Abstract
The growth of yeast cells to high densities at low, but constant, oxygen concentrations is difficult because the cells themselves respire oxygen; hence, as cell mass increases, so does oxygen consumption. To circumvent this problem, we have designed a system consisting of a computer-controlled gas flow train that adjusts oxygen concentration in the gas flow to match cellular demand. It does this by using a proportional-integral-differential algorithm in conjunction with a three-way valve to mix two gases, adjusting their proportions to maintain the desired oxygen concentration. By modeling yeast cell yields at intermediate to low oxygen concentrations, we have found that cellular respiration declines with oxygen concentration, most likely because of a decrease in the expression of genes for respiratory proteins. These lowered rates of oxygen consumption, together with the gas flow system described here, allow the growth of yeast cells to high densities at low oxygen concentrations. This system can also be used to grow cells at any desired oxygen concentration and for regulated shifts between oxygen concentrations.
Oxygen is an important environmental signal for most organisms (2). It determines whether energy generation occurs primarily through oxidative phosphorylation or glycolysis. The former is more efficient but also generates reactive oxygen species, which can damage cells. Consequently, cells require enzymes for the appropriate mode of energy metabolism and for protection from reactive oxygen species. In the yeast Saccharomyces cerevisiae, the expression of a large number of proteins is affected by oxygen tension (15). Most of these proteins function in respiration, in other processes that use oxygen (e.g., the synthesis of sterols, heme, or unsaturated fatty acids), or in the oxidative stress response. Some of these proteins are expressed optimally in air, some are expressed optimally under anaerobic conditions, and some are expressed optimally at intermediate microaerophilic oxygen tensions (2, 12, 15). The effects of oxygen on the expression of many of these proteins are exerted through transcription of their genes. Those genes that are expressed optimally in air have been designated aerobic genes; those genes that are expressed optimally under microaerophilic or anaerobic conditions have been designated hypoxic genes.
Most genetic studies of oxygen-regulated gene expression in yeast have been done with cells grown either aerobically or anaerobically, or with mutant strains deficient in heme biosynthesis. These studies have implicated heme and a few transcription factors in the expression of aerobic and hypoxic genes but have not revealed how yeast cells actually sense oxygen. Recently, we began to address this question by examining the effects of oxygen concentration on the expression of several genes that encode proteins of the terminal portion of the respiratory chain (3). In these studies, oxygen concentration was varied by sparging fermentor cultures with gases of different fixed oxygen concentrations. Cells were grown until oxygen demand exceeded supply, as indicated by a decrease in the dissolved-oxygen (DO) concentration in the culture. They were then harvested for RNA isolation and assessment of gene expression by Northern blot analysis. The dose-response curves obtained from these studies revealed that expression of these genes switches on or off at very low oxygen thresholds (0.5 or 1 μM O2). Although the fermentor system used for these studies performed adequately, cell yields from cultures grown at or near these oxygen thresholds were very low (3). Unfortunately, these low yields preempted biochemical studies aimed at elucidating the molecular basis for either oxygen sensing or oxygen thresholds in these cells.
Biochemical studies generally require substantial amounts of cellular material. Because of this, very little biochemistry has been done with yeast cells grown at the low oxygen concentrations (i.e., 0.5 to 1 μM O2) that serve as thresholds for the above-mentioned genes. Indeed, such studies would require the growth of yeast cells to high density at low, but constant, limiting oxygen tensions. This is difficult because cells themselves consume oxygen; hence, as cell mass increases, so does oxygen consumption. The system described here was designed to circumvent this problem. It is a computer-controlled feedback system for gas flow that allows oxygen concentration in the gas entering the fermentor to increase with cellular demand. It does this by mixing two gases, adjusting their proportions to maintain a desired fixed oxygen concentration.
MATERIALS AND METHODS
Strains and media.
