Abstract
Respiratory quinones were used as biomarkers to study bacterial community structures in activated sludge reactors used for enhanced biological phosphate removal (EBPR). We compared the quinone profiles of EBPR sludges and standard sludges, of natural sewage and synthetic sewage, and of plant scale and laboratory scale systems. Ubiquinone (Q) and menaquinone (MK) components were detected in all sludges tested at molar MK/Q ratios of 0.455 to 0.981. The differences in MK/Q ratios were much larger when we compared different wastewater sludges (i.e., raw sewage and synthetic sewage) than when we compared sludges from the EBPR and standard processes or plant scale and laboratory scale systems. In all sludges tested a Q with eight isoprene units (Q-8) was the most abundant quinone. In the MK fraction, either tetrahydrogenated MK-8 or MK-7 was the predominant type, and there was also a significant proportion of MK-6 to MK-8 in most cases. A numerical cluster analysis of the profiles showed that the sludges tested fell into two major clusters; one included all raw sewage sludges, and the other consisted of all synthetic sewage sludges, independent of the operational mode and scale of the reactors and the phosphate accumulation. These data suggested that Q-8-containing species belonging to the class Proteobacteria (i.e., species belonging to the beta subclass) were the major constituents of the bacterial populations in the EBPR sludge, as well as in standard activated sludge. Members of the class Actinobacteria (gram-positive bacteria with high DNA G+C contents) were the second most abundant group in both types of sludge. The bacterial community structures in activated sludge processes may be affected more by the nature of the influent wastewater than by the introduction of an anaerobic stage into the process or by the scale of the reactors.
Activated sludge processes with changing anaerobic-oxic (AO) or anaerobic-aerobic conditions have been used successfully for phosphate removal from wastewater. It is typical in the enhanced biological phosphate removal (EBPR) process that the sludge releases Pi (with concomitant uptake of wastewater organic carbon) in the anaerobic phase and takes up Pi rapidly in the aerobic stage. The Pi removed from wastewater is accumulated as a form of polyphosphate (polyP) in the sludge bacteria. Therefore, which phylogenetic and taxonomic groups of bacteria are responsible for phosphate removal and polyP accumulation has been and is still a subject of major concern for understanding the EBPR mechanism and the control of this process (for reviews see references 28 and 31).
Previous studies performed with conventional cultural methods suggested that Acinetobacter species predominate and/or play an important role in the EBPR process (6, 11, 12, 33, 45). However, none of Acinetobacter isolates tested exhibited characteristics that are consistent with the biochemical model of this process (13, 37). Some other species of bacteria were also isolated and identified as possible phosphate removers in the anaerobic-aerobic system (5, 18, 29, 33, 46, 47). Recently, a new polyP-accumulating gram-positive bacterium named Microlunatus phosphovorus was isolated from an EBPR process (36). To our knowledge, this bacterium is the first organism to exhibit Pi release and uptake in response to changing anaerobic-aerobic stages at the pure-culture level. Nevertheless, it is difficult to reconstruct bacterial community structures in activated sludge by studying only cultural information because of the well-known biases of culture-dependent methods, which may apply to only 1 to 15% of the total population of the sludge (1) and may provide misleading information about community structure (48).
In recent years, attempts have been made to describe bacterial communities in activated sludge systems by using non-culture-dependent chemotaxonomic and molecular methods (3, 4, 7, 15–19, 23, 24, 34, 43, 48, 49), and these approaches have provided data which contradict the previous results obtained with laboratory cultivation methods. Immunofluorescence (7) and quinone-profiling studies (17, 18) have indicated that the numbers of Acinetobacter cells are low in the EBPR process, as well as in the standard activated sludge process. Similar results were obtained with two molecular approaches (4, 49). One of these approaches, rRNA-targeted in situ hybridization, showed that members of the beta subclass of the Proteobacteria and gram-positive bacteria with high DNA G+C contents (now classified as the class Actinobacteria [44]) were numerically abundant in the EBPR system (49). The other approach involved PCR cloning and sequencing of environmental 16S rRNA genes, and this approach also showed that members of the beta subclass of the Proteobacteria were the major population constituents in this system (4).
Although rRNA approaches have become common in this area of study, the information obtained with these methods is still uncertain and somewhat different depending on the method used. This may be due in part to technical problems specific to the molecular methods, including problems with DNA retrieval, PCR bias, hybridization efficiency, and imposed selection of the retrieved or target sequences. For example, quinone pattern analyses have shown that partially saturated menaquinones (MKs), which are good biomarkers of the class Actinobacteria, usually constitute more than 20% of the total quinone content in plant scale sewage sludge (19, 23, 24), whereas 16S ribosomal DNA (rDNA) clone library approaches appear to underestimate the numbers of these bacteria (4, 43).
Previously, we used the quinone profile method to characterize bacterial community structures in the EBPR process as noted above (17) because of its simplicity and high reproducibility as a culture-independent technique. However, our previous research had a weakness; namely, a laboratory scale anaerobic-aerobic system fed with synthetic wastewater was the only system studied. Therefore, this study was designed to reexamine bacterial community structures in both plant scale and laboratory scale activated sludge reactors for EBPR by using respiratory quinone profiles. We report here that there were small differences in quinone profiles (i.e., community structures) between the EBPR and standard activated sludge systems. The usefulness of quinone profiling as a non-culture-dependent tool for quantitative evaluation of population shifts over time and space is also discussed.
