Significance
Pontocerebellar hypoplasia (PCH) is a collection of rare neurodevelopmental disorders that affect brain development. Identifying PCH-associated genes and disease mechanisms is crucial for improving diagnosis and developing treatment options. Here, we present FAM91A1 as a unique PCH-associated gene since zebrafish depleted of FAM91A1 display similar developmental issues as those of TBC1D23, which is a known PCH gene. The complex structure between FAM91A1 and TBC1D23 illustrates how the two proteins interact with each other. This interaction is required for protein trafficking from endosomes and strongly correlates with the risk to develop PCH-like symptoms in zebrafish. Our study contributes to our understanding of PCH and identifies impaired endosomal trafficking as a convergent mechanism for many PCH subtypes.
Keywords: membrane trafficking, endosomal trafficking, Golgi, pontocerebellar hypoplasia, neuronal development
Abstract
Pontocerebellar hypoplasia (PCH) is a group of rare neurodevelopmental disorders with limited diagnostic and therapeutic options. Mutations in WDR11, a subunit of the FAM91A1 complex, have been found in patients with PCH-like symptoms; however, definitive evidence that the mutations are causal is still lacking. Here, we show that depletion of FAM91A1 results in developmental defects in zebrafish similar to that of TBC1D23, an established PCH gene. FAM91A1 and TBC1D23 directly interact with each other and cooperate to regulate endosome-to-Golgi trafficking of KIAA0319L, a protein known to regulate axonal growth. Crystal structure of the FAM91A1–TBC1D23 complex reveals that TBC1D23 binds to a conserved surface on FAM91A1 by assuming a Z-shaped conformation. More importantly, the interaction between FAM91A1 and TBC1D23 can be used to predict the risk of certain TBC1D23-associated mutations to PCH. Collectively, our study provides a molecular basis for the interaction between TBC1D23 and FAM91A1 and suggests that disrupted endosomal trafficking underlies multiple PCH subtypes.
Pontocerebellar hypoplasia (PCH) is a group of rare autosomal recessive neurological diseases that lead to microcephaly, intellectual disability, and various developmental disorders (1–3). Currently, clinical diagnosis of PCH relies on cranial MRI, clinical manifestations, and whole exome sequencing (3, 4). However, treatment options for PCH are limited to supportive care, which has poor efficacy and significant impact on patients and their families (2, 5). So far, more than 20 PCH-associated genes have been identified, and many of them are involved in RNA processing (2). In addition, several PCH-associated genes, including TBC1D23 (6, 7), VPS51 (8), VPS53 (9), and CHMP1A (10), are all involved in protein trafficking from endosomes. Currently, it is unclear how mutations of these genes contribute to the pathogenesis of PCH, which prevent the development of effective therapeutic strategies.
Proteins entering the endosomal network are either delivered to lysosomes for degradation, transported to the trans-Golgi network (TGN), or recycled back to the plasma membrane for reuse (11–15). Endosomal trafficking is an important cellular process that plays a significant role in protein sorting, localization, and regulation of intracellular signaling, ultimately contributing to proper cellular functions (16, 17). In addition to PCH, dysregulation of this pathway can contribute to the development of a wide range of diseases, including Alzheimer’s disease (18–20), Parkinson’s disease (21, 22), and cancer (23–25). Recent studies have identified multiple critical regulators of endosome-to-TGN trafficking, including TBC1D23, Golgi tethering proteins golgin-97 and golgin-245, and the FAM91A1 complex (composed of FAM91A1, WDR11, and C17orf75) (26). TBC1D23 encompasses an N-terminal TBC domain followed by a Rhodanese-like domain, and a C-terminal PH domain. The TBC and Rhodanese-like domains of TBC1D23 interact with TGN-localized golgin-97/245, and the PH domain binds to the endosomal-vesicle-localized WASH complex, thereby promoting the protein trafficking from endosomes to the TGN (27, 28). The exact functions of the FAM91A1 complex are not fully understood yet; however, recent studies have revealed that the FAM91A1 complex also facilitates protein trafficking from endosomes to the TGN (29). The FAM91A1 complex directly interacts with TBC1D23 via the N terminus of FAM91A1 and the loop connecting the Rhodanese-like and PH domains from TBC1D23 (26). Surprisingly, TBC1D23 and the FAM91A1 complex play an overlapping but not identical role in promoting protein endosome-to-TGN trafficking. For instance, depletion of TBC1D23, but not that of FAM91A1, impairs the trafficking of TGN46 (30), a transmembrane protein shuttling between the TGN and cell surface via endosomes (26). It is necessary to determine the molecular details of the interaction between FAM91A1 and TBC1D23 in order to dissect their common and unique functions.
Recently, multiple individuals diagnosed with PCH carry homozygous mutations in the TBC1D23 gene, which often encode a protein with a truncated C-terminal domain (6, 7). Interestingly, in addition to these deletion mutants, many missense mutations have been found in the TBC1D23. Currently, it is unclear whether these mutations pose any risk of developing PCH for their carriers. Intriguingly, mutations in the FAM91A1 complex subunit WDR11 have been also identified in patients from various families with “PCH-like” symptoms, including intellectual disability, microcephaly, and cerebellar hypoplasia (31). Deletion of both WDR11 alleles in mice led to mid-gestation embryonal lethality or significant growth retardation, phenotypes much severe than PCH (32). Thus, it is still uncertain whether mutations in WDR11 or FAM91A1 are causative factors in the development of PCH, as definitive evidence is yet to be established.
To investigate the relationship between the FAM91A1 complex, TBC1D23, and PCH, we performed a multidisciplinary study of biochemical, structural, cellular, and zebrafish models. We show that depletion of FAM91A1 in zebrafish results in similar developmental defects as that of TBC1D23, recapitulating some key features of PCH patients. The crystal structure of the FAM91A1–TBC1D23 complex reveals that the TBC1D23 peptide adopts a Z-shaped conformation and binds to a conserved surface on FAM91A1. Importantly, the FAM91A1–TBC1D23 interaction is required for endosome-to-TGN trafficking of KIAA0319L, a protein participating in axon guidance, and brain development in zebrafish. Overall, our work expands the list of PCH-associated genes and provides a unique way to predict the risk of PCH based on the FAM91A1–TBC1D23 interaction.
Results
Depletion of FAM91A1 in Zebrafish Leads to Similar Phenotypes as that of TBC1D23.
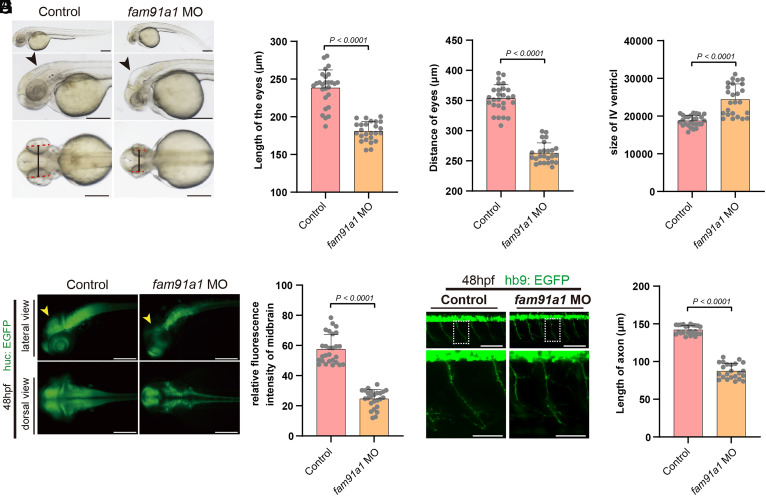
The following observations raised the possibility that FAM91A1 and TBC1D23 may play analogous roles during development. First, FAM91A1 directly interacts with TBC1D23, and both proteins function in endosomal trafficking (26). Second, homozygous mutations in WDR11 have been identified in patients with PCH-like symptoms (31). To test our hypothesis, we utilized a splice-blocking morpholino (MO) technology to knock down FAM91A1 expression in zebrafish and observed a significant reduction in FAM91A1 mRNA levels by over 60% following injection of 5.0 ng of MO (SI Appendix, Fig. S1A). Depletion of FAM91A1 resulted in significantly smaller eyes and shorter distances between the eyes compared with the control, similar to that of TBC1D23 (7, 26, 28) (Fig. 1 A–C). Moreover, depletion of FAM91A1 led to an enlarged IV ventricle relative to control, analogous to that of TBC1D23 (Fig. 1D).
