Abstract
A nitroalkane-oxidizing enzyme was purified to homogeneity from Neurospora crassa. The enzyme is composed of two subunits; the molecular weight of each subunit is approximately 40,000. The enzyme catalyzes the oxidation of nitroalkanes to produce the corresponding carbonyl compounds. It acts on 2-nitropropane better than on nitroethane and 1-nitropropane, and anionic forms of nitroalkanes are much better substrates than are neutral forms. The enzyme does not act on aromatic compounds. When the enzyme reaction was conducted in an 18O2 atmosphere with the anionic form of 2-nitropropane as the substrate, acetone (with a molecular mass of 60 Da) was produced. This indicates that the oxygen atom of acetone was derived from molecular oxygen, not from water; hence, the enzyme is an oxygenase. The reaction stoichiometry was 2CH3CH(NO2)-CH3 + O2→2CH3COCH3 + 2HNO2, which is identical to that of the reaction of 2-nitropropane dioxygenase from Hansenula mrakii. The reaction of the Neurospora enzyme was inhibited by superoxide anion scavengers in the same manner as that of the Hansenula enzyme. Both of these enzymes are flavoenzymes; however, the Neurospora enzyme contains flavin mononucleotide as a prosthetic group, whereas the Hansenula enzyme contains flavin adenine dinucleotide.
Nitro compounds are useful as solvents and fuels and are widely used in the chemical industry. Because nitro groups can be easily converted to other functional groups, they are useful as intermediate compounds in chemical synthesis. However, many nitro compounds are known to be toxic. 2-Nitropropane is used commercially as a solvent, even though it is a known mutagen in bacteria and a powerful hepatocarcinogen in rats (2, 5, 13).
Nitro compounds are synthesized not only in the chemical industry but also by various organisms. Many antibiotics, e.g., chloramphenicol and azomycin, contain nitro groups, and many leguminous plants produce nitro toxins such as 3-nitro-1-propionic acid and 3-nitro-1-propanol (19).
Several enzymological studies have been done on the metabolism of nitro compounds. In particular, biochemical oxidation of nitroalkanes has been studied in some detail. Thus far, two different kinds of nitroalkane-oxidizing flavoenzymes, nitroalkane oxidase (6) and 2-nitropropane dioxygenase (7, 9), have been purified to homogeneity and characterized. Nitroalkane oxidase was isolated from Fusarium oxysporum and catalyzes the denitrification of various nitroalkanes as follows: R1—CH(NO2)—R2 + O2 + H2O → R1—CO—R2 + HNO2 + H2O2. The enzyme contains flavin adenine dinucleotide (FAD) as a prosthetic group. A notable feature of this enzyme is that some portion of the purified enzyme contains a modified FAD, which we assume is produced during a catalytic side reaction in vivo. The modified form of FAD is labile in the presence of oxygen when it is dissociated from the protein moiety (11). Recently, it was identified as 5-nitrobutyl-FAD (3). The reaction mechanism of nitroalkane oxidase was also analyzed (4).
The other nitroalkane-oxidizing enzyme, 2-nitropropane dioxygenase, has been isolated from the yeast Hansenula mrakii (9). This enzyme also contains FAD as a prosthetic group. The reaction proceeds as follows: 2CH3CH(NO2)CH3 + O2→2CH3COCH3 + 2HNO2. This enzyme is unique in that the oxygen atoms of the dioxygen molecule are split and separately incorporated into two molecules of the substrates (8). This is in striking contrast to most other dioxygenases, which incorporate both oxygen atoms into a single molecule of the substrate. We have determined the primary structure of 2-nitropropane dioxygenase from H. mrakii (17) and found that there are several proteins in the GenBank and EMBL databases that are homologous to this enzyme, e.g., proteins encoded by YJR149w of Saccharomyces cerevisiae (accession no. Z49649; similarity, 28.3%) and yrpB of Bacillus subtilis (U93875; similarity, 22.2%).
