Abstract
Bacteria which were β-d-galactosidase and β-d-glucuronidase positive or expressed only one of these enzymes were isolated from environmental water samples. The enzymatic activity of these bacteria was measured in 25-min assays by using the fluorogenic substrates 4-methylumbelliferyl-β-d-galactoside and 4-methylumbelliferyl-β-d-glucuronide. The enzyme activity, enzyme induction, and enzyme temperature characteristics of target and nontarget bacteria in assays aimed at detecting coliform bacteria and Escherichia coli were investigated. The potential interference of false-positive bacteria was evaluated. Several of the β-d-galactosidase-positive nontarget bacteria but none of the β-d-glucuronidase-positive nontarget bacteria contained unstable enzyme at 44.5°C. The activity of target bacteria was highly inducible. Nontarget bacteria were induced much less or were not induced by the inducers used. The results revealed large variations in the enzyme levels of different β-d-galactosidase- and β-d-glucuronidase-positive bacteria. The induced and noninduced β-d-glucuronidase activities of Bacillus spp. and Aerococcus viridans were approximately the same as the activities of induced E. coli. Except for some isolates identified as Aeromonas spp., all of the induced and noninduced β-d-galactosidase-positive, noncoliform isolates exhibited at least 2 log units less mean β-d-galactosidase activity than induced E. coli. The noncoliform bacteria must be present in correspondingly higher concentrations than those of target bacteria to interfere in the rapid assay for detection of coliform bacteria.
Indicators of pollution (e.g., coliforms, fecal coliforms, and Escherichia coli) are traditionally used for monitoring the microbiological safety of water supplies and recreational water. Several techniques for detection of coliforms and E. coli are based on enzymatic hydrolysis of fluorogenic or chromogenic substrates for β-d-galactosidase and β-d-glucuronidase (9, 20). Current methods of recovery are usually culture based, and the analysis time is 18 to 24 h. In addition to enzymatic activity, these techniques use growth at appropriate temperatures in the presence of inhibitors, combined with demonstration of enzymatic activity, to selectively detect target bacteria.
Rapid methods which require less than 6 h and are based on chromogenic, fluorogenic, or chemiluminogenic substrates for detection of coliforms, fecal coliforms, or E. coli have been described (1–3, 10, 27, 28). These rapid assays are based on the assumption that β-d-galactosidase and β-d-glucuronidase are markers for coliforms and E. coli, respectively. However, when the incubation time is 1 h or less, growth is not a selective step, and all β-d-galactosidase-positive or β-d-glucuronidase-positive microorganisms in a water sample contribute to the activity measured. At low initial concentrations of target bacteria (i.e., E. coli and total coliforms), increasing the preincubation time to 5 to 6 h did not result in a predominance of target bacteria compared to nontarget bacteria (28).
The β-d-galactosidase or β-d-glucuronidase activity calculated per cultivable coliform or fecal coliform bacterium in environmental samples can be 1 to 2 log units higher than the activity per induced E. coli cell in pure culture (11, 26). The presence of active, noncultivable bacteria can be one reason for this. Studies of survival (7, 24, 25) and disinfection (26) of E. coli have shown that loss of cultivability does not necessarily result in a loss of β-d-galactosidase activity. The presence of false-positive bacteria can be another reason.
β-d-Galactosidase has been found in numerous microorganisms, including gram-negative bacteria (e.g., strains belonging to the Enterobacteriaceae, Vibrionaceae, Pseudomonadaceae, and Neisseriaceae), several gram-positive bacteria, yeasts, protozoa, and fungi (17, 29). β-d-Glucuronidase is produced by most E. coli strains and also by other members of the Enterobacteriaceae, including some Shigella and Salmonella strains and a few Yersinia, Citrobacter, Edwardia, and Hafnia strains. Production of β-d-glucuronidase by Flavobacterium spp., Bacteroides spp., Staphylococcus spp., Streptococcus spp., anaerobic corynebacteria, and Clostridium has also been reported (12).
High numbers of false-positive bacteria in sewage and contaminated water have been revealed by enumeration of β-d-galactosidase- and β-d-glucuronidase-positive CFU on nonselective agar supplemented with fluorogenic or chromogenic substrates (11, 28). Whether the activity from nontarget organisms can be neglected in a rapid assay depends on the number of nontarget organisms compared with the number of target bacteria and also on the level of their enzyme activity. Plant and algal biomass must be present at high concentrations to interfere in rapid bacterial β-d-galactosidase and β-d-glucuronidase assays (8).
