Abstract
After spiking anoxic sediment slurries of three acidic oligotrophic lakes with either HgCl2 at 1.0 μg/ml or CH3HgI at 0.1 μg/ml, both mercury methylation and demethylation rates were measured. High mercury methylation potentials were accompanied by high demethylation potentials in the same sediment. These high potentials correlated positively with the concentrations of organic matter and dissolved sulfate in the sediment and with mercury levels in fish. Adjustment of the acidic sediment pH to neutrality failed to influence either the methylation or the demethylation rate of mercury. The opposing methylation and demethylation processes converged to establish similar Hg2+-CH3Hg+ equilibria in all three sediments. Because of their metabolic dominance in anoxic sediments, mercury methylation and demethylation in pure cultures of sulfidogenic, methanogenic, and acetogenic bacteria were also measured. Sulfidogens both methylated and demethylated mercury, but the methanogen tested only catalyzed demethylation and the acetogen neither methylated nor demethylated mercury.
Mercury from nonpoint atmospheric sources reaches even pristine lakes and, after its biomethylation, accumulates in fish. This process is especially pronounced in oligotrophic acidic lakes (19–21). The determination of mercury levels at or above the 1-μg/g regulatory limit for fish has led to the introduction of economically damaging sport fishing restrictions on wide areas of Canada and the midwestern United States (19). Similar contamination of fish by mercury was recently reported in the Pine Barrens region of southern New Jersey (14). The Pine Barrens is a partially wooded, sparsely populated area straddling the Kirkwood-Cohansee aquifer (10). Due to its natural and water resources, it enjoys some measure of ecological protection, and the discovery of mercury contamination of fish in this relatively pristine area was particularly disturbing. No point sources of mercury discharge were identified, and the mercury contamination is believed to be mainly of atmospheric origin, related to distant incinerators and the burning of fossil fuels. Clearly, the balance of microbial mercury methylation and demethylation activities in the lakes is critical, since only the hydrophobic methylmercury accumulates in fish to levels that require regulatory attention (19, 21). Inhibitor experiments clearly tied the methylation of inorganic mercury in anoxic aquatic sediments to the activities of certain sulfidogenic bacteria (8, 9, 12). The types of bacteria that demethylate methylmercury in sediments were found to be more diverse, and their activities could not be demonstrated unequivocally in pure culture (15). The factors that influence the balance of the opposing methylation and demethylation processes and are thus responsible for the overall environmental methylmercury concentrations are as yet insufficiently understood (20, 21). For these reasons, we conducted mercury methylation and demethylation experiments in sediments of some affected Pine Barrens lakes, attempting to correlate these activities with the prevailing environmental parameters, such as sulfate and sulfide concentrations, pH, and sediment organic matter level. In an effort to interpret our results, the mercury transformation potentials of sulfidogenic, methanogenic, and acetogenic bacteria in pure culture were also reexamined.
MATERIALS AND METHODS
Collection, processing, and incubation of sediments.
Sediment cores were collected from lakes in the Pine Barrens area of southern New Jersey from May to November 1996. Atlantic City Reservoir, Batsto Lake, and East Creek Lake were selected for sampling because, in an earlier survey, some fish from these lakes exceeded the 1-μg/g regulatory limit for mercury (12). The three lakes were similar in character, all being retained by artificial dams at their outflow. They are relatively shallow and have mostly sandy sediments and moderately acidic (pH 5.5 to 6.0) water that is dark because of humic substances. All three lakes are relatively pristine, being situated in wooded nonresidential areas, but are accessible by paved roads. None has received sewage discharges.
