Abstract
A bacterial biosensor for benzene, toluene, and similar compounds has been constructed, characterized, and field tested on contaminated water and soil. The biosensor is based on a plasmid incorporating the transcriptional activator xylR from the TOL plasmid of Pseudomonas putida mt-2. The XylR protein binds a subset of toluene-like compounds and activates transcription at its promoter, Pu. A reporter plasmid was constructed by placing the luc gene for firefly luciferase under the control of XylR and Pu. When Escherichia coli cells were transformed with this plasmid vector, luminescence from the cells was induced in the presence of benzene, toluene, xylenes, and similar molecules. Accurate concentration dependencies of luminescence were obtained and exhibited K1/2 values ranging from 39.0 ± 3.8 μM for 3-xylene to 2,690 ± 160 μM for 3-methylbenzylalcohol (means ± standard deviations). The luminescence response was specific for only toluene-like molecules that bind to and activate XylR. The biosensor cells were field tested on deep aquifer water, for which contaminant levels were known, and were able to accurately detect toluene derivative contamination in this water. The biosensor cells were also shown to detect BETX (benzene, toluene, and xylene) contamination in soil samples. These results demonstrate the capability of such a bacterial biosensor to accurately measure environmental contaminants and suggest a potential for its inexpensive application in field-ready assays.
Many different species of soil- and water-borne bacteria have adapted to the presence of xenobiotic organic molecules in their environment by developing the capacity to use such compounds as carbon sources (7). These microbes have evolved networks of enzymes by which complex organic compounds are broken down into metabolic intermediates. In many instances, transcription of the genes encoding the enzymes that participate in these degradation pathways is regulated so that expression of these catabolic enzymes is dramatically enhanced by the presence of the compounds that they degrade. This type of transcriptional control is achieved by the interaction of transcriptional activator proteins with specific gene promoters. These proteins contain a DNA binding domain, a transcriptional activation domain, and a recognition domain (18). Organic molecules bind to the recognition domain and induce a conformational change in the protein that results in enhanced interaction of the DNA binding domain with specific promoter sequences. This interaction triggers the formation of a complex on the promoter DNA consisting of the transcriptional activator, ς-factor 54, integration host factor, and the RNA polymerase (19, 20). The complex effectively initiates transcription of the genes encoding the catabolic enzymes that lie directly downstream of the promoter sequence.
One such system is found in the TOL plasmid pWWO carried by the toluene-degrading soil microbe Pseudomonas putida mt-2 (3, 4, 28). In the TOL operon, the binding of toluene and related compounds to the transcriptional activator XylR results in an activating interaction between XylR and a promoter sequence (termed Pu) in the upper region of the operon (reviewed in reference 19). This interaction triggers transcription of the genes involved in converting toluene to benzoate. A second transcriptional activator, XylS, binds benzoate and activates transcription at a second promoter site (Pm) further downstream in the operon. The genes downstream of the Pm promoter encode the enzymes responsible for breakdown of benzoate to Krebs cycle intermediates. Thus, the enzymatic degradation of toluene is greatly increased precisely when the bacteria are exposed to toluene. A number of transcriptional activator proteins, in addition to XylR and XylS, with specificity for other common organic contaminants have been described for other bacterial species. These include DmpR (23, 24), a phenol-specific transcriptional activator, and NahR (21, 29), which is activated by salicylate, a product of naphthalene catabolism.
The discovery of transcriptional activators and their corresponding promoter sequences has made possible the development of bacterial biosensors of organic pollutants. A biosensor of this type can be engineered by placing a reporter gene, such as those encoding β-galactosidase or luciferase, under the control of the transcriptional activator. In the presence of an organic contaminant, the transcriptional activator becomes operative and transcription of the reporter gene is enhanced. The resulting increase in reporter gene product is then detected by measuring the activity of the reporter enzyme. Thus, under appropriate conditions, a direct correlation between organic contaminant concentration and reporter enzyme activity can be established. This potential for development of bacterial biosensors has been demonstrated for naphthalene (8, 17). The gene for bacterial luciferase (lux) was randomly inserted by transposon mutagenesis into the nahG gene of a naphthalene catabolic plasmid in Pseudomonas fluorescens. The resulting mutant cells showed a good correlation between naphthalene exposure and bioluminescence. Initial reports have also described biosensors for benzene and benzoate and their derivatives using the Pu and Pm promoters and XylR and XylS transcriptional activators linked to the luciferase or β-galactosidase reporter genes (5, 11). Other known transcriptional activators could be used to make biosensors of this type for a wide variety of organic pollutants. Sensitive and accurate biosensors would be useful alternatives to current chemical analysis methods for organic contaminants because of their low cost and their adaptability to field analysis or in situ monitoring.
In this paper, we report the construction, laboratory characterization, and environmental sample testing of a specific biosensor that detects toluene and related compounds when placed in Escherichia coli. A reporter plasmid in which expression of the luc gene for firefly luciferase was placed directly under the control of the Pu promoter and the xylR transcriptional activator of P. putida mt-2 was constructed. Cells harboring this construct detected toluene and specific derivatives with high sensitivity and quantitative accuracy in both contaminated water and soil samples.
MATERIALS AND METHODS
Construction of the biosensor plasmid.
