Abstract
Pancreatic cancer is one of the leading causes of cancer deaths, with pancreatic ductal adenocarcinoma (PDAC) being the most common subtype. Advanced stage diagnosis of PDAC is common, causing limited treatment opportunities. Gemcitabine is a frequently used chemotherapeutic agent which can be used as a monotherapy or in combination. However, tumors often develop resistance to gemcitabine. Previous studies show that the proto-oncogene PIM kinases (PIM1 and PIM3) are upregulated in PDAC compared to matched normal tissue and are related to chemoresistance and PDAC cell growth. The PIM kinases are also involved in the PI3K/AKT/mTOR pathway to promote cell survival. In this study, we evaluate the effect of the novel multikinase PIM/PI3K/mTOR inhibitor, AUM302, and commercially available PIM inhibitor, TP-3654. Using five human PDAC cell lines, we found AUM302 to be a potent inhibitor of cell proliferation, cell viability, cell cycle progression, and phosphoprotein expression, while TP-3654 was less effective. Significantly, AUM302 had a strong impact on the viability of gemcitabine-resistant PDAC cells. Taken together, these results demonstrate that AUM302 exhibits antitumor activity in human PDAC cells and thus has the potential to be an effective drug for PDAC therapy.
Introduction
Pancreatic cancer is the seventh leading cause of cancer deaths worldwide and the third leading cause of cancer deaths in the United States and its predicated to become the second leading cause of death by 2030 [1, 2]. Pancreatic cancer has a poor prognosis, with a 5-year survival rate of just 12% [1, 3]. This low survival rate is caused by several factors, of which perhaps the most important is the prevalence of late-stage diagnoses, with 80% of patients having locally advanced or metastatic pancreatic cancer at the time of diagnosis [4]. Current treatment options include surgical resection, chemotherapy, and radiotherapy [4]. However, only 10%-20% of patients are eligible for curative resection [5] and tumors often show resistance to chemotherapy and radiotherapy [6, 7]. Gemcitabine is a widely used chemotherapeutic agent against locally advanced and metastatic pancreatic cancer [8–10]. Although pancreatic cancer is most receptive to gemcitabine than other anticancer agents, many patients develop resistance within weeks of starting the treatment [11]. Multiple studies showed that reactivation and/or deregulation of SHH, PI3K/AKT, MEK, WNT, and NOTCH signaling pathways impact cell cycle and apoptosis and, in combination with disruption of gemcitabine metabolism leads to development of gemcitabine resistance [12–18]. Recent studies showed that remodeling of gemcitabine metabolism pathway and targeting apoptotic machinery provide promising results [8, 19–21].
Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, is genetically heterogeneous driven by mutations in oncogenes and tumor suppressor. K-Ras mutations are major drivers of PDAC development and progression and are identified in 90% of PDAC cases [22–25]. Mutations in tumor-suppressor genes such as CDKN2A, TP53, or SMAD4, and in oncogenes ERBB2 and EGFR, and in signaling pathways genes and genes regulating metabolism accelerate the formation and progression of pancreatic lesions [23, 24, 26, 27]. Dysregulation of multiple signaling pathways allows tumor cells to resist cell death, increase angiogenesis, invasion and metastasis, modify metabolism to nutrient- and oxygen-deficient environment, and remodel tumor-promoting immune response [25, 28–30].
Proviral integration site for moloney murine leukemia virus kinases (PIMs) are serine/threonine kinases that promote cell survival by regulating the cell cycle, cell proliferation, apoptosis, and transcription [31–33]. The PIM family consists of three members, PIM1, PIM2, and PIM3, from which PIM1 and PIM3 have been shown to be upregulated in solid cancers, while PIM2 mostly in hematological cancers [33–36]. PIM1 is upregulated in primary pancreatic tumor tissue compared to matched normal tissue in PDAC patients due to hypoxic environment and has been identified as prognostic marker [37, 38]. A study by Li and colleagues found PIM3 abundantly expressed in pancreatic cancer tissue but not in normal pancreatic tissue [32]. The overexpression of the PIM kinase family is related to chemotherapy and radiotherapy resistance with PIM3 expression specifically acting as a prognostic indicator related to poor patient survival [39]. PIM1 increases the stability of c-Myc through phosphorylation and together they promote cell cycle progression [40]. Multiple studies demonstrated that selective PIM inhibitors reduce phosphorylation levels of ribosomal protein S6 and thus, modulate the translation potential of numerous cancer cell lines [41–45]. PIM1 and PIM3 can phosphorylate pro-apoptotic BAD at Ser-112 to deactivate it and thereby promote cancer cell survival and progression [40]. Importantly, studies show that inhibition of PIM1 or PMI3 in PDAC cells reduces growth, invasion, and chemosensitizes the cells to gemcitabine treatment, respectively [37, 46]. Furthermore, PIM kinases interact with the PI3K/AKT/mTOR pathway to drive cancer cell proliferation and survival [31, 47]. The regulation of mTOR signaling by PIM can also affect mTOR outputs such as S6 kinase affecting cell growth and metabolism [48]. Consequently, PIM kinases are appropriate targets for cancer therapy through the use of PIM kinase inhibitors.
TP-3654 is a second generation small-molecule PIM kinase inhibitor that has been studied in vitro in several cancers, including pancreatic cancer, and is currently being used in a Phase I first-in-human study in patients with advanced solid tumors [49, 50]. The compound AUM302 is a novel triple PIM/PI3K/mTOR inhibitor that has been shown to induce apoptosis and decrease cell viability in prostate cancer [51]. Co-targeting of PIM and PI3K/AKT/mTOR pathways may be a useful approach as these kinases share several downstream targets such as p21, p27, and BAD [47]. Importantly, PI3K pathway has been implicated as one of the mechanisms of gemcitabine resistance in PDAC and targeting its activity provided potential path to sensitize the cells to gemcitabine [13, 52–56]. The aim of this study is to determine the efficacy of AUM302 in comparison with TP-3654 and gemcitabine in inhibiting pancreatic cancer cell lines growth. Here, we show that AUM302, a novel triple kinase PIM/PI3K/mTOR inhibitor, decreases proliferation of pancreatic cancer cell lines in vitro. Moreover, we showed that AUM302 sensitized pancreatic cancer cells’ response to gemcitabine treatment.
Materials and methods
Cell lines and compounds
PDAC cell lines BxPC-3 (CRL-1687), Capan-2 (HTB-80), MIA PaCa-2 (CRL-1420), PANC-1 (CRL-1469), and Hs766T (HTB-134) were purchased from ATCC (Manassas, VA) in 2020 and 2021. BxPC-3 cells were maintained in RPMI-1640 medium and Capan-2 cells in McCoy’s medium. MIA PaCa-2, Hs766T and PANC-1 cells were maintained in DMEM medium. All media were supplemented with 10% FBS and 1% penicillin/streptomycin. MIA PaCa-2-Gemcitabine (MIA PaCa-2 GemR) resistant cells were a gift from the laboratory of Dr. Lee M. Graves (Department of Pharmacology, School of Medicine, the University of North Carolina at Chapel Hill, Chapel Hill, NC, USA). (MIA PaCa-2 GemR) cells were grown in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin and 50 nM gemcitabine. All cells were maintained at 37°C and 5% CO2. All experiments were performed on cells with passage range between 5 and 29. TP-3654, GDC-0941, BEZ235, and gemcitabine were purchased from Selleck Chemicals (Houston, TX) and AUM302 was provided by AUM Biosciences. TP-3654, AUM302, GDC-0941, BEZ235 and gemcitabine were suspended in DMSO.
Cell viability assay
BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cells were seeded at 1 x 10³ cells per well in 100 μl of appropriate media in 96-well plate format. Twenty-four hours post seeding, cells were treated with DMSO (vehicle) and variable concentrations of TP-3654, AUM302, GDC-0941, BEZ235 and gemcitabine for 72 hours. MIA PaCa-2 GemS and MIA PaCa-2 GemR cell lines were seeded at 1 x 10³ cells per well in 100 μl of appropriate media in 96-well plate format, treated with 10 nM, 100 nM, or 1 μM of TP-3654 or AUM302 for 72 hours. Cell viability was analyzed using the Cell Titer-Glo luciferase assay system (Promega; Madison, WI), according to the manufacturer’s protocol, and a SpectraMax M3 plate reader (Molecular Devices; San Jose, CA). The IC50 values were calculated using GraphPad Prism for Windows version 10.0.2 (GraphPad Software) [57–60].
Cell proliferation assay
BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cells were seeded at 7.5 x 10⁴ cells per well in 2 ml of appropriate media in 6-well plate format. Twenty-four hours after seeding, cells were treated with DMSO (vehicle) or TP-3654 (10 nM and 100 nM) or AUM302 (10 nM and 100 nM). MIA PaCa-2 GemR cell line was seeded as mentioned above, and then treated with 10 nM, 100 nM, or 1μM of TP-3654 or AUM302. Cell count was determined using the Z-Series Coulter Counter (Beckman Coulter; Indianapolis, IN) after 24, 48, and 72 hours of treatment. Each experiment was performed in triplicate. The measurement of the control (cells with medium and DMSO) was defined as 100% and the results from other measurements were calculated accordingly [57–60].
Cell cycle assay
BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cells were seeded at 7.5 x 10⁴ cells per well in 2 ml of appropriate media in 6-well plate format. Twenty-four hours after seeding, cells were treated with DMSO (vehicle) or TP-3654 (100 nM) or AUM302 (100 nM). Cells were stained with propidium iodide and analyzed by FACS analysis using Cytoflex PC after 24, 48, and 72 hours of treatment. Each experiment was performed in triplicate. The measurement of the control (cells with medium and DMSO) was defined as 100% and the results from other measurements were calculated accordingly [59, 60].
Western blot analysis
BxPC-3, Capan-2, MIA PaCa-2, PANC-1, Hs766T, MIA PaCa-2 GemR pancreatic cancer cells were seeded at 7.5 x 10⁴ cells per well in 2 ml of appropriate media in 6-well plate format. Twenty-four hours post seeding, the first five cell lines were treated with DMSO (vehicle) or TP-3654 (10 and 100 nM) or AUM302 (10 and 100 nM) for 72 hours. MIA PaCa-2 GemR cells were treated with 10 nM, 100 nM, and 1 μM of TP-3654 or AUM302 for 72 hours. Cells were lysed in Laemmli buffer and total protein extracts were subjected to electrophoresis in 4–20% or 10% tris-glycine gels. The proteins were then transferred to a nitrocellulose membrane, blocked in 5% non-fat milk in 1 x TTBS buffer, and developed with appropriate antibodies. Protein bands were detected using an enhanced chemiluminescence detection kit using Azure c400 (Azure Biosystems). Densitometry analysis of western blots was performed using FIJI software [61].
