Abstract
Childhood cancer and birth defects are leading causes of childhood mortality, and studies suggest that birth defects increase pediatric cancer risk. The Gabriella Miller Kids First Pediatric Research Program (Kids First) seeks to alleviate these conditions by building an expansive resource of genetic and clinical data from patients with pediatric cancer and birth defects and their families. This article describes the data and support provided by the Kids First Data Resource Center and the Kids First Data Resource Center Data Resource Portal, which enables the public to review Kids First studies and request access to individual data. The Kids First Portal contains data from more than 34 000 participants and connects with CAVATICA (Seven Bridges Genomics, Inc, now part of Velsera), a cloud-based analysis and sharing platform. Researchers have used Kids First data to investigate a variety of cancers and further funding opportunities are available. The Kids First Portal is a unique resource that unites pediatric cancer and birth defects to uncover their genetic etiology and improve patients’ lives.
Birth defects are the leading cause of mortality during the first year of life and remain among the top 10 causes of mortality throughout childhood. Childhood cancer is the leading cause of disease-related death among children 5 to 18 years of age (1). Epidemiologic studies suggest that the presence of structural birth defects is associated with an increased risk of developing a variety of childhood cancers (2-4). These associations suggest a shared etiology, but studies examining the shared genetic origins of birth defects and cancer have been limited in part due to the rarity of these diagnoses and the resulting low power for the discovery of genetic associations in small sample sizes. The Gabriella Miller Kids First Pediatric Research Program (Kids First) is named in honor of Gabriella Miller, who was diagnosed with an inoperable brain tumor and died in 2013 at the age of 10. Two weeks before she died, Gabriella called upon our elected officials to “stop talking and start doing.” The Gabriella Miller Kids First Research Act was developed through the tireless advocacy of Gabriella and her family and was signed into law in April 2014, 6 months after Gabriella’s untimely death. This bill authorized a pediatric research initiative through the National Institutes of Health (NIH) to empower genetic discoveries by building a large resource of genetic and clinical data from patients and families afflicted with childhood cancer or birth defects and focused on stimulating collaborative research within the pediatric community. With support and management from the NIH Common Fund, program leaders across the NIH are collaborating to develop and grow this resource. At the National Cancer Institute, Kids First is an important element of the Childhood Cancer Data Initiative Data Ecosystem, a cornerstone of the Childhood Cancer Data Catalog, and it is aligned with the National Cancer Institute’s Division of Cancer Control and Population Science’s priority for data strategies.
Kids First has ambitious goals to collect, generate, and curate large amounts of genotype and phenotype data from children and families afflicted by childhood cancer or birth defects and to make these data available and accessible to researchers, patients, and health-care professionals. A key component of this effort is the Kids First Data Resource Center (DRC) Data Resource Portal (Kids First Portal), which facilitates access and analysis of data collected and generated through Kids First. Through this publicly available portal, users can review available studies, data types, and curated clinical data (including phenotypes) and request access to individual-level “omics” data. The data within the Kids First Portal is 1 of the largest pediatric data resources of its kind and a unique resource of both pediatric cancers and birth defects data. The Kids First Portal will eventually provide access to more than 56 000 genomes from 41 000 patient and family participants from 70 childhood cancer and birth defect cohorts (5). Currently, the resource contains data from more than 34 000 participants, amounting to 1.8 petabytes of data.
The Kids First Portal includes 9 Kids First cancer cohorts, 20 Kids First birth defect cohorts, and 2 cohorts with both cancer and birth defects as of August 2023 (Table 1). Childhood cancer cohorts include (Table 2) neuroblastoma (Genotype and Phenotype Database [dbGaP] phs001436), enchondromatoses (dbGaP phs001987), germ cell tumors (dbGaP phs002322), leukemia (dbGaP phs002187, phs002330, phs002276, phs001738), sarcomas (dbGaP Ewing sarcoma phs001228 and osteosarcoma phs001714), and studies investigating multiple pediatric cancer histologies (dbGaP phs001683 and phs001846). Additionally, studies from the Pediatric Brain Tumor Atlas, the Therapeutically Applicable Research To Generate Effective Treatments (TARGET) program (TARGET-Acute Myeloid Leukemia and TARGET-Neuroblastoma) (dbGaP AML phs000465 and neuroblastoma phs000467), and Project Open DIPG (diffuse intrinsic pontine glioma) are included. Sequencing data types vary by cancer study, but the resource includes whole-genome, whole-exome, and RNA sequencing data (Figure 1 and Table 2). Both germline and somatic data may be available, with harmonized data products that allow for cross-study comparisons. Other sequencing types, including proteomic and long-read sequencing data, may also be available in the future for some studies. Demographic information about sex, race, and ethnicity is available for a majority of participants, as well (Figure 2). A variety of study designs were employed by Kids First cancer studies, including proband only, trios (mother, father, and proband), duos (parent and proband), and trio plus (mother, father, and other afflicted family members) (Table 2). Rich, curated clinical data include information about observed phenotypic abnormalities (Supplementary Figure 1, available online).
