Significance
The IL-1 family member IL-38 has been characterized primarily as an antiinflammatory cytokine for systemic diseases. Here, we describe a central role of IL-38 in driving intestinal stem cell differentiation through the upregulation of WNT3a and IL-1β. Our findings reveal a dual role of IL-38 in regulating intestinal functions; a) in resting conditions, IL-38 maintains intestinal homeostasis, driving WNT3a production and organoid budding, whereas b) in highly inflamed conditions, IL-38 contributes to proper recovery, by exerting antiinflammatory activities. Thus, we demonstrate a pivotal role of IL-38 in driving tissue turnover and maintenance of homeostasis in intestinal health.
Keywords: inflammation, interleukin, gastroenterology
Abstract
The IL-1 Family member IL-38 has been characterized primarily as an antiinflammatory cytokine in human and mouse models of systemic diseases. Here, we examined the role of IL-38 in the murine small intestine (SI). Immunostaining of SI revealed that IL-38 expression partially confines to intestinal stem cells. Cultures of intestinal organoids reveal IL-38 functions as a growth factor by increasing organoid size via inducing WNT3a. In contrast, organoids from IL-38-deficient mice develop more slowly. This reduction in size is likely due to the downregulation of intestinal stemness markers (i.e., Fzd5, Ephb2, and Olfm4) expression compared with wild-type organoids. The IL-38 binding to IL-1R6 and IL-1R9 is still a matter of debate. Therefore, to analyze the molecular mechanisms of IL-38 signaling, we also examined organoids from IL-1R9-deficient mice. Unexpectedly, these organoids, although significantly smaller than wild type, respond to IL-38, suggesting that IL-1R9 is not involved in IL-38 signaling in the stem cell crypt. Nevertheless, silencing of IL-1R6 disabled the organoid response to the growth property of IL-38, thus suggesting IL-1R6 as the main receptor used by IL-38 in the crypt compartment. In organoids from wild-type mice, IL-38 stimulation induced low concentrations of IL-1β which contribute to organoid growth. However, high concentrations of IL-1β have detrimental effects on the cultures that were prevented by treatment with recombinant IL-38. Overall, our data demonstrate an important regulatory function of IL-38 as a growth factor, and as an antiinflammatory molecule in the SI, maintaining homeostasis.
The cytokine IL-38 belongs to the IL-1 family (1). It is encoded by the Il1f10 gene and is expressed in infiltrating immune cells of colonic samples from patients with inflammatory bowel disease (IBD), particularly CD19+ B cells (2) and CD123+ cells (3). IL-38 is mostly studied as an antiinflammatory cytokine (1, 4) and exerts protective functions in several organs, suggesting that this protein mainly functions in the maintenance of organ homeostasis (1). As demonstrated by van de Veerdonk et al., IL-38 has biological effects in immune cells that resemble IL-36Ra functions, downregulating the levels of IL-22, IL-17, and IFNγ in Candida-stimulated Th17 cells (5). A recent study by The et al. also demonstrated that IL-38 dampens aortic valve calcification, probably dependent on IL-1R9 (encoded by Il1rapl1 gene), inhibiting Nucleotide-binding oligomerization domain-containing protein-like receptor (NLR) family pyrin domain containing 3 (NLRP3) and IL-1β maturation (6). Moreover, Il1f10-deficient mice exhibit higher levels of inflammation when exposed to DSS treatment compared to wild-type mice and express higher levels of NLRP3 and caspase-1 (7). Regarding IBD, IL-38 is expressed in colon tissue of ulcerative colitis patients compared to healthy controls (8) and reduces inflammation in the colon (2).
Although most studies demonstrate that IL-38 has antiinflammatory properties by, for example, blocking IL-1 b maturation, the biological functions of IL-38 are more complex than only inhibiting the inflammatory response and some in vivo studies demonstrate that this cytokine favors disease progression. For instance, as shown in Huard et al. (9), IL-38 ablation in mice ameliorates autoimmune encephalomyelitis leading to the downregulation of inflammation markers like Mertk, Tnfa, Ptgs2, Tgfb1, and Tgfb2 (9). Additionally, experiments performed on mouse and A431 human epidermoids cancer cell line reveal that IL-38 has a protumorigenic function and can stimulate cancer cell proliferation using an IL-1R6-dependent mechanism (10). Moreover, as regards keratinocytes differentiation, Mermoud et al. (11) have recently demonstrated that IL-38 is expressed in primary keratinocytes as well as in the N/TERT1 keratinocyte cell line. Notably, the overexpression of IL-38 promotes the differentiation of these cells, inhibiting proliferation (11).
Another debated aspect of IL-38 biology relates to the identity of its receptor. As previously proposed by van de Veerdonk et al. (5), IL-38 binds IL-36R (IL-1R6) which is encoded by the Il1rl2 gene. Other evidence regarding the interaction of IL-38 with IL-1R6 comes from Mora et al. (12), who demonstrated that IL-38 binds either IL-1R6 or IL-1R9 (12). Additionally, pulldown experiments performed by Zhou et al. (10) support the IL-38–IL-1R6 interaction. IL-38 may also bind IL-1R1 and the affinity of interaction depends on the truncation of the N-terminal amino acids of IL-38 (13).
Given the emerging importance of a therapeutic role for IL-38 (14–16) and the function of this cytokine in the intestine (2, 7, 8, 17), here we analyzed the effects of IL-38 in intestinal stem cells (ISCs) in organoid cultures derived from wild-type and Il1f10 mice. Moreover, to better dissect the mechanism of action of IL-38 in the gut, we employed Il1rapl1- deficient mice and observed that IL-1R9 is not essential for a response to IL-38. Additionally, silencing experiments highlight the role of IL-1R6 in IL-38 signaling. Moreover, we presented that IL-38 induces low concentrations of IL-1β. This finding, confirmed in Il1r1 and Nlrp3- deficient mice, reveals that homeostatic levels of IL-1β are beneficial for organoids.
Results
Il1f10-Deficient SI Shows Lower Expression of Intestinal Stem Cell Markers.
Located in the intestinal crypts, ISCs represent the precursor cells of the entire intestine (18). These cells regulate homeostasis of the intestine, producing soluble factors that affect the fate of the intestine. The crypts are divided into two zones: the stem cell zone and the transient applying zone (TA). The stem cell zone contains the crypt base columnar cells that act as ISCs, as well as Paneth cells that protect and feed the ISCs (19). The TA zone is composed by lineage-committed stem cells, which rapidly divide and differentiate; the mature cell zone is populated by large numbers of epithelial cells (19). To determine these areas, cell markers have been identified to show differentiated and undifferentiated cells. Historically, the most important marker of ISCs is LGR5 (18, 20–22), a G-protein-coupled receptor and a target of the WNT pathway (23). As demonstrated by Carmon et al., LGR5 binds R-spondins for enhancing the activation of the WNT/β-catenin signaling in WNT-responsive cells (24). The WNT/β-catenin axis is fundamental for crypt homeostasis and maintains ISCs in an undifferentiated state (21); however, WNT/β-catenin signaling can also drive the differentiation of ISCs in Paneth cells (25).