Statistical analysis
The analysis was performed using appropriate statistical test with a value of p < 0.05 considered significant. This analysis was performed using GraphPad Prism for Windows version 10.0.2 (GraphPad Software).
Results
AUM302 is a potent inhibitor of pancreatic cancer cell lines growth in vitro
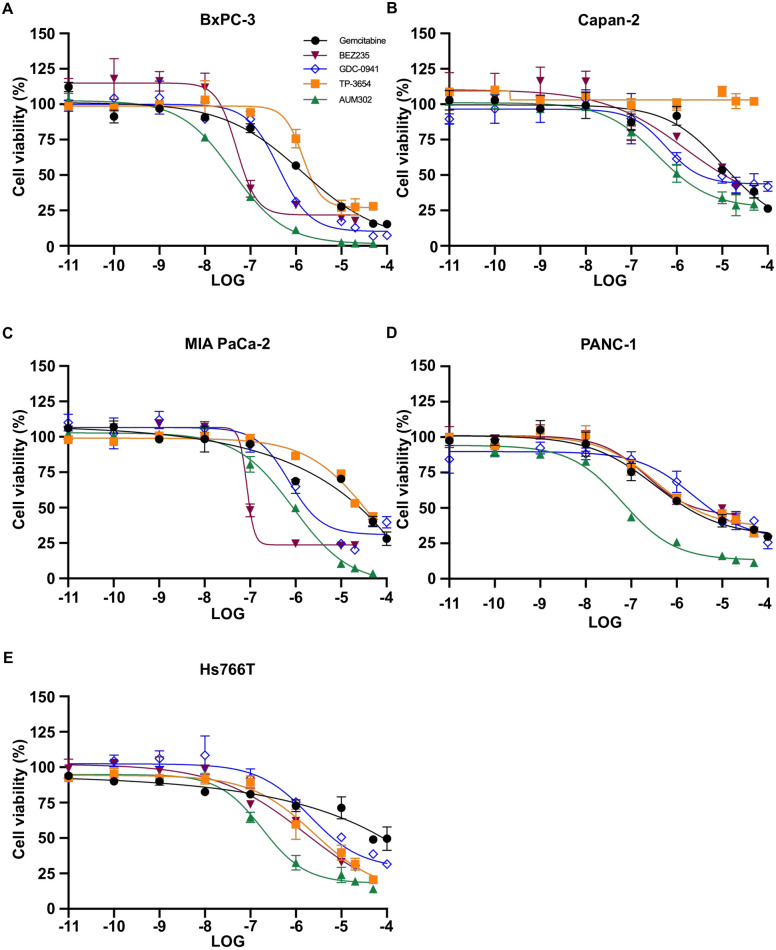
To assess the efficacy of gemcitabine, TP-3654, and AUM302 on the viability of pancreatic cancer cell lines, we performed cell proliferation and growth assays using BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cells treated with variable concentrations of these compounds for 72 hours. Furthermore, we treated these cell lines with two PI3K/mTOR inhibitors, GDC-0941 and BEZ235, to establish whether AUM302 or TP-3654, triple PIM/PI3K/mTOR inhibitors have higher efficacy of growth inhibition than dual-inhibitors [62–64]. We determined IC50 values using the Cell Titer-Glo assay (Fig 1 and Table 1). Our results showed that all five compounds inhibit the viability of tested pancreatic cancer cell lines. However, as shown in Table 1, the AUM302 compound has more favorable IC50 values as compared to gemcitabine, GDC-0941, and BEZ235 in BxPC-3, Capan-2, PANC-1, and Hs766T pancreatic cancer cell lines with AUM302 compound being even more effective than TP-3654. Only in MIA PaCa-2 cell line (Fig 1C, Table 1) are IC50 values for BEZ235 and GDC-0941 lower than AUM302. Further studies compared the effectiveness of two triple PIM/PI3K/mTOR inhibitors: TP-3654 and AUM302.
Fig 1. Gemcitabine, BEZ235, GDC-0941, TP-3654, and AUM302 inhibit viability of multiple pancreatic cancer cell lines.
Pancreatic cancer cell lines BxPC-3 (A), Capan-2 (B), MIA PaCa-2 (C), PANC-1 (D), and Hs766T (E) were treated with variable concentrations of gemcitabine or BEZ235 or GDC-0941 or TP-3654 or AUM302 twenty-four hours after seeding. Cells were treated with test compounds for 72 hours and cell viability was measured using Cell Titer-Glo. Each experiment was performed in triplicate and the results are shown as mean ±SD (N = 3).
Table 1. Experimental analysis of IC50 values of gemcitabine, BEZ235, GDC-0941, TP-3654, and AUM302 tested in BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cell lines.
| Cell line | Gemcitabine | BEZ235 | GDC-0941 | TP-3654 | AUM302 |
|---|---|---|---|---|---|
| BxPC-3 | 1375 ± 1.59 | 49.96 ± 1.29 | 418.8 ± 1.18 | 1418 ± 1.38 | 41.12 ± 1.07 |
| Capan-2 | 11080 ± 1.99 | 1517 ± 12.42 | 576 ± 1.44 | Unstable | 376 ± 1.25 |
| MIA PaCa-2 | 5.40E+09* | 84.84* | 713.4 ± 1.29 | 40390 ± 3.46 | 891.2 ± 1.16 |
| PANC-1 | 341.1 ± 1.38 | 214.6 ± 1.32 | 2308 ± 1.60 | 333.4 ± 1.38 | 65.6 ± 1.17 |
| Hs766T | 1.57E+12* | 1653 ± 3.38 | 2021 ± 1.52 | 2494 ± 1.71 | 182.2 ± 1.16 |
The values are expressed in nanomolar concentrations as mean with ± standard error.
*—standard error of mean (SEM) LogIC50 value > 12.
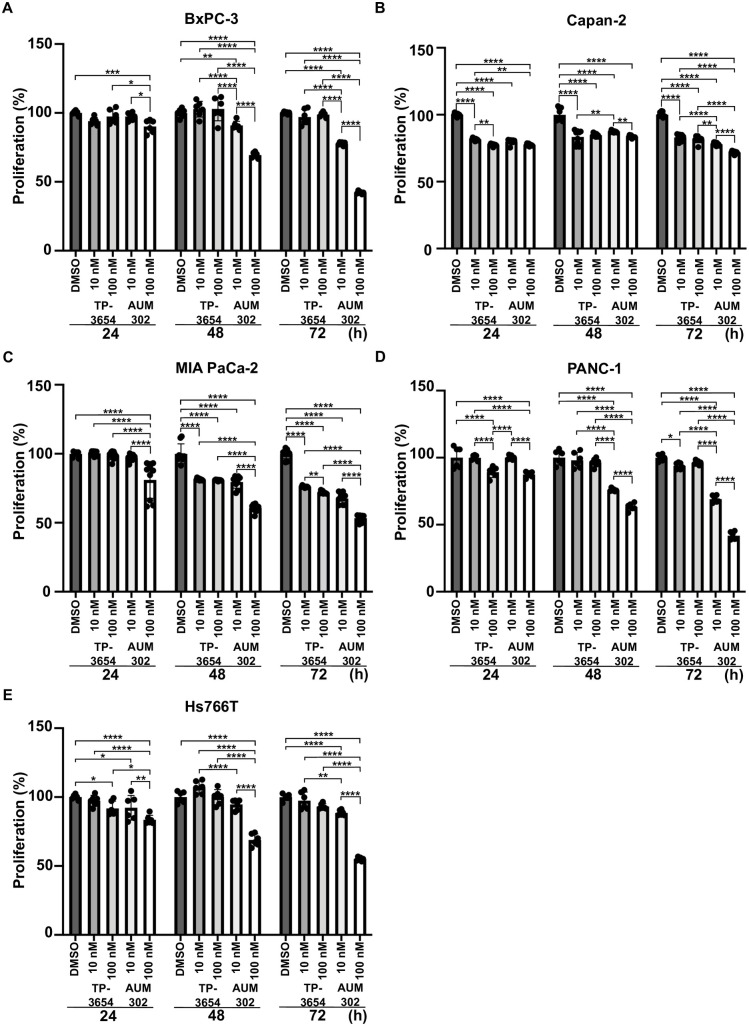
To examine the impact of TP-3654 and AUM302 on the growth of pancreatic cancer cell lines, we performed cell proliferation assay using previously tested pancreatic cancer cell lines. As shown in Fig 2, the two compounds, each tested at 10 nM and 100 nM, significantly inhibited proliferation and growth of pancreatic cancer cells over the course of three-day treatment in comparison to DMSO (vehicle)-treated cells. Additionally, analysis showed that AUM302 demonstrated a robust inhibitory effect, not only in comparison with vehicle but also in comparison to TP-3654 treatment in all tested pancreatic cancer cell lines (Fig 2).
Fig 2. AUM302 inhibits the proliferation of multiple pancreatic cancer cell lines.
The following pancreatic cancer cell lines, BxPC-3 (A), Capan-2 (B), MIA PaCa-2 (C), PANC-1 (D), and Hs766T (E), were treated with DMSO or TP-3654 (10 nM and 100 nM), or AUM302 (10 nM and 100 nM). Cell count was determined 24, 48, and 72 hours after treatment using a cell counter. The measurement of the control (cells with DMSO) was defined as 100%. Data represent mean ±SD (N = 6). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 calculated with two-way ANOVA.