MATERIALS AND METHODS
Sludge samples.
All of the activated sludge samples tested are listed in Table 1. Four EBPR sludge samples were collected from a main aeration basin used for the AO process in the Shinjiko-tobu sewage treatment plant (24) in Matsue, Japan, from July to December 1992; these samples were designated P-AO1, P-AO2, P-AO3, and P-AO4. Plant scale standard activated sludge, designated P-St, was collected from a sewage treatment plant in Chiba Prefecture, Japan. All plant sludge samples were placed in sterile polyethylene bottles, transported to the laboratory at −20°C, and stored at −80°C until analysis. For comparison, activated sludge cultivated in our laboratory was used. The laboratory system, which consisted of four jar fermentors with temperature and dissolved oxygen controllers, was seeded with standard sludge from the Chiba plant and was operated on a fill-and-draw basis with a 24-h batch cycle as described previously (18). The level of mixed liquor suspended solids (MLSS) was adjusted to 700 to 900 mg/liter every day. The reactors were fed either with raw sewage taken from the plant or with synthetic sewage (15, 23) at a biological oxygen demand loading rate of 220 to 300 mg/g of MLSS/day. The synthetic sewage was composed of (per liter of tap water) 3.0 g of Polypeptone (Daigo, Tokyo, Japan), 3.0 g of meat extract (Kyokuto, Tokyo, Japan), 3.0 g of anhydrous sodium acetate, 1.0 g of (NH4)2SO4, 1.0 g of KH2PO4, 1.0 g of K2HPO4, 0.2 g of MgCl2 · 6H2O, and 0.1 g of CaCl2 · 2H2O (pH 7.0); it was diluted with tap water at a given biological oxygen demand loading rate before feeding. The four reactors were operated as an AO system with raw sewage (L-AO/RS), an AO system with synthetic sewage (L-AO/SS), a standard system with raw sewage (L-St/RS), and a standard system with synthetic sewage (L-St/SS). After 5 weeks of acclimation, sludge samples were removed from the reactors and analyzed.
TABLE 1.
Sludges tested and their phosphorus and quinone contents
| Sludge sample | Source | Type of process | Wastewater type | Phosphorus content (μmol/mg of MLSS)a | Quinone content (nmol/mg of MLSS)
|
MK/Q ratio | ||
|---|---|---|---|---|---|---|---|---|
| Q | RQ | MK | ||||||
| P-AO1 | Shinjiko plant (July 1992) | AO | Raw sewage | 1.94 | 0.396 | 0.017 | 0.388 | 0.981 |
| P-AO2 | Shinjiko plant (August 1992) | AO | Raw sewage | 1.58 | 0.442 | 0.027 | 0.399 | 0.903 |
| P-AO3 | Shinjiko plant (October 1992) | AO | Raw sewage | 1.43 | 0.470 | 0.016 | 0.433 | 0.922 |
| P-AO4 | Shinjiko plant (December 1992) | AO | Raw sewage | 0.91 | 0.501 | 0.002 | 0.417 | 0.833 |
| P-St | Chiba plant | Standard | Raw sewage | 0.67 | 0.474 | 0.005 | 0.454 | 0.958 |
| L-AO/RS | Laboratory | AO | Raw sewage | 1.67 | 0.486 | NDb | 0.464 | 0.954 |
| L-St/RS | Laboratory | Standard | Raw sewage | 0.62 | 0.597 | ND | 0.497 | 0.833 |
| L-AO/SS | Laboratory | AO | Synthetic sewage | 1.94 | 0.734 | ND | 0.379 | 0.517 |
| L-ST-SS | Laboratory | Standard | Synthetic sewage | 0.58 | 0.887 | ND | 0.404 | 0.455 |
Sum of the phosphorus contents of the acid-soluble, lipid, alkali-soluble, hot-acid-soluble, and residual fractions.
ND, not detected.
Extraction and fractionation of quinones.
Sludge was harvested by centrifugation (12,000 × g, 10 min), washed with 50 mM phosphate buffer (pH 6.8) containing 1 mM ferricyanide, and resuspended in this buffer by using a total volume of 10 ml. Quinones were extracted three times with 2.5 volumes of a chloroform-methanol mixture (2:1, vol/vol), evaporated in a vacuum, and reextracted three times with n-hexane–water (1:1, vol/vol). Then, the crude quinone extract in n-hexane was concentrated and applied to a Sep-Pak Plus Silica column (Waters Corp., Milford, Mass.). MK and ubiquinone (Q) fractions were eluted with 20 ml of n-hexane–diethyl ether (98:2, vol/vol) and then with 20 ml of n-hexane–diethyl ether (90:10, vol/vol), respectively. The presence of MKs and Qs in both fractions was confirmed by silica gel thin-layer chromatography and UV light detection prior to high-performance liquid chromatography (HPLC) assays. Detailed information concerning the procedure used for quinone extraction and fractionation has been given elsewhere (19, 24).
Identification and quantification of quinones.
