Abstract
Penicillium vietnamense sp. nov. was isolated from Nha Trang Bay, Vietnam in June 2017. It is phylogenetically distinct from the sister species of Penicillium section Charlesia series Indica based on multi-locus sequence typing results using internal transcribed spacer, large subunit ribosomal RNA, β-tubulin, calmodulin, and RNA polymerase II second largest subunit regions. It showed strong growth on Czapek yeast autolysate agar at 37 °C, a strong acid production on Creatine sucrose agar, and produced short stipes, small vesicles, and subglobose to globose conidia delicately roughened with very short ridges. As the first novel marine fungi species described from Vietnam and discovered in a unique environment, the data could be significant for understanding the taxonomy and geographical distribution of marine fungi in tropical coastal systems such as Vietnam.
Keywords: DNA sequencing, marine fungi, morphology, phylogeny, scanning electron microscopy
1. Introduction
Historically, Johann Heinrich Friedrich Link firstly introduced the generic name Penicillium, meaning “brush” and classified the genus in the order Eurotiales within the family Trichocomaceae in 1809 [1]. It is now more than 200 years with many changes in the name and number of species. Penicillium was then redefined as part of the family Aspergillaceae [2] and contained 354 accepted species from a current systematic study [3]. Penicillium now become one of the most common fungi with global distribution in diverse environments and has a big economic and social impact [3]. Current reviews have shown that marine-derived Penicillium has provided numerous valuable pharmaceuticals with a variety of biological activities such as antimicrobial, cytotoxic, and anticancer properties [4–8]. Its species are identified based on the combination of morphological features including colony pattern, conidiophore structure, and sclerotia production, and molecular characteristics with sequence of internal transcribed spacer (ITS) region and additional markers such as large subunit ribosomal RNA (LSU) [9], β-tubulin (BenA), calmodulin (CaM) [10] and the RNA polymerase II second largest subunit (RPB2) [3].
In our current study on fungi diversity and community composition of the surface coastal marine and deeper waters at Nha Trang Bay and Van Phong Bay in Vietnam, a collection of marine fungi strains have been isolated and identified, which belong to 3 phyla, 5 subdivisions, 7 classes, 12 orders, 17 families, 22 genera and at least 40 species [11]. Among 29 identified fungal species, 12 and 28 species were new records in a global marine source and in a marine ecosystem of Vietnam, respectively.
This study describes one of the unidentified Penicillium strains in our collection isolated from Nha Trang Bay, located in the province of Khanh Hoa, the southern maritime area of Central Vietnam. The Bay has had the Hon Mun marine-protected area since 2001, with nine islands covering approximately 16,000 ha [12]. The living condition here is ideal for most marine organisms in the tropics such as warm water, the temperature from 23 °C in January up to 28 °C in May–June, and salinity from 3.2% to 3.4% [13,14]. The Bay has high diverse ecosystems with unique coral reefs linked to the open sea and attached to many valuable marine micro- and macro-organisms with diverse ecological functions and valuable bioactive compounds [13–17]. With marine fungi from Nha Trang Bay screened morphologically and molecularly, we decided that our fungus represents an undescribed species, which is described and illustrated here as Penicillium vietnamense sp. nov.
2. Materials and methods
2.1. Isolation of marine fungi
The deep waters (DW) at the depth of 40–47 m (N: 12°18′31′′, E: 109°31′67′′), pH 7.0, the temperature of 22 °C and salinity of 3.44%, were collected from Nha Trang Bay, Vietnam on June 1, 2017. Fungi were isolated within 3 h of collection using the membrane filtration technique [18]. Briefly, the DW samples were diluted into concentrations at 10−1, 10−2, and 10−3. Subsequently, diluted water samples (15 ml, triplicate) were filtered through a sterile 0.45 μm cellulose esters membrane (MilliporeSigma, Burlington, Massachusetts, USA). These membranes were then placed on solid media plates of Sabouraud Dextrose Agar (SDA) (Thermo Fisher Scientific, Waltham, Massachusetts, USA) supplemented with antibiotics (0.075% streptomycin sulfate and 0.05% ampicillin) (Thermo Fisher Scientific) to suppress bacterial growth. The plates were incubated at 25 °C for 2–3 days and examined daily for the growth of fungi. Fungal colonies that developed were subcultured onto fresh Potato Dextrose Agar (PDA) (Thermo Fisher Scientific) and incubated at 25 °C for 7 days to allow fungal growth. The isolate DW14M was deposited in Vietnam Type Culture Collection (VTCC), Microbiological Culture Collection at Nha Trang University (NTU), and Nha Trang Institute of Technology Innovation and Application (NITIA), Vietnam with allotted no. VTCC 930029, NTU DW14M, and NITIA DW14M, respectively.
2.2. Morphological analysis
The isolate DW14M was cultured on PDA and then transferred to Malt extract agar (MEA, Oxoid), Czapek yeast autolysate agar (CYA), Yeast extract sucrose agar (YESA), Czapek yeast autolysate agar with 5% NaCl (CYAS), Czapek’s agar (CZ), Oatmeal Agar (OA), and Creatine sucrose agar (CREA) for morphological analysis [3]. Plates were incubated at 25 °C in the dark for 7 days, and plates with CYA were additionally incubated at 30 °C and 37 °C in the dark for 7 days. After incubation, diameters, the density of sporulation, obverse and reverse colony colors, and the existence of soluble pigments were recorded. Fungal morphological characterization was identified by using a light microscope (Eclipse 80i, Nikon, Tokyo, Japan) and a scanning electron microscope (SEM, S-4800, Hitachi, Tokyo, Japan) [3,19]. With 85% lactic acid and 99% ethanol, fixed specimen images were acquired [20].