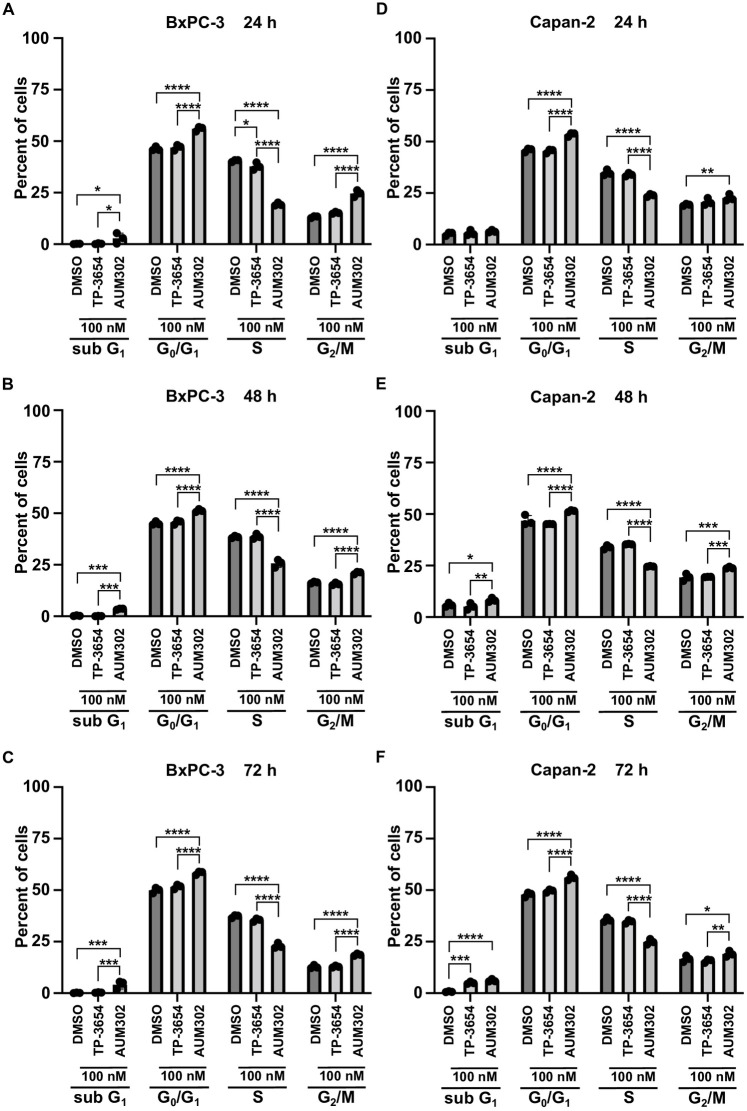
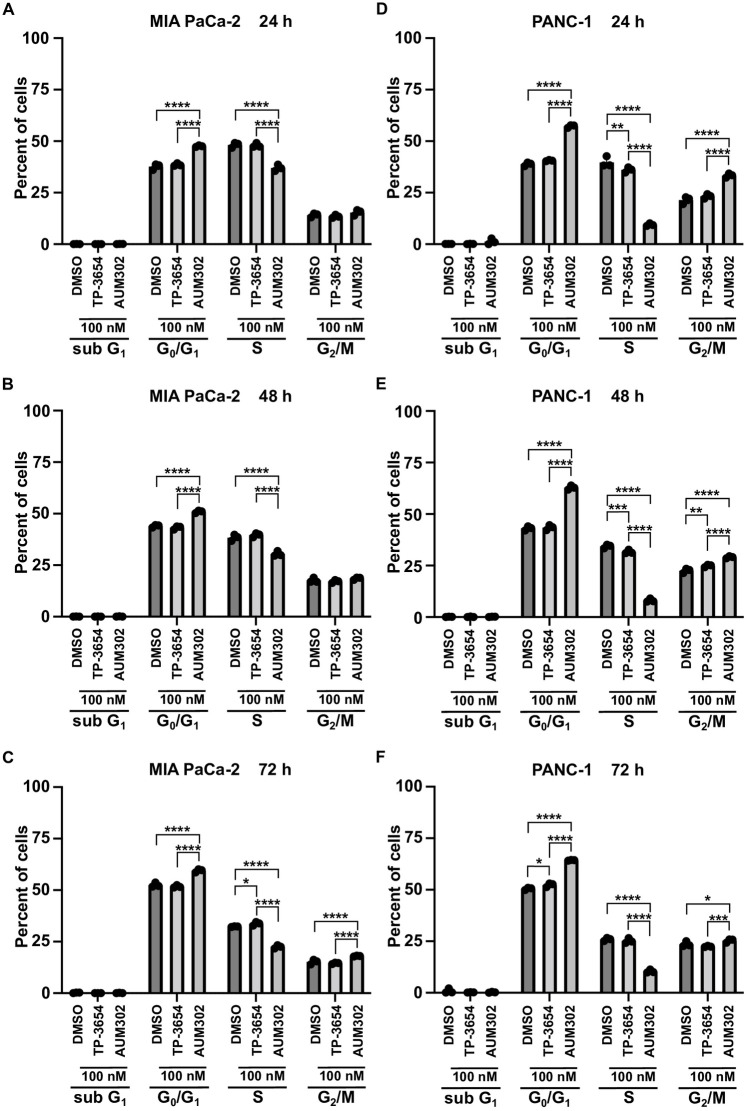
AUM302 alters cell cycle progression of pancreatic cancer cell lines
PIM kinases and PI3K/mTOR signaling pathways have been shown to play an important role in the regulation of cell cycle progression in multiple cancers, including pancreatic [18, 65–71]. Thus, we evaluated the impact of TP-3654 and AUM302 on cell cycle progression. We treated BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cell lines over three days with vehicle or 100 nM of test compounds and then analyzed cell cycle using flow cytometry. Our results demonstrated that TP-3654 at tested concentration (Figs 3–5) does not significantly affect the cell cycle progression of tested pancreatic cancer cell lines. In contrast, AUM302 was able to increase the cell number in G0/G1 phases, G2/M, and decrease the number of cells within the S-phase as compared to vehicle-treated cells. Notably, in the case of AUM302 there was a significant modification in the number of cells within aforementioned phases, in comparison to not only vehicle-treated cells but also to cells treated with TP-3654 compound. Importantly, in BxPC-3 and Capan-2 pancreatic cancer cell lines, treatment with AUM302 compound increased the number of cells within subG1 population, suggesting that this compound may induce apoptosis (Fig 3). These data suggest that treatment with AUM302 compound alters the cell cycle of pancreatic cancer cell lines and can additionally induce cell apoptosis.
Fig 3. AUM302 changes the cell cycle profile of BxPC-3 and Capan-2 pancreatic cancer cell lines.
Cells were treated with DMSO or TP-3654 (100 nM) or AUM302 (100 nM) for 24 (A and D), 48 (B and E), and 72 hours (C and F). Cells were stained with propidium iodide and analyzed by FACS analysis. Data are represented as mean ±SD, N = 3, *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 calculated with two-way ANOVA.
Fig 5. AUM302 changes the cell cycle profile of Hs766T pancreatic cancer cell line.
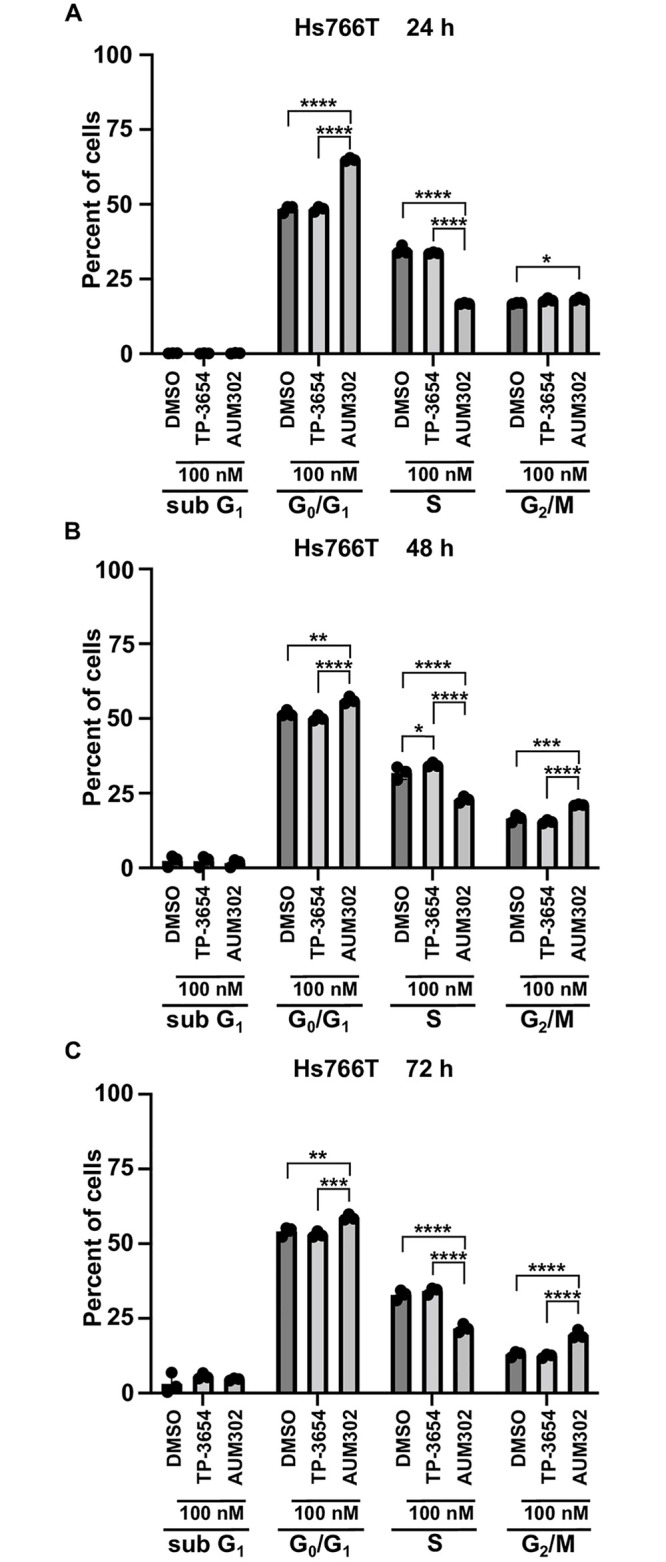
Cells were treated with DMSO or TP-3654 (100 nM) or AUM302 (100 nM) for 24 (A), 48 (B), and 72 hours (C). Cells were stained with propidium iodide and analyzed by FACS analysis. Data are represented as mean ±SD, N = 3, *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 calculated with two-way ANOVA.
Fig 4. AUM302 changes the cell cycle profile of MIA PaCa-2 and PANC-1 pancreatic cancer cell lines.
Cells were treated with DMSO or TP-3654 (100 nM) or AUM302 (100 nM) for 24 (A and D), 48 (B and E), and 72 hours (C and F). Cells were stained with propidium iodide and analyzed by FACS analysis. Data are represented as mean ±SD, N = 3, *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 calculated with two-way ANOVA.