Yeast strain JM43 (MATα leu2-3,112 his4-580 trp1-289 ura3-52) (6) was used for all of the studies described here. Cells were grown in a semisynthetic medium, SSG (containing, per liter, 3 g of Bacto yeast extract, 10 g of galactose, 0.8 g of NH4SO4, 1 g of KH2PO4, 0.5 g of NaCl, 0.7 g of MgSO4 · 7H2O, 5 μg of FeCl2, and 0.4 g of CaCl2), supplemented with amino acids and uracil at 40 μg/ml, 0.1% (vol/vol) Tween 80, 20 μg of ergosterol per ml, and 350 ppm of Dow Corning FG-10 Silicone antifoam.
Growth conditions.
Precultures were grown aerobically in a shaker (200 rpm) at 28°C to mid-logarithmic phase. The resulting cultures were inoculated into a New Brunswick BioFloIIc fermentor, with a working volume of 3.5 liters, and grown to early or mid-log phase before harvesting. Temperature, pH, and agitation were maintained at 28°C, 5.0, and 300 rpm, respectively. Oxygen concentration in the fermentor was either regulated or unregulated. In the latter case, gases of fixed oxygen concentration were used and cultures were harvested when the oxygen tension dropped below 80% of the initial value. When oxygen concentration was regulated, oxygen was monitored with an oxygen sensor as described below. For both types of experiments, cultures were inoculated with enough cells to allow 5 to 7 doublings in cell mass before harvesting. At the time of harvest, cells were quick-chilled through several feet of coiled copper tubing immersed in a salted ice bath, collected by centrifugation, and washed.
Measurement and control of DO.
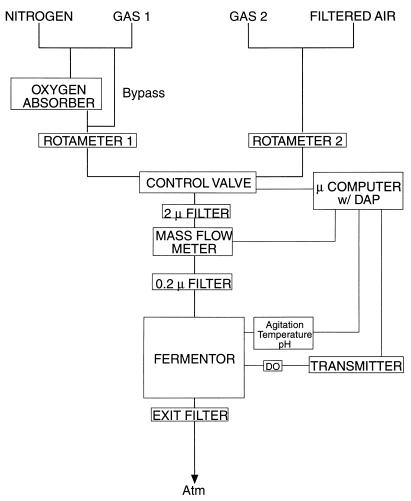
A block diagram of the gas flow train and control system is shown in Fig. 1. Either nitrogen or a process gas (gas 1) enters the flow train on the left while air or a second process gas (gas 2) enters on the right. Gas flow in both the right and left arms is set with Cole Palmer 150-mm rotameters (model no. L-03294-26), selected by a solenoid control valve, and monitored with a mass flow meter. Two in-line filters (2 and 0.2 μm) protect the mass flow meter and ensure sterility in the fermentor vessel. To reduce time delays, which can cause stability problems with regulation by the proportional-integral-differential (PID) algorithm, the length of tubing between the fermentor vessel and the solenoid valve was made as short as possible. The gases used for the experiments described here are oxygen-free nitrogen (Matheson), which contains less than 0.5 ppm of oxygen; filtered house air; and process gases, which are mixtures of Certified Standard gases made up in oxygen-free nitrogen. The process gases are attached to the gas flow train through two semiautomatic changeover manifolds (model no. 5201; Matheson).
FIG. 1.
Gas flow train and control system. A solenoid control valve allows either gas 1 or gas 2 to enter the fermentor, where oxygen tension is monitored with a polarographic DO sensor; the signal is amplified at the transmitter and sent to a microcomputer with a DAP. The solenoid valve is under computer control and may be programmed for a constant duty cycle, to maintain a particular oxygen concentration, or to shift oxygen concentrations. Gas 1 is usually either the low-oxygen gas or nitrogen, while gas 2 is usually either the high-oxygen gas or air. Gas flow is adjusted with the rotameters and monitored by the mass flow meter. Atm, atmosphere.
Oxygen-free nitrogen and air are used to calibrate the DO sensor (12 mm; Ingold, Wilmington, Mass.). For anaerobic experiments, nitrogen is passed through an Oxyclear oxygen absorber (LabClear, Oakland, Calif.) in order to reduce its residual oxygen level to below 50 ppb. At the end of each experiment, the zero calibration of the DO sensor is checked with a chemical zeroing gel (Ingold).
