Abstract
Nigrospora is a monophyletic genus belonging to Apiosporaceae. Species in this genus are phytopathogenic, endophytic, and saprobic on different hosts. In this study, leaf specimens with disease symptoms were collected from host plants from the Shandong Peninsula, China. The fungal taxa associated with these leaf spots were studied using morphology and phylogeny based on ITS, TEF1, and TUB2 gene regions. In this article, we report on the genus Nigrospora with N. gorlenkoana, N. oryzae, N. osmanthi, N. rubi, and N. sphaerica identified with 13 novel host associations including crops with economic importance such as bamboo and Chinese rose.
Keywords: Ascomycota, morphology, multi-gene phylogeny, new host records, Xylariales
1. Introduction
The genus Nigrospora Zimm. (Apiosporaceae, Xylariales, and Sordariomycetes) was established to accommodate N. panici Zimm. [1,2]. Nigrospora species are cosmopolitan, filamentous, dematiaceous taxa, with a diverse host range including crops with economic importance [2,3]. Species of this genus are pathogens, endophytes, and saprobes of various hosts [4,5]; Table 1 presents a list of reported N. occurrences including disease incidents. These studies emphasize N. oryzae and N. sphaerica as the most frequently reported pathogens of Nigrospora.
Table 1.
Occurrences of Nigrospora species on different hosts and their nutritional relationship.
| Causative agent | Nutritional relationship | Disease | Host | Region | References |
|---|---|---|---|---|---|
| N. lacticolonia | Pathogenic | Reddish brown spots | Hylocereus polyrhizus | Malaysia | Kee et al. [32] |
| N. musae | Endophytic | NA | Musa accuminata | Australia | Brown et al. [33] |
| N. oryzae | Pathogenic | Lint rot | Gossypium hirsutum | Alabama | Palmateer et al. [34] |
| N. oryzae | Pathogenic | Stem blight | Brassica juncea | India | Sharma et al. [35] |
| N. oryzae | Pathogenic | Leaf spot | Aloe vera | China | Zhai et al. [36] |
| N. oryzae | Pathogenic | Leaf spot | Dendrobium candidum | China | Wu et al. [37] |
| N. oryzae | Pathogenic | Brown/black spot disease | Actinidia deliciosa | China | Li et al. [38] |
| N. oryzae | Pathogenic | Leaf spots | Gossypium hirsutum | China | Zhang et al. [28] |
| N. oryzae | Pathogenic | Leaf spots | Poa pratensis | Canada | Zheng et al. [39] |
| N. oryzae | Pathogenic | Foliar and cane rot | Arundo donax | France, Crete, Cyprus, Italy, Morocco, and Spain | Widmer et al. [27] |
| N. oryzae | Pathogenic | Leaf spot | Pearl millet | Iran | Kalati et al. [40] |
| N. oryzae | Pathogenic | Leaf spot | Phoenix dactylifera | Iraq | Abass [41] |
| N. oryzae | Endophytic | NA | Emblica officinalis | India | Rathod et al. [42] |
| N. oryzae | Endophytic | NA | Artemisia sp. | China and Canary Islands | Cosoveanu [43] |
| N. oryzae | Saprobic | NA | Musa acuminate | Hong Kong and Australia | Brown et al. [33] |
| N. osmanthi | Pathogenic | Leaf blight | Stenotaphrum secundatum | Tropics and sub tropics and China | Mei et al. [44] |
| N. osmanthi | Pathogenic | Leaf blight | Ficus pandurata | China | Liu et al. [45] |
| N. sacchari | Endophytic | NA | Bauhinia phoenicea | India | Raviraja et al. [46] |
| N. sphaerica | Pathogenic | Leaf blight | Sesamum indicum | China | Zhao et al. [47] |
| N. sphaerica | Pathogenic | Leaf blight | Saccharum | China | Cui et al. [48] |
| N. sphaerica | Pathogenic | Leaf blight | Camellia sinensis | India | Dutta et al. [49] |
| N. sphaerica | Pathogenic | Shot hole disease | Morus alba | India and China | Chen et al. [50], Arunakumar et al. [51] |
| N. sphaerica | Pathogenic | Leaf spots, twigs, and shoot blight | Vaccinium corymbosum | Buenos Aires, Entre Ríos | Wright et al. [31] |
| N. sphaerica | Pathogenic | Leaf and stem black spot disease | Phoenix dactylifera | Iraq | Abass [41], Abass et al. [52] |
| N. sphaerica | Pathogenic | Reddish brown spots | Hylocereus polyrhizus | Malaysia | Kee et al. [32] |
| N. sphaerica | Pathogenic | Black end and squirter disease | Musa sp. | Australia | Allen [53], Simmonds [54] |
| N. sphaerica | Pathogenic | Leaf spots | Actinidia sp. | China | Chen et al. [55] |
| N. sphaerica | Pathogenic | Postharvest rot | Actinidia sp. | China | Li et al. [56] |
| N. sphaerica | Pathogenic | Leaf spots | Lagenaria siceraria | Georgia | Li et al. [57] |
| N. sphaerica | Pathogenic | Leaf blight | Camellia sinensis | China | Liu et al. [58] |
| N. sphaerica | Pathogenic | Leaf blight | Cunninghamia lanceolata | China | Xu et al. [59] |
| N. sphaerica | Pathogenic | Leaf spots | Kinnow Mandarin | Pakistan | Alam et al. [60] |
| N. sphaerica | Pathogenic | Leaf spots | Phoenix dactylifera | Pakistan | Alam et al. [61] |
| N. sphaerica | Pathogenic | Leaf spots | Mangiferra indica | India | Pandey et al. [62] |
| N. sphaerica | Endophytic | NA | Artemisia sp. | China | Cosoveanu [43] |
| Nigrospora. sp. | Endophytic | NA | Azadirachta indica | Southwest China | Wu et al. [63] |
NA: not applicable.
Species of Nigrospora harbor a great potential in bioactive secondary metabolite production. N. sphaerica is a rich source of secondary metabolites such as bioactive compounds with antileukemic (tested on HL60 and K562 cell lines), antileishmanial, and antifungal activities [6]. An endophytic Nigrospora species isolated from Moringa oleifera root produced a few important bioactive secondary metabolites under in vitro conditions, including griseofulvin, dechlorogriseofulvin, and mellein with antifungal activity [7]. A new hydroanthraquinone derivative and new azaphilones produced by Nigrospora sp. YE3033 was reported to be successful in inhibiting influenza viral strain of A/Puerto Rico/8/34 (H1N1) [8].
Species delimitation in Nigrospora was previously based on morphological characters [9], but it was found that some key morphological characters such as conidial dimensions overlap between phylogenetically distinct species [3]. To address this issue, a polyphasic approach, combining both morphology and molecular phylogeny, is necessary. A recent study reassessing Nigrospora species by Wang et al. [3] sequenced previously introduced Nigrospora species from their herbarium materials. Further, they affirmed the placement of the genus in Apiosporaceae (Xylariales) based on multi-locus molecular phylogeny (internal transcribed spacer (ITS), translation elongation factor 1-α (TEF1) and β-tubulin (TUB2) gene regions) [3]. In their study, the new species N. aurantiaca Mei Wang & L. Cai, N. bambusae Mei Wang & L. Cai, N. camellia-sinensis Mei Wang & L. Cai, N. chinensis Mei Wang & L. Cai, N. guilinensis Mei Wang & L. Cai, N. hainanensis Mei Wang & L. Cai, N. lacticolonia Mei Wang & L. Cai, N. osmanthi Mei Wang & L. Cai, N. pyriformis Mei Wang & L. Cai, N. rubi Mei Wang & L. Cai, N. vesicularis Mei Wang & L. Cai and N. zimmermanii Crous. were introduced. N. vietnamensis Hol.-Jech. was transferred to Arthrinium and synonymized under Arthrinium vietnamensis (Hol.-Jech.) Mei Wang & L. Cai. based on the multigene phylogenetic analyses [3].
Shandong Peninsula, the target site of this study, is bordered by the Bohai Sea to the North and Yellow Sea to the Southeast. The fungal ecology in this region would be an interesting aspect to study. This study focuses on Nigrospora species associated with leaf spots on forest plants. It also aims to provide molecular data for the genus to support molecular phylogeny based species identification. Furthermore, novel host associations of Nigrospora are identified and potential threats on forest plant species and crops with economic importance are predicted.