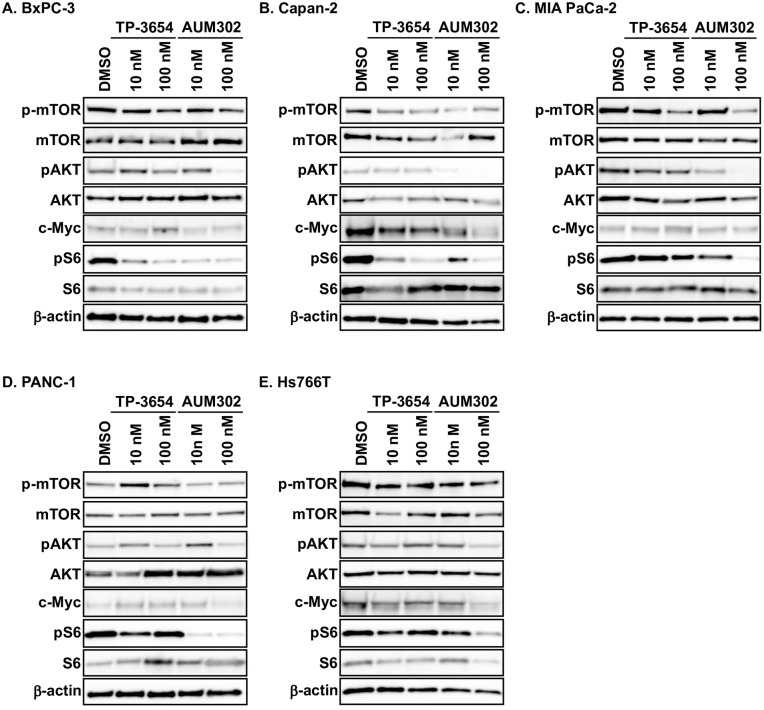
TP-3654 and AUM302 alter the cell signaling pathways controlled by PIM kinases and PI3K/mTOR signaling pathway
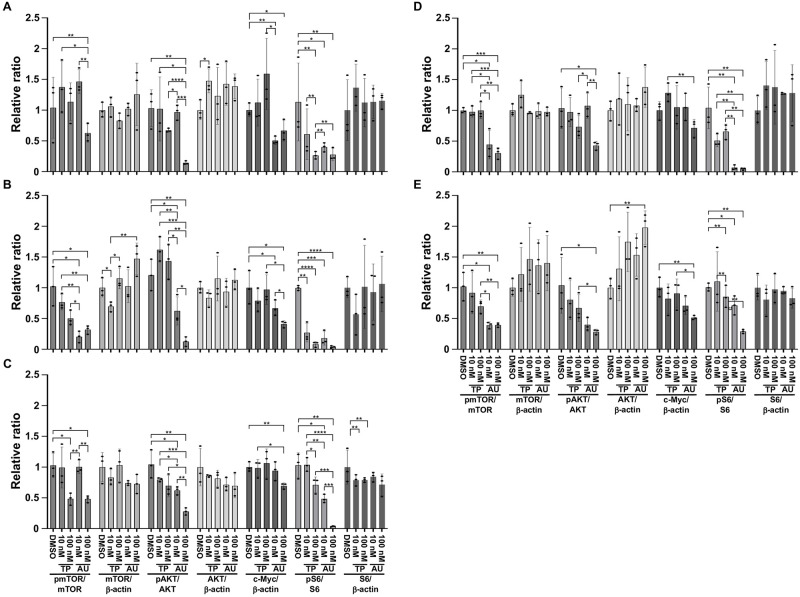
PIM kinases have been shown to positively modulate gene expression in the cell cycle and inhibit apoptosis by directly and indirectly regulating multiple targets such as c-Myc, BAD, and P21 [40, 72–75]. Furthermore, studies showed that inhibitors of PIM kinases reduce the phosphorylation status of ribosomal protein S6 in multiple cancer types [76]. In addition, myriad publications showed that the PI3K/mTOR pathway plays an essential role in the tumorigenesis of numerous cancers, including pancreatic cancer [16, 77–79]. Thus, we decided to investigate the ability of TP-3654 and AUM302 to alter the expression levels of the components of these pathways in pancreatic cancer cell lines. We treated BxPC-3, Capan-2, MIA PaCa-2, PANC-1, and Hs766T pancreatic cancer cell lines with DMSO (vehicle) and tested compounds at 10 nM or 100 nM, collecting the cells for western blot analysis at 24h. As shown in Fig 6, TP-3654 and AUM302 alter the expression of analyzed proteins. However, the AUM302 compound has the most negative effect on the phosphorylation of mTOR, AKT, and S6 kinases (Figs 6 and 7) while exerting minimal impact on the level of appropriate total proteins. Additionally, AUM302 significantly inhibited the levels of c-Myc compared to DMSO- and TP-3654 treatments. Thus far, we have demonstrated that AUM302 more effectively inhibits the proliferation of pancreatic cancer cells. Importantly, in contrast to TP-3654, AUM302 significantly modified the progression of the cell cycle and induced apoptosis. It has been shown that PI3K plays a vital role in the development of chemoresistance to gemcitabine in pancreatic cancer, and inhibition of PI3K activity can reverse this resistance [52, 80–83]. Hence, we decided to determine whether treatment with AUM302 inhibits the growth of gemcitabine-resistant pancreatic cancer cells.
Fig 6. AUM302 inhibits the cell signaling pathways regulated by PIM kinases and PI3K/mTOR pathway.
BxPC-3 (A), Capan-2 (B), MIA PaCa-2 (C), PANC-1 (D), and Hs766T (E) cells were treated with DMSO (vehicle) or TP-3654 (10 and 100 nM) or AUM302 (10 and 100 nM) for 24 hours.
Fig 7. Densitometry analysis of western blots results of protein regulated by PIM and PI3K/mTOR pathway.
BxPC-3 (A), Capan-2 (B), MIA PaCa-2 (C), PANC-1 (D), and Hs766T (E) cells were treated with DMSO (vehicle) or TP-3654 (10 and 100 nM) or AUM302 (10 and 100 nM) for 24 hours. Each experiment was performed in triplicate and the results are shown as mean ±SD (N = 3). Densitometry analysis was performed using FIJI software [61]. Statistical analysis was performed using the Student’s test followed by an analysis of the normal distribution (Tukey’s test). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
AUM302 inhibits growth of gemcitabine-resistant pancreatic cancer cells
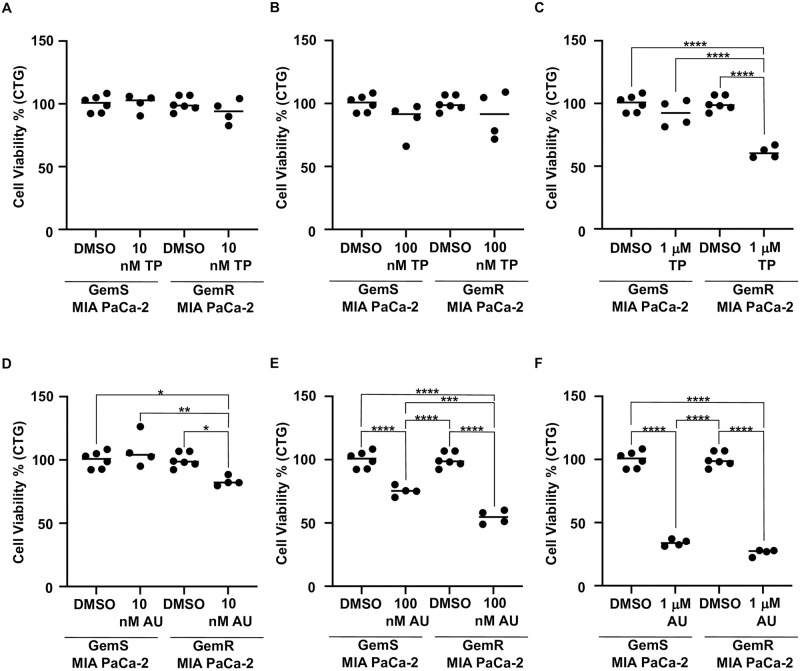
To assess the ability of TP-3654 and AUM302 to inhibit the growth of gemcitabine resistant pancreatic cancer cells we employed MIA PaCa-2 Gemcitabine Resistant (MIA PaCa-2 GemR) cell line. MIA PaCa-2 GemR cell line was continually grown in media supplemented with 50 nM gemcitabine. First, we grew MIA PaCa-2 GemS and MIA PaCa-2 GemR cell lines in the presence of TP-3654 and AUM302 at 10 nM, 100 nM, and 1 μM. We assessed the cell viability using Cell Titer-Glo 72 hours post-treatment. As shown in Fig 8, TP-3654 at 10 nM and 100 nM did not affect the growth of MIA PaCa-2 GemS and MIA PaCa-2 GemR cells (Fig 8A and 8B). The significant decrease in MIA PaCa-2 GemR cells viability was shown when cells were treated with 1 μM TP-3654 (Fig 8C). In contrast, even low doses of AUM302 (10 nM and 100 nM) significantly inhibited viability of MIA PaCa-2 GemR (Fig 8D and 8E). The effect was even more pronounced when cells were treated with 1 μM AUM302 (Fig 8F).
Fig 8. AUM302 and TP-3654 decrease the cell viability of MIA PaCa-2 gemcitabine-resistant (GemR) cell line.
MIA PaCa-2 GemR cell line was treated with 10 nM, 100 nM, or 1 μM of TP-3654 (A, B, & C) or AUM302 (D, E, & F) twenty-four hours after seeding. Cells were treated with test compounds for 72 hours and cell viability was measured using Cell Titer-Glo. Data represents mean ±SD (N = 6 and N = 4). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 calculated with two-way ANOVA.