Fig. 1.
FAM91A1 is required for zebrafish neuronal development and brain growth. (A) Bright-field images of zebrafish embryos of control and fam91a1 MO zebrafish based on head and body morphology at 48 hpf (hours post fertilization). The dotted red line indicates eye length, the black solid line indicates eye distance, and the black arrows indicate the fourth ventricle. Control: control MO injection; fam91a1 MO: fam91a1 MO injection. All injections are performed at one cell stage of the development (Scale bar, 100 μm). (B) The length of eyes of embryos in control and fam91a1 MO zebrafish at 48 hpf. Experiments were repeated three times. Each dot represents one zebrafish and at least 20 zebrafish were used per group. Statistical comparisons were performed using an unpaired t test and data are presented as mean ± SD. (C) The distance of eyes of embryos in control and fam91a1 MO zebrafish at 48 hpf. Experiments were repeated three times. Each dot represents one fish, and at least 20 fish were used per group. Statistical comparisons were performed using an unpaired t test and data are presented as mean ± SD. (D) The size of the fourth ventricle of control and fam91a1 MO zebrafish at 48 hpf. Each dot represents one fish, and at least 20 zebrafish were used per group. The data are presented as mean ± SD. P values were calculated using unpaired t test. Experiments were repeated three times. (E) HuC (green) expression in Tg [HuC: GFP] transgenic zebrafish. Arrows indicate midbrain. Top, lateral views; bottom, dorsal views (Scale bar, 100 μm). (F) The midbrain area fluorescence intensity of embryos in control and fam91a1. The fluorescence intensity in the midbrain area of the fam91a1 MO group is approximately half that of the control group. Each dot represents one fish, and at least 20 zebrafish were used per group. The data are presented as mean ± SD. P values were calculated using unpaired t test. Experiments were repeated three times. (G) Morphology of CaP axons from embryos at 48 hpf that were injected control or fam91a1 MO. All injections are performed at one cell stage of the Tg [Hb9: GFP]ml2 transgenic zebrafish embryos. Lateral views and enlarged views are shown (Scale bar, 20 μm). (H) Statistical results of the length of CaP axons in embryos treated as in G. For each group, more than 20 axons from 6 Tg [Hb9: GFP]ml2 transgenic zebrafish embryos are scored. The data are presented as mean ± SD. P values were calculated using unpaired t test. Experiments were repeated three times.
To further investigate the role of FAM91A1 in neuronal development, we used the HuC-GFP transgenic line, which expresses an early marker of pan-neuronal cells (33, 34). The fam91a1 MO group displayed a significantly reduced fluorescence intensity in midbrain when compared to the control group (Fig. 1 E and F). Specifically, the midbrain size of the fam91a1 MO group was reduced by approximately 60% when compared to the control (Fig. 1F). Using the Tg [Hb9: GFP]ml2 transgenic line that labels the motor neurons in zebrafish, we found that the fam91a1 MO group displayed abnormal CaP motor neuron morphology compared to control zebrafish (Fig. 1 G-H). The average length of the CaP axon in the fam91a1 MO group was only 67% of that in control zebrafish (Fig.1H). These phenotypes were highly similar to those of tbc1d23 MO reported by us and others. Thus, using three different zebrafish models, we demonstrate that depletion of FAM91A1 caused similar defects as that of TBC1D23, indicating that FAM91A1 and TBC1D23 may cooperate to regulate brain development and neuron formation.
Crystal Structure of the FAM91A1–TBC1D23 Complex.
Previous studies showed that residues 514–558 of TBC1D23 (TBC1D23514-558) were responsible for contacting the N terminus of FAM91A1 encompassing residues 1–328 (FAM91A1N) (26). We further truncated TBC1D23514-558 into two smaller fragments, TBC1D23514-538 and TBC1D23539-558, and presented that TBC1D23514-538, but not TBC1D23539-558, directly interacted with FAM91A1N (SI Appendix, Fig. S2). To understand the binding mechanism between TBC1D23 and FAM91A1, we tried to crystallize FAM91A1N together with multiple peptides derived from TBC1D23. After screening many different constructs and conditions, we successfully produced diffraction-quality crystals using a chimera construct encoding TBC1D23514-543 followed by a 3 × GGS linker and FAM91A1N. The complex structure was solved by molecular replacement using the FAM91A1N AlphaFold2 model and refined to a resolution of 2.51 Å (SI Appendix, Fig. S3 and Table S1).
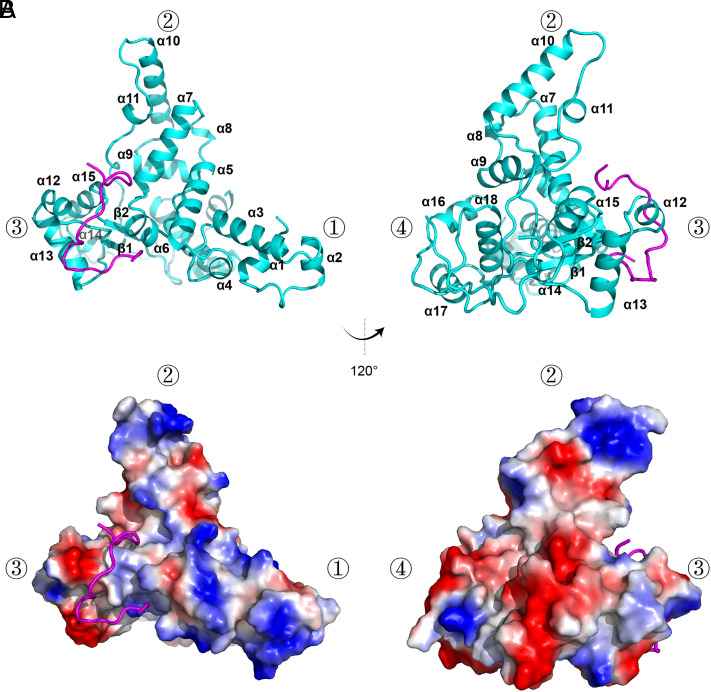
In the complex structure, all FAM91A1 residues, except for residues 236–251, are visible in the electron density. Residues 236–251 likely form a structurally flexible loop. The DALI search (35) did not identify any structure bearing significant similarity with that of FAM91A1N, indicating that FAM91A1N adopts a novel fold. FAM91A1N encompasses 18 alpha-helices and 2 beta-strands, forming a triangular pyramid-like structure. α1–6 form the first cone of the triangular pyramid, α7–11 form the second cone, α12–15 and β1–2 form the third cone, and α16–18 form the fourth cone. TBC1D23518-538 assumes a Z-shape and binds to the surface between the first, second, and third cones (Fig. 2 A and B).
Fig. 2.
Crystal structure of FAM91A1 in complex with TBC1D23. (A) Crystal structure of FAM91A1N in complex with the TBC1D23 fragment, shown in two orientations rotated 120° with respect to each other. Cyan: FAM91A1N; purple: TBC1D23 peptide. The four vertices of the triangular pyramid-like structure formed by FAM9A11-328 are indicated by ①, ②, ③, and ④, respectively. (B) Electrostatic potential surface of the complex of FAM91A1 with TBC1D23 peptide (ribbon diagram). Blue: positive potential; red: negative potential. The complex is shown in the same orientation as that of the above molecule in A.