Oxygen concentration in the fermentor is monitored with the DO sensor. The signal from this electrode is amplified with an Ingold 4300 transmitter, and a scaled, 4- to 20-mA signal is sent to the MicroStar Data Acquisition Processor (DAP) model 1200/2. Analog data from the mass flow meter (type 0258C 0 to 5 V; MSK) and the BioFloIIc fermentor (0 to 1 V) are also sent to the DAP, where they are digitized, averaged where appropriate, and sent on to a microcomputer (model 333P; Dell) for display and logging to disk. The DAP contains an 80186 microprocessor with firmware for data processing and control, including the PID algorithm, and digital output lines that are used to control the solenoid control valve (model no. 8320B174Q; ASCO). The solenoid control valve operates at 120 V and is optically isolated from its 0 to 5 V control signal with an SSR OPTO22 120D25 relay. It is switched to gas 1 or gas 2, depending on the signal from the DAP. When oxygen concentration is regulated, the valve is switched to gas 1 for some fraction (0 to 100%) of the 1-s duty cycle and to gas 2 for the remainder of the 1-s duty cycle. The DAP is controlled by scripts (instruction sets) sent to it by the microcomputer. Source code is available upon request.
The oxygen concentration (in μM) in the fermentor vessel was calculated from the measured DO level and based on oxygen solubility in the growth medium at 28°C, the ambient barometric pressure, and the pressure in the fermentor vessel (i.e., differential pressure) by using the following formula: O2 = [(bi − bo)/(bs − bo) × f] × cO2. The value bi is the measured output (in bits) from the dissolved oxygen sensor at time i, bo is the measured output in the absence of oxygen, bs is the measured output in air-saturated growth medium, f is the amplification factor (0 to 1) of the oxygen meter, and cO2 is the concentration of O2 (in μM) in air-saturated medium at the measured pressure (i.e., ambient plus differential pressure). The osmotic pressure of the supplemented SSG growth medium used here was 184 mosM. The differential pressure in the fermentor vessel at the 4-liters/min sparge rate used for these experiments was extremely low (less than 0.4% of an atmosphere) and therefore had little effect on the value of cO2. The value for bo was obtained for each experiment by equilibration of the oxygen sensor in either O2-free nitrogen or zeroing gel. The lowest value obtained from these treatments was used in all cases. The value for DO concentration in air-saturated growth medium in our laboratory in Boulder, Colo., was typically between 195 and 197 μM.
RNA isolation and hybridization.
Total RNA was isolated from washed cells essentially as described (7). RNA samples were separated on 0.22 M formaldehyde-agarose gels in a morpholinepropanesulfonic acid (MOPS)-formaldehyde buffer (0.02 M MOPS, 0.04 M Na acetate, 0.008 M EDTA, 0.22 M formaldehyde) and transferred to Nytran or Nytran Plus membranes (Schleicher and Schuell). Approximately 30 μg of total RNA was loaded per lane; loading was adjusted to give equal signals for hybridization to the ACT1 gene. DNA probes were prepared by random-primer labeling of double-stranded DNA fragments with [α-32P]dCTP (Dupont NEN). Probes were a 500-bp PstI fragment for COX5a, a 370-bp AccI-BglII fragment for COX5b, and a 520-bp StyI fragment for ACT1. Hybridization and stringency washes were performed as described previously (13). Signal intensity was quantitated with an AMBIS radioanalytic imaging system. For quantitation of transcripts, signals were normalized to that for ACT1 mRNA.
Miscellaneous.
Statistical analysis, modeling, and data plots were done with MATHCAD (MathSoft, Cambridge, Mass.). Cell density was monitored by measuring turbidity with a Klett meter fitted with a no. 54 green filter.
RESULTS
Computer control at intermediate and low oxygen tensions.