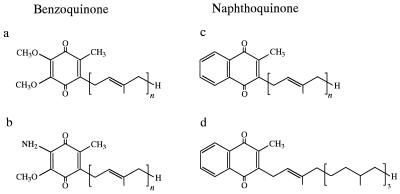
Quinone components were separated and identified by reverse-phase HPLC and photodiode array detection with internal and external standard quinones. The analytical system used has been described previously (24). Silver ion-modified thin-layer chromatography (20) and mass spectroscopy (8) performed with a Shimadzu model QP-2000 mass spectrometer were also used as supplementary tools for quinone analysis. Standard Qs and phylloquinone (K1) were obtained from Sigma Chemical Co. (St. Louis, Mo.). MK standards were prepared from known species of bacteria (15, 19). Below, Qs, rhodoquinones (RQs), and MKs with n isoprene units in their side chains are designated Q-n, RQ-n, and MK-n, respectively (Fig. 1 shows chemical structures). Partially hydrogenated MKs are designated MK-n(Hx), where x indicates the number of hydrogen atoms saturating the side chain.
FIG. 1.
Four quinone structural classes found in activated sludge. (a) Q-n. (b) RQ-n. (c) MK-n. (d) K1.
Numerical analysis.
To indicate dissimilarities among sludge samples on the basis of quinone profiles, a dissimilarity index (D) was calculated by the overlap method as described previously (19). This parameter was defined as follows:
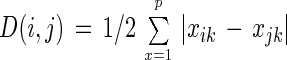 |
where Σxik = Σxjk = 100 and xik and xjk indicate the percentages of quinone homolog k in samples i and j, respectively.
Another parameter, the microbial divergence index based on quinone profiles (MDq) (23, 26), was also used to show the degree of microbial divergence of sludge. This parameter was defined as follows:
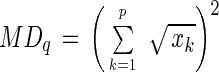 |
where xk ≥ 0.001, and xk indicates the molar ratio of quinone homolog k to the total quinone content.
Calculation of D and MDq values, tabulation of D value matrix data, and construction of a dendrogram for clustering were performed with a program written by us (27) by using the Microsoft Visual Basic programming system. The algorithm of the neighbor-joining method (41) was used for dendrogram construction. This method has been shown to give more accurate topography of dendrograms than the group average linkage method (23), which was used previously for clustering quinone patterns (19).
Other analytical methods.
Total cell counts were determined by epifluorescence microscopy performed with membrane filtration and staining with 4′,6-diamidino-2-phenylindole as described previously (39, 49). The phosphorus compounds in sludge were extracted and fractionated by the method of Langen et al. (32) with small modifications, and the acid-hydrolyzed Pi in each fraction was measured colorimetrically as described previously (25). MLSS and volatile solids in MLSS (VSS) were determined by using standard methods (2).
RESULTS
Phosphorus and quinone contents.
The phosphorus and quinone contents of all sludges tested, together with information on the processes from which the sludges were collected, are shown in Table 1. All of the EBPR sludges from the plants and the laboratory except sample P-AO4 contained high amounts of phosphorus (range, 1.35 to 1.82 μmol per mg of MLSS), whereas the phosphorus contents of the standard sludges were 0.58 to 0.67 μmol/mg of MLSS. The phosphorus contents of the EBPR sludges corresponded to 4.6 to 6.1% of the dry weight. More than 50% of the total phosphorus contents of the EBPR sludges was included in the alkali- and hot acid-soluble polyP fractions (data not shown). These data suggested that all of the EBPR sludges tested except P-AO4 had actually worked as phosphate removal systems.
All of the sludges tested contained both Qs and MKs at MK/Q molar ratios of 0.455 to 0.981. The plant sludges also contained much lower but appreciable amounts of RQs, which are derivatives of Qs. We observed no marked differences in quinone contents and MK/Q ratios between the EBPR and standard sludges. On the other hand, there were significant differences in MK/Q ratios between the sludges loaded with raw sewage and synthetic sewage.
Relationships between biomass and quinone contents.
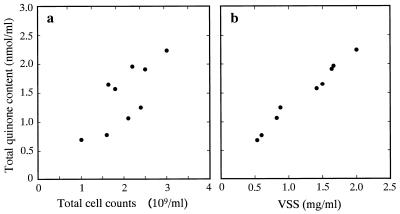
The total cell counts in the aeration basins as measured by fluorescence microscopy ranged from 1.0 × 109 to 3.0 × 109 cells/ml. The concentrations of MLSS determined were 700 to 2,430 mg/liter, and VSS accounted for 72 to 91% of the MLSS (data not shown). There was a positive relationship among the total quinone concentration, the total cell count, the MLSS content, and the VSS content in the aeration basins. Examples of the relationships between the total quinone concentration and the total cell count or VSS content are shown in Fig. 2; the correlation coefficients (r) determined for the former and latter relationships were 0.765 and 0.990, respectively. The regression equations derived from these relationships indicated that 1 nmol of quinones corresponded to 1.3 × 109 cells and 0.8 mg of VSS. The weaker direct relationship between quinone content and total cell count was probably due to the effect of dispersion and dilution of sludge flocs during cell counting.
FIG. 2.
Relationships between total quinone contents and total cell counts (a) or VSS (b) in aeration basins. The correlation coefficients (r) for the former and latter relationships are 0.765 and 0.990, respectively (n = 9).
Quinone composition.
The quinone compositions of all sludges tested, as determined by HPLC, are summarized in Table 2. In the Q fraction, Q-8 predominated, Q-10 was the second most common type, and Q-9 and other homologs were minor components in all test sludges. Also, Q-8 was the most abundant type in the total quinone (35 to 50% of the total). Small proportions of RQs, mainly RQ-8, as confirmed by mass spectrometry (M+ at m/z 712), were detected in all plant scale sludges. In the MK fraction, either MK-7 or MK-8(H4) predominated and there were significant proportions of MK-6 and MK-8 in the plant scale sludges. In the laboratory sludges, MK-8(H4) was predominant and MK-7 was the second most common type.