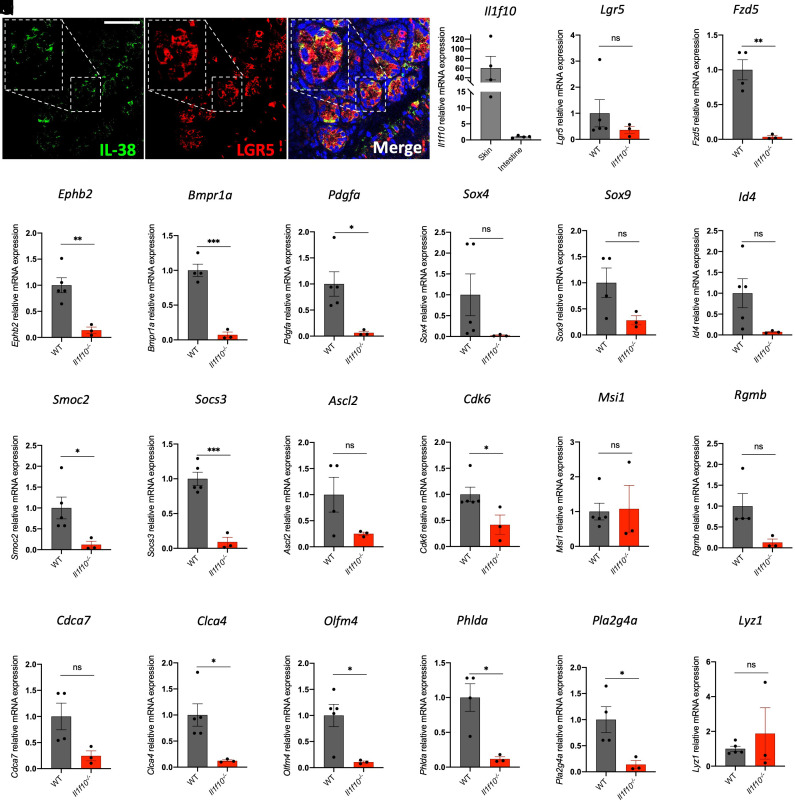
IL-38 is an antiinflammatory cytokine that can protect the organism challenged with highly inflammatory triggers, as recently observed in dextran sodium sulfate colitis or aortic calcification models (6, 7). We sought to confirm whether IL-38 is expressed in the mouse intestine (8) and if the IL-38 protein is expressed in the small intestine (SI) at the level of the stem cell crypts. As shown in Fig. 1A and SI Appendix, Fig. S1G, IL-38 staining colocalizes with LGR5 staining, revealing that IL-38 is expressed in stem cell crypts and suggesting a role of this cytokine in ISCs. IL-38 gene (Il1f10) expression in the SI is markedly lower when compared to a high Il1f10-expressing tissue like skin, probably because intestinal IL-38 expression is specifically localized in a small SI stem cell crypt subpopulation (Fig. 1B).
Fig. 1.
IL-38 is essential for intestinal crypt gene expression. (A) Staining of IL-38 (green) and LGR5 (red) in wild-type SI. An enlargement of one intestinal crypt is shown in dashed frame. (Scale bar, 50 μm.) (B) gene expression of Il1f10 in skin and intestine of wild type mice. (C–V) gene expression analysis of Lgr5 (C), Fzd5 (D), Ephb2 (E), Bmpr1a (F), Pdgfa (G), Sox4 (H), Sox9 (I), Id4 (J), Smoc2 (K), Socs3 (L), Ascl2 (M), Cdk6 (N), Msi1 (O), Rgmb (P), Cdca7 (Q), Clca4 (R), Olfm4 (S), Phlda (T), Pla2g4a (U), and Lyz1 (V) in SIs of wild-type and Il1f10-deficient mice. Comparisons between WT and Il1f10 knockout samples were performed using Student t test. *P < 0.05; **P < 0.01, ***P < 0.001, ns = not significant. Mean ± SEM.
To confirm the putative contribution of IL-38 to homeostasis of ISCs, we analyzed the intestinal histology and compared wild type with Il1f10−/− mice. Longitudinal histological sections shown in SI Appendix, Fig. S1 A–F did not reveal overt structural differences between the two experimental groups in healthy conditions. Therefore, we decided to analyze whether some intestinal fundamental pathways are transcriptionally impaired in the knockouts compared to the wild type. First, we measured the level of expression of stem cell niche markers (taken from refs. 26 and 27) in SIs samples from wild-type and Il1f10-deficient mice. Among the markers analyzed, Fzd5, Ephb2, Bmpr1a, Clca4, Pdgfa, Olfm4, Smoc2, Socs3, Cdk6, Phlda, and Pla2g4a are significantly down-regulated in Il1f10-deficient mice compared to wild type (Fig. 1 C–W), demonstrating the pivotal role of IL-38 in intestinal stem cell niche gene expression. These findings are further corroborated by western blot of intestinal extracts against LGR5. As shown in SI Appendix, Fig. S1 H and I, LGR5 expression is significantly reduced in Il1f10-deficient intestines compared to wild type. We then decided to analyze the expression levels of other genes involved in intestinal regeneration (28, 29) such as Clu, Ly64, Anxa8, Ereg, Rxna, Il33, Tacstd2, and Mlsn in wild-type intestines compared to Il1f10−/−-deficient ones. Among these transcripts, only Ly64 resulted up-regulated in IL-38 knockout samples, suggesting for an impairment of intestinal barrier functions due to IL-38 loss (SI Appendix, Fig. S2 A–H).
Additionally, we decided to analyze the level of expression of several IL-encoding genes, to see whether IL-38 deficiency leads to an impairment in ILs expression. As reported in SI Appendix, Fig. S3 A–E, Il36a, Il36b, Il36g, Il36rn, and Il1b (encoding, respectively, for IL-36α, IL-36β, IL-36γ and IL-36RA and IL-1β) are not differentially expressed in wild-type intestines when compared to Il1f10 knockout. We also measured the intracellular levels of IL-β in the intestines from wild-type and Il1f10-deficient mice and we did not observe statistical differences between the two experimental groups (SI Appendix, Fig. S3F). To confirm that IL-38 does not influence the expression of IL-36 genes, we injected 1 μg of recombinant moue IL-38 (3 to 152 residues) in wild-type mice every day for 15 d. At the end of the experiment, we collected the intestines from Phosphate buffer saline (PBS)-injected controls (Vehicle) and from IL-38-injected animals and we measured the expression of Il36a, Il36b, Il36g, and Il36rn. As reported in SI Appendix, Fig. S4 A–D, IL-38 injection did not affect the expression of these genes, suggesting that, at least in this context, IL-38 does not induce IL-36 gene expression.
IL-38 Is a Growth Factor in Intestinal Organoids.
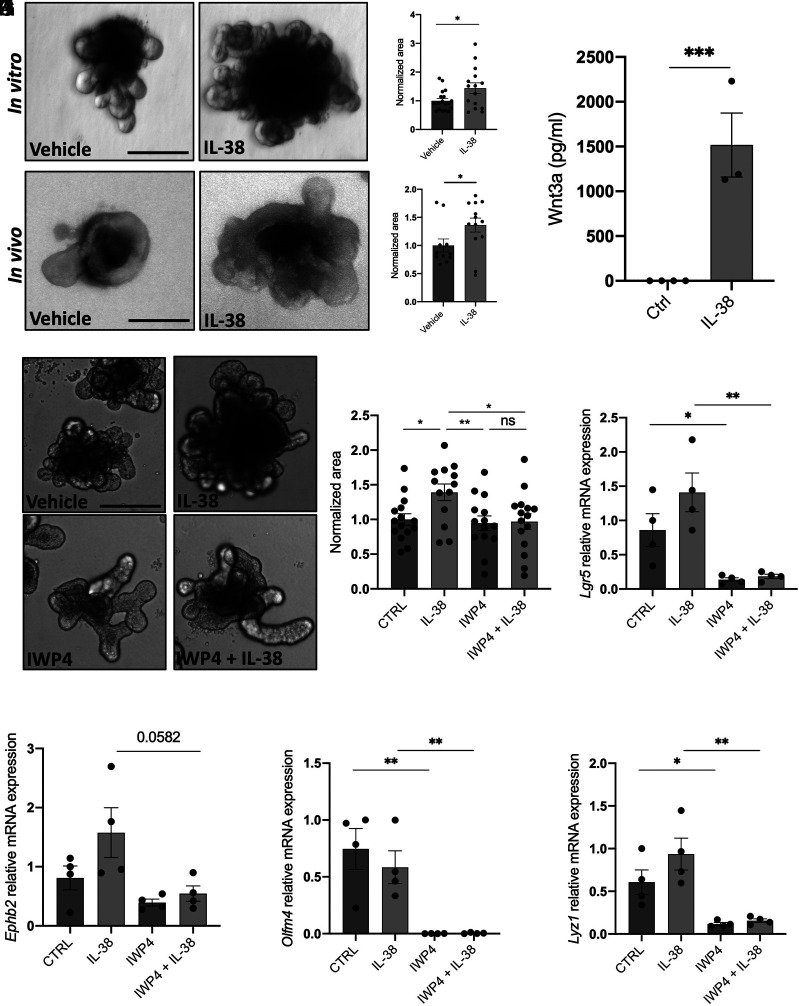
To test the functions of IL-38 ex vivo, we generated organoid cultures from wild-type SI. Organoids were stimulated with recombinant mouse IL-38 (3 to 152 residues) at three different concentrations. Interestingly, 2 ng/mL and 200 ng/mL did not affect organoid size (SI Appendix, Fig. S4 E–H) (P = 0.2314 and 0.9996, respectively), whereas 20 ng/mL of IL-38 increased the size of organoids (Fig. 2 A and B) (P = 0.0239). Additionally, we injected 1 μg recombinant mouse IL-38 in wild-type mice every day for 15 d and collected SI to generate organoid cultures. Notably, organoids obtained from IL-38-injected mice are larger than vehicle-injected mice (P = 0.0459), confirming that IL-38 can function as a growth factor in healthy in vivo intestines (Fig. 2 C and D).