Before the discovery of the above two nitroalkane-oxidizing enzymes, Little (12) partially purified a nitroalkane-oxidizing enzyme from Neurospora crassa and suggested that it was a nitroalkane oxidase, although its stoichiometry was not completely consistent with that of nitroalkane oxidase. In the present study, we purified a nitroalkane-oxidizing enzyme from N. crassa to homogeneity and showed that it is 2-nitropropane dioxygenase. This is the first 2-nitropropane dioxygenase purified to homogeneity from organisms other than yeast and is the first flavin mononucleotide (FMN)-dependent 2-nitropropane dioxygenase.
MATERIALS AND METHODS
Chemicals.
2-Nitropropane, 1-nitropropane, and nitroethane were purchased from Nacalai Tesque (Kyoto, Japan). All other nitro compounds were from Aldrich Chemical Co. (Milwaukee, Wis.). DEAE-Toyopearl, carboxymethyl (CM)-Toyopearl, and butyl-Toyopearl (for hydrophobic chromatography) were obtained from Toso Co. (Tokyo, Japan). The anionic forms of nitroalkanes, which undergo extraordinarily slow reprotonation (15), were obtained by addition of an equimolar amount of KOH to the neutral forms as described previously (9).
Enzyme and protein assays.
Nitropropane-oxidizing activity was assayed with 20 mM 2-nitropropane (anionic form) as a substrate. The reaction was carried out at 30°C in a final volume of 1 ml of Britton-Robinson’s buffer (pH 6.5) (a mixture of 16 mM phosphoric acid, 16 mM acetic acid, and 16 mM boric acid, whose pH is adjusted with NaOH), and the amount of nitrite was determined with sulfanilamide and N-(1-naphthyl)ethylenediamine (7). One unit of enzyme activity was defined as the amount of enzyme that catalyzes the formation of 1 μmol of nitrite per min. The protein concentrations were measured with a Bio-Rad protein assay kit. The stoichiometry of the oxidation of the anionic form of 2-nitropropane by the nitroalkane-oxidizing enzyme was examined at 30°C for 5 min by using a mixture containing 20 μmol of anionic 2-nitropropane in 1 ml of Britton-Robinson’s buffer (pH 6.5).
Purification of nitroalkane-oxidizing enzyme.
All the experiments described below were carried out between 0 and 4°C unless otherwise specified.
(i) Cultivation of N. crassa.
Mycelia of N. crassa ATCC 10337 were aerobically grown at 29°C for 24 h in 600 ml of Vogel’s minimal medium (18) containing 2% glucose and 0.5% yeast extract in a 2-liter Sakaguchi flask with reciprocal shaking (150 strokes per min). The mycelia grown in five flasks were subsequently transferred to 100 liters of fresh medium and cultivated at 29°C for 38 h. They were then collected and washed with distilled water. The yield of mycelia was about 4 kg (wet weight).
(ii) Preparation of crude extract.
The washed mycelia (1.5 kg [wet weight]) were suspended in 6 liters of 100 mM potassium phosphate buffer (pH 7.2) and disrupted with a Dyno-Mill (Willy A; Bachofen, Basel, Switzerland) at a flow rate of about 6 liters per h with glass beads (diameter, 0.15 to 0.3 mm). The suspension was centrifuged at 12,000 × g for 30 min, and the supernatant solution was used as an enzyme source.
(iii) Ammonium sulfate fractionation.
Ammonium sulfate was added to the supernatant solution to a final concentration of 1.6 M, and the precipitate was removed by centrifugation. Ammonium sulfate was added to the supernatant solution to a concentration of 2.4 M. The precipitate collected by centrifugation was dissolved in 1 liter of 20 mM potassium phosphate buffer (pH 7.2), and dialyzed 1,000-fold against the same buffer. The pH of the dialyzed enzyme solution was adjusted to 11 with NaOH. Ammonium sulfate was added to a concentration of 1.6 M, and the precipitate was removed by centrifugation. The ammonium sulfate concentration in the supernatant solution was brought to 2.4 M. The precipitate was collected by centrifugation and dissolved in 175 ml of 20 mM potassium phosphate buffer (pH 7.2).
