Abstract
Background
Pulmonary cryptococcosis (PC) contributes to the ongoing global disease burden in human immunodeficiency virus (HIV)-negative populations. Since some PC patients are misdiagnosed under existing diagnostic guidelines, new diagnostic markers are needed to improve diagnostic accuracy and therapeutic efficacy and reduce disease risk.
Methods
Our previously established sphingolipidomic approach was employed to explore the use of serum sphingolipids (SPLs) in diagnosing HIV-negative patients with PC. A clinical cohort of PC, pulmonary aspergillosis (PA), and tuberculosis (TB) patients and healthy controls was assessed to identify SPL biomarkers.
Results
A total of 47 PC, 27 PA, and 18 TB patients and 40 controls were enrolled. PC and TB patients had similar clinical features, laboratory test results and radiological features, excluding plural effusion. The serum ceramide [Cer (d18:1/18:0)] level showed a significant increase in PC patients compared to controls and PA and TB patients (P<0.05). Cer (d18:1/18:0) was identified as a specific diagnostic biomarker for PC. The optimal cut-off value of greater than 18.00 nM showed a diagnostic sensitivity of 76.60% and a specificity of 95.00% and better distinguished PC patients from PA and TB patients. Furthermore, the serum Cer (d18:1/18:0) level gradually decreased after 3 and 6 months of treatment, suggesting the prediction potential for therapeutic efficacy of this biomarker. In addition, Cer (d18:1/18:0) analysis presented a higher sensitivity than the cryptococcal antigen (CrAg) assay.
Conclusions
This is the first study to report the use of the SPL Cer (d18:1/18:0) as a serum biomarker for diagnosing Cryptococcus spp. infection in HIV-negative patients.
Keywords: Pulmonary cryptococcosis (PC), sphingolipid (SPL), Cer (d18:1/18:0), diagnostic biomarker
Highlight box.
Key findings
• The key finding is the use of the sphingolipid (SPL) Cer (d18:1/18:0) could work as a serum biomarker for diagnosing pulmonary cryptococcosis (PC) in human immunodeficiency virus (HIV)-negative patients.
What is known and what is new?
• According to current research, it is difficult to distinguish the clinical symptoms of HIV-negative cryptococcosis patients from other diseases. In addition, the diagnosis of PC patients mainly relies on methods such as lung tissue biopsy, culture and cryptococcal antigen (CrAg) assay, but each method has its limitations.
• We found the use of the SPL Cer (d18:1/18:0) could work as a serum biomarker for diagnosing cryptococcosis in HIV-negative patients. The optimal cut-off value of greater than 18.00 nM showed a diagnostic sensitivity of 76.60% and a specificity of 95.00%.
What is the implication, and what should change now?
• We believe that Cer (d18:1/18:0) can serve as a serum diagnostic marker for HIV negative PC patients.
Introduction
Cryptococcus neoformans (C. neoformans) and Cryptococcus gattii (C. gattii) are the two primary etiological factors underlying cryptococcosis. In Europe and America, cryptococcosis attributable to C. neoformans predominantly manifests in human immunodeficiency virus (HIV)-positive individuals, whereas infections induced by C. gattii are more frequently observed in immunocompetent patients, including those who are HIV-negative (1,2). However, C. neoformans are more frequently observed in HIV-negative patients in China (3). Pulmonary cryptococcosis (PC) is the most prevalent form of cryptococcosis and ranks as the third most common infectious fungal lung disease. Globally, approximately 1 million cases of cryptococcosis are reported annually, resulting in an estimated 625,000 deaths. The mortality rate for this disease ranges from 20% to 70% (4,5). PC is a frequently encountered life-threatening pulmonary mycosis that affects both individuals with compromised immune systems (including those with HIV) and individuals with competent immune function (5-7). Recent investigations have demonstrated a rising incidence of PC in HIV-negative patients, attributed to the growing population with chronic underlying conditions such as haematological malignancies, diabetes, pulmonary tuberculosis (TB), as well as those undergoing bone marrow or organ transplantation and receiving immunosuppressants (5,8,9). Furthermore, certain study has reported susceptibility to Cryptococcus infection even among individuals without underlying health issues (10).
The majority of patients with PC typically present with non-specific symptoms such as cough and expectoration, accompanied by ambiguous imaging manifestations. Consequently, the disease can easily be confused with and misdiagnosed as other pulmonary conditions, including pulmonary aspergillosis (PA), TB, lung cancer, and bacterial pneumonia (11,12). For instance, it has been reported that approximately 51.9% of South African miners tested for PC were mistakenly diagnosed with TB (13). Such misdiagnosis can lead to severe consequences as PC necessitates distinct treatment strategies compared to diseases with similar symptomatology. Therefore, it becomes imperative to explore novel diagnostic biomarkers that can facilitate early detection of PC, prevent the dissemination of cryptococcal infection, and reduce PC-related mortality.