Fig. 2.
IL-38 works as a growth factor stimulating WNT3a secretion. (A) Representative pictures and (B) measurement of wild-type organoids treated with 20 ng/mL IL-38 for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between control and IL-38-treated organoids were performed using Student t test. (C) Representative pictures and (D) measurements of wild-type organoids obtained from vehicle- and IL-38-injected mice. PSB or 1 μg recombinant murine IL-38 was intraperitoneally injected every day for 15 d. Each dot represents an individual organoid, organoid cultures were produced from 5 different mice per condition and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between control and IL-38-treated organoids were performed using Student t test. (E) WNT3a expression in the supernatants in wild-type organoids treated with 20 ng/mL IL-38 for 6 d. Comparisons between control and IL-38-treated organoids were performed using Student t test. (F) Representative pictures and (G) measurement of wild-type organoids treated with vehicle, 20 ng/mL IL-38, 5 μM IWP4, and 5 μM IWP4 +20 ng/mL IL-38. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between the four experimental groups were performed using one-way ANOVA. (H–K) Gene expression of Lgr5 (H), Ephb2 (I), Olfm4 (J), and Lyz1 (K) in wild-type organoids treated with vehicle, 20 ng/mL IL-38, 5 μM IWP4, and 5 μM IWP4 +20 ng/mL IL-38. *P < 0.05; **P < 0.01, ***P < 0.001, ns = not significant. Mean ± SEM.
WNT3a is the major soluble factor produced by Paneth cells that drives ISCs differentiation (24). To understand how IL-38 affects organoid growth, we measured the levels of WNT3a in organoids. As shown in Fig. 2F, we observed a significant increase with IL-38 stimulation (1,500-fold increase, P < 0.001) (Fig. 2E). To confirm whether the increase of organoid size depends on IL-38-induced WNT3a secretion, we treated organoids with the WNT inhibitor IWP4 (30) either with or without IL-38 stimulation. As expected, IL-38 stimulated organoid growth (P = 0.0082), whereas IWP4 did not affect organoid size compared to vehicle yet changed the organoid shape (Fig. 2 F and G). Moreover, organoids treated with IL-38 and IWP4 combination do not show significant differences when compared to either vehicle (P = 0.9962) or IWP4-treated (P = 9988) organoids but are significantly smaller compared to IL-38-treated organoids (Fig. 2 F and G). Furthermore, we observed the same phenotype in organoids treated with IWP4 and IL-38+IWP4 (Fig. 2 F and G). These data suggest that IL-38 stimulates organoid growth by inducing WNT3a production and that these effects are dependent on the WNT pathway. To confirm the actual inhibition of WNT pathway, we also measured the expression of WNT and stemness marker genes and we observed that Lgr5, Lyz1, and Olfm4 are significantly down-regulated in IWP4-treated organoids compared to control and IL-38-treated organoids (Fig. 2 H–K).
Il1f10-Deficient (Il1f10−/−) Organoids Show Growth Defects.
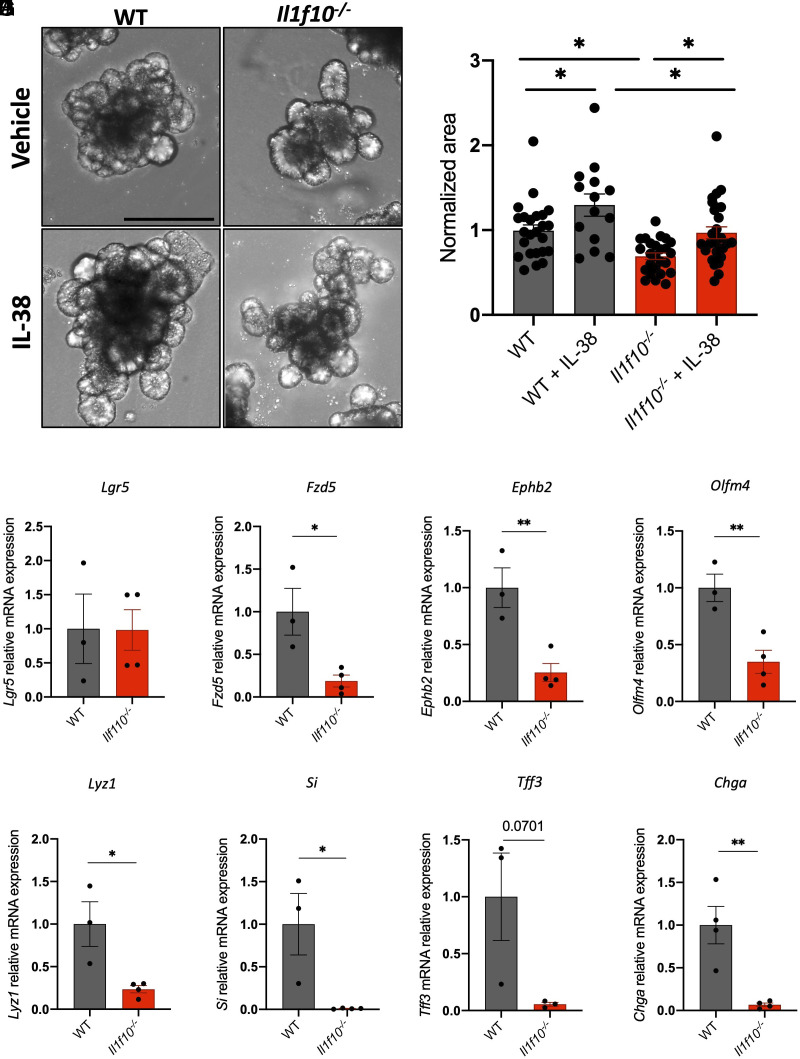
Organoid cultures from wild-type and IL-38-deficient mice (Il1f10−/−) were evaluated for growth. IL-38-deficient organoids are significantly smaller than wild-type organoids (P = 0.0135) and, as expected, 20 ng/mL recombinant IL-38 significantly increased the size of either wild-type (P = 0.0497) or IL-38-deficient organoids (0.0210) (Fig. 3 A and B).
Fig. 3.
Il1f10-deficient organoids show growth defects. (A) Representative pictures and (B) measurements of wild-type and Il1f10−/− organoids treated either with vehicle or 20 ng/mL IL-38 for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between the four experimental groups were performed using one-way ANOVA. (C–J) Gene expression analysis of Lgr5 (C), Fzd5 (D), Ephb2 (E), Olfm4 (F), Lyz1 (G), Si (H), Tff3 (I), and Chga (J) in wild-type and Il1f10−/− organoids. Comparisons between WT and Il1f10 knockout samples were performed using Student t test. *P < 0.05; **P < 0.01, ****P < 0.0001. Mean ± SEM.
Interestingly, we observed a marked downregulation of stemness markers in Il1f10 knockout organoids compared to wild type. Lgr5 does not show significant differences between Il1f10−/− and wild-type organoids, whereas Fzd5, Ephb2, and Olfm4 are significantly down-regulated in Il1f10-deficient organoids when compared to wild type (P = 0.9763, 0.0208, 0.0076, and 0.0091, respectively) (Fig. 3 C–F).
To further characterize Il1f10−/− organoids, we measured the levels of expression of markers of other cell types such as Lyz1 (Paneth cells), Si (absorptive cells), Tff3 (goblet cells), and Chga (goblet cells) (31). Notably, all these markers were down-regulated in Il1f10-deficient organoids (P = 0.0195, 0.0217, 0.0701, and 0.0055, respectively), suggesting that the lack of IL-38 fundamentally affects the differentiation of ISC through several intestinal cell types (Fig. 3 G–J).
Il1rapl1-Deficient (Il1rapl1−/y) Organoids Show Growth Defects.
IL-38 functions are determined by its interaction with its putative receptors IL-1R6 and IL-1R9 (5, 10, 12, 13).