(iv) Butyl-Toyopearl hydrophobic column chromatography.
Ammonium sulfate was added to the enzyme solution to a final concentration of 1.6 M at 4°C. The supernatant solution obtained by centrifugation was applied to a butyl-Toyopearl column (3 by 24 cm). The column was washed with 500 ml of 20 mM potassium phosphate buffer (pH 7.2) containing 1.6 M ammonium sulfate, and the elution was carried out with a linear gradient of ammonium sulfate (700 ml of 20 mM potassium phosphate buffer [pH 7.2] containing 1.6 M ammonium sulfate in the mixing chamber and 700 ml of 20 mM potassium phosphate buffer [pH 7.2] in the reservoir). Fractions (11 ml) were collected at a flow rate of 50 ml/h. The enzyme was eluted at an ammonium sulfate concentration of about 0.4 M. The active fractions were pooled, dialyzed against 20 mM potassium phosphate buffer (pH 7.2) and concentrated by ultrafiltration.
(v) DEAE-Toyopearl column chromatography.
The enzyme solution was loaded onto a DEAE-Toyopearl column (5.6 by 20 cm). The column was washed with 1.5 liters of 20 mM potassium phosphate buffer (pH 7.2), and the enzyme was eluted with a 3-liter linear gradient of 0 to 0.3 M KCl in 20 mM potassium phosphate buffer (pH 7.2). Fractions (10 ml) were collected at a flow rate of 100 ml/h. Active fractions were collected, and the enzyme was precipitated with ammonium sulfate at a final concentration of 2.4 M. The precipitate was dissolved in 12.5 ml of 20 mM potassium phosphate buffer (pH 7.2), and dialyzed 1,000-fold against the same buffer.
(vi) CM-Toyopearl column chromatography.
The pH of the enzyme solution was adjusted to 6.5, and the solution was applied to a CM-Toyopearl column (1.2 by 25 cm). The enzyme was eluted by washing the column with 20 mM potassium phosphate buffer (pH 6.5). Fractions (5 ml) were collected at a flow rate of 50 ml/h. Active fractions were pooled and concentrated by ultrafiltration. This is a negative chromatographic step in which contaminating proteins are effectively removed by binding to the column.
(vii) MonoQ HR column chromatography.
The pH of the enzyme solution was adjusted to 7.2, and the solution was applied to a MonoQ HR 10/10 column. The column was washed with 30 ml of 20 mM potassium phosphate buffer (pH 7.2). The elution was carried out with a 100-ml linear gradient of 0 to 0.3 M NaCl in 20 mM potassium phosphate buffer (pH 7.2) at a flow rate of 3 ml/min. Active fractions were collected and supplemented with 1.6 M ammonium sulfate.
(viii) Phenyl-Superose HR column chromatography.
The enzyme solution was loaded onto a phenyl-Superose HR 5/5 column, and the elution was carried out with a 20-ml linear gradient of 1.6 to 0 M ammonium sulfate in 20 mM potassium phosphate buffer (pH 7.2) at a flow rate of 0.5 ml/min. Active fractions were pooled and stored in 20 mM potassium phosphate buffer (pH 7.2) at −70°C.
Analytical methods.
Oxygen consumption during the 2-nitropropane oxidation was determined with an oxygen electrode. Acetone was detected spectrophotometrically with 3-methyl-2-benzothiazolonehydrazone hydrochloride at 300 nm (14). Hydrogen peroxide was determined by a previously described method (16). The amounts of FAD and FMN were determined by reversed-phase high-performance liquid chromatography (Cosmosil-5C18-AR column [4.6 by 150 mm] [Nacalai Tesque, Kyoto, Japan]; solvent, methanol–10 mM NaH2PO4 [pH 5.5] [35:65, vol/vol]).