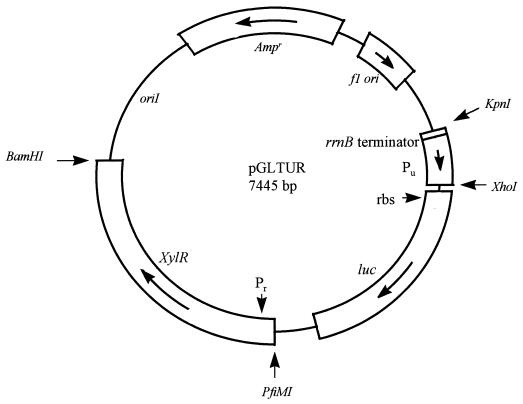
The starting vector for engineering of the toluene biosensor plasmid was a previously constructed plasmid, pGLTMRS, made for the detection of benzoate (21a). The benzoate plasmid was prepared from the commercially available luciferase reporter plasmid pGL2 (Promega, Madison, Wis.). An E. coli rrnB terminator sequence and the Pm promoter sequence were PCR amplified from E. coli and P. putida mt-2, respectively. They were ligated together at a PstI restriction site. The fused sequence was inserted at the KpnI and XhoI restriction sites in the multiple cloning region of pGL2, just upstream of the promoterless luc gene, with the Pm promoter positioned adjacent to the luc gene. The xylS and xylR genes were PCR amplified as a single amplicon from P. putida mt-2 and inserted at the BamHI digestion site of the pGL2 vector. The resulting pGLTMRS construct was used to prepare the toluene biosensor plasmid, pGLTUR. The Pu promoter was amplified from DNA purified from P. putida mt-2 by PCR using the forward primer 5′-CCAACTGCAGGGAAAGCGCGATGAAC-3′ containing a PstI site and a reverse primer, 5′-CCAGCTCGAGGACTCCAGGCGTAACG-3′, containing an XhoI site. The Pu sequence and pGLTMRS plasmid DNA were cleaved with PstI and XhoI, ligated, and transformed into competent E. coli DH5α cells (Life Technologies, Gaithersburg, Md.). The xylR gene, including its promoter (Pr), was also PCR amplified using the forward primer 5′-GCGCCAACCTATGGATTTTAATGTGGGCTGCTTGGT-3′ containing a PfiMI site and the reverse primer 5′-CGCGGATCCTTTTCACACAACCTGGGGCG-3′ containing a BamHI site. The xylR amplicon and the plasmid with the Pu insert were digested with PfiMI plus BamHI. The two were ligated together and then transformed into DH5α cells to produce the final pGLTUR plasmid construct (see Fig. 1).
FIG. 1.
Plasmid map of the pGLTUR biosensor construct. Important features of the toluene biosensor are indicated, including the location and orientation of the Pu promoter, the E. coli rrnB transcription terminator sequence, the luc luciferase gene, the Pr promoter, and the xylR transcriptional activator gene. Restriction sites used to insert Pu and xylR are shown for reference. rbs, ribosome binding site.
Induction of luciferase by toluene and its derivatives.
Single colonies of DH5α cells harboring the pGLTUR plasmid were grown in 10 ml of Luria-Bertani (LB) medium containing 100 μM ampicillin at 37°C. Cells were allowed to grow in log phase (6 to 8 h) to an optical density at 600 nm (OD600) of between 0.2 and 1.0. Similar luminescence responses were observed within this growth density range, but above values of 1.0, luminescence responses began to diminish. After this growth period, the cell culture was diluted to an OD600 of 0.2 with LB medium, and luciferase transcription was induced by mixing 0.9 ml of the diluted culture with 0.9 ml of LB medium containing increasing amounts of toluene or its derivative compounds. Incubation was done in 2.0-ml glass vials sealed with Teflon septa to avoid loss of the volatile organic compound. The cultures were incubated at 37°C for 30 min and then cooled on ice for 10 min. Luciferase activity was measured as described previously (22). Briefly, 75 μl of the induced culture was lysed by addition of 25 μl of 4× lysis buffer (100 mM Tris · HCl [pH 7.8], 32 mM NaH2PO4, 8 mM dithiothreitol, 8 mM CDTA, 4% [vol/vol] Triton-X 100, and 200 μg of polymyxin B sulfate per ml). Twenty-five microliters of the lysed cell solution was added to 25 μl of a combined 4× concentrate of luciferase substrates A and B (Analytical Luminescence Laboratories, Ann Arbor, Mich.) to give a final 2× concentration of each substrate. The luminescence was read immediately after substrate addition for 45 s in a Turner TD-20e luminometer.
Determination of organic compound concentrations in cell cultures.
