Abstract
Purpose:
To perform automated segmentation of corneal nerves and other structures in corneal confocal microscopy (CCM) images of the subbasal nerve plexus (SNP) in eyes with ocular surface diseases (OSD).
Methods:
A deep learning-based two-stage algorithm was designed to perform segmentation of SNP features. In the first stage, to address applanation artifacts, a generative adversarial network enabled deep network was constructed to identify 3 neighboring corneal layers on each CCM image: epithelium, SNP, and stroma. This network was trained/validated on 470 images of each layer from 73 individuals. The segmented SNP regions were further classified in the second stage, by another deep network into: background, nerve, neuroma, and immune cells. Twenty-onefold cross-validation was used to assess the performance of the overall algorithm on a separate dataset of 207 manually segmented SNP images from 43 OSD patients.
Results:
For the background, nerve, neuroma, and immune cell classes, the Dice similarity coefficients (DSC)s of the proposed automatic method were 0.992, 0.814, 0.748, and 0.736, respectively. The performance metrics for automatic segmentations were statistically better or equal as compared to human segmentation. Additionally, the resulting clinical metrics had good to excellent intraclass correlation coefficients (ICC)s between automatic and human segmentations.
Conclusion:
The proposed automatic method can reliably segment potential CCM biomarkers of OSD onset and progression with accuracy on par with human gradings in real clinical data, which frequently exhibited image acquisition artifacts. To facilitate future studies on OSD, we made our dataset and algorithms freely available online as an open-source software package.
Keywords: CCM, Ocular Surface Disease, Dry Eye Disease, Neuropathic Corneal Pain
Introduction
The cornea, through its 7000 nerve terminals per square millimeter, is the most densely innervated tissue in the body.1 These nerves, in addition to their sensory role, are critical for maintaining the homeostasis of the ocular surface. Specifically, they serve as the fundamental input channel to a dynamic circuitry known as the lacrimal function unit (LFU), which regulates blinking and lacrimation and maintains the structural and functional integrity of the ocular surface.2
Naturally, disruptions to the nerves and/or their connections may result in an inappropriate response to environmental stressors culminating in ocular surface damage or disease. For this reason, ocular surface diseases (OSD), such as dry eye disease (DED) and neuropathic corneal pain (NCP), frequently present with corneal nerve anomalies.3,4 In response, patients experience behavioral changes and report increased or decreased levels of corneal sensitivity. For diagnostic purposes, clinicians cannot rely on functional data alone as it has been, thus far, insufficient for objective disease differentiation and progression analysis.5,6 As a result, clinicians struggle to (a) identify a patient’s position on the neurosensorial spectrum, (b) prescribe appropriate therapies for the underlying OSD, and (c) assess improvement or decline in the patient’s condition.
Direct visualization of the corneal nerves and their surroundings may prove fruitful in solving these challenges. Corneal nerve abnormalities, neuromas, and immune cells in the subbasal nerve plexus (SNP) layer of the cornea have been observed in eyes with OSD.7–9 Similar to advances in the treatment of retinal diseases,10 one may potentially use these structural features to stratify OSD patients into subgroups that reflect specific pathophysiological mechanisms and use these subgroupings for better identification and to develop rapid, objective, and personalized treatment plans for OSD patients.
Corneal confocal microscopy (CCM), also known as in vivo confocal microscopy, is an emerging approach for the visualization of corneal nerves in clinics.11 However, the avalanche of data generated by CCM imaging systems (consisting of up to hundreds of images per eye) is often too large, costly, and time-consuming to be comprehensively evaluated manually by expert scientists and clinicians. Moreover, the subjective nature of human image analysis is difficult to consistently reproduce, quantify, and compare with other groups. Thus, in recent years, there have been efforts to utilize machine learning to segment features of interest in CCM images.12–17 Despite several exciting advances, there is a need for a tool that can segment and analyze the nerves, neuromas, and immune cells with accuracy on par with human grading on low-quality clinical images, including those with applanation artifacts,18 which can impact the performance of segmentation algorithms and clinical metrics.
Therefore, we propose a comprehensive and robust AI-based technology to attain fast and objective measurements of nerves, neuromas, and immune cells in CCM images of the SNP from different subjects. The overall AI algorithm will be implemented in two steps: (1) we segment and remove applanation artifacts, and (2) we detect nerves and other structures in the unmarred portion of the image. Subsequently, we use these segmentations to automatically calculate clinical metrics, which can be applied at the corneal vortex and other (optimally central) cornea locations. To facilitate future studies on OSD, we have made the algorithms resulting from this study and the corresponding annotated dataset freely available online as an open-source software package.
Materials and Methods
Dataset
CCM images were captured from controls and OSD patients at the Foster Center for Ocular Immunology at the Duke Eye Center, under the IRB protocol 00095121. All eyes were imaged with the Heidelberg Retina Tomograph 3 with the Rostock Cornea Module (Heidelberg Engineering, GmbH, Heidelberg, Germany). Volume imaging with 15 frames per second was captured at depths of 0–100 μm to visualize the epithelium, SNP, and stroma. Additionally, sequence imaging with 15 frames per second was captured at a depth of 50–80 μm to visualize a broader region of the SNP in the central cornea and around the corneal whorl. The height and width of the CCM images were 384 pixels, with a resolution of 1.04 μm/pixel.
We had two mutually exclusive datasets: the applanation artifact removal network (AAR-Net) dataset and the subbasal nerve plexus network (SNP-Net) dataset. To avoid the algorithm being trained and tested on the same patient, these datasets did not share any patients. The AAR-Net dataset patients were exclusively used for training, whereas the SNP-Net dataset patients were used for both training and testing (via cross-validation).
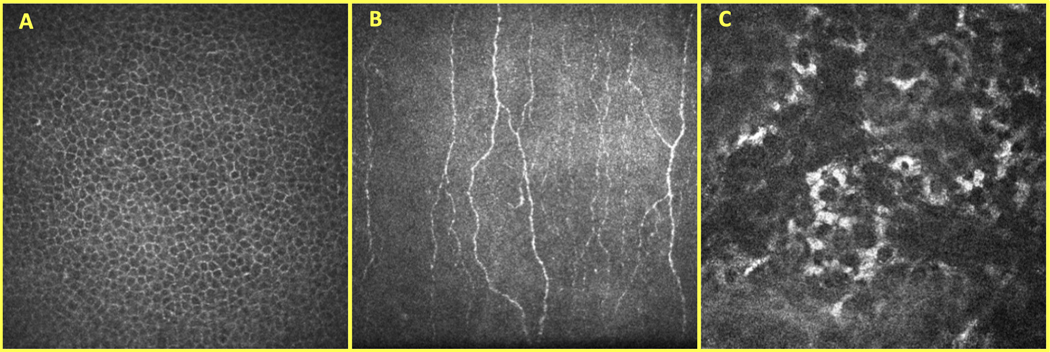
The AAR-Net dataset included 470 epithelium, 470 SNP, and 470 stroma images from 117 eyes (64 controls; 53 OSD) and 73 individuals (40 controls; 33 OSD). The OSD eyes included patients with DED, NCP, neurotrophic keratitis, infectious keratitis, and keratouveitis. One to ten non-identical images that did not present applanation artifacts were selected per eye from each corneal layer. Figure 1 shows an example of CCM images of the epithelium (A), SNP (B), and stroma (C).
Figure 1.
Corneal confocal microscopy (CCM) images of the epithelium (A), subbasal nerve plexus (SNP) (B), and stroma (C) layers of the cornea.