2.3. DNA extraction, PCR, and sequencing
The isolate DW14M was grown on PDA and incubated at 25 °C for 3–5 days. Mycelia were harvested, lyophilized, and crushed in a mortar with a pestle using liquid nitrogen to a fine powder. Genomic DNA from the isolate was extracted using the EZ-10 Spin Column Plant Genomic DNA Miniprep Kit (Bio Basic, Canada) as directed by the manufacturer. The extracted DNA samples were stored at −20 °C until use for PCR. Five regions, internal transcribed spacer (ITS), large subunit ribosomal RNA (LSU), β-tubulin (BenA or tub2), calmodulin (CaM), and RNA polymerase II second largest subunit (RPB2), were amplified with the primer pairs in Table 1. The PCR method was performed as described previously [17,26] with some modifications. Briefly, one microliter of DNA (∼25 ng) was added to a 50 μl reaction volume containing 1 μl of Taq polymerase (Bioline, Memphis, Tennessee, USA), 10 μl of 5X MyTaq reaction buffer, 35 μl of distilled deionized water, and 10 pmoles of each primer. The PCR program was run for the initial denaturation step at 93 °C for 3 min, followed by 35 cycles of 0.5 min at 93 °C, 0.5 min at 55–60 °C, and 0.5 min at 72 °C, and a final extension at 72 °C for 5 min. The amplified products were separated on a 1% (w/v) agarose gel stained with ethidium bromide and visualized under a UV transilluminator.
Table 1.
Phylogenetic loci and PCR primers used in the present study.
| Locus | Primer name | Direction | Primer sequence (5′–3′) | Reference |
|---|---|---|---|---|
| Internal transcribed spacer (ITS) | ITS1-F_KYO2 | Forward | TAG AGG AAG TAA AAG TCG TAA | [21] |
| ITS4-R | Reverse | TCC TCC GCT TAT TGA TAT GC | [22] | |
| Large subunit ribosomal RNA (LSU) | NL1 | Forward | GCA TATC AAT AAG CGG AGG AAA AG | [23] |
| NL4 | Reverse | GG TCC GTG TTT CAA GAC GG | ||
| β-tubulin (BenA) | Bt2a | Forward | GGT AAC CAA ATC GGT GCT GCT TTC | [24] |
| Bt2b | Reverse | ACC CTC AGT GTA GTG ACC CTT GGC | ||
| Calmodulin (CaM) | CF1 | Forward | GCC GAC TCT TTG ACY GAR GAR | [10] |
| CF4 | Reverse | TTT YTG CAT CAT RAG YTG GAC | ||
| RNA polymerase II second largest subunit (RPB2) | 5Feur | Forward | GAY GAY CGK GAY CAY TTC GG | [25] |
| 7CReur | Reverse | CCC ATR GCY TGY TTR CCC AT |
PCR products were purified using the QIAquick PCR purification Kit (Qiagen, Hilden, Germany), and then Sanger sequenced at Macrogen (Korea) in both directions with the same PCR primers as described above using Big Dye terminator in a 3730xl DNA Analyzer (Applied Biosystems, Waltham, Massachusetts, USA). The DNA sequences generated in this study are deposited in GenBank under the accession numbers MT102836 (ITS), MT209882 (LSU), MT230561 (BenA), ON209438 (CaM), and MT222288 (RPB2) to the isolate DW14M.
2.4. Phylogenetic analysis
DNA sequences of the isolate DW14M obtained in this study and reference sequences available in GenBank (Table 2) were used for sequence analysis at the National Center for Biotechnology Information (NCBI) using BLASTn (http://www.ncbi.nlm.nih.gov/BLAST). DNA sequences were aligned using ClustalW [27], and regions with gaps were removed using BioEdit. Model selection was used to determine the best fit model with the lowest Bayesian Information Criterion score for the Maximum-likelihood, Minimum-evolution, and Neighbor-joining method [28], which was then used to construct a phylogenetic tree using the MEGA X program [26]. The robustness of the tree topology was tested by bootstrap analysis with 1000 re-samplings [29]. The evolutionary distances were computed using the Tamura 3-parameter method [30] and Tamura–Nei method [31].
Table 2.
Details of the strains used in phylogenetic and identity analyses.
| Species | Strain | Sequence accession numbers |
||||
|---|---|---|---|---|---|---|
| ITS | LSU | BenA | CaM | RPB2 | ||
| Penicillium vietnamense | VTCC 930029 = DW14M | MT102836 | MT209882 | MT230561 | ON209438 | MT222288 |
| Penicillium chermesinum | CBS 231.81 | AY742693 | MH873092 | KJ834441 | AY741728 | JN406581 |
| Penicillium lunae | PPRI 25881 | MK450725 | MK598746 | MK451088 | MK451660 | MK450863 |