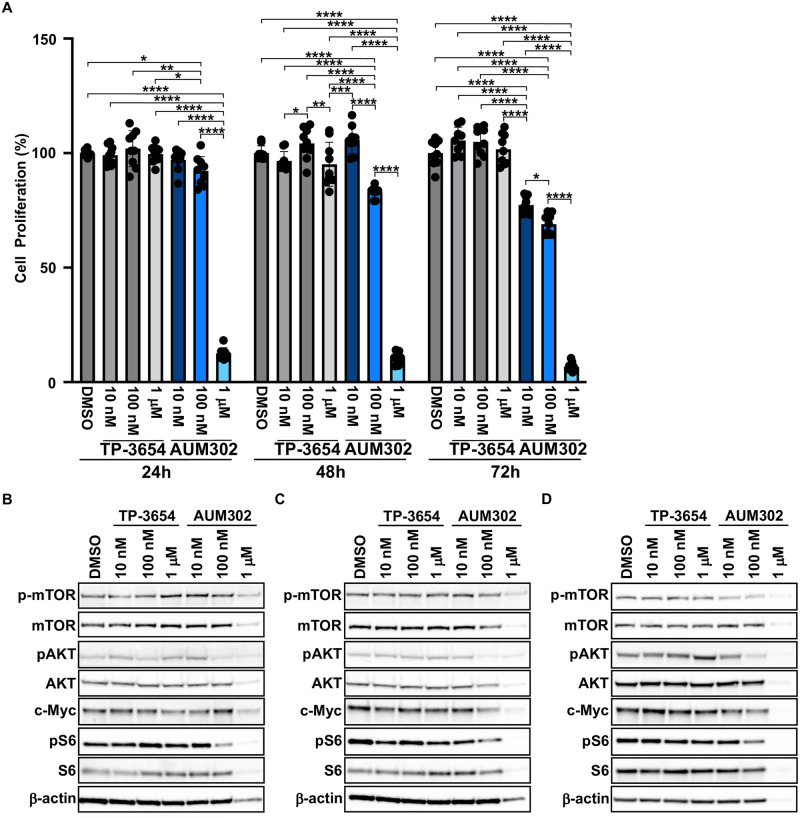
The effect of TP-3654 and AUM302 was additionally tested on the proliferation of MIA PaCa-2 GemR cell line over three days (Fig 9A). MIA PaCa-2 GemR cells were treated with at DMSO (vehicle) and 10 nM, 100 nM, or 1 μM of TP-3654 and AUM302 and cells were collected and counted 24, 48, and 72 hours post-treatment. As shown in Fig 9A, TP-3654 treatment did not affect the growth of MIA PaCa-2 GemR cells. In contrast, AUM302 reduces the growth of MIA PaCa-2 GemR cells at 72 hours at all tested concentration. Notably, treatment with 1 μM AUM302 almost completely inhibited the growth of MIA PaCa-2 GemR after 24 hours of treatment (Fig 9B). Furthermore, protein analysis was completed on MIA PaCa-2 GemR cells treated with DMSO, or TP-3654 or AUM302 at 10 nM, 100 nM, or 1 μM concentration for 24, 48, and 72 hours before total protein extraction. Western blot analysis demonstrated that components of the PI3K/AKT/mTOR signaling pathway are significantly inhibited with AUM302 compared to DMSO- and TP-3654-treated cells (Fig 9B–9D and S1–S3 Figs). Importantly, the inhibitory effects of AUM302 (100 nM and 1 μM) on the phosphorylation levels of mTOR, AKT, and S6 were noted already at 24-hour timepoint. In addition, the levels of non-phosphorylated counterparts of these proteins were downregulated after the treatment with 1μM AUM302 compared to other treatments. While treatment with TP-3654 resulted with significantly lower inhibition of the phosphorylation of mTOR, AKT, and S6, and downregulation of the total protein levels. Furthermore, we assessed the levels of c-Myc, and we showed that AUM302 at 1 μM concentration was able to significantly reduce its levels at three tested time points.
Fig 9. AUM302 inhibits the proliferation of MIA PaCa-2 gemcitabine-resistant (GemR) cells and activity of multiple signaling pathways.
(A) MIA PaCa-2 GemR cells were treated with 10 nM, 100 nM, and 1 μM of TP-3654 or AUM302. Cell count was determined 24, 48, and 72 hours after treatment using a cell counter. The measurement of the control (cells with DMSO) was defined as 100%. Data represent mean ±SD (N = 9). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 calculated with two-way ANOVA. (B—D) MIA PaCa-2 GemR cells were treated with DMSO (vehicle) or TP-3654 or AUM302 (10 nM, 100 nM, and 1 μM) for 24 (B), 48 (C), and 72 (D) hours and analyzed with western blot.
Discussion
Pancreatic ductal adenocarcinoma (PDAC) is a lethal cancer often resistant to the widely used chemotherapeutic agent gemcitabine. Currently, limited options exist for the treatment of PDAC. The surgery followed by adjuvant chemotherapy with FOLFIRINOX is available only to 10–15% of PDAC patients and provides an overall survival rate of four and a half years [84–86]. PDAC multidrug treatment provides between two to six months of benefit for patients with locally advanced or metastasis compared to single compound treatment [84, 85]. Consequently, studying PDAC molecular pathways gives rise to targeted therapies that may be a promising approach to treating pancreatic cancer [87]. Targeting PIM kinases has become a novel cancer therapeutic approach [88]. PIM inhibitors such as AZD1208, PIM447, and TP-3654 [16] are used against different cancers and have all entered the clinical stage [89]. A phase I dose-escalation study on 35 patients with solid tumors found that AZD1208 induced no functional response, although the PIM kinases were inhibited [90]. TP-3654 has improved potency, and clinical findings suggest it can treat patients with heavily pretreated, relapsed, and resistant solid tumors. There is a strong rationale to investigate PIM inhibitors in combination with PI3K/AKT/mTOR pathway inhibitors, as their interactions drive cancer cell proliferation and cell survival [31].
PI3K inhibitors are a targeted therapy with limited success in treating PDAC but promising results when combined during pre-clinical studies [16]. Initially, pan-PI3K or PI3K/mTOR inhibitors such as GDC-0941 or BEZ235 were developed and tested in preclinical models. Studies showed that simultaneous inhibition of PI3K with GDC-0941 and MEK or PORCN inhibitors provides a synergistic effect [91–93]. Other studies showed BEZ235 enhanced response to the chemotherapy and antiangiogenic or pan-histone deacetylase inhibitors in PDAC treatment [94, 95]. However, resistance to PI3K inhibitors is possible, and PIM overexpression was found to be related to such resistance [96]. Furthermore, PIM mimics the effects of AKT, leading to cell cycle progression, cell survival, and growth [97]. mTOR is a link between the PIM and PI3K pathways, also responsible for cell survival. These pathways are so intertwined in cancer that a multikinase inhibitor may be an efficient approach.
Here, we investigated using a triple kinase PIM/PI3K/mTOR inhibitor, AUM302, compared to TP-3654 and gemcitabine in PDAC cell lines. Initially, we compared the efficacy of two known dual PI3K/mTOR inhibitors, GDC-0941 and BEZ235, with triple PIM/PI3K/mTOR inhibitors, TP-3654 and AUM302, to reduce the viability of PDAC cell lines. Our results showed AUM302 had lower IC50 values in four tested cell lines (BxPC-3, Capan-2, PANC-1, and Hs766T) than other treatments. MIA PaCa-2 cell line was more susceptible to tested dual inhibitors than triple ones. It has been shown that the downregulation of PIM1 by shRNA in MIA PaCa-2 cell lines decreases proliferation [98]. However, the treatment in the context of PI3K/mTOR inhibitors may not result in more efficient viability inhibition (Fig 1, Table 1). AUM302 potently inhibited growth in PDAC cell lines, generating IC50 values in the low nanomolar range and significantly decreased cell viability. TP-3654 had IC50 values in the micromolar range and was less effective in altering the viability of PDAC cells, and gemcitabine even less so. Follow-up experiments comparing the impact of TP-3654 and AUM302 on cell proliferation and cell cycle showed that AUM302 is a more potent inhibitor of proliferation and blocker of the cell cycle progression than TP-3654 (Figs 2–5). Furthermore, AUM302 consistently decreased the number of cells in the S-phase and increased in G2/M phase compared to TP-3654 treated cells and control. This may be due to inhibition of PIM1 activity, which regulates normal cell cycle progression, particularly at the G1/S checkpoint [99]. The effect of AUM302 may be due to reduced expression of transcription factors like c-Myc, regulating cellular metabolism and protein translation through mTOR and AKT, regulating apoptosis through BAD, and decreasing the phosphorylation and activity of the ribosomal protein S6 [100]. Our results demonstrated that 24-hour treatment with AUM302 inhibited the phosphorylation of mTOR, AKT, and S6, while the total levels of the appropriate proteins were almost unchanged (Figs 6 and 7). In contrast, TP-3654 did not have a significant effect or demonstrated minimal impact on the expression levels of these proteins. In addition, we showed that the levels of c-Myc, an effector of the PI3K/mTOR pathway, were significantly decreased upon AUM302 compared to other treatments. This observation agrees with the previous studies demonstrating the downregulation of c-Myc levels upon inhibition of PI3K/mTOR in several cancer types [101–103]. Importantly, PIM1 was shown to phosphorylate c-Myc and increase its stability [40]. Thus, the downregulation of c-Myc in our model could be due to inhibition of kinase activity of PIM and an increase in c-Myc degradation.
We further investigated the effect of AUM302 and TP-3654 on gemcitabine-resistant cells using the MIA PaCa-2 gemcitabine-resistant cell line. Treatment with TP-3654 had no significant impact on the cell viability of gemcitabine-resistant cells until a concentration of 1μM, AUM302 did have a considerable effect at 10 nM, 100 nM, and 1 μM (Figs 8 and 9). Similarly, AUM302 significantly decreased cell proliferation of gemcitabine-resistant cells compared to vehicle- and TP-3654-treated cells. This may be due to AUM302 reducing the phosphorylation and thus the activity of mTOR, AKT, and S6 and decreasing the levels of c-Myc over three-day treatment (Fig 1 and S1–S3 Figs).
This is the first in vitro study demonstrating that AUM302 is an effective inhibitor of the PIM/PI3K/mTOR pathways and decreases PDAC cell viability. Its efficacious multikinase properties make it an advantageous approach to cancer therapy compared to kinase inhibitors like TP-3654 or dual PI3K/mTOR inhibitors. Notably, the compound can overcome gemcitabine resistance in PDAC cells. AUM302 is a potent inhibitor of PDAC cell growth in vitro, and our results suggest a clinical benefit in future research. However, PDAC is characterized by great genetic intra- and inter-heterogeneity [104]. In addition, treatment outcome also depends on the interaction between tumor cells and the microenvironment. In the case of PDAC, extensive fibrosis with little vascularization limits the drug’s efficacy. Thus, to fully assess the effectiveness of AUM302, studies using models mirroring in vivo characteristics of PDAC, such as chemically-induced animal models and genetically engineered mice, patient-derived organoids, and xenografts, should be performed [105, 106].
