Abstract
To establish a high-quality, easy-to-use, and effective risk prediction model for hepatic encephalopathy, to help healthcare professionals with identifying people who are at high risk of getting hepatic encephalopathy, and to guide them to take early interventions to reduce the occurrence of hepatic encephalopathy. Patients (n = 1178) with decompensated cirrhosis who attended the First Affiliated Hospital of Guangxi University of Chinese Medicine between January 2016 and June 2022 were selected for the establishment and validation of a nomogram model for risk prediction of hepatic encephalopathy. In this study, we screened the risk factors for the development of hepatic encephalopathy in patients with decompensated cirrhosis by univariate analysis, LASSO regression and multifactor analysis, then established a nomogram model for predicting the risk of getting hepatic encephalopathy for patients with decompensated cirrhosis, and finally performed differentiation analysis, calibration analysis, clinical decision curve analysis and validation of the established model. A total of 1178 patients with decompensated cirrhosis who were hospitalized and treated at the First Affiliated Hospital of Guangxi University of Chinese Medicine between January 2016 and June 2022 were included for modeling and validation. Based on the results of univariate analysis, LASSO regression analysis and multifactor analysis, a final nomogram model with age, diabetes, ascites, spontaneous peritonitis, alanine transaminase, and blood potassium as predictors of hepatic encephalopathy risk prediction was created. The results of model differentiation analysis showed that the AUC of the model of the training set was 0.738 (95% CI 0.63–0.746), while the AUC of the model of the validation set was 0.667 (95% CI 0.541–0.706), and the two AUCs indicated a good discrimination of this nomogram model. According to the Cut-Off value determined by the Jorden index, when the Cut-Off value of the training set was set at 0.150, the sensitivity of the model was 72.8%, the specificity was 64.8%, the positive predictive value was 30.4%, and the negative predictive value was 91.9%; when the Cut-Off value of the validation set was set at 0.141, the sensitivity of the model was 69.7%, the specificity was 57.3%, the positive predictive value was 34.5%, and the negative predictive value was 84.7%. The calibration curve and the actual events curve largely overlap at the diagonal, indicating that the prediction with this model has less error. The Hosmer–Lemeshow test for goodness of fit was also applied, and the results showed that for the training set, χ2 = 1.237587, P = 0.998, and for the validation set, χ2 = 31.90904, P = 0.0202, indicating that there was no significant difference between the predicted and actual observed values. The results of the clinical decision curve analysis showed that the model had a good clinical benefit, compared with the two extreme clinical scenarios (all patients treated or none treated), and the model also had a good clinical benefit in the validation set. This study showed that aged over 55 years, complications of diabetes, ascites, and spontaneous bacterial peritonitis, abnormal glutamate aminotransferase and abnormal blood potassium are independent risks indicators for the development of hepatic encephalopathy in patients with decompensated cirrhosis. The nomogram model based on the indicators mentioned above can effectively and conveniently predict the risk of developing hepatic encephalopathy in patients with decompensated cirrhosis. The nomogram model established on this study can help clinical healthcare professionals to timely and early identify patients with high risk of developing hepatic encephalopathy.
Subject terms: Risk factors, Gastrointestinal diseases
Introduction
Hepatic encephalopathy (HE) is an extremely serious complication of cirrhosis, and is the most common cause of death among various liver diseases. Hepatic encephalopathy has a great impact on patients' quality of life. Therefore, anticipatory interventions for those at risk for hepatic encephalopathy are particularly important. The personal experience of healthcare professionals is still the main basis for the assessment and identification of hepatic encephalopathy in clinical practice, making the diagnosis and treatment of hepatic encephalopathy significantly limited. If the risk of hepatic encephalopathy can be accurately predicted, if patients gets timely and early intervention, the progression of them getting hepatic encephalopathy can be stopped. However, the risk factors for the development of hepatic encephalopathy are not uniformly reported worldwide, the predictive ability of existed risk prediction models for hepatic encephalopathy is still unknown and failed to meet clinical needs. Therefore, this study screened the risk factors for the development of hepatic encephalopathy in patients with decompensated cirrhosis by univariate analysis, LASSO regression and multifactor analysis, and established a nomogram model for the prediction of risk of getting hepatic encephalopathy. The differentiation analysis, calibration analysis, clinical decision curve analysis and validation of the established model were also performed.
Methods
General information
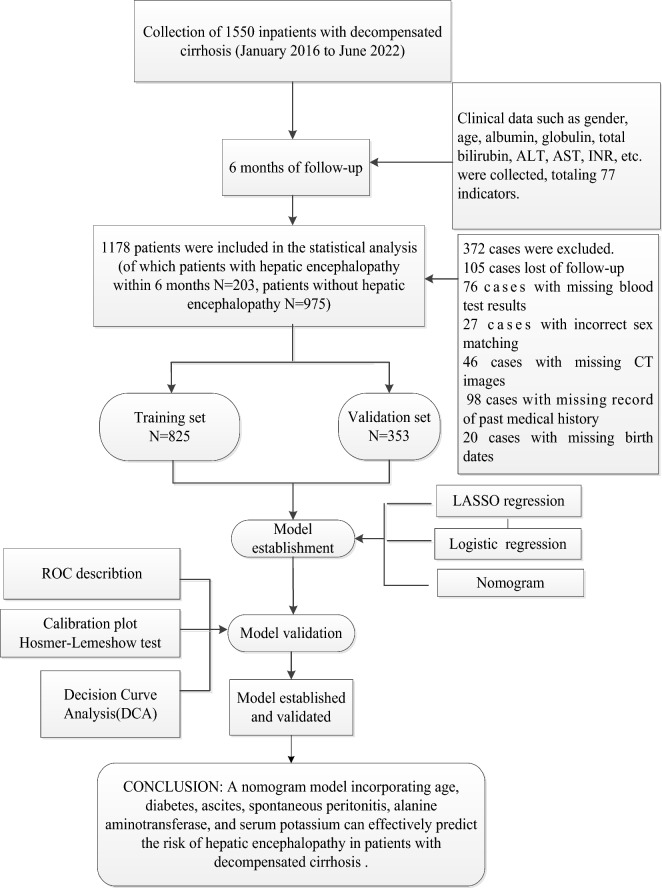
This study is a retrospective cohort study. We identified 77 relevant indicators of decompensated cirrhosis. We collected data from a total of 1550 inpatients with decompensated cirrhosis who attended the First Affiliated Hospital of Guangxi University of Chinese Medicine from January 2016 to June 2022, and these patients were followed up and screened for six months. Of these 1550 patients, 372 were not included in the final analysis due to missing follow-up, missing or incorrect data, and/or missing indicators, the total missing rate was 24%. The final sample size evaluated for the modeling analysis was 1178. All patients received routine treatment, including treatment of the cause (antiviral, alcohol cessation, etc.); use of lactulose, probiotics, etc. to keep the patient's bowel movements soft with a frequency of 1–2 times per day during the follow-up period; and treatment of their complications, respectively. Among these 1178 patients, 203 patients developed hepatic encephalopathy within six months of follow-up and 975 patients did not develop hepatic encephalopathy. A flow diagram of the study design is shown in Fig. 1.
Figure 1.
Flow diagram of the study.
Ethics statement
This study was reviewed and approved by The Ethics Committee of the First Affiliated Hospital of Guangxi University of Chinese Medicine (approval No.: 2022-080-02). All methods of this study were performed in accordance with the relevant guidelines and regulations. All subject researchers have pledged to follow the principle of confidentiality. All data and information collected were used for this study only. Informed consents were obtained from all subjects.
Inclusion criteria
age ≥ 18 years.
Meet the diagnostic criteria of decompensated cirrhosis1.
No current overt hepatic encephalopathy.
No previous history of hepatic encephalopathy.
Exclusion criteria
Missing more than 10% of data.
Imaging or pathological biopsy findings of liver cancer.
Combined severe systemic diseases or drug addicts who have difficulty quitting.
Pregnant or lactating women.
Combined pulmonary or other organ infections or gastrointestinal bleeding at admission.
Combined sepsis, shock from various causes, etc. at admission.
Combined acquired immunodeficiency virus (HIV) infection, syphilis spirochete infection.
Altered consciousness caused by concomitant psychiatric disease, metabolic encephalopathy, toxic encephalopathy and craniosynostosis.
History of laparotomy within 4 weeks prior to admission.
Record of alcohol consumption throughout the follow-up period.
Statistical analysis
Data grouping
Random numbers were generated using R software, and patients with decompensated cirrhosis included in the study were randomly divided into a training set(70%) and a validation set(30%). The training set was set to construct a risk prediction model, and the validation set was set to verify the accuracy of the prediction model.
Statistical descriptions
Analysis of variance was performed for the 77 selected relevant indicators in the training and validation sets. Between-group comparisons were made between patients’ data in the hepatic encephalopathy and non-hepatic encephalopathy groups. Among these indicators, continuous variables such as white blood cell count (WBC), were compared between groups using one-sample independent t tests or Wilcoxon rank sum tests, expressed as Mean ± SD or Median (P25, P75). Categorical variables, such as smoking history, hypertension, diabetes mellitus, presence of gastrointestinal bleeding, presence of ascites, presence of cardiovascular disease, were compared between groups by Pearson's chi-square test or Fisher test for rates and expressed as frequencies (percentages). Statistically significant difference was set at P < 0.05.
Handling of missing values
Multiple interpolation of missing data was performed using SPSS software. Most of the traditional methods for handling missing values use median or mean for interpolation. Multiple interpolation deals with missing values by using other variables given in the dataset, fitting the missing values by iteration and pre-defined matrix construction models, and then using the fitted predicted values to multiply fill the missing values of this variable. This method gives a higher accuracy of the missing value alternatives.
Model establishment and demonstration
(1) Determination of independent risk factors Least absolute shrinkage and selection operator (LASSO) regression was performed using the "glmnet" package of R software. LASSO regression is a linear regression that avoids overfitting by imposing a penalty on the magnitude of the model coefficients. Some of the variables derived from the LASSO regression mightnot be significantly correlated with the results in the multi-factor logistic regression analysis.
(2) Establishment of the model After screening the predictor variables by LASSO regression, the variables with P < 0.1 were used as predictors, and the risk prediction model was constructed by binary logistic regression using the glm function in R software.
(3) Presentation of the model In order to visualize the weights of each predictor and to make the established risk prediction model more convenient and concise for clinical application, the "rms" package of R software was used to build a nomographic plot based on the results of the multi-factor logistic model by using the lrm function and nomogram function.