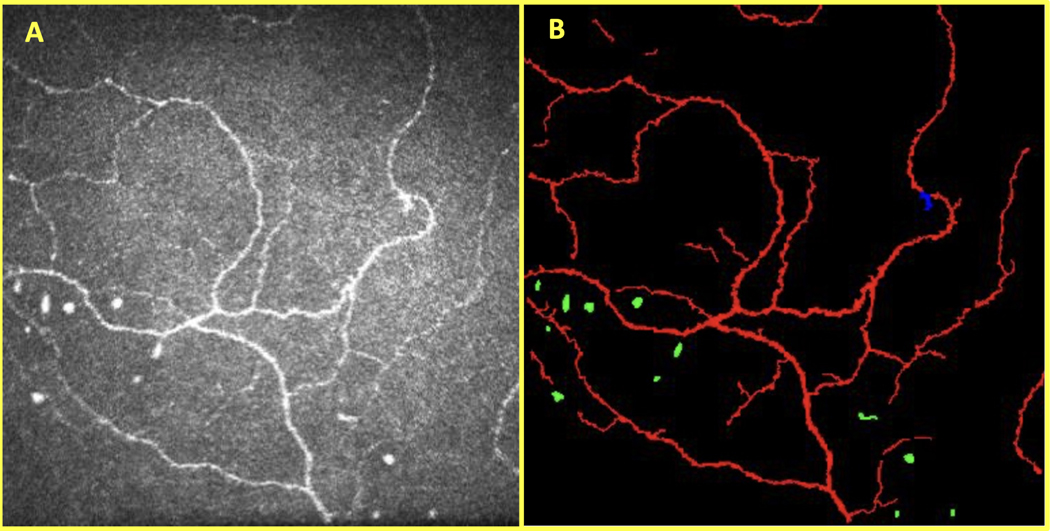
The SNP-Net dataset included 69 eyes (45 DED; 24 NCP) from 43 OSD patients. All eyes came from DED suspects who were imaged sequentially at the Duke Eye Center. Three SNP images were randomly selected per eye (207 images total); one image was selected from the corneal whorl (if available) and the other images were from other SNP locations in the central cornea. Next, a trained reader (Reader 1, Z.Z.Z.) used a custom MATLAB program to segment structures in the SNP. In addition to its multi-class capabilities, this program enabled the reader to perform more detail-oriented segmentations than possible with ACCMetrics.19 The reader manually segmented nerves, presumed neuromas, and presumed immune cells. We defined these structures according to a review done by Chinnery et al.20 and a literature search. Nerves consisted of thin, elongated, and branch-like structures that were brighter than the background. Presumed neuromas were defined as an asymmetrical thickening of the nerve or a sudden swelling across the nerve.11,21–24 They were usually amorphous and hyperreflective as compared to the surrounding nerves and tissue. Finally, presumed immune cells in the central cornea had four primary appearances: (1) bright, round/globular structures (i.e., non-dendritic immune cells), (2) bright, crescent-shaped structures (i.e., immature dendritic cells), (3) bright, short, serpentine structures (i.e., mature dendritic cells), and (4) bright, large trapezoidal-shaped structures with multiple branches (i.e., mature dendritic cells) .25–28 Figure 2 shows an example of the manual segmentation of the target features on a CCM image.
Figure 2.
CCM image of the SNP (A) and its corresponding manual segmentation (B) with nerves (red), neuromas (blue), and immune cells (green).
A subset of the patients from the AAR-Net dataset was also used to train (but not test) SNP-Net. Specifically, we obtained SNP images containing corneal compression artifacts from these patients. This dataset included 25 images from 11 eyes (5 controls; 6 OSD) and 7 patients (3 controls; 4 OSD). The corneal compression artifacts were manually segmented out (Reader 1, Z.Z.Z) of the image such that they could be used for training without incorporating the other regions of the image.
SNP Segmentation Algorithm
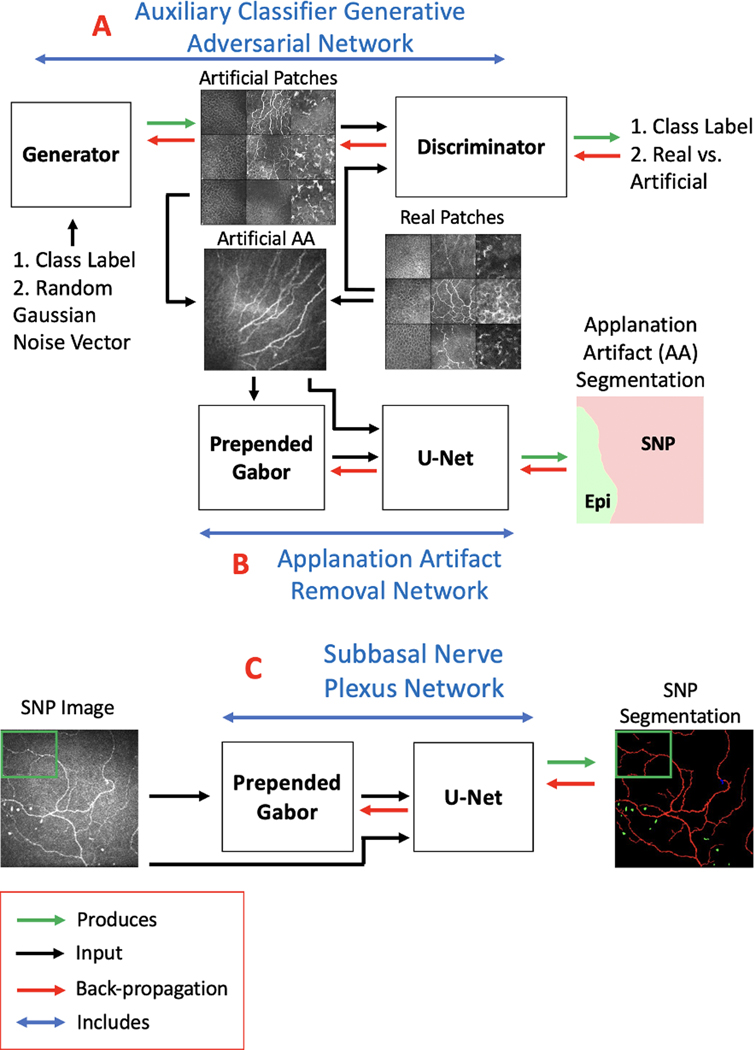
The schematic in Figure 3 outlines the core steps in our segmentation algorithm. The proposed method consisted of two convolutional neural networks (CNNs) that operated in sequential stages. In the first stage, the AAR-Net (Figure 3.B) was developed to segment three corneal layers and remove stroma and epithelium layers from an image. For the second stage, the SNP-Net (Figure 3.C) was developed to segment structures such as nerve, neuromas, and immune cells in the SNP layer.
Figure 3.
Schematic for the proposed corneal confocal microscopy image analysis method. The subbasal nerve plexus (SNP) segmentation algorithm includes: (A) an auxiliary classifier generative adversarial network (AC-GAN), (B) an applanation artifact removal network (AAR-Net), and (C) an SNP segmentation network (SNP-Net).