Supporting information
Each experiment was performed in triplicate and the results are shown as mean ±SD (N = 3). Densitometry analysis was performed using FIJI software [61]. Statistical analysis was performed using the Student’s test followed by an analysis of the normal distribution (Tukey’s test). *p<0.05; **p<0.01; ***p<0.001.
(PDF)
Each experiment was performed in triplicate and the results are shown as mean ±SD (N = 3). Densitometry analysis was performed using FIJI software [61]. Statistical analysis was performed using the Student’s test followed by an analysis of the normal distribution (Tukey’s test). *p<0.05; **p<0.01; ***p<0.001.
(PDF)
Each experiment was performed in triplicate and the results are shown as mean ±SD (N = 3). Densitometry analysis was performed using FIJI software [61]. Statistical analysis was performed using the Student’s test followed by an analysis of the normal distribution (Tukey’s test). *p<0.05; **p<0.01; ***p<0.001.
(PDF)
(PDF)
Acknowledgments
We would like to thank Research Flow Cytometry Core in the Department of Pathology, Stony Brook University for assistance with data analysis.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
The work was supported by a grant from the National Institutes of Health awarded to ABB (DK124342). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rawla P, Sunkara T, Gaduputi V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J Oncol. 2019;10(1):10–27. Epub 2019/03/06. doi: 10.14740/wjon1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw Open. 2021;4(4):e214708. Epub 2021/04/08. doi: 10.1001/jamanetworkopen.2021.4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. Epub 2022/01/13. doi: 10.3322/caac.21708 . [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Zeng L, Chen Y, Lian G, Qian C, Chen S, et al. Pancreatic Cancer Epidemiology, Detection, and Management. Gastroenterol Res Pract. 2016;2016:8962321. Epub 2016/03/05. doi: 10.1155/2016/8962321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillen S, Schuster T, Meyer Zum Buschenfelde C, Friess H, Kleeff J. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010;7(4):e1000267. Epub 2010/04/28. doi: 10.1371/journal.pmed.1000267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng S, Pottler M, Lan B, Grutzmann R, Pilarsky C, Yang H. Chemoresistance in Pancreatic Cancer. Int J Mol Sci. 2019;20(18). Epub 2019/09/14. doi: 10.3390/ijms20184504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seshacharyulu P, Baine MJ, Souchek JJ, Menning M, Kaur S, Yan Y, et al. Biological determinants of radioresistance and their remediation in pancreatic cancer. Biochim Biophys Acta Rev Cancer. 2017;1868(1):69–92. Epub 2017/03/03. doi: 10.1016/j.bbcan.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amrutkar M, Gladhaug IP. Pancreatic Cancer Chemoresistance to Gemcitabine. Cancers (Basel). 2017;9(11). Epub 2017/11/17. doi: 10.3390/cancers9110157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsujimoto A, Sudo K, Nakamura K, Kita E, Hara R, Takayama W, et al. Gemcitabine plus nab-paclitaxel for locally advanced or borderline resectable pancreatic cancer. Sci Rep. 2019;9(1):16187. Epub 2019/11/09. doi: 10.1038/s41598-019-52486-x TOWA and Teijin Pharma, and a consulting role for Ono Pharmaceutical, outside the submitted work. Other authors declare no conflict of interest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–703. Epub 2013/10/18. doi: 10.1056/NEJMoa1304369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koltai T, Reshkin SJ, Carvalho TMA, Di Molfetta D, Greco MR, Alfarouk KO, et al. Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma: A Physiopathologic and Pharmacologic Review. Cancers (Basel). 2022;14(10). Epub 2022/05/29. doi: 10.3390/cancers14102486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia Y, Xie J. Promising molecular mechanisms responsible for gemcitabine resistance in cancer. Genes Dis. 2015;2(4):299–306. Epub 2015/07/30. doi: 10.1016/j.gendis.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Huang W, Wang X, Zhang J, Tao T, Zheng Y, et al. Hsa-miR-3178/RhoB/PI3K/Akt, a novel signaling pathway regulates ABC transporters to reverse gemcitabine resistance in pancreatic cancer. Mol Cancer. 2022;21(1):112. Epub 2022/05/11. doi: 10.1186/s12943-022-01587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansari D, Ohlsson H, Althini C, Bauden M, Zhou Q, Hu D, et al. The Hippo Signaling Pathway in Pancreatic Cancer. Anticancer Res. 2019;39(7):3317–21. Epub 2019/07/03. doi: 10.21873/anticanres.13474 . [DOI] [PubMed] [Google Scholar]
- 15.Avila JL, Kissil JL. Notch signaling in pancreatic cancer: oncogene or tumor suppressor? Trends Mol Med. 2013;19(5):320–7. Epub 2013/04/03. doi: 10.1016/j.molmed.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehra S, Deshpande N, Nagathihalli N. Targeting PI3K Pathway in Pancreatic Ductal Adenocarcinoma: Rationale and Progress. Cancers (Basel). 2021;13(17). Epub 2021/09/11. doi: 10.3390/cancers13174434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortazavi M, Moosavi F, Martini M, Giovannetti E, Firuzi O. Prospects of targeting PI3K/AKT/mTOR pathway in pancreatic cancer. Crit Rev Oncol Hematol. 2022;176:103749. Epub 2022/06/22. doi: 10.1016/j.critrevonc.2022.103749 . [DOI] [PubMed] [Google Scholar]
- 18.Murthy D, Attri KS, Singh PK. Phosphoinositide 3-Kinase Signaling Pathway in Pancreatic Ductal Adenocarcinoma Progression, Pathogenesis, and Therapeutics. Front Physiol. 2018;9:335. Epub 2018/04/20. doi: 10.3389/fphys.2018.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiki Y, Hirota S, Horii A. Attempts to remodel the pathways of gemcitabine metabolism: Recent approaches to overcoming tumours with acquired chemoresistance. Cancer Drug Resist. 2020;3(4):819–31. Epub 2020/10/12. doi: 10.20517/cdr.2020.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minami K, Shinsato Y, Yamamoto M, Takahashi H, Zhang S, Nishizawa Y, et al. Ribonucleotide reductase is an effective target to overcome gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells with dual resistant factors. J Pharmacol Sci. 2015;127(3):319–25. Epub 2015/04/04. doi: 10.1016/j.jphs.2015.01.006 . [DOI] [PubMed] [Google Scholar]
- 21.Thummuri D, Khan S, Underwood PW, Zhang P, Wiegand J, Zhang X, et al. Overcoming Gemcitabine Resistance in Pancreatic Cancer Using the BCL-X(L)-Specific Degrader DT2216. Mol Cancer Ther. 2022;21(1):184–92. Epub 2021/10/21. doi: 10.1158/1535-7163.MCT-21-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giri B, Sethi V, Dudeja V, Banerjee S, Livingstone A, Saluja A. Genetics of pancreatic cyst-cancer progression: standing on the shoulders of giants. Curr Opin Gastroenterol. 2017;33(5):404–10. Epub 2017/07/07. doi: 10.1097/MOG.0000000000000382 . [DOI] [PubMed] [Google Scholar]
- 23.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–3 e9. Epub 2012/01/10. doi: 10.1053/j.gastro.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen H, Lundy J, Strickland AH, Harris M, Swan M, Desmond C, et al. KRAS G12D Mutation Subtype in Pancreatic Ductal Adenocarcinoma: Does It Influence Prognosis or Stage of Disease at Presentation? Cells. 2022;11(19). Epub 2022/10/15. doi: 10.3390/cells11193175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30(4):355–85. Epub 2016/02/18. doi: 10.1101/gad.275776.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers N, Gerard C, Lemaigre FP, Jacquemin P. Differential impact of the ERBB receptors EGFR and ERBB2 on the initiation of precursor lesions of pancreatic ductal adenocarcinoma. Sci Rep. 2020;10(1):5241. Epub 2020/04/07. doi: 10.1038/s41598-020-62106-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JT, Wang YP, Yin M, Lei QY. Metabolism remodeling in pancreatic ductal adenocarcinoma. Cell Stress. 2019;3(12):361–8. Epub 2019/12/14. doi: 10.15698/cst2019.12.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elebo N, Fru P, Omoshoro-Jones J, Patrick Candy G, Nweke EE. Role of different immune cells and metabolic pathways in modulating the immune response in pancreatic cancer (Review). Mol Med Rep. 2020;22(6):4981–91. Epub 2020/11/12. doi: 10.3892/mmr.2020.11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin C, Yang G, Yang J, Ren B, Wang H, Chen G, et al. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Mol Cancer. 2020;19(1):50. Epub 2020/03/04. doi: 10.1186/s12943-020-01169-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Zheng Y, Yang F, Zhu L, Zhu XQ, Wang ZF, et al. The molecular biology of pancreatic adenocarcinoma: translational challenges and clinical perspectives. Signal Transduct Target Ther. 2021;6(1):249. Epub 2021/07/06. doi: 10.1038/s41392-021-00659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathi A, Kumar D, Hasan GM, Haque MM, Hassan MI. Therapeutic targeting of PIM KINASE signaling in cancer therapy: Structural and clinical prospects. Biochim Biophys Acta Gen Subj. 2021;1865(11):129995. Epub 2021/08/30. doi: 10.1016/j.bbagen.2021.129995 . [DOI] [PubMed] [Google Scholar]
- 32.Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66(13):6741–7. Epub 2006/07/05. doi: 10.1158/0008-5472.CAN-05-4272 . [DOI] [PubMed] [Google Scholar]
- 33.Mukaida N, Wang YY, Li YY. Roles of Pim-3, a novel survival kinase, in tumorigenesis. Cancer Sci. 2011;102(8):1437–42. Epub 2011/04/27. doi: 10.1111/j.1349-7006.2011.01966.x . [DOI] [PubMed] [Google Scholar]