RESULTS
Purification of nitroalkane-oxidizing enzyme.
The maximal nitroalkane-oxidizing activity was found in N. crassa mycelia grown for 38 h in minimal Vogel’s medium (18) with nitrate as the sole nitrogen source. The enzyme was purified 2,500-fold with an 8.3% yield from the mycelia (Table 1). The purified enzyme migrated as a single protein band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The specific activity of the purified enzyme was about 25 times higher than that of 2-nitropropane dioxygenase from H. mrakii when the anionic form of 2-nitropropane was used as a substrate (7).
TABLE 1.
Purification of the nitroalkane-oxidizing enzyme from N. crassa
| Step | Total activity (U) | Total amt of protein (mg) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Cell extract | 240,000 | 60,000 | 4.0 | 1 | 100 |
| (NH4)2SO4 (pH 7.2) | 160,000 | 18,000 | 9.0 | 2.2 | 67 |
| (NH4)2SO4 (pH 11.0) | 140,000 | 2,600 | 54 | 14 | 58 |
| Butyl-Toyopearl | 92,000 | 900 | 100 | 25 | 38 |
| DEAE-Toyopearl | 24,000 | 100 | 240 | 60 | 10 |
| CM-Toyopearl | 23,000 | 50 | 460 | 120 | 9.6 |
| MonoQ | 21,000 | 9 | 2,300 | 580 | 8.8 |
| Phenyl-Superose | 20,000 | 2 | 10,000 | 2,500 | 8.3 |
The observation by Little (12) that the nitropropane-oxidizing enzyme of N. crassa is stable at alkaline pH was particularly useful. Most contaminating proteins were denatured by incubation at pH 11 for 1 h, and a sixfold purification was achieved in this single step.
Molecular weight of the enzyme.
The subunit molecular weight of 2-nitropropane dioxygenase was estimated to be approximately 40,000 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gel filtration with a GS 520H-Shodex column showed that the native enzyme has a molecular weight of about 70,000. It follows that the 2-nitropropane dioxygenase of N. crassa is probably a dimer.
Substrate specificity and kinetic properties.
Nitroalkanes in aqueous solution are in a state of equilibrium between a protonated neutral form and a nonprotonated anionic form. The protonation of anionic forms of nitroalkanes is quite slow (15); for example, the half-life of the protonation of anionic 2-nitropropane is 104 min under the assay conditions we used (9). Therefore, we examined the substrate specificity of the enzyme with various anionic forms of nitro compounds (Table 2). 2-Nitropropane was the preferred substrate for this enzyme. It also showed a high activity on 1-nitropropane, nitroethane, and 3-nitro-2-pentanol. Aromatic nitro compounds such as nitrobenzene and 2-nitrobenzoic acid were not substrates.
TABLE 2.
Substrate specificity of the nitroalkane-oxidizing enzyme from N. crassa
| Nitro compound | Relative activitya |
|---|---|
| 2-Nitropropane | 100 |
| 1-Nitropropane | 21 |
| Nitroethane | 27 |
| 2-Nitroethanol | 13 |
| 3-Nitro-2-butanol | 14 |
| 3-Nitro-2-pentanol | 20 |
| 3-Nitropropionic acid | 12 |
| 1-Nitropentane | 3 |
| Nitrocyclohexane | 2 |
The enzyme activity was assayed by measuring nitrite production with 20 mM anionic nitro compounds as substrates under the conditions described in Materials and Methods. The following nitro compounds were inert: 2-methyl-2-nitropropanol, nitrobenzene, 2-nitrobenzoic acid, 2-nitrophenol, 4-nitrocatechol, and 2,4-dinitrotoluene.