Both AAR-Net (Figure 3.B) and SNP-Net (Figure 3.C) are patch-based networks, which included a trainable, prepended Gabor network and a U-Net backbone. Patch-based networks29 are helpful when constrained by a small dataset; the network will have fewer weights to train and a greater quantity of images to train on. We prepended a trainable Gabor filtering network as they are well-suited for identifying elongated structures such as corneal nerves.30 In this Gabor network, the only trainable parameters were the frequency and orientation. As such, the addition of a Gabor network facilitated training on a small dataset without substantially increasing the number of network parameters. The U-Net backbone is frequently utilized for medical image segmentation.31 It had four identical encoder blocks, a bottleneck block, four identical decoder blocks, and a prediction block. Every encoder block consisted of a “double convolution” followed by a max pooling layer. A double convolution included a convolution layer, a batch normalization layer,32 and a ReLU non-linearity repeated twice. The bottleneck block was equivalent to the encoder block but without the max pooling layer. Every decoder block consisted of a concatenation between the outputs of a transposed convolution layer and the corresponding encoder block, followed by a double convolution. At the end, the prediction block consisted of a convolution layer and a SoftMax operation, which normalized the final outputs to probability values ranging from 0 to 1. This probability map, p, had the same dimensionality as the input image, but with one channel for every class.
The loss functions of AAR-Net and SNP-Net utilized pixel-wise cross-entropy loss, defined as
where was the binary manual segmentation map of each class. AAR-Net was further optimized by performing inverse class weighting33 and edge weighting34 on the pixel-wise cross-entropy loss. The AAR-Net’s loss was defined as
Similarly, SNP-Net’s loss was defined as
Finally, the AAR-Net received supplemental training via image examples (Figure 4.A2–C2) generated from an auxiliary classifier generative adversarial network (AC-GAN).35 The AC-GAN consisted of two neural networks trained in opposition to one another: the generator and discriminator (Figure 3.A). The generator took a class label (one of the three corneal layers) and a random Gaussian noise vector as inputs and output an artificial image. It had one fully connected layer followed by three sequential convolutional layers. Between each layer, it had batch normalization, leaky ReLU non-linearity, and nearest-neighbor up-sampling. The final output was normalized via a Tanh operation. The discriminator took either a real training image () or an artificial image () as input and output probability distributions over sources (real versus artificial) and class labels. It had four sequential convolutional layers followed by two parallel fully connected layers. Between each layer, it had leaky ReLU non-linearity, dropout, and batch normalization. One fully connected layer’s output was normalized via a sigmoid operation whereas the other was normalized via a SoftMax operation. These output probability distributions were then converted to cross-entropy loss terms and , respectively. The generator’s loss was defined as
and calculated for the artificial images. The discriminator’s loss was defined as
but it was calculated for both real and artificial images.
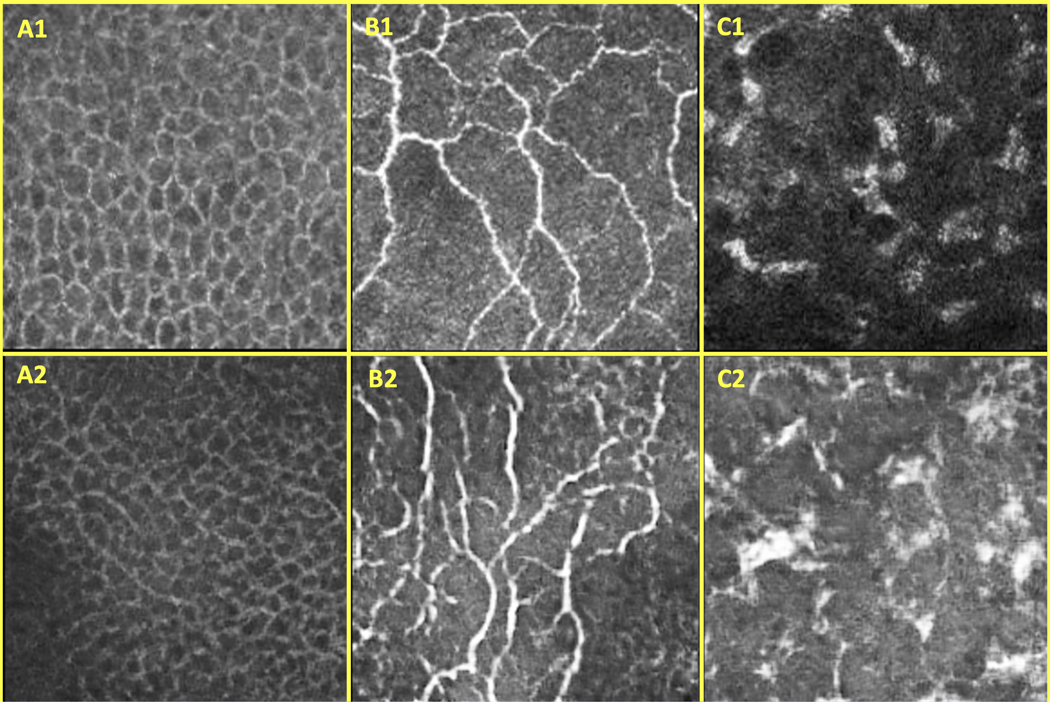
Figure 4.
A set of real CCM images (1) and a set of AC-GAN-generated CCM images (2) of the epithelium (A), SNP (B), and stroma (C)
Training and Validation
AAR-Net was designed and trained to identify different corneal layers on singular images, a capability beyond previous works that could identify only one layer per image.36 To improve the generalizability of AAR-Net, instead of solely relying on traditional image augmentations (i.e., rotation, translation, cropping, and scaling), we included an AC-GAN to synthesize a diverse set of realistic CCM images for training.35 While training AAR-Net, the AC-GAN produced one artificial image for every real image in our training dataset, which consisted of 1230 manually labeled images (410 of each class). The remaining 180 images (60 of each class) were used for validation. Given this curated dataset, we randomly extracted patches (1/4 of total image size) from the single-class areas of the real images. These extracted image patches, along with the ones generated by the trained AC-GAN, were used in the training of the AAR-Net. Every patch had a random probability of being merged with a patch from a different corneal layer. These merged patches were intensity equalized and then smoothed and stitched along a randomly generated junction line. The ground truth segmentation masks were based on both the original cornea layer classes and the location of the junction line. Through this process, we created innumerable permutations of marred images to train AAR-Net. Ultimately, image regions containing stroma or epithelium, as identified by AAR-net, were not trained, validated, or tested by SNP-Net.
SNP-Net was trained to reproduce the manual segmentations and to ignore corneal compression artifacts. We trained it on randomly extracted patches (1/16 of total image size) from the applanation artifact-free regions of the SNP images. Every patch had a random probability of being merged with patches of segmented images containing corneal compression artifacts. Regions that were replaced with a corneal compression artifact were then labeled as containing background class.
The AAR and SNP networks’ weights were randomly initialized using He initialization.37 Adam stochastic gradient descent,38 with learning rates of 0.005 and 0.0005, was used to optimize the weights by minimizing and in the first and second stages, respectively. The learning rates had a decay rate of 0.5 every 20 epochs. Additionally, L2 weight regularization was applied with a factor of 1e-8. The networks were trained with a fixed batch size for a fixed number of epochs. The validation dice similarity coefficient was monitored on the validation set for the last 40 epochs, and the weights corresponding to the highest Dice similarity coefficient were retained as the final network weights that were then used for testing. For AAR-Net, a batch size of 20 and 80 epochs were used, whereas for SNP-Net, a batch size of 80 and 100 epochs were used.