Evaluation and validation of the model
The risk prediction model for patients with decompensated cirrhosis getting hepatic encephalopathy built based on the training set was evaluated in terms of its discriminative efficacy, consistency test and clinical benefit. The model was validated in the validation set.
(1) Evaluation of the model ROC (receiver operator charteristics), area under curve (AUC), concordance index (C-index), sensitivity, specificity, positive predictive value (PPR), and risk prediction model were used to evaluate the model. The area under the ROC curve reflects the discriminative power of the model. The risk prediction model is considered to be having good discriminatory ability if the area under the ROC curve was greater than 0.7. On the contrary, when the area under the ROC curve was close or even equals to 0.5, the risk prediction model would be considered to be having low diagnostic value.
(2) Validation of the model In this study, we used the bootstrap resampling method, the Hosmer–Lemeshow test, the ROC curve, the area under the ROC curve, and the calibration curve to measure and validate the model. The clinical benefit of the model was evaluated using decision curve analysis.
This study used Excel software for data entry and SPSS 26.0 and R 3.6.3 software for statistical analysis of the data. All P values were two-sided tests, P < 0.05 indicates that the differences are statistically significant unless otherwise stated.
Results
Sample size estimation and general information of the included patients
We calculated the minimum sample size required for modeling in this study as 323 cases based on R2 and 344 cases based on C-index. According to the sample size estimation, we included 1178 patients with decompensated cirrhosis were finally included in the analysis, including 203 patients who developed hepatic encephalopathy within six months and 975 patients who did not. 849(72.1%) of the 1178 patients in the sample were male, 329(27.9%) were female. 585 patients(49.7%) were < 55 years of age, 593 patients(50.3%) were over 55 years of age. 195 patients(16.6%) had ascites; 410 patients(34.8%) had infection; 193 patients(16.4%) had diabetes mellitus; as shown on Table 1.
Table 1.
General information of the included patients.
| Variables (categorical variables) | N (%)/Mean ± SD/Median (P25, P75) | Variables (continuous variables) | N (%)/Mean ± SD/Median (P25, P75) |
|---|---|---|---|
| Occurrence of hepatic encephalopathy | HGB | 126 [101;145] | |
| No | 975 (82.8%) | MCV | 92.1 [85.3;97.0] |
| Yes | 203 (17.2%) | MCH | 30.7 [27.8;32.4] |
| Sex | PLT | 126 [72.0;186] | |
| Male | 849 (72.1%) | MPV | 9.90 [9.10;10.6] |
| Female | 329 (27.9%) | PCT | 0.12 [0.07;0.18] |
| Age (years) | K | 3.80 [3.54;4.10] | |
| < 55 years old | 585 (49.7%) | Na | 140 [138;142] |
| ≥ 55 years old | 593 (50.3%) | Cl | 104 [102;106] |
| History of diabetes | P | 1.01 [0.89;1.13] | |
| No | 985 (83.6%) | Mg | 0.83 [0.76;0.90] |
| Yes | 193 (16.4%) | Ca | 2.18 [2.06;2.29] |
| History of smoking | TG | 0.94 [0.70;1.36] | |
| No | 939 (79.7%) | CHOL | 4.09 [3.30;4.98] |
| Yes | 239 (20.3%) | HDL | 1.17 [0.92;1.43] |
| History of alcohol consumption | APOA1 | 1.24 [0.98;1.49] | |
| No | 847 (71.9%) | APOB | 0.78 [0.58;0.97] |
| Yes | 331 (28.1%) | VLDL | 0.43 [0.32;0.62] |
| History of cerebrovascular disease | LDL | 2.33 [1.72;3.09] | |
| No | 1142 (96.9%) | LDH | 188 [158;235] |
| Yes | 36 (3.06%) | CK | 107 [70.0;165] |
| History of cardiovascular disease | AHBDH | 150 [125;188] | |
| No | 1115 (94.7%) | IgA | 2.67 [1.91;3.36] |
| Yes | 63 (5.35%) | IgG | 15.6 [12.8;19.5] |
| Hypertension | IgM | 1.22 [0.81;1.78] | |
| No | 980 (83.2%) | PT | 14.4 [13.2;16.6] |
| Yes | 198 (16.8%) | PTA | 83.0 [64.0;99.0] |
| Ascites | INR | 1.12 [1.01;1.35] | |
| No | 983 (83.4%) | APTT | 41.0 [37.4;45.7] |
| Yes | 195 (16.6%) | TT | 17.5 [16.6;18.7] |
| Gastrointestinal bleeding | FIB | 2.55 [1.96;3.14] | |
| No | 1154 (98.0%) | D-Dimer | 0.55 [0.25;2.14] |
| Yes | 24 (2.04%) | TP | 66.7 [61.5;71.8] |
| Spontaneous bacterial peritonitis | ALB | 37.9 [31.8;43.0] | |
| No | 1092 (92.7%) | GLO | 28.2 [24.6;32.9] |
| Yes | 86 (7.30%) | AVSG | 1.38 [1.02;1.69] |
| infection | PA | 156 [79.0;223] | |
| No | 768 (65.2%) | CHE | 5768 [3425;8271] |
| Yes | 410 (34.8%) | TBIL | 20.3 [13.5;35.1] |
| Biliary tract disorders | DBIL | 6.60 [4.10;14.5] | |
| No | 964 (81.8%) | IBIL | 13.4 [9.10;20.3] |
| Yes | 214 (18.2%) | TBA | 18.4 [5.70;55.6] |
| Hepatorenal syndrome (HRS) | AKP | 83.0 [65.0;117] | |
| No | 1166 (99.0%) | GGT | 37.0 [23.0;75.8] |
| Yes | 12 (1.02%) | ALT | 29.0 [19.0;44.0] |
| Hepatitis B | AST | 35.0 [27.0;52.8] | |
| No | 193 (16.4%) | ADA | 19.0 [14.0;27.2] |
| Yes | 985 (83.6%) | AFU | 28.0 [23.0;36.0] |
| Hepatitis C | BUN | 4.61 [3.70;5.78] | |
| No | 1127 (95.7%) | CREA | 72.0 [61.0;84.0] |
| Yes | 51 (4.33%) | UA | 309 [245;380] |
| Variables (continuous variables) | CysC | 0.88 [0.72;1.12] | |
| WBC (× 109/L) | 5.10 [3.80;6.50] | CO2 | 24.2 [22.3;26.1] |
| NEUT | 2.81 [2.10;3.80] | SOD | 151 [126;174] |
| NEUP | 58.5 (11.9) | CRP | 10.0 [4.50;22.5] |
| LYT | 0.14 [0.08;0.25] | Ammo | 40.1 [23.4;57.0] |
| LYP | 2.90 [1.60;4.90] | GLU | 5.03 [4.50;5.97] |
| RBC(× 109/L) | 4.26 [3.45;4.88] |
N (%): number of cases (percentage); Mean ± SD: mean ± standard deviation; Median: median; P25: 25th percentile; P75: 75th percentile.
Comparsion of the training and validation sets
1178 patients were randomly divided into a training set and a validation set in the ratio of 70% and 30%, where the training set n = 826 and the validation set n = 352 (Table 2). The training set included 128 patients who heve hepatic encephalopathy, accounting for 15.5% of the group; the validation set included 75 patients who have hepatic encephalopathy, accounting for 21.3% of the group. Statistical analysis was performed on the general data, and the results shown that except for the neutrophil ratio (NEUT), absolute neutrophil value (NEUP), immunoglobulin M (IgM), and a-L-amyloidase (AFU), the differences between the two groups were not statistically significant (P > 0.05) for all variables. This indicates that the indicators in the training and validation sets are evenly distributed, which can effectively avoid the conclusion bias.
Table 2.
Baseline table for training set-validation set grouping.