TABLE 2.
Quinone compositions of sludgesa
| Quinone homolog | mol% in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plant sludges
|
Laboratory sludges
|
||||||||
| P-AO1 | P-AO2 | P-AO3 | P-AO4 | P-St | L-AO/RS | L-St/RS | L-AO/SS | L-St/SS | |
| Benzoquinones | |||||||||
| Q-6 | 0.26 | 0.35 | 0.30 | 0.55 | 0.38 | 0.29 | 0.21 | 0.21 | 0.23 |
| Q-7 | 0.69 | 0.65 | 0.80 | 0.67 | 0.82 | 0.68 | 0.98 | 1.45 | 1.46 |
| Q-8 | 36.14 | 38.60 | 37.28 | 39.75 | 35.14 | 36.41 | 38.55 | 47.22 | 50.43 |
| Q-9 | 2.80 | 2.74 | 4.09 | 3.01 | 3.32 | 3.22 | 4.03 | 3.11 | 4.06 |
| Q-10 | 9.03 | 8.41 | 8.40 | 10.05 | 10.81 | 10.05 | 10.56 | 12.88 | 12.43 |
| Q-11 | 0.20 | 0.10 | 0.11 | 0.26 | 0.28 | 0.15 | 0.14 | 0.02 | 0.08 |
| RQ-7 | 0.08 | 0.06 | |||||||
| RQ-8 | 2.06 | 3.06 | 1.69 | 0.20 | 0.51 | 0.60 | |||
| Naphthoquinones | |||||||||
| K1 | 0.54 | 0.11 | 0.31 | 0.31 | 0.10 | 0.16 | 0.15 | 0.02 | 0.07 |
| MK-5 | 0.41 | 0.36 | 0.40 | 0.59 | 0.24 | 0.26 | 0.23 | 0.03 | |
| MK-6 | 5.24 | 4.21 | 5.20 | 5.10 | 2.52 | 5.85 | 4.66 | 3.26 | 2.64 |
| MK-7 | 11.91 | 9.21 | 8.70 | 7.90 | 7.45 | 10.22 | 9.02 | 8.23 | 9.09 |
| MK-8 | 8.90 | 5.90 | 7.31 | 5.34 | 9.55 | 6.68 | 8.58 | 2.01 | 1.98 |
| MK-9 | 0.44 | 0.56 | 0.40 | 0.42 | 0.51 | 0.42 | 0.41 | 0.18 | 0.21 |
| MK-10 | 1.40 | 2.86 | 1.74 | 1.20 | 1.44 | 1.01 | 0.76 | 0.39 | 0.58 |
| MK-11 | 0.34 | 0.20 | 0.33 | 0.28 | 0.38 | 0.22 | 0.39 | 0.09 | 0.11 |
| MK-12 | 0.25 | 0.18 | 0.19 | 0.18 | 0.21 | 0.12 | 0.14 | 0.01 | |
| MK-7(H2) | 0.15 | 0.57 | 0.19 | 0.75 | 1.35 | 0.12 | 0.56 | ||
| MK-8(H2) | 3.36 | 1.84 | 2.45 | 1.76 | 4.01 | 2.65 | 4.41 | 0.14 | 0.81 |
| MK-8(H4) | 11.83 | 12.99 | 12.19 | 16.90 | 11.98 | 13.47 | 9.36 | 19.03 | 14.02 |
| MK-8(H6) | 0.03 | 0.03 | 0.02 | 0.01 | 0.03 | 0.08 | 0.07 | 0.01 | |
| MK-9(H2) | 0.77 | 1.20 | 2.00 | 1.48 | 4.21 | 2.43 | 2.33 | 0.21 | 0.23 |
| MK-9(H4) | 1.92 | 2.03 | 3.14 | 2.66 | 2.25 | 1.91 | 1.21 | 1.33 | 1.31 |
| MK-9(H6) | 0.18 | 0.12 | 0.09 | 0.06 | 0.02 | 0.03 | 0.01 | 0.01 | |
| MK-9(H8) | 0.78 | 3.58 | 2.64 | 0.52 | 2.45 | 2.95 | 3.21 | 0.21 | 0.21 |
| MK-10(H2) | 0.02 | 0.02 | 0.02 | 0.03 | 0.01 | ||||
The contents of unidentified quinone homologs, which accounted for 0.2 to 1.2% of the total quinone contents, are not included, and the percentages of identified quinone homologs were recalculated to give a total value of 100%.
Numerical analysis.
The differences in quinone profiles among the samples tested were evaluated quantitatively by calculating the D and MDq values (Table 3). The D values when the sludges were compared ranged from 5.1 to 24.2%. The D values were relatively low (5.1 to 13.8%) for the sludges from the same wastewater type (i.e., sludges with raw sewage or with synthetic sewage), independent of the mode of operation (EBPR versus standard), the scale of the reactors (plant scale versus laboratory scale), and the phosphate accumulation. The values increased to 14.3 to 24.2% (mostly >20%) for the raw sewage and synthetic sewage sludges. The MDq values were similar (range, 12.4 to 13.8) for the raw sewage sludges, whereas the MDq values were lower (7.9 to 8.3) for the synthetic sewage sludges, regardless of the mode of operation and the scale of the reactors.
TABLE 3.