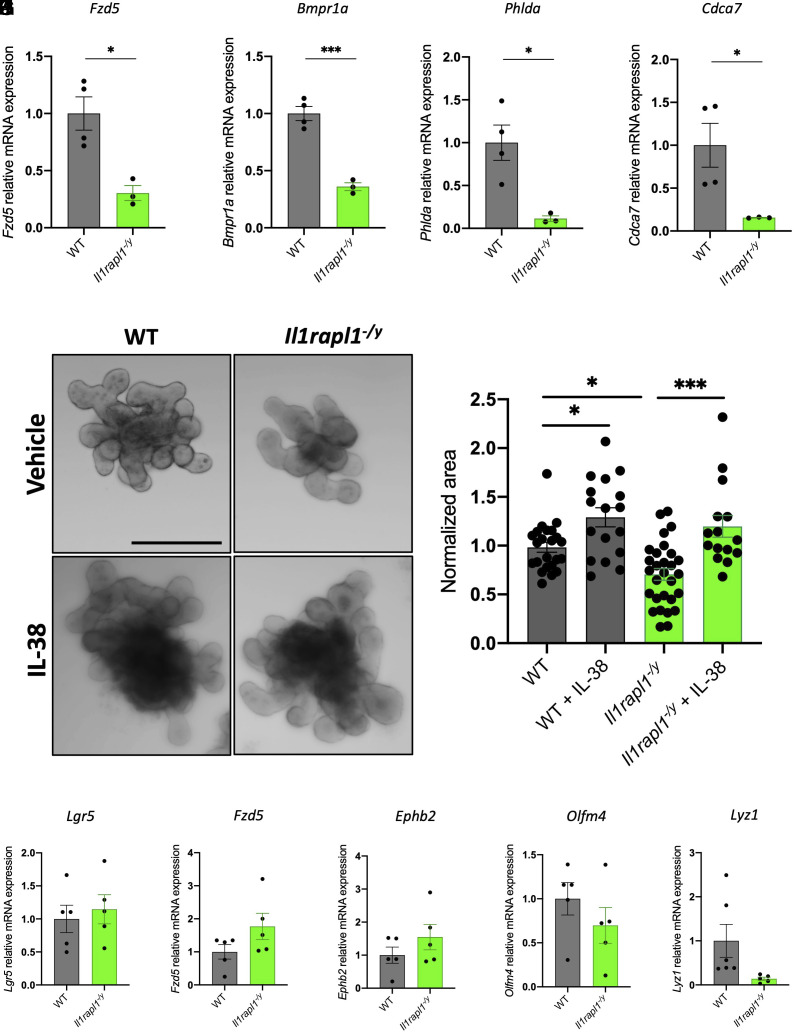
Using RNA extracted from wild-type and Il1rapl1-deficient SIs, we demonstrated that, among all the transcripts analyzed in Fig. 1, Fzd5, Bmpr1a, Cdca7, and Phlda are significantly down-regulated in Il1rapl1-deficient intestines compared to wild-type controls (Fig. 4 A–D). In contrast, other genes are not differentially expressed between wild-type and Il1rapl1-deficient intestines, whereas Clca4 was significantly up-regulated (SI Appendix, Fig. S5 A–O). These observations suggest that IL-1R9 is involved in the correct intestinal stem cell niche homeostasis, but its absence impacts stemness markers less than IL-38 deficiency.
Fig. 4.
Il1rapl1-deficient organoids show growth defects. (A–D) Gene expression analysis of Fzd5 (A), Bmpr1a (B), Phlda (C), and Cdca7 (D) in SI of wild-type and Il1rapl1-deficient mice. Comparisons between WT and Il1rapl1 knockout samples were performed using Student t test. (E) Representative pictures and (F) measurements of wild-type and Il1rapl1-deficient organoids treated either with vehicle or 20 ng/mL IL-38 for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between the four experimental groups were performed using one-way ANOVA. (G–K) Gene expression analysis of Lgr5 (G), Fzd5 (H), Ephb2 (I), Olfm4 (J), and Lyz1 (K) in wild-type and Il1rapl1-deficient organoids. Comparisons between WT and Il1rapl1 knockout samples were performed using Student t test. *P < 0.05; **P < 0.01. Mean ± SEM.
Next, we cultured organoids derived from wild-type and Il1rapl1-deficient mice and we observed that Il1rapl1 knockout organoids are significantly smaller than wild type–derived organoids (P = 0.0249), highlighting the importance of IL-1R9 in intestinal stem cell niche homeostasis (Fig. 4 E and F). Moreover, IL-38 stimulation of organoid cultures revealed that both wild-type and Il1rapl1-deficient cultures (P = 0.0001) respond to stimulation. In fact, we observed a significant increase of organoid size in IL-38-treated wild-type (P = 0.0328) and Il1rapl1-deficient organoids (P = 0.0001) compared to untreated controls (Fig. 4 E and F). In addition, RT-qPCR for Lgr5, Fzd5, Ephb2, Olfm4, and Lyz1 revealed no differences between wild-type and Il1rapl1 KO organoids (Fig. 4 G–K). These data demonstrate that IL-1R9 is not required for IL-38 biological activities in intestinal organoids and that another receptor is involved in IL-38-signaling.
IL-1R6 Is Involved in IL-38 Signaling in the Intestinal Crypt.
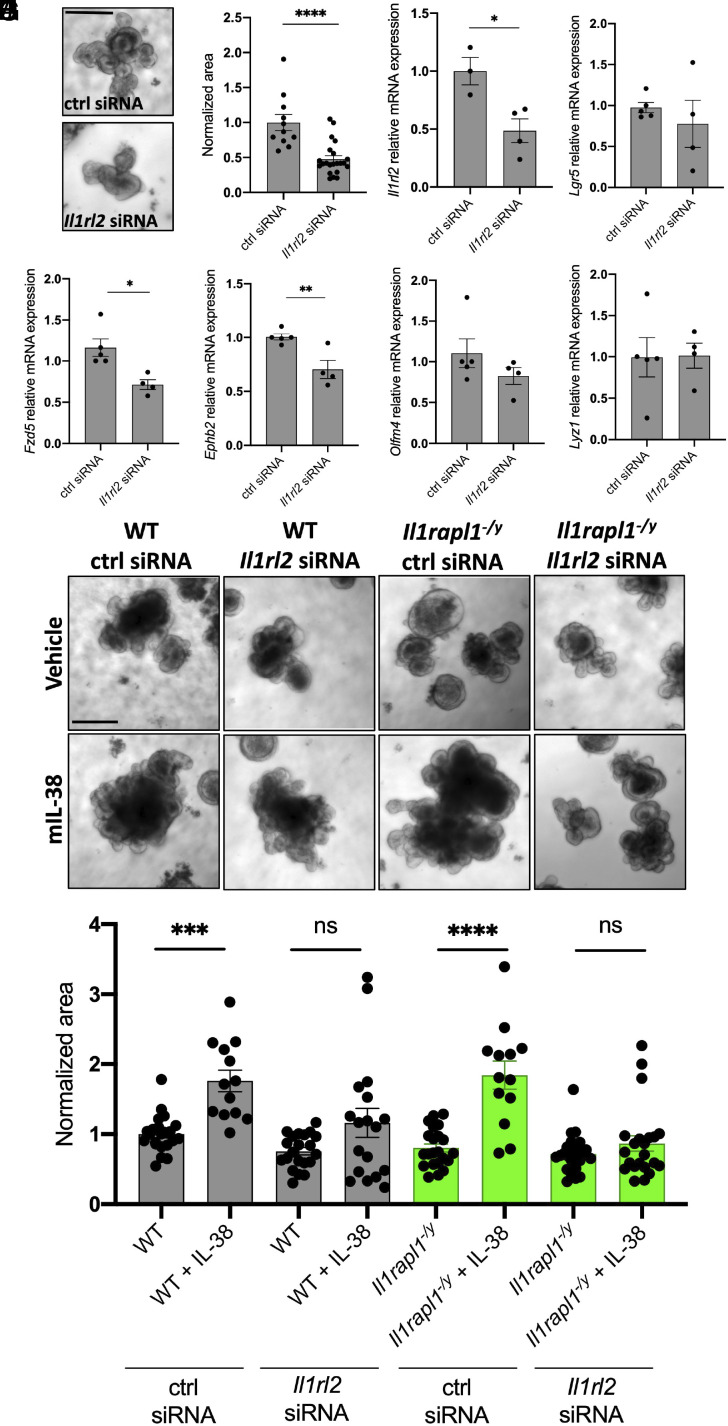
IL-1R6 is encoded by the Il1rl2 gene. In order to understand whether the lack of this receptor has a detrimental effect on organoid growth, we used Il1rl2 siRNAs on wild-type organoids. As shown in Fig. 5 A and B, Il1rl2 silencing determines a significant reduction of organoid size (P < 0.0001), suggesting an important role of IL-1R6 in this process. After validating Il1rl2 RNA silencing by RT-qPCR (Fig. 5C) (P = 0.0218), we measured the expression levels of Lgr5, Fzd5, Ephb2, Olfm4, and Lyz1. Differently to what we observed with Il1rapl1-deficient organoids, Il1rl2 silencing resulted in a significant downregulation of Fzd5 and Ephb2 (P = 0.0113 and 0.0074, respectively), without significantly affecting the expression of the other markers analyzed (Fig. 5 D–H).