Table 1.
Number of Kids First studies per major condition
| Major condition | Kids First studies, a No. |
|---|---|
| Cancer | 9 |
| Birth defect | 20 |
| Cancer and birth defect | 2 |
Other programs’ studies can also be explored through the Kids First Portal, including the Pediatric Brain Tumor Atlas, Therapeutically Applicable Research To Generate Effective Treatments, and Project Open DIPG [diffuse intrinsic pontine glioma]. Kids First = Gabriella Miller Kids First Pediatric Research Program; Kids First Portal = Kids First Data Resource Center Data Resource Portal.
Table 2.
Kids First cancer studies
| Cancer type, study focus | dbGaP study accession | Data types available a | Study designs b | No. of probands/study participants |
|---|---|---|---|---|
| Acute lymphoblastic leukemia, co-occurring Down syndrome | phs002330 | RNA sequencing, whole-genome sequencing | Proband only, trio, duo | 1777/2233 |
| Acute lymphoblastic leukemia, T-cell | phs002276 | RNA sequencing, whole-genome sequencing, whole-exome sequencing | Proband only | 1358/1358 |
| Enchondromatoses | phs001987 | Whole-genome sequencing | Proband only, trio, duo | 75/211 |
| Ewing sarcoma | phs001228 | Whole-genome sequencing | Proband only, trio | 469/1157 |
| Germ cell tumors | phs002322 | Whole-genome sequencing | Trio, duo | 130/356 |
| Hematopoietic malignancies, familial | phs001738 | Whole-genome sequencing | Trio plus | 0/185c |
| Myeloid malignancies | phs002187 | RNA sequencing, whole-genome sequencing | Proband only | 408/408 |
| Neuroblastoma | phs001436 | Linked-read whole-genome sequencing, RNA sequencing, whole-genome sequencing, whole-exome sequencing | Proband only, trio, duo | 609/1681 |
| Osteosarcoma | phs001714 | Whole-genome sequencing, whole-exome sequencing | Proband only | 129/129 |
| Pan-cancer, co-occurring birth defects | phs001846 | Whole-genome sequencing | Proband only, trio, duo | 1453/1777 |
| Pan-cancer, familial | phs001683 | Whole-genome sequencing | Proband only, trio | 247/718 |
Not all data types are available for all participants. dbGaP = Genotype and Phenotype Database; Kids First = Gabriella Miller Kids First Pediatric Research Program.
Multiple designs are possible in a study and include proband only (no family member comparator), trio (mother, father, and proband), duo (1 parent and proband), and trio plus (mother, father, and other afflicted family members).
Due to the nature of this familial study design, a proband designation was not assigned to these participants because multiple participants in a single family have leukemia-affected status; 79/185 participants in this study are affected by leukemia.
Figure 1.
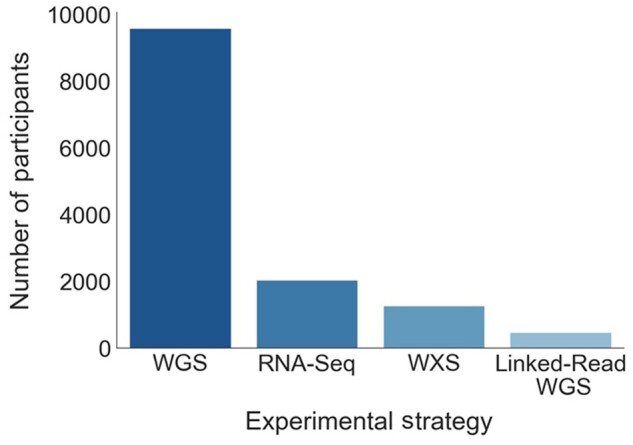
Available data types for Gabriella Miller Kids First Pediatric Research Program cancer study participants. This figure shows the number of individuals with each available data type, by experimental sequencing strategy. RNA-Seq = RNA sequencing; WGS = whole-genome sequencing; WXS = whole-exome sequencing; Linked-Read WGS = Linked-read whole-genome sequencing using the 10× Genomics Chromium platform.