The lake sediments were collected at the deepest (3 to 4 m) portions of the lakes by using a Wildco (Saginaw, Mich.) corer with acrylic liners. The 5- by 20-cm cores were immediately sealed into their liners without air pockets, transported to the laboratory, and placed, within 3 h of collection, in an anaerobic chamber (PACE 6500; Labline Instruments, Melrose Park, Ill.) with an atmosphere of 5% H2, 5% CO2, and 90% N2. All subsequent operations were performed within this chamber. The sediment cores were pooled, slurried with deaerated lake water, and passed through a no. 18 sieve (1-mm-diameter openings) to remove vegetation, stones, and woody debris. The resulting slurries had approximately 350 mg of sediment per ml. The sediments were divided into two batches. One batch was adjusted to pH 7.0, using NaOH and an electronic pH meter. The other batch was left at its original pH. The two batches were subdivided once more and spiked with either 1.0 of μg HgCl2 or 0.1 μg of CH3HgI per ml. With continuous stirring and the use of a tilting repeater dispenser (Kontes, Vineland, N.J.), 20-ml amounts of the spiked slurries, containing about 7 g of sediment each, were placed into 50-ml anaerobic vials. The vials were sealed with butyl rubber stoppers and aluminum crimp seals. They were subsequently incubated at 27°C, approximating the maximum summer temperature of the lakes. Abiotic (autoclaved) sediment controls were included.
Gas samples from the sediment of Atlantic City Reservoir were collected during the coring process. The gas bubbles rising from the disturbed sediment were captured by means of a submerged inverted funnel. Connected to the funnel was a 60-ml disposable plastic syringe (Becton Dickinson, Franklin Lakes, N.J.) that could be closed with a stainless steel valve (Popper & Sons, New Hyde Park, N.Y.). The captured gas was drawn into the syringe, and the valve was closed prior to the disconnection of the funnel. The gas sample was analyzed within 24 h of collection.
Pure-culture experiments.
Desulfovibrio desulfuricans LS was isolated in our laboratory (8). D. desulfuricans ND-132, selected for its high Hg2+ methylation activity, was kindly donated by C. Gilmore (Academy of Natural Sciences of Philadelphia). The purity of the cultures was ascertained by repeated single-colony isolations from agar shake tubes in media B and E (16). The identification of the cultures was verified by their fluorescence under UV illumination (due to desulfoviridin), their morphology, and their substrate utilization range (16). Methanococcus maripaludis ATCC 4300 (2) was kindly donated by the laboratory of W. B. Whitman, University of Georgia, Athens. It was maintained and pregrown for experiments in ATCC medium 1439 (2) under an atmosphere of 80% H2 and 20% CO2. The acetogen Eubacterium limosum ATCC 8486 (2) was kindly donated by the laboratory of L. Young, Rutgers University, New Brunswick, N.J. It was grown in ATCC medium 1019 under an atmosphere of 30% CO2 and 70% N2. For mercury methylation and demethylation experiments, the sulfidogens were pregrown in low-sulfate medium D with pyruvate as the carbon and energy source (16). The methanogen and the acetogen were grown in their recommended ATCC media (2). Anaerobic vials (50 ml) containing 20 ml of the appropriate medium were spiked with either 1.0 μg of HgCl2 or 0.1 μg of CH3HgI per ml, inoculated at 5% (vol/vol) with 2-day-old precultures, and incubated at 37°C with slow reciprocal shaking. Methylation and demethylation experiments were always conducted simultaneously with matched cell suspensions. Unless an inhibitor was present, the initial inoculum increased 15- to 20-fold during incubation.
Specific inhibitors of sulfidogens (sodium molybdate, 2 mM) and methanogens (bromoethane sulfonate [BES], 0.5 mM), employed in earlier sediment experiments (8, 9, 15), were added in some of our pure-culture incubations to verify their effects on mercury methylation and demethylation. Strict anaerobic processes were observed, just as in the sediment experiments. Abiotic (uninoculated) controls were included.
Analytical procedures.