Although indirectly involved in the SNP segmentation pipeline, the AC-GAN had to be trained as well. The AC-GAN was trained on the same dataset of 1410 manually labeled images as the AAR-Net. We randomly extracted patches (1/4 of total image size) from the single-class areas of the images and used them to train the AC-GAN. The network’s weights were randomly initialized using a normal distribution. Adam stochastic gradient descent, with a learning rate of 0.002 and decay rate of 0.5 every 100 epochs, was used to optimize the weights. Over 400 epochs, and with a batch size of 20, the parameters of the generator were fine-tuned to produce realistic, labeled CCM images from an implicit distribution derived from a sampling of the real data distribution.
Testing
During testing, for each SNP image, a trained AAR-Net was used to predict the marred regions of the image (i.e., the regions containing stroma or epithelium). AAR-Net performed patch-based segmentation29 on the SNP-Net dataset. A weighted amalgamation of the patch’s probability maps was used to produce global probability maps from which the arguments of the maxima were found. In total, 5 AAR-Net models were trained to completion. For each SNP image, the mean of the five AAR-Net outputs was used as the final segmentation.
In the second stage, the SNP-Net performed patch-based segmentation on the applanation artifact-free regions of the images. A weighted amalgamation of the patch’s probability maps was used to produce global probability maps from which the arguments of the maxima were found. In total, 5 SNP-Net models were trained to completion for each cross-validation group. For each SNP image, the mean of the five SNP-Net outputs was used as the final segmentation.
Post-processing
Post-processing techniques were implemented on the SNP-Net segmentation outputs. (1) To prevent objects with neuroma and immune cell class mixtures, we assigned all pixels within a given mixed object to the majority class. (2) To minimize the occurence of objects with nerve and immune cell class mixtures and improve the segmentation of mature immune cells, we assigned all pixels to the immune cell class if it was the majority class (3) Non-background classes that were engulfed by another non-background class were reassigned to the surrounding class. (4) Very small non-background class objects were reassigned to the background class.
Performance Metrics
To make a quantitative evaluation of automated segmentation results, we calculated the overlap with the manual segmentation. Specifically, we used the performance metric, Dice similarity coefficient (DSC), for each class. This metric ranges from 0 to 1, with 1 showing perfect overlap and 0 indicating no overlap. The DSC was calculated as
where TP was the number of true positives (in pixels), TN was the number of true negatives (in pixels), FP was the number of false positives (in pixels), and FN was the number of false negatives (in pixels) of the given class for predicted classification of the SNP images. The DSC was weighted by the number of positive occurrences of each class in the manually segmented images.39
We also calculated the sensitivity and specificity of each class. Sensitivity is a measure of the ability to correctly identify regions containing the class of interest, whereas specificity is a measure of the ability to correctly identify regions not containing the class of interest. The sensitivity and specificity were calculated as
The sensitivity and specificity were weighted by the number of positive occurrences and negative occurrences, respectively, of each class in the manually segmented images.39
Finally, we used the one-sided Wilcoxon signed-rank test to determine the statistical significance of the observed differences between automated segmentation and manual segmentation.
Cross Validation
We used twenty-onefold cross validation to evaluate the performance of the SNP-Net on all 207 images to avoid any selection bias and to ensure independence between the training, validation, and testing sets. The 43 patients were randomly divided into 21 groups, each consisting of 3–4 eyes. Eyes from the same patient were placed in the same group to avoid the algorithm being trained on one eye and tested on the other eye. As such, correlation between eyes from the same patient would not influence the performance of the algorithm. Nineteen groups were placed in the training set, one group was placed in the validation set, and the remaining group was placed in the testing set. The groups were rotated such that each group was used once for validation and once for testing. Therefore, a different network (trained to completion 5 times and averaged) was used for testing in each group, resulting in 21 different networks used for testing to enable testing on all available data.
Clinical Metrics
Some of the previous publications in this field15,17 have compared their model performance to ACCMetrics,19 which calculates corneal nerve fiber density, corneal nerve branch density, corneal nerve fiber length, and corneal nerve fiber tortuosity from manual segmentations. These clinical metrics are not specifically designed to be used at the corneal vortex, which comprises ~1/3 of our SNP-Net dataset. Clinical metrics such as nerve density, average nerve thickness, average nerve segment tortuosity, junction point density, neuroma density, and immune cell density, however, can be applied at both the cornea vortex and other locations. As such, we will use these metrics and the resulting intraclass correlation coefficient (ICC)40 to measure the agreement between automatic and manual segmentations.
The nerve density is defined as the μm2 of nerve in a 160,000 μm2 region of SNP. The average nerve thickness is defined as the total nerve area (μm2) divided by the total nerve path length (μm). The (weighted) average nerve segment tortuosity is the absolute curvature, as defined by Scarpa et al., averaged across nerve segments.41 The junction point density is defined as the number of nerve junctions in a 160,000 μm2 region of SNP. The neuroma density is defined as the number of neuromas in a 160,000 μm2 region of SNP. The immune cell density is defined as the number of immune cells in a 160,000 μm2 region of SNP.
Implementation and Distribution
The proposed method was implemented in Python using the PyTorch (1.11.0) library on cloud with an Intel Xeon Platinum 8259CL CPU and one NVIDIA Tesla P100 GPU. All code for the method proposed in this paper, along with the annotated datasets, are available at https://github.com/ZaneZemborain/SNPNETv1
Results
Qualitative Analysis
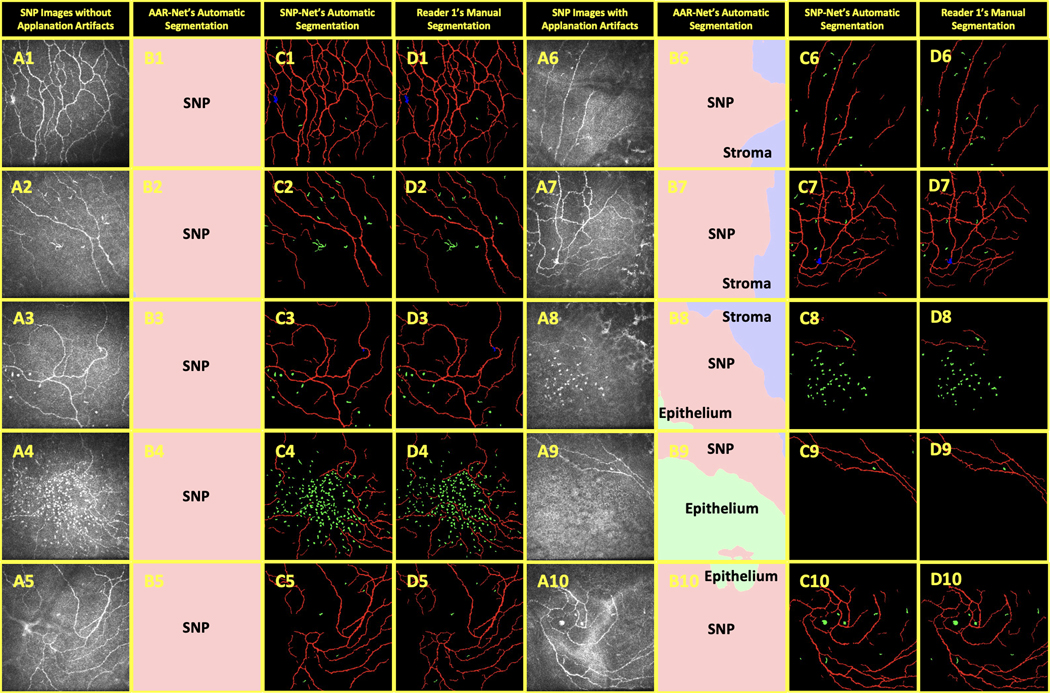
The SNP images in our dataset consisted of examples both with (Figure 5.A6–A10) and without (Figure 5.A1–A5) real applanation artifacts. Thirty-three of the 207 SNP images evaluated by AAR-Net were found to have non-SNP regions occupying more than 5% of the image; we considered these images to have applanation artifacts. AAR-Net identified and removed these regions with stroma (Figure 5.B6–B8) and with epithelium (Figure 5.B8–B10).