Interaction between FAM91A1 and TBC1D23.
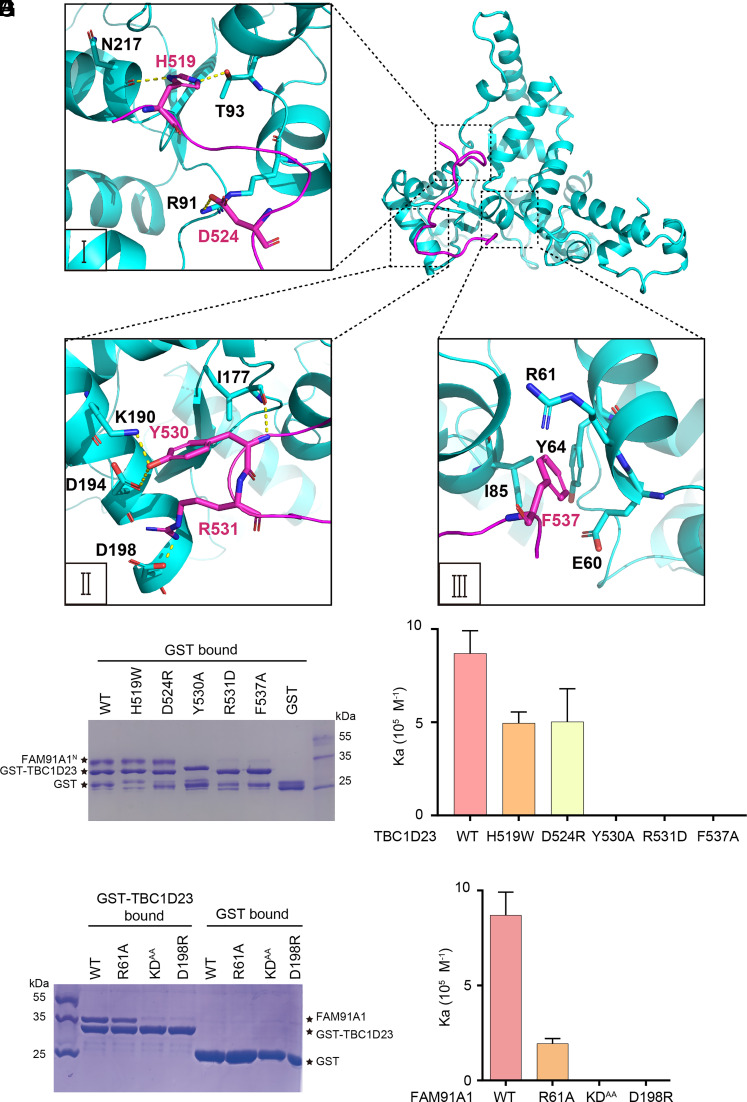
The interaction between FAM91A1 and TBC1D23 involves multiple hydrogen bonds, salt bridges, and Van der Waal forces, resulting in a buried surface area of ~1,003 Å2. According to our structure, TBC1D23518–538 can be divided into three segments to interact with FAM91A1. The first segment of TBC1D23 consists of residues 518–524 and contacts FAM91A1 predominately via hydrogen bonds. The side chain of TBC1D23H519 forms hydrogen bonds with the main chain oxygen of N217 and the side chain of T93 in FAM91A1. TBC1D23D524 forms a salt bridge and a hydrogen bond with FAM91A1R91 (Fig. 3A). The second segment of TBC1D23 comprises residues 525–531 and interacts with FAM91A1 by forming hydrogen bonds and a salt bridge. The side chain of TBC1D23Y530 forms two hydrogen bonds with FAM91A1K190 and FAM91A1D194. TBC1D23R531 contacts FAM91A1D198 by forming a salt bridge and a hydrogen bond (Fig. 3B). In contrast to the first and second segment, the third segment of TBC1D23 (comprising residues 532–538) interacts with FAM91A1 mainly through hydrophobic interactions (Fig. 3C). Most notably, TBC1D23F537 is buried in a hydrophobic groove formed by side chains of E60, R61, Y64, and I85 of FAM91A1 (Fig. 3C).
Fig. 3.
Molecular basis of TBC1D23 recognition by FAM91A1. (A) Detailed interaction between FAM91A1 (cyan) and the first segment of TBC1D23 (purple). Yellow dotted line: hydrogen bonds. (B) Detailed interaction between FAM91A1 (cyan) and the second segment of TBC1D23 (purple). Yellow dotted line: hydrogen bonds. (C) Detailed interaction between FAM91A1 (cyan) and the third segment of TBC1D23 (purple). (D) GST pull-down assays were performed with GST–TBC1D23514-558, or GST, and purified FAM91A1N wild-type (WT) or the indicated mutants. Shown is the Coomassie blue-stained SDS-PAGE gel of bound samples. (E) The affinity between FAM91A1N and TBC1D23514-538 WT or mutants in a buffer containing 100 mM HEPES, pH 7.5, 200 mM NaCl, determined by ITC. Association constants (Ka) were shown as mean ± SD. (F) GST pull-down assays were performed with GST–TBC1D23514-558 WT and purified FAM91A1N WT or the indicated mutants. Shown is the Coomassie blue-stained SDS-PAGE gel of bound samples. (G) The affinity between TBC1D23514-538 WT and FAM91A1N WT or mutants in a buffer containing 100 mM HEPES, pH 7.5, 200 mM NaCl, determined by ITC. The association constants (Ka) are presented as mean ± SD.
To verify our structure, we mutated multiple FAM91A1-contacting residues in TBC1D23 and examined their binding to FAM91A1 using GST pull-down assays. In contrast with TBC1D23 wild type (WT), TBC1D23Y530A, TBC1D23R531D, and TBC1D23F537A completely lost the ability to contact FAM91A1 (Fig. 3D). The results were further confirmed by the isothermal titration calorimetry (ITC) experiments, which showed that TBC1D23 WT bound FAM91A1N with an affinity of 1.15 ± 0.16 µM, whereas no binding was detected for these three mutants (Fig. 3E). Furthermore, TBC1D23H519W and TBC1D23D524R slightly decreased the affinity toward FAM91A1N (Fig. 3E).
Similarly, we mutated multiple FAM91A1 residues that are involved in contacting TBC1D23: R61A, K190A/D194A (KDAA), and D198R. Both the KDAA and D198R mutants completely abolished the binding to TBC1D23 in GST-pulldown and ITC assays (Fig. 3 F and G). Furthermore, the R61A mutants decreased the binding by >80% (Fig. 3G). Sequence alignment using proteins from various model organisms, including human (Homo sapiens), mouse (Mus musculus), zebrafish (Danio rerio), tropical clawed frog (Xenopus tropicalis), fruit fly (Drosophila melanogaster), and nematode (Caenorhabditis elegans) indicate that key TBC1D23 and FAM91A1 residues involved in the interactions are highly conserved, suggesting that the interaction between FAM91A1 and TBC1D23 is conserved across different species (SI Appendix, Fig. S4).