Quinone profile-based D and MDq values for the sludges tested
| Sample |
D value (%) compared with:
|
MDq | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P-AO1 | P-AO2 | P-AO3 | P-AO4 | P-St | L-AO/RS | L-St/RS | L-AO/SS | ||
| 13.2 | |||||||||
| P-AO2 | 9.7 | 13.5 | |||||||
| P-AO3 | 7.3 | 6.8 | 13.6 | ||||||
| P-AO4 | 12.6 | 9.8 | 10.0 | 12.5 | |||||
| P-St | 10.5 | 13.4 | 10.0 | 13.8 | 13.8 | ||||
| L-AO/RS | 7.4 | 8.3 | 6.2 | 9.4 | 12.1 | 13.0 | |||
| L-St/RS | 10.5 | 10.9 | 8.6 | 12.5 | 8.5 | 8.3 | 12.7 | ||
| L-AO/SS | 24.2 | 21.3 | 22.9 | 14.3 | 23.6 | 21.0 | 19.6 | 7.9 | |
| L-St/SS | 22.9 | 20.0 | 20.7 | 15.1 | 22.1 | 22.3 | 20.0 | 5.1 | 8.3 |
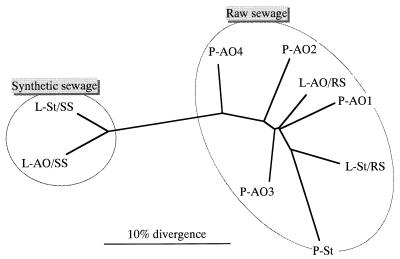
Based on the D matrix data shown in Table 3, a dendrogram grouping the sludges tested was constructed by using the algorithm of the neighbor-joining method (Fig. 3). The sludges tested were divided into two major clusters, one of which consisted of all of the EBPR and standard sludges containing raw sewage and one of which included all laboratory sludges fed with synthetic sewage. Thus, the topology of the dendrogram was independent of the capacity of sludge for phosphate accumulation. Within the raw sewage cluster, the plant and laboratory sludges overlapped each other.
FIG. 3.
Dendrogram grouping the sludges tested based on D value matrix data. The dendrogram was constructed by using the algorithm of the neighbor-joining method. The two major clusters for raw sewage sludges and synthetic sewage sludges are surrounded by lines.
When the quinone profile data reported previously for standard and EBPR sludges fed with the synthetic sewage (18, 23) were incorporated into the numerical analysis, all of these sludges fell into the synthetic sewage cluster described above at a dissimilarity level of less than 13% (data not shown).
Assignment of different quinone producers to phylogenetic taxa.
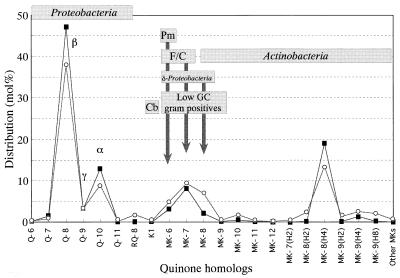
The distributions of different quinone homologs in the EBPR sludges (based on the data in Table 1) are illustrated in Fig. 4, and these quinones were assigned to phylogenetic taxa (bacteria that may have been present) on the basis of the available chemotaxonomic information. Q-8 was assigned to the beta subclass of the Proteobacteria and some members of the gamma subclass; Q-9 was assigned members of the gamma subclass, such as members of the genera Acinetobacter and Pseudomonas; and Q-10 was assigned to the alpha subclass of the Proteobacteria (50). RQ-8 might have been derived from the second quinone component of certain members of the beta subclass, such as Brachymonas (21) and Zoogloea (22) species. MKs with short isoprene units (MK-6 to MK-8) were assigned to the Flavobacterium-Cytophaga group (35, 38), the gram-positive bacteria with low G+C contents (9), the genus Planctomyces (42), and the delta subclass of the Proteobacteria (9, 10), and MKs with longer side chains or partially hydrogenated chains were assigned to the class Actinobacteria. Partially saturated MKs are distributed in some species of the sulfate-reducing proteobacteria (10). However, since these MKs are limited to MKs with short isoprene units (less than seven units long), most of the partially hydrogenated MKs found in the sludges could be assigned to the class Actinobacteria.
FIG. 4.
Distribution of different quinone homologs in EBPR sludges and assignment of these quinones to their possible sources (phylogenetic groups of bacteria). Symbols: ○, average data for the plant EBPR sludges with high phosphate contents (P-AO1, P-AO2, and P-AO3); ▪, data for the laboratory EBPR sludge fed with synthetic sewage (L-AO/SS). Cb, cyanobacteria; F/C, Flavobacterium-Cytophaga cluster in the Cytophaga-Bacteroides phylum. Pm, Planctomyces.
DISCUSSION
Advances in microbial chemotaxonomy based on quinone patterns (8, 9) provide a basis for the idea that quinone profiling of an environmental sample yields insight into the proportions of species with different homolog types in the total population. One of the major problems that complicate the assessment of in situ bacterial populations by quinone profiling is that the quinone contents of bacterial cells vary among taxa or in response to environmental stress. A previous study showed that the ratio of total quinones to biomass as measured by membrane lipid phosphate was different under different environmental conditions (14). In this study, however, we found that the total quinone content was highly correlated with the biomass, as indicated by the VSS data for activated sludge systems (Fig. 2). Another research group has also demonstrated that there is a positive relationship between quinone content and biomass in soil (40). Biomarker approaches, including quinone profiling, in general suffer from another problem, namely, that only information concerning culturable bacteria can be considered when a community structure is interpreted, even if numerous unknown bacteria are present. Nevertheless, this is not a problem in cluster analyses of microbial communities based on numerically processed quinone data.