| Penicillium cuddlyae | PREM 623302 | MK951942 | MN388754 | MK951835 | MK951908 | MN418450 |
| Penicillium indicum | CBS 115.63 | AY742699 | AY742699 | EU427263 | AY741744 | JN406640 |
| Penicillium charlesii | CBS 304.48 | AF033400 | JX091508 | AY741754 | JN121486 | |
| Penicillium fellutanum | CBS 229.81 | AF033399 | KJ834450 | AY741753 | JN121460 | |
| Penicillium coffeae | CBS 119387 | AY742702 | KJ834443 | AY741747 | JN121436 | |
| Penicillium phoeniceum | CBS 249.32 | KC411711 | KJ834483 | AY741729 | JN406597 | |
| Penicillium costaricense | CBS 140998 | KT887873 | KT887834 | KT887795 | MN969173 | |
| Penicillium eremophilum | CBS 123361 | GU733341 | KY709170 | KY611931 | KY611970 | |
| Aspergillus oryzae | NRRL 447 | EF661560 | EF661483 | EF661506 | EF661438 | |
| Penicillium sp. | CBS 140613 | KX961204 | KX961227 | KX961262 | KX961286 | |
| Penicillium sp. | CBS 140614 | KX961205 | KX961228 | KX961263 | KX961288 | |
| Penicillium sp. | CGMCC 3.18173 | KX961206 | KX961229 | KX961264 | KX961287 | |
| Penicillium chermesinum | CMV011D8 | MK450679 | MK451202 | MK451596 | MK450829 | |
3. Results
3.1. Phylogenetic analysis
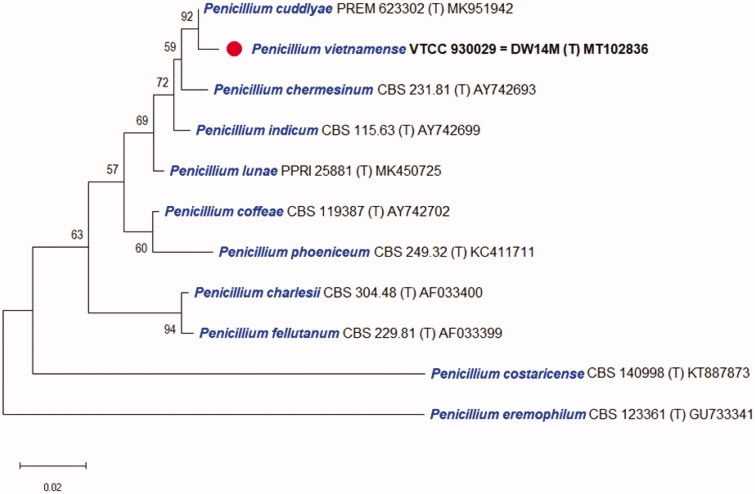
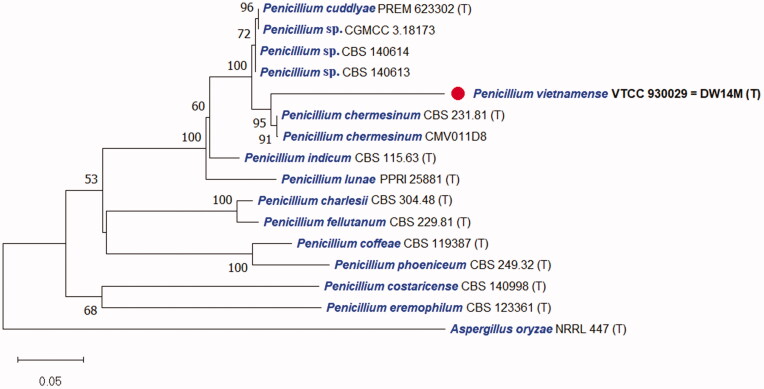
The DNA sequences of the new species were registered in the GenBank database of the NCBI (MT102836 for rDNA–ITS, MT209882 for LSU, MT230561 for BenA, ON209438 for CaM, and MT222288 for RPB2) (Table 2). The target strain DW14M was positioned using the sequences of the ITS region (Figure 1) and the combined sequences of ITS, BenA, CaM, and RPB2 regions (Figure 2). Based on these regions, the target strain was compared with other strains available in the NCBI database. The phylogenetic trees were constructed following the bootstrap analysis of 1000 replicates, and DW14M was the most similar to the type strains of Penicillium chermesinum, Penicillium cuddlyae, Penicillium lunae, and Penicillium indicum within the Penicillium section Charlesia series Indica [32]. In addition, BLASTn search was performed against the strain DW14M. ITS and RPB2 sequence analyses showed that DW14M was the most similar to the type strain of P. cuddlyae with 99.7% and 97.7% sequence identity, respectively (Table 3). LSU sequence analysis showed that DW14M was the most similar to the type strain of P. lunae with 99.5% sequence identity (Table 3). BenA and CaM gene sequence analyses showed that DW14M was the most similar to the type strain of P. chermesinum with 99.1% and 98.8% sequence identity, respectively (Table 3). Sequence analysis with a combination of four DNA regions (ITS, BenA, CaM, RPB2) (Table 2) showed that DW14M was the most similar to P. chermesinum with 91% sequence identity, P. cuddlyae with 90%, P. indicum with 88%, and P. lunae with 86% (data not shown).
Figure 1.
Phylogenetic tree based on the neighbor-joining analysis of the ITS sequences for Penicillium species classified in the Charlesia section. Penicillium eremophilum as the most related to the Charlesia section was included as an outgroup. Bootstrap analysis was performed with 1000 replications with values of at least 50% indicated at the nodes. The bar indicates the number of substitutions per position. Each branch indicates taxon name, strain name and GenBank accession number sequentially. (T) indicates the type strain of the species. The isolate in this study is marked as ( ).
).
Figure 2.
Phylogenetic tree based on the neighbor-joining analysis of the combined ITS, BenA, CaM, and RPB2 dataset for Penicillium species classified in the Charlesia section. Penicillium eremophilum was added as the most related species to the Charlesia section. Aspergillus oryzae was included as an outgroup. Bootstrap analysis was performed with 1000 replications with values of at least 50% indicated at the nodes. The bar indicates the number of substitutions per position. Each branch indicates taxon name and strain name sequentially. (T) indicates the type strain of the species. The isolate in this study is marked as ( ).
).
Table 3.