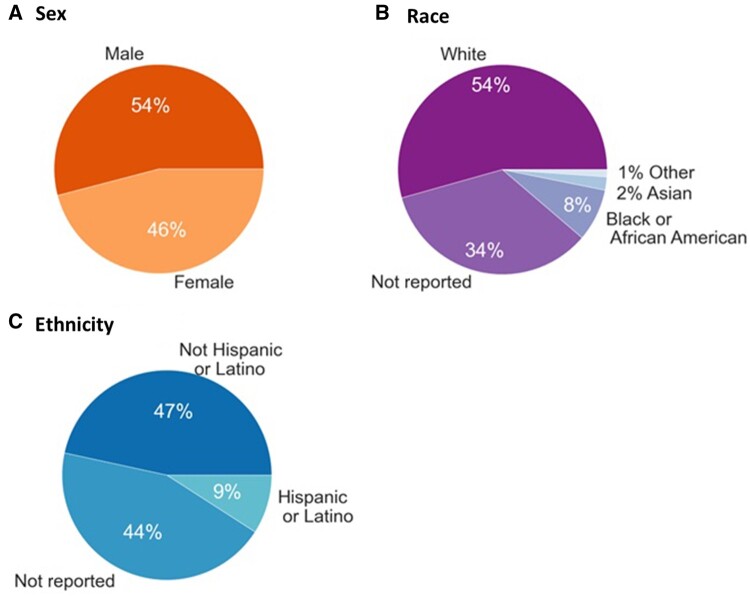
Figure 2.
Gabriella Miller Kids First Pediatric Research Program (Kids First) cancer study participant demographics. A, Reported sex of Kids First cancer study participants. B, Reported race of Kids First cancer study participants. The “Other” category represents individuals of American Indian or Alaska Native descent (0.5%), individuals of more than 1 race (0.5%), or individuals of Native Hawaiian or Other Pacific Islander descent (0.1%). C, The reported ethnicity of Kids First cancer study participants.
The Kids First data align with findable, accessible, interoperable, reusable (FAIR) data principles and undergo extensive harmonization to enable cross-cohort analysis. Clinical and phenotypic data are harmonized using community-based ontologies and standards, and the DRC is developing genomic harmonization methods for standardized characterization of genetic variation and somatic mutations in tumor samples. The availability and harmonization of pediatric cancer data with birth defect data will enable researchers to explore shared phenotypes and features across cancer and birth defect cohorts and discover both individually associated and common genetic pathways.
Support resources for investigators
Investigators using the Kids First Portal have access to a multitude of other resources to assist them in their research (Supplementary Table 1, available online). For example, the Kids First Portal allows users to explore data to identify relevant datasets for analyses and to build virtual cohorts by combining different studies or grouping participants with shared phenotypic or genetic features (see portal screenshot for an example of query building, Supplementary Figure 2, available online). Through the Kids First Portal, researchers can be directed to data repositories (eg, dbGaP) to request access to individual-level data. When access is granted, datasets can be seamlessly integrated into CAVATICA (Seven Bridges Genomics, Inc, now part of Velsera), a cloud-based analysis platform that provides a secure environment for analysis and collaboration. Kids First datasets are already in the cloud and, after investigators have been granted access, they do not need to download or upload them. Within CAVATICA, investigators can develop and share workflows to analyze the data and upload their own data for additional analysis. The Kids First Portal’s Variant Explorer enables easy querying of summary-level information about germline genetic variants identified in Kids First participants by variant type, gene, frequency, pathogenicity, and other factors. The Kids First Portal also connects with PedcBioPortal, an instance of cBioPortal for Cancer Genomics, allowing for the review, visualization, and analysis of Kids First somatic data within a browser-based tool.
Guidance for the use of the Kids First Portal and its associated tools is provided in the form of FAQs, YouTube step-by-step guides, a recorded workflow creation and maintenance course, and written tutorials for searching Kids First datasets and using CAVATICA. Additionally, monthly Kids First DRC office hours provide investigators with access to members of the Kids First DRC’s data operations and bioinformatics teams and a venue in which to ask questions about Kids First datasets and the use of the Kids First Portal and to get help with data analysis. The latest links and scheduling information about these tools can be found under the “Resources” tab on the Kids First Portal.