The Km and kcat values of the enzyme were determined for several nitro compounds (Table 3). Anionic as well as neutral forms of nitroalkanes were used for these experiments. The enzyme had lower Km values and higher kcat values for the anionic forms of nitroalkanes. The best substrate for this enzyme was the anionic form of 2-nitropropane. Glucose oxidase and d-amino acid oxidase also catalyze the oxidation of the anionic form of 2-nitropropane as a side reaction (7). However, the reactions are much less efficient than those of the nitroalkane oxygenases from H. mrakii and N. crassa (Table 3).
TABLE 3.
Comparison of the kinetic parameters for anionic and neutrala nitroalkanes
| Substrate | Anionic form
|
Neutral form
|
||
|---|---|---|---|---|
| Km (mM) | kcat (s−1) | Km (mM) | kcat (s−1) | |
| 2-Nitropropane dioxygenase (N. crassa) | ||||
| 2-Nitropropane | 3.1 | 6,400 | 33 | 160 |
| Nitroethane | 6.0 | 2,000 | 23 | 32 |
| 1-Nitropropane | 8.3 | 1,300 | 20 | 35 |
| 2-Nitropropane dioxygenase (H. mrakii) | ||||
| 2-Nitropropane | 1.6 | 290 | ||
| Glucose oxidase | ||||
| 2-Nitropropane | 17 | 0.026 | ||
| d-Amino acid oxidase | ||||
| 2-Nitropropane | 11 | 0.23 | ||
Various concentrations of anionic nitroalkanes were incubated with the enzyme at pH 6.5. The reactions for the neutral forms were carried out in 100 mM potassium phosphate buffer (pH 8.0) as described previously (9).
Stoichiometry of the reaction.
The stoichiometry of the 2-nitropropane oxidation was deduced by measuring the amount of oxygen consumed and the amounts of nitrite, acetone, and hydrogen peroxide produced. The amount of oxygen consumed (215 nmol) was approximately half the amounts of nitrite (380 nmol) and acetone (420 nmol) produced; no hydrogen peroxide was formed in the reaction. The data are consistent with the following equation: 2CH3CH(NO2)CH3 + O2 → 2CH3COCH3 + 2HNO2.
Enzymatic incorporation of 18O2 into acetone.
Oxidation of 2-nitropropane by the N. crassa enzyme was carried out in an 18O2 atmosphere, and the reaction mixture was analyzed by gas chromatography-mass spectrometry. The mass spectrum of the acetone peak showed a parent peak of m/e = 60 corresponding to [18O]acetone. A parent peak of m/e = 60 was also observed for acetone produced with 2-nitropropane dioxygenase from H. mrakii as described previously (8). This result and the reaction stoichiometry described above indicate that two atoms of molecular oxygen were incorporated into the carbonyl groups of two molecules of acetone. Thus, this enzyme can be designated 2-nitropropane dioxygenase in the same manner as the enzyme from H. mrakii.
Prosthetic group.
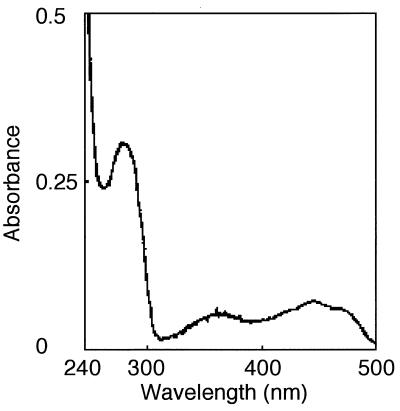
The absorption spectrum of the enzyme showed maxima at 276, 380, and 445 nm (Fig. 1), suggesting that the enzyme contains a flavin moiety, like other 2-nitropropane-oxidizing enzymes from H. mrakii and F. oxysporum. To analyze the prosthetic group, the enzyme solution was incubated at 80°C for 20 min to denature the protein moiety, cooled, and centrifuged to remove the denatured protein. The supernatant solution containing the dissociated prosthetic group was subjected to reversed-phase chromatography. The prosthetic group was identified as FMN based on its retention time. The quantitative analysis revealed the presence of 2 mol of FMN per mol of subunit.