2. Materials and methods
2.1. Sample collection, isolation, and herbarium specimens
Leaf specimens from various plants with leaf spot symptoms were collected from Shandong Peninsula, China and brought to the laboratory in paper bags. Symptomatic leaves with leaf spots were selected and cut into approximately 2 × 2 mm pieces composed of both the diseased and healthy leaf tissue areas. The leaf pieces were surface sterilized by washing with 1% sodium hypochlorite for 30 s, 70% ethanol for 30 s, and finally, three times in sterilized water prior to culturing on potato dextrose agar (PDA) (1/4 PDA) and incubated at 25 °C. Hyphal tips of growing mycelia from leaf tissues on PDA were carefully picked up with a sterile toothpick and transferred onto fresh PDA plates to obtain pure cultures.
Morphological characters were observed and photographed using an Axio Imager Z2 photographic microscope (Carl Zeiss Microscopy, Oberkochen, Germany) and measurements were made with ZEN PRO 2012 software (Carl Zeiss Microscopy). Fifty conidial measurements were taken per isolate and cultures were allowed to grow until they completely covered a 90 mm petri dish to measure growth rate. The growth rate was calculated as the mean of two perpendicular measurements.
Voucher specimens were deposited in the herbarium collection of Beijing Academy of Agricultural and Forestry Sciences (JZBH) and all the cultures were deposited at the culture collections of Beijing Academy of Agricultural and Forestry Sciences (JZB), China and Kunming Institute of Botany (KUMCC), China. Following Jayasiri et al. [10], Faces of Fungi (FOF) numbers were acquired.
2.2. Dna extraction, PCR amplification, and sequencing
Fungal mycelia grown on PDA for 4–7 d were scraped off and collected. Genomic DNA was extracted using a modified CTAB protocol described in Guo et al. [11]. The following loci are amplified with the primer pairs given in Table 2. Polymerase chain reactions (PCR) were conducted in an Applied Biosystems C1000 TouchTM Thermal Cycler with the following PCR conditions for ITS, TEF1, and TUB2 regions [12]: initial denaturation for 3 min at 95 °C followed by 34 cycles of denaturation for 30 s at 95 °C and 30 s of annealing and 1 min elongation at 72 °C, and a final extension for 10 min at 72 °C. The annealing temperatures were as follows: 58 °C for both ITS and TUB2, and 52 °C for TEF1. The PCR reaction mixture was composed of 0.3 µL of TaKaRa Ex-Taq DNA polymerase (TaKaRa, Beijing, China), 2.5 µL of 10x Ex-Taq buffer (TaKaRa), 3.0 µL of dNTPs (TaKaRa), 1 µL of genomic DNA, 1 µL of each primer, and 16.2 µL of double-distilled H2O. The PCR products were visualized on 1% agarose gel followed by ethidium bromide staining, under UV light using a GelDoc XR + Molecular Imager (Bio-Rad, Hercules, CA, USA). Sequencing of PCR products was done by Beijing Biomed Gene Technology Co., Ltd, Beijing, China.
Table 2.
Primers used in the study, with sequences and references.
| Gene abbreviation | Definition | Primer | Sequence (5′-3′) | References |
|---|---|---|---|---|
| ITS1-5.8S-ITS2 | Internal transcribed spacer | ITS 4 | TCCTCCGCTTATTGATATGC | White et al. [12] |
| ITS 5 | GGAAGTAAAAGTCGTAACAAGG | |||
| TEF 1 | Partial translation elongation factor 1- α | TEF1-728F | CATCGAGAAGTTCGAGAAGG | Carbone et al. [64] |
| EF-2 | GGA(G/A)GTACCAGT(G/C)ATCATGTT | O’Donnell et al. [65] | ||
| TUB2 | β-Tubulin | BT-2F | AACATGCGTGAGATTGTAAGT | O’Donnell et al. [66] |
| BT-4R | TAGTGACCCTTGGCCCAGTTG |
2.3. Sequence alignment and phylogenetic analyses
Sequence chromatograms were checked with Chromas version 2.6.6 (Technelysium Pty Ltd., South Brisbane, Australia) and low-quality regions were trimmed prior to sequence alignments. Consensus sequences were generated for the TUB2 gene region using DNAStar version 5.1 (DNASTAR, Inc. Madison, WI, USA). All the sequences generated in this study were analyzed using the BLASTn searches in the GenBank. Reference sequences were obtained from GenBank referring to recently published relevant phylogenies and are listed in Table 3 [3]. Individual data sets of ITS, TEF1, and TUB2 were aligned using the default settings of the MAFFT version 7 webserver [13]. The alignments were manually edited further discarding leading or trailing gaps and concatenated in the following order, ITS, TEF1, and TUB2 using BioEdit version 7.0.5.2 (Department of Microbiology, North Carolina State University, NC, USA) [14]. Phylogenetic analyses of the aligned data were based on maximum likelihood (ML), Maximum parsimony (MP), and Bayesian posterior probabilities (BYPP) analyses.
Table 3.
Strains of the Nigrospora species and related GenBank accession numbers of taxa included in this study.
| Taxa | Culture collection Numbera,b | Hostc | GenBank Accession numbersd |
||
|---|---|---|---|---|---|
| ITS | TUB2 | TEF1 | |||
| N. aurantiaca | CGMCC 3.18130* = LC 7302 | Nelumbo sp. (leaf) | KX986064 | KY019465 | KY019295 |
| N. aurantiaca | LC 7034 | Musa paradisiaca | KX986093 | KY019598 | KY019394 |
| N. bambusae | CGMCC 3.18327* = LC 7114 | Bamboo (leaf) | KY385307 | KY385319 | KY385313 |
| N. bambusae | LC 7244 | Bamboo (leaf) | KY385306 | KY385320 | KY385314 |
| N. bambusae | LC 7245 | Bamboo (leaf) | KY385305 | KY385321 | KY385315 |
| N. camelliae-sinensis | LC 2710 | Castanopsis sp. | KX985957 | KY019484 | KY019310 |
| N. camelliae-sinensis | LC 3287 | Camellia sinensis | KX985975 | KY019502 | KY019323 |
| N. camelliae-sinensis | LC 3496 | Camellia sinensis | KX985985 | KY019510 | KY019327 |
| N. camelliae-sinensis | CGMCC 3.18125* = LC 3500 | Camellia sinensis | KX985986 | KY019460 | KY019293 |
| N. camelliae-sinensis | LC 6684 | Camellia sinensis | KX986046 | KY019570 | KY019449 |
| N. chinensis | LC 2696 | Lindera aggregata | KX985947 | KY019474 | KY019424 |
| N. chinensis | LC 3493 | Camellia sinensis | KX985984 | KY019509 | KY019434 |
| N. chinensis | LC 4433 | Castanopsis sp. | KX986013 | KY019536 | KY019436 |
| N. chinensis | LC 4558 | Unknown host plant | KX986020 | KY019543 | KY019441 |
| N. chinensis | CGMCC 3.18127* = LC 4575 | Machilus breviflora | KX986023 | KY019462 | KY019422 |
| N. chinensis | LC 4660 | Quercus sp. | KX986026 | KY019548 | KY019445 |
| N. chinensis | LC 6631 | Camellia sinensis | KX986043 | KY019569 | KY019448 |
| N. chinensis | LC 6851 | Unknown host plant | KX986049 | KY019579 | KY019450 |
| N. gorlenkoana | CBS 480.73* | Vitis vinifera | KX986048 | KY019456 | KY019420 |
| N. gorlenkoana | JZB 3230001 | Cirsium setosum** | MN495939 | MN549381 | MN544645 |
| N. guilinensis | LC 7301 | Vitis vinifera | KX986063 | KY019608 | KY019404 |
| N. guilinensis | CGMCC 3.18124* = LC 3481 | Nelumbo sp. (stem) | KX985983 | KY019459 | KY019292 |
| N. hainanensis | CGMCC 3.18129* = LC 7030 | Musa paradisiaca (leaf) | KX986091 | KY019464 | KY019415 |
| N. hainanensis | LC 6979 | Musa paradisiaca (leaf) | KX986079 | KY019586 | KY019416 |
| N. hainanensis | LC 7031 | Musa paradisiaca (leaf) | KX986092 | KY019597 | KY019417 |
| N. hainanensis | LC 7042 | Musa paradisiaca (leaf) | KX986094 | KY019599 | KY019418 |