At appropriate time intervals, the methylmercury contents of replicate vials were extracted in their entirety for analysis, using the procedure of Longbottom et al. (13). This procedure converts all of the methylmercury in the sample to monomethylmercury. Monomethylmercury is the principal alkylmercury species present in sediments and fish (21). Dimethylmercury, if detectable at all, is present only in trace amounts. Sediment samples were extracted immediately after being spiked, as well as after appropriate incubation periods. Methylation and demethylation were stopped by injecting 2.0-ml aliquots of a 1.0 M CuSO4 solution into the serum vials containing sediment slurry or culture suspension. After solvent extraction and cleanup, monomethylmercury levels were measured with a Hewlett-Packard model 5890 gas chromatograph equipped with a model AT-35 macrobore capillary column (0.53-mm internal diameter, 15 m long; Alltech, Deerfield, Ill.). Operating conditions were as follows: 95:5 Ar-CH4 (vol/vol) carrier gas (Matheson Gas Products, East Rutherford, N.J.) delivered at 35 ml/min, injector at 150°C, oven at 100°C, and electron capture detector at 250°C. Monomethylmercury peak areas (retention time, 1.25 min) were recorded by a Hewlett-Packard 3392A integrator that had been calibrated with monomethylmercury (CH3HgI) standards (American Tokyo Kasei, Inc., Portland, Oreg.). The detection limit for monomethylmercury was 1 ng/ml of sediment slurry or culture suspension.
In sediment experiments, the number of time points analyzed did not allow the replication of every point, but several time points in each experiment were analyzed in triplicate. The standard deviations for these time points are shown as error bars in the figures below. Similar standard errors need to be assumed for the nonreplicated time points that have no error bars. In the pure-culture experiments, the time points were not replicated.
The collected gas from the Atlantic City Reservoir sediment was analyzed for its methane content by using a model 1200 gas partitioner (Fisher Co., Springfield, N.J.) operated at 50°C with 30 ml of helium carrier per min. The sample volumes analyzed were 250 μl, and the instrument was calibrated with methane gas standards (Fisher Co.).
Organic matter content was determined as weight loss on ignition (600°C, overnight) of dried (100°C) sediment samples. Acid-labile sulfide was determined iodometrically in 50-g (wet weight) sediment samples (1). For sulfate measurements, sediments were vigorously agitated after addition of an equal volume of distilled water and then centrifuged. Sulfate in the clear supernatant was determined gravimetrically after precipitation with barium (1). Organic-matter and sulfide concentrations are reported on a dry-sediment basis, while the concentration of sulfate, which is assumed to be in the dissolved state, is reported per milliliter of pore water in the undiluted sediments (see Table 1).
TABLE 1.
Physicochemical characteristics of three Pine Barrens lakes with relevance to mercury methylation and demethylation
| Lake | pH | Conc of:
|
||
|---|---|---|---|---|
| Organic matter (%) | Sulfate (μg/ml of pore water) | Sulfide (acid labile; μg/g)a | ||
| Atlantic City Reservoir | 6.0 | 31.0 | 67 | 86 |
| Batsto Lake | 6.0 | 24.0 | 30 | 69 |
| East Creek Lake | 5.5 | 25.8 | 44 | 75 |
Per gram of dry sediment.
Each experiment was repeated at least once, and results were considered valid if the time curves were reproduced within the error limits.
RESULTS AND DISCUSSION
Mercury methylation and demethylation in lake sediments.
Lake sediment characteristics of potential relevance to methylation and demethylation of mercury are listed in Table 1. Free sulfide was not detectable in the sediments, and gas bubbles rising to the surface during core sampling were indicative of methanogenic activity in all three lake sediments. Gas evolution during coring was particularly abundant in Atlantic City Reservoir. The gas collected at this location contained 72.4% methane by volume. The organic matter and sulfate concentrations were the highest in the Atlantic City Reservoir sediment and somewhat lower in the East Creek Lake and Batsto Lake sediments. The differences in pH and in acid-labile sulfide values were slight.
