WT levels.
Figure 4. RNA-seq analysis reveals that human H2A.Z.1 restores the expression of more than 84% of misregulated genes in h2a.z plants back to the WT levels.
(A) Correlation of DESeq2 normalized transcript counts between WT and h2a.z + AtHTA11 plants (left), WT and h2a.z + HsH2A.Z.1 plants (middle), and between WT and h2a.z plants (right). Color represents log2 fold change of normalized counts in sample vs WT. (B) Histogram representing total number of misexpressed genes in mutant and transgenic plants versus WT (C) Upset plot of differentially expressed genes across h2a.z + AtHTA11, h2a.z + HsH2A.Z.1, and h2a.z plants compared to WT. Differential expression is defined by |L2FC| > 0.6 and padj < 0.05.
Finally, we also examined the average H2A.Z enrichment in WT across the bodies of genes either downregulated or upregulated in h2a.z plants and discovered that the genes upregulated in h2a.z have higher H2A.Z levels compared to downregulated genes (Supplemental Figure 4). These results suggest that H2A.Z enrichment at gene bodies is negatively correlated with gene expression, which is consistent with the results from previous studies [13].
Taken all together, our results suggest that human H2A.Z.1 rescues the molecular defects of h2a.z plants at a remarkably high level even though the amino acid sequences between Arabidopsis and human N-terminal ends are significantly different (Figure 1B).
N-terminal truncated Arabidopsis H2A.Z proteins rescue the majority of severe phenotypic defects in h2a.z plants
Because human H2A.Z.1 recapitulates the normal function of Arabidopsis H2A.Zs in transgenic plants so efficiently in spite of the divergent N-terminal sequences among these proteins (Figure 1B), we hypothesized that post-translational modifications of the key amino acid residues in the N-terminal tail of Arabidopsis H2A.Zs are not critical for the overall function of H2A.Z in development. Since the PTMs on plant H2A.Z have only begun to be documented [29], instead of replacing the individual key residues within the N-terminus, we decided to employ a more drastic approach of testing our hypothesis by truncating the amino acids of the N-terminal tail of Arabidopsis HTA11 to various lengths. We engineered four different AtHTA11 constructs, truncated for 7, 13, 21, or 28 amino acids at the N-terminus (see Figure 1B for reference), and transformed them into h2a.z +/− plants to assess their ability to rescue the mutant defects.
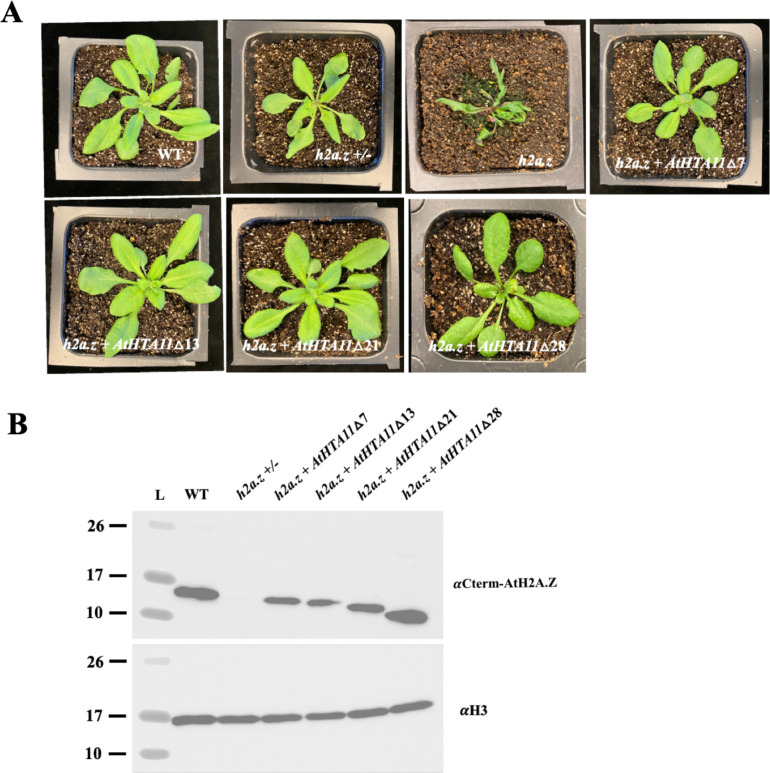
Phenotypic analysis of h2a.z plants carrying either of the four truncated AtHTA11 constructs revealed that these plants morphologically closely resembled WT plants (Figure 5A). We have also shown that the truncated AtHTA11 proteins are successfully deposited into chromatin (Figure 5B). Based on these results we concluded that, surprisingly, the N-terminal tail of Arabidopsis H2A.Z appears to be dispensable for its major functions in plant development.