Fig. 5.
Il1rl2 silencing negatively affects organoid growth. (A) Representative pictures and (B) measurements of wild-type organoids treated with control siRNA or Il1rl2 siRNA for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between control siRNA and Il1rl2 siRNA organoids were performed using Student t test. (C–H) Gene expression analysis of Il1rl2 (C), Lgr5 (D), Fzd5 (E), Ephb2 (F), Olfm4 (G), and Lyz1 (H) in wild-type organoids treated with either control siRNA or Il1rl2 siRNA for 6 d. Comparisons between control siRNA and Il1rl2 siRNA organoids were performed using Student t test. (I) Representative pictures and (J) measurements of wild-type and Il1rapl1-deficient organoids treated with control siRNA, Il1rl2 siRNA, vehicle, or IL-38 for 6 d. Each dot represents an individual organoid; organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between the eight experimental groups were performed using one-way ANOVA. *P < 0.05; **P < 0.01; ****P < 0.0001. Mean ± SEM.
Next, we sought to assess whether organoids can respond to IL-38 even in the absence of IL-1R6. To do so, we incubated wild-type organoids with either control siRNA or Il1rl2 siRNA and we stimulated them for 6 d with IL-38. As observed in Fig. 5 I and J, recombinant IL-38 significantly increases the organoid size in control siRNA-treated organoids (P < 0.0001), but we could not see significant differences between Il1rl2-silenced organoids treated with vehicle or with recombinant IL-38 (P > 0.999), suggesting that IL-38 increases organoid growth through IL-1R6. To better evaluate the role of IL-1R6 and IL-1R9 in IL-38 biological activity, we silenced Il1rl2 in Il1rapl1-deficient organoids. As previously observed in Fig. 4 E and F, Il1rapl1-deficient organoids indeed respond to IL-38 (P < 0.0001); however, upon silencing with Il1rl2 siRNA, IL-38 did not stimulate growth (P > 0.999) (Fig. 5 I and J). These data demonstrate that IL-38 signaling depends on IL-1R6, whereas IL-1R9 plays a separate role in intestinal organoid homeostasis.
Low Concentrations of IL-1β Stimulate Organoid Growth.
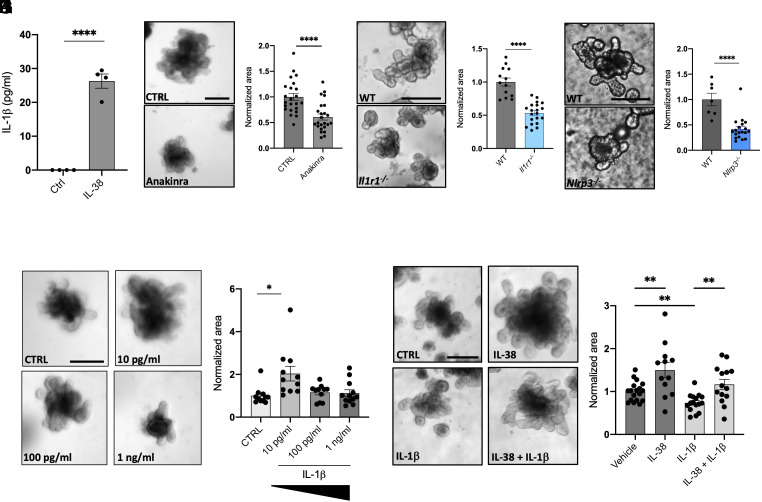
Together with the induction of WNT3a in organoid supernatants (as observed in Fig. 2), IL-38 induces a low but significant concentration of IL-1β (about 25 pg/mL; P < 0.0001) (Fig. 6A), suggesting a possible role of this cytokine in the correct homeostasis of organoids. To test the importance of IL-1β in intestinal organoid cultures, we first decided to block IL-1 signaling by treating wild-type organoids with 10 μg/mL Anakinra (a commercially available antagonist of IL-1 receptor) for 6 d. Anakinra significantly reduces organoids size (P < 0.0001), suggesting the importance of IL-1 signaling for the proper development of intestinal cells (Fig. 6 B and C). To confirm this result, we generated cultures of Il1r1 knockout organoids. As observed in Fig. 6 D and E, Il1r1-deficient organoids are significantly smaller than wild type (P < 0.0001), recapitulating with a genetic model what we previously observed with Anakinra treatment (Fig. 6 B–E). Similarly, Nlrp3 knockout organoids, that do not produce NLRP3, fundamental for inflammasome formation thus for the conversion of pro-IL-1β into mature IL-1β (32), show a marked reduction in size when compared to wild type (P < 0.0001) (Fig. 6 F and G). These data demonstrate a central role of IL-1β in organoid growth.
Fig. 6.
Low concentrations of IL-1β induce organoid growth. (A) IL-1β expression in the supernatants in wild-type organoids treated with 20 ng/mL IL-38 for 6 d. Comparisons between control and IL-38-treated organoids were performed using Student t test. (B) Representative pictures and (C) measurements of wild-type organoids treated with 10 μg/mL Anakinra for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between control and Anakinra-treated organoids were performed using Student t test. (D) Representative pictures and (E) measurement of wild-type and Il1r1−/− organoids. Each dot represents an individual organoid; organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between control and Il1r1 knockout organoids were performed using Student t test. (F) Representative pictures and (G) measurement of wild type and Nlrp3−/− organoids. Each dot represents an individual organoid; organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between control and Nlrp3 knockout organoids were performed using Student t test. (H) Representative pictures and (I) measurements of wild-type organoids treated with 10 pg/mL, 100 pg/mL and 1 ng/mL IL-1β for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between the four experimental groups were performed using one-way ANOVA. (J) Representative pictures and (K) measurements of wild-type organoids treated with vehicle, 20 ng/mL IL-38, 10 ng/mL IL-1β, and 20 ng/mL IL-38 +10 ng/mL IL-1β for 6 d. Each dot represents an individual organoid, organoid cultures were produced from 3 different mice and plated in three wells for each experimental condition. (Scale bar: 200 μm.) Comparisons between the four experimental groups were performed using one-way ANOVA. *P < 0.05; **P < 0.01; ****P < 0.0001. Mean ± SEM.
Since we observed that IL-38 induces low concentrations of IL-1β (Fig. 6A), we decided to test the response of organoids to different concentrations of this cytokine. As also observed by Katsura et al. in lung organoids (33), low concentrations (10 pg/mL) of IL-1β stimulate organoid growth (P = 0.0076) (Fig. 6 H and I). Notably, higher concentrations do not affect organoid size possibly because of IL-1β-dependent cytotoxicity (Fig. 6 H and I). The highest IL-1β concentration used (10 ng/mL), on the other hand, probably inducing cell death in the organoids, significantly reduced their size (P = 0.0485) (Fig. 6 J and K). The protective functions of IL-38 against high levels of IL-1β are confirmed by treating wild-type organoids with 20 ng/mL IL-38 and 10 ng/mL IL-1β. Organoids treated with 20 ng/mL IL-38 and 10 ng/mL IL-1β are significantly larger than 10 ng/mL IL-1β-treated organoids (P = 0.0028) and do not appear significantly different to vehicle organoids (P = 0.5736) (Fig. 6 J and K).
Another confirmation of the protective role of IL-38 was obtained from LPS-treated cultures. LPS exerts detrimental effects on organoid growth (34) (SI Appendix, Fig. S6 A and B). Therefore, we tested three concentrations of LPS (10, 100, and 1,000 ng/mL) and we observed a nonsignificant trend of reduction in organoid size upon 10 ng/mL LPS treatment compared to untreated organoids (P = 0.982), whereas 100 and 1,000 ng/mL significantly reduced organoid size compared to untreated controls (P = 0.0027 and 0.0002, respectively) (SI Appendix, Fig. S6 A and B). To test the protective effects of IL-38, we treated organoids with 100 ng/mL LPS with or without the presence of 20 ng/mL IL-38. While LPS decreases organoid size (P = 0.0139) and IL-38 alone increases size (P = 0039), IL-38 rescues LPS-dependent negative effects (P = 0.0113) (SI Appendix, Fig. S6 C and D).