BLASTn search against the strain DW14M (VTCC 930029) compared with the closest relatives and type strains in Genbank based on the ITS, LSU, BenA, CaM, and RPB2 sequences.
| DNA sequence name | GenBank accession No. | Closest relatives and type strains (T) | GenBank accession no. | Identity (%) | Coverage (%) |
|---|---|---|---|---|---|
| ITS | MT102836 | Penicillium sp. CGMCC 3.18173 | KX961206 | 99.5 | 99.8 |
| Penicillium sp. CBS 140614 | KX961205 | 99.5 | 99.8 | ||
| Penicillium sp. CBS 140613 | KX961204 | 99.5 | 99.8 | ||
| Penicillium cuddlyae PPRI 26355 (T) | NR168823 | 99.7 | 97.6 | ||
| Penicillium lunae PPRI 25881 (T) | NR168788 | 97.8 | 99.8 | ||
| Penicillium indicum NRRL 3387 (T) | NR121311 | 98.2 | 94.2 | ||
| Penicillium chermesinum CBS 231.81 (T) | AY742693 | 98.0 | 94.2 | ||
| LSU | MT209882 | Penicillium lunae PPRI 25881 (T) | MK598746 | 99.5 | 100 |
| Penicillium gerundense CBS 179.81 (T) | MH873084 | 99.0 | 100 | ||
| Penicillium chermesinum NRRL 2048 (T) | AY742693 | 99.1 | 99.5 | ||
| Penicillium indicum NRRL 3387 (T) | AY742699 | 99.0 | 99.5 | ||
| Penicillium cuddlyae PPRI 26355 (T) | NG067917 | 100 | 95.7 | ||
| BenA | MT230561 | Penicillium chermesinum CMV011D8 | MK451202 | 99.1 | 100 |
| Penicillium chermesinum A1S4-D39 | KJ767035 | 99.1 | 100 | ||
| Penicillium chermesinum A1S4-D3 | KJ767033 | 99.1 | 100 | ||
| Penicillium chermesinum CBS 231.81 (T) | KJ834441 | 99.1 | 91.4 | ||
| Penicillium cuddlyae CMV016A6 (T) | MK951835 | 95.7 | 94 | ||
| CaM | ON209438 | Penicillium chermesinum NRRL 2048 (T) | AY741728 | 98.8 | 100 |
| Penicillium cuddlyae CMV016A6 (T) | MK951908 | 97.6 | 97.9 | ||
| RPB2 | MT222288 | Penicillium chermesinum CMV011D8 | MK450829 | 99.7 | 99.8 |
| Penicillium cuddlyae CMV016A6 (T) | MN418450 | 97.7 | 97.7 | ||
| Penicillium chermesinum CBS 231.81 (T) | MN969111 | 99.7 | 94 |
3.2. Morphological feature
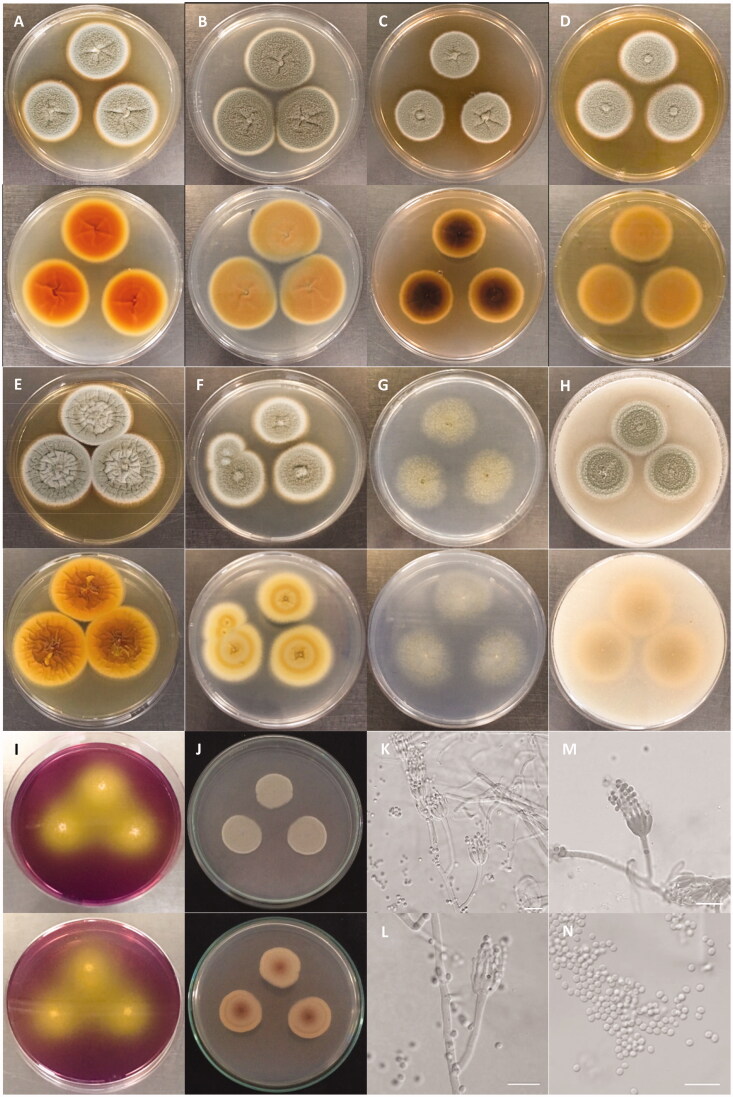
The colony of various plates and the photomicrographs of morphological structures of the DW14M strain are shown in Figures 3 and 4. The detailed fungal morphological descriptions are in the Taxonomy section. Distinct morphological features between P. vietnamense and its related species are summarized in Table 4.
Figure 3.
Characteristics of Penicillium vietnamense (VTCC 930029 = DW14M) grown on different media. Colony on (A) CYA 25 °C; (B) CYA 30 °C; (C) CYA 37 °C; (D) MEA 25 °C; (E) YESA 25 °C; (F) CYAS 25 °C; (G) CZ 25 °C; (H) OA 25 °C; (I) CREA 25 °C; (J) PDA 25 °C (top: obverse, bottom: reverse). (K–M) Conidiophores; (N) Conidia (Scale bar = 10 μm in (K)–(N)).