Figure 5. N-terminal truncations of AtHTA11 largely rescue h2a.z defects.
(A) 2.5 week old plants were grown under long-day conditions and individually photographed. (B) Western-blot analysis using 10ul of leaf chromatin extracts from wild type (WT), h2a.z +/− plant, and T2 h2a.z plants with four different truncated Arabidopsis HTA11 constructs probed with Arabidopsis H2A.Z specific antibody that recognizes the C-terminal end of the protein (top panel), and with H3 antibody that was used as a loading control (bottom panel).
DISCUSSION
Human H2A.Z proteins rescue morphological and molecular defects of h2a.z plants at a remarkably high level
We originally hypothesized that human H2A.Zs would not be able to rescue all phenotypic defects of h2a.z plants since the functionally significant amino acid residues found at the N-terminal tails of these proteins are very different in sequence (Figure 1B). However, we found that human H2A.Z.1, when expressed in h2a.z plants, complemented not only all morphological abnormalities of mutant plants but also rescued the molecular defects caused by the loss of endogenous H2A.Zs at a remarkably high level. On the other hand, human H2A.Z.2.2 had an opposite effect when expressed in plants and may interfere with the function of the endogenous H2A.Zs. Even though the mechanism of this obstruction is not known, human H2A.Z.2.2, in the form of an inducible construct, could potentially be used to temporally disrupt endogenous H2A.Z functions in plants.
Overall, these results clearly demonstrated that there is an incredibly high degree of functional conservation between Arabidopsis and human H2A.Z histones and raise an interesting question about the ability of Arabidopsis H2A.Zs to rescue any defects caused by the loss of H2A.Zs in human cells, or in other model organisms.
The N-terminal tail of Arabidopsis H2A.Z is not essential for its major developmental functions
To explain the finding that human H2A.Zs are highly functional in Arabidopsis, we contemplated two possible scenarios, which are not mutually exclusive: 1) Post-translational modifications (PTMs) of amino acids at the N-terminal end of Arabidopsis H2A.Zs are important for their overall function and plant enzyme machinery that deposits those modifications can correctly recognize key residues within the N-terminal tails of human H2A.Zs to properly modify them, 2) PTMs of the key amino acid residues at the N-terminal tail of Arabidopsis H2A.Zs are not important for their function and, therefore, human H2A.Zs may function in plants as efficiently as endogenous H2A.Zs regardless of their ability to be post-translationally modified by plant machinery.
Since PTMs of amino acids at the N-terminal tail of Arabidopsis H2A.Zs are not well characterized [29], we utilized a more aggressive approach to test the second hypothesis by transforming four different N-terminally truncated constructs of Arabidopsis HTA11 into h2a.z +/− plants and assessing their ability to rescue the phenotypic defects of h2az plants. Surprisingly, all four constructs successfully recapitulated morphological defects, including Δ28 AtHTA11, which is practically an N-terminal tailless H2A.Z, indicating that the N-terminal end of Arabidopsis H2A.Z is not essential to support normal development. At first, these results were quite unexpected. However, a recent study in yeast has demonstrated that it is not the N-terminal tail but rather the specific set of conserved amino acids at the C-terminus that contributes to the unique functions of H2A.Z, at least in this organism [30]. Indeed, all Arabidopsis and human H2A.Zs, except for a few residues that are missing in HsH2A.Z.2.2, possess these conserved amino acids, while they are absent in Arabidopsis HTA2 histone (Supplemental Figure 5).