The main objective of this study was to investigate the enzyme characteristics of β-d-galactosidase- and β-d-glucuronidase-positive bacteria isolated from environmental water samples and to evaluate the potential influence of false-positive bacteria in rapid assays for coliform bacteria or E. coli in water. The effect of temperature on enzyme activity and on the interference of nontarget bacteria in the rapid assays was investigated as an important factor.
(Some of the results were presented at the 97th General Meeting of the American Society for Microbiology 1997, Miami Beach, Fla., 4 to 8 May 1997.)
MATERIALS AND METHODS
Isolation of β-d-galactosidase- and β-d-glucuronidase-positive environmental bacteria.
Cultivable bacteria were recovered from sewage effluent (after primary treatment), polluted river water, or coastal water after membrane filtration (type GSWP Millipore filter; diameter, 47 mm; pore size, 0.22 μm) by growing them on tryptic soy agar (TSA) (Difco Laboratories) supplemented with 50 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma Chemical Co.) per liter and 50 mg of 4-methylumbelliferyl-β-d-glucuronide (MUGlu) (Diagnostic Chemicals Ltd.) per liter for 48 h at 35°C or for 72 h at 20°C. β-d-Galactosidase- and/or β-d-glucuronidase-positive bacteria were isolated by randomly picking green colonies (β-d-galactosidase positive) and/or fluorescent colonies (β-d-glucuronidase positive) from the agar plates. The colonies were purified by streaking them onto new agar and were identified on the basis of the results of Gram staining, the oxidase test, and the catalase test, as well as the API 20E and API 20NE tests (gram-negative rods) and the ID 32 Staph or ID 32 Strep test (gram-positive cocci) (bioMérieux). The isolates identified by the API systems as Aeromonas spp. and gram-positive rods were not further differentiated.
Preparation of induced and noninduced cells.
The isolated pure cultures were grown in 10% tryptic soy broth without dextrose (TSB) (Difco Laboratories) supplemented with 0.6 g of isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma Chemical Co.) per liter to achieve β-d-galactosidase induction. To achieve β-d-glucuronidase induction, TSB supplemented with 0.2 g of methyl-β-d-glucuronide (MetGlu) (Sigma Chemical Co.) per liter was used unless otherwise specified. TSB without IPTG or MetGlu was used to grow noninduced cells. The cultures were grown at 35 or 20°C depending on their temperature tolerance until the bacterial count was approximately 108 cells per ml (stationary phase, obtained after 1 to 4 days). Some strains showed weak growth, and the bacterial count was only 106 to 107 cells per ml after 4 days of incubation.
Cell counts.
The numbers of CFU in bacterial suspensions were determined by using membrane filtration (type GSWP Millipore filter; diameter, 47 mm; pore size, 0.22 μm) and TSA (Difco Laboratories) after 1 to 4 days of incubation at 35 or 20°C (depending on temperature tolerance and growth rates of the cultures).
The total numbers of cells (acridine orange direct counts [AODC]) in bacterial suspensions were determined directly by epifluorescence microscopy by using the standard acridine orange direct procedure of Hobbie et al. (16). Aliquots were preserved in 0.5% (wt/vol) (final concentration) formaldehyde. The AODC values below are the mean counts obtained with 200 cells or 10 microscopic fields of view. Agglomerated cell suspensions were treated for 1 min with an ultrasonic liquid processor (model VC 50; Sonics & Materials Inc.) before AODC were determined.
β-d-Galactosidase assay.
The β-d-galactosidase assay was performed by using a modification of the method described by Fiksdal et al. (10). Each bacterial culture was diluted in phosphate-buffered saline (pH 7.3) and filtered through a 0.2-μm-pore-size 47-mm-diameter polycarbonate filter (Poretics). The filter was then placed in a 250-ml flask containing 20 ml of phosphate buffer (PB) (pH 7.3) supplemented with 0.2 g of 4-methylumbelliferyl-β-d-galactoside (MUGal) (Diagnostic Chemicals Ltd.) per liter, 0.2 g of sodium laurylsulfate (Sigma Chemical Co.) per liter, and 0.1% nutrient broth (Difco Laboratories). The flasks were incubated in a shaking water bath at 44.5°C unless otherwise specified, and the fluorescence intensities of sample aliquots (2.5 ml of sample and 100 μl of 10 M NaOH) were measured every 5 min for 25 min with a Sequia Turner model 450 fluorometer with excitation at 365 nm and emission at 440 nm. At least two replicates were analyzed. Enzymatic activity, determined by a least-squares linear regression, was measured by determining the amount of methylumbelliferyl (MU) released per minute per CFU. Enzymatic activity is reported below as activity per total count in suspensions of Aerococcus viridans, Bacillus spp., and two unidentified gram-negative rods when cell agglomeration caused the difference between the CFU and the total count to be more than 0.5 log unit.