| N. lacticolonia | CGMCC 3.18123* = LC 3324 | Camellia sinensis | KX985978 | KY019458 | KY019291 |
| N. lacticolonia | LC 7009 | Musa paradisiaca (leaf) | KX986087 | KY019594 | KY019454 |
| N. musae | CBS 319.34* | Musa paradisiaca (fruit) | KX986076 | KY019455 | KY019419 |
| N. musae | LC 6385 | Camellia sinensis | KX986042 | KY019567 | KY019371 |
| N. oryzae | LC 6761 | Oryza sativa | KX986056 | KY019574 | KY019376 |
| N. oryzae | LC 7297 | Nelumbo sp. (leaf) | KX985936 | KY019605 | KY019400 |
| N. oryzae | LC 2693 | Neolitsea sp. | KX985944 | KY019471 | KY019299 |
| N. oryzae | LC 2707 | Rhododendron simiarum | KX985954 | KY019481 | KY019307 |
| N. oryzae | LC 4338 | Camellia sp. | KX986008 | KY019532 | KY019349 |
| N. oryzae | LC 4961 | Pittosporum illicioides | KX986031 | KY019553 | KY019358 |
| N. oryzae | LC 5243 | Submerged wood | KX986033 | KY019555 | KY019360 |
| N. oryzae | LC 6923 | Oryza sativa L. | KX986051 | KY019581 | KY019383 |
| N. oryzae | JZB 3230002 | Phyllostachys nigra** | MN495940 | – | MN544639 |
| N. oryzae | JZB 3230003 | Rudbeckia hirta** | MN495941 | – | MN544640 |
| N. oryzae | JZB 3230004 | Scirpus sp.** | MN495942 | MN549382 | MN544641 |
| N. osmanthi | CGMCC 3.18126* = LC 4350 | Osmanthus sp. | KX986010 | KY019461 | KY019421 |
| N. osmanthi | LC 4487 | Hedera nepalensis | KX986017 | KY019540 | KY019438 |
| N. osmanthi | JZB 3230005 | Rosa chinensis** | MN495943 | MN549383 | MN508179 |
| N. osmanthi | JZB 3230006 | Rosa chinensis** | MN495944 | MN549384 | MN508180 |
| N. osmanthi | JZB 3230007 | Phragmites australis** | MN495945 | MN549385 | MN508181 |
| N. osmanthi | JZB 3230008 | Cirsium setosum** | MN495946 | MN549386 | MN508182 |
| N. osmanthi | JZB 3230009 | Phyllostachys nigra** | MN495947 | MN549387 | MN508183 |
| N. osmanthi | JZB 3230010 | Phyllostachys nigra** | MN495948 | MN549388 | MN508184 |
| N. osmanthi | JZB 3230011 | Rudbeckia hirta** | MN495949 | MN549389 | MN508185 |
| N. pyriformis | CGMCC 3.18122* = LC 2045 | Citrus sinensis | KX985940 | KY019457 | KY019290 |
| N. pyriformis | LC 2688 | Lindera aggregate | KX985941 | KY019468 | KY019297 |
| N. pyriformis | LC 2694 | Rubus reflexus | KX985945 | KY019472 | KY019300 |
| N. pyriformis | LC 3099 | Camellia sinensis | KX985971 | KY019498 | KY019322 |
| N. pyriformis | LC 3292 | Camellia sinensis | KX985976 | KY019503 | KY019324 |
| N. rubi | CGMCC 3.18326* = LC 2698 | Rubus sp. | KX985948 | KY019475 | KY019302 |
| N. rubi | JZB 3230012 | Fraxinus sp.** | MN495950 | – | MN544646 |
| N. sphaerica | LC 7312 | Nelumbo sp. (leaf) | KX985935 | KY019618 | KY019414 |
| N. sphaerica | LC 7298 | Nelumbo sp. (leaf) | KX985937 | KY019606 | KY019401 |
| N. sphaerica | LC 2840 | Harpullia longipetala | KX985965 | KY019492 | KY019318 |
| N. sphaerica | LC 3477 | Camellia sinensis | KX985982 | KY019508 | KY019326 |
| N. sphaerica | LC 4264 | Rhododendron arboretum | KX985993 | KY019517 | KY019334 |
| N. sphaerica | LC 4307 | Rhododendron arboretum | KX986005 | KY019529 | KY019346 |
| N. sphaerica | LC 5901 | Submerged wood | KX986034 | KY019556 | KY019361 |
| N. sphaerica | LC 6294 | Camellia sinensis | KX986044 | KY019565 | KY019369 |
| N. sphaerica | LC 6996 | Musa paradisiaca (leaf) | KX986085 | KY019592 | KY019390 |
| N. sphaerica | JZB 3230013 | Cirsium setosum** | MN495951 | MN549390 | MN544642 |
| N. sphaerica | JZB 3230014 | Phragmites australis ** | MN495952 | MN549391 | MN544643 |
| N. sphaerica | JZB 3230015 | Fraxinus sp.** | MN495953 | MN549392 | MN544644 |
| Nigrospora sp. 1 | LC 2725 | Symplocos zizyphoides | KX985960 | KY019487 | KY019313 |
| Nigrospora sp. 1 | LC 4566 | Lithocarpus sp. | KX986022 | KY019545 | KY019354 |
| Nigrospora sp. 2 | LC 6704 | Camellia sinensis | KX986047 | KY019571 | KY019373 |
| N. vesicularis | LC 0322 | Unknown host plant | KX985939 | KY019467 | KY019296 |
| N. vesicularis | CGMCC 3.18128* = LC 7010 | Musa paradisiaca (leaf) | KX986088 | KY019463 | KY019294 |
| N. zimmermanii | CBS 167.26 | Unknown | KY385308 | KY385318 | KY385312 |
| N. zimmermanii | CBS 290.62* | Saccharum officinarum (leaf) | KY385309 | KY385317 | KY385311 |
| N. zimmermanii | CBS 984.69 | Saccharum officinarum (leaf) | KY385310 | KY385322 | KY385316 |
| Arthrinium obovatum | LC 4940 | KY494696 | KY705166 | KY705095 | |
| Arthrinium malaysianum | CBS 102053 | NR120273 | KF144988 | KF145030 | |
CGMCC: China General Microbiological Culture Collection, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; CBS: Culture Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; JZB: Beijing Academy of Agriculture and Forestry Sciences Culture Collection, China; LC: working collection of Lei Cai, housed at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
,*Ex-type culture.
,**Novel host associations.
ITS: internal transcribed spacer region (ITS1-5.8S-ITS2); TUB2: β-tubulin; TEF1: translation elongation factor 1-α.
Sequences generated in this study are in bold type face.
ML analysis was performed using RAxML-HPC2 on XSEDE version 8.2.8 (San Diego Supercomputer Center, CA, USA) [15,16] in the CIPRES Science Gateway platform [17] using GTR + CAT model of evolution. MP analysis was performed in PAUP version 4.0b10 (Sinauer Associates, Sunderland, MA, USA) [18], with the heuristic search option. Ambiguous regions in the alignment were excluded from the analyses, and gaps were treated as missing data. The stability of generated trees was evaluated by 1000 random bootstrap replicates. Maxtrees was set to 1000 and branches of zero length were collapsed and all multiple parsimonious trees were saved. Descriptive tree statistics for parsimony (tree length [TL], consistency index [CI], retention index [RI], relative consistency index [RCI], and homoplasy index [HI]) were calculated. Differences between the trees inferred under different optimality criteria were evaluated with Kishino–Hasegawa tests (KHT) [19].
Bayesian analysis was executed in MrBayes version 3.1.2 [20] through Markov Chain Monte Carlo (MCMC) sampling to calculate the posterior probabilities (PP) [18,21]. Partitioning of data was initially done by locus and then the parameters of the nucleotide substitution models for every partition were selected independently using MrModeltest version 2.3 [22] under the Akaike information criterion (AIC) executed in PAUP version 4.0b10. The models GTR + G for ITS and HKY + I + G for TEF1 and TUB2 were set for their respective genes in the analysis. Six Markov chains were run in parallel for 3 million generations with trees being sampled at every 1000th generation. Twenty-five percent of the trees were discarded representing the burn-in phase. Generated trees were used to calculate the PP in the majority rule consensus tree. The resulting trees were viewed in FigTree version 1.4.0 (Institute of Evolutionary Biology, University of Edinburgh, UK) [23] and annotated in Adobe Illustrator CC 2017 version 21.0.0 (Adobe Systems Incorporated, Seattle, WA). All the sequence data generated in this study were deposited in NCBI GenBank (Table 3). The sequence alignment generated in this study was deposited in TreeBase under the accession number of 25396.