Currently, the clinical diagnostic techniques employed for PC primarily comprise lung tissue biopsy, direct microscopic examination of stained infected body fluids, and culture of respiratory samples such as sputum, bronchoalveolar lavage fluid (BALF), pleural effusion, and lung tissue (14). Recently, the detection of cryptococcal antigen (CrAg) in serum using lateral flow assay (LFA) and latex agglutination (LA) has emerged as the most rapid and widely adopted diagnostic method for PC. However, it has been observed that both LFA and LA exhibit lower sensitivity in HIV-negative patients compared to HIV-positive individuals. Specifically, in cases of disseminated disease, the serum CrAg sensitivity for both LA and LFA in HIV-positive individuals exceeded 94%, whereas in HIV-negative patients, the sensitivity was only 78.1% and 82.6% for LA and LFA, respectively (15). Additionally, CrAg testing in lung aspirates has demonstrated its diagnostic value for PC (16), but concerns persist regarding its routine application in BALF (17). Moreover, the relationship between the CrAg titre and therapeutic outcomes remains unclear, further contributing to various limitations in CrAg testing. Therefore, the development of an alternative or auxiliary non-invasive, efficient, rapid, sensitive, and specific method or biomarker for the diagnosis of PC is highly desired.
Sphingolipids (SPLs) and their metabolites serve as essential structural constituents of the cell membrane and participate in various signal transduction processes and biological functions, including cell growth, differentiation, senescence, and programmed cell death. Disruptions in SPL metabolism are closely related to heart disease, diabetes, neurodegenerative diseases, tumours, chronic obstructive pulmonary disease, cystic fibrosis, and other respiratory infections (18,19). Previous study has indicated that the level of the SPL Sa (d16:0) in serum can serve as a biomarker for the rapid diagnosis of Talaromyces marneffei infection in HIV-negative patients (20), suggesting the involvement of SPL in fungal infectious diseases.
In light of these considerations, this study aimed to explore novel biomarkers for diagnosing PC through sphingolipidomic analysis of serum. We collected a clinical cohort comprising patients with PC, PA, and TB, and healthy controls. The serum SPL components were analysed using liquid chromatography-mass spectrometry (LC-MS) to identify specific markers that can facilitate the accurate diagnosis of PC. We present this article in accordance with the STARD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-125/rc).
Methods
Study design and participants
The participants for this study were recruited from two medical institutions: the First Affiliated Hospital of Guangzhou Medical University (Guangzhou, China) and the First Affiliated Hospital of Guangxi Medical University (Guangxi, China). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. 2019-26). All participants agreed with and signed the consent form before enrolment. The recruitment process strictly adhered to the guidelines set forth by the Chinese Center for Disease Control and Prevention (CDC) and the World Health Organization (WHO). Prior to sample collection, written informed consent was obtained from all participants in accordance with the principles outlined in the Declaration of Helsinki. The clinical data and serum samples of patients diagnosed with PC, PA, and TB, as well as healthy controls were collected for analysis in this study.
Inclusion criteria for patients with PC
Patients are enrolled when they are diagnosed with PC after pathology or culture. The inclusion and exclusion criteria for patients diagnosed with PC adhered to the guidelines for the diagnosis and management of invasive fungal diseases (21). The inclusion criteria were as follows: (I) age ≥18 years; (II) confirmed negative HIV test; (III) first onset of the disease; (IV) provision of signed informed consent; and (V) presence of clinical and/or imaging manifestations indicative of PC, supported by microbiological examination or histopathological examination results meeting any of the following conditions: (I) identification of Cryptococcus in or cryptococcal growth from samples of blood, pleural effusion (consistent with a clinically determined site of infection), or smears of diseased lung tissue specimens collected under sterile conditions. (II) Detection of Cryptococcus in or cryptococcal growth from pus smears of the extrapulmonary infection site, collected under sterile conditions from patients with disseminated infection. (III) Identification of granulomatous lesions through histopathological examination of specimens collected from diseased tissue via bronchoscopy, lung puncture, or surgery, accompanied by corresponding tissue inflammation and confirmation of cryptococcus through special staining. All the patients we included were patients with simple PC, and the patients had no neurological symptoms at the time of admission and follow-up.