In order to grow cells to high densities at low, but constant, oxygen concentrations, we developed a computer-controlled gas flow system (Fig. 1) that mixes two gases, adjusting their proportions to maintain the desired oxygen concentration. This allows oxygen concentration in the gas flow to increase with cellular demand (i.e., cell mass). With this computer-controlled system, two process gases may be mixed to give any desired oxygen concentration in the feed gas.
In the simplest case, program control of the solenoid valve (Fig. 1) is set for a constant duty cycle, which may employ one process gas or the other, or any combination of the two. As an example, we compared cell growth obtained with a fixed duty cycle, mixing air and nitrogen to give ∼100 μM O2 in the saturated culture media, with cell growth in which the duty cycle was regulated to maintain the oxygen concentration at 100 μM. In the culture with a fixed duty cycle, a decline in oxygen concentration was apparent at 6 h and this concentration dropped to 80% of its initial value at 16 h, at which time the experiment was terminated. In the culture with the regulated duty cycle, the duty cycle began to change by 6 h to accommodate increased cellular demand for oxygen. This culture was terminated at the same time point (i.e., 16 h) as the unregulated culture, so that mRNA levels could be compared. The cell growth rates in both cultures were equivalent. Moreover, the cell yields at the time of harvest in both cultures were comparable (Table 1). However, the regulated experiment could have continued for several more hours because the air fraction of the process gas had only increased from 50 to 65% of the maximum for the duty cycle at the time of harvest. Hence, the regulated culture could have produced more cell mass.
TABLE 1.
Growth and yield for cells grown at different oxygen tensions with or without regulation of oxygen
| O2 concn (μM) | Type of culturea | Growth rateb | Yield (g [wet weight]/liter) |
|---|---|---|---|
| 100 | U | 2.7 | 1.4 |
| 100 | R | 2.6 | 1.4 |
| 1 | U | 2.6 | 0.08 |
| 1 | R | 2.6 | 1.4 |
U, unregulated; R, regulated.
Growth rate is given as doubling time in hours.
We also compared the growth rates of regulated and unregulated cultures at 1 μM O2, which is two orders of magnitude below the oxygen concentration used for the experiment described above. For this experiment, two different feed gas mixtures were used—one containing 0.1% oxygen, the other 1.0% oxygen—to keep the oxygen concentration near 1 μM. In the unregulated culture, cells were maintained in exponential growth until the oxygen concentration dropped to 80% of its initial value, at which time they were harvested. The regulated culture was grown for 16 h and then harvested. Although the signal-to-noise ratio was higher at these lower oxygen concentrations, the gas flow train successfully maintained its initial oxygen concentration in the regulated culture to within 10% of its initial set point. As in the first experiment, the growth rates in both cultures were equivalent (Table 1), but, unlike in the first experiment, oxygen became limiting (i.e., declined to 80% of its initial concentration) at very low cell densities in the unregulated culture. This resulted in an extremely low cell yield (0.08 g [wet weight] per liter) (Table 1). In contrast, the yield from the regulated culture was significantly higher because this culture could be grown for a longer period of time before oxygen concentration declined. In this experiment, cells were harvested at 16 h so that mRNA levels could be compared to those in the experiment described above. The cell yield was 1.4 g (wet weight) per liter, the same yield obtained after 16 h in the regulated culture grown at 100 μM O2 (Table 1). Higher yields than this are possible because, as in the culture grown at 100 μM O2, the fraction of the duty cycle for the higher-oxygen gas was still low (i.e., in the range of 20 to 40%) at the time of cell harvest.