β-d-Glucuronidase assay.
The β-d-glucuronidase assay was performed like the β-d-galactosidase assay, except that the filters were placed in flasks containing 17 ml of PB (pH 6.4), 3 ml of a MUGlu solution (50 mg of MUGlu in 50 ml of PB supplemented with 1 drop of Triton X-100), and 0.1% nutrient broth (Difco Laboratories).
Measurement of induced and noninduced enzyme levels.
The levels of enzymatic activity per cell in cell suspensions, cultivated in the presence and absence of inducer, were determined by performing the 25-min enzyme assays described above at 44.5°C.
Temperature dependence.
The levels of enzymatic activity per cell of induced cell suspensions were determined by performing the 25-min enzyme assays described above at 25, 35, and 44.5°C.
RESULTS
β-d-Galactosidase activity of environmental isolates. (i) Members of the Enterobacteriaceae.
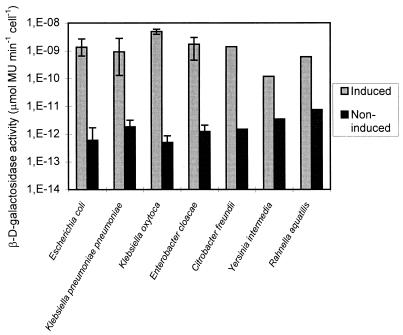
The β-d-galactosidases of all seven environmental isolates identified as E. coli were induced by IPTG. The levels of activity per induced cell varied between 7 × 10−10 and 3 × 10−9 μmol of MU min−1 CFU−1 and were 3 to 4 log units higher than the levels of activity per noninduced cell, which varied between 9 × 10−14 and 2 × 10−12 μmol of MU min−1 CFU−1. The β-d-galactosidases of other bacterial isolates belonging to the Enterobacteriaceae, including Klebsiella pneumoniae subsp. pneumoniae, Klebsiella oxytoca, Enterobacter cloacae, Citrobacter freundii, Yersinia intermedia, and Rahnella aquatilis isolates, were also inducible, and the β-d-galactosidase activities of these organisms were similar to the activities of E. coli (Fig. 1).
FIG. 1.
β-d-Galactosidase activities of different members of the Enterobacteriaceae isolated from environmental water samples and grown in the presence or absence of inducer (IPTG). The broad bars indicate the means, and the error bars indicate the maximum and minimum activities of isolates identified as members of the same species (seven E. coli isolates, six K. pneumoniae subsp. pneumoniae isolates, two K. oxytoca isolates, and two Enterobacter cloacae isolates).
(ii) Isolates that are not members of the Enterobacteriaceae.
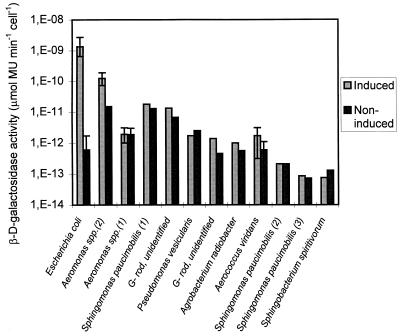
All of the bacterial isolates that did not belong to the Enterobacteriaceae showed no or only slight (≤1-log unit) induction by IPTG (Fig. 2). The maximum levels of activity of nontarget isolates (except Aeromonas sp. type 2 isolates) were at least 2 log units lower than the mean value obtained for E. coli (Fig. 2). If the IPTG-induced enzyme levels observed in this study reflect environmental conditions, a CFU ratio for nontarget and target bacteria of 101 to 104 is necessary for nontarget bacteria to affect the β-d-galactosidase rapid assay results. The results presented in Fig. 1 and 2 represent isolates that produce positive β-d-galactosidase reactions on TSA supplemented with X-Gal (green colonies). Other isolates that produce weakly positive β-d-galactosidase reactions were also investigated, but the levels of activity after growth in TSB supplemented with IPTG were below the detection limit (<10−14 μmol of MU min−1 cell−1). Two of these isolates were identified as A. viridans strains.
FIG. 2.
β-d-Galactosidase activities of different β-d-galactosidase-positive bacteria isolated from environmental water samples and grown in the presence or absence of inducer (IPTG). The broad bars indicate the means, and the error bars indicate the maximum and minimum activities of isolates identified as members of the same species (seven E. coli isolates, three Aeromonas sp. type 2 isolates, four Aeromonas sp. type 1 isolates, and two A. viridans isolates). G−, gram negative. Numbers in parentheses indicate bacterial types.
Temperature dependence of β-d-galactosidase activity.