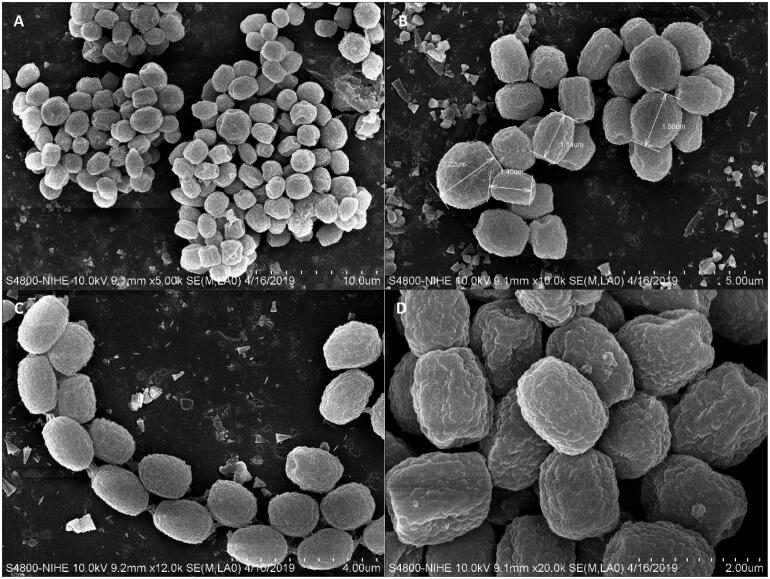
Figure 4.
(A–D) Conidial morphology of Penicillium vietnamense (VTCC 930029 = DW14M) using scanning electron microscopy. Scale bar = 10 μm in (A). Scale bar = 5 μm in (B). Scale bar = 4 μm in (C). Scale bar = 2 μm in (D).
Table 4.
Morphological comparison between Penicillium vietnamense and its closest species.
| Morphological features | Penicillium vietnamense | Penicillium chermesinum | Penicillium cuddlyae | Penicillium indicum | Penicillium lunae |
|---|---|---|---|---|---|
| Type strain | VTCC 930029 = NTU DW14M = NITIA DW14M | NRRL 2048 = CBS 231.81 | PPRI 26355 = CMV016A6 | CBS 115.63 = NRRL 3387 | PPRI 25881 = CMV006E6 |
| Colonies on CYA 7 days, 25/30/37 °C | 33–35/37–39/27–28 mm | Growth at 37 °C | 24–26/31–33/19–21 mm | Growth at 37 °C | 34–36/28–29 mm/no growth |
| Colonies on CYA with 5% NaCl (CYAS) 7 days, 25 °C | 32–35 mm | 19–20 mm | 33–35 mm | ||
| Colonies on CREA 7 days, 25 °C | 19–21 mm, strong acid production | 12–14 mm, no acid production | |||
| Sclerotia production | No | No | No | Yes | No |
| Conidiophores | Monoverticillate, miner proportion biverticillate | Strictly monoverticillate with no branched structures observed | Monoverticillate | Almost entirely monoverticillate and only occasionally showing a branch | Monoverticillate, miner proportion biverticillate |
| Stipes | Smooth-walled, 14–31 × 1.5–3.1 μm | Smooth-walled, mostly 20–40 (−50) × 2.0–2.5 µm | Smooth-walled, 20–45 × 2–3 μm | Smooth or finely roughened, 50–100 × 2.0–2.5 μm | Smooth-walled, 13–60 × 2–3 (−3.5) μm |
| Vesicle | 3.5–4.0 μm wide | 4.0–4.5 µm wide | 5–6 μm wide | 5–7 μm; metulae two when present, 18–30 × 2–3(−3.5) μm | |
| Phialides | Ampulliform, usually 8–15 per vesicle, 9 − 11 × 1.7–2.8 μm | Sterigmata crowded, often incurved, usually 10–15 per vesicle, 6–8 × 2.0–2.5 µm | Ampulliform, 10–20 per vesicle, 8–10 × 2–3 μm (9 ± 0.7 × 2.6 ± 0.2) | Sterigmata mostly in compact clusters, up to 12 or 15 per vesicle, 7–9 × 2.0–2.5 µm | Ampulliform, 10–20 per vesicle, (7.5) 8–10 × 2–3 μm (8.8 ± 0.8 × 2.5 ± 0.4); average length metula/phialide 2.5 µm |
| Conidia | Smooth-walled (light microscopy), delicately roughened with very short ridges (scanning electron microscopy), subglobose to globose, 1.9–2.5 × 2.1–2.7 µm | Smooth, elliptical, 2.0–2.5 × 1.5–2.0 µm, appearing faintly green in mass. | Smooth-walled, ellipsoid, often almost appearing cylindrical, 2–3 × 2.5–2 μm (2.5 ± 0.2 × 1.8 ± 0.2), average length/width = 0.73, n = 54 | Smooth, elliptical to subglobose, 2.0–2.5 (−3.0) µm, appearing slightly green under the microscope | Smooth-walled, subglobose to broadly ellipsoid, 2–3 (−3.5) × 1.5–2 (−2.5) μm (2.2 ± 0.4 × 1.8 ± 0.2), average width/length = 1.2, n = 70 |
| Reference | This study | [1] | [33] | [34,35] | [36] |
3.3. Taxonomy
Penicillium vietnamense V.D. Nguyen & T.T. Pham, sp. nov. (Figures 3 and 4)
Typus: VTCC 930029 = NTU DW14M = NITIA DW14M
Mycobank: MB840587
Etymology: The name refers to the country where the type specimen was collected (Vietnam)
DNA barcodes: ITS MT102836, LSU MT209882, BenA MT230561, CaM ON209438, and RPB2 MT222288.
Colony diameter (mm), 7 days, 25 °C (unless stated otherwise): CYA 33–35; CYA 30 °C 37–39; CYA 37 °C 27–28; MEA 32–33; YESA 40–41; CYAS 32–35; CREA 19–21; CZ 30–32; OA 28–30; PDA 38–42.