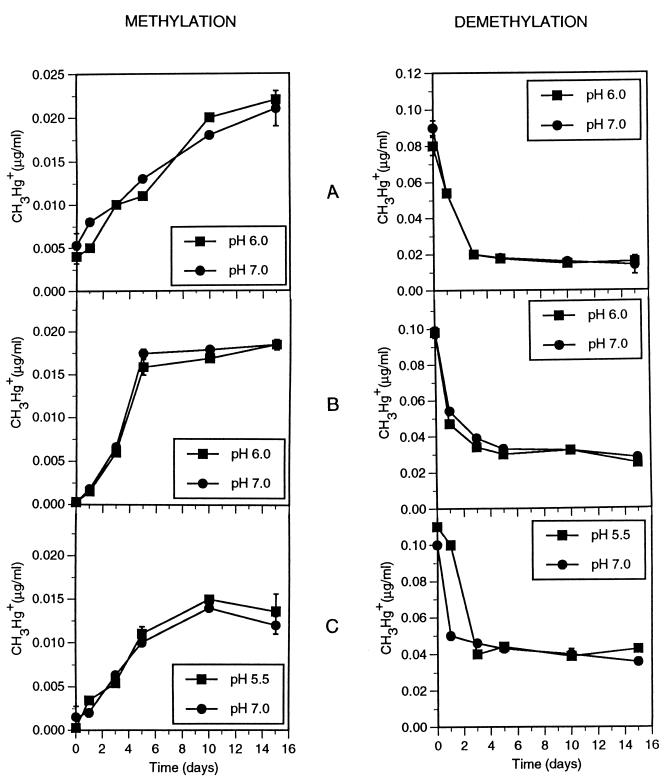
When the lake sediment slurries were spiked with 1.0 μg of HgCl2 per ml and were incubated for 15 days, 15 to 22 ng of methylmercury per ml was formed (Fig. 1). Initial synthesis was rapid and approached equilibrium after 5 to 10 days. At time zero, only the Atlantic City Reservoir sediment slurry contained a detectable amount of methylmercury (4 ng/ml). When spiked with 0.1 μg of methylmercury per ml, the same sediments showed a rapid decrease in methylmercury concentration to 16 to 40 ng/ml. Again, most of this decrease occurred in the first 5 days of incubation.
FIG. 1.
Methylation and demethylation of mercury in sediments of Atlantic City Reservoir (A), Batsto Lake (B), and East Creek Lake (C) spiked with either 1.0 μg of HgCl2 or 0.10 μg of CH3HgI per ml. In methylation experiments, triplicate samples were analyzed on days 0, 5, and 15; in demethylation experiments, triplicate samples were analyzed on days 0, 10, and 15. The error bars (±1 standard deviation) were omitted when smaller than the symbols. For the rest of the time points, only single samples were analyzed, but similar standard errors must be assumed. Note that the CH3Hg+ concentration scales for the mercury methylation and demethylation experiments differ.
Both the mercury methylation and demethylation activities were most extensive in the Atlantic City Reservoir sediment, where equilibrium concentrations of 18 to 22 ng of methylmercury per ml were attained during the experimental period, regardless of the initial spiking material used. Both the methylation and demethylation processes were slightly less extensive in the Batsto Lake and East Creek Lake sediments. In these two lakes, a complete methylmercury concentration equilibrium was not attained during the 15-day incubation period, although a trend toward such an equilibrium was discernable. The high mercury methylation and demethylation activities in Atlantic City Reservoir sediments appeared to correlate positively with the level of organic matter in the sediment, the concentration of dissolved sulfate in the pore water (Table 1), and the concentration of mercury in fish (14). The parameters and activity levels in the other two lakes were too similar to each other to reveal any trend.
Largemouth bass (Micropterus salmoides) from Atlantic City Reservoir were found to contain 3.0 to 8.9 μg of mercury/g, while the same species from Batsto Lake contained 0.7 to 1.3 μg/g (14). This fish species was not obtained from East Creek Lake, but the chain pickerel (Esox niger), which occupies a trophic level similar to that of the largemouth bass, contained 0.8 to 2.8 μg of mercury/g. Since practically all of the mercury found in fish is present in the form of methylmercury, mercury methylation rates may be expected to correlate with elevated mercury levels in fish (21).
Previously, the level of organic matter was found to be positively correlated with mercury methylation activity in estuarine sediments (5), and this correlation appears to exist also in the case of the three freshwater lakes investigated here. Virtually all mercury methylation in anoxic aquatic sediments has been tied to the activity of sulfidogens (8, 9, 11). Yet, because of the reaction of H2S with Hg2+ to form HgS, which is virtually unavailable for methylation, sulfate concentrations of estuarine sediments were found to correlate inversely with mercury methylation activity (3, 9). At the much lower sulfate concentrations of freshwater lakes, addition of sulfate to the 200 mM level was found to stimulate Hg2+ methylation (11, 12). The sulfate concentrations measured by us in the Pine Barrens lakes were only slightly higher, and thus a positive correlation of sulfate levels with mercury methylation in these lakes was not unexpected.