Our previous studies have shown that the quinone patterns of municipal sewage activated sludges are similar to each other (15–19, 23, 24). The general quinone profiles of these sludges are as follows: (i) Qs and MKs are present at MK/Q ratios of 0.6 to 1.0 in most cases (but the MK/Q ratios are 0.3 to 0.6 for synthetic sewage sludges); (ii) Q-8 is the most abundant type of quinone; (iii) the proportion of Q homologs decreases in the order Q-8 > Q-10 > Q-9 > other Qs; and (iv) the predominant MK type is either MK-7, MK-8, or MK-8(H4) [in some cases, MK-6 or MK-9(H8)]. The results of this study are consistent with the general information noted above and also support our previous data obtained for laboratory scale EBPR activated sludge with synthetic sewage (17).
In view of the present data, together with the previous findings, it is clear that most of the bacteria in both EBPR and standard activated sludge systems are members of the Proteobacteria and that Q-8-producing species (i.e., species belonging to the beta subclass) are the most abundant species (>30% of the total population) (Fig. 4). Some species belonging to the gamma subclass, such as the enterobacterial species, also contain Q-8 as a major quinone. However, the contribution of these bacteria to the Q-8-producing population may be negligible, because demethylmenaquinone, another indicator of the enterobacteria, did not occur in appreciable amounts in any sludge. The low Q-9 contents in all sludges suggest that Acinetobacter and Pseudomonas species, which are representatives of the gamma subclass, constitute minor populations (less than 4% of the total population). Members of the alpha subclass of the Proteobacteria may constitute about 10% of the total population, as judged from the Q-10 content. These results agree well with the results of rRNA in situ hybridization and 16S rDNA clone library studies, all of which have indicated that members of the beta subclass of the Proteobacteria are predominant and minor populations of Acinetobacter strains occur in EBPR and standard systems (4, 30, 43, 48, 49). The finding that MKs with long isoprene units (n ≤ 10) and with partially saturated chains were present at relatively high levels (>18%) suggests that the Actinobacteria is the most abundant phylogenetic group next to the beta subclass of the Proteobacteria. A high proportion of members of the Actinobacteria in an EBPR process has also been revealed by rRNA-targeted oligonucleotide probing (49), whereas 16S rRNA clone library studies failed to detect this phylogenetic group as a major population component (4, 43). It has been suggested that the polyP-accumulating bacterium M. phosphovorus, which contains MK-9(H4) as its sole quinone (36), is only a minor component of the populations in EBPR sludges if it is present at all, in view of the low MK-9(H4) contents of these sludges (less than 3%). A similar result was obtained by dual staining of EBPR sludge with rRNA-targeted probes and a polyP-specific fluorescent dye (30). The MK profiles of the sludges also suggest that bacteria containing MK-6 to MK-8 (e.g., bacteria in the Cytophaga-Flavobacterium group, Planctomyces strains, and/or gram-positive bacteria with low G+C contents) are present at significant levels in both EBPR and standard processes. The presence of these phylogenetic groups in the EBPR process was demonstrated by a 16S rDNA clone library study (4). In view of the quinone profiles of the EBPR sludge, it is necessary to consider a number of species of at least two phylogenetic groups, the Proteobacteria and the Actinobacteria, as possible phosphate removers.
Numerical analyses of quinone data indicated that there were small differences in the profiles of EBPR and standard activated sludges fed with the same type of wastewater (Table 3). The levels of dissimilarity between the two processes, as indicated by the D values, were 5.1 to 13.8%. Previously, it has been shown that seasonal variations in the D values in a sewage activated sludge treatment plant are less than 20%, if the plant is operated under normal conditions (24). Similar variations in D values have also been found among sewage sludges in different plants which are operating under good conditions (23). Therefore, the dissimilarities between the EBPR and standard processes for the same wastewater type may correspond to (or be smaller than) the dissimilarities found seasonally in the same plant or the dissimilarities among different sewage treatment plants. In contrast, the levels of dissimilarity between the sludges loaded with different types of wastewater (i.e., natural sewage versus synthetic sewage) were higher (mostly >20%), independent of the operational mode, the scale of the reactors, and the phosphate accumulation. The neighbor-joining dendrogram based on the D value matrix data indicated that the sludges tested fell into two major clusters depending on the type of wastewater (i.e., there was a raw sewage cluster and a synthetic sewage cluster). Within the raw sewage cluster, the EBPR and standard sludges or the plant scale and laboratory scale sludges overlapped each other. These findings suggest that the introduction of an anaerobic stage into the aerobic process results in no more significant population shift than changes in the quality of wastewater. Also, the scale of reactors may have no or little effect on the community structure as long as the quality and loading rate of wastewater are constant.
The natural and synthetic sewage EBPR sludges were almost identical in their qualitative quinone patterns (Fig. 4). However, the quantitative quinone profiles of the two groups of sludge differed significantly, as indicated by the D and MDq values. This may be explained by the possibility that similar genera or species of phosphate-accumulating bacteria play the major role in both EBPR systems but the proportions of the individual populations in the total population differ in the two systems. Alternatively, it is possible to speculate that the same quinone-containing species but different species of phosphate-accumulating bacteria are present in the two EBPR systems; this would affect the whole community structure differentially, resulting in different levels of microbial divergence in the two systems. Although phosphate removal can be effectively achieved by the laboratory scale synthetic sewage EBPR process, as well as by the plant scale system, it is our view that the former system should not be considered a model of the latter with respect to bacterial population structure.