Colony characteristics: On CYA 25 °C, 7 days. Colonies nearly circular, low, radially sulcate, raised centrally; margins low, narrow (1 mm), white, entire; mycelia white to gray; texture floccose; sporulation sparse, conidial color en masse gray-green; soluble pigments absent; exudates clear to light brown; reverse orange with light yellow margins (Figure 3(A)). On CYA 30 °C, 7 days: Colonies similar to those on CYA 25 °C, 7 days but larger scale, exudates clear; reverse light brown (Figure 3(B)). On CYA 37 °C, 7 days: Colonies similar to those on CYA 25 °C, 7 days but smaller scale, exudates light brown; reverse blackish brown (Figure 3(C)). On MEA 25 °C, 7 days: Colonies nearly circular, low, plain, concentrically sulcate, raised centrally; margins low, narrow (1–2 mm), white, entire; mycelia white to gray; texture floccose; sporulation sparse, conidial color en masse gray-green; soluble pigments absent; exudates clear; reverse light brown with light yellow margins (Figure 3(D)). On YESA 25 °C, 7 days: Colonies nearly circular, low, radially and concentrically sulcate, raised centrally; margins low, narrow (1 mm), white, entire; mycelia gray, bright gray at center; texture velutinous and floccose; sporulation sparse, conidial color en masse gray-green; soluble pigments absent; exudates clear to light brown; reverse orange to pale yellow (Figure 3(E)). On CYAS 25 °C, 7 days: Colonies nearly circular, concentrically sulcate, raised centrally; margins low, wide (2–3 mm), white, entire; mycelia gray; texture floccose; sporulation sparse, conidial color en masse gray-green; soluble pigments absent; exudates clear; reverse milk-white with white margins (Figure 3(F)). On CZ 25 °C, 7 days: Colonies nearly circular, raised centrally; margins absent, entire; mycelia light gray; texture floccose; sporulation sparse, conidial color en masse yellowish green; soluble pigments absent; exudates clear; reverse white (Figure 3(G)). On OA 25 °C, 7 days: Colonies nearly circular, concentrically sulcate, raised centrally; margins low, wide (2–3 mm), light white, entire; mycelia gray; texture floccose; sporulation sparse, conidial color en masse gray-green; soluble pigments absent; exudates clear; no sclerotia; reverse milk-white to yellowish orange with white margins (Figure 3(H)). On CREA 25 °C, 7 days: Colonies moderate growth, acid production strongly present with color reaction from purple to yellow (Figure 3(I)). On PDA 25 °C, 7 days: Colonies nearly circular, concentrically sulcate, low, plain; margins low, narrow (1 mm), white, 2/3 entire; mycelia gray-green; texture floccose; soluble pigments absent; exudates clear; reverse light yellow, reddish-brown at the center (Figure 3(J)).
Micromorphology: Conidiophores monoverticillate, miner proportion biverticillate; stipes smooth-walled, 13.5–31.1 × 1.5–3.1 μm; vesicle 3.5–4.0 μm wide; phialides ampulliform, usually 8–15 per vesicle, 9.3–10.8 × 1.7–2.8 μm; conidia smooth-walled (light microscopy), delicately roughened with very short ridges (scanning electron microscopy), subglobose to globose, joined into chains, 1.9–2.5 × 2.1–2.7 μm.
Type strain: VTCC 930029 = NTU DW14M = NITIA DW14M, isolated from the deep waters (N: 12°18′31′′, E: 109°31′67′′) at the depth of 40–47 m, pH 7.0, the temperature of 22 °C and salinity of 3.44% in Nha Trang Bay, Khanh Hoa province, Vietnam, June 1, 2017, collector V.D. Nguyen. The culture is preserved in Vietnam Type Culture Collection (VTCC) in Hanoi, Vietnam, as well as in Microbiological Culture Collections at Nha Trang University (NTU) and Nha Trang Insitute of Technology Innovation and Application (NITIA) in Nha Trang city, Khanh Hoa province, Vietnam. Molecular markers for the species are MT102836 for rDNA-ITS, MT209882 for large subunit ribosomal RNA, MT230561 for β–tubulin, ON209438 for calmodulin, and MT222288 for RNA polymerase II second largest subunit.
Note: BLAST search against a type reference sequence dataset placed the new species in the Penicillium section Charlesia series Indica [32]. An ITS-based single phylogeny and a multigene phylogeny based on ITS, BenA, CaM, and RPB2 resolve P. vietnamense as sister to P. chermesinum, P. indicum, and the recently described species P. cuddlyae [33] and P. lunae [36]. All markers LSU, ITS, BenA, CaM, and RPB2 distinguish these species. Morphologically, P. vietnamense is the only of the five that can produce subglobose to globose (vs ellipsoid to subglobose) conidia delicately roughened with very short ridges under scanning electron microscopy [37]. It usually displays shorter stipes (14–31 vs up to 40–100 µm), smaller vesicle (3.5–4 vs 4–7 μm wide), and longer phialides (9–11 µm vs 6–8 (P. chermesinum), 7–9 (P. indicum), 8–10 µm (P. cuddlyae and P. lunae) ). While P. lunae can not grow on CYA at 37 °C after 7 days [36], P. cuddlyae can grow with colony size at 19–21 mm [33] and the new species can even grow more rapidly at 27–28 mm in the same condition. Penicillium cuddlyae displays weak growth and no acid production on CREA but the new species has moderate growth and strong acid production. Sclerotia are produced in P. indicum only, but not in the remaining four species.
Additional strains studied: Penicillium spp. strains CGMCC 3.18173, CBS 140614, and CBS 140613, China, Beijing, indoor air, May 2014, Chen, A.J., Sun, B.D., Houbraken, J., Frisvad, J.C., Yilmaz, N., Zhou, Y.G. and Samson, R.A. (unpublished data but available on NCBI Nucleotide), which shared the most similar to DW14M with 99.5% identity (ITS) (Table 3), distinguished with DW14M on the phylogenetic tree based on combined ITS, BenA, CaM, and RPB2 regions and thus they can belong to P. cuddlyae or a novel species (Figure 2).