| Variables | Total N = 1178 |
Validation set N = 352 |
Training set N = 826 |
P value |
|---|---|---|---|---|
| History of diabetes | 0.406 | |||
| No | 985 (83.6%) | 289 (82.1%) | 696 (84.3%) | |
| Yes | 193 (16.4%) | 63 (17.9%) | 130 (15.7%) | |
| Smoking history | 0.762 | |||
| No | 939 (79.7%) | 283 (80.4%) | 656 (79.4%) | |
| Yes | 239 (20.3%) | 69 (19.6%) | 170 (20.6%) | |
| History of alcohol consumption | 0.364 | |||
| No | 847 (71.9%) | 260 (73.9%) | 587 (71.1%) | |
| Yes | 331 (28.1%) | 92 (26.1%) | 239 (28.9%) | |
| Cardiovascular disease | 0.927 | |||
| No | 1115 (94.7%) | 334 (94.9%) | 781 (94.6%) | |
| Yes | 63 (5.35%) | 18 (5.11%) | 45 (5.45%) | |
| Hypertension | 0.777 | |||
| No | 980 (83.2%) | 295 (83.8%) | 685 (82.9%) | |
| Yes | 198 (16.8%) | 57 (16.2%) | 141 (17.1%) | |
| Cerebrovascular disorders | 0.519 | |||
| No | 1142 (96.9%) | 339 (96.3%) | 803 (97.2%) | |
| Yes | 36 (3.06%) | 13 (3.69%) | 23 (2.78%) | |
| Gastrointestinal bleeding | 1.000 | |||
| No | 1154 (98.0%) | 345 (98.0%) | 809 (97.9%) | |
| Yes | 24 (2.04%) | 7 (1.99%) | 17 (2.06%) | |
| Spontaneous bacterial peritonitis | 0.769 | |||
| No | 1092 (92.7%) | 328 (93.2%) | 764 (92.5%) | |
| Yes | 86 (7.30%) | 24 (6.82%) | 62 (7.51%) | |
| Biliary tract disorders | 0.811 | |||
| No | 964 (81.8%) | 290 (82.4%) | 674 (81.6%) | |
| Yes | 214 (18.2%) | 62 (17.6%) | 152 (18.4%) | |
| Co-infection | 0.230 | |||
| No | 768 (65.2%) | 220 (62.5%) | 548 (66.3%) | |
| Yes | 410 (34.8%) | 132 (37.5%) | 278 (33.7%) | |
| Hepatorenal syndrome | 0.527 | |||
| No | 1166 (99.0%) | 350 (99.4%) | 816 (98.8%) | |
| Yes | 12 (1.02%) | 2 (0.57%) | 10 (1.21%) | |
| Ascites | 0.094 | |||
| No | 983 (83.4%) | 304 (86.4%) | 679 (82.2%) | |
| Yes | 195 (16.6%) | 48 (13.6%) | 147 (17.8%) | |
| Hepatitis B | 0.797 | |||
| No | 193 (16.4%) | 56 (15.8%) | 137 (16.6%) | |
| Yes | 985 (83.6%) | 298 (84.2%) | 687 (83.4%) | |
| Hepatitis C | 0.494 | |||
| No | 1127 (95.7%) | 336 (94.9%) | 791 (96.0%) | |
| Yes | 51 (4.33%) | 18 (5.08%) | 33 (4.00%) | |
| Age (years) | 0.499 | |||
| < 55 years old | 585 (49.7%) | 169 (48.0%) | 416 (50.4%) | |
| ≥ 55 years old | 593 (50.3%) | 183 (52.0%) | 410 (49.6%) | |
| Sex | 0.308 | |||
| Male | 849 (72.1%) | 246 (69.9%) | 603 (73.0%) | |
| Female | 329 (27.9%) | 106 (30.1%) | 223 (27.0%) | |
| GLU | 5.03[4.50;5.97] | 5.00[4.53;5.77] | 5.04[4.49;6.03] | 0.550 |
| WBC | 5.10[3.80;6.50] | 4.90[3.70;6.12] | 5.20[3.80;6.60] | 0.136 |
| NEUT | 2.81[2.10;3.80] | 2.70[2.00;3.70] | 2.90[2.10;3.90] | 0.023 |
| LYT | 0.14[0.08;0.25] | 0.14[0.08;0.25] | 0.14[0.08;0.25] | 0.921 |
| NEUP | 58.5 (11.9) | 57.0 (12.1) | 59.1 (11.9) | 0.007 |
| LYP | 2.90[1.60;4.90] | 3.15[1.63;4.80] | 2.87[1.56;4.90] | 0.394 |
| HGB | 126 [101;145] | 126 [99.0;147] | 126 [102;144] | 0.765 |
| MCV | 92.1[85.3;97.0] | 92.0[85.5;96.0] | 92.2[85.3;97.3] | 0.609 |
| MCH | 30.7[27.8;32.4] | 30.6[27.9;32.0] | 30.7[27.7;32.6] | 0.368 |
| PLT | 126 [72.0;186] | 130 [76.0;185] | 125 [70.0;186] | 0.382 |
| MPV | 9.90[9.10;10.6] | 9.80[9.00;10.6] | 9.90[9.20;10.7] | 0.106 |
| RBC | 4.26[3.45;4.88] | 4.22[3.35;4.88] | 4.29 [3.46;4.89] | 0.542 |
| PCT | 0.12 [0.07;0.18] | 0.12 [0.08;0.18] | 0.12 [0.07;0.18] | 0.540 |
| K | 3.80 [3.54;4.10] | 3.80 [3.57;4.10] | 3.80 [3.53;4.10] | 0.968 |
| Na | 140 [138;142] | 140 [138;142] | 140 [137;142] | 0.286 |
| Cl | 104 [102;106] | 104 [102;107] | 104 [101;106] | 0.339 |
| P | 1.01 [0.89;1.13] | 1.01 [0.88;1.14] | 1.01 [0.89;1.13] | 0.647 |
| Mg | 0.83 [0.76;0.90] | 0.84 [0.76;0.90] | 0.83 [0.76;0.90] | 0.482 |
| Ca | 2.18 [2.06;2.29] | 2.19 [2.05;2.28] | 2.18 [2.06;2.29] | 0.920 |
| TG | 0.94 [0.70;1.36] | 0.94 [0.70;1.36] | 0.95 [0.70;1.36] | 0.873 |
| CHOL | 4.09 [3.30;4.98] | 4.09 [3.29;5.06] | 4.10 [3.30;4.97] | 0.930 |
| HDL | 1.17 [0.92;1.43] | 1.17 [0.90;1.43] | 1.17 [0.94;1.43] | 0.497 |
| APOA1 | 1.24 [0.98;1.49] | 1.23 [0.96;1.49] | 1.25 [0.99;1.50] | 0.615 |
| APOB | 0.78 [0.58;0.97] | 0.78 [0.58;0.97] | 0.77 [0.58;0.97] | 0.794 |
| VLDL | 0.43 [0.32;0.62] | 0.43 [0.32;0.61] | 0.43 [0.32;0.62] | 0.845 |
| LDL | 2.33 [1.72;3.09] | 2.36 [1.75;3.09] | 2.30 [1.69;3.08] | 0.506 |
| LDH | 188 [158;235] | 187 [157;228] | 189 [158;237] | 0.626 |
| AHBDH | 150 [125;188] | 145 [125;187] | 151 [126;188] | 0.506 |
| IgA | 2.67 [1.91;3.36] | 2.67 [1.88;3.32] | 2.66 [1.92;3.38] | 0.647 |
| IgG | 15.6 [12.8;19.5] | 15.6 [12.5;19.5] | 15.7 [12.9;19.5] | 0.464 |
| IgM | 1.22 [0.81;1.78] | 1.13 [0.76;1.68] | 1.26 [0.85;1.84] | 0.001 |
| PT | 14.4 [13.2;16.6] | 14.3 [13.1;16.7] | 14.4 [13.3;16.5] | 0.509 |
| PTA | 83.0 [64.0;99.0] | 83.5 [63.0;100] | 82.0 [64.0;98.0] | 0.508 |
| INR | 1.12 [1.01;1.35] | 1.12 [1.00;1.36] | 1.13 [1.01;1.34] | 0.472 |
| APTT | 41.0 [37.4;45.7] | 40.9 [37.6;45.5] | 41.0 [37.3;45.8] | 0.875 |
| TT | 17.5 [16.6;18.7] | 17.6 [16.6;18.8] | 17.5 [16.5;18.7] | 0.524 |
| D-Dimer | 0.55 [0.25;2.14] | 0.62 [0.24;2.38] | 0.48 [0.25;2.01] | 0.300 |
| TP | 66.7 [61.5;71.8] | 66.6 [61.5;71.2] | 66.8 [61.5;71.9] | 0.571 |
| ALB | 37.9 [31.8;43.0] | 38.0 [31.7;42.9] | 37.9 [31.8;43.0] | 0.949 |
| GLO | 28.2 [24.6;32.9] | 28.4 [24.6;32.2] | 28.2 [24.5;33.2] | 0.674 |
| AVSG | 1.38 [1.02;1.69] | 1.40 [1.06;1.70] | 1.37 [1.02;1.68] | 0.579 |
| PA | 156 [79.0;223] | 156 [81.8;225] | 155 [79.0;222] | 0.746 |
| CHE | 5768 [3425;8271] | 5646 [3485;8083] | 5865 [3399;8273] | 0.881 |
| TBIL | 20.3 [13.5;35.1] | 20.0 [13.4;35.2] | 20.4 [13.6;35.1] | 0.521 |
| DBIL | 6.60 [4.10;14.5] | 6.50 [4.07;14.1] | 6.70 [4.10;14.6] | 0.588 |
| IBIL | 13.4 [9.10;20.3] | 13.3 [8.88;19.8] | 13.5 [9.20;20.4] | 0.444 |
| TBA | 18.4 [5.70;55.6] | 18.3 [6.20;56.8] | 18.4 [5.40;54.7] | 0.768 |
| AKP | 83.0 [65.0;117] | 82.0 [63.0;111] | 83.0 [65.2;118] | 0.266 |
| GGT | 37.0 [23.0;75.8] | 37.0 [22.0;75.2] | 38.0 [23.0;76.5] | 0.419 |
| ALT | 29.0 [19.0;44.0] | 29.0 [19.0;43.0] | 29.0 [20.0;44.0] | 0.472 |
| AST | 35.0 [27.0;52.8] | 35.0 [26.0;50.0] | 35.0 [27.0;54.0] | 0.428 |
| ADA | 19.0 [14.0;27.2] | 19.0 [13.0;27.0] | 19.0 [14.0;27.8] | 0.300 |
| AFU | 28.0 [23.0;36.0] | 28.0 [21.1;35.0] | 29.0 [23.0;36.0] | 0.037 |
| FIB | 2.55 [1.96;3.14] | 2.55 [1.95;3.16] | 2.55 [1.96;3.13] | 0.938 |
| BUN | 4.61 [3.70;5.78] | 4.65 [3.80;5.97] | 4.60 [3.69;5.70] | 0.296 |
| CREA | 72.0 [61.0;84.0] | 72.0 [59.0;87.2] | 72.0 [62.0;83.0] | 0.593 |
| UA | 309 [245;380] | 306 [240;378] | 311 [247;381] | 0.432 |
| CysC | 0.88 [0.72;1.12] | 0.88 [0.71;1.11] | 0.89 [0.72;1.13] | 0.549 |
| CO2 | 24.2 [22.3;26.1] | 24.4 [22.4;26.0] | 24.2 [22.1;26.1] | 0.658 |
| SOD | 151 [126;174] | 150 [123;175] | 151 [127;174] | 0.453 |
| CK | 107 [70.0;165] | 106 [69.0;153] | 108 [70.2;172] | 0.353 |
| CRP | 10.0 [4.50;22.5] | 10.0 [4.48;23.6] | 10.1 [4.50;22.1] | 0.532 |
| Ammo | 40.1 [23.4;57.0] | 40.0 [23.7;57.8] | 40.1 [23.4;56.4] | 0.820 |
Comparison of hepatic encephalopathy and non-hepatic encephalopathy grouping
As shown in Table 3, among the 1178 included patients, 203 had hepatic encephalopathy and 975 did not have hepatic encephalopathy within six months of following up. The morbidity rate was 17.23%. The differences of the following variables between the hepatic encephalopathy and non-hepatic encephalopathy groups were statistically significant (P < 0.05) : age, diabetes mellitus (DM), ascites, gastrointestinal bleeding, spontaneous bacterial peritonitis, blood potassium (K), alkaline transaminase (AKP), glutamic aminotransferase (AST), and glutamic alanine transaminase (ALT) .
Table 3.