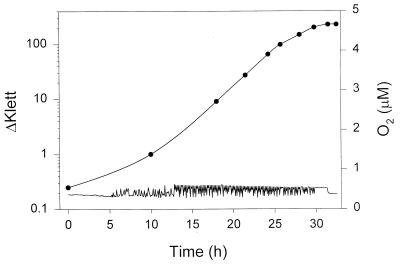
To illustrate the usefulness of the regulated system for growing cells to high densities at low oxygen concentrations, we did the experiment whose results are shown in Fig. 2. Here, we used two feed gas mixtures (0.05 and 0.5% oxygen) to keep the oxygen concentration at 0.44 μM and allowed cells to grow to stationary phase. The oxygen concentration was invariant for the first 5 h, after which the culture began to consume appreciable amounts of oxygen and the solenoid control valve began to regulate the oxygen concentration. For the duration of the experiment, the oxygen concentration was maintained at 0.44 ± 0.1 μM (mean ± standard deviation). The cell yield in this experiment was 7.04 g (wet weight) per liter. It is nearly two orders of magnitude higher than yields that could be obtained in unregulated cultures grown at this oxygen concentration.
FIG. 2.
Cell growth at low oxygen concentration. Yeast cells were grown in the fermentor at an oxygen concentration of 0.44 μM by using feed gas mixtures of 0.05 and 0.5% oxygen. Cell growth was monitored by taking 5-ml samples and assaying culture density turbidometrically with a Klett colorimeter equipped with a green (no. 54) filter. The difference in turbidity between the culture and the uninoculated growth medium is expressed as a difference in Klett units (ΔKlett) (upper trace) and is plotted on the left ordinate. The concentration of oxygen in the culture medium (lower trace) is plotted in 5-min-interval increments on the right ordinate. The oxygen concentration in the culture throughout the entire experiment was 0.44 ± 0.1 μM (mean ± standard deviation). The cells were harvested and weighed after cessation of growth.
Modeling cell growth and oxygen consumption.
Cell yields can be estimated from the oxygen balance equation:
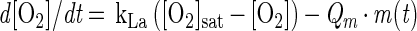 |
1 |
where KLa is the gas-liquid mass transfer coefficient, [O2]sat is the oxygen concentration supplied by the feed gas, [O2] is the oxygen concentration in the fermentor, Qm is the specific respiration rate, and m(t) is the cell mass in grams at time t. Under our experimental conditions, kLa is 40 h−1, Qm is 100 μM h−1g−1 at moderate to high oxygen concentrations (i.e., 100 to 200 μM O2 [air]) and 30 μM h−1g−1 at low oxygen concentrations (i.e., 1 μM O2), and m(t) increases exponentially with doubling times of 2.6 to 3.0 h, depending on the experiment.
Under steady-state conditions, where oxygen concentration is constant, we can calculate the maximal cell mass yield (in grams), mass yield as a function of time, and the variation in the duty cycle necessary to maintain a constant oxygen concentration from equation 1. Maximal cell mass yield, Mmax, calculated from the limiting condition, when the feed gas is entirely the second, higher-oxygen, gas, is given by:
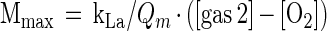 |
2 |
where [gas 2] is the concentration of oxygen in the second, higher-oxygen, gas. With one feed gas (i.e., unregulated conditions in which oxygen tension decreases as cell mass increases), equation 1 becomes:
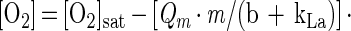 |
3 |
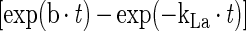 |
where b is [ln(2)/mass doubling time] and t is time (in hours). From equation 3 it is possible to calculate oxygen concentration in the fermentor as a function of time.
When d[O2]/dt in equation 1 is set equal to zero, the equation can be used to model cell growth and respiration under steady-state conditions. In contrast, equation 3 can be used to model cell growth and respiration under conditions where oxygen tension decreases with time. Given the cell growth rate, cell yield, and oxygen concentration in the fermentor, we have used these equations to calculate the specific respiration rate as 100 μM O2/h/g for cultures grown in 100 or 200 μM O2 and as 30 μM O2/h/g for cultures grown at 1 μM O2. This reduction in specific respiration rate at reduced oxygen concentrations has been observed previously (8a, 10) and is significant because it leads to a lack of proportionality between cell yield and oxygen concentration; this, in turn, leads to an increase in cell yield in low-oxygen cultures. By using the appropriate specific respiration rate (above), the cell yields from regulated cultures can be calculated to be considerably higher than the cell yields from unregulated cultures. For example, if growth were allowed to continue until the feed gas is entirely the high-oxygen gas, cell yields from regulated cultures would be 7- and 30-fold higher than from unregulated cultures grown in 100 μM and 1 μM O2, respectively.