It is difficult to imagine that the N-terminal tail, carrying many conserved amino acids known to be the substrates for PTMs, has no significance for H2A.Z biology in plants. For instance, Arabidopsis HTA11 contains 11 lysines at the N-terminus, which are known targets of numerous modifications and plants do possess enzymatic machinery capable of recognizing and modifying these residues. Therefore, it is reasonable to assume that many of the AtHTA11 lysines would be modified at some point during development. The question then arises, what role do modified amino acids at the N-terminus play in H2A.Z biology that we have not observed at the phenotypic level? One possibility is that the putatively modified residues may fine-tune H2A.Z function in response to diverse external and internal stimuli, such as the exposure of plants to various abiotic stresses. This scenario, together with the complete identification of post-translationally modified amino acids at the N-terminus of Arabidopsis H2A.Zs, remains to be tested and characterized in the near future.
MATERIALS and METHODS
Plant material, growth conditions, and transformation
Arabidopsis thaliana of the Columbia (Col-0) ecotype was used as the wild-type reference. Seedlings were grown either in soil or on half-strength Murashige and Skoog (MS) media agar plates [31], in growth chambers at +20°C under a 16h light/8 hour dark cycle. All plasmids used for plant transformation were introduced into Agrobacterium tumefaciens GV3101 strain by electroporation. T0 plants were transformed with plasmids using the floral dip method [32]. Primary transgenic plants were selected on half-strength MS media agar plates containing 25 mg/L BASTA and 100 mg/L timentin, and then transferred to soil.
Production of h2a.z plants using CRISPR methodology
To generate CRISPR-induced h2a.z null mutant plants we utilized an egg cell-specific promoter-controlled Cas9 plasmid that contains two single guide RNAs, which can target two separate genes at the same time [33]. HTA9 and HTA11 were targeted first since hta9/hta11 double mutants have easily observable phenotypes. We sequenced the regions around the target sites using DNA samples isolated from T2 transgenic plants with h2a.z phenotypes and confirmed that these plants were indeed double homozygous mutants for hta9 and hta11. All mutations were simple base pair insertions causing a frame shift in the coding sequence leading to premature stop codons (Suppl. Figure 6). We then identified plants that were hta9/hta11 null without the Cas9 transgene and transformed them with the same egg cell-specific promoter-controlled Cas9 plasmid, this time targeting the HTA8 gene. After DNA regions around HTA8 target sites were sequenced, we identified new primary transgenic plants that were homozygous for hta9 and hta11 and heterozygous for HTA8, herein named h2a.z +/− plants. The HTA8 CRISPR allele was a simple base pair insertion causing a frame shift in the coding sequence (Suppl. Figure 6). We then identified h2a.z +/− plants that segregated out the Cas9 transgene and those were used for all transformation experiments. Eventually, we identified h2a.z null mutants among h2a.z +/− progeny plants and confirmed by Sanger sequencing that these plants indeed had mutations in all three H2A.Z genes.
Plasmid DNA constructs
To produce all constructs that were driven by the native HTA11 promoter we first cloned 1855 bp of the AtHTA11 promoter sequence (1855 bp upstream from the AtHTA11 ATG start codon), with added NcoI restriction site at the 3’ end, into gateway-compatible pENTR-D/TOPO plasmid (Invitrogen). We then cloned 482 bp of the AtHTA11 terminator sequence (482 bp downstream from the AtHTA11 stop codon) into the same pENTR plasmid that contained the AtHTA11 promoter just downstream of the NcoI restriction site. Various PCR-generated AtHTA11 constructs and synthesized and plant codon-optimized human H2A.Zs were then cloned into this plasmid using NcoI restriction site. All constructs were verified by sequencing and eventually subcloned into pMDC123 gateway destination plasmid [34] using the LR clonase II enzyme in LR recombination reaction (Invitrogen). To produce 35S::HTA2 construct we first cloned HTA2 gene sequence into pENTR-D/TOPO plasmid (Invitrogen). After the correct sequence of HTA2 was verified by Sanger sequencing, the construct was subcloned into pEG100 gateway destination plasmid [35] using the LR clonase II enzyme in LR recombination reaction (Invitrogen).
cDNA production and real-time RT-PCR (qRT-PCR)
Total RNA was isolated from third and fourth pair of leaves from 35S::HTA2 transgenic h2a.z +/− plants using the RNeasy plant mini kit (Qiagen). 2 μg of total RNA was converted into cDNA with LunaScript RT SuperMix kit (New England Biolabs). The cDNAs were used as templates for real-time PCR and ran on StepOnePlus real-time PCR system (Applied Biosystems) using SYBR Green as a detection reagent. The PP2A gene (AT1G13320) was used as the endogenous gene expression control [36], while primers specific for the 35S::HTA2 transgene were used to detect its expression in three individual T1 h2a.z +/− plants.