Analysis of differences between the hepatic encephalopathy and non-hepatic encephalopathy groups.
| Variables | Total (N = 1178) | Non-HE (N = 975) | HE (N = 203) | OR | P value |
|---|---|---|---|---|---|
| Gender | 0.27 (0.44) | 0.28 (0.45) | 0.23 (0.43) | 0.80 [0.52;1.25] | 0.324 |
| Age (years) | 0.50 (0.50) | 0.46 (0.50) | 0.69 (0.47) | 2.57 [1.72;3.84] | < 0.001 |
| Smoking history | 0.21 (0.40) | 0.20 (0.40) | 0.23 (0.43) | 1.22 [0.78;1.91] | 0.385 |
| History of alcohol consumption | 0.29 (0.45) | 0.28 (0.45) | 0.34 (0.47) | 1.30 [0.87;1.94] | 0.207 |
| Cerebrovascular disease | 0.03 (0.16) | 0.03 (0.17) | 0.02 (0.15) | 0.81 [0.24;2.78] | 0.742 |
| Cardiovascular disease | 0.05 (0.23) | 0.05 (0.22) | 0.06 (0.24) | 1.19 [0.54;2.62] | 0.664 |
| Diabetes mellitus | 0.16 (0.36) | 0.14 (0.34) | 0.27 (0.45) | 2.39 [1.53;3.73] | < 0.001 |
| Hypertension | 0.17 (0.38) | 0.16 (0.37) | 0.20 (0.40) | 1.29 [0.80;2.08] | 0.29 |
| Ascites | 0.18 (0.38) | 0.15 (0.36) | 0.34 (0.47) | 2.89 [1.89;4.41] | < 0.001 |
| Gastrointestinal bleeding | 0.02 (0.14) | 0.02 (0.12) | 0.05 (0.21) | 3.07 [1.12;8.46] | 0.03 |
| Co-infection | 0.34 (0.47) | 0.33 (0.47) | 0.38 (0.49) | 1.22 [0.83;1.81] | 0.317 |
| Biliary tract disorders | 0.18 (0.39) | 0.18 (0.38) | 0.23 (0.42) | 1.37 [0.87;2.16] | 0.178 |
| Spontaneous bacterial peritonitis | 0.08 (0.26) | 0.05 (0.21) | 0.23 (0.42) | 5.90 [3.43;10.1] | < 0.001 |
| Hepatorenal syndrome | 0.01 (0.11) | 0.01 (0.10) | 0.02 (0.15) | 2.37 [0.60;9.28] | 0.216 |
| Hepatitis B | 0.83 (0.37) | 1.00 (0.00) | 0.04 (0.20) | 1.5 [0.78;3.53] | 0.987 |
| Hepatitis C | 0.04 (0.20) | 0.04 (0.18) | 0.06 (0.24) | 1.84 [0.84;4.04] | 0.130 |
| WBC | 5.47 (2.55) | 5.51 (2.64) | 5.27 (2.00) | 0.96 [0.89;1.04] | 0.341 |
| NEUT | 3.33 (2.15) | 3.36 (2.24) | 3.16 (1.57) | 0.95 [0.86;1.05] | 0.341 |
| NEUP | 59.1 (11.9) | 59.1 (11.9) | 59.0 (11.5) | 1.00 [0.98;1.01] | 0.895 |
| LYT | 0.19 (0.20) | 0.19 (0.20) | 0.19 (0.16) | 0.90 [0.34;2.41] | 0.836 |
| LYP | 3.77 (3.45) | 3.76 (3.51) | 3.83 (3.11) | 1.01 [0.95;1.06] | 0.817 |
| RBC | 4.16 (1.02) | 4.18 (1.02) | 4.06 (1.03) | 0.89 [0.74;1.07] | 0.225 |
| HGB | 122 (29.0) | 122 (29.1) | 121 (28.3) | 1.00 [0.99;1.01] | 0.725 |
| MCV | 90.1 (11.9) | 89.8 (11.9) | 91.7 (11.5) | 1.01 [1.00;1.03] | 0.092 |
| MCH | 29.6 (4.60) | 29.5 (4.63) | 30.2 (4.41) | 1.04 [0.99;1.08] | 0.106 |
| PLT (× 109/L) | 135 (82.3) | 135 (82.4) | 131 (81.4) | 1.00 [1.00;1.00] | 0.604 |
| MPV | 9.97 (1.32) | 9.96 (1.28) | 10.0 (1.52) | 1.05 [0.91;1.21] | 0.539 |
| PCT | 0.13 (0.08) | 0.13 (0.08) | 0.13 (0.08) | 1.04 [0.09;11.5] | 0.975 |
| K | 3.83 (0.50) | 3.84 (0.50) | 3.74 (0.49) | 0.65 [0.45;0.96] | 0.028 |
| Na | 139 (4.04) | 139 (4.10) | 139 (3.67) | 1.02 [0.97;1.07] | 0.544 |
| Cl | 104 (4.33) | 104 (4.32) | 104 (4.41) | 1.01 [0.97;1.06] | 0.672 |
| P | 1.01 (0.21) | 1.02 (0.21) | 1.00 (0.20) | 0.70 [0.28;1.74] | 0.439 |
| Mg | 0.82 (0.11) | 0.82 (0.11) | 0.82 (0.12) | 1.24 [0.23;6.70] | 0.806 |
| Ca | 2.17 (0.17) | 2.17 (0.17) | 2.15 (0.18) | 0.52 [0.18;1.53] | 0.233 |
| TG (mmol/L) | 1.17 (0.95) | 1.16 (0.95) | 1.19 (0.95) | 1.03 [0.86;1.24] | 0.714 |
| HDL | 1.18 (0.41) | 1.19 (0.41) | 1.14 (0.42) | 0.77 [0.49;1.22] | 0.264 |
| APOA1 | 1.22 (0.40) | 1.23 (0.40) | 1.17 (0.42) | 0.67 [0.42;1.06] | 0.089 |
| APOB | 0.79 (0.30) | 0.79 (0.31) | 0.80 (0.29) | 1.06 [0.57;1.95] | 0.864 |
| LDL | 2.44 (1.06) | 2.45 (1.07) | 2.40 (0.98) | 0.95 [0.80;1.14] | 0.612 |
| VLDL | 0.53 (0.43) | 0.53 (0.43) | 0.55 (0.44) | 1.12 [0.76;1.64] | 0.563 |
| LDH | 205 (78.3) | 203 (68.7) | 217 (117) | 1.00 [1.00;1.00] | 0.072 |
| CK | 139 (120) | 142 (125) | 127 (90.1) | 1.00 [1.00;1.00] | 0.189 |
| IgA | 2.79 (1.29) | 2.78 (1.25) | 2.84 (1.50) | 1.04 [0.90;1.19] | 0.639 |
| IgG | 16.6 (5.50) | 16.6 (5.46) | 16.6 (5.72) | 1.00 [0.97;1.03] | 0.975 |
| IgM | 1.47 (0.96) | 1.45 (0.90) | 1.56 (1.20) | 1.11 [0.93;1.33] | 0.262 |
| PT | 15.3 (2.97) | 15.3 (2.97) | 15.3 (3.03) | 1.01 [0.95;1.07] | 0.793 |
| PTA | 81.2 (22.4) | 81.2 (22.2) | 81.0 (23.4) | 1.00 [0.99;1.01] | 0.928 |
| AHBDH | 164 (63.1) | 162 (55.9) | 172 (92.7) | 1.00 [1.00;1.00] | 0.125 |
| INR | 1.22 (0.32) | 1.22 (0.31) | 1.23 (0.32) | 1.12 [0.63;2.00] | 0.708 |
| APTT | 42.3 (7.65) | 42.2 (7.58) | 43.2 (8.00) | 1.02 [0.99;1.04] | 0.156 |
| TT | 17.8 (2.11) | 17.9 (2.10) | 17.7 (2.15) | 0.96 [0.87;1.06] | 0.388 |
| FIB | 2.63 (0.93) | 2.62 (0.92) | 2.71 (0.99) | 1.10 [0.91;1.34] | 0.317 |
| D-Dimer | 1.77 (3.02) | 1.81 (3.13) | 1.57 (2.34) | 0.97 [0.90;1.04] | 0.411 |
| ALB | 37.1 (7.39) | 37.1 (7.35) | 36.6 (7.63) | 0.99 [0.96;1.01] | 0.403 |
| TP | 66.4 (8.07) | 66.5 (8.13) | 66.0 (7.76) | 0.99 [0.97;1.02] | 0.506 |
| GLO | 29.4 (7.19) | 29.4 (7.22) | 29.4 (7.06) | 1.00 [0.98;1.03] | 0.908 |
| AVSG | 1.35 (0.45) | 1.35 (0.44) | 1.33 (0.46) | 0.89 [0.58;1.35] | 0.578 |
| CHE | 5976 (2934) | 6006 (2940) | 5813(2907) | 1.00 [1.00;1.00] | 0.493 |
| TBIL | 34.5 (50.5) | 33.4 (50.0) | 40.1 (52.8) | 1.00 [1.00;1.01] | 0.18 |
| DBIL | 16.5 (35.3) | 15.7 (34.7) | 20.9 (38.2) | 1.00 [1.00;1.01] | 0.138 |
| TBA | 44.2 (63.9) | 43.1 (63.9) | 50.3 (63.9) | 1.00 [1.00;1.00] | 0.243 |
| IBIL | 18.2 (17.6) | 17.8 (17.6) | 19.9 (17.6) | 1.01 [1.00;1.02] | 0.219 |
| AKP(μ/L) | 102 (68.4) | 99.2 (63.2) | 116 (90.9) | 1.00 [1.00;1.01] | 0.018 |
| GGT | 79.6 (173) | 78.0 (176) | 88.3 (152) | 1.00 [1.00;1.00] | 0.544 |
| AST(μ/L) | 52.8 (78.4) | 49.6 (62.1) | 70.2 (136) | 1.00 [1.00;1.00] | 0.025 |
| ADA | 21.6 (10.5) | 21.3 (10.3) | 23.1 (11.7) | 1.02 [1.00;1.03] | 0.075 |
| AFU | 30.2 (10.5) | 30.0 (10.5) | 31.1 (10.5) | 1.01 [0.99;1.03] | 0.27 |
| CREA(umol/L) | 75.4 (26.4) | 75.1 (24.8) | 77.1 (34.0) | 1.00 [1.00;1.01] | 0.449 |
| BUN | 5.20 (2.89) | 5.25 (3.00) | 4.94 (2.17) | 0.96 [0.89;1.03] | 0.273 |
| UA | 319 (107) | 320 (109) | 316 (97.6) | 1.00 [1.00;1.00] | 0.682 |
| CysC | 1.02 (0.78) | 1.02 (0.78) | 1.06 (0.80) | 1.06 [0.86;1.30] | 0.582 |
| CO2 | 24.1 (3.22) | 24.0 (3.19) | 24.1 (3.36) | 1.01 [0.95;1.07] | 0.771 |
| SOD | 148 (35.4) | 148 (35.1) | 147 (37.2) | 1.00 [0.99;1.00] | 0.559 |
| CRP | 16.2 (19.7) | 16.0 (18.5) | 17.5 (25.3) | 1.00 [0.99;1.01] | 0.442 |
| Ammo | 40.9 (22.4) | 40.5 (22.5) | 42.7 (22.2) | 1.00 [1.00;1.01] | 0.325 |
| PA | 155 (82.3) | 155 (81.3) | 150 (87.5) | 1.00 [1.00;1.00] | 0.491 |
| ALT(μ/L) | 46.6 (109) | 40.5 (42.5) | 79.7 (257) | 1.00 [1.00;1.01] | 0.004 |
| CHOL | 4.18 (1.34) | 4.20 (1.36) | 4.11 (1.22) | 0.95 [0.82;1.10] | 0.479 |
| GLU | 5.80 (2.70) | 5.84 (2.74) | 5.63 (2.46) | 0.97 [0.90;1.05] | 0.426 |
Assignment of variables in the model for risk prediction of hepatic encephalopathy
A total of 1 dependent variable (occurrence of hepatic encephalopathy) and 77 independent variables were included in this study. Table 4 assigned values to the variables individually and converted the corresponding variables to categorical variables, including the assignment of dichotomous variables and the handling of dummy variables of multicategorical variables. The occurrence of HE was set as the dependent variable Y. The independent continuous variables (X) such as white blood cell count, platelet count, hemoglobin, glutamate aminotransferase, blood creatinine, blood urea nitrogen, urea, glucose, total cholesterol, triglycerides, total bile acids, albumin, were still included in the model analysis as numerical variables.