Although access to Kids First data is free of charge, there are fees for running computational workflows in CAVATICA. To help lower the barrier for use of Kids First data for analysis, Kids First has developed the Cloud Credits Pilot program, which provides credits for accessing cloud computing data analysis resources for reasonable proposals analyzing Kids First data. Kids First also supports investigators through a variety of funding opportunity announcements. Expansion of data available through Kids First is supported by funding opportunities (eg, PAR-23-035) for X01 sequencing grants, which allow investigators to propose established cancer or birth defect cohorts for sequencing at a Kids First–supported sequencing center. After sequencing, the data are returned to investigators, and then shared with the community through the Kids First Portal. Cohorts proposed for sequencing must allow for data sharing, and those with the broadest sharing criteria are prioritized. Some NIH institutes and centers are joining the effort to support small project R03 grants for Kids First data analysis. This funding opportunity (PAR-23-075) supports the analysis of Kids First data with other non–Kids First datasets, including those available on the Kids First Portal, or an investigator’s own data, which can be uploaded to CAVATICA and stored for a fee to conduct joint analysis. In addition, an NIH Common Fund funding opportunity supports R03 grants for enhancing the value and utility of datasets (RFA-RM-23-003) available on the Kids First Portal by collecting or processing new data types (eg, environmental exposures, deep phenotyping, medical imaging), creating new tools or workflows for the analysis of Kids First data or curating and annotating available data, among other activities. Investigators can sign up for the Kids First listserv or follow the program on Facebook, Twitter (@kidsfirstDRC), YouTube (@kidsfirstdataresourcecente8800) or GitHub to keep abreast of information regarding the release of new datasets, funding opportunity announcements, or workflows.
Examples of projects using Kids First data
Studies funded by Kids First are already underway to discover genetic variants associated with childhood cancer and birth defects using Kids First data. At the end of 2022, a total of 489 dbGaP access applications were approved for the use of data within Kids First studies, 212 of which were for cancer studies. Cancer researchers are using Kids First data to better understand the genetic basis of pediatric cancers and cancer-associated disorders that cause structural birth defects (Supplementary Table 2, available online) (6). Investigators are taking advantage of the inclusion of both patient and family genome sequences in the Kids First data to discover de novo alterations, inherited alterations, and structural variants contributing to the etiology of Ewing sarcoma (R03CA218733; dbGaP phs001228), T-cell lymphoblastic leukemia (R03CA256550; dbGaP phs002276), and cancer-associated disorders such as Ollier disease and Maffucci syndrome (R03CA256535; dbGaP phs001987). Available Kids First genetic trio data are being used to enhance the accuracy of algorithms to detect transposable element insertion in childhood cancers (R03CA249364). Furthermore, DNA and RNA sequencing data are being used to discover structural variants in cancers of the nervous system (R03CA230366) and somatic genome rearrangements in brain tumors (R03CA246228). The power of discovering the shared etiology of childhood cancers and developmental disorders is evidenced by recent discoveries that chromosome 16p11.2 deletions in neuroblastoma are also associated with neurodevelopmental disorders such as autism spectrum disorder and birth defects (7) (dbGaP phs001436) and that rare missense variants in the HIF1A, VHL, and IDH1 genes may underlie phenotypic abnormalities in Ollier disease and Maffucci syndrome (8) (dbGaP phs001987). Additionally, Kids First investigators are enhancing the utility of the Kids First resource by contributing new types of accompanying data, developing new tools or workflows for data analysis, and enriching phenotypic data, among other activities (Supplementary Table 2, available online). Separately, researchers outside of Kids First have accessed Kids First cancer datasets to develop new methods and tools (9,10), reveal new avenues of exploration (11), and empower new cancer discoveries (12).
Limitations and future directions
Although the Kids First Portal provides investigators access to a unique and valuable dataset, it is not without its limitations. As a pediatric cancer resource, the availability of samples and participants is constrained by the rarity of pediatric cancer diagnoses and the availability of high-quality samples for existing cohorts. Therefore, the number of cancer cases with data available on the portal is smaller than other adult data resources (eg, the National Cancer Institute Genomic Data Commons Data Portal). Additionally, data types other than whole-genome sequencing are available only for a subset of Kids First cancer studies (Table 2 and Figure 1). These issues will be addressed by the continued expansion of Kids First sequencing data availability and the addition of new data types. Along with the expansion of genomic and RNA sequencing datasets, future Kids First studies may also incorporate additional “omics” (eg, proteomics and long-read sequencing), imaging, or environmental exposure data. Careful harmonization of Kids First data is a key component of the Kids First Portal, and the addition of new data types will provide new challenges and opportunities for the Kids First Portal and pediatric cancer research.