Quinone profiling has gained general acceptance as a biomarker approach for characterizing in situ bacterial communities but has received less attention as a culture-independent tool than molecular techniques that now enjoy widespread use in wastewater microbiology and microbial ecology. Certainly, the quinone profile method is inferior to the rRNA approaches for resolving phylogenetic taxa. However, since this biomarker method is a direct chemical analysis method for environmental lipids, it provides higher reproducibility and reliability without any bias based on extraction and identification of the molecules. It also provides quantitative data useful for grouping whole microbial populations in situ. A neighbor-joining dendrogram inferred from quinone-based D value matrix data is useful for quantitative evaluation of microbial population shifts over time and space, as reported here. A combination of the quinone profile method with molecular and ecophysiological techniques should provide better understanding of the EBPR process.
REFERENCES
- 1.Amann R L, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2540 Solids. In: Eaton A D, Clesceri L S, Greenberg A E, editors. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C: American Public Health Association; 1995. pp. 2-53–2-58. [Google Scholar]
- 3.Auling G, Pilz F, Busse H-J, Karrasch S, Streichan M, Shön G. Analysis of the polyphosphate-accumulating microflora in phosphorus-eliminating, anaerobic-aerobic activated sludge systems by using diaminopropane as a biomarker for rapid estimation of Acinetobacter spp. Appl Environ Microbiol. 1991;57:3583–3592. doi: 10.1128/aem.57.12.3585-3592.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludge from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodisch K E U, Joyner S J. The role of microorganisms other than Acinetobacter in biological phosphate removal in activated sludge process. Water Sci Technol. 1983;15:117–125. [Google Scholar]
- 6.Buchan L. The location and nature of accumulated phosphorus in seven sludges from activated sludge plants which exhibited enhanced phosphorus removal. Water S A (Preteria) 1981;7:1–7. [Google Scholar]
- 7.Cloete T E, Steyn P L. A combined membrane filter-immunofluorescent technique for the in situ identification and enumeration of Acinetobacter. Water Res. 1988;22:961–969. [Google Scholar]
- 8.Collins M D. Analysis of isoprenoid quinones. Methods Microbiol. 1985;18:329–366. [Google Scholar]
- 9.Collins M D, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins M D, Widdel F. Respiratory quinones of sulphate-reducing and sulphur-reducing bacteria: a systematic investigation. Syst Appl Microbiol. 1986;8:8–18. [Google Scholar]
- 11.Deinema M H, Habets L H A, Scholten J, Turkstra E, Webers H A A M. The accumulation of polyphosphate in Acinetobacter spp. FEMS Microbiol Lett. 1980;9:275–279. [Google Scholar]
- 12.Fuhs G W, Chen M. Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater. Microb Ecol. 1975;2:119–138. doi: 10.1007/BF02010434. [DOI] [PubMed] [Google Scholar]
- 13.Groenestijn J W, Zuidema M, van de Worp J J M, Deinema M H, Zehnder A J B. Influence of environmental parameters on polyphosphate accumulation in Acinetobacter sp. Antonie Leeuwenhoek. 1989;55:67–82. doi: 10.1007/BF02309620. [DOI] [PubMed] [Google Scholar]
- 14.Hedrick D B, White D C. Microbial respiratory quinones in the environment. I. A sensitive liquid chromatographic method. J Microbiol Methods. 1986;5:243–254. [Google Scholar]
- 15.Hiraishi A. Respiratory quinone profiles as tools for identifying different bacterial populations in activated sludge. J Gen Appl Microbiol. 1988;34:39–56. [Google Scholar]
- 16.Hiraishi A. Isoprenoid quinone profiles for identifying and classifying microorganisms in the environment. In: Hattori T, Ishida Y, Maruyama Y, Morita R Y, Uchida A, editors. Recent advances in microbial ecology. Tokyo, Japan: Japan Scientific Societies Press; 1989. pp. 663–668. [Google Scholar]
- 17.Hiraishi A, Masamune K, Kitamura H. Characterization of the bacterial population structure in an anaerobic-aerobic activated sludge system on the basis of respiratory quinone profiles. Appl Environ Microbiol. 1989;55:897–901. doi: 10.1128/aem.55.4.897-901.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraishi A, Morishima Y. Capacity for polyphosphate accumulation of predominant bacteria in activated sludge showing enhanced phosphate removal. J Ferment Bioeng. 1990;69:368–371. [Google Scholar]
- 19.Hiraishi A, Morishima Y, Takeuchi J. Numerical analysis of lipoquinone patterns in monitoring bacterial community dynamics in wastewater treatment systems. J Gen Appl Microbiol. 1991;37:57–70. [Google Scholar]
- 20.Hiraishi A, Shin Y K, Sugiyama J. Rapid profiling of bacterial quinones by two-dimensional thin-layer chromatography. Lett Appl Microbiol. 1992;14:170–173. [Google Scholar]
- 21.Hiraishi A, Shin Y K, Sugiyama J. Brachymonas denitrificans gen. nov., sp. nov., an aerobic chemoorganotrophic bacterium which contains rhodoquinones, and evolutionary relationships of rhodoquinone producers to bacterial species with various quinone classes. J Gen Appl Microbiol. 1995;41:99–117. [Google Scholar]
- 22.Hiraishi A, Shin Y K, Sugiyama J, Komagata K. Isoprenoid quinones and fatty acids of Zoogloea. Antonie Leeuwenhoek. 1992;61:231–236. doi: 10.1007/BF00584229. [DOI] [PubMed] [Google Scholar]
- 23.Hiraishi A, Ueda Y. Evaluation of microbial population structures of synthetic-wastewater activated sludge and plant-scale sewage sludge on the basis of respiratory quinone profiles. Jpn J Water Treat Biol. 1997;33:137–149. . (In Japanese.) [Google Scholar]
- 24.Hiraishi A, Ueda Y, Ishihara J, Mori T. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol. 1996;42:457–469. [Google Scholar]
- 25.Hiraishi A, Yanase A, Kitamura H. Polyphosphate accumulation by Rhodobacter sphaeroides under different environmental conditions with special emphasis on the effect of external phosphate concentrations. Bull Jpn Soc Microb Ecol. 1991;6:25–32. [Google Scholar]
- 26.Hu H-I. Kinetic and ecological studies on aerobic submerged biofilter for wastewater treatment. Ph.D. thesis. Yokohama, Japan: Yokohama National University; 1993. . (In Japanese.) [Google Scholar]
- 27.Iwasaki, M., and A. Hiraishi. Unpublished data.