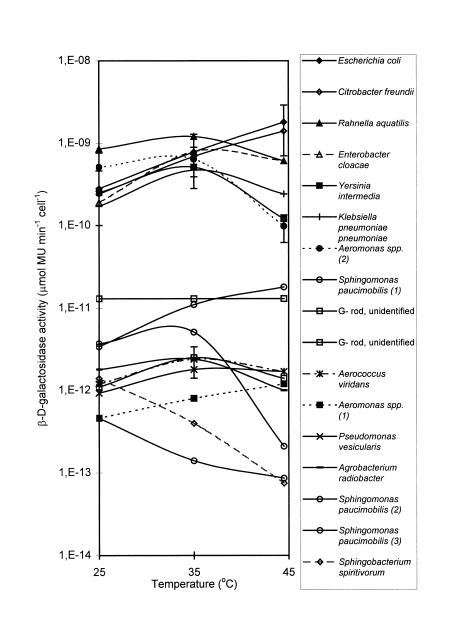
When the β-d-galactosidase activities of E. coli at 44.5, 35, and 25°C were compared, the maximum activity was observed at 44.5°C (Fig. 3). Other coliforms exhibited varying temperature dependence for this enzyme. The responses of C. freundii (Fig. 3) and some of the isolates identified as K. pneumoniae subsp. pneumoniae and Enterobacter cloacae (data not shown) were similar to the response of E. coli. The β-d-galactosidases of Enterobacter cloacae, K. pneumoniae subsp. pneumoniae, Y. intermedia, and R. aquatilis (Fig. 3) were not stable at 44.5°C, and the activity at this temperature was less than the activity at 35°C.
FIG. 3.
Induced β-d-galactosidase activities at 25, 35, and 44.5°C of different β-d-galactosidase-positive environmental isolates. The datum points are means, and the error bars indicate standard deviations for isolates identified as members of the same species (three E. coli isolates, three Aeromonas sp. type 2 isolates, and two A. viridans isolates). G−, gram negative. Numbers in parentheses indicate bacterial types.
At 44.5°C the enzyme activities of all but three of the other noncoliform β-d-galactosidase-positive bacteria were unstable (Fig. 3). Aeromonas spp. were differentiated on the basis of their β-d-galactosidase temperature characteristics. The isolates with a stable enzyme at 44.5°C were designated type 1, and the isolates with an unstable enzyme at 44.5°C were designated type 2. Similarly, isolates identified as Sphingomonas paucimobilis with different enzyme temperature dependence characteristics were differentiated and designated types 1, 2, and 3 (Fig. 3). However, the two isolates with an unstable enzyme at 44.5°C showed poor growth in API tests, and the identification of these organisms may be questioned.
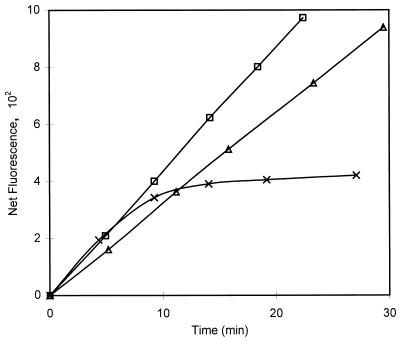
The combined effects of different temperatures and time on substrate hydrolysis by Aeromonas spp. (type 2) are shown in Fig. 4. While the production of MU (net fluorescence) was nearly linear during the 30-min assay period at 35 and 25°C, the fluorescence reached a plateau value after approximately 10 min at 44.5°C, demonstrating that enzyme destabilization occurred. The enzymatic activity measured by the 25-min assay, therefore, decreased compared to activities at lower temperatures. Type 2 Aeromonas spp. isolates had higher activities than type 1 isolates, and their activities were similar to or higher than the activities of E. coli at 25 and 35°C. However, at 44.5°C the activities of type 2 isolates were more than 1 log unit lower than the activity of E. coli (Fig. 3). Therefore, the interference of Aeromonas spp. type 2 in a rapid assay for coliform bacteria can be reduced by increasing the assay temperature from 35 to 44.5°C.
FIG. 4.
Plot of net fluorescence versus time (β-d-galactosidase activity) for Aeromonas spp. (type 2) at 25°C (▵), 35°C (□), and 44.5°C (×).
β-d-Glucuronidase activity of environmental isolates.