Chromatin extraction and protein gel blotting
To probe chromatin-associated proteins by western blotting we used the chromatin extraction protocol described by Luo and colleagues [37], with following modifications: 1) 0.5 grams of rosette leaves from each sample were ground in liquid nitrogen and homogenized in 5 ml of Honda buffer, 2) Homogenates were filtered through 70 μm cell strainer and centrifuged at 1,500g at + 4°C degree for 20 minutes. The chromatin fractions from each sample were resuspended in 80–100 μl of 1x Laemmli’s sample buffer. Western blotting was performed as previously described [38], using 1:1000 dilutions of following primary antibodies: N-terminal Arabidopsis H2A.Z antibody [22], in-house C-terminal Arabidopsis H2A.Z antibody, and human H2A.Z antibody (Abcam, ab188314). All blots were incubated with ECL detection reagents for 2 minutes (Thermo Scientific) and scanned for chemiluminescence signal using ChemiDoc MP imaging system instrument (BioRad).
Chromatin immunoprecipitation (ChIP) with Arabidopsis and human H2A.Z antibodies
ChIP experiments were performed in biological duplicates on WT, h2a.z + AtHTA11, and on h2a.z + HsH2A.Z.1 leaf tissue (third and/or fourth pair of rosette leaves) as described previously [39], with following modifications: 1) For each sample, 0.5 grams of leaf tissue were used, 2) ground samples were lysed in 600 μl of buffer S, 3) around 400 μl of the slurry were transferred to 0.6 ml tubes and sonicated in the Bioruptor (Diagenode) at + 4°C for 1 hour on the “high” setting with sonication intervals set to 45 seconds on/15 seconds off, 4) The sonicated lysates were centrifuged at 20,000×g for 10 min at + 4°C and 300 μl of the supernatants were transferred to a fresh 5 ml tube where 2.7 ml of buffer F was added and mixed well, 5) 50 μl of this mixture was saved as the input sample while 1.5 ml was used for immunoprecipitation with specific antibodies, 6) Arabidopsis-specific H2A.Z antibody [22] and human-specific H2A.Z antibody (Abcam, ab188314) were added to the diluted lysates at a final concentration of ~2 μg/ml and incubated overnight at + 4°C with rocking, 7) 25 μl of washed Protein A dynabeads were added to each sample and incubated at + 4°C for 2 hours with rocking, 8) immunoprecipitated DNA fragments were purified using Qiagen Minelute kit and eluted in 14 μl of elution buffer. Eluted DNA samples were quantified using Picogreen reagents.
ChIP-seq library preparation, sequencing, and data analysis
ChIP-seq libraries were prepared starting with ~1000 picograms of ChIP or input DNA samples using the ThruPlex DNA-seq kit (Takara) according to the manufacturer’s instructions. Libraries were pooled together and sequenced using paired-end 150 nt reads on an Illumina NovaSeq 6000 instrument. Reads were trimmed of adapter content using Trimgalore [40] and mapped to the Arabidopsis thaliana Col-PEK genome assembly [41] using Bowtie2 [42] with the following parameters: --local --very-sensitive --no-mixed --no-discordant --phred33 -I 10 -X 700. Aligned reads were converted to the BAM file format and quality filtered using Samtools [43]. Duplicate reads were removed using Picard markDuplicates [44]. For visualization, deduplicated BAM files of each genotype were converted to bedgraph files using bedtools bamtobed followed by genomecov [45]. Samples were then normalized to the lowest read depth and converted to BigWig using bedGraphToBigWig [46]. Finally, the signal from each sample was expressed as the log2 ratio relative to the corresponding input using bigWigCompare with –psuedocount = 1 to avoid 0/x [47]. Profile plots and heatmaps of the resulting bigwig files were generated using Deeptools [47]. Mapped fragments overlapping with genes or peaks were counted using featureCounts [48]. Gene counts were then analyzed for differential enrichment using DESeq2 [49]. DESeq2 results were visualized using ggplot2 package in R [50].