Table 4.
Variable assignment.
| Variable name | Variable name | Variable value representation and meaning |
|---|---|---|
| Y | Whether hepatic encephalopathy occurs | No = 0, Yes = 1 |
| X1 | Sex | Male = 0, Female = 1 |
| X2 | Age (years) | < 55 years old = 0, ≥ 55 years old = 1 |
| X3 | Marital status | X3-1 = 0:unmarried, X3-2 = 1:married, X3-3 = 2:other |
| X4 | Smoking history | X4 = 1: Yes; X4 = 0: No |
| X5 | History of alcohol consumption | X5 = 1: Yes; X5 = 0: No |
| X6 | Cerebrovascular disease | X6 = 1: Yes; X6 = 0:No |
| X7 | Cardiovascular disease | X7 = 1: Yes; X7 = 0: No |
| X8 | Diabetes | X8 = 1: Yes; X8 = 0: No |
| X9 | History of hypertension | X9 = 1: Yes; X9 = 0: No |
| X10 | Ascites | X10 = 1: Yes; X10 = 0: No |
| X11 | Gastrointestinal bleeding | X11 = 1: Yes; X11 = 0: No |
| X12 | Infection | X12 = 1: Yes; X12 = 0: No |
| X13 | Biliary tract disorders | X13 = 1: Yes; X13 = 0: No |
| X14 | Spontaneous bacterial peritonitis | X14 = 1: Yes; X14 = 0: No |
| X15 | Hepatorenal syndrome | X15 = 1: Yes; X15 = 0: No |
| X16 | Hepatitis B | X16 = 1: Yes; X16 = 0: No |
| X17 | Hepatitis C | X17 = 1: Yes; X17 = 0: No |
| X18 | WBC | Numeric variables |
| X19 | NEUT | Numeric variables |
| X20 | LYT | Numeric variables |
| X21 | LYP | Numeric variables |
| X22 | RBC | Numeric variables |
| X23 | HGB | Numerical variables |
| X24 | MCV | Numerical variables |
| X25 | MCH | Numerical variables |
| X26 | PLT | Numerical variables |
| X27 | MPV | Numerical variables |
| X28 | PCT | Numerical variables |
| X29 | K | Numerical variables |
| X30 | Na | Numerical variables |
| X31 | Cl | Numerical variables |
| X32 | P | Numerical variables |
| X33 | Mg | Numerical variables |
| X34 | Ca | Numerical variables |
| X35 | TG | Numerical variables |
| X36 | HDL | Numerical variables |
| X37 | APOA1 | Numerical variables |
| X38 | APOB | Numerical variables |
| X39 | LDL | Numerical variables |
| X40 | VLDL | Numerical variables |
| X41 | LDH | Numerical variables |
| X42 | CK | Numerical variables |
| X43 | IgA | Numerical variables |
| X44 | IgG | Numerical variables |
| X45 | IgM | Numerical variables |
| X46 | PT | Numerical variables |
| X47 | PTA | Numerical variables |
| X48 | AHBDH | Numerical variables |
| X49 | INR | Numerical variables |
| X50 | APTT | Numerical variables |
| X51 | TT | Numerical variables |
| X52 | FIB | Numerical variables |
| X53 | D-Dimer | Numerical variables |
| X54 | ALB | Numerical variables |
| X55 | TP | Numerical variables |
| X56 | GLO | Numerical variables |
| X57 | AVSG | Numerical variables |
| X58 | CHE | Numerical variables |
| X59 | TBIL | Numerical variables |
| X60 | DBIL | Numerical variables |
| X61 | TBA | Numerical variables |
| X62 | IBIL | Numerical variables |
| X63 | AKP | Numerical variables |
| X64 | GGT | Numerical variables |
| X65 | AST | Numerical variables |
| X66 | ADA | Numerical variables |
| X67 | AFU | Numerical variables |
| X68 | CREA | Numerical variables |
| X69 | BUN | Numerical variables |
| X70 | UA | Numerical variables |
| X71 | CysC | Numerical variables |
| X72 | CO2 | Numerical variables |
| X73 | SOD | Numerical variables |
| X74 | CRP | Numerical variables |
| X75 | Ammo | Numerical variables |
| X76 | PA | Numerical variables |
| X77 | ALT | Numerical variables |
| X78 | CHOL | Numerical variables |
| X79 | GLU | Numerical variables |
Univariate logistic regression analysis
The occurrence of hepatic encephalopathy was used as the dependent variable Y, and all candidate predictors based on 826 patients in the training set were used as independent variables. Univariate logistic regression analysis was performed to screen the potential predictors. The results shown that the following variables were considered to be the potential predictors and were entered into the regression equation (Table 5): age, serum alkaline transaminase (AKP), glutamic aminotransferase (ALT), adenosine deaminase (ADA), glutamic aminotransferase (AST), diabetes mellitus, serum lactate dehydrogenase (LDH), apolipoprotein A, serum potassium, red blood cell volume (MCV), ascites, spontaneous bacterial peritonitis, and gastrointestinal bleeding.
Table 5.
Results of univariate logistic regression analysis.
| Variables | Β | SD | OR | 95% CI | Z | P |
|---|---|---|---|---|---|---|
| Age | 0.943 | 0.20524 | 2.569 | 1.718–3.841 | 4.597 | 0 |
| ADA | 0.015 | 0.00861 | 1.015 | 0.998–1.033 | 1.781 | 0.075 |
| AST | 0.002 | 0.00103 | 1.002 | 1–1.004 | 2.234 | 0.025 |
| ALT | 0.004 | 0.00141 | 1.004 | 1.001–1.007 | 2.865 | 0.004 |
| AKP | 0.003 | 0.00118 | 1.003 | 1–1.005 | 2.367 | 0.018 |
| History of diabetes mellitus | 0.871 | 0.22696 | 2.389 | 1.531–3.727 | 3.837 | 0 |
| Serum lactate dehydrogenase | 0.002 | 0.00108 | 1.002 | 1–1.004 | 1.801 | 0.072 |
| Apolipoprotein A | − 0.405 | 0.23813 | 0.667 | 0.418–1.064 | − 1.7 | 0.089 |
| Serum potassium | − 0.428 | 0.19476 | 0.652 | 0.445–0.955 | − 2.196 | 0.028 |
| Red blood cell volume | 0.014 | 0.00847 | 1.014 | 0.998–1.031 | 1.685 | 0.092 |
| Ascites | 1.061 | 0.21522 | 2.889 | 1.895–4.406 | 4.93 | 0 |
| Spontaneous bacterial peritonitis | 1.775 | 0.27639 | 5.903 | 3.434–10.147 | 6.424 | 0 |
| Gastrointestinal bleeding | 1.122 | 0.51694 | 3.072 | 1.115–8.46 | 2.171 | 0.03 |
Predictors screening
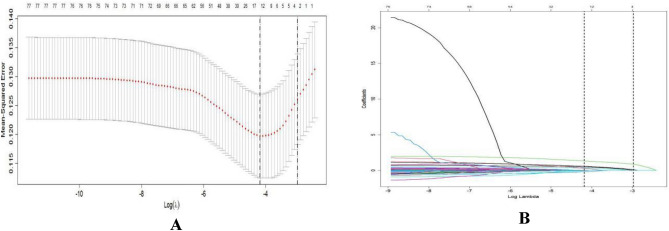
LASSO regression was performed using the "glmnet" package in R software. All independent variables were screened using LASSO regression, and the adjustment parameter λ was validated using the ten-fold crossover method. Conversely, if the regression coefficient is not zero, it indicates that the variable is strongly associated with the occurrence of HE in patients with cirrhosis. The two dashed lines indicate lambda.min, which represents the value of λ corresponding to the smallest error and which can correspond to the least number of predictor variables, and lambda.1se, which represents the value of λ for the most streamlined model within one standard error of lambda.min. All the independent variables in this study were screened by LASSO regression, and finally 15 variables with non-zero regression coefficients were output at lambda.min. These 15 variables were listed below: age, sex, diabetes mellitus (DM), spontaneous bacterial peritonitis, ascites, gastrointestinal bleeding, serum α-hydroxybutyrate dehydrogenase (α-HBDH), white blood cells ( WBC), red blood cell volume (MCV), serum potassium (K), prothrombin time (TT), serum alkaline phosphatase (AKP), alanine transaminase (ALT), adenosine deaminase (ADA), and plasma ammonia (Ammo). The above 15 predictor variables were included in a multifactorial logistic regression analysis, and six fo these 15 variables showed statistically significant differences (P < 0.05) (Fig. 2A, B, Table 6).
Figure 2.
LASSO regression diagram.
Table 6.