Figure 5.
Comparison of automatic and manual segmentations of OSD biomarkers on CCM images. The 207 SNP-Net dataset images included images without applanation artifacts (A1-A5) and with applanation artifacts (A6-A10). The applanation artifacts (or lack thereof) were segmented by AAR-Net (B1-B10) as containing epithelium (light green), SNP (pink), and stroma (light blue). The SNP-Net and Reader 1 segmentations of background (black), nerves (red), neuromas (blue), and immune cells (green) are shown in C1-C10 and D1-D10, respectively.
The automatic segmentations from SNP-Net (Figure 5.C1–C10) closely replicated the manual segmentations from Reader 1 (Figure 5.D1–D10). SNP-Net successfully segmented large clusters of immune cells, such as in Figure 5.C4 and Figure 5.C8. It achieved similar success with the mature dendritic cell in Figure 5.C2. Additionally, it correctly identified clear neuromas such as in Figure 6.C1, Figure 6.C3, and Figure 6.C7. Finally, SNP-Net correctly segmented the corneal compression artifact as background (Figure 6.C5).
Figure 6.
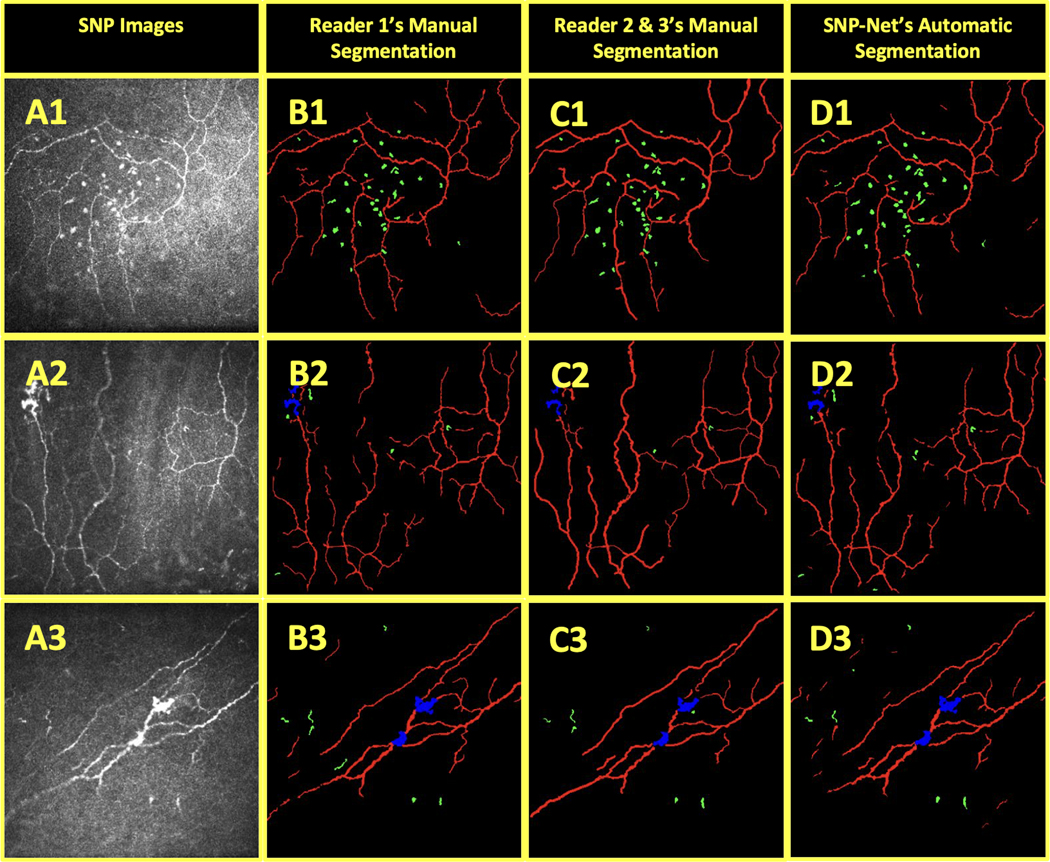
Comparison of different graders’ manual segmentations of OSD biomarkers on CCM images with automatic segmentation. The background (black), nerves (red), neuromas (blue), and immune cells (green) in SNP images (A) were independently segmented by Reader 1 (B) and Readers 2 & 3 (C). These images were also segmented by SNP-Net (D).
Quantitative Analysis
The performance metrics of the proposed SNP-Net are shown in Table 1. In this table, the average and standard deviation of the DSC, sensitivity, and specificity were calculated for all 207 SNP-Net dataset images.
Table 1.
Average performance metrics (mean ± SD) for the proposed SNP-Net as compared to Reader 1 for the 207 SNP-Net dataset images.
| Background | Nerve | Neuroma | Immune Cell | |
|---|---|---|---|---|
| DSC | 0.992 ± 0.004 | 0.814 ± 0.042 | 0.748 ± 0.222 | 0.736 ± 0.104 |
| Sensitivity | 0.991 ± 0.004 | 0.829 ± 0.054 | 0.780 ± 0.266 | 0.789 ± 0.152 |
| Specificity | 0.839 ± 0.050 | 0.992 ± 0.004 | 0.9997 ± 0.0009 | 0.9991 ± 0.0012 |
The clinical metrics of the proposed SNP-Net segmentations, as compared to Reader 1’s manual segmentations, are shown in Table 2. In this table, the average and standard deviation of the nerve density, average nerve thickness, average nerve segment tortuosity, junction point density, neuroma density, and immune cell density were calculated for all 207 SNP-Net dataset images.
Table 2.
Average clinical metrics (mean ± SD) and intraclass correlation coefficient (ICC) for the proposed SNP-Net as compared to Reader 1 for the 207 SNP-Net dataset images.
| Nerve Density (μm2/160,000μm2) | Average Nerve Thickness (μm) | Average Nerve Segment Tortuosity | Junction Point Density (count/160,000μm2) | Neuroma Density (count/160,000μm2) | Immune Cell Density (count/160,000μm2) | |
|---|---|---|---|---|---|---|
| SNP-Net | 6579 ± 3498 | 2.8 ± 0.3 | 18.0 ± 7.4 | 19.1 ± 20.1 | 0.8 ± 1.5 | 14.0 ± 21.1 |
| Reader 1 | 6342 ± 3450 | 2.5 ± 0.4 | 18.9 ± 7.6 | 25.0 ± 22.3 | 0.6 ± 1.1 | 13.6 ± 22.2 |
| ICC | 0.99 | 0.93 | 0.82 | 0.98 | 0.74 | 0.99 |
Inter-reader analysis
The SNP images were also manually segmented by the combined efforts of two additional trained readers (Readers 2 & 3, N.S.A. & M.S.). Reader 2 performed the initial segmentation of the images and then the more experienced Reader 3 made corrections. In total, they segmented 43 images or one image per patient. Reader 2 and Reader 3 completed this task independent of Reader 1. Figure 6 provides examples of their manual segmentation set side by side with those from Reader 1 and the SNP-Net segmentation.