Saturated solutions of toluene and its derivative compounds were made by mixing 4 ml of LB medium with 4 ml of organic compound for 1 h at room temperature on a platform shaker. The solutions were then centrifuged for 5 min at 3,000 × g to separate the organic and aqueous phases. Dilutions of the saturated LB medium in the aqueous phase were made by addition of fresh LB medium and were used to induce luciferase transcription in the biosensor cells. To determine the concentrations of toluene and its derivatives in the saturated LB media, solutions containing the 1% (wt/vol) NaCl of LB medium, without the tryptone and yeast extract, were saturated with organic compound in parallel with the LB medium solutions. The concentration of the organic compound in the 1% saline solutions was determined by measuring the UV absorption at λmax and calculating the concentration from the extinction coefficient for each compound at λmax. Extinction coefficients for all the compounds tested were obtained from literature values measured in ethyl alcohol or methanol (9, 26, 27). The saturation concentration of the organic compound in LB medium was taken to be the same as that of the 1% NaCl solution. This assumption was verified by directly measuring the saturation concentration of toluene in LB medium and 1% NaCl by high-pressure liquid chromatography (HPLC) analysis. Saturated concentrations of toluene in each solution were made as described above and were analyzed on an SD-200 HPLC system (Rainin Instrument, Ridgefield, N.J.) using a Rainin Microsorb-mv C18 analytical column. Elution from the column was monitored at 250 nm with a Rainin Dynamax UV-DII absorbance detector. LB medium components were eluted from the column by isocratic flow in 50% acetonitrile–0.1% trifluoroacetic acid in H2O for 5 min. The mobile phase was then increased to 100% acetonitrile–0.1% trifluoroacetic acid in 1 min and run isocratically at 100% acetonitrile for 7 min. Toluene eluted as a single peak at 9.5 min. By comparing the peak area for toluene in LB medium or 1% NaCl solutions with the peak area for known amounts of toluene in ethanol, the saturation concentrations of toluene (means ± standard deviations) were found to be 5.2 ± 0.1 mM (n = 3) in LB medium and 4.82 ± 0.06 mM (n = 3) in 1% NaCl. Thus, the solubility of toluene is the same, within 10%, in LB medium or 1% NaCl. These values correlate with the toluene saturation concentrations determined by UV spectroscopy (6.1 ± 0.9 mM; n = 19). The values compare well with the published value for toluene saturation in water (5.8 mM [10]).
Testing of contaminated water.
Deep aquifer water from well OS-13, known to have BETX (benzene, toluene, and xylene) contamination, from the Baca Street site in Santa Fe, N. Mex., was obtained with permission from the Underground Storage Tank Bureau of the State of New Mexico Environmental Department. Three well volumes of water were purged before samples were taken. Sample water was filtered through a 0.2-μm-pore-size filter and stored in vials with Teflon septa at 4°C. When stored in this manner, no change in toluene contaminant concentration was observed for 1 week. Longer storage periods were not evaluated. Water was tested by adding 850 μl of OS-13 sample to 284 μl of 4× concentrate of LB medium, 216 μl of LB medium, and 450 μl of DH5α cells harboring the pGLTUR plasmid in LB medium at a cell density of 0.4 OD600. The cells were incubated for 1 h at 37°C, and then luciferase activity was measured as described above. Samples containing known concentrations of toluene in the 216-μl portion of LB medium were tested in parallel with 850 μl of deionized, distilled laboratory water in place of well water to generate a standard curve. As a control, water from an adjacent well (well USTB-2), which had no BETX contamination, was tested in the same manner. Standard curve data were fitted to the Hill equation, and the concentrations of toluene or toluene equivalents in the well water samples were calculated from the standard curve.
Testing of contaminated soil.
Contaminated soil was obtained from the DP road site at Los Alamos National Laboratory. This site, a former vehicle fueling and maintenance area, was known to be contaminated with BETX materials and was in the process of being remediated. The soil was placed in tightly sealing glass bottles immediately after excavation and stored at 4°C until assayed. Similar results were obtained for 1 month when the soil was stored in this manner. Uncontaminated soil from the same site was also obtained and used as a control. Thirty grams of soil samples were extracted with 30 ml of ethyl alcohol by being mixed on a platform shaker for 2 to 3 h at room temperature. Samples were centrifuged at 13,000 × g for 1 min to pellet the soil. Fifty microliters of the ethyl alcohol extraction supernatant was added to 850 μl of LB medium and 900 μl of cells at a cell density of 0.2 OD600. Parallel samples containing known toluene concentrations in the 850-μl portion of LB medium were also tested by using 50 μl of pure ethyl alcohol to obtain a standard curve. The data were fitted to the Hill equation, and the concentration of toluene or toluene equivalents in the ethyl alcohol extract was calculated from the standard curve. The 50 μl of ethyl alcohol extract inhibited toluene-induced luminescence by 40% (data not shown). This was caused entirely by the ethyl alcohol and not by any other component of the extract. To account for ethyl alcohol inhibition in the unknown samples, 50 μl of ethyl alcohol was added to the samples used to generate the standard curve. Thus, unknown and control samples contained the same amount of ethyl alcohol, and any error resulting from addition of ethyl alcohol to the biosensor cells was accounted for.
RESULTS AND DISCUSSION
Construction of the toluene biosensor.
A map of the pGLTUR plasmid is shown in Fig. 1. The position of the Pu promoter was 43 bp upstream of the luc gene at the XhoI site. The promoter fragment spanned 325 bp of the Pu region of the TOL plasmid including the upstream regulatory sequences for XylR binding, the ntr/nif −24 and −12 promoter sequences, the transcription initiation site, and 90 bp of the xylA gene (toluene oxygenase) (12, 16). The xylR structural gene sequence was placed 318 bp directly downstream of the luc gene in the same orientation as luc at the PfiMI site. It consisted of 2,335 bp spanning the autorepressor region upstream of xylR (1, 6, 14) and the entire open reading frame of xylR (13, 15, 25). The rrnB sequence was positioned between the Ampr gene and the luc gene at the KpnI site to decrease background luminescence. When DH5α cells were transformed with this vector containing the Pu promoter, luciferase reporter, and xylR activator together on the same plasmid, consistent luminescence signals were observed in response to toluene and derivative compounds.
Characterization of the toluene biosensor.