Multifactorial logistic regression analysis based on LASSO regression.
| Variables | β | SD | OR | 95% CI | Z | P |
|---|---|---|---|---|---|---|
| Age | 0.815 | 0.22239 | 2.26 | 1.461–3.494 | 3.666 | 0 |
| History of diabetes mellitus (DM) | 0.504 | 0.25423 | 1.656 | 1.006–2.725 | 1.983 | 0.047 |
| Ascites | 0.706 | 0.24217 | 2.025 | 1.26–3.255 | 2.914 | 0.004 |
| Spontaneous bacterial peritonitis (SBP) | 1.58 | 0.30707 | 4.856 | 2.66–8.865 | 5.146 | 0 |
| ALT, U/L | 0.005 | 0.00142 | 1.005 | 1.002–1.007 | 3.216 | 0.001 |
| K, mmol/L | − 0.414 | 0.2065 | 0.661 | 0.441–0.99 | − 2.007 | 0.045 |
Establishment and demonstration of the model
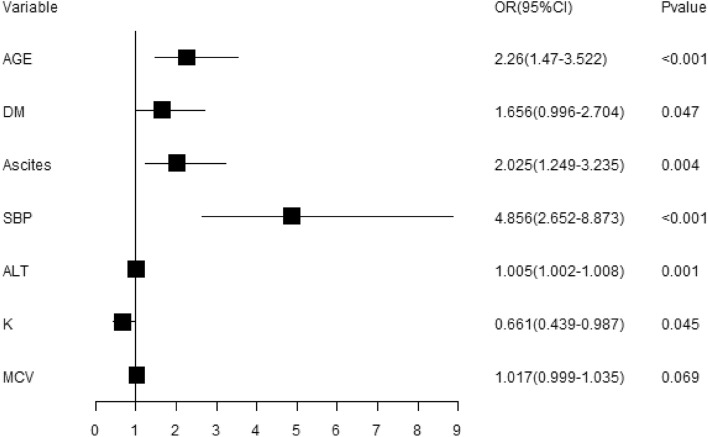
The risk prediction model for the occurrence of HE in patients with decompensated cirrhosis was established based on the above six predictors (risk factors), and the OR values obtained after incorporating the model are shown in Fig. 3, in which the combination of SBP or not had the greatest effect on the occurrence of hepatic encephalopathy, and the risk of getting hepatic encephalopathy in decompensated cirrhosis patients with SBP was 4.856 times higher than that in patients without SBP (2.66, 8.865); the risk of getting hepatic encephalopathy in decompensated cirrhosis patients older than 55 years of age was 2.26 times higher than in patients not older than 55 years (1.461, 3.494); the risk of getting hepatic encephalopathy in decompensated cirrhosis patients with a history of diabetes mellitus was 1.656 times higher than in patients without a history of diabetes mellitus (1.006, 2.725); the risk of getting hepatic encephalopathy in decompensated cirrhosis patients with ascites was 2.025 times higher than in patients without ascites (1.26, 3.255); the risk of getting hepatic encephalopathy increased incrementally with the increasing serum glutamate values in decompensated cirrhosis patients, with an OR value of 1.005 (1.002, 1.007).
Figure 3.
OR (95% CI) forest plot of predictors.
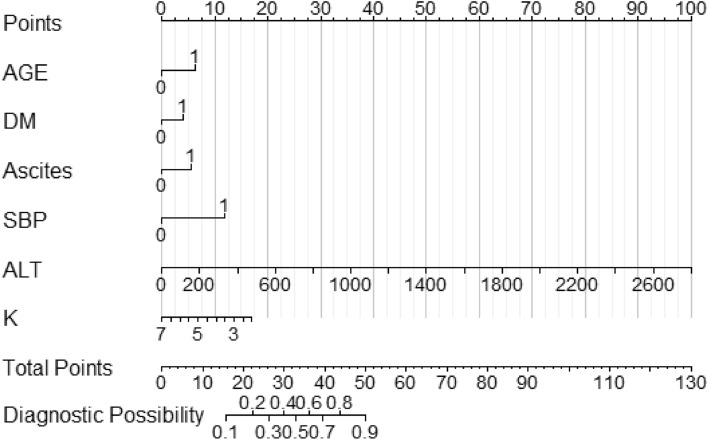
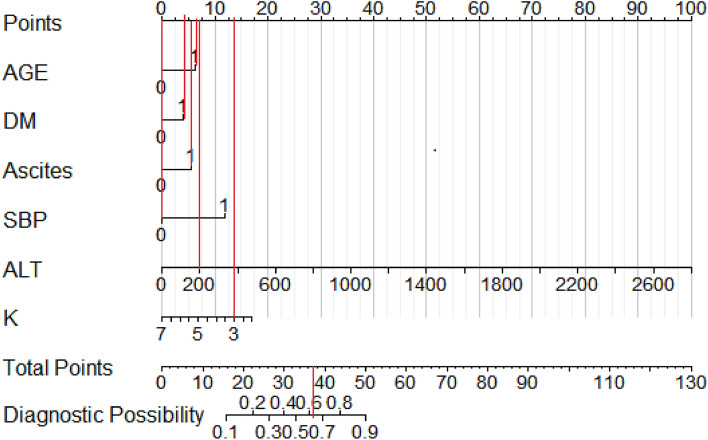
In order to visualize the weight of each predictor in the model and to visualize the model for clinical application, we used R software to construct a nomogram to demonstrate the model. The scores and risks of each predictor are shown in Fig. 4, Tables 7, and 8. The higher the total score, the higher the risk of developing hepatic encephalopathy.
Figure 4.
Nomogram for predicting the risk of getting hepatic encephalopathy in patients with decompensated cirrhosis. Note: AGE: age; DM: history of diabetes mellitus; SBP: spontaneous bacterial peritonitis; ALT: glutamic aminotransferase; K: serum potassium concentration; 1 = yes; 0 = no.
Table 7.
Total scores of the predictors and their corresponding probability of diagnosis of hepatic encephalopathy.
| Total scores | 16 | 22 | 26 | 30 | 33 | 36 | 39 | 44 | 50 |
|---|---|---|---|---|---|---|---|---|---|
| Probability of diagnosis | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 |
Table 8.
Scores for each predictor in the nomogram.
| Predictor | AGE | DM | Ascites | SBP | ALT (U/L) | K (mmol/L) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | S | A | S | A | S | A | S | A | S | A | S | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.0 | 17 | |
| 1 | 6 | 1 | 4 | 1 | 6 | 1 | 12 | 200 | 7 | 2.5 | 15 | |
| 400 | 14 | 3.0 | 14 | |||||||||
| 600 | 21 | 3.5 | 12 | |||||||||
| 800 | 29 | 4.0 | 10 | |||||||||
| 1000 | 36 | 4.5 | 9 | |||||||||
| 1200 | 43 | 5.0 | 7 | |||||||||
AGE: age; DM: history of diabetes mellitus; SBP: spontaneous bacterial peritonitis; ALT: glutamic aminotransferase; K: serum potassium concentration; A: assignment; S: score.
The following example (see Fig. 5) illustrates the clinical application of the nomogram model for predicting the risk of getting hepatic encephalopathy: for example, a decompensated cirrhosis patients is 60 years old (6 points), he/she has diabetes mellitus (4 points), he is found to have ascites (6 points), he does not have spontaneous bacterial peritonitis (0 points), and laboratory tests suggested the value of his glutamic aminotransferase is 200 U/L (7 points), the value of his serum potassium concentration is 3.0 mmol /L (14 points). Then, this patient had a final score of 37, and his probability of developing hepatic encephalopathy would be greater than 0.6. That is to say, this patient had a high probability (risk) of developing hepatic encephalopathy, suggesting the need for timely and early intervention by medical personnel to reduce his risk of developing hepatic encephalopathy.
Figure 5.
Example of the application of the established nomogram. Note: AGE: age; DM: history of diabetes mellitus; SBP: spontaneous bacterial peritonitis; ALT: glutamic aminotransferase; K: serum potassium concentration; 1 = yes; 0 = no.
Evaluation and validation of the model
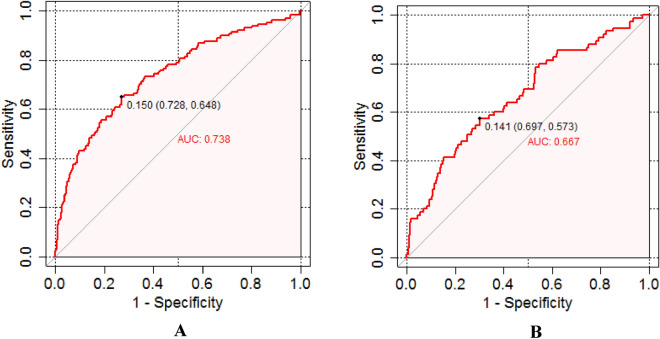
Distinguishability
The distinguishability of the model was evaluated using ROC (receiver operator charteristics) curves (as shown in Fig. 6A, B), and the results showed that the AUC of the model was 0.738 (95% CI 0.63–0.746) in the training set (as shown in Fig. 6A) and 0.667 (95% CI 0.541–0.706) (as shown in Fig. 6B), and the AUCs of two sets indicated that the nomogram model was greatly differentiated. According to the Cut-Off value determined by the Jorden index, when the Cut-Off value of the training set was taken as 0.150, the sensitivity of the model was 72.8%, the specificity was 64.8%, the PPV was 30.4%, and the NPV was 91.9%; when the Cut-Off value of the validation set was taken as 0.141, the sensitivity of the model was 69.7%, the specificity was 57.3%, the PPV was 34.5%, and the NPV was 84.7%.
Figure 6.
ROC curves of the training set (A) and validation set (B). Note: The area under the ROC curve for the model is 0.738 and 0.667 for the training and validation sets, respectively.