FIG. 1.
Absorption spectrum of 2-nitropropane dioxygenase from N. crassa.
Inhibitors.
The inhibitory effects of various compounds on the enzyme activity were examined (Table 4). The oxidation of anionic 2-nitropropane was inhibited by various superoxide scavengers. The addition of superoxide dismutase inhibited the enzyme completely. Other O2− scavengers such as cytochrome c and nitroblue tetrazolium also acted as inhibitors. These results suggest that O2− is generated as an essential intermediate during the oxidation of anionic nitroalkanes by 2-nitropropane dioxygenase from N. crassa. The addition of thiol compounds (2-mercaptoethanol, dithiothreitol, and glutathione) and a thiol-modifying reagent (iodoacetate) resulted in a marked decrease in the enzyme activity. EDTA did not affect the enzyme activity.
TABLE 4.
Effect of various compounds on the oxidation of 2-nitropropane catalyzed by 2-nitropropane dioxygenase from N. crassa
| Additive | Concn (mM) | Relative activitya (%) |
|---|---|---|
| None | 100 | |
| Superoxide dismutase | 100 U | 0 |
| Cytochrome c | 0.04 | 34 |
| Nitroblue tetrazolium | 2.5 | 10 |
| EDTA | 1 | 95 |
| Iodoacetate | 1 | 0 |
| Dithiothreitol | 1 | 0 |
| 2-Mercaptoethanol | 1 | 2 |
| Glutathione (reduced form) | 5 | 28 |
The enzyme was assayed at 30°C for 5 min, as described in Materials and Methods, in a mixture containing the indicated concentration of each compound and 20 μmol of anionic 2-nitropropane in 1 ml of Britton-Robinson’s buffer (pH 6.5).
DISCUSSION
We have purified and characterized a nitroalkane-oxidizing enzyme from N. crassa. Previously, Little (12) suggested that the nitroalkane-oxidizing enzyme from N. crassa is an oxidase and that the oxygen atom of the product is derived from water, not from molecular oxygen in the reaction. However, the present data clearly shows that the enzyme we purified from N. crassa is not an oxidase but an oxygenase, although we cannot exclude the possibility that the enzyme shown by Little is produced by the fungus in addition to the enzyme we purified. However, Little’s failure to detect hydrogen peroxide as a product of the enzyme reaction (12) strongly suggests that this enzyme was also an oxygenase.
We previously found 2-nitropropane dioxygenase in the yeast H. mrakii (7). Thus, this enzyme is probably widely distributed in eukaryotic microorganisms. It has also been reported that the stoichiometry of the 2-nitropropane oxidation catalyzed by the nitroalkane-oxidizing enzyme partially purified from Streptomyces is identical to that of the oxidation catalyzed by 2-nitropropane dioxygenase (1). However, the direct incorporation of oxygen atoms into the substrate from molecular oxygen has not yet been confirmed, and it is not known whether this enzyme is a flavoenzyme.
2-Nitropropane dioxygenase purified in the present study is similar to 2-nitropropane dioxygenase from H. mrakii in several respects: both of them are flavoenzymes, their reaction stoichiometry is identical, anionic forms of nitroalkanes are much better as substrates than their neutral counterparts, and both enzyme reactions are inhibited by superoxide anion radical scavengers. These findings suggest that the reactions probably proceed by the same mechanism.