RNA extraction, RNA-seq library preparation, sequencing, and data analysis
Total RNA was isolated from the following plants: WT, h2a.z, h2a.z + AtHTA11, and h2a.z + HsH2A.Z.1. Five individual leaves (third or fourth pair of rosette leaves) were collected from 5 different plants of WT, h2a.z + AtHTA11, and h2a.z + HsH2A.Z.1 genotypes. The leaves were ground in liquid nitrogen and powder was resuspended in 2 ml of RLT buffer from RNeasy Plant Mini kit (Qiagen). For each sample, three equal aliquots of 400 μl of this resuspension were further processed to extract total RNA, according to the manufacturer’s instructions. For h2a.z plants, aboveground tissue from three different plants were harvested and individually processed to extract total RNA following the manufacturer’s recommendations. Total RNA was DNase-treated (Ambion) and quantified using Nanodrop One (Thermo Scientific). 100 nanograms for each of the three replicates from every sample were then used as a starting material to generate RNA-seq libraries following the Universal RNA-seq with NuQuant protocol (Tecan). Libraries were pooled together and sequenced using paired-end 150 nt reads on an Illumina NovaSeq 6000 instrument. Reads were trimmed of adapter content using Trimgalore [40]. Trimmed reads were mapped to the Arabidopsis thaliana TAIR10 genome assembly, converted to the BAM file format, and sorted by coordinate using STAR [51]. Aligned reads were indexed and quality filtered using Samtools [43]. Mapped fragments overlapping with exons were counted using featureCounts [48]. Gene counts were then analyzed for differential enrichment using DESeq2 [49]. Significantly differentially expressed genes were defined as those having an adjusted p-value less than 0.05 and an absolute fold change greater than 1.5. DESeq2 results were visualized using ggplot2 and upsetR in R [50].
Supplementary Material
FUNDING
This work was supported by funding from the National Institutes of Health (R01GM134245) to RBD. DHH was also supported by an NIH training grant (T32GM008490).
Data availability
All sequencing data are in the process of submission to the NCBI GEO database. Accession numbers will be added to an updated version of this manuscript.
REFERENCES
- 1.Talbert PB, Henikoff S. Histone variants at a glance. J Cell Sci. 2021;134(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martire S, Banaszynski LA. The roles of histone variants in fine-tuning chromatin organization and function. Nat Rev Mol Cell Biol. 2020;21(9):522–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foroozani M, Holder DH, Deal RB. Histone Variants in the Specialization of Plant Chromatin. Annu Rev Plant Biol. 2022;73:149–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreienbaum C, Paasche LW, Hake SB. H2A.Z’s ‘social’ network: functional partners of an enigmatic histone variant. Trends Biochem Sci. 2022;47(11):909–20. [DOI] [PubMed] [Google Scholar]
- 5.Borg M, Jiang D, Berger F. Histone variants take center stage in shaping the epigenome. Curr Opin Plant Biol. 2021;61:101991. [DOI] [PubMed] [Google Scholar]
- 6.Lewis TS, Sokolova V, Jung H, Ng H, Tan D. Structural basis of chromatin regulation by histone variant H2A.Z. Nucleic acids research. 2021;49(19):11379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic acids research. 2012;40(21):10719–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7(12):1121–4. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular cell. 2012;48(5):723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou BO, Wang SS, Xu LX, Meng FL, Xuan YJ, Duan YM, et al. SWR1 complex poises heterochromatin boundaries for antisilencing activity propagation. Molecular and cellular biology. 2010;30(10):2391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112(5):725–36. [DOI] [PubMed] [Google Scholar]
- 12.Rosa M, Von Harder M, Cigliano RA, Schlogelhofer P, Mittelsten Scheid O. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. The Plant cell. 2013;25(6):1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS genetics. 2012;8(10):e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.March-Diaz R, Reyes J. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Molecular plant. 2009;2(4):565–77. [DOI] [PubMed] [Google Scholar]
- 15.Giaimo BD, Ferrante F, Herchenrother A, Hake SB, Borggrefe T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin. 2019;12(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colino-Sanguino Y, Clark SJ, Valdes-Mora F. The H2A.Z-nucleosome code in mammals: emerging functions. Trends Genet. 2022;38(5):516. [DOI] [PubMed] [Google Scholar]
- 17.Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, et al. Histone variant H2A.Z is required for early mammalian development. Current biology : CB. 2001;11(15):1183–7. [DOI] [PubMed] [Google Scholar]