Gene expression with the regulated system.
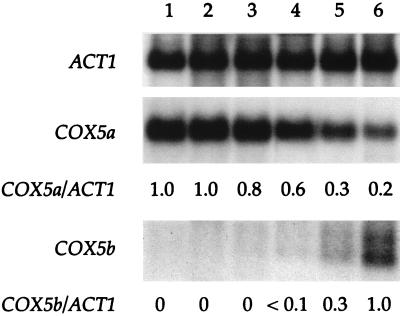
The utility of this fermentor system for studies of oxygen-regulated gene expression is illustrated by the experiment whose results are shown in Fig. 3. Here, the effects of oxygen concentration on the expression of ACT1, COX5a, and COX5b are shown. ACT1 encodes actin, COX5a encodes the aerobic isoform (Va) of cytochrome c oxidase subunit V, and COX5b encodes the hypoxic isoform (Vb) of cytochrome c oxidase subunit V. The ACT1 gene, whose expression is not oxygen sensitive, was used as a control. COX5a is optimally expressed in air (200 μM O2), while COX5b is optimally expressed under anaerobic conditions. The results shown in Fig. 3 allow us to determine the oxygen thresholds for expression of these two genes. The mRNA level from COX5a declines precipitously at oxygen concentrations of 0.5 μM or below. This agrees with the 0.5 μM O2 threshold we determined previously for unregulated cultures (3). In contrast, the mRNA level from COX5b increases at oxygen concentrations of 0.2 μM or below, indicating that the threshold for switching on this gene is between 0.5 and 0.2 μM O2. Previous attempts to define this threshold in unregulated cultures have been unsuccessful (3) because of low cell yields. The high cell yields produced in the regulated cultures now make possible studies aimed at determining the biochemical bases for these low oxygen thresholds for gene expression.
FIG. 3.
Effects of oxygen on the expression of COX5a and COX5b. Northern blot of total RNA isolated from cells grown in 200 μM O2 (air) (lane 1), 50 μM O2 (lane 2), 5 μM O2 (lane 3), 0.5 μM O2 (lane 4), 0.2 μM O2 (lane 5), and oxygen-free N2 (lane 6). The RNA was hybridized with probes specific for COX5a, COX5b, and ACT1, as described in Materials and Methods. The ACT1 gene was probed as a control for RNA load. The levels of COX5a and COX5b mRNAs, normalized to the ACT1 mRNA, are presented as decimal percentages relative to their levels in air (for COX5a) or nitrogen (for COX5b).
System stability and regulation.
A PID-algorithm-regulated system may be hard to tune for stability in the face of rapid shifts or transients and tight regulation at set points. Because tightly regulated systems may not be stable to large transients, we have designed the control program to approach the desired oxygen tension by operating at the theoretical duty cycle for that oxygen tension until one of two criteria is met: either the error signal becomes less than a specified value, or the time allowed to reach the set point is exceeded. Once one of the above criteria is met, the program changes from the programmed, theoretical duty cycle to PID control of the duty cycle. This is a modified type of PID control inasmuch as the error signal increases or decreases the duty cycle of the solenoid valve, which will change the proportion of oxygen in the gas flow.