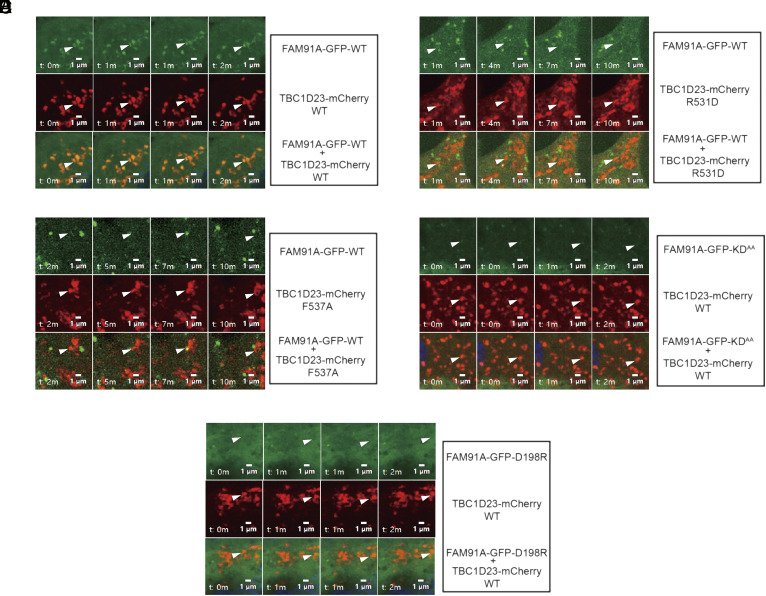
To further investigate the relevance of FAM91A1–TBC1D23 interaction in vivo, we performed live-cell time-lapse imaging to determine the subcellular localization of FAM91A1 and TBC1D23 (Fig. 4). GFP-FAM91A1 WT and mCherry-TBC1D23 WT formed punctate structures in live cells, with extensive colocalization (Fig. 4A). However, the colocalization was lost when TBC1D23–R531D or F537A was used (Fig. 4 B and C). Unlike FAM91A1 WT, the FAM91A1 mutants (KDAA or D198R) were dispersed throughout the cells and no longer formed puncta (Fig. 4 D and E). To investigate whether these puncta of TBC1D23 represent endosomal or Golgi localization, we employed time-lapse imaging to investigate their colocalization with early endosome (FYVE) (36), late endosome (CD63) (37), and trans-Golgi (GRIP: a region encompassing amino acids 688 to 767 of golgin-97) (38). Our experiments revealed that GFP-TBC1D23 strongly colocalized with mCherry-GRIP, rather than mCherry-2 × FYVE or mCherry-CD63, resembling the pattern seen with endogenous TBC1D23 (26) (SI Appendix, Fig. S5 A–D). Furthermore, the FAM91A1-binding deficient mutants (Y530A, R531D, and F537A) still exhibited significant colocalization with GRIP, indicating that interacting with FAM91A1 is not required for the TGN localization of TBC1D23 (SI Appendix, Fig. S5 E–H). Altogether, our studies demonstrate that the interaction between FAM91A1 and TBC1D23 is essential for their colocalization in cells.
Fig. 4.
Time-lapse chasing of FAM91A1-GFP and TBC1D23-mCherry colocalization events. (A) Time-lapse chasing of FAM91A1-GFP with TBC1D23-mCherry WT, R531D, and F537A in COS7 cells. For each group, the Top shows FAM91A1-GFP images, the Middle shows TBC1D23-mCherry images, and the Bottom shows the merged images of FAM91A1-GFP and TBC1D23-mCherry. The experiment was repeated independently three times (Bars, 1 µm). (B) Time-lapse chasing of FAM91A1-GFP with TBC1D23-mCherry R531D in COS7 cells. For each group, the Top shows FAM91A1-GFP images, the Middle shows TBC1D23-mCherry images, and the Bottom shows the merged images of FAM91A1-GFP and TBC1D23-mCherry. The experiment was repeated independently three times (Bars, 1 µm). (C) Time-lapse chasing of FAM91A1-GFP with TBC1D23-mCherry F537A in COS7 cells. For each group, the Top shows FAM91A1-GFP images, the Middle shows TBC1D23-mCherry images, and the Bottom shows the merged images of FAM91A1-GFP and TBC1D23-mCherry. The experiment was repeated independently three times (Bars, 1 µm). (D) Time-lapse chasing of TBC1D23-mCherry with FAM91A1-GFP KDAA, in COS7 cells. For each group, the Top shows FAM91A1-GFP images, the Middle shows TBC1D23-mCherry images, and the Bottom shows the merged images of FAM91A1-GFP and TBC1D23-mCherry. The experiment was repeated independently three times (Bars, 1 µm). (E) Time-lapse chasing of TBC1D23-mCherry with FAM91A1-GFP D198R, in COS7 cells. For each group, the Top shows FAM91A1-GFP images, the Middle shows TBC1D23-mCherry images, and the Bottom shows the merged images of FAM91A1-GFP and TBC1D23-mCherry. The experiment was repeated independently three times (Bars, 1 µm).
The FAM91A1–TBC1D23 Interaction Is Critical for KIAA0319L Endosome-to-Golgi Trafficking.
Given that both TBC1D23 and the FAM91A1 complex are known to regulate protein trafficking from endosome to the TGN (26, 29), we sought to determine whether the FAM91A1–TBC1D23 interaction was required for the process. We chose KIAA0319L as our model cargo, which is transported to the TGN in an FAM91A1-dependent manner (29, 39, 40). Furthermore, the intracellular transport of KIAA0319L also depends on golgin-97 (39), a TBC1D23-interacting protein. By conducting internalization assays using an antibody against the extracellular region of KIAA0319L (41), we confirmed that KIAA0319L was transported from endosomes to the Golgi after internalization. KIAA0319L predominantly localized to the plasma membrane 2 min after internalization (SI Appendix, Fig. S6A). The colocalization between KIAA0319L and early endosome marker, EEA1, substantially decreased from 30 min to 60 min; in contrast, its colocalization with the Golgi increased, indicating that internalized KIAA0319L undergoes endosome-to-Golgi trafficking, similar to classical cargo proteins, such as CI-MPR (SI Appendix, Fig. S6).
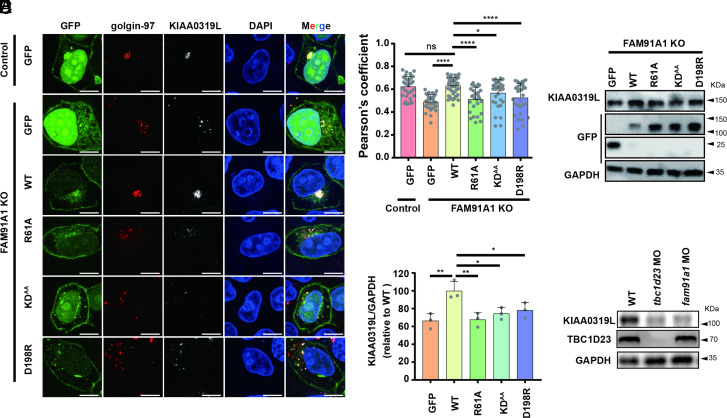
We generated TBC1D23 or FAM91A1 polyclonal knockout (KO) HeLa and HEK293T cells using CRISPR/Cas9 technology, which had a knockout efficiency of ~ 80% (SI Appendix, Fig. S7A). From these polyclonal KO cells, we obtained one monoclonal cell (KO-2) that was completely depleted of TBC1D23 or FAM91A1 (SI Appendix, Fig. S7B). Consistent with previous studies (29), FAM91A1 KO cells displayed a dispersed distribution of KIAA0319L relative to control cells. The colocalization between KIAA0319L and the TGN marker golgin-97 was significantly lower than control cells, as determined by immunofluorescence (SI Appendix, Fig. S7 C and D). To test whether the interaction between FAM91A1 and TBC1D23 was required for KIAA0319L trafficking, FAM91A1 KO cells were reexpressed with FAM91A1 WT and the TBC1D23-interacting deficient mutants (R61A, KDAA, D198R) (Fig. 5 A and B). FAM91A1 WT, but not the empty vector, efficiently rescued KIAA0319L missorting, as indicated by the colocalization of KIAA0319L and golgin-97 (Fig. 5 A and B). None of the three mutants supported the proper trafficking of KIAA0319L, suggesting that the FAM91A1–TBC1D23 interaction is required for the proper trafficking of KIAA0319L. Missorted proteins are often delivered to lysosomes for degradation. Indeed, we found that deletion of FAM91A1 led to decreased protein levels of KIAA0319L (Fig. 5 C and D). FAM91A1 WT, but none of the mutants, restored the protein levels of KIAA0319L (Fig. 5 C and D).