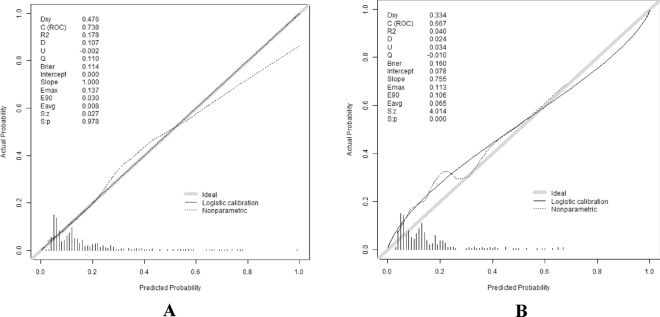
Calibration
Bootstrap sampling method was used to perform the calibration. Patients in the training and validation sets were repeatedly sampled 1000 times, respectively, and the calibration curves were plotted after validation. The horizontal coordinate indicates the likelihood of developing hepatic encephalopathy in patients with decompensated cirrhosis, and the vertical coordinate indicates the actual event occurrence. The further the calibration curve deviates from the diagonal, the greater the error (as shown in Fig. 7A, B). The Hosmer–Lemeshow test for goodness of fit was also applied, and the results showed that χ2 = 1.237587, P = 0.998 in the training set, χ2 = 31.90904, P = 0.0202 in the validation set, indicating that there was no significant difference between the predicted and actual observed values.
Figure 7.
Calibration curves of the training set (A) and validation set (B). Note: The X-axis is the predicted probability of developing hepatic encephalopathy in patients with decompensated cirrhosis, and the Y-axis is the actual probability of developing hepatic encephalopathy in patients with decompensated cirrhosis. The diagonal dashed line indicates a perfect prediction, while the solid line indicates the actual corrected prediction.
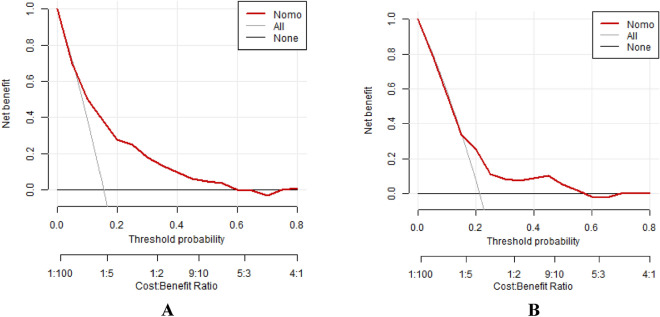
Clinical decision curve analysis
We used clinical decision curve analysis (DCA) to assess the net benefit of the model in clinical application. As shown in (Fig. 8A, B), the results of the DCA show that the model has good clinical benefit in both the training and validation sets when compared to two extreme clinical scenarios (all patients received treatment or none of them received).
Figure 8.
Calibration curves of the training set (A) and validation set (B). Note: The horizontal axis indicates that no patient received treatment after the application of the model, with a net benefit of 0. The diagonal line indicates that all patients received treatments.
Discussion
Hepatic encephalopathy is a complex disease with a wide range of etiologies and varying degrees of severity of morbidity. The use of appropriate measurement tools to assess the risk of getting hepatic encephalopathy can help to develop targeted interventions to reduce the occurrence of hepatic encephalopathy, which is important to improve patients’ quality of life and reduce the burden of medical care. Therefore, the development of high-quality risk prediction tools has become the focus of research on the prevention and treatment of hepatic encephalopathy. In recent years, scholars in various countries have constructed various risk prediction models for the development of hepatic encephalopathy based on the characteristics of the local population and epidemiological data. However, those risk prediction models constructed are diverse, the predictive indicators incorporated in each model are not consistent, the assessment contents and applicable population are not uniform, resulting in a certain gap between the prediction results and the real situation.
Risk prediction model studies aim to estimate the probability of an event occurring in an individual and can be divided into diagnostic models (presence or absence of a disease or symptom) and prognostic models (whether a specific outcome will occur in the future)2. The common metrics used to evaluate predictive models are the degree of discrimination and calibration, and if a good degree of discrimination is available, it indicates that the predictive model can accurately distinguish high-risk population with different risks. AUC value of 0.50 indicates that the model has predictive power but poor discrimination, 0.51–0.70 indicates that the model has low discrimination, 0.71–0.90 indicates good discrimination, and higher than 0.90 indicates high discrimination3. Sensitivity reflects the ability to correctly detect positive diagnosis patients, also known as the true positive rate, and specificity reflects the ability to correctly determine people who are actually disease-free as true negatives, known as the true negative rate4.
The risk prediction models can be divided into traditional statistical algorithm models and machine learning algorithm models according to the model building method. Traditional statistical algorithmic models are mathematical models based on statistical analysis of risk factors, i.e., the probability of disease occurrence is calculated by constructing mathematical models in which factors that can independently predict the occurrence of an event are selected as predictors. The most common models are logistic regression and Cox proportional risk regression models. Takikawa5 applied logistic regression analysis to construct a predictive model for the risk of developing hepatic encephalopathy, and the findings suggest that advanced age, prolonged prothrombin time, and high total serum bilirubin can be used as risk predictors for the development of hepatic encephalopathy. Although the specificity of this study was very high, its sensitivity was low, indicating that the inclusion of the above factors alone was not sufficient to predict the development of hepatic encephalopathy. In 2019, Labenz6 used history of minimal hepatic encephalopathy, history of hepatic encephalopathy, C-reactive protein, albumin, MELD score, serum interleukin 6 (IL-6) as predictors to establish a prediction model to validate the predictive value of IL-6 to identify the occurrence of hepatic encephalopathy, and the results showed that the predictive performance was substantially improved (AUC of 0.931).
In contrast to the logistic regression model, the Cox proportional risk regression model uses survival outcome and survival time as dependent variables, allowing simultaneous analysis of the effects of numerous factors on survival to study the incidence at different time points. Tapper7 used demographic, clinical, laboratory, and pharmacological data to construct a predictive model for the risk of developing hepatic encephalopathy based on the Cox proportional risk regression model, and The final prediction model consisted of four predictors: albumin, bilirubin, statin usage and non-selective β-blocker usage.The model was validated using bootstrapping and obtained an AUC of 0.73, indicated a hi gh degree of discrimination.
In this study, we chose to use logistic regression analysis to construct our analytical model, as opposed to opting for Cox's proportional risk model for several reasons: (1) the purpose of our study was to examine the impact of specific risk factors on a dichotomous outcome variable (whether or not a specific event occurs) rather than the impact on survival time. Therefore, we considered that logistic regression was more appropriate for our study, whereas the Cox proportional risk model was more appropriate for survival analysis. (2) Our dataset did not contain information on survival times, nor did we record the start and end times of observations for individuals, so it’s hard to perform analyses using the Cox proportional risk model. (3) Some of the independent variables in our dataset are categorical or ordinal, whereas the Cox's proportional risk model requires the independent variables to be continuous or dichotomous. If we convert these variables to dichotomous variables, we may lose some information and precision. Logistic regression, on the other hand, can handle multi-categorical or sequential variables and only requires dummy variable coding8,9. (4) There are some independent variables in our data set that may not meet the basic assumption of the Cox's proportional risk model, i.e., the assumption of equal proportional risk. This means that the impact of the independent variable on the outcome variable changes over time. If the equal proportional risk assumption does not hold, the results of the Cox's proportional risk model will lose their explanatory power. Logistic regression, on the other hand, does not require this assumption and is more flexible and robust.
In this study, we refered to the prevailing practice of the popular clinical prediction models10–29 and chose the logistic regression method to develop a nomogram model for predicting the risk of developing hepatic encephalopathy in patients with decompensated cirrhosis, and the model results were as described above. The results showed that the model performed well in terms of differentiation, calibration and clinical applicability and can be used in clinical practice.
The results obtained in this study suggested that age, diabetes mellitus (DM), ascites, spontaneous bacterial peritonitis (SBP), abnormal glutamate aminotransferase (ALT), and abnormal blood potassium (K) are risk factors (predictors) for the development of hepatic encephalopathy. These predictors were finally entered into the subsequent analysis and were used to build the nomogram model.
We included both potassium and sodium in the possible risk factors, and after statistical analysis, the difference in potassium was statistically significant (P < 0.05), while the difference in sodium was not statistically significant (P > 0.05), so we finally included potassium as an independent factor in the nomogram model for risk prediction of hepatic encephalopathy. It is worth noting that potassium is the major intracellular cation involved in maintaining electrolyte homeostasis and acid–base balance inside and outside the cell. Hypokalemia is defined as a serum potassium concentration less than 3.5mmol/L, which is commonly found in patients with liver cirrhosis, especially when combined with ascites, diuresis, vomiting, and diarrhea30. Hypokalemia may lead to metabolic alkalosis, which in turn promotes the occurrence of hepatic encephalopathy. Sodium, on the other hand, is the major extracellular cation involved in maintaining body fluid volume and osmolality. The impact of blood sodium abnormalities on hepatic encephalopathy is unclear. In light of this, we have also included a number of relevant studies in the Discussion section that support the rationale for choosing potassium rather than sodium as an independent factor in the development of hepatic encephalopathy in cirrhosis30–34.
In this study, the ROC curve was used to evaluate the predictive ability of the model, and the area under the ROC curve was calculated to evaluate the model performance. The accuracy of the model was evaluated by plotting the calibration curve. The clinical benefit of the model was evaluated using decision curve analysis (DCA). DCA is a method to evaluate prediction models by calculating the net clinical benefit. The results of the DCA showed that the risk prediction model established in this study had good clinical benefit in both the training set and the validation set when compared with two extreme clinical scenarios (i.e., all patients were treated or none of them were treated). This further validated the good performence and high value of this model in practical clinical work.
This study has some limitations. This is a single-center study, the sample size and the representativeness of the sample might be insufficient. Our study was conducted from 2016 to 2022, and we initially collected 1550 patients, and finally, after rigorous inclusion–exclusion screening, the final sample size was 1178, which was much larger than the minimum sample size requirement of constructing a risk prediction model (323 patients)35. Therefore, we considered that the results of our study can be applied well in clinical practices. Although this prediction model has been set up with a validation set for internal validation, an external validation with a larger sample size and multiple centers would be helpful to demonstrate the feasibility of this model in order to better generalize it. We intend to conduct more multicenter investigations to improve the sample's representativeness and applicability of the study results in future studies.
In conclusion, this study showed that age over 55 years, diabetes, ascites, spontaneous bacterial peritonitis, abnormal glutamate aminotransferase, and abnormal blood potassium concentration are independent risk factors(predictors) for the development of hepatic encephalopathy in patients with cirrhosis, and these six indicators are very meaningful for identifying the risk of developing hepatic encephalopathy in patients with decompensated cirrhosis. The risk prediction nomogram model based on the above risk factors can effectively and conveniently predict the risk of developing hepatic encephalopathy in patients with decompensated cirrhosis. This model can help clinical healthcare professionals to timely and early identify patients at high risk of developing hepatic encephalopathy, so as to intervene early and prevent the disease progression in time.