Discussion
Several studies highlight the role of interleukins in regulating intestinal crypt functions. For instance, IL-22 protects ISCs from immune-mediated tissue damage and promotes intestinal epithelial regeneration (35, 36). Additionally, IL-22, by activating the STAT3 pathway and inhibiting WNT and Notch signaling, suppresses cell differentiation and intestinal stem cell self-renewal (37). However, a study of human intestinal organoids has recently highlighted the role of IL-22 in the maturation of Paneth cells (38). Similar effects on cell differentiation were observed by Deng et al. in their report on IL-10, which leads to inhibition of the WNT pathway and the subsequent depletion of ISCs (39). On the other hand, the chronic exposure of IL-1β induces the expression of stem cell markers like Bmi1, Lgr5, c-Myc, β-catenin, and Nanog in the IEC-18 rat epithelial cell line, showing a putative role of this cytokine in stem cell homeostasis (40). Additionally, IL-33, deriving from pericryptal fibroblasts, drives intestinal differentiation, inducing the expansion of secretory cells (41).
In the present studies, we analyzed the role of IL-38, in order to better define its functions in the gut, including the downstream molecular mechanisms that IL-38 activates to maintain the homeostasis of intestinal tissue. We know that IL-38 reduces intestinal inflammation, and the lack of this cytokine hampers gut recovery after DSS-induced injury (2, 7, 8). Therefore, we focused on the stem cell compartment in intestinal crypt. We demonstrated that exogenous IL-38 stimulation induced the activation of WNT pathway triggering the secretion of the agonist WNT3a, which resulted in the increase of organoid size and budding. On the other hand, genetic ablation of IL-38 resulted in the downregulation of several stem cell markers (Fzd5, Ephb2, Olfm4). These markers are usually related to stemness and WNT activity (42–44). In addition, IL-38 deficiency also severely dampened the expression of markers of Paneth cells (Lyz1), absorptive cells (Si), and goblet cells (Tff3 and Chga). These observations revealed the central role of IL-38 in maintaining the homeostasis of ISCs. In its role in intestinal homeostasis, IL-38 determines the differentiation towards several intestinal cell types, thus explaining the importance of this cytokine in the correct recovery of intestinal injuries.
In order to identify the molecular mechanisms of IL-38 in the intestines, we studied IL-1R9 and IL-1R6, which were identified as putative receptors of IL-38 (5, 10, 12, 13). Intestines taken from IL-38-deficient mice show a significant downregulation of Fzd5, Ephb2, Bmpr1a, Pdgfa, Smoc2, Socs3, Cdk6, Clca4, Olfm4, Phlda, and Pla2g4a compared to wild type. On the other hand, we could not see differences in the expression of regenerative genes and IL-36 transcripts in wild type compared to Il1f10-deficient mice. This observation highlights the fact that IL-38 ablation specifically impairs stem cell niche–related expression. Conversely, Fzd5, Bmpr1a, Phlda, and Cdca7 are the only stem cell niche markers that appeared significantly downregulated in IL-1R9-deficient mice compared to wild type. Fzd5 encodes for a Frizzled5 receptor for WNT ligands and crypt proliferation requires it and/or Frizzled8 (45). BMP signaling is essential for driving the correct differentiation of ISCs, and an increasing gradient of BMP from the crypt to the top of the villus determines the proper zonation of intestinal epithelium (46). Among the several receptors of BMP, the one encoded by Bmpr1a is important because it regulates the phosphorylation of SMAD1/5/8 in the intestine (26). Phlda marks epithelial stem cells (47). Cdca7 is another marker of ISCs (26). The downregulation of these four transcripts in Il1rapl1−/y intestines reveals a general disorganization of the intestinal tissue in these mice, but not as dramatic as shown in Il1f10 knockouts. Moreover, we observed that, using the IL-1R9-deficient mouse, the absence of IL-1R9 does not affect IL-38-dependent increase of organoid size, although IL-1R9 deficiency per se gives rise growth defects. These findings exclude IL-1R9 as a coreceptor for IL-38, in the intestinal crypt compartment. On the other hand, IL-1R6 silencing resulted in a significant downregulation of organoid size and expression of two stem cell markers (Fzd5 and Ephb2), and the lack of IL-1R6 also blocks the responsiveness of organoids to recombinant IL-38 (Fig. 5). We conclude that IL-1R6 works as IL-38 receptor in the gut stem cell niche compartment.
Another controversial aspect of IL-38 is its nature as a pro- or antipathological cytokine. In most diseased conditions, IL-38 exerts an antiinflammatory function, yet it can induce detrimental effects (10, 48). As regards colorectal cancer, IL-38 was reported to inhibit the ERK pathway and thus suppresses cell migration and proliferation (49). Therefore, given its dual role, IL-38 can be considered as a homeostatic factor that balances the intestine according to its needs. This may also explain why IL-38 works only in a precise concentration range. Upon testing of three different concentrations of IL-38 (2, 20, and 200 ng/mL), only 20 ng/mL induced organoid growth, whereas 2 ng/mL was not sufficient to trigger the IL-38-dependent mechanisms. On the other hand, 200 ng/mL could have saturated the whole receptors necessary for IL-1 signaling, thus inhibiting the effects of IL-1 family members downstream to IL-38. The homeostatic nature of IL-38 was further validated in response to IL-1β stimulation. In fact, in healthy conditions, IL-38 induces low levels of IL-1β (about 20 pg/mL) that helped organoids growth. These findings are in line with the discovery of detectable yet low resting circulating concentrations of IL-1β in healthy patients (50) that are likely necessary to keep the biological systems in a steady state. However, based on our observations in intestinal organoids challenged by high concentrations of IL-1β or LPS, IL-38 exerts a protective signal that restores the normal conditions and acts as an antiinflammatory cytokine.
All in all, we can conclude that IL-38, by signaling via binding IL-1R6 but not IL-1R9, favors stem cell renewal and differentiation by inducing WNT3a and regulating the expression of crypt-related genes, acting as a growth factor. In this context, IL-38 induces a homeostatic release of IL-1β (at pg/mL level) that supports growth. On the other hand, in inflamed conditions, IL-38 heals injuries by returning the system to the normal state, again exerting prohomeostatic functions. These observations allow us to conclude that, in the intestine, IL-38 should not be categorized as a pro- or antiinflammatory cytokine, as in different contexts, it can either induce or repress IL-1β to maintain intestinal homeostasis. These findings are remarkably important if we consider the emerging therapeutic potential of IL-38 (14–16). Our findings revealed that, comparably with what was observed with IL-37, another enigmatic IL-1 family member (51), IL-38 exerts its functions on organoids at a specific concentration. IL-37 and IL-38 are mostly described as antiinflammatory cytokines (52, 53) and recent studies demonstrated that they both can inhibit trained immunity (54–57). In conclusion, our results highlight the importance of IL-38 not only as a growth factor but also as a cytokine that, working only in a specific concentration range, can be a potent modulator of intestinal inflammation in human therapies.
Furthermore, these data highlight the commercial potential of IL-38 as a supplement of commercially available kits for organoids growth: The addition of IL-38 in these kits would increase the culturing efficiency, accelerating growth and crypt budding.
Materials and Methods
Intestinal Crypt Isolation and Organoid Culture.
Intestinal segments were collected from wild-type, Il1f10-deficient, Il1rapl1-deficient, and Nlrp3 -deficient mice and were flushed in cold PBS. Two-cm long Intestinal pieces were cut longitudinally and washed vigorously for 20 times to remove mucus and debris. Subsequently, tissues were incubated with Gentle Cell Dissociation Reagent (Stemcell Technologies) in a rocking platform for 20 min at room temperature. Cells were removed and intestinal pieces were resuspended in PBS +0.1% BSA and pipetted up and down 10 times. The supernatant was passed through a 70-μm strainer into a 50-mL tube and centrifuged at 290 × g for 5 min at 2 to 8 °C. Cell pellets were resuspended in DMEM/F-12 with 15 mM HEPES and centrifuged at 200 × g for 5 min at 2 to 8 °C. Isolated crypts in a 50:50 mixture of IntestiCult™ Organoid Growth Medium and Matrigel® and 20 μL of this solution was transferred to each well of a preheated 24-well plate. The plate was then incubated at 37 °C for 15 min and 550 μL of complete IntestiCult™ Organoid Growth Media was added to each well and changed every 3 d. Organoids were generally passaged every 7/8 d and kept in culture for no more than 3 passages.