Fig. 5.
FAM91A1 residues interacting with TBC1D23 are required for endosome-to-Golgi trafficking of KIAA0319L. (A) Subcellular location of KIAA0319L in HeLa cells. The FAM91A1 KO cells were transfected with GFP, GFP-tagged FAM91A1 WT, R61A, KDAA, or D198R respectively. Cells were then incubated with antibodies against KIAA0319L (gray) and golgin-97 (red) (Scale bar, 10 μm). (B) Colocalization analysis between KIAA0319L and golgin-97 in A. Each dot represents Pearson’s correlation coefficients from one cell. Data are presented as mean ± SD, and P values were calculated using one-way ANOVA and Tukey’s multiple comparisons tests. Ns: not significant, *P < 0.05, 0.001 < ****P < 0.0001; the figure is representative of n = 3 independent experiments with similar results. (C) Immunoblot of whole-cell extracts showing that knockout of FAM91A1 decreased the total protein level of KIAA0319L in HEK293T cells. Transient expression of WT, but not these mutants or GFP vector, rescued the reduction. (D) The relative abundance of KIAA0319L compared to GAPDH was quantified in (E) and compared to the WT group. Data are presented as mean ± SD, and P values were calculated using one-way ANOVA and Tukey’s multiple comparisons tests. Ns: not significant, *P < 0.05, 0.05 < **P < 0.001; (E) Immunoblot of entire zebrafish tissue extracts showing that injection of tbc1d23 MO or fam91a1 MO decreased the protein level of KIAA0319L.
Similar to FAM91A1, knockout of TBC1D23 also altered the cellular distribution of KIAA0319L and decreased its protein level (SI Appendix, Fig. S8 A–D). These defects could be restored by TBC1D23 WT but not by the mutants deficient for FAM91A1 binding (Y530A, R531D, or F537A) (SI Appendix, Fig. S8 A–D). Depletion of TBC1D23 could lead to the missorting of KIAA0319L and enhanced degradation by the lysosome. Indeed, we found that the diminished KIAA0319L level could be partially rescued by the treatment of the lysosome inhibitor chloroquine (SI Appendix, Fig. S8 E and F). Similar to mammalian cells, the protein level of KIAA0319L was significantly decreased in both tbc1d23 and fam91a1 MO zebrafish (Fig. 5E). Since FAM91A1 and TBC1D23 knockout cells or mutant-expressing rescued HeLa cells only moderately impair endosome-to-Golgi trafficking of KIAA0319L, it is likely that the transport of KIAA0319L is regulated by additional proteins or trafficking routes.
To further confirm our conclusion, we took advantage of the previously developed mitochondria recruitment assay and localized TBC1D23 to the mitochondria’s outer membrane (42). TBC1D23 WT was able to redirect KIAA0319L from Golgi to mitochondria, while the empty vector or the FAM91A1-binding deficient mutants (Y530A, R531D, and F537A) failed to promote the translocation (SI Appendix, Fig. S9 A and B). Likewise, targeting TBC1D23 to mitochondria also leads to the redistribution of CI-MPR to the same organelle. The redistribution was absent in the cells expressing empty vector or the FAM91A1-binding deficient mutants (Y530A, R531D, and F537A) (SI Appendix, Fig. S9 C and D). Collectively, our results suggest that the FAM91A1–TBC1D23 interaction is necessary for the efficient transport of KIAA0319L and perhaps other cargo proteins from endosomes to the TGN.
The FAM91A1–TBC1D23 Interaction Predicts the Risk of PCH Due to TBC1D23 Mutations.
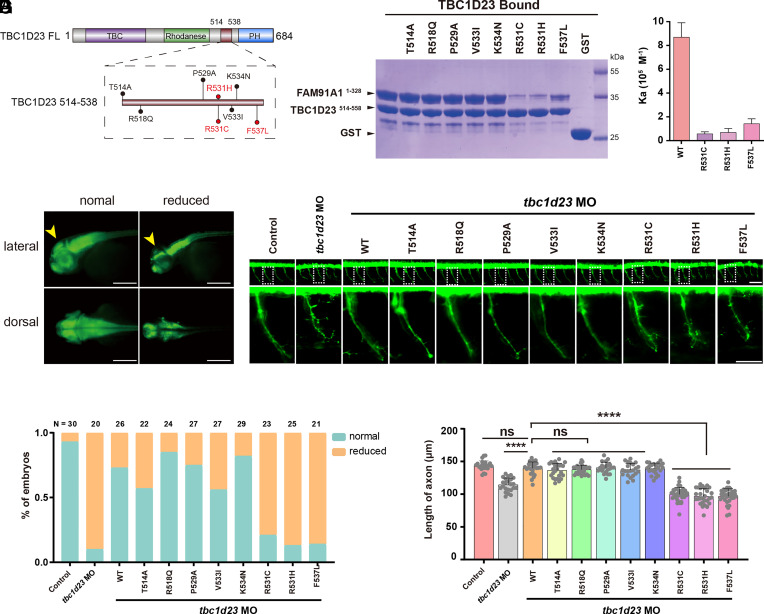
So far, our results showed that FAM91A1 and TBC1D23 coordinately regulated neuronal development, suggesting that disruption of the interaction between FAM91A1 and TBC1D23 could contribute to the development of PCH. To test this hypothesis, we conducted a comprehensive analysis of gene mutations across multiple databases, including gnomAD (43) and TCGA, and found 8 missense mutants (with occurrence ≥ 2) in the FAM91A1-contacting region of TBC1D23 (Fig. 6A). Using GST-pulldown assays, we found that TBC1D23 R531C, R531H, and F537L significantly reduced the FAM91A1 binding compared to WT, while other mutants (T514A, R518Q, P529A, V533I, and K534N) did not alter the binding (Fig. 6B). ITC experiments showed that the affinity between TBC1D23 R531C, R531H, and F537L and FAM91A1 was about 6.6%, 7.9%, and 16.4% that of WT, respectively (Fig. 6C).
Fig. 6.
The binding of FAM91A1 with TBC1D23 predicts the risk of PCH caused by TBC1D23 mutations. (A) Schematic representation of human TBC1D23 mutations within the FAM91A1 binding region, reported in the gnomAD and TCGA databases. (B) GST pull-down assays performed with GST-TBC1D23514–558 WT, mutants or GST, and purified FAM91A1N WT. Shown is the Coomassie blue-stained SDS-PAGE gel of bound samples. (C) ITC assays determining the affinity between TBC1D23514–538 WT or mutants and FAM91A1N. Association constants (Ka) are shown as mean ± SD. (D) HuC (green) expression in Tg [HuC: GFP] transgenic zebrafish. Classification of zebrafish embryos based on the expression level of HuC (elavl3) at 48 hpf. Reduced: decreased midbrain size. Top: lateral views; bottom: dorsal views (Scale bar, 100 μm). (E) Percentage of embryos in each group as defined in D. N presents the number of embryos used for statistical analysis. All experiments were performed in three times. (F) Morphology of CaP axons in Tg[Hb9:GFP]ml2 transgenic zebrafish at 48 hpf. Bottom: the enlarged views are shown at the Top. Control: control MO injection; tbc1d23 MO: tbc1d23 MO injection; tbc1d23 MO + WT: tbc1d23 MO and human TBC1D23 WT mRNA coinjection; tbc1d23 MO + T514A: tbc1d23 MO and TBC1D23 T514A mRNA coinjection; tbc1d23 MO + R518Q: tbc1d23 MO and TBC1D23 R518Q mRNA coinjection; tbc1d23 MO + P529A: tbc1d23 MO and TBC1D23 P529A mRNA coinjection; tbc1d23 MO + V533I: tbc1d23 MO and TBC1D23 V533I mRNA coinjection; tbc1d23 MO + K534N: tbc1d23 MO and TBC1D23 K534N mRNA coinjection; tbc1d23 MO + R531C: tbc1d23 MO and TBC1D23 R531C mRNA coinjection; tbc1d23 MO + R531H: tbc1d23 MO and TBC1D23 R531H mRNA coinjection; tbc1d23 MO + F537L: tbc1d23 MO and TBC1D23 F537L mRNA coinjection. All injections were performed at the one-cell stage of the Tg [Hb9: GFP]ml2 transgenic zebrafish embryos (Scale bar, 20 μm). (G) Statistical results of the length of CaP axons in embryos were treated as in F. For each group, ~20 to 30 axons from 6 to 12 Tg [Hb9: GFP]ml2 transgenic zebrafish embryos are scored. Experiments were repeated three times. ****P < 0.001, ns: not significant. P-values were calculated using one-way ANOVA, and Tukey’s multiple comparisons test. All experiments were performed in three times.