3. Results
3.1. Phylogenetic analysis
The combined ITS, TEF1, and TUB2 gene data set comprised 64 sequences from Nigrospora including isolates from this study. Arthrinium malaysianum (CBS 102053) and Arthrinium obovatum (LC 4940) were considered as outgroup taxa (Figure 1). The combined alignment of three gene regions was analyzed and the best scoring RAxML tree is shown in Figure 1 with a final ML optimization likelihood value of −9176.491460. The matrix had 605 distinct alignment patterns, with 8.57% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.209857, C = 0.308100, G = 0.240726, and T = 0.241318; substitution rates AC = 0.968792, AG = 2.885236, AT = 0.956737, CG = 0.911966, CT = 4.642164, and GT = 1.000000; proportion of invariable sites I = 0.401481; gamma distribution shape parameter α = 0.808089. The MP analysis with combined ITS, TEF1, and TUB2 gene data comprised 1344 total characters including gaps, of which 759 characters were constant, 498 characters were parsimony-informative, while 87 variable characters are parsimony-uninformative. In the most parsimonious tree, TL = 1621, CI = 0.570, RI = 0.907, RCI = 0.517, and HI = 0.430. The Bayesian analysis resulted in 15,000 trees after 3,000,000 generations. All trees (ML, MP, and BYPP) were similar in topology and did not differ significantly (data not shown). At the generic level, relationships are in agreement with the previous study based on multi-gene phylogeny [3]. Our phylogenetic analyses resulted in 18 clades corresponding to species in Nigrospora similar to the study conducted by Wang et al. [3]. Isolates from this study clustered within five clades corresponding to known species and thus confirmed their identities.
Figure 1.
Multilocus phylogenetic tree based on the combined ITS, TEF1, and TUB2 sequences alignment generated from a maximum likelihood phylogenetic analysis. Bootstrap support values for ML, MP (> 70%), and posterior probabilities (> 0.9) are given at the nodes (ML/MP/PP). The tree is rooted with Arthrinium malaysianum (CBS 102053) and Arthrinium obovatum (LC 4940). (*indicates the ex-type isolates).
3.2. Taxonomy
Nigrospora Zimm., Centbl. Bakt. ParasitKde, Abt. I 8:220 (1902),
Synonym: Khuskia H.J. Huds., Trans. Br. mycol. Soc. 46:358 (1963),
Nigrospora gorlenkoana Novobr., Nov. sist. Niz. Rast. 9:180 (1972),
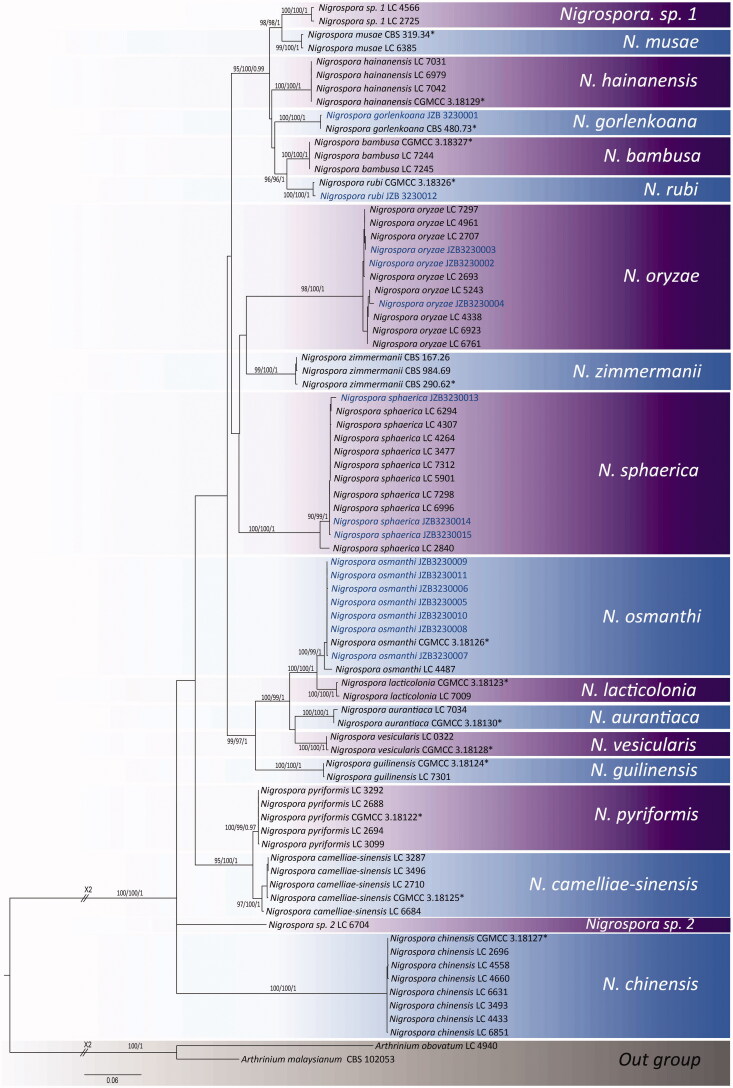
Facesoffungi number: FoF 06595 (Figure 2).
Figure 2.
Nigrospora gorlenkoana (JZB 3230001). (a and b) Appearance of leaf spots on the host substrate; (c and d) Upper view (c) and reverse view (d) of culture on PDA; (e) Conidia on aerial mycelia on PDA; (f) Mature conidia; (g–i) Mature conidia attached to conidiogenous cells. Scale bars f, g = 20 μm, h, and i = 10 μm.
Pathogenic or saprobic on leaves of Cirsium setosum (Willd.) Besser ex M.Bieb (Asteraceae). Asexual morph: Hyphae smooth, branched, septate, and hyaline. Conidiophores mostly reduced to conidiogenous cells. Conidiogenous cells 6.9–10 × 4.2–8 μm diam. (= 8.4 × 6 μm, n = 30), monoblastic, solitary, discrete, determinate, doliiform to ampulliform, and pale brown. Conidia 10.3–14 × 13.3–17.2 μm diam. (= 12.5 × 15.2 μm, n = 50), solitary, globose or oblate, dark brown to black, shiny, sparse, discrete on aerial mycelia, and smooth-walled. Sexual morph: Undetermined.
Culture characteristics – Colonies on PDA, reach 9 cm diam. after 5 d at 25 °C, circular shaped, entire margined, floccose with aerial mycelium, surface initially white, turning grayish when mature and reverse initially white, turning smoke gray when mature.
Material examined – China, Shandong Peninsula, on living leaves of Cirsium setosum, 07 October 2017, Yuanyuan Hao (JZBH 3230001), living culture JZB 3230001, and KUMCC 19-0222.
Leaf spot symptoms – Leaf spots irregularly scattered and composed of a dark brown circular outer ring with a light brown inner ring, margined by apparently healthy leaf tissues.
Notes – Based on the phylogenetic analysis of combined ITS, TEF1, and TUB2 sequence data of Nigrospora species (Figure 1), our strain Nigrospora gorlenkoana (JZB 3230001) clustered with the ex-type strain of N. gorlenkoana (CBS 480.73) with strong bootstrap support and Bayesian probabilities (100% ML, 100% MP, and 1.00 BYPP) (Figure 1). The base pair difference comparison of ITS, TEF1, and TUB2 gene regions between our strain (JZB 3230001) and ex-isotype strain of N. gorlenkoana (CBS 480.73) reveal less than 1% difference and the two specimens share similar morphological characters confirming both strains are conspecific. In contrast to the ex-type strain (CBS 480.73), an equatorial slit on conidia was not observed in our strain (JZB 3230001) [3]. Nigrospora gorlenkoana has not frequently been identified as a plant pathogen and it was previously reported to be isolated from leaves and fruits of Vitis vinifera [3]. This is the first report of Nigrospora gorlenkoana from Cirsium setosum.
Nigrospora oryzae (Berk. & Broome) Petch, J. Indian Bot. Soc. 4:24 (1924),
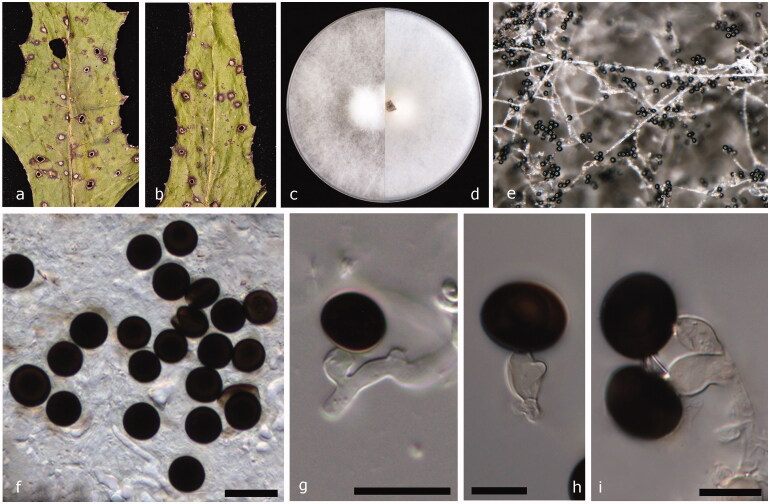
Facesoffungi number: FoF 06596 (Figure 3).
Figure 3.
Nigrospora oryzae (JZB 3230004). (a and b) Appearance of leaf spots on the host substrate; (c and d) Upper view (c) and reverse view (d) of culture on PDA; (e) Surface view of the colony on PDA; (f) Colony on PDA; (g–k) Mature conidia attached to conidiogenous cells. Scale bars f, g = 20 μm, h, and i = 10 μm.