4. Discussion
With four DNA sequences (ITS, CaM, BenA, and RPB2), phylogenetic analysis was processed to study the relationship of Penicllium section Charlesia [3,32]. The construction of the phylogenetic tree was done with two kinds of versions: a single ITS region and a combination of ITS, BenA, CaM, and RPB2 regions. The result for individual and combined markers showed a close relationship of P. vietnamense with P. chermesinum, P. cuddlyae, P. lunae, and P. indicum as confirmed by results of Neighbor-joining phylogenetic trees (Figures 1 and 2). It has been known that ITS works well as an official barcode for placing a Penicillium species into one of the 32 sections but only sometimes provide a species identification [3,32]. However, in this study, ITS is good enough for distinguishing all closely related species within the section Charlesia and with P. eremophilum as the most similar species to the section [32]. The phylogenetic tree based on the combination with additional markers such as BenA, CaM, and RPB2 agrees well with that based on single marker ITS but provides better resolution. The results also confirm the molecular phylogeny of Penicllium section Charlesia with new series and species included recently [32]. In this study, P. vietnamense is added as the 10th member of the section Charlesia which includes the series Costaricensia (Penicillium costaricense), series Fellutana (Penicillium charlesii and Penicillium fellutanum), series Indica (P. chermesinum, P. cuddlyae, P. indicum, P. lunae, P. vietnamense), and series Phoenicea (Penicillium coffeae and Penicillium phoeniceum).
Along with molecular characteristics, Penicillium species could be distinguished according to the morphology of the conidiophores, stipes, vesicle, phialides, and conidia, as well as growth temperature [3]. In the result of comparison with Penicillium sister species, P. vietnamense was morphologically similar to all other species within the section Charlesia series Indica. The colonies grow moderately fast or spread with conidial color en masse dull green or gray-green. The conidiophores are mostly monoverticillate, conspicuously vesiculate, and smooth. The stipes are smooth-walled, mostly 2–3 μm wide. The vesicle ranges from 3.5 to 7 μm wide. The phialides are mostly ampulliform, 10–20 per vesicle, ranging 6–11 × 2–3 μm. The conidia are subglobose to ellipsoidal, smooth-walled under light microscopy, mostly 1.5–3 μm in size.
However, not like the section Charlesia series Indica sister species, P. vietnamense has a unique conidiophore structure that composes of subglobose to globose conidia delicately roughened with very short ridges under scanning electron microscopy, as well as shorter stipes (14–31 µm), smaller vesicle (3.5–4.0 μm) but longer phialides (9–11 µm). It grows rapidly on CYA at 37 °C, whereas others can grow moderately and P. lunae not. It also displays a strong acid production on CREA while this activity is not found or not determined in others yet. White to cream sclerotia are produced in P. indicum, but this structure was not observed in P. vietnamense and its other sister species.
On extrolite profile, P. chermesinum is reported to produce costaclavin [38], potential ribotoxin proteins [39], chermesinones and terphenyllins [40], penicilliumolides and PR-toxins [41], and chermesins [42]. Some of them come from diverse marine origins such as mangrove endophytic fungus [40] and marine algal-derived endophytic fungus [42] in the South China Sea. As the most similar species to P. chermesinum and the first novel marine fungi species described from Vietnam, P. vietnamense is expected to express a unique extrolite profiling and ecological function in tropical marine ecosystems.
The roles of marine fungi are diverse but their ecological functions are still open questions and unsolved problems, resulting in a lack of understanding of their ecology [43]. Especially, the diversity of marine fungi in tropical coastal systems such as Vietnam has been not fully discovered yet. Our current studies have shown that marine fungi at Nha Trang Bay and Van Phong Bay in Vietnam belong to at least 40 species, of which 29 species have been identified and several species are likely novel [11]. Among unidentified species, the strain DW14M isolated from deep waters at Nha Trang Bay is described in this study as a novel Penicillium species. The strain DW14M has shown an ability to degrade protein with moderate protease activity but no activities of amylase, phytase, cellulase, and chitinase were monitored (data not shown). Other Penicillium isolates from marine sediments collected in Vietnam were found to express diverse secondary metabolites [44], thus novel species become an invaluable source of novel active secondary metabolites possessing various biological activities. In addition, along with Candida, Aspergillus, and Cladosporium, Penicillium was among the most dominant genera in both surface coastal marine and deeper waters at Nha Trang Bay, which suggested the contribution of marine saprotrophic ascomycetes fungi to the degradation of organic materials in marine ecosystems [11]. Therefore, the present study contributes to a comprehensive understanding of the taxonomy, phylogenetics, morphology, and geographical distribution of Penicillium species in tropical coastal systems such as Vietnam.
Acknowledgment
The authors acknowledge Tran Chau Loan at Nha Trang University (NTU) for technical support.