We have previously shown that the knockdown of TBC1D23 in zebrafish significantly decreased the expression of HuC in mature neurons and resulted in smaller midbrains (27, 28). To test the importance of the FAM91A1–TBC1D23 interaction in the midbrain development, we coinjected mRNA encoding TBC1D23 WT or mutants together with tbc1d23 MO. TBC1D23 WT and mutants with normal binding toward FAM91A1 (T514A, R518Q, P529A, V533I, and K534N) effectively rescued the reduced HuC expression and smaller midbrains (Fig. 6 D and E). However, TBC1D23 mutants (R531C, R531H, and F537L) defective in FAM91A1 binding were unable to restore the defects (Fig. 6 D and E). In the second assay, we took advantage of previous observations that depletion of TBC1D23 significantly reduced the axonal length of the CaP motor neurons. Importantly, we found that the abnormalities in the CaP motor neurons could be rescued by TBC1D23 WT or mutants with normal binding to FAM91A1, but not the mutants defective in binding to FAM91A1 (Fig. 6 F and G). Taken together, these results demonstrate that the interaction between TBC1D23 and FAM91A1 is critical for zebrafish neuronal development and brain growth. The pathogenic TBC1D23 mutations identified by our study could be used in genetic counseling and to predict the risk of PCH.
Discussion
Recent studies have identified WDR11 mutations in patients with PCH-like symptoms, including cerebellar hypoplasia and intellectual disability, suggesting that WDR11 could be a novel PCH-associated gene, although more clinical evidence is needed (31). Here, we demonstrate that the depletion of FAM91A1, which tightly associates with WDR11, causes similar phenotypes as that of TBC1D23 and recapitulates some important features of PCH. Furthermore, the complex structure between FAM91A1N and a TBC1D23 peptide illustrates the binding mechanism between the two proteins. Importantly, the interaction between FAM91A1 and TBC1D23 is informative to predict the risk of PCH associated with TBC1D23 mutations. Overall, our studies suggest that FAM91A1 and likely WDR11 represent unique PCH-associated genes and provide a new method to predict the risk of PCH.
In addition to the FAM91A1 complex and TBC1D23, mutations in at least three other genes regulating endosomal trafficking, VPS51 (8), VPS53 (9), and CHMP1A (10) have been linked with PCH. CHMP1A is primarily involved in the formation of multivesicular bodies and targeting of endosomal proteins to lysosomes (44). VPS51 and VPS53 are part of the GARP and EARP complexes, respectively, and also regulate retrograde transport of endosomes to the TGN (45, 46). Interestingly, individuals with mutations in VLDLR, a transmembrane receptor that undergoes endosome-to-plasma membrane recycling, exhibit clinical features that largely overlap with those of PCH patients (47). We show that loss of TBC1D23 and FAM91A1 significantly reduces the protein levels of KIAA0319L in cells and zebrafish, possibly due to impaired transport of KIAA0319L by TBC1D23 and FAM91A1. Moreover, KIAA0319L is highly conserved across multiple eukaryotic taxa and is known to regulate the growth of neuronal axons and dendritic spines during neuronal development (48–50). Further investigations are needed to determine whether the loss of KIAA0319L contributes to the development of PCH.
One of the outstanding questions in the endosomal trafficking field is how the FAM91A1 complex regulates protein endosome-to-TGN trafficking. Although FAM91A1 and TBC1D23 directly interact with each other (26) and cooperate to regulate the trafficking of some cargo proteins, such as KIAA0319L in this study, they appear to have distinct functions. For instance, Shin et al showed that the endosome-to-TGN transport of TGN46 required TBC1D23 but not the FAM91A1 complex (26). By establishing the interaction mechanism between FAM91A1 and TBC1D23, our studies allow to precisely separate their functions through specific mutations and lay a foundation to further illustrate their common and unique functions.
Methods
Antibodies and Plasmids.
DNA constructs and antibodies used in this paper were listed in SI Appendix, Tables S2 and S3, respectively.
Cloning, Expression, and Purification.
The TBC1D23–FAM91A1 chimera construct was cloned in the PGEX 4 T-1 vector, resulting an N-terminal cleavable GST tag, followed by TBC1D23 (residues 514–543), a 3 × GGS linker, and FAM91A1 (residues 1–328). The chimera construct was transformed into the BL21(DE3) cells, and the protein was expressed at 18 °C overnight after the addition of 0.5 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). The cells were harvested in lysis buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 1 mM DTT, and 1 mM PMSF), lysed by sonication, and cleared by centrifugation at 18,000g. The cleared supernatant was passed through glutathione-Sepharose beads and washed with lysis buffer. The bound proteins were subjected to tobacco etch virus (TEV) protease cleavage in a buffer containing 20 mM Tris pH 7.5, 200 mM NaCl, and 1 mM DTT. The elution was collected, concentrated, and further purified by anion-exchange and size exclusion chromatography (Superdex 200 column, GE Healthcare) in buffer containing 20 mM Tris, pH 7.5, 200 mM NaCl, and 0.5 mM TCEP.
Crystallization, Data Collection, and Structural Determination.
Crystals of the TBC1D23–FAM91A1 chimera protein were produced using the hanging-drop vapor-diffusion technique at 16 °C. The chimera protein was concentrated at 5 mg/mL and mixed with equal amounts of reservoir buffer containing 0.03 M Diethylene glycol, 0.03 M Triethylene glycol, 0.03 M Tetraethylene glycol, 0.03 M Pentaethylene glycol, 0.1 M PH8.5 Tris (base), 20% v/v Ethylene glycol, and 10% w/v PEG 8000. The crystals were cryo-protected in the reservoir buffer supplmented with 20% glycerol prior to their rapid freezing in liquid nitrogen. X-ray diffraction data were collected at Shanghai Synchrotron Radiation Facility beamline BL02U1, and the data collection statistics can be found in SI Appendix, Table S1.
The initial model of the FAM91A1–TBC1D23 complex was solved by molecular replacement using the FAM91A1 structure predicted by Alphafold2 as the searching model (51, 52). Model building and crystallographic refinement were conducted with COOT and PHENIX software (53, 54), respectively. The final FAM91A1–TBC1D23 complex structure was solved at 2.51 Å (SI Appendix, Table S1) and deposited into the Protein Data Bank with the accession entry of 8JJ9.
ITC.