Basionym: Monotospora oryzae Berk. & Broome, J. Linn. Soc., Bot. 14: 99 (1873) [1875]
≡ Khuskia oryzae H.J. Huds., Trans. Br. mycol. Soc. 46(3): 358 (1963)
≡ Apiospora oryzae (H.J. Huds.) Arx, Gen. Fungi Sporul. Cult., Edn 2: 129 (1974).
Pathogenic or saprobic on leaves of Scirpus sp. (Cyperaceae). Asexual morph: Hyphae smooth, branched, septate, hyaline or pale brown. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 8.6–14 × 6.4–11.9 μm diam. ( = 11.18 × 7.98 μm, n = 30), aggregated in clusters on hyphae, monoblastic, determinate, ampulliform or doliiform, and hyaline to pale brown. Conidia 9.0–13.2 × 12.6–15.8 μm diam. ( = 10.95 × 14 μm, n = 50), formed abundantly, solitary, globose or oblate, dark brown to black, shiny, smooth, and aseptate. Sexual morph: Undetermined.
Culture characteristics – Colonies on PDA reach 9 cm diam. in 6 d at 25 °C, circular, entire margined, floccose, filiform, surface and reverse initially white, becoming dark gray, or black toward the center with age.
Material examined – China, Shandong Peninsula, on living leaves of Scirpus sp., October 7 2017, Yuanyuan Hao (JZBH 3230004), living culture JZB 3230004, and KUMCC 19-0225.
Leaf spot symptoms – Randomly scattered and elliptical shaped leaf spots are composed of dark brick, slightly dispersed outer halo with light brown inner core, and margined by healthy leaf tissues.
Other materials examined – China, Shandong Peninsula, on living leaves of Phyllostachys nigra (Lodd. ex Lindl.) Munro (Poaceae), October 7 2017, Yuanyuan Hao (JZBH 3230002), living culture JZB 3230002, KUMCC 19-0223; China, Shandong Peninsula, on living leaves of Rudbeckia hirta L. (Asteraceae), October 7 2017, Yuanyuan Hao (JZBH 3230003), living culture JZB 3230003, and KUMCC 19-0224.
Notes – Nigrospora gorlenkoana and N. oryzae are reported to have the same synonym of Basisporium gallarum in Mycobank. However in our phylogenetic analysis, N. oryzae and N. gorlenkoana are placed in two distinct clades. Khuskia oryzae was introduced as the teleomorph of N. oryzae. The multi-gene phylogeny generated herein indicates that our strains of Nigrospora oryzae form a strongly supported lineage (98% ML, 100% MP, and 1.00 BYPP) in N. oryzae cluster (Figure 1). Base pair comparison of ITS, TEF1, and TUB2 gene regions between our strain (JZB 3230004) and reference strain of N. oryzae (LC 5243) reveal less than 1% difference. The morphological characters, such as conidiogenous cells, conidial dimensions, and culture characteristics also overlap confirming that the two strains are the same species [3]. This is the first time N. oryzae has been reported from Scirpus sp., which is an aquatic grass-like plant species, Phyllostachys nigra commonly known as black bamboo and Rudbeckia hirta, a garden plant belongs to the sunflower family.
Nigrospora osmanthi Mei Wang, F. Liu, P.W. Crous & L. Cai. Persoonia 39:135 (2017),
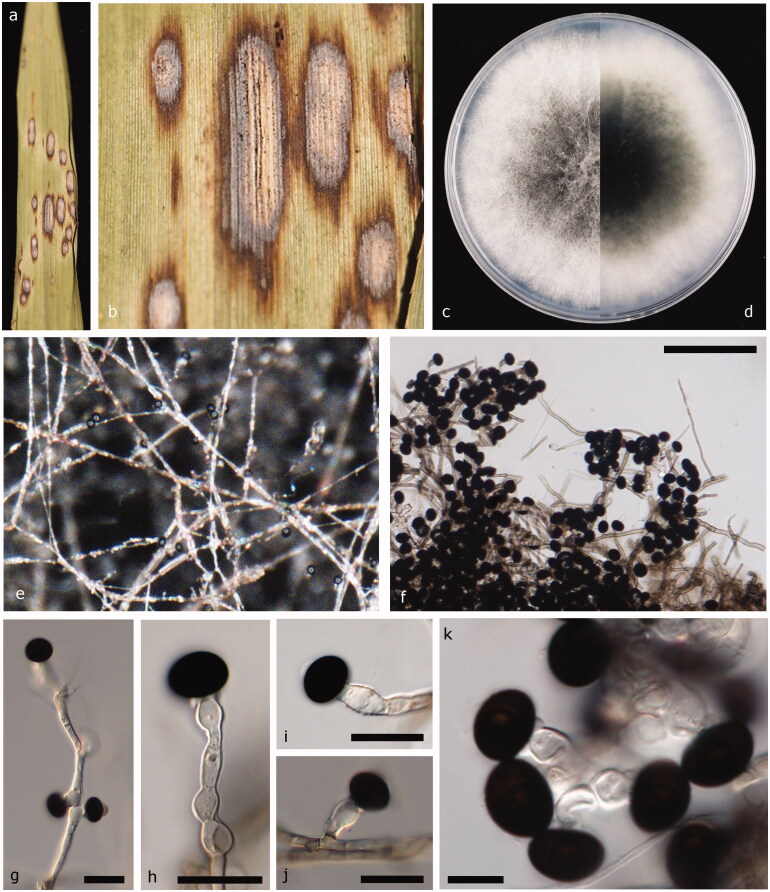
Facesoffungi number: FoF 06597 (Figure 4).
Figure 4.
Nigrospora osmanthi (JZB 3230011). (a and b) Appearance of leaf spots on host substrate; (c) Enhanced view of leaf spot on the host substrate; (d and e) Upper view (c) and reverse view (d) of culture on PDA; (f) Colony on PDA; (g–i) Mature conidia attached to conidiogenous cells. Scale bars f = 50 μm, g–i = 10 μm.
Pathogenic or saprobic on leaves of Rudbeckia hirta L. Asexual morph: Hyphae smooth, branched, septate, hyaline, or pale brown. Conidiophores mostly reduced to conidiogenous cells. Conidiogenous cells 6.8–12.6 × 5.3–7.4 μm diam. ( = 9.3 × 6.3 μm, n = 30), discrete, solitary, monoblastic, determinate, ampulliform to subglobose, straight or curved, hyaline. Conidia 9–11.5 × 12.5–14.6 μm diam. (= 10 × 13.2 μm, n = 50), discrete on aerial mycelia, solitary, globose or oblate, dark brown to black, shiny, smooth-walled, and aseptate. Sexual morph: Undetermined.
Culture characteristics – Colonies on PDA reach 9 cm diam. in 5 d at 25 °C, circular, entire margined, flat with aerial mycelium, floccose, filiform, surface initially white turning dark gray when mature and reverse initially white, and turning leek green when mature.
Material examined - China, Shandong Peninsula, on living leaves of Rudbeckia hirta L., 07 October 2017, Yuanyuan Hao (JZBH 3230011), living culture JZB 3230011, KUMCC 19-0229.
Leaf spot symptoms and characters – Irregularly scattered and free-form shaped leaf spots are composed of dark brown outer border with light brown inner core, margined by apparently healthy leaf tissues.
Other materials examined – China, Shandong Peninsula, on living leaves of Cirsium setosum, October 7 2017, Yuanyuan Hao (JZBH 3230008), living culture JZB 3230008, KUMCC 19-0227; China, Shandong Peninsula, on living leaves of Phyllostachys nigra, October 07 2017, Yuanyuan Hao (JZBH 3230009), living culture JZB 3230009, KUMCC 19-0228; China, Shandong Peninsula, on living leaves of Phragmites australis (Cav.) Trin. ex Steud. (Poaceae), October 7 2017, Yuanyuan Hao (JZBH 3230007), living culture JZB 3230007; China, Shandong Peninsula, on living leaves of Rosa chinensis Jacq. (Rosaceae), October 7 2017, Yuanyuan Hao (JZBH 3230005), living culture JZB 3230005, and KUMCC 19-0226.