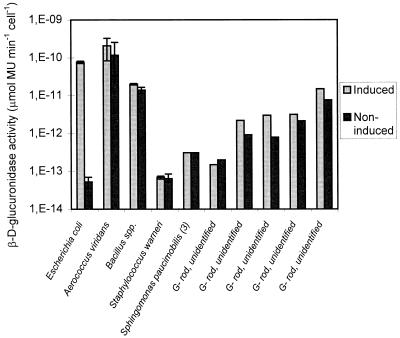
The β-d-glucuronidase of E. coli was successfully induced by MetGlu. Supplementing the growth medium with 0.1, 0.2, and 0.4 g of inducer per liter increased the β-d-glucuronidase activity of E. coli by factors of 1.4 × 102, 1.5 × 103, and 2.2 × 103, respectively. The other β-d-glucuronidase-positive bacteria were not induced or were only slightly induced (<1 log unit) by 0.2 g of MetGlu per liter (Fig. 5). The activities of false-positive gram-negative isolates and of three different gram-positive isolates identified as Staphylococcus warneri strains were 0.7 to 3 log units lower than the activity of induced E. coli. If the Met-Glu-induced enzyme levels observed in this study reflect environmental conditions, the bacterial concentrations of these nontarget bacteria must be correspondingly higher than those of target bacteria to interfere in a rapid β-d-glucuronidase assay. However, two isolates identified as Bacillus spp. and four isolates identified as A. viridans showed enzyme levels similar to those of induced E. coli. Therefore, the interference of these organisms cannot be neglected when the numbers of target and nontarget bacteria in environmental waters are similar. Other isolates (such as Pseudomonas pickettii), which produce positive β-d-glucuronidase reactions on TSA supplemented with MUGlu, were also investigated. The activities of these organisms after growth in TBS supplemented with MetGlu were below the detection limit (<10−14 μmol of MU min−1 cell−1).
FIG. 5.
β-d-Glucuronidase activities of different β-d-glucuronidase-positive bacteria isolated from environmental water samples and grown in the presence or absence of inducer (MetGlu). The broad bars indicate means, and the error bars indicate the maximum and minimum activities of isolates identified as members of the same species (two E. coli isolates, three Staphylococcus warneri isolates, two Bacillus sp. isolates, and four A. viridans isolates). G−, gram negative. (3), type 3.
Temperature dependence of β-d-glucuronidase.
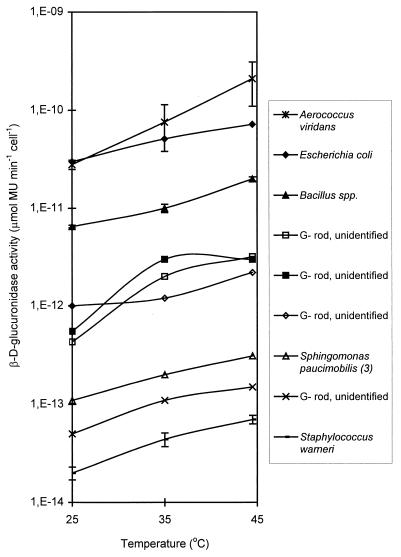
All of the nontarget β-d-glucuronidase-positive bacteria that were tested exhibited stable enzyme activity at 44.5°C (Fig. 6). However, all of these organisms except the isolates identified as Staphylococcus warneri were unable to grow at this temperature. Our results indicate that an increase in the assay temperature cannot be used to reduce nontarget interference in the rapid β-d-glucuronidase assay, as shown by the Bacillus spp. and A. viridans data (Fig. 6).
FIG. 6.
Induced β-d-glucuronidase activities at 25, 35, and 44.5°C of different β-d-glucuronidase-positive environmental isolates. The datum points are means, and the error bars indicate standard deviations for isolates identified as members of the same species (two Bacillus sp. isolates, three Staphylococcus warneri isolates, and four A. viridans isolates). G−, gram negative. (3), type 3.
Sensitivity limit of the rapid enzyme assays. (i) β-d-Galactosidase assay.
To obtain a detectable signal at 44.5°C after the bacterial concentration was increased 25-fold by filtration, a concentration of induced pure-culture E. coli cells of 103 CFU/100 ml before filtration was required. For other bacteria the following detection limits were observed: other coliforms, 103 to 104 CFU/100 ml; Aeromonas spp. (type 2), 104 CFU/100 ml; Aeromonas spp. (type 1), 106 CFU/100 ml; different gram-negative rods, 105 to 108 CFU/100 ml; and A. viridans, 106 to 108 cells/100 ml.
(ii) β-d-Glucuronidase assay.
For nonfiltered samples the following detection limits were observed when the filtration procedure was similar to the filtration procedure used in the β-d-galactosidase-assay: E. coli, A. viridans, and Bacillus spp., 104 to 105 cells/100 ml; Staphyloccus warneri, 107 to 108 CFU/100 ml; and different gram-negative rods, 105 to 108 CFU/100 ml.