Inclusion criteria for patients with PA and TB
Patients with PA and TB were required to meet the following criteria: (I) age ≥18 years; (II) confirmed negative HIV test; (III) first onset of PA or TB and prior to treatment; (IV) fulfilling the criteria for the diagnosis of PA or TB, as specified in the guidelines for the diagnosis and management of invasive fungal diseases (21) and the guidelines for the diagnosis and treatment of TB (22), respectively; and (V) providing signed informed consent.
Inclusion criteria for healthy controls
Healthy controls were also recruited from the First Affiliated Hospital of Guangzhou Medical University (Guangzhou, China). Healthy controls were selected to match the age and sex of the patient groups and were free of documented pulmonary infectious, chronic or malignant diseases.
Serum sample collection and processing
Serum samples were collected from patients prior to treatment, as well as at 3 months and 6 months after enrolment. The collection procedure followed routine clinical operation guidelines and was conducted by trained nurses. Blood was collected using separation tubes and allowed to coagulate at room temperature for 30 min. Subsequently, the samples were centrifuged at 3,000 rpm for 5 min to obtain serum, which was then stored at −80 ℃ until SPL extraction.
SPL extraction
SPLs were extracted from serum using a three-step extraction procedure previously described (23). Briefly, 20 µL of serum, 0.75 mL of chloroform (CHCl3)/methanol (MeOH) solvent (1:2, v/v), and 10 µL of a 2.5 µM SPL internal standard (Avanti Polar Lipids Inc., AL, USA) were combined in a borosilicate glass tube. The mixture was sonicated for 30 s at room temperature and incubated at 48 ℃ for 12 h to extract total lipids. Then, alkaline hydrolysis was performed for 2 h by adding 75 µL of 1 M potassium hydroxide. After neutralization with acetic acid, the supernatant was transferred to a new bottle, and the residue underwent sequential extraction with 1 mL of CHCl3/MeOH solvent (2:1, v/v) solvent and a mixed solvent containing 0.4 mL of CHCl3/MeOH (1:2, v/v), 1 mL of CHCl3 and 2 mL of water (H2O). Finally, the extract was combined, dried under a nitrogen flow, and redissolved with 100 µL of MeOH for sphingolipidomic analysis.
Ultra-high-pressure liquid chromatography (UHPLC)-triple-quadrupole (QQQ)-mass spectrometer (MS) analysis
Sphingolipidomic analysis of the serum samples was conducted using an optimized UHPLC-MS approach (24). Chromatographic separation was performed using an Agilent 1290 Infinity UHPLC system (Agilent, Santa Clara, CA, USA) equipped with an Agilent Eclipse Plus C18 column (100 mm × 2.1 mm, 1.8 µm). The mobile phase consisted of (I) MeOH/H2O/formic acid (FA) (60:40:0.2, v/v/v) and (II) MeOH/isopropanol (IPA)/FA (60:40:0.2, v/v/v), both containing 10 mM ammonium acetate. An Agilent 6460 QQQ MS operated in multiple reaction monitoring (MRM) mode and positive ionization mode was used for quantifying SPLs.
CrAg assay
Quantitative detection of CrAg in serum samples was performed using LFA according to the manufacturer’s instructions. In brief, 40 µL of the specimen was placed on the test strip, and the readings were obtained using an immune-quantitative analyser after a 10-min incubation. A reading of ≥8 indicated a positive result for cryptococcosis.
Statistical analysis
Measurement data were presented as the means with standard deviations (SD), while count data were expressed as frequencies or percentages. For continuous data that were normally distributed, the t-test was used to compare the means of two groups, while the Mann-Whitney test was employed for non-normally distributed data. To assess the association between two categorical variables, such as sex and the presence of symptoms, the Fisher’s exact test was applied. Statistical analysis was performed using SPSS software (v25.0; IBM, Armonk, NY, USA), with a P value less than 0.05 considered statistically significant.
The raw data were acquired and processed using Agilent MassHunter Quantitative Analysis B.09.00 software. Subsequently, the processed data were imported into SIMCA 17 software (Sartorius, Gottingen, Germany) for multivariate statistical analysis. Orthogonal partial least squares discriminate analysis (OPLS-DA) was carried out to explore the differentially expressed SPLs responsible for discriminating among the different groups. The goodness of fit of the OPLS-DA model was assessed by the R2Y and Q2Y values, and cumulative values of R2Y and Q2Y close to 1 demonstrated an excellent model. SPLs with variable importance in projection (VIP) values greater than 1.00 were considered potential diagnostic biomarkers. GraphPad Prism 9 software (GraphPad Software, CA, USA) was employed to construct the receiver operating characteristic (ROC) curve and heatmap plot, as well as to perform all additional statistical analyses. The ROC curve was plotted by sensitivity (true positive rate) against 1−specificity (false positive rate) for all possible cut-off values, and the area under the ROC curve (AUC) was used to reflect the diagnostic efficacy of the potential biomarkers. A higher AUC value, closer to 1, indicates a better overall diagnostic performance of the biomarker.