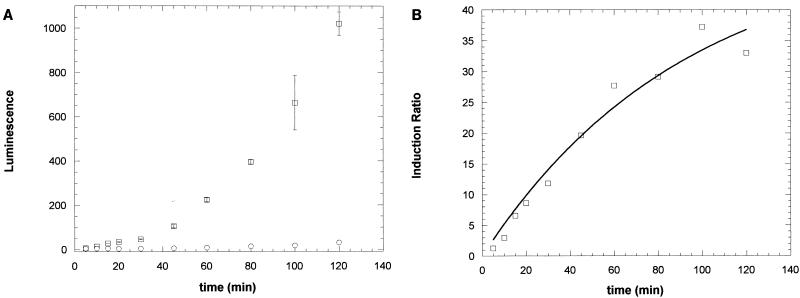
The time course of luciferase activity in DH5α cells harboring the pGLTUR plasmid is shown in Fig. 2A. Cells were exposed to 250 μM 3-xylene and were assayed for luciferase activity at the times indicated. Luminescence in exposed cells increased rapidly from a relative luminescence of 5.85 U at 5 min to 1,020 U at 120 min. Luminescence from unexposed cells increased much more slowly from 4.68 to 30.9 U over the same period as a result of cell growth. To determine the rate of induction of luciferase activity independently of the contribution of cell growth to that rate, the ratio of luminescence from induced versus uninduced cells was determined (Fig. 2B). The luciferase synthesis rate was treated as a pseudo-first-order reaction, and the induction ratio was fitted to a first-order rate equation. From the fit, the rate constant (mean ± standard deviation) was found to be 0.011 min−1 ± 0.003, yielding a half-life of 64 min and a maximal induction ratio of 50.5 ± 10.0. This rate is about twice that reported previously for xylR-activated transcription from the Pu promoter in E. coli grown in LB medium (1). Thus, relatively rapid induction of luciferase activity occurred in DH5α cells containing the pGLTUR plasmid. Testing of toluene and other xylR effectors was completed with 30-min induction periods in the linear portion of the time response. Testing of environmental samples was completed by using 60-min induction periods to increase sensitivity.
FIG. 2.
Kinetics of induction of the toluene biosensor by 3-xylene. (A) Rate of luminescence increase (arbitrary luminescence units) in DH5α cells harboring the toluene biosensor plasmid in nonexposed cells (○) or after exposure to 250 μM 3-xylene (□). Luminescence was measured as described in Materials and Methods. Error bars represent the standard deviations from three replicates of each time point. The rate of cell growth was the same for 3-xylene-exposed and control cell suspensions (data not shown). (B) Kinetics of the ratio of luminescence from 3-xylene-exposed and -unexposed cells in panel A. The line represents the nonlinear least-squares fit of the data to the first-order rate equation y = ymax (1 − e−kt), in which y is the ratio of luminescence from 3-xylene-exposed versus -unexposed cells at time t, ymax is the maximal value of the induction ratio at infinite time, and k is the rate constant. A ymax value of 50.5 ± 10.0 and a k value of 0.011 ± 0.003 min−1 were obtained from the curve fit.
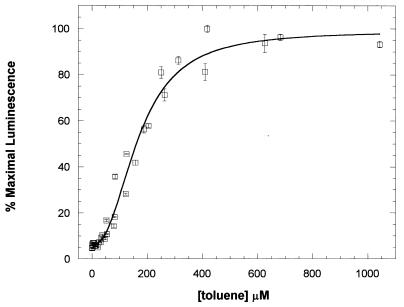
The response of the biosensor cell suspensions to toluene is shown in Fig. 3. Results were very reproducible between experiments, as shown by the small amount of scatter in the data. The luminescence exhibited a sigmoidal dependence on toluene concentration and was best fit by the Hill equation. A nonlinear least-squares fit of the data gave a K1/2 of 169 ± 9.6 μM (mean ± standard deviation), an napp of 2.3 ± 0.3, and a maximal luminescence increase of 16-fold. The limit of toluene detection was between 10 and 20 μM (0.92 to 1.84 ppm). The source of the observed cooperativity is not known, but it was observed with all the toluene derivatives tested that were potent activators (see Table 1). The cooperativity could result from allosteric interactions between XylR monomers in an oligomeric complex. It has previously been suggested that XylR may act as a cooperative oligomer to activate transcription from Pu (2). Alternatively, the cooperativity may have resulted from the way in which the pGLTUR vector was constructed. The xylR gene is directly downstream of the luciferase gene and in the same orientation (Fig. 1). Thus, transcriptional activation at Pu could also increase the transcription of xylR. However, transcription of the xylR gene is inhibited by XylR protein at its promoter, Pr (1, 14). Therefore, one would predict the opposite effect, that transcription of xylR would decrease as XylR protein increased until a steady-state level of XylR protein is reached.
FIG. 3.
The toluene concentration dependence of luminescence from toluene biosensor cells. Luminescence from DH5α cells harboring the pGLTUR plasmid was measured at the concentrations of toluene indicated, as described in Materials and Methods. Data from three separate experiments were normalized for maximal luminescence and combined. The error bars represent the standard deviations of three replicates of each sample within the same experiment. The line represents the nonlinear least-squares fit of the data to the Hill equation, y = {(ymax − ymin)[toluene]n/(K1/2n + [toluene]n)} + ymin, in which y is the percentage of maximal luminescence at a given concentration of toluene, ymax is the percentage of maximal luminescence at the saturating concentration of toluene (100%), and ymin is the percentage of maximal luminescence at a toluene concentration of zero. K1/2 is the concentration of toluene at which half-maximal effect is observed, and n is the Hill coefficient, napp. Values of 169 ± 9.6 μM for K1/2 and 2.3 ± 0.3 for napp were obtained from the curve fit.