Acknowledgements
This study is founded by the Guangxi Natural Science Foundation (project No.: AB22035076); the natural science foundation of china (NSFC) (project No.: 82260899); the Innovation Project of Guangxi Graduate Education (YCBXJ2021013, YCSW2022343); the Guangxi Natural Science Foundation (project No.: 2020GXNSFAA297070). We thank Mr. William Dowding, an Australian biologist, for his help and advice. We also thank those who have helped to make this work possible and better.
Author contributions
This study was completed in collaboration among all authors. Y.C. and H.G.C. conceived, designed and coordinated the study. Y.C., H.L., H.G.C., W.M., M.D.W., W.M.G., Z.J.H., H.J.J., L.X.R., Z.R.Z., X.J.C., Y.F. and C.C. enrolled the participants and performed the study. W.M., H.L. and H.G.C. carried out the date analyses. H.G.C. and H.L. drafted the manuscript. Y.C., H.L. and H.G.C. contributed equally to this work and therefore should be considered co-first authors. All authors have contributed to, seen and approved the final manuscript.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hepatic encephalopathy in chronic liver disease: 2014 practice guideline by the European Association for the Study of the Liver and the American Association for the Study of Liver Diseases. J. Hepatol., 61(3), 642–659 (2014). [DOI] [PubMed]
- 2.Moons KGM, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015;162(1):W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 3.Verbakel JY, Steyerberg EW, Uno H, et al. ROC curves for clinical prediction models part 1. ROC plots showed no added value above the AUC when evaluating the performance of clinical prediction models. J. Clin. Epidemiol. 2020;126:207–216. doi: 10.1016/j.jclinepi.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Muñoz Díaz B, Martínez De La Iglesia J, Romero-Saldaña M, et al. Development of predictive models for nutritional assessment in the elderly. Public Health Nutr. 2021;24(3):449–456. doi: 10.1017/S1368980020002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takikawa Y, Endo R, Suzuki K, et al. Prediction of hepatic encephalopathy development in patients with severe acute hepatitis. Dig. Dis. Sci. 2006;51(2):359–364. doi: 10.1007/s10620-006-3138-7. [DOI] [PubMed] [Google Scholar]
- 6.Labenz C, Toenges G, Huber Y, et al. Raised serum Interleukin-6 identifies patients with liver cirrhosis at high risk for overt hepatic encephalopathy. Aliment. Pharmacol. Ther. 2019;50(10):1112–1119. doi: 10.1111/apt.15515. [DOI] [PubMed] [Google Scholar]
- 7.Tapper EB, Parikh ND, Sengupta N, et al. A risk score to predict the development of hepatic encephalopathy in a population-based cohort of patients with cirrhosis. Hepatology. 2018;68(4):1498–1507. doi: 10.1002/hep.29628. [DOI] [PubMed] [Google Scholar]
- 8.Xue X, Kim MY, Gaudet MM, et al. A comparison of the polytomous logistic regression and joint cox proportional hazards models for evaluating multiple disease subtypes in prospective cohort studies. Cancer Epidemiol. Biomark. Prev. 2013;22(2):275–285. doi: 10.1158/1055-9965.EPI-12-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant SW, Hickey GL, Head SJ. Statistical primer: Multivariable regression considerations and pitfalls. Eur. J. Cardiothorac. Surg. 2019;55(2):179–185. doi: 10.1093/ejcts/ezy403. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Ju S, Li X, et al. Prediction of minimal hepatic encephalopathy by using an radiomics nomogram in chronic hepatic schistosomiasis patients. PLoS Negl. Trop. Dis. 2021;15(10):e0009834. doi: 10.1371/journal.pntd.0009834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin X, Zhang F, Guo H, et al. A nomogram to predict the risk of hepatic encephalopathy after transjugular intrahepatic portosystemic shunt in cirrhotic patients. Sci. Rep. 2020;10(1):9381. doi: 10.1038/s41598-020-65227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi K, Huang Y, Zhang Q, et al. A dynamic nomogram to predict transplant-free mortality in patients with hepatitis B-related cirrhosis and overt hepatic encephalopathy. Int. Immunopharmacol. 2022;108:108879. doi: 10.1016/j.intimp.2022.108879. [DOI] [PubMed] [Google Scholar]
- 13.Li SW, Wang K, Yu YQ, et al. Psychometric hepatic encephalopathy score for diagnosis of minimal hepatic encephalopathy in China. World J. Gastroenterol. 2013;19(46):8745–8751. doi: 10.3748/wjg.v19.i46.8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malinchoc M, Kamath PS, Gordon FD, et al. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology. 2000;31(4):864–871. doi: 10.1053/he.2000.5852. [DOI] [PubMed] [Google Scholar]
- 15.Luo S, Zhou ZM, Guo DJ, et al. Radiomics-based classification models for HBV-related cirrhotic patients with covert hepatic encephalopathy. Brain Behav. 2021;11(2):e01970. doi: 10.1002/brb3.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aicart-Ramos M, Ibáñez-Samaniego L, Hernández-Gea V, et al. A simple nomogram identifies patients with refractory ascites with worse outcome after transjugular intrahepatic portosystemic shunt. Eur. J. Gastroenterol. Hepatol. 2021;33(1S Suppl 1):e587–e593. doi: 10.1097/MEG.0000000000002179. [DOI] [PubMed] [Google Scholar]
- 17.Chen JF, Weng WZ, Huang M, et al. Derivation and validation of a nomogram for predicting 90-day survival in patients with HBV-related acute-on-chronic liver failure. Front. Med. (Lausanne) 2021;8:692669. doi: 10.3389/fmed.2021.692669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao F, Zhang Q, Liu Y, et al. Nomogram prediction of individual prognosis of patients with acute-on-chronic hepatitis B liver failure. Dig. Liver Dis. 2019;51(3):425–433. doi: 10.1016/j.dld.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 19.García-Rodríguez MT, Pértega-Díaz S, López-Calviño B, et al. Nomogram and validity of a model for predicting malnutrition in patients on liver transplant lists. Dig. Dis. Sci. 2018;63(7):1952–1961. doi: 10.1007/s10620-018-5079-3. [DOI] [PubMed] [Google Scholar]
- 20.Gao F, Li X, Wan G, et al. Development and external validation of a prognostic nomogram for acute decompensation of chronic hepatitis B cirrhosis. BMC Gastroenterol. 2018;18(1):179. doi: 10.1186/s12876-018-0911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang RR, Gu HQ, Wei YY, et al. Development and validation of a prognostic model for one-year survival of cirrhosis patients with first-ever spontaneous bacterial peritonitis. J. Clin. Transl. Hepatol. 2021;9(5):647–654. doi: 10.14218/JCTH.2021.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan YP, Wang AJ, Zhang W, et al. Development and validation of a nomogram for predicting overall survival in cirrhotic patients with acute kidney injury. World J. Gastroenterol. 2022;28(30):4133–4151. doi: 10.3748/wjg.v28.i30.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue R, Yang J, Wu J, et al. Novel prognostic models for predicting the 180-day outcome for patients with hepatitis-b virus-related acute-on-chronic liver failure. J. Clin. Transl. Hepatol. 2021;9(4):514–520. doi: 10.14218/JCTH.2021.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirano H, Saito M, Yano Y, et al. Chronic Liver Disease Questionnaire would be a primary screening tool of neuropsychiatric test detecting minimal hepatic encephalopathy of cirrhotic patients. Hepatol. Res. 2015;45(9):994–1003. doi: 10.1111/hepr.12442. [DOI] [PubMed] [Google Scholar]
- 25.Zhong BY, Wang WS, Shen J, et al. Single-centre retrospective training cohort using artificial intelligence for prognostic prediction of encephalopathy, mortality, and liver dysfunction after early TIPS creation. Cardiovasc. Intervent. Radiol. 2021;44(10):1597–1608. doi: 10.1007/s00270-021-02907-0. [DOI] [PubMed] [Google Scholar]
- 26.Bianchi G, Marchesini G, Vilstrup H, et al. Hepatic amino-nitrogen clearance to urea-nitrogen in control subjects and in patients with cirrhosis: A simplified method. Hepatology. 1991;13(3):460–466. doi: 10.1002/hep.1840130313. [DOI] [PubMed] [Google Scholar]
- 27.Xiao N, Liu L, Zhang Y, et al. A practical nomogram based on serum interleukin-6 for the prognosis of liver failure. Front. Med. (Lausanne) 2022;9:1035699. doi: 10.3389/fmed.2022.1035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou YF, Xu Y, Ding YF, et al. Novel nomogram model for predicting 6-week mortality in liver cirrhosis patients with acute upper gastrointestinal bleeding. J. Dig. Dis. 2022;23(8–9):516–526. doi: 10.1111/1751-2980.13137. [DOI] [PubMed] [Google Scholar]
- 29.Kim JM, Jo YY, Na SW, et al. The predictors for continuous renal replacement therapy in liver transplant recipients. Transplant. Proc. 2014;46(1):184–191. doi: 10.1016/j.transproceed.2013.07.075. [DOI] [PubMed] [Google Scholar]
- 30.Rudler M, Weiss N, Bouzbib C, Thabut D. Diagnosis and management of hepatic encephalopathy. Clin. Liver Dis. 2021;25(2):393–417. doi: 10.1016/j.cld.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Jiménez JV, Carrillo-Pérez DL, Rosado-Canto R, et al. Electrolyte and acid-base disturbances in end-stage liver disease: A physiopathological approach. Dig. Dis. Sci. 2017;62(8):1855–1871. doi: 10.1007/s10620-017-4597-8. [DOI] [PubMed] [Google Scholar]
- 32.Tapper EB, Parikh ND. Diagnosis and management of cirrhosis and its complications: A review. JAMA. 2023;329(18):1589–1602. doi: 10.1001/jama.2023.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Praharaj DL, Anand AC. Clinical implications, evaluation, and management of hyponatremia in cirrhosis. J. Clin. Exp. Hepatol. 2022;12(2):575–594. doi: 10.1016/j.jceh.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alukal JJ, John S, Thuluvath PJ. Hyponatremia in cirrhosis: An update. Am. J. Gastroenterol. 2020;115(11):1775–1785. doi: 10.14309/ajg.0000000000000786. [DOI] [PubMed] [Google Scholar]
- 35.Riley RD, Ensor J, Snell KIE, et al. Calculating the sample size required for developing a clinical prediction model. BMJ. 2020;368:m441. doi: 10.1136/bmj.m441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.