Notes – Based on the phylogenetic analysis of combined ITS, TEF1, and TUB2 sequence data of Nigrospora species (Figure 1), our strains of N. osmanthi (JZB 3230005, JZB 3230006, JZB 3230007, JZB 3230008, JZB 3230009, JZB 3230010, and JZB 3230011) form a strongly supported lineage (100% ML, 99% MP, and 1.00 BYPP) with the ex-type strain N. osmanthi (CGMCC 3.18126) (Figure 1). The base pair comparison shows 100% similarity in all three gene regions of ITS, TEF1, and TUB2 between our strain (JZB 3230011) and ex-type strain (CGMCC 3.18126). The two specimens share similar morphological characters except for culture characteristics where our strain (JZB 3230011) has an entire margin and reference strain (CGMCC 3.18126) has a lobate margin [3]. This is the first time N. osmanthi has been isolated from Rudbeckia hirta L., Cirsium setosum, which is a Chinese herb, Phyllostachys nigra, Phragmites australis which is a perennial grass species found in wetlands, and Rosa chinensis.
Nigrospora rubi Mei Wang, F. Liu, P.W. Crous & L. Cai. Persoonia 39:135 (2017),
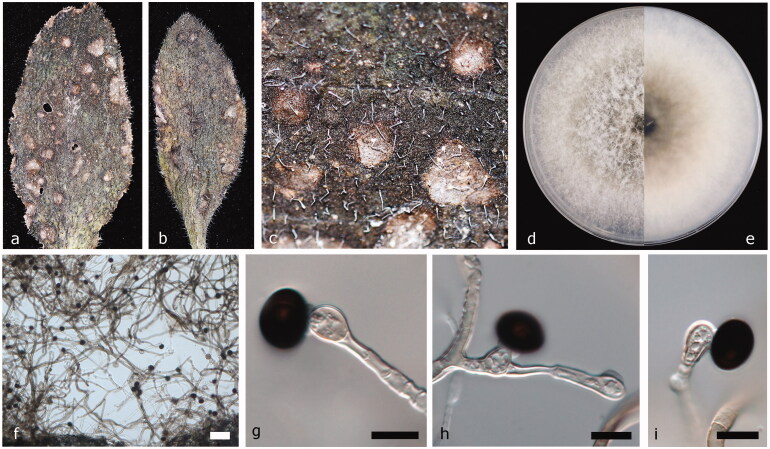
Facesoffungi number: FoF 06598 (Figure 5).
Figure 5.
Nigrospora rubi (JZB 3230012). (a and b) Appearance of leaf spots on host substrate; (c) Enhanced view of leaf spot on the host substrate; (d and e) Upper view (c) and reverse view (d) of culture on PDA; (f) Surface view of the colony on PDA; (g) Colony on PDA (h, j, and k) Mature conidia attached to conidiogenous cells; (i) Mature conidia. Scale bars g = 100 μm, h = 10 μm, i = 20 μm, j, and k = 10 μm.
Pathogenic or saprobic on leaves of Fraxinus sp. (Oleaceae). Asexual morph: Hyphae smooth, branched, septate, and hyaline. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 5.2–7.4 × 6.6–7.3 μm diam (= 6.7 × 6.9 μm, n = 30), clustered on hyphae, unbranched, ampulliform, short, and squat pale brown. Conidia 7.9–10.7 × 10–12.1 μm diam. ( = 9.58 × 11.17 μm, n = 50), solitary, spherical or subglobose, black, shiny, smooth, and aseptate. Sexual morph: Undetermined.
Culture characteristics – Colonies on PDA reach 9 cm diam. after 6 d at 25 °C, circular, entire margined, velvety to lanose, surface initially white, becoming dark olive-green to gray with age and reverse initially white, and turning leek green when mature.
Material examined – China, Shandong Peninsula, on living leaves of Fraxinus sp., October 7 2017, Yuanyuan Hao (JZBH 3230012), living culture JZB 3230012, and KUMCC 19-0242.
Leaf spot symptoms and characters – Irregularly scattered and free-form shaped leaf spots are composed of dark brick outer border with light brown inner core, margined by healthy leaf tissues.
Notes – Based on multi-locus molecular phylogeny, our isolate of N. rubi (JZB 3230012) forms a strongly supported lineage (100% ML, 100% MP, and 1.00 BYPP) with N. rubi as the type species (CGMCC 3.18326) (Figure 1) and the base pair comparison between these two strains exhibit 100% similarity in ITS and 98.8% similarity in TEF1 gene region. The TUB2 gene sequence could not be obtained for our strain (JZB 3230012). The conidial measurements were slightly larger (11.5 × 16.5 μm) in type specimen (CGMCC 3.18326), compared to our strain (JZB 3230012, 9.58 × 11.17 μm) [3]. The culture characteristics slightly deviate in color; the ex-type culture (CGMCC 3.18326) was initially white, becoming black with age and reverse smoke-gray in patches, where our strain shows initially white surface becoming dark olive-green to gray with age and initially white reverse turning leek green when mature (JZB 3230012). Nigrospora rubi has been previously isolated from Rubus species [3]. This is the first time N. rubi has been isolated from Fraxinus sp.
Nigrospora sphaerica (Sacc.) E.W. Mason, Trans. Br. Mycol. Soc. 12: 158 (1927),
Facesoffungi number: FoF 06599 (Figure 6).
Figure 6.
Nigrospora sphaerica (JZB 3230015). (a and b) Appearance of leaf spots on host substrate; (c) Enhanced view of leaf spot on the host substrate; (d and e) Upper view (c) and reverse view (d) of culture on PDA; (f) Surface view of the colony on PDA; (g) Colony on PDA; (h) Mature conidia. (i and j) Mature conidia attached to conidiogenous cells. Scale bars g = 50 μm, h = 10 μm, i = 20 μm, and j = 10 μm.
Basionym: Trichosporum sphaericum Sacc., Michelia 2 (no. 8): 579 (1882).
Pathogenic or saprobic on leaves of Fraxinus sp. Asexual morph: Hyphae smooth, branched, septate, hyaline, or pale brown. Conidiophores mostly reduced to conidiogenous cells. Conidiogenous cells 9.5–16.5 × 7.4–9.8 μm diam. ( = 12.7 × 8.4 μm, n = 30), discrete, monoblastic, determinate, unbranched, and ampulliform to subglobose hyaline to pale brown. Conidia 11.5–15.7 × 13.3–19.6 μm diam. ( = 14 × 16.7 μm, n = 50), sparse, discrete, globose or subglobose, black, shiny, smooth, and aseptate. Sexual morph: Undetermined.
Culture characteristics – Colonies on PDA reach 9 cm diam. in 5 d at 25 °C, circular, entire margined, floccose or suede-like texture, surface initially white, becoming dark gray with age and reverse initially white, and turning smoke gray when mature.
Material examined – China, Shandong Peninsula, on living leaves of Fraxinus sp., October 7 2017, Yuanyuan Hao (JZBH 3230015), living culture JZB 3230015, and KUMCC 19-0232.
Leaf spot symptoms – Leaf spots irregularly scattered and free-form shaped, composed of dark brick outer border with light brown inner core, and margined by healthy leaf tissues.
Other materials examined – China, Shandong Peninsula, on living leaves of Cirsium setosum, October 7 2017, Yuanyuan Hao (JZBH 3230013), living culture JZB 3230013, KUMCC 19-0230; China, Shandong Peninsula, on living leaves of Phragmites australis, October 7 2017, Yuanyuan Hao (JZBH 3230014), living culture JZB 3230014, and KUMCC 19-0231.
Notes – Nigrospora sphaerica is identified as a widely distributed plant pathogen on a diverse range of host species worldwide. Since the DNA sequence data of N. sphaerica type specimen was not available, Wang et al. [3] determined a collection of Nigrospora isolates from their study as N. sphaerica by comparing morphological characters of vesicular structures and conidial dimensions to the original description. In combined phylogenetic analysis, our isolates of N. sphaerica (JZB 3230013, JZB 3230014, and JZB 3230015) clustered with strong bootstrap support and posterior probability values (90% ML, 99% MP, and 1.00 BYPP). Less than 1% base pair difference was observed in base pair comparison of ITS, TEF1, and TUB2 gene regions between our strain (JZB 3230015) and reference N. sphaerica (LC 6996) strain. Also, similar morphologies were observed between the two strains confirming these two strains as conspecific. This is the first time N. sphaerica has been isolated from Fraxinus sp., Cirsium setosum and Phragmites australis.
4. Discussion
This study illustrates five different Nigrospora species isolated from various hosts in Shandong Peninsula, China. Nigrospora gorlenkoana, N. oryzae, N. osmanthi, N. rubi and N. sphaerica are reported from this study. Thirteen novel host associations (Table 3) were revealed on hosts such as Fraxinus sp., Phragmites australis, Scirpus sp. and including economically important plant varieties, such as Cirsium setosum, Phyllostachys nigra, Rosa chinensis, and Rudbeckia hirta.
Nigrospora is a monophyletic genus in Apiosporaceae (Xylariales) [3]. The phylogenetic construction of the DNA sequences of combined ITS, TEF1, and TUB2 gene regions provide robust confirmation and resolution for species delimitation by separating different species of the genus with high bootstrap support (Figure 1).