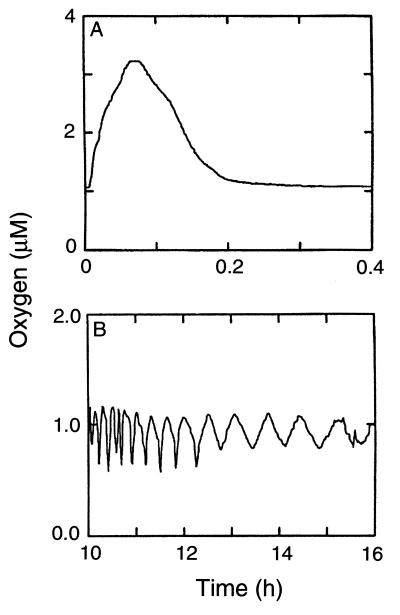
Figure 4A shows the response of the system to transient introduction of a gas of higher oxygen concentration. The system was equilibrated with 0.1% oxygen, and regulation was started with the second gas (1% oxygen) fully on. Within 5 min, the DAP script for PID control reversed the rise in oxygen tension, and within 10 min it returned it to the control value. Figure 4B shows oscillations at low oxygen concentration when the set point is just above the actual oxygen concentration for gas 1 (Fig. 1); this oscillation is reduced after the culture begins to consume oxygen. Such oscillations are common with simple PID control (4, 14); further adjustment of the three parameters for low oxygen concentrations might improve the regulation. Alternatively, a predictor model (14) or a feed-forward model (11) could be used at low oxygen concentrations.
FIG. 4.
System responses. (A) Response time of the modified PID controller. The system was equilibrated with gas 1 prior to initiation of computer control; regulation was started with gas 2 fully on. (B) Oscillations in oxygen tension when the set point is just above the actual oxygen concentration for gas 1. Initially, there is little oxygen consumption because the density of the cell culture is low. Later, as the culture begins to consume more oxygen, the size of the oscillation in oxygen concentration is reduced.
Other uses.
This is a versatile system that was set up initially for use at intermediate and low oxygen concentrations. It has also been used to grow cells in atmospheric and even hyperbaric oxygen (data not shown). In addition, it has been used to shift cells from one oxygen concentration to another. The shift in oxygen concentration within the fermentor is typically 95% complete within 4 min; this allows changes in gene expression to be monitored after a rapid shift in conditions.
DISCUSSION
Several strategies have been used to control DO concentration in fermentor cultures of various microbes (4, 5, 8, 10, 11, 14). These vary in the type of control algorithm used and in the parameters that are regulated. The easiest parameters to regulate are agitation within the fermentor vessel and total gas flow into the vessel. Consequently, the most commonly used controllers have relied on varying the agitation rate or on varying both the agitation rate and the rate of inlet gas flow. Both strategies regulate oxygen concentration by altering the agitation or sparge rate, both of which modify the gas-liquid mass transfer coefficient, kLa. Alterations in the agitation or sparge rate concomitantly alter the fluid flow and shear characteristics within the fermentor (9). Because DO probes may be sensitive to shear, alterations in shear can exacerbate measurement problems at the low oxygen levels that approach the limits of probe sensitivity. The regulation of agitation and/or sparge rate is therefore not useful for studies aimed at growing cells at very low oxygen concentrations. In addition, yeast cells grown at low oxygen concentrations require added unsaturated fatty acids (usually supplied as Tween 80) and ergosterol (1). These supplements increase foaming in the medium, which can be very difficult to suppress when the agitation or sparge rate increases.
Our solution to the problem of growing yeast cells at low but constant oxygen concentrations is to mix two gases at constant agitation and gas flow levels. Because the DO probe may be sensitive to changes in fluid flow, especially at the limits of probe sensitivity, we chose to maintain constant agitation and gas flow. This minimizes variations in gas-liquid transfer rates, foaming, and oxygen probe sensitivity problems. The proportion of the two gases is adjusted to maintain oxygen tension near the set point by using a PID-control algorithm. Although this strategy is similar to that used by Chen et al. (4), our system regulates at lower oxygen tensions and permits higher cell yields. Our system also allows one to start a fermentation with a constant gas mixture before switching to PID control; this avoids instabilities that are characteristic of many PID systems responding to large changes in set point (4). Regulation with this system is good at intermediate oxygen concentrations and satisfactory at low oxygen concentrations. Tighter regulation at low oxygen concentrations might be achieved by using a feed-forward algorithm similar to that used by Smith et al. (11).