Table 3 shows the segmentation performance of the Readers 2 & 3 versus Reader 1 and SNP-Net versus Reader 1 for these 43 images. The SNP-Net achieved a higher (or statistically equal) average DSC, sensitivity, and specificity for the background, nerve, and immune cell classes.
Table 3.
Average performance metrics (mean ±SD) for Readers 2 & 3 and the proposed SNP-Net segmentation as compared to Reader 1 for the 43 images.
| Background | Nerve | Neuroma | Immune Cell | ||
|---|---|---|---|---|---|
| DSC | Readers 2 & 3 | 0.988 ± 0.005 | 0.721 ± 0.089 | 0.766 ± 0.200 | 0.711 ± 0.145 |
| SNP-Net | 0.993 ± 0.003 | 0.823 ± 0.034 | 0.726 ± 0.245 | 0.740 ± 0.087 | |
| P-Value | <0.001** | <0.001** | 0.858 | 0.017* | |
| Sensitivity | Readers 2 & 3 | 0.986 ± 0.007 | 0.762 ± 0.093 | 0.721 ± 0.219 | 0.713 ± 0.190 |
| SNP-Net | 0.992 ± 0.003 | 0.839 ± 0.042 | 0.722 ± 0.285 | 0.809 ± 0.153 | |
| P-Value | <0.001** | <0.001** | 0.509 | <0.001** | |
| Specificity | Readers 2 & 3 | 0.766 ± 0.093 | 0.986 ± 0.007 | 0.9999 ± 0.0002 | 0.9995 ± 0.0009 |
| SNP-Net | 0.846 ± 0.040 | 0.992 ± 0.004 | 0.9998 ± 0.0004 | 0.9993 ± 0.0012 | |
| P-Value | <0.001** | <0.001** | 0.957 | 0.997 | |
P-values were calculated using a one-sided Wilcoxon signed-rank test.
A p-value less than 0.05 is denoted with
and a p-value less than 0.01 is denoted with.
Discussion
In this paper, we presented a deep learning-based method for automated segmentation of corneal nerves and other structures in CCM images of the SNP in eyes with OSD. We showed that the proposed automatic method could reliably segment potential biomarkers of OSD with accuracy on par with human gradings in real clinical data.
In an ideal practice, the operator of the CCM will acquire unmarred images of the layer of interest. However, in real clinical practice, the operator may frequently position the microscope’s object plane such that it intersects two or more adjacent layers of the cornea, resulting in the applanation artifacts. Even with careful imaging, the operator may be unable to avoid producing this artifact as CCM imaging involves pressing a naturally curved cornea against a flat surface. This applanation can be exacerbated by thin layers, excessive curvature of the cornea, and a lack of cooperation from patients. Thus, CCM images targeted at imaging the SNP layer frequently contain the neighboring epithelium and stroma layers (16% of the cases in our study). As such, a preprocessing step was needed to identify the marred image regions containing the undesired adjacent layers and exclude them from the analysis. We proposed to achieve this by utilizing the AAR-Net, which labels each pixel of an image as belonging to one of the three corneal layers. Subsequently, only the regions of the image that belong to the SNP layer will be processed by the SNP-Net and used to calculate clinical metrics.
SNP-Net’s segmentation of nerve, neuromas, and immune cells enables quantification of nerve density, average nerve thickness, average nerve segment tortuosity, junction point density, neuroma density, and immune cell density. The ICC’s between the automatic and manual segmentations indicate good to excellent reliability. These metrics can be used by our lab and others to perform objective disease differentiation, progression analysis, and treatment evaluation at the corneal vortex and other locations. Additionally, thse automatically generated metrics are expected to facilitate investigations to determine the clinical significance of neuromas and immune cells in ocular surface diseases.
It should also be noted that subjectivity is inherent to manual segmentation. Research groups may use different criteria, but, even with the same criteria and training, readers may differ in their interpretation of the images. For example, in Figure 6.A2–D2, Reader 1 used a slightly more conservative approach in terms of nerve thickness, visibility, and connections than Readers 2 & 3. Similarly, Readers 2 & 3 appear to have more conservatively labeled objects as immune cells than Reader 1. Since the automatic segmentation was trained on Reader 1’s gradings, it is reasonable to expect that it objectively replicates Reader 1’s style of grading.
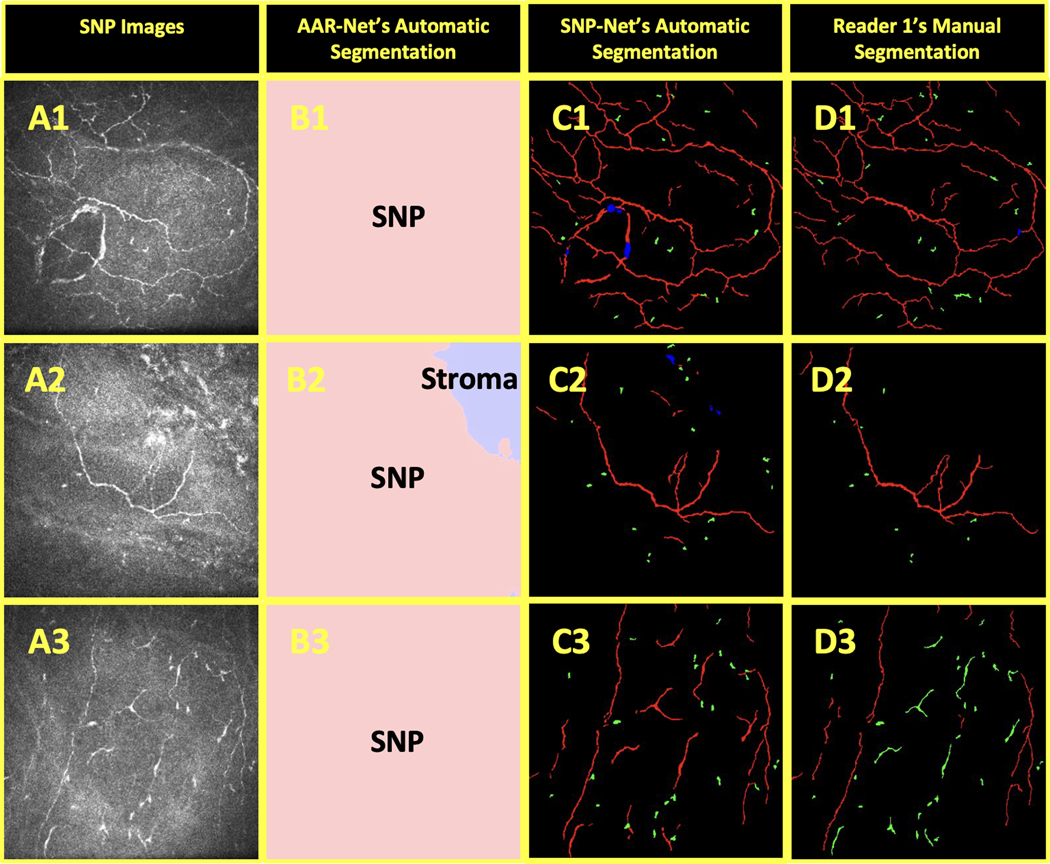
The limitations of this study include its retrospective design and relatively small dataset.. Furthermore, as with any other deep learning-based image analysis method, the proposed algorithms may not perform as well on images of participants that fall outside the target population and imaging protocol. For example, SNP-Net may perform suboptimally when applied on images from participants with other corneal diseases or dystrophies. In Figure 7.C1, SNP-Net incorrectly segmented anterior basement membrane corneal dystrophy in the lower lefthand corner of the figure. Other potential limitations include the proper labeling of nerve structures as normal (i.e., corneal nerve penetration) or pathologic (i.e., neuroma or micro-neuroma). This has been studied in a recent systematic review,20 which observed a significant overlap in the literature terminology used to describe healthy and abnormal nerve features and argued that the terms ‘neuromas’ and ‘micro-neuromas’ could potentially be physiological epithelial nerve penetration sites. We approached this issue with caution and strove to properly differentiate neuromas and micro-neuromas (i.e., presumed neuromas) from nerve perforation sites, but this remains a potential source of error. Additionally, there is a lack of compelling evidence for the correlations between CCM images and the actual corneal histology, which might lead to a miscategorization of the segmented structures.