DISCUSSION
Interference of nontarget organisms in a rapid assay depends on the number of nontarget organisms relative to the number of target bacteria. Previous studies demonstrated that different ratios between β-d-galactosidase-positive bacteria and coliform bacteria occur in sewage, river water, and coastal water (11). However, the individual enzyme activities of target and nontarget organisms are also important. Differences in the enzyme stabilities of target and nontarget bacteria should generally be considered when it is desirable to reduce the signal of nontarget bacteria. In this study the enzyme levels and enzyme temperature characteristics of different environmental bacteria were investigated. The β-d-galactosidases (18) and β-d-glucuronidases (23) of target bacteria (coliforms and E. coli) are both inducible, and enzyme levels were studied in the absence and presence of inducer.
β-d-Galactosidase temperature dependence.
The temperature of the β-d-galactosidase assay was previously optimized by using environmental E. coli isolates (26). Consistent with the results of this study, the E. coli isolates showed stable enzyme activity at 44.5°C, but the critical temperature when the enzyme was no longer stable varied (26). The enzyme activities of most of the nontarget bacteria were reduced when the assay temperature was increased from 35 to 44.5°C, indicating that a similar increase in assay temperature should reduce the contribution of nontarget bacteria in the β-d-galactosidase assay. Such an increase can also reduce the activity of some of the coliform bacteria (e.g., Y. intermedia), but the reductions were in general found to be not important.
Of the nontarget bacteria, only one isolate identified as Sphingomonas paucimobilis and the Aeromonas sp. type 1 isolates had higher activities at 44.5°C than at 35°C. However, the activity of the Aeromonas type 1 isolates was 3 log units lower than the activity of E. coli at 44.5°C, and interference by this organism therefore depends on high bacterial numbers.
The reported temperature dependence of different β-d-galactosidase-positive environmental isolates can be used to evaluate the dominant types of β-d-galactosidase-positive bacteria in environmental samples. Sewage samples showed higher activity at 44.5°C than at 35°C, demonstrating that organisms with stable β-d-galactosidase at 44.5°C were dominant (26). If the temperature optimum of a sample is low, the β-d-galactosidase activity most probably is caused by nontarget organisms.
Interference of false-positive bacteria in β-d-galactosidase assays.
At 44.5°C, the enzyme activities of all β-d-galactosidase-positive nontarget bacteria were 1 to 4 log units lower than the activity of induced E. coli. Depending on the bacterial concentration, low levels of interference can be expected with most of the gram-negative rods, as well as the gram-positive cocci, because of their low enzyme levels or unstable enzymes at 44.5°C. These results are consistent with results reported by other workers (6) for possible interference of lactose-fermenting marine vibrios in a β-d-galactosidase assay for detection of coliforms.
In the 25-min rapid assays, enzymatic activity is measured as the amount of MU released per minute by a least-squares linear regression. A nonlinear fluorescence-versus-time curve sometimes occurred when environmental samples were analyzed at 44.5°C (results not shown). High numbers of Aeromonas sp. type 2 cells which produced a plateau fluorescence value after approximately 10 min or other microorganisms responding similarly could have caused the nonlinearity. Interference by this type of nontarget bacteria can therefore be reduced by 10 to 15 min of preincubation of the sample at 44.5°C before measurements are made.
The enzyme activity of most of the nontarget bacteria increased when the assay temperature was decreased to 35°C. At this temperature nontarget bacteria isolated from natural water samples showed a positive chemiluminescent response at a concentration of 102 to 106 CFU/ml when Colicult medium supplemented with cefsulodin (28) was used. The cell concentrations of Aeromonas spp. (103 to 104 CFU/ml) which produced positive responses were 1 to 2 log units higher than the cell concentrations of Sphingomonas paucimobilis and Bacillus spp. (102 CFU/ml); i.e., these data were similar to the difference in the enzyme activities of Sphingomonas paucimobilis (1) and Aeromonas spp. (1) observed in this study at 35°C.
Interference of false-positive bacteria in β-d-glucuronidase assays.
The β-d-glucuronidases of all nontarget bacteria were stable at 44.5°C. Therefore, in contrast to the β-d-galactosidase assay, an assay temperature of 44.5°C cannot be used to reduce nontarget interference. The enzyme activities of isolates identified as Staphylococcus warneri and of some of the gram-negative bacteria were low, indicating that the interference of these organisms in environmental samples can be neglected, except at high bacterial concentrations (107 to 108 CFU/100 ml). However, interference by nontarget bacteria with high β-d-glucuronidase activities (e.g., A. viridans and Bacillus spp. [this study], enterococci [14], anaerobic Corynebacterium strains [5], clostridia [14], and Bacteroides species [14]) cannot be neglected if these organisms are present at concentrations similar to the concentrations of the target bacteria.