Because of the known association of low pH with mercury accumulation in fish (21), the effects of pH on the balance of mercury methylation and demethylation were the subject of several studies. Ramlal et al. (17) and Steffan et al. (18) found that demethylation was relatively insensitive to pH changes but that acidification of the anoxic lake sediments to pH 5.0 or lower essentially stopped methylmercury synthesis. The authors demonstrated a decrease in Hg2+ availability for methylation in the acidified sediments. The postulated mechanisms were HgS formation by acid-mobilized H2S and increased Hg2+ adsorption. Of course, these results did not explain the accumulation of mercury in fish at low pH.
The unresolved state of the pH effect on the balance of mercury methylation and demethylation prompted us to reexamine this question by using Pine Barrens lake sediments. To avoid the possibility of H2S mobilization, we adjusted the naturally acidic (pH 5.5 to 6.0) lake sediments upward only to pH 7.0. Neither methylation nor demethylation rates were affected by this modest pH adjustment (Fig. 1). Our results confirm the opinion (17, 18, 21) that mercury accumulation in fish at low pH is not caused directly by stimulation of the methylation or inhibition of the demethylation process.
Methylation and demethylation of mercury by pure cultures of strictly anaerobic sediment bacteria.
Historically, the identification of sediment bacteria responsible for mercury methylation and demethylation followed a two-step process. Sediments were treated with a selective inhibitor of sulfate reduction (molybdate) or methanogenesis (BES), and the effects of these treatments on mercury methylation or demethylation were measured. Subsequently, pure cultures of the purportedly involved bacteria were isolated and their mercury methylation or demethylation potentials were confirmed. This approach tied over 95% of the mercury methylation in anoxic sediments to the activities of sulfidogens (8, 9), and the process was verified in pure cultures of D. desulfuricans (8). By a similar approach, Oremland et al. (15) investigated bacterial populations involved in demethylation of monomethylmercury. On the basis of inhibition experiments, they concluded that in anoxic freshwater sediments, both methanogens and sulfidogens participated in methylmercury demethylation, but in estuarine and hypersaline sediments, only the contributions of sulfidogens were significant. However, these authors had little success in verifying methylmercury demethylation in pure cultures of sulfidogens and methanogens. While their sediment samples typically decomposed 35 to 70% of the added methylmercury, their pure cultures decomposed less than 3% of the radiolabeled methylmercury, which contained 2% radiochemical impurities. Assuming that these inconclusive results were due to culture selection or test conditions, we embarked on reexamining the ability of strict anaerobes to decompose methylmercury in pure culture.
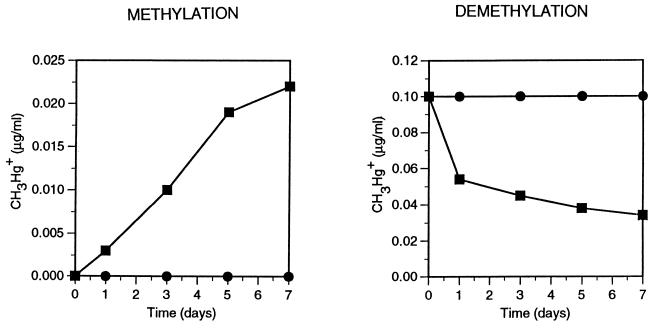
Figure 2 shows mercury methylation and demethylation in pure cultures of D. desulfuricans LS. Intentionally presented on the same scale as the sediment experiments of Fig. 1, a comparison shows that the mercury methylation and demethylation activities of this sulfidogen very closely resemble those measured in lake sediments. The addition of 2 mM molybdate completely inhibited both the methylation and demethylation activities, supporting the results of specific-inhibitor approaches used in earlier sediment experiments (8, 9, 15). A comparison of the relative mercury methylation and demethylation rates of D. desulfuricans LS with those of D. desulfuricans ND 138 (data not shown) indicated that these activities may vary independently. While the demethylation activity of D. desulfuricans ND 138 was almost identical to that of D. desulfuricans LS, the mercury methylation activity of the former was four times higher than that of strain LS. It is relevant that D. desulfuricans ND 138 was selected from numerous isolates for its high net methylmercury synthesis activity.