- 34.Li YY, Mukaida N. Pathophysiological roles of Pim-3 kinase in pancreatic cancer development and progression. World J Gastroenterol. 2014;20(28):9392–404. Epub 2014/07/30. doi: 10.3748/wjg.v20.i28.9392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Xiu J, Ren C, Yu Z. Protein kinase PIM2: A simple PIM family kinase with complex functions in cancer metabolism and therapeutics. J Cancer. 2021;12(9):2570–81. Epub 2021/04/16. doi: 10.7150/jca.53134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellon M, Nicot C. Targeting Pim kinases in hematological cancers: molecular and clinical review. Mol Cancer. 2023;22(1):18. Epub 2023/01/25. doi: 10.1186/s12943-023-01721-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Allsop SA, Witherspoon SM, Snider JL, Yeh JJ, Fiordalisi JJ, et al. The oncogenic kinase Pim-1 is modulated by K-Ras signaling and mediates transformed growth and radioresistance in human pancreatic ductal adenocarcinoma cells. Carcinogenesis. 2011;32(4):488–95. Epub 2011/01/26. doi: 10.1093/carcin/bgr007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiser-Erkan C, Erkan M, Pan Z, Bekasi S, Giese NA, Streit S, et al. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2008;7(9):1352–9. Epub 2008/08/19. doi: 10.4161/cbt.7.9.6418 . [DOI] [PubMed] [Google Scholar]
- 39.Liang C, Yu XJ, Guo XZ, Sun MH, Wang Z, Song Y, et al. MicroRNA-33a-mediated downregulation of Pim-3 kinase expression renders human pancreatic cancer cells sensitivity to gemcitabine. Oncotarget. 2015;6(16):14440–55. Epub 2015/05/15. doi: 10.18632/oncotarget.3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27(35):4809–19. Epub 2008/04/29. doi: 10.1038/onc.2008.123 . [DOI] [PubMed] [Google Scholar]
- 41.Keeton EK, McEachern K, Dillman KS, Palakurthi S, Cao Y, Grondine MR, et al. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014;123(6):905–13. Epub 20131220. doi: 10.1182/blood-2013-04-495366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadav AK, Kumar V, Bailey DB, Jang BC. AZD1208, a Pan-Pim Kinase Inhibitor, Has Anti-Growth Effect on 93T449 Human Liposarcoma Cells via Control of the Expression and Phosphorylation of Pim-3, mTOR, 4EBP-1, S6, STAT-3 and AMPK. Int J Mol Sci. 2019;20(2). Epub 20190116. doi: 10.3390/ijms20020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia PD, Langowski JL, Wang Y, Chen M, Castillo J, Fanton C, et al. Pan-PIM kinase inhibition provides a novel therapy for treating hematologic cancers. Clin Cancer Res. 2014;20(7):1834–45. Epub 20140128. doi: 10.1158/1078-0432.CCR-13-2062 . [DOI] [PubMed] [Google Scholar]
- 44.Iqbal A, Eckerdt F, Bell J, Nakano I, Giles FJ, Cheng SY, et al. Targeting of glioblastoma cell lines and glioma stem cells by combined PIM kinase and PI3K-p110alpha inhibition. Oncotarget. 2016;7(22):33192–201. doi: 10.18632/oncotarget.8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118(3):693–702. Epub 20110531. doi: 10.1182/blood-2010-12-323022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D, Cobb MG, Gavilano L, Witherspoon SM, Williams D, White CD, et al. Inhibition of oncogenic Pim-3 kinase modulates transformed growth and chemosensitizes pancreatic cancer cells to gemcitabine. Cancer Biol Ther. 2013;14(6):492–501. Epub 2013/06/14. doi: 10.4161/cbt.24343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aziz AUR, Farid S, Qin K, Wang H, Liu B. PIM Kinases and Their Relevance to the PI3K/AKT/mTOR Pathway in the Regulation of Ovarian Cancer. Biomolecules. 2018;8(1). Epub 2018/02/07. doi: 10.3390/biom8010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meja K, Stengel C, Sellar R, Huszar D, Davies BR, Gale RE, et al. PIM and AKT kinase inhibitors show synergistic cytotoxicity in acute myeloid leukaemia that is associated with convergence on mTOR and MCL1 pathways. Br J Haematol. 2014;167(1):69–79. Epub 2014/07/01. doi: 10.1111/bjh.13013 . [DOI] [PubMed] [Google Scholar]
- 49.Wu CP, Li YQ, Chi YC, Huang YH, Hung TH, Wu YS. The Second-Generation PIM Kinase Inhibitor TP-3654 Resensitizes ABCG2-Overexpressing Multidrug-Resistant Cancer Cells to Cytotoxic Anticancer Drugs. Int J Mol Sci. 2021;22(17). Epub 2021/09/11. doi: 10.3390/ijms22179440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dutta A, Nath D, Yang Y, Le BT, Rahman MF, Faughnan P, et al. Genetic ablation of Pim1 or pharmacologic inhibition with TP-3654 ameliorates myelofibrosis in murine models. Leukemia. 2022;36(3):746–59. Epub 2021/11/07. doi: 10.1038/s41375-021-01464-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luszczak S, Simpson BS, Stopka-Farooqui U, Sathyadevan VK, Echeverria LMC, Kumar C, et al. Co-targeting PIM and PI3K/mTOR using multikinase inhibitor AUM302 and a combination of AZD-1208 and BEZ235 in prostate cancer. Sci Rep. 2020;10(1):14380. Epub 2020/09/03. doi: 10.1038/s41598-020-71263-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okuno K, Xu C, Pascual-Sabater S, Tokunaga M, Han H, Fillat C, et al. Berberine Overcomes Gemcitabine-Associated Chemoresistance through Regulation of Rap1/PI3K-Akt Signaling in Pancreatic Ductal Adenocarcinoma. Pharmaceuticals (Basel). 2022;15(10). Epub 2022/10/28. doi: 10.3390/ph15101199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y, Wu H, Xiong J, Gou S, Cui J, Peng T. miR-222-3p-containing macrophage-derived extracellular vesicles confer gemcitabine resistance via TSC1-mediated mTOR/AKT/PI3K pathway in pancreatic cancer. Cell Biol Toxicol. 2022. Epub 2022/08/17. doi: 10.1007/s10565-022-09736-y . [DOI] [PubMed] [Google Scholar]
- 54.Liu K, Geng Y, Wang L, Xu H, Zou M, Li Y, et al. Systematic exploration of the underlying mechanism of gemcitabine resistance in pancreatic adenocarcinoma. Mol Oncol. 2022;16(16):3034–51. Epub 2022/07/11. doi: 10.1002/1878-0261.13279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y, Xu W, Yang Y, Zhang Z. miRNA-93-5p Promotes Gemcitabine Resistance in Pancreatic Cancer Cells by Targeting the PTEN-Mediated PI3K/Akt Signaling Pathway. Ann Clin Lab Sci. 2021;51(3):310–20. Epub 2021/06/25. . [PubMed] [Google Scholar]
- 56.Zhou HY, Yao XM, Chen XD, Tang JM, Qiao ZG, Wu XY. Mechanism of metformin enhancing the sensitivity of human pancreatic cancer cells to gem-citabine by regulating the PI3K/Akt/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(23):10283–9. Epub 2019/12/17. doi: 10.26355/eurrev_201912_19666 . [DOI] [PubMed] [Google Scholar]
- 57.Bialkowska AB, Crisp M, Bannister T, He Y, Chowdhury S, Schurer S, et al. Identification of small-molecule inhibitors of the colorectal cancer oncogene Kruppel-like factor 5 expression by ultrahigh-throughput screening. Mol Cancer Ther. 2011;10(11):2043–51. Epub 2011/09/03. doi: 10.1158/1535-7163.MCT-11-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bialkowska AB, Du Y, Fu H, Yang VW. Identification of novel small-molecule compounds that inhibit the proproliferative Kruppel-like factor 5 in colorectal cancer cells by high-throughput screening. Mol Cancer Ther. 2009;8(3):563–70. Epub 2009/02/26. doi: 10.1158/1535-7163.MCT-08-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Wang C, de Sabando AR, Cole HL, Huang TJ, Yang J, et al. The Novel Small-Molecule SR18662 Efficiently Inhibits the Growth of Colorectal Cancer In Vitro and In Vivo. Mol Cancer Ther. 2019;18(11):1973–84. Epub 2019/07/31. doi: 10.1158/1535-7163.MCT-18-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruiz de Sabando A, Wang C, He Y, Garcia-Barros M, Kim J, Shroyer KR, et al. ML264, A Novel Small-Molecule Compound That Potently Inhibits Growth of Colorectal Cancer. Mol Cancer Ther. 2016;15(1):72–83. Epub 2015/12/02. doi: 10.1158/1535-7163.MCT-15-0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. Epub 20120628. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18(1):26. Epub 20190219. doi: 10.1186/s12943-019-0954-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao HF, Wang J, Shao W, Wu CP, Chen ZP, To ST, et al. Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: current preclinical and clinical development. Mol Cancer. 2017;16(1):100. Epub 20170607. doi: 10.1186/s12943-017-0670-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51(18):5522–32. doi: 10.1021/jm800295d . [DOI] [PubMed] [Google Scholar]
- 65.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68(13):5076–85. Epub 2008/07/03. doi: 10.1158/0008-5472.CAN-08-0634 . [DOI] [PubMed] [Google Scholar]
- 66.Vadlakonda L, Pasupuleti M, Pallu R. Role of PI3K-AKT-mTOR and Wnt Signaling Pathways in Transition of G1-S Phase of Cell Cycle in Cancer Cells. Front Oncol. 2013;3:85. Epub 2013/04/19. doi: 10.3389/fonc.2013.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharya N, Wang Z, Davitt C, McKenzie IF, Xing PX, Magnuson NS. Pim-1 associates with protein complexes necessary for mitosis. Chromosoma. 2002;111(2):80–95. Epub 2002/07/12. doi: 10.1007/s00412-002-0192-6 . [DOI] [PubMed] [Google Scholar]
- 68.Iadevaia V, Caldarola S, Biondini L, Gismondi A, Karlsson S, Dianzani I, et al. PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene. 2010;29(40):5490–9. Epub 2010/07/20. doi: 10.1038/onc.2010.279 . [DOI] [PubMed] [Google Scholar]
- 69.Cen B, Mahajan S, Zemskova M, Beharry Z, Lin YW, Cramer SD, et al. Regulation of Skp2 levels by the Pim-1 protein kinase. J Biol Chem. 2010;285(38):29128–37. Epub 2010/07/29. doi: 10.1074/jbc.M110.137240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quan J, Zhou L, Qu J. Knockdown of Pim-3 suppresses the tumorigenicity of glioblastoma by regulating cell cycle and apoptosis. Cell Mol Biol (Noisy-le-grand). 2015;61(1):42–50. Epub 2015/03/31. . [PubMed] [Google Scholar]