- 28.Jenkins D, Tandori V. The applied microbiology of enhanced biological phosphate removal—accomplishments and needs. Water Res. 1991;25:1471–1478. [Google Scholar]
- 29.Kavanaugh R G, Randall C W. Bacterial populations in a biological nutrient removal plant. Water Sci Technol. 1994;29(7):25–34. [Google Scholar]
- 30.Kawaharasaki, M., H. Tanaka, T. Kanagawa, and K. Nakamura. Personal communication.
- 31.Kortstee G J J, Appeldoorn K J, Bonting C F C, van Niel E W J, van Veen H W. Biology of polyphosphate-accumulating bacteria involved in enhanced biological phosphorous removal. FEMS Microbiol Rev. 1994;15:137–153. doi: 10.1111/j.1574-6976.1994.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 32.Langen P, Liss E, Lohmann K. Art, Bildung und Umsatz der Polyphosphate der Hefe. Collq Int Cent Natl Rech Sci. 1960;106:603–612. [Google Scholar]
- 33.Lötter L H, Murphy M. The identification of heterotrophic bacteria in an activated sludge plant, with particular reference to polyphosphate accumulation. Water S A (Pretoria) 1985;11:179–184. [Google Scholar]
- 34.Manz W, Wagner M, Amann R, Schleifer K-H. In situ characterisation of the microbial consortia active in two wastewater treatment plants. Water Res. 1994;28:1715–1723. [Google Scholar]
- 35.Nakagawa Y, Yamasato K. Phylogenetic diversity of the genus Cytophaga revealed by 16S rRNA sequencing and menaquinone analysis. J Gen Microbiol. 1993;139:1155–1161. doi: 10.1099/00221287-139-6-1155. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura K, Hiraishi A, Yoshimi Y, Kawaharasaki M, Masuda K, Kamagata Y. Microlunatus phosphovorus gen. nov., sp. nov., a new gram-positive polyphosphate-accumulating bacterium isolated from activated sludge. Int J Syst Bacteriol. 1995;45:17–22. doi: 10.1099/00207713-45-1-17. [DOI] [PubMed] [Google Scholar]
- 37.Ohtake H, Takahashi K, Tsuzaki Y, Toda K. Uptake and release of phosphate by a pure culture of Acinetobacter calcoaceticus. Water Res. 1985;19:1587–1594. [Google Scholar]
- 38.Oyaizu H, Komagata K. Chemotaxonomic and phenotypic characterization of the strains of species in the Flavobacterium-Cytophaga complex. J Gen Appl Microbiol. 1981;27:57–107. [Google Scholar]
- 39.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 40.Saito, K., A. Katayama, and K. Fujie. Personal communication.
- 41.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Sittig M, Schlesner H. Chemotaxonomic investigation of various prosthecate and/or budding bacteria. Syst Appl Microbiol. 1993;16:92–103. [Google Scholar]
- 43.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stackebrandt E, Rainey F A, Ward-Rainey N L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 45.Stephenson T. Acinetobacter, its role in biological phosphate removal. In: Ramadori R, editor. Biological phosphate removal from wastewaters. Oxford, United Kingdom: Pergamon Press; 1987. pp. 313–316. [Google Scholar]
- 46.Streichan M, Golecki J R, Schön G. Polyphosphate-accumulating bacteria from sewage plants with different processes for biological phosphorus removal. FEMS Microbiol Ecol. 1990;73:113–124. [Google Scholar]
- 47.Surech N, Warburg R, Timmerman M, Wells J, Coccia M, Robert M F, Halvorson H O. New strategies for the isolation of microorganisms responsible for phosphate accumulation. Water Sci Technol. 1985;17(11/12):43–56. [Google Scholar]
- 48.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner M, Erhart R, Manz, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokota A, Akagawa-Matsushita M, Hiraishi A, Katayama Y, Urakami T, Yamasato K. Distribution of quinone systems in microorganisms: gram-negative eubacteria. Bull Jpn Fed Culture Collection. 1992;8:136–171. [Google Scholar]