FIG. 2.
Methylation and demethylation of mercury in pure cultures of D. desulfuricans LS spiked with either 1.0 μg of HgCl2 or 0.10 μg of CH3HgI per ml (▪), and the same in the presence of 2.0 mM sodium molybdate (•). For ease of comparison, the presentation matches the sediment experiments of Fig. 1 except for the time scale.
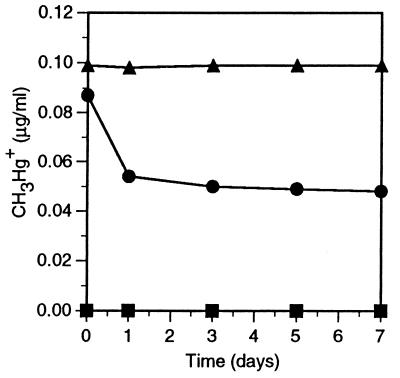
Figure 3 shows that M. maripaludis was unable to methylate inorganic mercury but demethylated methylmercury at a rate comparable to that of the sulfidogens. The presence of 0.5 mM BES completely stopped demethylation, again supporting the outcome of earlier sediment work with specific inhibitors (15). E. limosum, an acetogen, was unable to either methylate or demethylate mercury.
FIG. 3.
Methylation and demethylation of mercury in pure cultures of M. maripaludis spiked with either 1.0 μg of HgCl2 (▪) or 0.1 μg of CH3HgI (•) per ml, and demethylation of mercury by M. maripaludis in the presence of 0.5 mM BES (▴). The methylation and demethylation time curves of E. limosum (not shown) were identical to those of HgCl2 and BES, respectively.
Although earlier specific-inhibitor experiments have implicated sulfidogens and methanogens as being demethylators of methylmercury in sediments (15), the present study has led to the first conclusive report of methylmercury demethylation in pure cultures of these obligate anaerobes. It provides a necessary confirmation of the methylmercury demethylation potential attributed to these microorganisms in their anoxic-sediment environments. Oremland et al. (15) successfully reproduced methylmercury demethylation by pure cultures of facultative anaerobes such as Escherichia coli. Such microorganisms, which were active mainly in aerobic sediments, evolved 14CH4 from 14CH3Hg+, but little evolution of 14CH4 was measured in either freshwater or estuarine sediments under anoxic incubation conditions (15). In anoxic sediments, the principal gaseous product of 14CH3Hg+ decomposition was 14CO2, leading the authors to question the identity of the biochemical oxidative pathway which converts the methyl groups of monomethylmercury to carbon dioxide (15).
In our laboratory, the D. desulfuricans LS mercury methylation process was shown to be enzymatically catalyzed (6), with methylcobalamin serving as the methyl donor (4), and the acetyl coenzyme A synthase reaction was implicated in Hg2+ methylation (7). Running forward, this reaction seems to generate CH3Hg+ as an incidental side product. Running backward, the same reaction could release the methyl carbon as CO2. This proposition, while clearly only a hypothesis, is consistent with the results of Oremland et al. (15) concerning the participation of sulfidogens and methanogens as principal demethylators of methylmercury in anoxic sediments and also accounts for CO2 being the major product of methylmercury demethylation in such environments.
ACKNOWLEDGMENTS
We thank C. Gilmore (Academy of Natural Sciences of Philadelphia) for the culture of D. desulfuricans ND-132 and L. Young (Agbiotech Center, Rutgers University) for E. limosum and for the use of her gas partitioner for methane analysis. Our special thanks are expressed to R. England and B. Ruppert of the New Jersey Department of Environmental Protection for generously devoting time and facilities to our sediment collection needs.
This work was supported by the New Jersey Department of Environmental Protection, by the New Jersey Water Resources Institute, and by state funds.