TABLE 1.
Characteristics of induction of the toluene biosensor by toluene derivative compoundsa
| Compound | K1/2 (μM) | napp | Fold induction |
|---|---|---|---|
| Toluene | 169 ± 9.6 | 2.3 ± 0.3 | 16 |
| Benzene | 468 ± 17 | 3.2 ± 0.3 | 9.2 |
| 2-Xylene | 45.2 ± 5.4 | 1.9 ± 0.4 | 22 |
| 3-Xylene | 39.0 ± 3.8 | 1.8 ± 0.3 | 21 |
| 4-Xylene | 103 ± 7 | 2.0 ± 0.2 | 26 |
| 2-Chlorotoluene | 62.9 ± 6.4 | 1.9 ± 0.3 | 21 |
| 3-Chlorotoluene | 80.0 ± 2.1 | 2.8 ± 0.2 | 19 |
| 4-Chlorotoluene | 80.9 ± 1.6 | 3.8 ± 0.3 | 15 |
| 2-Methylbenzylalcohol | 2,690 ± 160 | 1.9 ± 0.2 | 2.2 |
| 3-Methylbenzylalcohol | 1,820 ± 170 | 2.4 ± 0.5 | 8.8 |
| 4-Methylbenzylalcohol | NDb | ||
| 2-Nitrotoluene | 610 ± 91 | 2.3 ± 0.7 | 4.3 |
| 3-Nitrotoluene | ∼1,100 | 1.2 ± 0.5 | 2.3 |
| 4-Nitrotoluene | ∼600 | 0.8 ± 0.3 | 2.9 |
The half-maximal induction values (K1/2), Hill coefficients (napp), and the increase in luminescence at saturating concentration are reported for several toluene derivatives. The data were obtained from titration curves for each compound as shown for toluene in Fig. 3 (see Materials and Methods). Data from at least three experiments were combined, and K1/2, napp, and induction values were obtained from fits of the combined data to the Hill equation as described in the legend to Fig. 3. The accuracy of the K1/2 values for 3- and 4-nitrotoluene suffered from the small amount of induction produced by these compounds resulting in curve fits that did not yield reasonable error values. The reported K1/2 values should be considered accurate within a factor of 2.
ND, no detectable induction up to 1,500 mM, the highest concentration assayed.
Several analogs of toluene were tested to define the substrate specificity and sensitivity of xylR in the pGLTUR construct. The concentration dependence of luminescence for each analog was measured over the full response range, as was done for toluene. The data are summarized in Table 1. Robust induction of luciferase activity was observed for xylenes and chlorotoluenes. All three isomers of xylene and chlorotoluene induced an ∼20-fold increase in luminescence and exhibited Hill coefficients generally near 2, values similar to those of toluene. The reporter was most sensitive to 3-xylene, with half-maximal binding occurring at 39.0 ± 3.8 μM (mean ± standard deviation) and a detection limit of ∼3 μM (0.4 ppm). Sensitivity to benzene was significantly less than those for toluene, xylenes, and chlorotoluenes. The K1/2 was 468 μM, a 12-fold decrease compared to that of 3-xylene. Moreover, the fold increase in luminescence at saturating concentrations was 9.2, approximately half that of 3-xylene. The methylbenzyl alcohols and nitrotoluenes were the weakest inducers of luciferase activity. Luminescence increased only two to four times at saturating concentrations of most isomers of these compounds. Values of K1/2 ranged from 600 to 1,000 μM for the nitrotoluenes to approximately 2 mM for 2- and 3-methylbenzylalcohol, reflecting a low affinity of these compounds for the effector binding pocket in XylR. Generally, the compounds with the lowest binding affinity for XylR also gave the smallest increases in luminescence at saturating concentrations. This result suggests that intermediate conformations of partially active XylR exist, somewhere between the fully active form when 3-xylene is bound and the inactive form when no inducer molecule is bound. It appears that molecules that bind with lower affinity generate this partially active conformation of XylR. Thus, XylR may exist in a continuum of conformations of varying activity depending on the affinity of the inducer molecule for the binding pocket. This is opposed to a situation in which XylR could exist in only two conformations, one fully active and the other inactive. Effector compounds would induce the fully active state at differing concentrations, depending on their affinity for the binding pocket.
These different responses to very similar compounds demonstrate the specificity of the pGLTUR-based biosensor. The specificity is also reflected by the number of compounds similar to toluene that do not induce luciferase activity. For example, 4-methylbenzylalcohol, 3-methylphenol, and benzoate did not increase luciferase activity, nor did the less-related compound trichloroethylene (data not shown). Thus, this biosensor was specific for toluene and a select number of its analogs. The data are consistent with and extend previous specificity measurements for XylR effectors (1).
Samples from contaminated sites often contain multiple xenobiotics. Thus, it is important to know how the toluene biosensor would respond to combinations of compounds. If compounds that induce luciferase activity bind the effector region of XylR at the same site and produce similar degrees of activation of XylR, then luminescence responses would be expected to be additive. If different compounds could bind XylR simultaneously at different sites or activate only partially, then synergistic or inhibitory effects could be observed. Benzene, toluene, and m-xylene were tested separately and in two-component mixtures for their ability to induce biosensor luminescence. Results from mixing components were compared with what would be predicted if the individual components acted in an additive manner (Table 2). Good agreement was found between the measured luminescence and the predicted luminescence in each of the two-component mixtures. Thus, it appears that these compounds are acting in an additive manner, suggesting that they bind to the same effector site on XylR and elicit similar responses.