Currently, there are 15 records of Nigrospora species in MycoBank and 16 in GenBank but sequence data are not available for Nigrospora aerophila, N. arundinacea, N. canescens, N. gallarum, N. gossypii, N. javanica, N. maydis, N. padwickii, and N. panici. Therefore, epitypification of these species must be carried out and further studies based on molecular phylogeny are needed on these species.
There are few studies conducted on the fungal ecology of the Shandong peninsula. A study on aquatic fungi in China revealed various fungal species isolated from different hosts from Shandong province; Arenariomyces trifurcata Höhnk, Buergenerula spartinae J. Kohlmerer & R.V. Gessner, Corollospora maritima Werderm., Dryosphaera navigans Jørg. Koch & E.B.G. Jones, Halosphaeriopsis mediosetigera (A.B. Cribb & J.W. Cribb) T.W. Johnson, Lignincola laevis Höhnk, Monosporascus cannonballus Pollack & Uecker, Natantispora retorquens (C.A. Shearer & J.L. Crane) J. Campb., J.L. Anderson & C.A. Shearer, Pleospora betae Björl., Pleospora spartinae (J. Webster & M.T. Lucas) Apinis & Chesters, Pleospora vitalbae (De Not.) Berl., Tetraploa aristata Berk. & Broome, Torula herbarum (Pers.) Link, Torpedospora radiata Meyers, Trichocladium achrasporum Meyers & R.T. Moore) M. Dixon ex Shearer & J.L. Crane, Zalerion maritimum (Linder) Anastasiou, Zalerion varium Anastasiou from driftwood; Ceriosporopsis halima Linder from bamboo; Passeriniella obiones (P. Crouan & H. Crouan) K.D. Hyde & Mouzouras from straw; Torpedospora radiata Meyers from drift bamboo as marine Ascomycetes [24], and Nia vibrissa R.T. Moore & S. P. Meyers from driftwood as marine Basidiomycetes [24], and Alternaria maritima G.K. Sutherl. from driftwood as marine Hyphomycetes [24]. Shandong province is also famous for economically important fungal resources, 182 taxa of wild edible and medicinal fungi belong to 39 families, and 80 genera are reported [25]. Agaricus silvaticus Schaeff., Agaricus silvicola (Vittad.) Peck, Ganoderma lingzhi Sheng H. Wu, Y. Cao & Y.C. Dai, Grifola frondosa (Dicks.) Gray, Lactarius deliciosus L., Lactarius subvellereus Peck, Perenniporia fraxinea (Bull.) Ryvarden, Pholiota adipose (Batsch) P. Kumm., Schizophyllum commune Fr., Suillus bovinus (L.) Roussel, Suillus granulatus (L.) Roussel, Xerocomellus chrysenteron (Bull.) Šutara, and Xerula radicata (Relhan) Dörfelt, are among edible fungi [25]. Further, Leptosphaeria agnita (Desm.) Ces. & De Not., L. dumetorum Niessl, L. eustomoides Sacc., and L. solani Romell ex Berl. were isolated from deadwood materials as saprophytic fungi from Shandong Peninsula [26]. There are no previous records on the occurrence of Nigrospora species from the Shandong peninsula.
Among the five Nigrospora species reported in this study, N. oryzae, N. osmanthi, and N. sphaerica were recorded frequently as pathogenic on a broader range of host plants (Table 1). Even though the pathogenic behavior of N. oryzae is prominent, in most cases it is identified as a weak pathogen [27,28]. Spore dispersal of Nigrospora is aided by the wind, rain splash and insect vectors [29] supporting a rapid spread of the disease. The presence of a sticky mucilaginous substance was observed on discharged spores [30]. It has been hypothesized that this mucilaginous substance facilitates adherence to the host substrate or to a vector, such as mites as a successful spore dispersal mechanism. Since Nigrospora infections occur easily on weakened or wounded plants, spore dispersal through vectors is an added advantage on disease establishment. Nigrospora sphaerica isolated from Blueberry (Vaccinium corymbosum) leaf spots, twigs and shoot blight was identified as a pathogen that penetrates the host plant through wounds caused by insects or abiotic frost damages [31]. Previously, it was believed that Nigrospora was limited to monocotyledonous hosts [9], but later studies revealed it can occur on a diverse range of hosts and the pathogenicity of Nigrospora alerts the concerns on agronomy and forestry management. Molecular phylogeny guided species identification would be essential in developing effective bio-control measures against these species. Here, we extend the known host range of five species in Nigrospora.
Acknowledgments
Authors would like to thank funding authorities for their support. Alan JL Phillips acknowledges the support from UID/MULTI/04046/2019 Research Unit grant from FCT, Portugal to BioISI.
Funding Statement
This project was funded by the Project of Regional collaborative innovation of Beijing Academy of Agriculture and Forestry Sciences [grant No. KJCX20170709].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that supports the findings of this study are openly available in GenBank and TreeBase public repositories. The GenBank accession numbers and the TreeBase submission number are given within the article.
References
- 1.Zimmerman A. Ueber einige an tropischen Kulturpflanzen beobachtete Pilze III. Zentralblatt Für Bakteriol Parasitenkd. 1902;8:216–221. [Google Scholar]
- 2.Hyde K, Norphanphoun C, Maharachchikumbura S, et al. Refined families of Sordariomycetes. Mycosphere. 2020;11(1):305–1059. [Google Scholar]
- 3.Wang M, Liu F, Crous PW, et al. Phylogenetic reassessment of Nigrospora: ubiquitous endophytes, plant and human pathogens. Persoonia. 2017;39(1):118–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rashmi M, Kushveer J, Sarma V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere. 2019;10(1):798–1079. [Google Scholar]
- 5.Sun X, Guo L-D, Hyde KD. Community composition of endophytic fungi in Acer truncatum and their role in decomposition. Fungal Divers. 2011;47(1):85–95. [Google Scholar]
- 6.Metwaly AM, Kadry HA, El-Hela AA, et al. Nigrosphaerin A a new isochromene derivative from the endophytic fungus Nigrospora sphaerica. Phytochem Lett. 2014;7:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao JH, Zhang YL, Wang LW, et al. Bioactive secondary metabolites from Nigrospora sp. LLGLM003, an endophytic fungus of the medicinal plant Moringa oleifera Lam. World J Microbiol Biotechnol. 2012;28(5):2107–2112. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SP, Huang R, Li FF, et al. Antiviral anthraquinones and azaphilones produced by an endophytic fungus Nigrospora sp. from Aconitum carmichaeli. Fitoterapia. 2016;112:85–89. [DOI] [PubMed] [Google Scholar]
- 9.Mason EW. On species of the genus Nigro-spora Zimmermann recorded on monocotyledons. Trans Br Mycol Soc. 1927;12(2–3):152–165. [Google Scholar]
- 10.Jayasiri SC, Hyde KD, Ariyawansa HA, et al. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74(1):3–18. [Google Scholar]
- 11.Guo LD, Hyde KD, Liew E. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000;147(3):617–630. [DOI] [PubMed] [Google Scholar]
- 12.White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols. Amsterdam, Netherlands: Elsevier; 1990. p. 315–322. [Google Scholar]
- 13.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 15.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57(5):758–771. [DOI] [PubMed] [Google Scholar]
- 16.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010 gateway computing environments workshop (GCE). Piscataway (NJ): IEEE; 2010. p. 1–8. [Google Scholar]
- 18.Swofford DL. PAUP: phylogenetic analysis using parsimony [Internet]. Vol. 42. Options. Sunderland (MA): Sinauer Associates; 2002. p. 294–307. [Google Scholar]
- 19.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989;29(2):170–179. [DOI] [PubMed] [Google Scholar]
- 20.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. [DOI] [PubMed] [Google Scholar]
- 21.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol. 1996;43(3):304–311. [DOI] [PubMed] [Google Scholar]
- 22.Nylander J. MrModeltest v2. Program distributed by the author. Evol Biol Cent Uppsala Univ. 2004;2:1–2. [Google Scholar]