ITC experiments were performed with a Microcal PEAQ-ITC instrument, following established protocols (55, 56). The proteins and peptides used in the experiments were first dialyzed to the ITC buffer (100 mM HEPES, pH 7.5, 200 mM NaCl). The TBC1D23 peptides (320 µM) were subsequently titrated into the FAM91A1N protein (20 µM) at 25 °C. Each experiment was repeated at least three times. The data obtained from the experiments were analyzed using Microcal PEAQ-ITC analysis software.
Pull-Down Experiments.
GST pull-down experiments were carried out as previous studies (57, 58). In short, 20 µg of GST or GST-tagged protein, 400 µg of bait protein, glutathione-sepharose beads (30 µL) was mixed in 0.5 mL of the pull-down buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 0.005% Triton-X100). After incubation at 4 °C for 1 h, the beads were washed three times with the pull-down buffer and separated on a 12% SDS-PAGE gel.
High-Resolution Time-Lapse Imaging.
High-resolution time-lapse imaging were carried out as previous studies (37). Briefly, HeLa cells were cultured in 24-well plates with glass bottoms (Corning) and then transfected with plasmids encoding FAM91A1-GFP and TBC1D23-mCherry. Twenty-four hours after transfection, images were collected by a confocal microscope (ScanR, Olympus) with a 100x oil objective. Images were taken every 30 s for a total of 30 min.
Generation of TBC1D23 and FAM91A1 Knockout and Rescued Cell Lines.
TBC1D23 and FAM91A1 knockout cells were generated according to previous studies (59, 60). The TBC1D23 target guide RNA 5′-CTGCCAACGTCGAGCGGCGA-3′ and the FAM91A1 target guide RNA 5′-GTTGCCGGCCAACGTGAGAC-3′ was inserted in the plasmid hSpCas9. HEK293T cells were seeded into 10-cm dishes for 24 h and transfected with hSpCas9 and helper plasmids (pRSV-Rev, pMDLg/pRRE, and pMD2.G) using liposomal transfection reagent (Yeasen, 40802ES03). Forty-eight hours later, the resultant virus was collected to infect HeLa and HEK293T cells, respectively. Cells infected with the virus were cultured in the full medium for 48 h, screened for two additional passages in a medium containing 2 µg/mL puromycin, and the knockdown efficiency was determined by immunoblotting. Monoclonal cells were diluted by dividing about 100 cells in a 96-well plate and replaced with fresh medium every 2 d for about 2 wk. Single colonies were expanded and selected by immunoblotting. TBC1D23 and FAM91A1 cDNAs used in this study were codon-optimized and was not recognized by the respective guide RNA.
Cell Culture, Immunofluorescence Staining, and Confocal Microscopy.
HEK293T, HeLa, and COS7 cell lines were cultured in DMEM medium supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. For immunofluorescence experiments, cells were preseeded at the bottom of the 24- or 12-well plates with glass slides and were transfected with Liposomal Transfection Reagent (Yeasen, 40802ES03) following the manufacturer’s instructions. After fixation with 4% formaldehyde in PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. The cells were then washed three times with PBS and blocked with 5% BSA in PBS for 1 h at room temperature. The primary antibody was diluted in 1% BSA in PBS and incubated with the cells overnight at 4 °C. The cells were washed three times with PBS and then incubated with the appropriate fluorescently labeled secondary antibody diluted in 1% BSA in PBS for 1 h at room temperature. After washing with PBS, the nuclei were stained with DAPI. Confocal images were acquired using either the Olympus FV-3000 or Olympus scanR confocal microscope and were analyzed using NIH ImageJ software. All experiments were repeated at least three times to ensure reproducibility.
KIAA0391L Internalization and Trafficking Assay.
KIAA0391L internalization and trafficking assays were performed according to the previous study (41). In brief, HeLa cells were initially incubated at 4 °C with anti-KIAA0319L antibodies (approximately 25 μg/mL) for 1 h. Subsequently, cells were subjected to three washes with 1 × PBS and then shifted to 37 °C to allow internalization. At 2-, 30-, and 60-min, cells were fixed using 4% PFA for 15 min. Following fixation, immunofluorescence staining was carried out to visualize the process of KIAA0319L trafficking All data presented are representative of three independent experiments.
Zebrafish, Morpholino, and mRNA Injections.
Zebrafish (D. rerio) experiments were conducted following the animal ethical committee of West China Hospital guidelines and standard procedures. Antisense morpholino oligonucleotides (MO) were purchased from GeneTools and were utilized following previously published protocols. The tbc1d23 MO (CTTCCCCTACAGCATCCGCCATTGC) was used in our previous study, which effectively knocked down the TBC1D23 expression. The sequence of fam91a1 MO (GAGAGAACGACTTACAGTCAGCAGC) was designed by GeneTools and verified by qPCR. Injections of MO and mRNAs were performed at the one-cell stage, with 5 ng of tbc1d23 MO and 5 ng of fam91a1 MO and/or 50–100 ng of mRNA administered per injection into the yolk and cell. The experiments were repeated at least three times to ensure reproducibility and the resulting samples were imaged using a focal microscope. Image analysis was conducted using NIH ImageJ software and ZEN 3.1 software.
Total RNA Isolation and Semiquantitative RT-PCR.
RNA isolation and semiquantitative RT-PCR were performed according to previously published methods (61). To extract RNA from zebrafish embryos, 50 embryos at 48 h postfertilization were collected, homogenized, and total RNA was isolated using the RNeasy Plant Mini Kit (FOREGRNE). The extracted RNA was reverse transcribed using the Prime Script Reverse-transcription PCR kit (TaKaRa DRR014A) to synthesize cDNA, which was then used as a template for PCR reactions using the Real Master Mix Kit (Roche).
Statistical Analysis.
All experiments involving zebrafish and cells were conducted a minimum of three times to ensure the accuracy and reliability of the results. Statistical analyses were performed using Prism 8.3.0 software (GraphPad) with one-way ANOVA and Tukey’s multiple comparisons tests. Significance was defined as P < 0.05 (*), P < 0.01 (**), or P < 0.001 (***), while nonsignificant findings were represented as “ns.” This approach ensured the validity and robustness of the statistical analyses and the accuracy of the reported results.
Supplementary Material
Appendix 01 (PDF)
Appendix 02 (PDF)
Acknowledgments
We thank members of our laboratory for helpful discussions. This work was supported by National Key Research and Development Program of China (2022YFA1105200), Natural Science Foundation of China (NSFC #92254302), NSF for Distinguished Young Scholars (#32125012), and NIH grant DK107733 to D.D.B.