TABLE 2.
Additivity of induction of the toluene biosensor by benzene, toluene, and 3-xylenea
| Compound(s) | % Maximum luminescence measured | % Maximum luminescence predicted |
|---|---|---|
| Benzene | 47 ± 3 | |
| Toluene | 51 ± 3 | |
| m-Xylene | 61 ± 8 | |
| Benzene + toluene | 68 ± 10 | 76 |
| Benzene + m-xylene | 79 ± 3 | 79 |
| Toluene + m-xylene | 89 ± 6 | 80 |
The effects of two-component mixtures were tested at 430 μM benzene, 120 μM toluene, and 22 μM 3-xylene as described in Materials and Methods. These concentrations were chosen to produce ∼50% maximal responses when added alone and ∼75% maximal responses when added in pairs. A standard curve of toluene, similar to the data of Fig. 3, was also done in parallel to determine the maximal response and the toluene equivalent response of benzene and 3-xylene. The toluene equivalent response represented the concentration of toluene equivalent to the luminescence response elicited by benzene or 3-xylene. This was determined by fitting the standard curve data to the Hill equation as described for Fig. 3 and calculating from the equation what toluene concentration would elicit the same response. The predicted percent luminescence was determined by adding the toluene equivalent concentrations for each pair and predicting from the toluene standard curve what luminescence response this sum of concentrations would elicit. The error range represents the standard deviations of duplicate samples. A representative data set from three separate determinations is shown.
Testing of contaminated water and soil with the toluene biosensor.
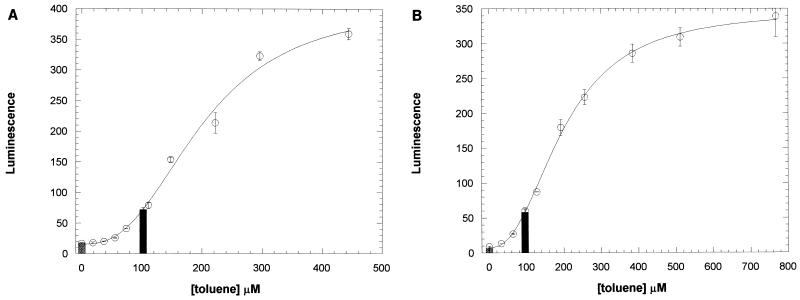
To determine the utility of the toluene biosensor in measuring actual environmental contamination, water samples of known contaminant concentration were tested and results of the biosensor assay were compared to the known concentrations. Deep aquifer water was obtained from contaminated and uncontaminated wells at the Baca Street site in Santa Fe, N. Mex. This site has been actively monitored by the Underground Storage Tank Bureau of the State of New Mexico Environmental Department for 3 years. One particular well, designated OS-13, was contaminated principally with benzene, toluene, xylenes, and ethylbenzene. Water from well OS-13 and from an uncontaminated well in the same area (USTB-2) was tested by using the pGLTUR-based biosensor. The results of a typical experiment are shown in Fig. 4A. Water from OS-13 gave a luminescence response of 72 ± 4 (mean ± standard deviation; n = 8) luminescence units. From the graph, one can see that this luminescence corresponded to ∼100 μM toluene. By calculating the contaminant concentration from the standard curve, a value of 102 ± 3.3 μM was obtained. Taking the dilution factor of the assay into account (see Materials and Methods), the final concentration of contaminants in the water was 217 ± 7.4 μM (20.0 ± 0.7 ppm) (means ± standard deviations). The average value from three separate tests was 215 ± 23.0 μM (19.8 ± 2.1 ppm). This concentration should be referred to as a toluene equivalent concentration because it could have resulted from 601 μM benzene or 50.1 μM 3-xylene or some combination of those compounds detected by the biosensor. K1/2 values from Table 1 give the relative sensitivity of the toluene biosensor for these other compounds. The toluene biosensor did not detect any contaminants in the uncontaminated well, USTB-2 (Fig. 4A). Furthermore, no quenching of the luminescence response to spiked samples containing 720 μM toluene was observed up to the highest concentration of OS-13 water added (47%) to the assay mixture.
FIG. 4.
Testing of contaminated water and soil by using the toluene biosensor. (A) Water from BETX-contaminated well OS-13 and from uncontaminated well USTB-2 at the Baca Street site in Santa Fe, N. Mex., was tested for toluene derivative contamination as described in Materials and Methods. The luminescence responses to water from the contaminated well (black bar) and the uncontaminated well (cross-hatched bar) are shown, as is the luminescence response to different known toluene concentrations (○). Error bars represent the standard deviations of three replicates of each sample. The standard curve was fit to the Hill equation as described for Fig. 3 (represented by the line), and the resulting equation was used to calculate the toluene equivalent concentrations of the well water (indicated by the positions of the bars on the x axis). (B) Soil from the DP road site at Los Alamos, N. Mex., was extracted with ethanol and tested for toluene derivative contamination as described in Materials and Methods. The luminescence responses of ethyl alcohol extracts from contaminated (black bar) and uncontaminated (cross-hatched bar) soil collected from the same site are shown, as is the luminescence response to different known toluene concentrations (○). Error bars represent the standard deviations of three replicates of each sample. The toluene equivalent concentrations were determined as for panel A.