Results
Characteristics of the study participants
A total of 47 patients with PC (27 in the test cohort and 20 in the validation cohort), 27 patients with PA, and 18 patients with TB, and 40 healthy controls (20 in the test cohort and 20 in the validation cohort) were enrolled in this study (Table 1). No significant differences in age or sex were observed among the groups.
Table 1. Participant characteristics by age and sex.
| Group | Cohort | Number of cases | Age, years | Sex (female), % |
|---|---|---|---|---|
| Control | Test | 20 | 34.85±8.89 | 50.00 |
| Validation | 20 | 33.80±8.64 | 50.00 | |
| PC | Test | 27 | 44.56±11.80 | 33.33 |
| Validation | 20 | 44.95±17.54 | 50.00 | |
| PA | 27 | 39.81±13.19 | 51.85 | |
| TB | 18 | 42.17±13.98 | 22.22 |
Age is shown as the mean ± standard deviation. Control, healthy control; PC, pulmonary cryptococcosis; PA, pulmonary aspergillosis; TB, pulmonary tuberculosis.
In clinical work, we found that the symptoms of PC were not typical; most patients only had cough, expectoration and other nonspecific symptoms, and it was difficult to distinguish PC patients from PA and TB patients. Therefore, we collected the clinical information of patients with PC, PA and TB, analysed the data, and found that it was indeed difficult for us to distinguish PC, PA and TB patients according to the clinical characteristics or imaging features of the patients (Table 2).
Table 2. Comparison of the clinical features of the PC, PA and TB patients.
| Clinical features | Baseline of PC | After 3–4 months PC treatment | PA | P value (baseline of PC vs. PA) | TB | P value (baseline of PC vs. TB) |
|---|---|---|---|---|---|---|
| Characteristics | ||||||
| Age (years) | 44.72±14.35 | – | 39.81±13.19 | 0.141 | 42.17±13.98 | 0.647 |
| Female | 19 (40.42) | – | 14 (51.85) | 0.341 | 4 (22.22) | 0.170 |
| Symptoms | ||||||
| Fever | 5 (12.2) | 0 | 4 (14.3) | 0.800 | 2 (11.1) | 0.906 |
| Cough | 24 (58.5) | 6 (31.6) | 27 (96.4) | <0.001 | 11 (61.1) | 0.853 |
| Expectoration | 19 (46.3) | 5 (26.3) | 25 (89.3) | <0.001 | 11 (61.1) | 0.296 |
| Sore throat | 2 (4.9) | 0 | 0 | 0.511 | 0 | 1.000 |
| Shortness of breath | 7 (17.1) | 3 (15.8) | 7 (25.0) | 0.421 | 5 (27.8) | 0.483 |
| Chest tightness/pain | 13 (31.7) | 6 (31.6) | 8 (28.6) | 0.781 | 6 (33.3) | 0.902 |
| Dyspnoea | 0 | 0 | 1 (3.6) | 0.406 | 1 (5.6) | 0.305 |
| Headache | 1 (2.4) | 0 | 0 | 1.000 | 0 | 1.000 |
| Dry rales | 1 (2.4) | 0 | 1 (3.6) | 1.000 | 0 | 1.000 |