- 71.Forshell LP, Li Y, Forshell TZ, Rudelius M, Nilsson L, Keller U, et al. The direct Myc target Pim3 cooperates with other Pim kinases in supporting viability of Myc-induced B-cell lymphomas. Oncotarget. 2011;2(6):448–60. Epub 2011/06/08. doi: 10.18632/oncotarget.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X, Song M, Kundu JK, Lee MH, Liu ZZ. PIM Kinase as an Executional Target in Cancer. J Cancer Prev. 2018;23(3):109–16. Epub 2018/10/30. doi: 10.15430/JCP.2018.23.3.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11(6):709–19. Epub 2000/01/08. doi: 10.1016/s1074-7613(00)80145-4 . [DOI] [PubMed] [Google Scholar]
- 74.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9(8):932–44. Epub 2007/07/24. doi: 10.1038/ncb1618 . [DOI] [PubMed] [Google Scholar]
- 75.Banerjee S, Lu J, Cai Q, Sun Z, Jha HC, Robertson ES. EBNA3C augments Pim-1 mediated phosphorylation and degradation of p21 to promote B-cell proliferation. PLoS Pathog. 2014;10(8):e1004304. Epub 2014/08/15. doi: 10.1371/journal.ppat.1004304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yi YW, You KS, Park JS, Lee SG, Seong YS. Ribosomal Protein S6: A Potential Therapeutic Target against Cancer? Int J Mol Sci. 2021;23(1). Epub 20211221. doi: 10.3390/ijms23010048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paplomata E ’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–66. Epub 2014/07/25. doi: 10.1177/1758834014530023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front Oncol. 2014;4:64. Epub 2014/05/02. doi: 10.3389/fonc.2014.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stanciu S, Ionita-Radu F, Stefani C, Miricescu D, Stanescu S, II, Greabu M, et al. Targeting PI3K/AKT/mTOR Signaling Pathway in Pancreatic Cancer: From Molecular to Clinical Aspects. Int J Mol Sci. 2022;23(17). Epub 2022/09/10. doi: 10.3390/ijms231710132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voutsadakis IA. Molecular predictors of gemcitabine response in pancreatic cancer. World J Gastrointest Oncol. 2011;3(11):153–64. Epub 2011/11/24. doi: 10.4251/wjgo.v3.i11.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Awasthi N, Kronenberger D, Stefaniak A, Hassan MS, von Holzen U, Schwarz MA, et al. Dual inhibition of the PI3K and MAPK pathways enhances nab-paclitaxel/gemcitabine chemotherapy response in preclinical models of pancreatic cancer. Cancer Lett. 2019;459:41–9. Epub 2019/06/04. doi: 10.1016/j.canlet.2019.05.037 . [DOI] [PubMed] [Google Scholar]
- 82.Arlt A, Gehrz A, Muerkoster S, Vorndamm J, Kruse ML, Folsch UR, et al. Role of NF-kappaB and Akt/PI3K in the resistance of pancreatic carcinoma cell lines against gemcitabine-induced cell death. Oncogene. 2003;22(21):3243–51. Epub 2003/05/23. doi: 10.1038/sj.onc.1206390 . [DOI] [PubMed] [Google Scholar]
- 83.Jung KH, Yan HH, Fang Z, Son MK, Lee H, Hong S, et al. HS-104, a PI3K inhibitor, enhances the anticancer efficacy of gemcitabine in pancreatic cancer. Int J Oncol. 2014;45(1):311–21. Epub 2014/05/14. doi: 10.3892/ijo.2014.2435 . [DOI] [PubMed] [Google Scholar]
- 84.Park W, Chawla A, O’Reilly EM. Pancreatic Cancer: A Review. JAMA. 2021;326(9):851–62. Epub 2021/09/22. doi: 10.1001/jama.2021.13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Principe DR, Underwood PW, Korc M, Trevino JG, Munshi HG, Rana A. The Current Treatment Paradigm for Pancreatic Ductal Adenocarcinoma and Barriers to Therapeutic Efficacy. Front Oncol. 2021;11:688377. Epub 2021/08/03. doi: 10.3389/fonc.2021.688377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang Y, Sohal DPS. Pancreatic Adenocarcinoma Management. JCO Oncol Pract. 2023;19(1):19–32. Epub 2022/09/23. doi: 10.1200/OP.22.00328 . [DOI] [PubMed] [Google Scholar]
- 87.Hosein AN, Dougan SK, Aguirre AJ, Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat Cancer. 2022;3(3):272–86. Epub 2022/03/31. doi: 10.1038/s43018-022-00349-2 . [DOI] [PubMed] [Google Scholar]
- 88.Asati V, Mahapatra DK, Bharti SK. PIM kinase inhibitors: Structural and pharmacological perspectives. Eur J Med Chem. 2019;172:95–108. Epub 2019/04/08. doi: 10.1016/j.ejmech.2019.03.050 . [DOI] [PubMed] [Google Scholar]
- 89.Chen Q, Wang Y, Shi S, Li K, Zhang L, Gao J. Insights into the Interaction Mechanisms of the Proviral Integration Site of Moloney Murine Leukemia Virus (Pim) Kinases with Pan-Pim Inhibitors PIM447 and AZD1208: A Molecular Dynamics Simulation and MM/GBSA Calculation Study. Int J Mol Sci. 2019;20(21). Epub 2019/11/02. doi: 10.3390/ijms20215410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cortes J, Tamura K, DeAngelo DJ, de Bono J, Lorente D, Minden M, et al. Phase I studies of AZD1208, a proviral integration Moloney virus kinase inhibitor in solid and haematological cancers. Br J Cancer. 2018;118(11):1425–33. Epub 2018/05/17. doi: 10.1038/s41416-018-0082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhong H, Sanchez C, Spitzer D, Plambeck-Suess S, Gibbs J, Hawkins WG, et al. Synergistic effects of concurrent blockade of PI3K and MEK pathways in pancreatic cancer preclinical models. PLoS One. 2013;8(10):e77243. Epub 20131009. doi: 10.1371/journal.pone.0077243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang H, Xu M, Li L, Grierson P, Dodhiawala P, Highkin M, et al. Concurrent HER or PI3K Inhibition Potentiates the Antitumor Effect of the ERK Inhibitor Ulixertinib in Preclinical Pancreatic Cancer Models. Mol Cancer Ther. 2018;17(10):2144–55. Epub 20180731. doi: 10.1158/1535-7163.MCT-17-1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong Z, Sepramaniam S, Chew XH, Wood K, Lee MA, Madan B, et al. PORCN inhibition synergizes with PI3K/mTOR inhibition in Wnt-addicted cancers. Oncogene. 2019;38(40):6662–77. Epub 20190807. doi: 10.1038/s41388-019-0908-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Awasthi N, Yen PL, Schwarz MA, Schwarz RE. The efficacy of a novel, dual PI3K/mTOR inhibitor NVP-BEZ235 to enhance chemotherapy and antiangiogenic response in pancreatic cancer. J Cell Biochem. 2012;113(3):784–91. doi: 10.1002/jcb.23405 . [DOI] [PubMed] [Google Scholar]
- 95.Venkannagari S, Fiskus W, Peth K, Atadja P, Hidalgo M, Maitra A, et al. Superior efficacy of co-treatment with dual PI3K/mTOR inhibitor NVP-BEZ235 and pan-histone deacetylase inhibitor against human pancreatic cancer. Oncotarget. 2012;3(11):1416–27. doi: 10.18632/oncotarget.724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Le X, Antony R, Razavi P, Treacy DJ, Luo F, Ghandi M, et al. Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer Discov. 2016;6(10):1134–47. Epub 2016/09/09. doi: 10.1158/2159-8290.CD-16-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Warfel NA, Kraft AS. PIM kinase (and Akt) biology and signaling in tumors. Pharmacol Ther. 2015;151:41–9. Epub 2015/03/10. doi: 10.1016/j.pharmthera.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu J, Xiong G, Cao Z, Huang H, Wang T, You L, et al. PIM-1 contributes to the malignancy of pancreatic cancer and displays diagnostic and prognostic value. J Exp Clin Cancer Res. 2016;35(1):133. Epub 20160905. doi: 10.1186/s13046-016-0406-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swords R, Kelly K, Carew J, Nawrocki S, Mahalingam D, Sarantopoulos J, et al. The Pim kinases: new targets for drug development. Curr Drug Targets. 2011;12(14):2059–66. Epub 2011/07/23. doi: 10.2174/138945011798829447 . [DOI] [PubMed] [Google Scholar]
- 100.Alvarado Y, Giles FJ, Swords RT. The PIM kinases in hematological cancers. Expert Rev Hematol. 2012;5(1):81–96. Epub 2012/01/26. doi: 10.1586/ehm.11.69 . [DOI] [PubMed] [Google Scholar]
- 101.Schild C, Wirth M, Reichert M, Schmid RM, Saur D, Schneider G. PI3K signaling maintains c-myc expression to regulate transcription of E2F1 in pancreatic cancer cells. Mol Carcinog. 2009;48(12):1149–58. doi: 10.1002/mc.20569 . [DOI] [PubMed] [Google Scholar]
- 102.Hsin IL, Shen HP, Chang HY, Ko JL, Wang PH. Suppression of PI3K/Akt/mTOR/c-Myc/mtp53 Positive Feedback Loop Induces Cell Cycle Arrest by Dual PI3K/mTOR Inhibitor PQR309 in Endometrial Cancer Cell Lines. Cells. 2021;10(11). Epub 20211027. doi: 10.3390/cells10112916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stengel S, Petrie KR, Sbirkov Y, Stanko C, Ghazvini Zadegan F, Gil V, et al. Suppression of MYC by PI3K/AKT/mTOR pathway inhibition in combination with all-trans retinoic acid treatment for therapeutic gain in acute myeloid leukaemia. Br J Haematol. 2022;198(2):338–48. Epub 20220425. doi: 10.1111/bjh.18187 . [DOI] [PubMed] [Google Scholar]
- 104.Gutierrez ML, Munoz-Bellvis L, Orfao A. Genomic Heterogeneity of Pancreatic Ductal Adenocarcinoma and Its Clinical Impact. Cancers (Basel). 2021;13(17). Epub 20210903. doi: 10.3390/cancers13174451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gundel B, Liu X, Lohr M, Heuchel R. Pancreatic Ductal Adenocarcinoma: Preclinical in vitro and ex vivo Models. Front Cell Dev Biol. 2021;9:741162. Epub 20211022. doi: 10.3389/fcell.2021.741162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mallya K, Gautam SK, Aithal A, Batra SK, Jain M. Modeling pancreatic cancer in mice for experimental therapeutics. Biochim Biophys Acta Rev Cancer. 2021;1876(1):188554. Epub 20210501. doi: 10.1016/j.bbcan.2021.188554 [DOI] [PMC free article] [PubMed] [Google Scholar]