- 23.Rambaut A. FigTree version 1.4.0. 2012. [cited 2020 April 05]. Available from http://tree.bio.ed.ac.uk/software/figtree/
- 24.Hu DM, Liu F, Cai L. Biodiversity of aquatic fungi in China. Mycology. 2013;4(3):125–168. [Google Scholar]
- 25.Wang J, Liu Y, Bau T. Evaluation of endangered status and conservation priority of macrofungi in Shandong Province. China Acta Ecol Sin. 2015;35(3):837–848. [Google Scholar]
- 26.Chi S, Yu H, Cai C, et al. New Chinese records of saprophytic Leptosphaeria from Shandong Peninsula. Mycosystema. 2013;32(2):208–215. [Google Scholar]
- 27.Widmer T, Kirk A, Kirk G, et al. Foliar and cane rot of Arundo donax caused by Nigrospora oryzae in Europe. Plant Dis. 2006;90(8):1107–1107. [DOI] [PubMed] [Google Scholar]
- 28.Zhang LX, Li SS, Tan GJ, et al. First report of Nigrospora oryzae causing leaf spot of cotton in China. Plant Dis. 2012;96(9):1379–1379. [DOI] [PubMed] [Google Scholar]
- 29.Wu PC, Tsai JC, Li FC, et al. Increased levels of ambient fungal spores in Taiwan are associated with dust events from China. Atmos Environ. 2004;38(29):4879–4886. [Google Scholar]
- 30.Webster J. Spore projection in the Hyphomycete Nigrospora sphaerica. New Phytol. 1952;51(2):229–235. [Google Scholar]
- 31.Wright ER, Folgado M, Rivera MC, et al. Nigrospora sphaerica causing leaf spot and twig and shoot blight on blueberry: a new host of the pathogen. Plant Dis. 2008;92(1):171–171. [DOI] [PubMed] [Google Scholar]
- 32.Kee YJ, Hafifi ABM, Huda-Shakirah AR, et al. First report of reddish brown spot disease of red-fleshed dragon fruit (Hylocereus polyrhizus) caused by Nigrospora lacticolonia and Nigrospora sphaerica in Malaysia. Crop Prot. 2019;122:165–170. [Google Scholar]
- 33.Brown KB, Hyde KD, Guest DI. Preliminary studies on endophytic fungal communities of Musa acuminata species complex in Hong Kong and Australia. Fungal Divers. 1998;1:27–51. [Google Scholar]
- 34.Palmateer AJ, McLean KS, van Santen E, et al. Occurrence of Nigrospora lint rot caused by Nigrospora oryzae on cotton in Alabama. Plant Dis. 2003;87(7):873–873. [DOI] [PubMed] [Google Scholar]
- 35.Sharma P, Meena PD, Chauhan JS. First report of Nigrospora oryzae (Berk. & Broome) Petch causing stem blight on Brassica juncea in India. J Phytopathol. 2013;161(6):439–441. [Google Scholar]
- 36.Zhai LF, Liu J, Zhang MX, et al. The first report of leaf spots in Aloe vera caused by Nigrospora oryzae in China. Plant Dis. 2013;97(9):1256–1256. [DOI] [PubMed] [Google Scholar]
- 37.Wu JB, Zhang CL, Mao PP, et al. First report of leaf spot caused by Nigrospora oryzae on Dendrobium candidum in China. Plant Dis. 2014;98(7):996–996. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Pan H, Chen MY, et al. First report of Nigrospora oryzae causing brown/black spot disease of kiwifruit in China. Plant Dis. 2018;102(1):243–243. [Google Scholar]
- 39.Zheng L, Shi F, Kelly D, et al. First report of leaf spot of Kentucky Bluegrass (Poa pratensis) caused by Nigrospora oryzae in Ontario. Plant Dis. 2012;96(6):909–909. [DOI] [PubMed] [Google Scholar]
- 40.Kalati TH, Jahani M, Zare R, et al. First report of Nigrospora leaf spot on Pennisetum americanum in Iran. J Plant Pathol. 2014;96(3):606. [Google Scholar]
- 41.Abass MH. Morphological, molecular and pathological study on Nigrospora oryzae and Nigrospora sphaerica, the leaf spot fungi of date palm abstract. Basra J Date Palm Res. 2014;13(1):26–38. [Google Scholar]
- 42.Rathod D, Dar M, Gade A, et al. Griseofulvin producing endophytic Nigrospora oryzae from Indian Emblica officinalis Gaertn: a new report. Austin J Biotechnol Bioeng. 2014;1(3):1–5. [Google Scholar]
- 43.Cosoveanu A. Fungi as endophytes in Chinese Artemisia spp.: juxtaposed elements of phylogeny, diversity and bioactivity. Mycosphere. 2016;7(2):102–117. [Google Scholar]
- 44.Mei SS, Wang ZY, Zhang J, et al. First report of leaf blight on Stenotaphrum secundatum caused by Nigrospora osmanthi in China. Plant Dis. 2019;103(7):1783–1783. [Google Scholar]
- 45.Liu J, Yang L, Miao P, et al. First report of leaf blight on Ficus pandurata caused by Nigrospora osmanthi in China. Plant Dis. 2019;103(10):2685–2685. [Google Scholar]
- 46.Raviraja NS, Maria GL, Sridhar KR. Antimicrobial evaluation of endophytic fungi inhabiting medicinal plants of the Western Ghats of India. Eng Life Sci. 2006;6(5):515–520. [Google Scholar]
- 47.Zhao H, Liu HY, Yang XS, et al. First report of Nigrospora leaf blight on sesame caused by Nigrospora sphaerica in China. Plant Dis. 2014;98(6):842–842. [DOI] [PubMed] [Google Scholar]
- 48.Cui YP, Wu B, Peng AT, et al. First report of Nigrospora leaf blight on sugarcane caused by Nigrospora sphaerica in China. Plant Dis. 2018;102(4):824–824. [Google Scholar]
- 49.Dutta J, Gupta S, Thakur D, et al. First report of Nigrospora leaf blight on tea caused by Nigrospora sphaerica in India. Plant Dis. 2015;99(3):417–417. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Xiang TT, Liu XY, et al. First report of Nigrospora sphaerica causing shot hole disease on mulberry in China. Plant Dis. 2018;102(1):245. [Google Scholar]
- 51.Arunakumar GS, Gnanesh BN, Supriya M, et al. First report of Nigrospora sphaerica causing shot hole disease on Mulberry in India. Plant Dis. 2019;103(7):1783. [Google Scholar]
- 52.Abass MH, Hameed MA, Ahmed AN. First report of Nigrospora sphaerica (Sacc.) Mason as a potential pathogen on date palm (Phoenix dactylifera L.). Can J Plant Pathol. 2013;35(1):75–80. [Google Scholar]
- 53.Allen R. Control of black-end and squirter diseases in bananas with benzimidazole and salicylanilide compounds. Aust J Exp Agric. 1970;10(45):490. [Google Scholar]
- 54.Simmonds JH. Banana leaf spot progress report. Queensl Agric J. 1933;39(1):21–40. [Google Scholar]
- 55.Chen Y, Yang X, Zhang A-F, et al. First report of leaf spot caused by Nigrospora sphaerica on kiwifruit in China. Plant Dis. 2016;100(11):2326. [Google Scholar]
- 56.Li L, Pan H, Liu YF, et al. First report of Nigrospora sphaerica causing kiwifruit postharvest rot disease in China. Plant Dis. 2018;102(8):1666. [Google Scholar]
- 57.Li YG, Huang MH, Sun LP, et al. Occurrence of leaf spot of calabash caused by Nigrospora sphaerica in Georgia. Plant Dis. 2016;100(7):1506–1506. [Google Scholar]
- 58.Liu YJ, Tang Q, Fang L. First report of Nigrospora sphaerica causing leaf blight on Camellia sinensis in China. Plant Dis. 2016;100(1):221. [Google Scholar]
- 59.Xu YM, Liu YJ. First report of Nigrospora sphaerica causing leaf blight on Cunninghamia lanceolata in China. Plant Dis. 2017;101(2):389. [Google Scholar]
- 60.Alam MW, Rehman A, Gleason ML, et al. First report of Nigrospora sphaerica causing leaf spot of Kinnow mandarin in Pakistan. J Plant Pathol. 2017;99(1):295. [Google Scholar]
- 61.Alam MW, Rehman A, Ahmad S, et al. First report of Nigrospora sphaerica causing leaf spot of Kinnow mandarin in Pakistan. J Plant Pathol. 2020;102(1):223–223. [Google Scholar]
- 62.Pandey A, Pandey S, Awasthi AK, et al. A new host record of Nigrospora sphaerica on Mangifera indica from Jabalpur, India. J Mycol Plant Pathol. 2013;43(2):255–256. [Google Scholar]
- 63.Wu SH, Chen YW, Shao SC, et al. Two new Solanapyrone analogues from the endophytic fungus Nigrospora sp. YB-141 of Azadirachta indica. Chem Biodivers. 2009;6(1):79–85. [DOI] [PubMed] [Google Scholar]
- 64.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556. [Google Scholar]
- 65.O’Donnell K, Kistler HC, Cigelnik E, et al. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA. 1998;95(5):2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that supports the findings of this study are openly available in GenBank and TreeBase public repositories. The GenBank accession numbers and the TreeBase submission number are given within the article.