Induction characteristics.
The β-d-galactosidases and β-d-glucuronidases (or the permease cascades bringing the substrate into the cells) of all nontarget environmental isolates were less inducible than the enzymes of coliforms and E. coli. Additional studies are needed to determine if this is true for a larger portion of environmental nontarget bacteria. However, this information can be used to improve the specificity of current presence-absence methods, including growth, for detection of coliforms and E. coli by the construction of a supplementary test. If parallel incubations with and without inducer in the presence of nutrients reveal that the activity of a sample is 2 to 3 log units higher with the inducer, this indicates that growing target bacteria cause the activity. If not, the activity is probably caused by either nongrowing but active target bacteria or nontarget organisms.
The β-d-galactosidase and β-d-glucuronidase of E. coli are effectively induced by the inducers IPTG (18) and MetGlu (23), respectively. However, other β-d-galactosidase- and β-d-glucuronidase-positive bacteria may require other inducers for maximum activity (4, 13, 19, 21). The measured enzyme activity depends both on the enzyme content and the permeability of the cell membrane; addition of IPTG to Pseudomonas sp. strain BAL-31 cells growing in rich medium induced an o-nitrophenyl-β-d-galactoside-hydrolytic activity which was detectable in cell extracts but cryptic in whole cells (15). The minor effect of inducers on nontarget bacteria which was observed in this study could have been caused by unsuitable inducers or by a lack of permeases or other transport systems in the cell membrane.
The environment of the bacteria affects the induction of enzyme production, the permeability of the cell membrane, the cell composition, and the bacterial cell size (22). The enzyme level in bacteria cultivated in a laboratory does not necessarily reflect the enzyme level in the same bacteria in the environment. However, the induced and noninduced enzyme levels of different environmental isolates are useful for evaluating the potential interference of nontarget bacteria in rapid enzyme assays under optimal and suboptimal environmental conditions.
Summary.
The results of this study indicate that rapid enzyme assays without a growth-selective phase should not generally be rejected when high numbers of nontarget bacteria are present in environmental water samples. The obvious advantage of the 25-min assays is the short assay time. The assays can be used for early warning of accidental pollution or for monitoring water quality for recreation or aquaculture. Several of the enzyme-positive nontarget bacteria isolated from environmental water samples had low enzyme levels, and the influence of these organisms can be neglected except at very high bacterial concentrations. However, some nontarget bacteria with high enzyme levels, especially high levels of β-d-glucuronidase, could interfere seriously if they are present at high enough concentrations.
Differences in the enzyme stabilities of target bacteria and nontarget bacteria should generally be considered for possibly reducing the signal of nontarget bacteria. In rapid assays not including growth, different bacteria may tolerate different concentrations of organic solvents and detergents (4, 15), and additional studies are necessary to evaluate the effect of destabilizers on the specificity of rapid assays.
ACKNOWLEDGMENTS
This work was supported by the Norwegian Research Council under the European Economic Area agreement with the Commission of the European Communities Environment Research and Development Program 1994–1996 (contract EV5V-CT93-0345) and by a Ph.D. fellowship from the Norwegian University of Science and Technology NTNU to I.T.
We thank Kari Ingeborg Flatås and Frode Width Gran for laboratory assistance.
REFERENCES
- 1.Apte S C, Batley G E. Rapid detection of sewage contamination in marine waters using a fluorometric assay. Sci Total Environ. 1994;141:175–180. [Google Scholar]
- 2.Apte S C, Davies C M, Peterson S M. Rapid detection of faecal coliforms in sewage using a colorimetric assay of β-d-galactosidase. Water Res. 1995;29:1803–1806. [Google Scholar]
- 3.Berg J D, Fiksdal L. Rapid detection of total and fecal coliforms in water by enzymatic hydrolysis of 4-methylumbelliferone-β-d-galactoside. Appl Environ Microbiol. 1988;54:2118–2122. doi: 10.1128/aem.54.8.2118-2122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Citti J E, Sandine W E, Elliker P R. β-Galactosidase of Streptococcus lactis. J Bacteriol. 1965;89:937–942. doi: 10.1128/jb.89.4.937-942.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlén G, Linde A. Screening plate method for detection of bacterial β-glucuronidase. Appl Microbiol. 1973;26:863–866. doi: 10.1128/am.26.6.863-866.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies C M, Apte S C, Peterson S M. Possible interference of lactose-fermenting marine vibrios in coliform β-d-galactosidase assays. J Appl Bacteriol. 1995;78:387–393. doi: 10.1111/j.1365-2672.1995.tb03423.x. [DOI] [PubMed] [Google Scholar]