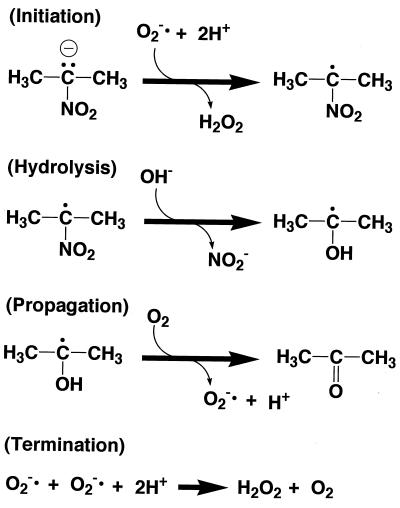
Kuo and Fridovich (10) described the reaction mechanism of a free-radical chain oxidation of 2-nitropropane initiated and propagated by a superoxide anion radical (Fig. 2) and claimed that their mechanism is applicable to the reaction catalyzed by 2-nitropropane dioxygenase (10). According to their mechanism, a hydroxide ion derived from water is incorporated into the 2-nitropropane radical at the hydrolysis step (Fig. 2). The hydrogen radical is removed by molecular dioxygen, and acetone is produced at the propagation step. Thus, the oxygen atom of acetone should be derived from water but not from molecular oxygen. However, this was not the case in our experiments.
FIG. 2.
Proposed reaction mechanism of free-radical chain oxidation of 2-nitropropane initiated and propagated by the superoxide anion radical (10).
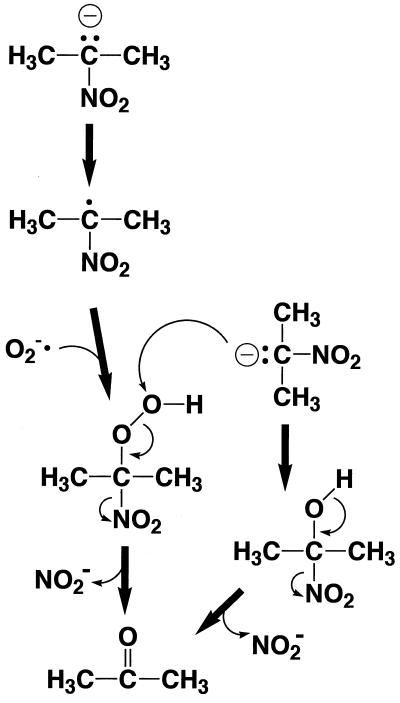
Therefore, we propose the mechanism shown in Fig. 3. An anionic form of 2-nitropropane is oxidized to form a 2-nitropropane radical, which reacts with a superoxide anion radical to produce a peroxide intermediate. This intermediate reacts with another molecule of an anionic form of 2-nitropropane, and consequently two nitrite ions are released to form two molecules of acetone. Two atoms of molecular oxygen are therefore incorporated into two molecules of acetone. The FMN of the N. crassa enzyme or the FAD of the H. mrakii enzyme probably participates in the formation of a 2-nitropropane radical and superoxide anion radical. This mechanism can explain the stoichiometry of the reaction, the direct incorporation of the oxygen atom from molecular oxygen, and the involvement of the superoxide anion radical as an essential intermediate.
FIG. 3.
Probable reaction mechanism of 2-nitropropane oxidation by 2-nitropropane dioxygenase.