Table 3 compares the results of State of New Mexico OS-13 water testing with the biosensor assay. Biosensor tests were conducted on water drawn on 4 June 1996. The nearest dates of testing by the State of New Mexico are shown for comparison. The toluene equivalent concentrations are calculated from the K1/2 values in Table 1, giving sums of 20.9 ppm on 6 March 1996 and 18.1 ppm on 7 October 1996. This trend of decreasing contaminant concentration with time had been observed for a 2-year period. The concentration of toluene equivalents of 19.8 ± 2.1 ppm (mean ± standard deviation) measured by the toluene biosensor assay compares closely to the state laboratory’s results. If one assumes a linear decrease with time, then on 4 June 1996 the toluene equivalent concentration should have been 20.4 ppm. Thus, the pGLTUR-based biosensor was able to measure toluene concentrations within 3% of those measured by conventional methods. These results demonstrate that this type of biosensor can give accurate measurements of BETX contaminants in aqueous environmental samples.
TABLE 3.
Comparison of contamination concentrations from OS-13 well watera
| Source | Date (mo/day/yr) | Compound(s) | ppm | Toluene equivalent (ppm)b |
|---|---|---|---|---|
| State of New Mexico | 3/6/96 | Benzene | 12 | 4.3 |
| Toluene | 7.4 | 7.4 | ||
| Ethylbenzene | 0.35 | 0 | ||
| Xylenes | 3.4 | 9.2 | ||
| Total | 20.9 | |||
| 10/7/96 | Benzene | 10.5 | 3.8 | |
| Toluene | 5.7 | 5.7 | ||
| Ethylbenzene | 0.6 | 0 | ||
| Xylenes | 3.2 | 8.6 | ||
| Total | 18.1 | |||
| This report | 6/4/96 | Toluene or equivalent | 13.0 ± 1.6 | 19.8 ± 2.1 |
The amount of contamination measured by the State of New Mexico using standard gas chromatography-mass spectroscopy methods is compared to the amounts measured by using the toluene biosensor. Results of the two tests by the State nearest to the data of the biosensor testing (6/4/96) are shown for comparison purposes. The biosensor results are averages of three separate determinations.
The toluene equivalent values were calculated by multiplying the parts per million for each compound by the ratio of its corresponding K1/2 to the K1/2 for toluene (Table 1). The State’s results did not distinguish between xylene isomers; therefore, the three K1/2 values were averaged in determining the toluene equivalent concentration. Ethylbenzene has been shown by others to activate XylR only weakly (1); therefore, it was considered not to contribute significant toluene equivalents.
Soil from a site at Los Alamos National Laboratory that was known to be contaminated with BETX materials was tested with the pGLTUR-based biosensor to determine its utility in detecting soil contamination. Samples were obtained immediately upon excavation, extracted with ethyl alcohol, and assayed for toluene equivalents as described in Materials and Methods. The results are shown in Fig. 4B. From the graph, one can see that 50 μl of ethyl alcohol extract containing soluble compounds from contaminated soil resulted in luminescence comparable to roughly 100 μM toluene, whereas assay of ethyl alcohol extracts from uncontaminated soil showed no increase in luminescence. From the standard curve, a value of 95.7 ± 3.3 μM (mean ± standard deviation; n = 4) toluene equivalents was obtained. Taking into account the dilution factor of the assay (see Materials and Methods), the final toluene equivalent concentration was 3.44 ± 0.12 mM. This corresponds to 317 ± 13 ppm toluene equivalents in the 30 g of extracted soil. These results demonstrate that the pGLTUR-based biosensor can be used to detect BETX contamination of soil. It should be noted that the sensitivity of the assay is limited by the amount of ethyl alcohol that can be added to the biosensor cell suspensions. In the assay, the ethyl alcohol concentration was 2.8%. Increasing this concentration resulted in high cell mortality and unacceptable loss of the luminescence signal. Thus, the concentration limit for detection of toluene in soil samples by this method is ∼30 ppm, compared to a detection limit of ∼1 ppm in aqueous samples (Fig. 3), because of the necessity to dilute the ethyl alcohol extract. This limit could be decreased by employing methods of concentrating ethyl alcohol extracts before dilution into the luciferase assay.
This study demonstrates the capabilities of a biosensor based on the transcriptional activator XylR to detect toluene and specific derivative compounds. By use of E. coli cells harboring the pGLTUR plasmid construct, BETX contamination in collected environmental samples was able to be accurately measured. Both aqueous and soil samples could be assayed. Detection in aqueous samples was limited by the binding affinity of the contaminant compound for XylR and ranged from 0.4 ppm for 3-xylene to 4 ppm for benzene. Detection in soil samples was limited by the ethyl alcohol concentration in soil extracts that the cells could tolerate. This limit was ∼30 times greater than for aqueous samples. The pGLTUR-based bacterial biosensor represents a fast, simple, and inexpensive alternative to conventional gas chromatographic and mass spectroscopic methods of BETX detection. The greatest advantage of this type of biosensor may be the ease with which it could be applied to field testing. Assay kits could readily be developed for accurate on-site BETX contamination analysis.
One potential problem with this method of testing of environmental samples is that components of the sample such as high salt, extremes of pH, or compounds toxic to E. coli could inhibit luciferase expression. However, no quenching was observed in the deep-well water samples and the ethanol-extracted soil examined here. In each environmental context, controls for quenching would have to be performed.
The development of this biosensor for toluene and its derivative compounds demonstrates the feasibility of constructing similar biosensors with specificity for other organic contaminants by using their corresponding transcriptional activators. Transfer of the biosensor plasmid from E. coli to other more environmentally hardy bacterial species could permit in situ testing of BETX-contaminated sites.