In Vivo Models.
All the mice used for this study belong to the C57BL/6 genetic strain. Mice were maintained in the same room in ventilated cages (40 to 60 air changes per hour) in 14 h light/10 h dark cycle. Cages are cleaned every 14 d and mouse health are daily checked by facility staff.
Animal protocols were approved by the University of Colorado Animal Care and Use Committee. Il1f10-deficient mice (GenBank accession number: NM_153077.2; Ensembl: ENSMUSG00000046845) were generated using CRISPR/Cas9 technology and were previously described in de Graaf et al. (7); Il1rapl1-deficient mice (GenBank accession number: NM_001160403.1; Ensembl: ENSMUSG00000052372) were generated by deleting Exon 3 using CRISPR/Cas9 technology (Cyagen Biosciences) and were previously described in The et al. (6); the Nlrp3 knockout mice (B6.129S6-Nlrp3tm1Bhk/J) were purchased from The Jackson Laboratories and previously described in Tengesdal et al. (58); the Il1r1 knockout mice (B6.129S7tm1lmx/J) was kindly provided by Shaikh M Atif.
We decided to use 8-wk-old mice for all the experiments performed. Male Il1f10−/−, Il1rapl1−/y, and Nlrp3−/− mice were used for this study. Il1r1−/− and their respective controls were female mice.
Protein Extraction and Western Blotting.
SIs were collected from wild-type, Il1f10-deficient mice. Tissues were lysed in RIPA buffer (Sigma) supplemented with protease and phosphatase inhibitors (Roche) and centrifuged at 13,000 g for 30 min at 4 °C, and the supernatants were obtained. Protein concentration was determined in the cleared supernatants using Bio-Rad protein assay (Bio-Rad Laboratories). Electrophoresis was performed on Mini-Protean TGX 4 to 20% gradient gels (Bio-Rad Laboratories) and blotted onto nitrocellulose 0.1-μm 145 membranes (GE Water & Process Technologies). Membranes were blocked in 5% rehydrated nonfat milk in TBS-Tween 0.5% for 1 h at room temperature. Primary antibody for LGR5 (R&D Systems) was used in combination with peroxidase-conjugated secondary antibodies. A primary antibody against β-Actin (Santa Cruz Biotechnology) was used to assess protein loading.
Mouse IL-38 Expression.
A recombinant expression plasmid was ordered from Twist Bioscience (San Francisco, CA) encoding a 6xHis-tagged Small Ubiquitin-Like Modifier (SUMO) followed by mouse IL-38 residues 3 to 152 within pET21b. A typical expression comprised 4 L of luria broth at 37 °C with ampicillin selection and induced at 0.6 ODs (600 nM) with isopropyl b-D-1-thiogalactopyranoside for 3 to 4 h. Soluble protein was lysed via sonication and applied to a Ni-affinity resin (Sigma) in Ni-A buffer (50 mM l Na2HPO4, pH 7, 500 mM NaCl, 10 mM imidazole) and eluted with Ni-B buffer (50 mM l Na2HPO4, pH 7, 500 mM NaCl, 400 mM imidazole). Elutions were dialyzed against Ni-A buffer for subsequent cleavage of the 6xHis-tagged SUMO via recombinant SUMO Protease produced in-house (also known as Ulp1p, UniProt accession A0A0L8VFW2). 6xHis-tagged Sumo was stripped via Ni-affinity, and the untagged IL-38 was concentrated for size-exclusion chromatography using a Superdex-75 column (Cytiva, 120 mL total bed volume) in final buffer (50 mM HEPES, pH 7, 150 mM NaCl). Fractions comprising IL-38 were concentrated and stored at −80 °C until further use.
RT-qPCR.
Total RNA was extracted from murine SI and from organoid cultures with TRIzol reagent. cDNA synthesis was performed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. qPCRs were performed in triplicate with SYBR Green Master Mix (Applied Biosystems) by means of QuantStudio 3 Real-Time PCR System (Applied Biosystem). Then, 18s was used as internal standard in each sample. The sequences of the primers used are listed in SI Appendix, Table S1. To reduce the number of mice to sacrifice, we used the same wild-type controls for either Il1f10 or Il1rapl1-deficient intestines.
Hematoxylin & Eosin Staining.
SI samples were included in paraffin and cut longitudinally. A regressive Hematoxylin and Eosin staining procedure was used. In this method, hematoxylin is applied to the slide followed by an acid differentiating solution to remove excess stain and clarify the nuclear detail. Eosin is then applied to the slide in order to stain the cytoplasm and surrounding structures.
Imaging.
Organoid stacks were taken with Olympus IX81 spinning disk after 6 d of culturing. Organoid size was measured using Fiji (ImageJ) software in pixel2; every measurement was normalized with the average of the control group for each experiment.
Immunofluorescence.
Mouse intestinal samples were collected and fixed in PBS with 4% paraformaldehyde (Sigma-Aldrich) in PBS. After dehydration, samples were embedded in paraffin molds for sectioning. Sections (5 µm) were obtained and transferred to glass microscope slides. Slides were deparaffinized with xylene and ethanol and permeabilized with 0.1%Tween in TBS solution. After antigen retrieval with citrate buffer, slides were blocked with normal donkey serum and incubated with primary antibodies against LGR5 (clone 803420, Catalog MAB8240, R&D Systems) and IL-38 (catalog ab180898, Abcam) dilution overnight at 4 °C. Alexa flour 647 anti-rat (catalog A48272, Invitrogen) and Alexa fluor 488 anti-rabbit at 1:100 (catalog 11008, Invitrogen) were used as secondary antibodies for 1 h at room temperature. Finally, slides were coverslipped in Mowiol mounting media with DAPI. Images were taken using Olympus FV1000 laser scanning confocal/CARS microscope. IL-38 antibody produces massive background probably due to possible cross-reactivity with other interleukins expressed in the intestine (i.e., IL-36a, IL-36b, IL-36g, and IL-36RA). For this reason, a reliable analysis of IL-38 staining can be performed only in comparison with an IL-38 null sample. We tried to use this IL-38 antibody for western blot, but, at least as regards intestinal samples, we could not obtain reliable results.
Cytokine Measurements.
Cytokine concentrations were measured by specific DuoSet ELISAs (WNT3a and IL-1β) according to the manufacturer’s instructions (R&D Systems). Commercially available ELISA kit from R&D Systems was used to detect IL-38 production but we could not obtain reliable results.
Il1rl2 Silencing.
Il1rl2 siRNAs (MBS8235553) and unspecific siRNA as negative control (MBS8241404) were purchased from MyBioSource and transfected with the siTran 2.0 siRNA transfection reagent following the manufacturer’s instructions. For transfection, we used 10 nM siRNAs for 24 h. Silencing efficiency was evaluated with RT-qPCR using the Il1rl2 primers listed in SI Appendix, Table S1.
Statistical Analysis.
Significance of differences was evaluated with Student’s t test or one-way ANOVA, where accordingly specified, using GraphPad Prism (GraphPad Software Inc.). Statistical significance was set at P < 0.05.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We are thankful to Jesper Falkesgaard Højen, Kibrom Meles Alula, Dominik Stich, Elizabeth Kaye, Radu Moldovan for their technical support. A special acknowledgment goes to Fabia Gamboni for her constant and careful support.