- 18.Lamaa A, Humbert J, Aguirrebengoa M, Cheng X, Nicolas E, Cote J, et al. Integrated analysis of H2A.Z isoforms function reveals a complex interplay in gene regulation. eLife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sales-Gil R, Kommer DC, de Castro IJ, Amin HA, Vinciotti V, Sisu C, et al. Non-redundant functions of H2A.Z.1 and H2A.Z.2 in chromosome segregation and cell cycle progression. EMBO Rep. 2021;22(11):e52061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonisch C, Schneider K, Punzeler S, Wiedemann SM, Bielmeier C, Bocola M, et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic acids research. 2012;40(13):5951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wratting D, Thistlethwaite A, Harris M, Zeef LA, Millar CB. A conserved function for the H2A.Z C terminus. J Biol Chem. 2012;287(23):19148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deal R, Topp C, McKinney E, Meagher R. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. The Plant cell. 2007;19(1):74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456(7218):125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi K, Park C, Lee J, Oh M, Noh B, Lee I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development (Cambridge, England). 2007;134(10):1931–41. [DOI] [PubMed] [Google Scholar]
- 25.March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio F, Reyes J. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. The Plant journal : for cell and molecular biology. 2008;53(3):475–87. [DOI] [PubMed] [Google Scholar]
- 26.Colino-Sanguino Y, Clark SJ, Valdes-Mora F. H2A.Z acetylation and transcription: ready, steady, go! Epigenomics. 2016;8(5):583–6. [DOI] [PubMed] [Google Scholar]
- 27.Law C, Cheung P. Expression of Non-acetylatable H2A.Z in Myoblast Cells Blocks Myoblast Differentiation through Disruption of MyoD Expression. The Journal of biological chemistry. 2015;290(21):13234–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie WF, Lei M, Zhang M, Tang K, Huang H, Zhang C, et al. Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(33):16641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartl M, Fussl M, Boersema PJ, Jost JO, Kramer K, Bakirbas A, et al. Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol Syst Biol. 2017;13(10):949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewis HT, Wang AY, Gaub A, Lau JJ, Stirling PC, Kobor MS. What makes a histone variant a variant: Changing H2A to become H2A.Z. PLoS genetics. 2021;17(12):e1009950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–97. [Google Scholar]
- 32.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant journal : for cell and molecular biology. 1998;16(6):735–43. [DOI] [PubMed] [Google Scholar]
- 33.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome biology. 2015;16(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, et al. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant journal : for cell and molecular biology. 2006;45(4):616–29. [DOI] [PubMed] [Google Scholar]
- 36.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant physiology. 2005;139(1):5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo YX, Hou XM, Zhang CJ, Tan LM, Shao CR, Lin RN, et al. A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. The EMBO journal. 2020;39(7):e102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sijacic P, Holder DH, Bajic M, Deal RB. Methyl-CpG-binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z-enriched regions in the Arabidopsis genome. PLoS genetics. 2019;15(8):e1008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, Xie L, Zhang Q, Ouyang W, Deng L, Guan P, et al. Integrative analysis of reference epigenomes in 20 rice varieties. Nat Commun. 2020;11(1):2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- 41.Hou X, Wang D, Cheng Z, Wang Y, Jiao Y. A near-complete assembly of an Arabidopsis thaliana genome. Molecular plant. 2022;15(8):1247–50. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England). 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.http://broadinstitute.github.io/picard/.
- 45.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England). 2010;26(6):841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics (Oxford, England). 2010;26(17):2204–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic acids research. 2014;42(Web Server issue):W187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England). 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickham H. ggplot2: Elegant graphics for data analysis. Springer Verlag. 2016. [Google Scholar]
- 51.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are in the process of submission to the NCBI GEO database. Accession numbers will be added to an updated version of this manuscript.