REFERENCES
- 1.Dhawale M R, Hornemann U. Nitroalkane oxidation by streptomycetes. J Bacteriol. 1979;137:916–924. doi: 10.1128/jb.137.2.916-924.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiala E S, Conaway C C, Mathis J E. Oxidative DNA and RNA damage in the livers of Sprague-Dawley rats treated with the hepatocarcinogen 2-nitropropane. Cancer Res. 1989;49:5518–5522. [PubMed] [Google Scholar]
- 3.Gadda G, Edmondson R D, Russell D H, Fitzpatrick P F. Identification of the naturally occurring flavin of nitroalkane oxidase from Fusarium oxysporum as a 5-isobutyl-FAD and conversion of the enzyme to the active FAD-containing form. J Biol Chem. 1997;272:5563–5570. doi: 10.1074/jbc.272.9.5563. [DOI] [PubMed] [Google Scholar]
- 4.Heasley C J, Fitzpatrick P F. Kinetic mechanism and substrate specificity of nitroalkane oxidase. Biochem Biophys Res Commun. 1996;225:6–10. doi: 10.1006/bbrc.1996.1122. [DOI] [PubMed] [Google Scholar]
- 5.Hite M, Skeggs H. Mutagenic evaluation of nitroparaffins in the Salmonella typhimurium/mammalian-microsome test and the micronucleus test. Environ Mutagen. 1979;1:383–389. doi: 10.1002/em.2860010411. [DOI] [PubMed] [Google Scholar]
- 6.Kido T, Hashizume K, Soda K. Purification and properties of nitroalkane oxidase from Fusarium oxysporum. J Bacteriol. 1978;133:53–58. doi: 10.1128/jb.133.1.53-58.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kido T, Soda K. Oxidation of anionic nitroalkanes by flavoenzymes, and participation of superoxide anion in the catalysis. Arch Biochem Biophys. 1984;234:468–475. doi: 10.1016/0003-9861(84)90294-7. [DOI] [PubMed] [Google Scholar]
- 8.Kido T, Soda K, Suzuki T, Asada K. A new oxygenase, 2-nitropropane dioxygenase of Hansenula mrakii. J Biol Chem. 1976;251:6994–7000. [PubMed] [Google Scholar]
- 9.Kido T, Tanizawa K, Inagaki K, Yoshimura T, Ishida M, Hashizume K, Soda K. 2-Nitropropane dioxygenase from Hansenula mrakii: re-characterization of the enzyme and oxidation of anionic nitroalkanes. Agric Biol Chem. 1984;48:2549–2554. [Google Scholar]
- 10.Kuo C F, Fridovich I. Free-radical chain oxidation of 2-nitropropane initiated and propagated by superoxide. Biochem J. 1986;237:505–510. doi: 10.1042/bj2370505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurihara T, Esaki N, Soda K, Ohishi N. A 340-nm chromophore of nitroalkane oxidase from Fusarium oxysporum. In: Yagi K, editor. Flavins and flavoproteins. Berlin, Germany: Walter de Gruyter; 1994. pp. 195–198. [Google Scholar]
- 12.Little H N. Oxidation of nitroethane by extracts from Neurospora. J Biol Chem. 1951;193:347–358. [PubMed] [Google Scholar]
- 13.Loefroth G, Nilsson L, Andersen J R. Structure-activity relationship of nitroalkane-induced mutagenicity in the Ames Salmonella assay. Prog Clin Biol Res. 1986;209:149–155. [PubMed] [Google Scholar]
- 14.Paz M A, Blumenfeld O O, Rajkind M, Henson E, Fureine H F, Gallop P M. Determination of carbonyl compounds with N-methyl benzothiazolone hydrazone. Arch Biochem Biophys. 1965;109:548–559. doi: 10.1016/0003-9861(65)90400-5. [DOI] [PubMed] [Google Scholar]
- 15.Porter D J, Bright H J. Mechanism of oxidation of nitroethane by glucose oxidase. J Biol Chem. 1977;252:4361–4370. [PubMed] [Google Scholar]
- 16.Snyder S H, Hendley E D. Sensitive fluorometric and radiometric assays for monoamine oxidase and diamine oxidase. Methods Enzymol. 1971;17B:741–746. [Google Scholar]
- 17.Tchorzewski M, Kurihara T, Esaki N, Soda Unique primary structure of 2-nitropropane dioxygenase from Hansenula mrakii. Eur J Biochem. 1994;226:841–846. doi: 10.1111/j.1432-1033.1994.00841.x. [DOI] [PubMed] [Google Scholar]
- 18.Vogel H J. Distribution of lysine pathways among fungi: evolutionary implications. Am Nat. 1964;98:435–446. [Google Scholar]
- 19.Williams M C, Barneby R C. The occurrence of nitro-toxins in North American Astragalus (Fabaceae) Brittonia. 1977;29:310–326. [Google Scholar]