The study of pediatric cancer and birth defects can be challenging because of the rarity of individual cancers and birth defects, but the Kids First Portal brings together these communities to accelerate the discovery of their shared genetic etiology and provide access to an expansive network of genetic, clinical, phenotypic, and multiomic data. Moreover, the availability of cloud-based data storage as well as collaborative analysis and data sharing tools (eg, CAVATICA and the Variant Explorer) remove the hurdles of moving large amounts of data between locations, expand the availability of these large datasets to a wider variety of institutions by reducing the cost burden of data storage and computation, and prevent data siloing by facilitating easy integration of data within and between diseases. The Kids First Portal is a unique and powerful resource for the study of pediatric cancer and birth defects, providing access to a diverse and growing collection of valuable and difficult-to-obtain data. The Kids First Portal will continue to facilitate critical insights into the etiology of these diseases and lead to treatments that improve the lives of thousands of children and their families.
Supplementary Material
Acknowledgements
We would like to thank the NIH Common Fund Program and the Gabriella Miller Kids First Pediatric Research Program working group and leadership team for their support, discussion, and helpful comments. We also thank the Kids First Data Resource Center for their insight into the Kids First Portal and their review of this manuscript. We also extend our deepest gratitude to Gabriella Miller and her family, whose advocacy efforts are continuing to accelerate pediatric cancer research.
Contributor Information
Alexandra Hudson, Center for Research Strategy, National Cancer Institute, Bethesda, MD, USA.
Marcia Fournier, Developmental Biology and Congenital Anomalies Branch, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, USA.
James Coulombe, Developmental Biology and Congenital Anomalies Branch, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, USA.
Danielle Daee, Genomic Epidemiology Branch, Division of Cancer Control and Population Sciences, National Cancer Institute, Bethesda, MD, USA.
Data availability
All data described in this article are available for access at the Kids First Portal (https://portal.kidsfirstdrc.org/login). Access to individual-level genetic data for all studies mentioned and future Kids First studies is available upon request through dbGaP. See Table 2 for the referenced dbGaP accession numbers.
Author contributions
Alexandra Hudson, PhD (Writing—original draft; Writing—review & editing), Marcia Fournier, PhD (Conceptualization; Writing—review & editing), James Coulombe, PhD (Conceptualization; Writing—review & editing), Danielle Daee, PhD (Conceptualization; Writing—original draft; Writing—review & editing).
Funding
This manuscript was supported by administrative funding from the NIH Common Fund Program.
Conflicts of interest
The authors have nothing to disclose.
References
- 1.Centers for Disease Control and Prevention. Centers for Disease Control: Leading Causes of Death and Injury. 2018. https://www.cdc.gov/injury/wisqars/LeadingCauses.html. Accessed April 11, 2022.
- 2. Johnson KJ, Lee JM, Ahsan K, et al. Pediatric cancer risk in association with birth defects: a systematic review. PLoS One. 2017;12(7):e0181246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daltveit DS, Klungsøyr K, Engeland A, et al. Cancer risk in individuals with major birth defects: large Nordic population based case-control study among children, adolescents, and adults. BMJ. 2020;371:m4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lupo PJ, Schraw JM, Desrosiers TA, et al. Association between birth defects and cancer risk among children and adolescents in a population-based assessment of 10 million live births. JAMA Oncol. 2019;5(8):1150-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institutes of Health, Office of Strategic Coordination, The Common Fund. The Gabriella Miller Kid's First Pediatric Research Program. 2022. https://commonfund.nih.gov/kidsfirst.
- 6. National Institutes of Health. RePORT Expenditures and Results module (RePORTER). 2022. https://reporter.nih.gov/ (accessed 2022).
- 7. Egolf LE, Vaksman Z, Lopez G, et al. Germline 16p11.2 microdeletion predisposes to neuroblastoma. Am J Hum Genet, 2019;105(3):658-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poll SR, Martin R, Wohler E, et al. Disruption of the HIF-1 pathway in individuals with Ollier disease and Maffucci syndrome. PLoS Genet. 2022;18(12):e1010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller DB, Piccolo SR.. trioPhaser: using Mendelian inheritance logic to improve genomic phasing of trios. BMC Bioinformatics. 2021;22(1):559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pedersen BS, Brown JM, Dashnow H, et al. Effective variant filtering and expected candidate variant yield in studies of rare human disease. NPJ Genom Med. 2021;6(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller DB, Piccolo SR.. A survey of compound heterozygous variants in pediatric cancers and structural birth defects. Front Genet. 2021;12:640242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malone CF, Dharia NV, Kugener G, et al. Selective modulation of a pan-essential protein as a therapeutic strategy in cancer. Cancer Discov. 2021;11(9):2282-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in this article are available for access at the Kids First Portal (https://portal.kidsfirstdrc.org/login). Access to individual-level genetic data for all studies mentioned and future Kids First studies is available upon request through dbGaP. See Table 2 for the referenced dbGaP accession numbers.