Footnotes
New Jersey Agricultural Experiment Station publication no. D-01408-01-97.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater, sect. 4. Washington, D.C: American Public Health Association; 1992. pp. 127–133. [Google Scholar]
- 2.American Type Culture Collection. Catalogue of bacteria and bacteriophages. 19th ed. 1996. p. 166. , 212, 514, 538. American Type Culture Collection, Rockville, Md. [Google Scholar]
- 3.Blum J E, Bartha R. Effect of salinity on methylation of mercury. Bull Environ Contam Toxicol. 1980;25:404–408. doi: 10.1007/BF01985546. [DOI] [PubMed] [Google Scholar]
- 4.Choi S-C, Bartha R. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1993;59:290–295. doi: 10.1128/aem.59.1.290-295.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi S-C, Bartha R. Environmental factors affecting mercury methylation in estuarine sediments. Bull Environ Contam Toxicol. 1994;53:805–812. doi: 10.1007/BF00196208. [DOI] [PubMed] [Google Scholar]
- 6.Choi S-C, Chase T, Jr, Bartha R. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1994;60:1342–1346. doi: 10.1128/aem.60.4.1342-1346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi S-C, Chase T, Jr, Bartha R. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1994;60:4072–4077. doi: 10.1128/aem.60.11.4072-4077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compeau G C, Bartha R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol. 1985;50:498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compeau G C, Bartha R. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl Environ Microbiol. 1987;53:261–265. doi: 10.1128/aem.53.2.261-265.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooley J. Natural sources of mercury in the Kirkwood Cohansey aquifer system of the NJ coastal plain. Draft report. Trenton: New Jersey Geological Survey; 1992. [Google Scholar]
- 11.Gilmore C C, Henry E A. Mercury methylation in aquatic systems affected by acid deposition. Environ Pollut. 1991;71:131–169. doi: 10.1016/0269-7491(91)90031-q. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore C C, Henry E A, Mitchell R. Sulfate stimulation of mercury methylation in freshwater sediments. Environ Sci Technol. 1992;26:2281–2287. [Google Scholar]
- 13.Longbottom J E, Dressman R C, Lichtenberg J J. Metals and other elements: gas chromatographic determination of methylmercury in fish, sediment, and water. J Assoc Off Anal Chem. 1973;56:1297–1303. [Google Scholar]
- 14.New Jersey Department of Environmental Protection. Preliminary assessment of total mercury concentrations in fish from rivers, lakes and reservoirs in New Jersey. Report no. 93-15F. Trenton: New Jersey Department of Environmental Protection; 1994. [Google Scholar]
- 15.Oremland R S, Culbertson C W, Winfrey M R. Methylmercury decomposition in sediments and bacterial cultures: involvement of methanogens and sulfate reducers in oxidative demethylation. Appl Environ Microbiol. 1991;57:130–137. doi: 10.1128/aem.57.1.130-137.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postgate J R. The sulphate-reducing bacteria. 2nd ed. Cambridge, United Kingdom: Cambridge University Press; 1984. p. 32. [Google Scholar]
- 17.Ramlal P S, Rudd J W M, Furutani A, Xun L. The effect of pH on methyl mercury production and decomposition in sediments. Can J Fish Aquat Sci. 1985;42:685–692. [Google Scholar]
- 18.Steffan R J, Korthals E T, Winfrey M R. Effects of acidification on mercury methylation, demethylation, and volatilization in sediments from an acid-susceptible lake. Appl Environ Microbiol. 1988;54:2003–2009. doi: 10.1128/aem.54.8.2003-2009.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain E B. Assessment of mercury contamination in selected Minnesota lakes and streams. Report to the Legislative Commission on Minnesota Resources. St. Paul: Minnesota Pollution Control Agency; 1989. [Google Scholar]
- 20.Watras C J, Morrison K A, Host J S. Concentration of mercury species in relationship to other site-specific factors in the surface waters of northern Wisconsin lakes. Limnol Oceanogr. 1995;40:556–565. [Google Scholar]
- 21.Winfrey M R, Rudd J W M. Environmental factors affecting the formation of methylmercury in low pH lakes. Environ Toxicol Chem. 1990;9:853–869. [Google Scholar]