Figure 7.
Examples of AAR-Net and SNP-Net errors in segmentation: anterior basement membrane corneal dystrophy was incorrectly segmented by SNP-Net (1), stroma in the upper righthand corner was only partially labeled as stroma by AAR-Net (2), and mature dendritic cells were mistaken for nerve by SNP-Net (3). The SNP images (A) were segmented by AAR-Net (B) as containing epithelium (light green), SNP (pink), and stroma (light blue). The background (black), nerves (red), neuromas (blue), and immune cells (green) in the SNP images were also segmented by SNP-Net (C) and Reader 1 (D).
Finally, it is constructive to study the major cases in which AAR-Net or SNP-Net failed to accurately segment features of interest. A failure by AAR-Net to segment the epithelium layer would result in SNP-Net erroneously segmenting the epithelial cell borders as nerve. Similarly, a failure to segment the stroma layer would result in SNP-Net erroneously segmenting the stomal cells as nerve, neuromas, and immune cells. The example of AAR-Net’s failure to completely segment the stroma, one of the worst segmentation performances in our dataset, is shown in the upper righthand corner of Figure 7.B2. Examples of notable SNP-Net errors include: (1) changes in nerve elevation near terminal nerve ends were sometimes incorrectly segmented as immune cells, (2) immune cells that overlaid nerve were sometimes mistaken for neuroma or vice versa, and (3) mature dendritic cells were often mistaken for nerve. Because the CCM images came from the central cornea, our SNP-Net training set had relatively few mature dendritic cell examples, which are more commonly found in the peripheral cornea.42 As such, the failure of SNP-Net in Figure 7.C3 is unsurprising. It is expected that a large training set will alleviate these types of errors. To further improve the accuracy of segmentation, as part of our ongoing work, we will extend the capabilities of the proposed SNP-Net by integrating the BiconNet connectivity principles,43 which have proven impactful for other image analysis applications.44
Conclusion
In conclusion, we have developed a fully automatic deep learning-based method to remove applanation artifacts and segment nerves and other structures on CCM images of the SNP in patients with OSD. Furthermore, we created an accompanying set of clinical metrics based on our automatic segmentations that can be applied at the cornea vortex and other locations. To our knowledge, it is the first study to perform multi-class segmentation of nerves, neuromas, and immune cells in clinical images and the first study to specifically address applanation artifacts. To promote future research in this area, we have made our annotated datasets and corresponding algorithms open-source and freely available online.
Funding
This work was supported by in part by the National Institutes of Health (U01 EY034687 and P30 EY005722), and Research to Prevent Blindness Unrestricted Grant to Duke University.
Footnotes
Victor L. Perez: Alcon (F); Heat Biologics (F); NIH (F); Brill (C); Dompe (C); Kala (C); Kiora (C); Novartis (C); Oyster Point Pharma (C); Trefoil (I)
References
- 1.Müller LJ, Marfurt CF, Kruse F, Tervo TM. Corneal nerves: structure, contents and function. Exp Eye Res 2003;76(5):521–42. (In eng). DOI: 10.1016/s0014-4835(03)00050-2. [DOI] [PubMed] [Google Scholar]
- 2.Stern ME, Beuerman RW, Fox RI, Gao J, Mircheff AK, Pflugfelder SC. The pathology of dry eye: the interaction between the ocular surface and lacrimal glands. Cornea 1998;17(6):584–9. (In eng). DOI: 10.1097/00003226-199811000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Phadatare SP, Momin M, Nighojkar P, Askarkar S, Singh KK. A Comprehensive Review on Dry Eye Disease: Diagnosis, Medical Management, Recent Developments, and Future Challenges. Advances in Pharmaceutics 2015;2015:704946. DOI: 10.1155/2015/704946. [DOI] [Google Scholar]
- 4.Treede RD, Jensen TS, Campbell JN, et al. Neuropathic pain: redefinition and a grading system for clinical and research purposes. Neurology 2008;70(18):1630–5. (In eng). DOI: 10.1212/01.wnl.0000282763.29778.59. [DOI] [PubMed] [Google Scholar]
- 5.Galor A, Moein HR, Lee C, et al. Neuropathic pain and dry eye. Ocul Surf 2018;16(1):31–44. (In eng). DOI: 10.1016/j.jtos.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yavuz Saricay L, Bayraktutar BN, Kenyon BM, Hamrah P. Concurrent ocular pain in patients with neurotrophic keratopathy. Ocul Surf 2021;22:143–151. (In eng). DOI: 10.1016/j.jtos.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moein HR, Akhlaq A, Dieckmann G, et al. Visualization of microneuromas by using in vivo confocal microscopy: An objective biomarker for the diagnosis of neuropathic corneal pain? Ocul Surf 2020;18(4):651–656. (In eng). DOI: 10.1016/j.jtos.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero-Moreno A, Liang H, Moreau N, et al. Corneal Nerve Abnormalities in Painful Dry Eye Disease Patients. Biomedicines 2021;9(10) (In eng). DOI: 10.3390/biomedicines9101424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson W, Chauhan SK, Dana R. Dry eye disease: an immune-mediated ocular surface disorder. Arch Ophthalmol 2012;130(1):90–100. (In eng). DOI: 10.1001/archophthalmol.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasti R, Allingham MJ, Mettu PS, et al. Deep learning-based single-shot prediction of differential effects of anti-VEGF treatment in patients with diabetic macular edema. Biomed Opt Express 2020;11(2):1139–1152. (In eng). DOI: 10.1364/boe.379150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruzat A, Qazi Y, Hamrah P. In Vivo Confocal Microscopy of Corneal Nerves in Health and Disease. Ocul Surf 2017;15(1):15–47. (In eng). DOI: 10.1016/j.jtos.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salahuddin T, Qidwai U. Evaluation of Loss Functions for Segmentation of Corneal Nerves. 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES)2021:533–537. [Google Scholar]
- 13.Wei S, Shi F, Wang Y, Chou Y, Li X. A Deep Learning Model for Automated Sub-Basal Corneal Nerve Segmentation and Evaluation Using In Vivo Confocal Microscopy. Translational Vision Science & Technology 2020;9(2):32-32. DOI: 10.1167/tvst.9.2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D, Huang F, M Khansari M, et al. Automatic corneal nerve fiber segmentation and geometric biomarker quantification. The European Physical Journal Plus 2020;135. DOI: 10.1140/epjp/s13360-020-00127-y. [DOI] [Google Scholar]