Funding Statement
This work was financially supported by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number [106-NN.02-2016.70] to T.T.P.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Biourge P. Les moissisures du groupe Penicillium link. La Cellule. 1923;33:7–331. [Google Scholar]
- 2.Samson RA, Yilmaz N, Houbraken J, et al. . Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70(1):159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visagie CM, Houbraken J, Frisvad JC, et al. . Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014;78:343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou X, Zhang X, Xue M, et al. . Recent advances in sorbicillinoids from fungi and their bioactivities (covering 2016-2021). J Fungi. 2022;8(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klas KR, Kato H, Frisvad JC, et al. . Structural and stereochemical diversity in prenylated indole alkaloids containing the bicyclo[2.2.2]diazaoctane ring system from marine and terrestrial fungi. Nat Prod Rep. 2018;35(6):532–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, Chen S, Pang X, et al. . Natural products from mangrove sediments-derived microbes: structural diversity, bioactivities, biosynthesis, and total synthesis. Eur J Med Chem. 2022;230:114117. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Su M, Song SJ, et al. . Marine-derived Penicillium species as producers of cytotoxic metabolites. Mar Drugs. 2017;15(10):329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Li Y, Zhang X, et al. . Structural diversity and biological activities of the cyclodipeptides from fungi. Molecules. 2017;22(12):2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson SW. 2000. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson RA, Pitt JI, editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam (The Netherlands): Harwood Academic Publishers; p. 163–178. [Google Scholar]
- 10.Peterson SW. Multilocus DNA sequence analysis shows that Penicillium biourgeianum is a distinct species closely related to P. brevicompactum and P. olsonii. Mycol Res. 2004;108(4):434–440. [DOI] [PubMed] [Google Scholar]
- 11.Pham TT, Dinh VK, Nguyen VD.. Biodiversity and enzyme activity of marine fungi with 28 new records from the tropical coastal ecosystems in Vietnam. Mycobiology. 2021;49(6):559–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Callaghan B. 2008. Vietnam: a collaborative approach to MPA development. Gland (Switzerland): The World Conservation Union. [Google Scholar]
- 13.Dung LD. Nha Trang Bay marine protected area, Vietnam: initial trends in coral structure and some preliminary linkages between these trends and human activities (2002–2005). Aquat Ecosyst Health Manag. 2009;12(3):249–257. [Google Scholar]
- 14.Girich EV, Yurchenko AN, Smetanina OF, et al. . Neuroprotective metabolites from Vietnamese marine derived fungi of Aspergillus and Penicillium genera. Mar Drugs. 2020;18(12):608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latypov YY. Some data on the composition and structure of coral communities in the Littoral and sublittoral in the province of Khanh Hoa, Vietnam. J Mar Sci Res Dev. 2014;4:1. [Google Scholar]
- 16.Nguyen VD, Le MH, Trang ST.. 2013. Application of probiotics from marine microbes for sustainable marine aquaculture development. In: Kim S-K, editor. Marine microbiology: bioactive compounds and biotechnological applications. Weinheim (Germany): Wiley; p. 307–349. [Google Scholar]
- 17.Pham TT, Ho THN, Nguyen VD.. Screening for bacteriocin-like antimicrobial activity against shrimp pathogenic vibrios and molecular identification of marine bacteria from otter clam Lutraria philippinarum. Thai J Vet Med. 2014;1:345–353. [Google Scholar]
- 18.Li L, Singh P, Liu Y, et al. . Diversity and biochemical features of culturable fungi from the coastal waters of Southern China. AMB Express. 2014;4:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitt JI. 1979. The genus Penicillium, and its teleomorphic states eupenicillium, and talaromyces. London (UK): Academic Press. [Google Scholar]
- 20.Choi DH, You YH, Lee IS, et al. . Penicillium ulleungdoense sp. nov. from Ulleung Island in Korea. Mycobiology. 2020;49(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toju H, Tanabe AS, Yamamoto S, et al. . High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One. 2012;7(7):e40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White TJ, Bruns T, Lee S.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky TJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press Inc.; p. 315–322. [Google Scholar]
- 23.Kurtzman CP, Robnett CJ.. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5' end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35(5):1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass NL, Donaldson GC.. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houbraken J, Spierenburg H, Frisvad JC.. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek. 2012;101(2):403–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Li M, et al. . MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, et al. . Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M.. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol. 1992;9:678–687. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Nei M.. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. [DOI] [PubMed] [Google Scholar]
- 32.Houbraken J, Kocsubé S, Visagie CM, et al. . Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crous PW, Wingfield MJ, Lombard L, et al. . Fungal planet description sheets: 951–1041. Persoonia. 2019;43:223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thom C. Cultural studies of species of Penicillium. USDA Bur Animal Ind Bull. 1910;118:1–107. [Google Scholar]
- 35.Sandhu DK, Sandhu RS.. A new species of Penicillium isolated from sputum. Can J Bot. 1963;41(8):1273–1274. [Google Scholar]
- 36.Crous PW, Carnegie AJ, Wingfield MJ, et al. . Fungal planet description sheets: 868–950. Persoonia. 2019;42:291–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez AT, Calvo MA, Ramirez C.. Scanning electron microscopy of Penicillium conidia. Antonie Van Leeuwenhoek. 1982;48(3):245–255. [DOI] [PubMed] [Google Scholar]
- 38.Agurell SL. Costaclavine from Penicillium chermesinum. Experientia. 1964;20(1):25–26. [DOI] [PubMed] [Google Scholar]
- 39.Hwu L, Cho C-J, Tzeanz S.. Nucleotide sequence and the action of ribotoxin gene (sar gene) of Penicillium isolates from Taiwan. Bot Bull Acad Sinica. 2001;42:101–107. [Google Scholar]
- 40.Huang H, Feng X, Xiao Z, et al. . Azaphilones and p-terphenyls from the mangrove endophytic fungus Penicillium chermesinum (ZH4-E2) isolated from the South China sea. J Nat Prod. 2011;74(5):997–1002. [DOI] [PubMed] [Google Scholar]
- 41.Darsih C, Prachyawarakorn V, Wiyakrutta S, et al. . Cytotoxic metabolites from the endophytic fungus Penicillium chermesinum: discovery of a cysteine-targeted Michael acceptor as a pharmacophore for fragment-based drug discovery, bioconjugation and click reactions. RSC Adv. 2015;5(86):70595–70603. [Google Scholar]
- 42.Liu H, Li X-M, Liu Y, et al. . Meroterpenoids with a drimane-type spirosesquiterpene skeleton from the marine algal-derived endophytic fungus Penicillium chermesinum EN-480. J Nat Prod. 2016;79(4):806–811. [DOI] [PubMed] [Google Scholar]
- 43.Amend A, Burgaud G, Cunliffe M, et al. . Fungi in the marine environment: open questions and unsolved problems. mBio. 2019;10(2):e01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le H, Do Q, Doan M, et al. . Chemical composition and biological activities of metabolites from the marine fungi Penicillium sp. isolated from sediments of Co To Island, Vietnam. Molecules. 2019;24(21):3830. [DOI] [PMC free article] [PubMed] [Google Scholar]