- 7.Davies C M, Apte S C, Peterson S M. β-d-Galactosidase activity of viable, non-culturable coliform bacteria in marine waters. Lett Appl Microbiol. 1995;21:99–102. doi: 10.1111/j.1472-765x.1995.tb01016.x. [DOI] [PubMed] [Google Scholar]
- 8.Davies C M, Apte S C, Peterson S M, Stauber J L. Plant and algal interference in bacterial β-d-galactosidase and β-d-glucuronidase assays. Appl Environ Microbiol. 1994;60:3959–3964. doi: 10.1128/aem.60.11.3959-3964.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton A D, Clesceri L S, Greenberg A E, Franson M A H, editors. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C: American Public Health Association; 1995. [Google Scholar]
- 10.Fiksdal L, Pommepuy M, Caprais M P, Midttun I. Monitoring of fecal pollution in coastal waters by use of rapid enzymatic techniques. Appl Environ Microbiol. 1994;60:1581–1584. doi: 10.1128/aem.60.5.1581-1584.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiksdal L, Tryland I, Nelis H. Rapid detection of coliform bacteria and influence of non-target bacteria. Water Sci Technol. 1997;35:415–418. [Google Scholar]
- 12.Frampton E W, Restaino L. Methods for Escherichia coli identification in food, water and clinical samples based on beta-glucuronidase detection. J Appl Bacteriol. 1993;74:223–233. doi: 10.1111/j.1365-2672.1993.tb03019.x. [DOI] [PubMed] [Google Scholar]
- 13.Hasan N, Durr I F. Induction of β-galactosidase in Lactobacillus plantarum. J Bacteriol. 1974;120:66–73. doi: 10.1128/jb.120.1.66-73.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawksworth G, Drasar B S, Hill M J. Intestinal bacteria and the hydrolysis of glycosidic bonds. J Med Microbiol. 1971;4:451–459. doi: 10.1099/00222615-4-4-451. [DOI] [PubMed] [Google Scholar]
- 15.Hidalgo C, Reyes J, Goldschmidt R. Induction and general properties of β-galactosidase and β-galactoside permease in Pseudomonas BAL-31. J Bacteriol. 1977;129:821–829. doi: 10.1128/jb.129.2.821-829.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams & Wilkins; 1994. [Google Scholar]
- 18.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 19.Jacox R F. Streptococcal β-glucuronidase. J Bacteriol. 1953;65:700–705. doi: 10.1128/jb.65.6.700-705.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manafi M, Kneifel W, Bascomb S. Fluorogenic and chromogenic substrates used in bacterial diagnostics. Microbiol Rev. 1991;55:335–348. doi: 10.1128/mr.55.3.335-348.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClatchy J K, Rosenblum E D. Induction of lactose utilization in Staphylococcus aureus. J Bacteriol. 1963;86:1211–1215. doi: 10.1128/jb.86.6.1211-1215.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 23.Novel G, Didier-Fichet M L, Stoeber F. Inducibility of β-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J Bacteriol. 1974;120:89–95. doi: 10.1128/jb.120.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nwoguh C E, Harwood C R, Barer M R. Detection of induced β-galactosidase activity in individual non-culturable cells of pathogenic bacteria by quantitative cytological assay. Mol Microbiol. 1995;17:545–554. doi: 10.1111/j.1365-2958.1995.mmi_17030545.x. [DOI] [PubMed] [Google Scholar]
- 25.Pommepuy M, Fiksdal L, Gourmelon M, Melikechi H, Caprais M P, Cormier M, Colwell R R. Effect of seawater on Escherichia coli β-d-galactosidase activity. J Appl Bacteriol. 1996;81:174–180. doi: 10.1111/j.1365-2672.1996.tb04496.x. [DOI] [PubMed] [Google Scholar]
- 26.Tryland, I., M. Pommepuy, and L. Fiksdal. Effect of chlorine on β-d-galactosidase activity of sewage bacteria and Escherichia coli. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 27.Van Poucke S O, Nelis H J. Development of a sensitive chemiluminometric assay for the detection of β-galactosidase in permeabilized coliform bacteria and comparison with fluorometry and colorimetry. Appl Environ Microbiol. 1995;61:4505–4509. doi: 10.1128/aem.61.12.4505-4509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Poucke S O, Nelis H J. Limitations of highly sensitive enzymatic presence-absence tests for detection of waterborne coliforms and Escherichia coli. Appl Environ Microbiol. 1997;63:771–774. doi: 10.1128/aem.63.2.771-774.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallenfels K, Weil R. The enzymes. VIII. New York, N.Y: Academic Press; 1972. β-Galactosidase; pp. 617–663. [Google Scholar]