ACKNOWLEDGMENTS
This work was conducted under the auspices of and was partially funded by the U.S. Department of Energy. Additional funding was received from the Department of Chemistry and Biochemistry, Brigham Young University.
We express thanks to Jerry Schoeppner and his colleagues at the Underground Storage Tank Bureau, State of New Mexico Environmental Department, for assistance in obtaining water samples and in providing results of contamination testing of water from Baca Street wells, Santa Fe, N. Mex. We also thank Thomas C. Terwilliger for helpful discussions.
REFERENCES
- 1.Abril M-A, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abril M-A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon. J Biol Chem. 1991;266:15832–15838. [PubMed] [Google Scholar]
- 3.Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- 4.Burlage R S, Hooper S W, Sayler G S. The TOL (pWWO) catabolic plasmid. Appl Environ Microbiol. 1989;55:1323–1328. doi: 10.1128/aem.55.6.1323-1328.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Fernandez S, Herrero M, Jakubzik U, Timmis K N. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 1993;130:41–46. doi: 10.1016/0378-1119(93)90344-3. [DOI] [PubMed] [Google Scholar]
- 6.Gomada M, Inouye S, Imaishi H, Nakazawa A, Nakazawa T. Analysis of an upstream regulatory sequence required for activation of the regulatory gene xylS in xylene metabolism directed by the TOL plasmid of Pseudomonas putida. Mol Gen Genet. 1992;233:419–426. doi: 10.1007/BF00265439. [DOI] [PubMed] [Google Scholar]
- 7.Harayama S, Timmis K N. Aerobic biodegradation of aromatic hydrocarbons by bacteria. In: Sigel H, Sigel A, editors. Metal ions in biological systems. Vol. 28. New York, N.Y: Marcel Dekker Inc.; 1992. pp. 99–156. [Google Scholar]
- 8.Heitzer A, Malachowsky K, Thonnard J E, Bienkowski P R, White D C, Sayler G S. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1994;60:1487–1494. doi: 10.1128/aem.60.5.1487-1494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirayama K. Handbook of ultraviolet and visible absorption spectra of organic compounds. New York, N.Y: Plenum Press; 1967. [Google Scholar]
- 10.Howard P H. Handbook of environmental fate and exposure data for organic chemicals. Chelsea, Mich: Lewis Publishers; 1989. p. 435. [Google Scholar]
- 11.Ikariyama Y, Nishiguchi S, Kobatake E, Aizawa M, Tsuda M, Nakazawa T. Luminescent biomonitoring of benzene derivatives in the environment using recombinant Escherichia coli. Sensors Actuators B. 1993;13–14:169–172. [Google Scholar]
- 12.Inouye S, Ebina Y, Nakazawa A, Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci USA. 1984;81:1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inouye S, Nakazawa A, Nakazawa T. Determination of the transcription initiation site and identification of the protein product of the regulatory gene xylR for xyl operons on the TOL plasmid. J Bacteriol. 1985;163:863–869. doi: 10.1128/jb.163.3.863-869.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inouye S, Nakazawa A, Nakazawa T. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc Natl Acad Sci USA. 1987;84:5182–5186. doi: 10.1073/pnas.84.15.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid for Pseudomonas putida. Gene. 1988;66:301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
- 16.Inouye S, Gomada M, Sangodkar U M, Nakazawa A, Nakazawa T. Upstream regulatory sequence for transcriptional activator xylR in the first operon of xylene metabolism on the TOL plasmid. J Mol Biol. 1990;216:251–260. doi: 10.1016/S0022-2836(05)80317-1. [DOI] [PubMed] [Google Scholar]
- 17.King J M H, Digrazia P M, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler G S. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1991;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 18.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;6:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 19.Marques S, Ramos J L. Transcriptional control of the Pseudomonas putida tol plasmid catabolic pathways. Mol Microbiol. 1993;9:923–929. doi: 10.1111/j.1365-2958.1993.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Martin J, Timmis K N, Delorenzo V. Coregulation by bent DNA: functional substitutions of the integration host factor site at ς(54)-dependent promoter Pu of the upper-tol operon by intrinsically curved sequences. J Biol Chem. 1994;269:22657–22662. [PubMed] [Google Scholar]
- 21.Schell M A, Wender P E. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J Bacteriol. 1986;166:9–14. doi: 10.1128/jb.166.1.9-14.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Schupp, J. M., and P. Keim. Unpublished results.
- 22.Schupp J M, Travis S E, Price L B, Shand R F, Keim P. Rapid bacterial permeabilization reagent useful for enzyme assays. BioTechniques. 1995;19:18–20. [PubMed] [Google Scholar]
- 23.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (dmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shingler V, Moore T. Sensing of aromatic compounds by the dmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spooner R A, Lindsay K, Franklin F C H. Genetic, functional and sequence analysis of the xylR and xylS regulatory genes of the TOL plasmid pWWO. J Gen Microbiol. 1986;132:1347–1358. doi: 10.1099/00221287-132-5-1347. [DOI] [PubMed] [Google Scholar]
- 26.Weast R C, editor. CRC handbook of chemistry and physics. Vol. 60. Boca Raton, Fla: CRC Press; 1980. p. C–161. [Google Scholar]
- 27.Weast R C, Grasselli J G, editors. Handbook of data on organic compounds. 2nd ed. Boca Raton, Fla: CRC Press; 1989. [Google Scholar]
- 28.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yen K M, Gunsalus I C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci USA. 1982;79:874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]