- 15.Williams BM, Borroni D, Liu R, et al. An artificial intelligence-based deep learning algorithm for the diagnosis of diabetic neuropathy using corneal confocal microscopy: a development and validation study. Diabetologia 2020;63(2):419–430. (In eng). DOI: 10.1007/s00125-019-05023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colonna A, Scarpa F, Ruggeri A. Segmentation of Corneal Nerves Using a U-Net-Based Convolutional Neural Network: First International Workshop, COMPAY 2018, and 5th International Workshop, OMIA 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16 – 20, 2018, Proceedings. 2018:185–192. [Google Scholar]
- 17.Setu MAK, Schmidt S, Musial G, Stern ME, Steven P. Segmentation and Evaluation of Corneal Nerves and Dendritic Cells From In Vivo Confocal Microscopy Images Using Deep Learning. Transl Vis Sci Technol 2022;11(6):24. (In eng). DOI: 10.1167/tvst.11.6.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petroll WM, Robertson DM. In Vivo Confocal Microscopy of the Cornea: New Developments in Image Acquisition, Reconstruction, and Analysis Using the HRT-Rostock Corneal Module. Ocul Surf 2015;13(3):187–203. (In eng). DOI: 10.1016/j.jtos.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Graham J, Dabbah MA, Petropoulos IN, Tavakoli M, Malik RA. An Automatic Tool for Quantification of Nerve Fibers in Corneal Confocal Microscopy Images. IEEE Trans Biomed Eng 2017;64(4):786–794. (In eng). DOI: 10.1109/tbme.2016.2573642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinnery HR, Rajan R, Jiao H, et al. Identification of presumed corneal neuromas and microneuromas using laser-scanning in vivo confocal microscopy: a systematic review. Br J Ophthalmol 2022;106(6):765–771. (In eng). DOI: 10.1136/bjophthalmol-2020-318156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal S, Kheirkhah A, Cavalcanti BM, et al. Autologous Serum Tears for Treatment of Photoallodynia in Patients with Corneal Neuropathy: Efficacy and Evaluation with In Vivo Confocal Microscopy. Ocul Surf 2015;13(3):250–62. (In eng). DOI: 10.1016/j.jtos.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goyal S, Hamrah P. Understanding Neuropathic Corneal Pain--Gaps and Current Therapeutic Approaches. Semin Ophthalmol 2016;31(1–2):59–70. (In eng). DOI: 10.3109/08820538.2015.1114853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieckmann G, Goyal S, Hamrah P. Neuropathic Corneal Pain: Approaches for Management. Ophthalmology 2017;124(11s):S34–s47. (In eng). DOI: 10.1016/j.ophtha.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morkin MI, Hamrah P. Efficacy of self-retained cryopreserved amniotic membrane for treatment of neuropathic corneal pain. Ocul Surf 2018;16(1):132–138. (In eng). DOI: 10.1016/j.jtos.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamel JT, Zhang AC, Downie LE. Corneal Epithelial Dendritic Cell Response as a Putative Marker of Neuro-inflammation in Small Fiber Neuropathy. Ocul Immunol Inflamm 2020;28(6):898–907. (In eng). DOI: 10.1080/09273948.2019.1643028. [DOI] [PubMed] [Google Scholar]
- 26.Cavalcanti BM, Cruzat A, Sahin A, Pavan-Langston D, Samayoa E, Hamrah P. In vivo confocal microscopy detects bilateral changes of corneal immune cells and nerves in unilateral herpes zoster ophthalmicus. The ocular surface 2018;16(1):101–111. (In eng). DOI: 10.1016/j.jtos.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vera LS, Gueudry J, Delcampe A, Roujeau JC, Brasseur G, Muraine M. In vivo confocal microscopic evaluation of corneal changes in chronic Stevens-Johnson syndrome and toxic epidermal necrolysis. Cornea 2009;28(4):401–7. (In eng). DOI: 10.1097/ICO.0b013e31818cd299. [DOI] [PubMed] [Google Scholar]
- 28.Zhivov A, Stave J, Vollmar B, Guthoff R. In vivo confocal microscopic evaluation of Langerhans cell density and distribution in the normal human corneal epithelium. Graefes Arch Clin Exp Ophthalmol 2005;243(10):1056–61. (In eng). DOI: 10.1007/s00417-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 29.Feng Z, Yang J, Yao L. Patch-based fully convolutional neural network with skip connections for retinal blood vessel segmentation. 2017 IEEE International Conference on Image Processing (ICIP) 2017:1742–1746. [Google Scholar]
- 30.Luan S, Zhang B, Zhou S, et al. Gabor Convolutional Networks. 2018. IEEE Winter Conference on Applications of Computer Vision (WACV)2018:1254–1262. [Google Scholar]
- 31.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015:234–241. [Google Scholar]
- 32.Sergey I, Christian S. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. PMLR:448–456. [Google Scholar]
- 33.Cui Y, Jia M, Lin T-Y, Song Y, Belongie S. Class-Balanced Loss Based on Effective Number of Samples2019. [Google Scholar]
- 34.Jurdi RE, Petitjean C, Cheplygina V, Abdallah F. A Surprisingly Effective Perimeter-based Loss for Medical Image Segmentation. MIDL2021. [Google Scholar]
- 35.Odena A, Olah C, Shlens J. Conditional Image Synthesis with Auxiliary Classifier GANs. In: Doina P, Yee Whye T, eds. Proceedings of the 34th International Conference on Machine Learning. Proceedings of Machine Learning Research: PMLR; 2017:2642–2651. [Google Scholar]
- 36.Hamrah P, Koseoglu D, Kovler I, Ben Cohen A, Soferman R. Deep Learning Convolutional Neural Network for the Classification and Segmentation of In Vivo Confocal Microscopy Images. Investigative Ophthalmology & Visual Science 2018;59(9):1733-1733. [Google Scholar]
- 37.He K, Zhang X, Ren S, Sun J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. 2015. IEEE International Conference on Computer Vision (ICCV)2015:1026–1034. [Google Scholar]
- 38.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. 2015. [Google Scholar]
- 39.Crum WR, Camara O, Hill DL. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans Med Imaging 2006;25(11):1451–61. (In eng). DOI: 10.1109/tmi.2006.880587. [DOI] [PubMed] [Google Scholar]
- 40.Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med 2016;15(2):155–63. (In eng). DOI: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scarpa F, Zheng X, Ohashi Y, Ruggeri A. Automatic Evaluation of Corneal Nerve Tortuosity in Images from In Vivo Confocal Microscopy. Investigative Ophthalmology & Visual Science 2011;52(9):6404–6408. DOI: 10.1167/iovs.11-7529. [DOI] [PubMed] [Google Scholar]
- 42.Hamrah P, Huq SO, Liu Y, Zhang Q, Dana MR. Corneal immunity is mediated by heterogeneous population of antigen-presenting cells. J Leukoc Biol 2003;74(2):172–8. (In eng). DOI: 10.1189/jlb.1102544. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Soltanian-Zadeh S, Farsiu S. BiconNet: An edge-preserved connectivity-based approach for salient object detection. Pattern Recogn 2022;121(C):11. DOI: 10.1016/j.patcog.2021.108231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Soltanian-Zadeh S, Chu KK, et al. Connectivity-based deep learning approach for segmentation of the epithelium in in vivo human esophageal OCT images. Biomed Opt Express 2021;12(10):6326–6340. (In eng). DOI: 10.1364/boe.434775. [DOI] [PMC free article] [PubMed] [Google Scholar]