This regulated system for growing yeast cells has been used reliably to grow cells at oxygen concentrations between 0.2 and 200 μM. Gene expression in the oxygen-regulated cultures is comparable to that for unregulated cultures. However, the regulated system supports higher cell yields at very low oxygen tensions. Indeed, it increases cell yield nearly two orders of magnitude, at oxygen concentrations below 1 μM, relative to those levels achievable in an unregulated system. The gas flow system described here is also useful for programmed shifts between different oxygen concentrations and is sufficiently versatile to allow a wide range of gas mixtures to be used, in conjunction with appropriate probes.
ACKNOWLEDGMENTS
This work was supported by GM 30228 (R.O.P.) from the National Institutes of Health and grant 4557 from the Tobacco Research Council.
REFERENCES
- 1.Andreasen A A, Stier T J B. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J Cell Comp Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- 2.Bunn H F, Poyton R O. Oxygen sensing and molecular adaptation to hypoxia. Physiol Res. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 3.Burke P V, Raitt D C, Allen L A, Kellogg E A, Poyton R O. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J Biol Chem. 1997;272:14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Tannahill A L, Shuler M L. Design of a system for the control of low dissolved oxygen concentrations: critical oxygen concentrations for Azotobacter vinelandii and Escherichia coli. Biotechnol Bioeng. 1985;27:151–155. doi: 10.1002/bit.260270208. [DOI] [PubMed] [Google Scholar]
- 5.Clark T, Hasketh T, Seddon T. Automatic control of dissolved oxygen tension via fermentor agitation speed. Biotechnol Bioeng. 1985;27:1507–1511. doi: 10.1002/bit.260271017. [DOI] [PubMed] [Google Scholar]
- 6.Cumsky M G, Ko C, Trueblood C E, Poyton R O. Two nonidentical forms of subunit V are functional in yeast cytochrome c oxidase. Proc Natl Acad Sci USA. 1985;82:2235–2239. doi: 10.1073/pnas.82.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elder R T, Loh E Y, Davis R W. RNA from yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci USA. 1983;80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosz R, Stephanopoulis G. Physiological, biochemical, and mathematical studies of micro-aerobic continuous ethanol fermentation by Saccharomyces cerevisiae. I. Hysteresis, oscillations, and maximum specific ethanol productivities in chemostat culture. Biotechnol Bioeng. 1990;36:1006–1019. doi: 10.1002/bit.260361006. [DOI] [PubMed] [Google Scholar]
- 8a.Kellogg, E. A., and P. V. Burke. Unpublished data.
- 9.Nishihawa M, Kato H, Hashimoto K. Heat transfer in aerated tower filled with non-Newtonian liquid. Ind Eng Process Des Dev. 1977;16:133–137. [Google Scholar]
- 10.Rogers P J, Stewart P R. Mitochondrial and peroxisomal contributions to the energy metabolism of Saccharomyces cerevisiae in continuous culture. J Gen Microbiol. 1973;79:205–217. doi: 10.1099/00221287-79-2-205. [DOI] [PubMed] [Google Scholar]
- 11.Smith J M, Davidson S W, Payne G F. Development of a strategy to control the dissolved concentration of oxygen and carbon dioxide at constant shear in a plant cell bioreactor. Biotechnol Bioeng. 1990;35:1088–1101. doi: 10.1002/bit.260351104. [DOI] [PubMed] [Google Scholar]
- 12.Stansfield I, Cliffe K R, Kelly S L. Chemostat studies of microsomal enzyme induction in Saccharomyces cerevisiae. Yeast. 1991;7:147–156. doi: 10.1002/yea.320070208. [DOI] [PubMed] [Google Scholar]
- 13.Trueblood C T, Poyton R O. Differential effectiveness of yeast cytochrome c oxidase subunit V genes results from differences in expression not function. Mol Cell Biol. 1987;7:3520–3526. doi: 10.1128/mcb.7.10.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi G, Hwang Y B, Chang H N, Lee K S. Computer control of cell mass concentration in continuous culture. Automatica. 1989;25:243–249. [Google Scholar]
- 15.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]