Author contributions
A.D. and C.A.D. designed research; A.D., M.M., J.A.-A., T.A., J.M.G., C.M., A.T., R.G., J.S.R., W.S.W., S.M.A., and C.A.D. performed research; A.D., J.S.R., S.M.A., S.L., E.Z.E., and C.A.D. contributed new reagents/analytic tools; A.D., M.M., J.A.-A., T.A., J.M.G., C.M., A.T., R.G., J.S.R., W.S.W., S.M.A., S.L., E.Z.E., D.M.d.G., and C.A.D. analyzed data; and A.D., D.M.d.G., and C.A.D. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: G.P., Universite de Geneve; and A.W., Goethe-University Frankfurt.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.de Graaf D. M., Teufel L. U., Joosten L. A. B., Dinarello C. A., Interleukin-38 in health and disease. Cytokine 152, 155824 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Xie C., et al. , Interleukin-38 is elevated in inflammatory bowel diseases and suppresses intestinal inflammation. Cytokine 127, 154963 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Fonseca-Camarillo G., Furuzawa-Carballeda J., Iturriaga-Goyon E., Yamamoto-Furusho J. K., Differential expression of IL-36 family members and IL-38 by immune and nonimmune cells in patients with active inflammatory bowel disease. Biomed. Res. Int. 2018, 5140691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garraud T., Harel M., Boutet M.-A., Le Goff B., Blanchard F., The enigmatic role of IL-38 in inflammatory diseases. Cytokine Growth Factor Rev. 39, 26–35 (2018). [DOI] [PubMed] [Google Scholar]
- 5.van de Veerdonk F. L., et al. , IL-38 binds to the IL-36 receptor and has biological effects on immune cells similar to IL-36 receptor antagonist. Proc. Natl. Acad. Sci. U.S.A. 109, 3001–3005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The E., et al. , Interleukin 38 alleviates aortic valve calcification by inhibition of NLRP3. Proc. Natl. Acad. Sci. U.S.A. 119, e2202577119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Graaf D. M., et al. , IL-38 gene deletion worsens murine colitis. Front. Immunol. 13, 840719 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno M., et al. , The anti-inflammatory and protective role of interleukin-38 in inflammatory bowel disease. J. Clin. Biochem. Nutr. 70, 64–71 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huard A., et al. , IL-38 ablation reduces local inflammation and disease severity in experimental autoimmune encephalomyelitis. J. Immunol. 206, 1058–1066 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Zhou H., et al. , Interleukin-38 promotes skin tumorigenesis in an IL-1Rrp2-dependent manner. EMBO Rep. 23, e53791 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mermoud L., et al. , IL-38 orchestrates proliferation and differentiation in human keratinocytes. Exp. Dermatol. 31, 1699–1711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mora J., et al. , Interleukin-38 is released from apoptotic cells to limit inflammatory macrophage responses. J. Mol. Cell Biol. 8, 426–438 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lin H., et al. , Cloning and characterization of IL-1HY2, a novel interleukin-1 family member. J. Biol. Chem. 276, 20597–20602 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Xu W.-D., et al. , IL-38, a potential therapeutic agent for lupus, inhibits lupus progression. Inflamm. Res. 71, 963–975 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Mercurio L., et al. , IL-38 has an anti-inflammatory action in psoriasis and its expression correlates with disease severity and therapeutic response to anti-IL-17A treatment. Cell Death Dis. 9, 1104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., et al. , Therapeutic effect of IL-38 on experimental autoimmune uveitis: Reprogrammed immune cell landscape and reduced Th17 cell pathogenicity. Invest. Ophthalmol. Vis. Sci. 62, 31 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Ma L., An C., Wise S. G., Bao S., The role of IL-38 in intestinal diseases–its potential as a therapeutic target. Front. Immunol. 13, 1051787 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker N., et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Gehart H., Clevers H., Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Haegebarth A., Clevers H., Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 174, 715–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T., et al. , Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Muñoz J., et al. , The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent “+4” cell markers. EMBO J. 31, 3079–3091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar K. K., Burgess A. W., Gulbis J. M., Structure and function of LGR5: An enigmatic G-protein coupled receptor marking stem cells. Protein Sci. 23, 551–565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmon K. S., Gong X., Lin Q., Thomas A., Liu Q., R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 11452–11457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Es J. H., et al. , Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381–386 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Qi Z., et al. , BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H., et al. , Loss of aryl hydrocarbon receptor suppresses the response of colonic epithelial cells to IL22 signaling by upregulating SOCS3. Am. J. Physiol. Gastrointest. Liver Physiol. 322, G93–G106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukonin I., et al. , Phenotypic landscape of intestinal organoid regeneration. Nature 586, 275–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuberger J., et al. , High Yap and Mll1 promote a persistent regenerative cell state induced by Notch signaling and loss of p53. Proc. Natl. Acad. Sci. U.S.A. 118, e2019699118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narytnyk A., et al. , Differentiation of human epidermal neural crest stem cells (hEPI-NCSC) into virtually homogenous populations of dopaminergic neurons. Stem Cell Rev. Rep. 10, 316–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong X.-Y., et al. , Lgr5 positive stem cells sorted from small intestines of diabetic mice differentiate into higher proportion of absorptive cells and Paneth cells in vitro. Dev. Growth Differ. 57, 453–465 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Paik S., Kim J. K., Silwal P., Sasakawa C., Jo E.-K., An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell. Mol. Immunol. 18, 1141–1160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsura H., Kobayashi Y., Tata P. R., Hogan B. L. M., IL-1 and TNFα contribute to the inflammatory niche to enhance alveolar regeneration. Stem Cell Rep. 12, 657–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naito T., et al. , Lipopolysaccharide from crypt-specific core microbiota modulates the colonic epithelial proliferation-to-differentiation balance. mBio 8, e01680-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanash A. M., et al. , Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity 37, 339–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindemans C. A., et al. , Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., et al. , Interleukin-22 regulates the homeostasis of the intestinal epithelium during inflammation. Int. J. Mol. Med. 43, 1657–1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He G.-W., et al. , Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation. Cell Stem Cell 29, 1333–1345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng F., et al. , Interleukin-10 expands transit-amplifying cells while depleting Lgr5+ stem cells via inhibition of Wnt and notch signaling. Biochem. Biophys. Res. Commun. 533, 1330–1337 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Wang L., et al. , Pro-inflammatory cytokine interleukin-1β promotes the development of intestinal stem cells. Inflamm. Res. 61, 1085–1092 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Mahapatro M., et al. , Programming of intestinal epithelial differentiation by IL-33 derived from pericryptal fibroblasts in response to systemic infection. Cell Rep. 15, 1743–1756 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Mah A. T., Yan K. S., Kuo C. J., Wnt pathway regulation of intestinal stem cells. J. Physiol. 594, 4837–4847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmberg J., et al. , EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125, 1151–1163 (2006). [DOI] [PubMed] [Google Scholar]
- 44.van der Flier L. G., Haegebarth A., Stange D. E., van de Wetering M., Clevers H., OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Miao Y., et al. , Next-generation surrogate Wnts support organoid growth and deconvolute frizzled pleiotropy in vivo. Cell Stem Cell 27, 840–851.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beumer J., et al. , BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38, 110438 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Sakthianandeswaren A., et al. , PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 71, 3709–3719 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Kinoshita F., et al. , Interleukin-38 promotes tumor growth through regulation of CD8+ tumor-infiltrating lymphocytes in lung cancer tumor microenvironment. Cancer Immunol. Immunother. 70, 123–135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang L., Zhang H., Zhao D., Hu H., Lu Z., Interleukin-38 suppresses cell migration and proliferation and promotes apoptosis of colorectal cancer cell through negatively regulating extracellular signal-regulated kinases signaling. J. Interferon Cytokine Res. 41, 375–384 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Ter Horst R., et al. , Host and environmental factors influencing individual human cytokine responses. Cell 167, 1111–1124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S., et al. , Extracellular forms of IL-37 inhibit innate inflammation in vitro and in vivo but require the IL-1 family decoy receptor IL-1R8. Proc. Natl. Acad. Sci. U.S.A. 112, 2497–2502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavalli G., Dinarello C. A., Suppression of inflammation and acquired immunity by IL-37. Immunol. Rev. 281, 179–190 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Dinarello C. A., The IL-1 family of cytokines and receptors in rheumatic diseases. Nat. Rev. Rheumatol. 15, 612–632 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Cavalli G., et al. , The anti-inflammatory cytokine interleukin-37 is an inhibitor of trained immunity. Cell Rep. 35, 108955 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Li S., et al. , Role of nuclear interleukin-37 in the suppression of innate immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 4456–4461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teufel L. U., et al. , Opposing effects of interleukin-36γ and interleukin-38 on trained immunity. Int. J. Mol. Sci. 24, 2311 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Graaf D. M., et al. , IL-38 prevents induction of trained immunity by inhibition of mTOR signaling. J. Leukoc. Biol. 110, 907–915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tengesdal I. W., et al. , Targeting tumor-derived NLRP3 reduces melanoma progression by limiting MDSCs expansion. Proc. Natl. Acad. Sci. U.S.A. 118, e2000915118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.