Author contributions
L.Z., H.D., and D.J. designed research; L.Z., H.D., Q.Y., Y.T., J.Z., P.L., and T.L. performed research; L.Z., Q.Y., P.L., S.Z., X.Y., D.D.B., and D.J. analyzed data; and L.Z., H.D., and D.J. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Structure factor and atomic coordinates data have been deposited in Protein Data Bank with accession code 8JJ9 (62). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Rudnik-Schoneborn S., Barth P. G., Zerres K., Pontocerebellar hypoplasia. Am. J. Med. Genet. C, Semin. Med. Genet. 166C, 173–183 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Rusch C. T., Bolsterli B. K., Kottke R., Steinfeld R., Boltshauser E., Pontocerebellar hypoplasia: A pattern recognition approach. Cerebellum 19, 569–582 (2020). [DOI] [PubMed] [Google Scholar]
- 3.van Dijk T., Baas F., Barth P. G., Poll-The B. T., What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet J. Rare Dis. 13, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Accogli A., Addour-Boudrahem N., Srour M., Diagnostic approach to cerebellar hypoplasia. Cerebellum 20, 631–658 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Ammann-Schnell L., Groeschel S., Kehrer C., Frolich S., Krageloh-Mann I., The impact of severe rare chronic neurological disease in childhood on the quality of life of families—A study on MLD and PCH2. Orphanet J. Rare Dis. 16, 211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanova E. L., et al. , Homozygous truncating variants in TBC1D23 cause pontocerebellar hypoplasia and alter cortical development. Am. J. Hum. Genet. 101, 428–440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin-Valencia I., et al. , Homozygous mutations in TBC1D23 lead to a non-degenerative form of pontocerebellar hypoplasia. Am. J. Hum. Genet. 101, 441–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gershlick D. C., et al. , A neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes. Hum. Mol. Genet. 28, 1548–1560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinstein M., et al. , VPS53 mutations cause progressive cerebello-cerebral atrophy type 2 (PCCA2). J. Med. Genet. 51, 303–308 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Mochida G. H., et al. , CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat. Genet. 44, 1260–1264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu Y., Zhao L., Billadeau D. D., Jia D., Endosome-to-TGN trafficking: Organelle–vesicle and organelle–organelle interactions. Front. Cell Dev. Biol. 8, 163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., et al. , Endosomal receptor trafficking: Retromer and beyond. Traffic 19, 578–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen P. J., Steinberg F., To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Yong X., Billadeau D. D., Jia D., All ways lead to Rome: Assembly of retromer on membranes with different sorting nexins. Signal Transduction Targeted Ther. 6, 139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yong X., et al. , Targeting endosomal recycling pathways by bacterial and viral pathogens. Front. Cell Dev. Biol. 9, 648024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong X., et al. , Mechanism of cargo recognition by retromer-linked SNX–BAR proteins. PLoS Biol. 18, e3000631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yong X., et al. , SNX27-FERM-SNX1 complex structure rationalizes divergent trafficking pathways by SNX17 and SNX27. Proc. Natl. Acad. Sci. U.S.A. 118, e2105510118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qureshi Y. H., et al. , The neuronal retromer can regulate both neuronal and microglial phenotypes of Alzheimer’s disease. Cell Rep. 38, 110262 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simoes S., et al. , Tau and other proteins found in Alzheimer’s disease spinal fluid are linked to retromer-mediated endosomal traffic in mice and humans. Sci. Transl. Med. 12, eaba6334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daly J. L., et al. , Multi-omic approach characterises the neuroprotective role of retromer in regulating lysosomal health. Nat. Commun. 14, 3086 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P. K., Muqit M. M. K., Parkinson’s: A disease of aberrant vesicle trafficking. Annu. Rev. Cell Dev. Biol. 36, 237–264 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Rahman A. A., Morrison B. E., Contributions of VPS35 mutations to Parkinson’s disease. Neuroscience 401, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka N., Kyuuma M., Sugamura K., Endosomal sorting complex required for transport proteins in cancer pathogenesis, vesicular transport, and non-endosomal functions. Cancer Sci. 99, 1293–1303 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei Y., et al. , RAB21 controls autophagy and cellular energy homeostasis by regulating retromer-mediated recycling of SLC2A1/GLUT1. Autophagy 19, 1070–1086 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X., et al. , A thiazole-derived oridonin analogue exhibits antitumor activity by directly and allosterically inhibiting STAT3. J. Biol. Chem. 294, 17471–17486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin J. J. H., Gillingham A. K., Begum F., Chadwick J., Munro S., TBC1D23 is a bridging factor for endosomal vesicle capture by golgins at the trans-Golgi. Nat. Cell Biol. 19, 1424–1432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W., et al. , Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH. Proc. Natl. Acad. Sci. U.S.A. 116, 22598–22608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., et al. , Structure of TBC1D23 N-terminus reveals a novel role for rhodanese domain. PLoS Biol. 18, e3000746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro Negredo P., Edgar J. R., Manna P. T., Antrobus R., Robinson M. S., The WDR11 complex facilitates the tethering of AP-1-derived vesicles. Nat. Commun. 9, 596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeffer S. R., Entry at the trans-face of the Golgi. Cold Spring Harb. Perspect. Biol. 3, a005272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haag N., et al. , Biallelic loss-of-function variants in WDR11 are associated with microcephaly and intellectual disability. Eur. J. Hum. Genet. 29, 1663–1668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y. J., et al. , WDR11-mediated Hedgehog signalling defects underlie a new ciliopathy related to Kallmann syndrome. EMBO Rep. 19, 269–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H. C., et al. , Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci. Lett. 279, 81–84 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Kim C. H., et al. , Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216, 109–112 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutateladze T. G., Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta 1761, 868–877 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu N., Liu K., Yang C., WDR91 specifies the endosomal retrieval subdomain for retromer-dependent recycling. J. Cell Biol. 221, e202203013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu L., Tai G., Wu M., Song H., Hong W., Multilayer interactions determine the Golgi localization of GRIP golgins. Traffic 7, 1399–1407 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Shin J. J. H., et al. , Spatial proteomics defines the content of trafficking vesicles captured by golgin tethers. Nat. Commun. 11, 5987 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer N. L., Chapman M. S., Adeno-associated virus (AAV) cell entry: Structural insights. Trends Microbiol. 30, 432–451 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pillay S., et al. , An essential receptor for adeno-associated virus infection. Nature 530, 108–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong M., Munro S., Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science 346, 1256898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gudmundsson S., et al. , Variant interpretation using population databases: Lessons from gnomAD. Hum. Mutat. 43, 1012–1030 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stauffer D. R., Howard T. L., Nyun T., Hollenberg S. M., CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell Sci. 114, 2383–2393 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Schindler C., Chen Y., Pu J., Guo X., Bonifacino J. S., EARP is a multisubunit tethering complex involved in endocytic recycling. Nat. Cell Biol. 17, 639–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Victoria F. J., et al. , Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol. Biol. Cell 21, 3386–3395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valence S., et al. , RELN and VLDLR mutations underlie two distinguishable clinico-radiological phenotypes. Clin. Genet. 90, 545–549 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Carrion-Castillo A., Franke B., Fisher S. E., Molecular genetics of dyslexia: An overview. Dyslexia 19, 214–240 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Dennis M. Y., et al. , A common variant associated with dyslexia reduces expression of the KIAA0319 gene. PLoS Genet. 5, e1000436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peschansky V. J., et al. , The effect of variation in expression of the candidate dyslexia susceptibility gene homolog Kiaa0319 on neuronal migration and dendritic morphology in the rat. Cereb. Cortex. 20, 884–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mirdita M., et al. , ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D, Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng L., et al. , Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis. Nat. Struct. Mol. Biol. 28, 1–12 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Yong X., et al. , Cryo-EM structure of the Mon1-Ccz1-RMC1 complex reveals molecular basis of metazoan RAB7A activation. Proc. Natl. Acad. Sci. U.S.A. 120, e2301725120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao J., et al. , Mechanism of inhibition of retromer transport by the bacterial effector RidL. Proc. Natl. Acad. Sci. U.S.A. 115, E1446–E1454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin J., et al. , Structural and mechanistic insights into secretagogin-mediated exocytosis. Proc. Natl. Acad. Sci. U.S.A. 117, 6559–6570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao L., et al. , Phosphorylation of SNX27 by MAPK11/14 links cellular stress-signaling pathways with endocytic recycling. J. Cell Biol. 220, e202010048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao L., et al. , SARS-CoV-2 spike protein harnesses SNX27-mediated endocytic recycling pathway. MedComm 2, 798–809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z., et al. , SCGN deficiency is a risk factor for autism spectrum disorder. Signal Transduction Targeted Ther. 8, 3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng H. Q., Zhang S. T., Jia D., Human FAM91A1 N terminal domain in complex with TBC1D23. Protein Data Bank (PDB). rcsb.org/structure/unreleased/8JJ9. Deposited 29 May 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Appendix 02 (PDF)
Data Availability Statement
Structure factor and atomic coordinates data have been deposited in Protein Data Bank with accession code 8JJ9 (62). All other data are included in the manuscript and/or supporting information.