| Moist rales | 6 (14.6) | 0 | 9 (32.1) | 0.083 | 3 (16.7) | 0.842 |
| Laboratory test | ||||||
| Erythrocyte sedimentation rate (mm/h) | 35.06±30.01 | 21.23±20.80 | 51.32±43.35 | 0.116 | 46.67±34.15 | 0.215 |
| White blood cell (×109/L) | 7.33±2.50 | 6.04±1.67 | 7.70±3.95 | 0.637 | 7.28±2.70 | 0.946 |
| Neutrophil (×109/L) | 4.81±2.25 | 3.34±1.34 | 5.31±3.76 | 0.488 | 4.78±2.81 | 0.969 |
| Lymphocyte (×109/L) | 1.67±0.74 | 2.00±0.49 | 1.52±0.60 | 0.371 | 1.67±0.67 | 0.974 |
| Haemoglobin (g/L) | 129.30±22.63 | 142.92±14.95 | 116.00±18.00 | 0.012 | 120.50±27.43 | 0.203 |
| Platelet (×109/L) | 265.86±110.45 | 214.85±35.10 | 253.07±113.73 | 0.642 | 279.33±76.76 | 0.641 |
| 1,3-ß-D-glucan ≥100 pg/mL | 6 (12.61) | 1 (8.3) | 5 (23.8) | 0.478 | 0 | 0.517 |
| Radiological features | ||||||
| Ground glass opacity | 8 (19.5) | 4 (16.7) | 4 (14.3) | 0.574 | 1 (5.6) | 0.170 |
| Interlobular thickening | 0 | 1 (4.2) | 0 | – | 0 | – |
| Cystic lesions | 2 (4.9) | 2 (8.3) | 5 (17.9) | 0.080 | 0 | 1.000 |
| Consolidation or patches | 36 (87.8) | 21 (87.5) | 26 (92.9) | 0.495 | 17 (94.4) | 0.437 |
| Plural effusion | 1 (2.4) | 0 | 6 (21.4) | 0.010 | 5 (27.8) | 0.003 |
| Small nodules | 32 (78.0) | 18 (75.0) | 14 (50.0) | 0.015 | 7 (38.9) | 0.003 |
Data are presented as number (percentage) or mean ± standard deviation. PC, patients with pulmonary cryptococcosis; PA, patients with pulmonary aspergillosis; TB, patients with pulmonary tuberculosis.
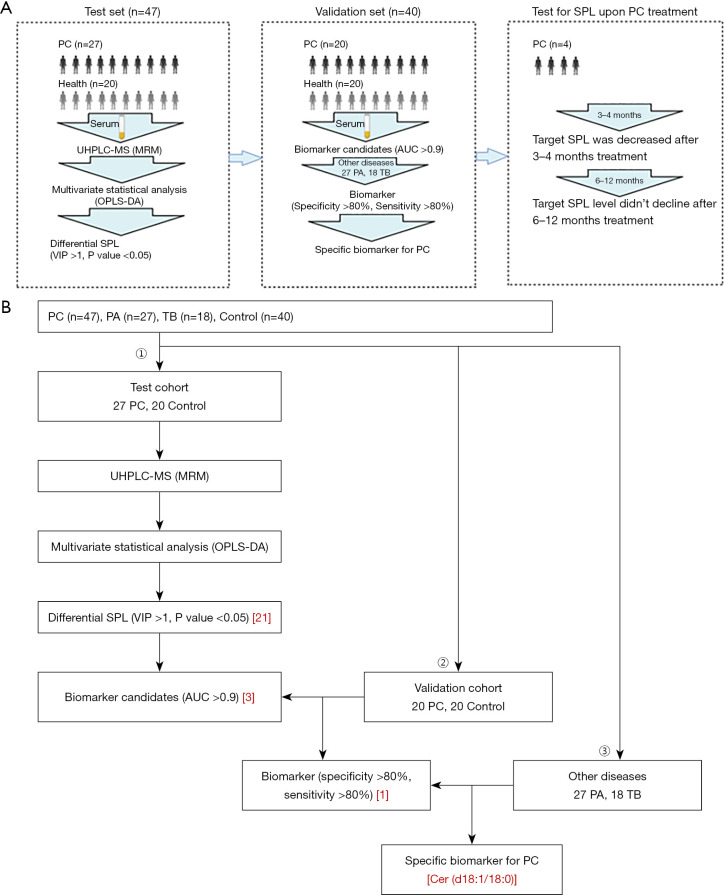
These patients were divided into different cohorts, including test and validation cohorts as well as a follow-up cohort. The serum SPLs were analysed step by step to obtain specific SPL markers for PC diagnosis (Figure 1A,1B).
Figure 1.
Flow chart. (A) Design of the study. From the establishment of a clinical cohort to the collection, treatment and analysis of serum and then to the follow-up of those patients upon PC treatment. (B) Scheme to screen specific diagnostic biomarkers for PC. PC, patients with pulmonary cryptococcosis; UHPLC-MS, ultra high performance liquid chromatography-mass spectrometry technology; MRM, multiple reaction monitoring; OPLS-DA, orthogonal partial least squares discriminate analysis; VIP, variable importance in the projection; AUC, area under curve; PA, patients with pulmonary aspergillosis; TB, patients with pulmonary tuberculosis; SPL, sphingolipid.
Comparison of serum SPLs between different groups of participants
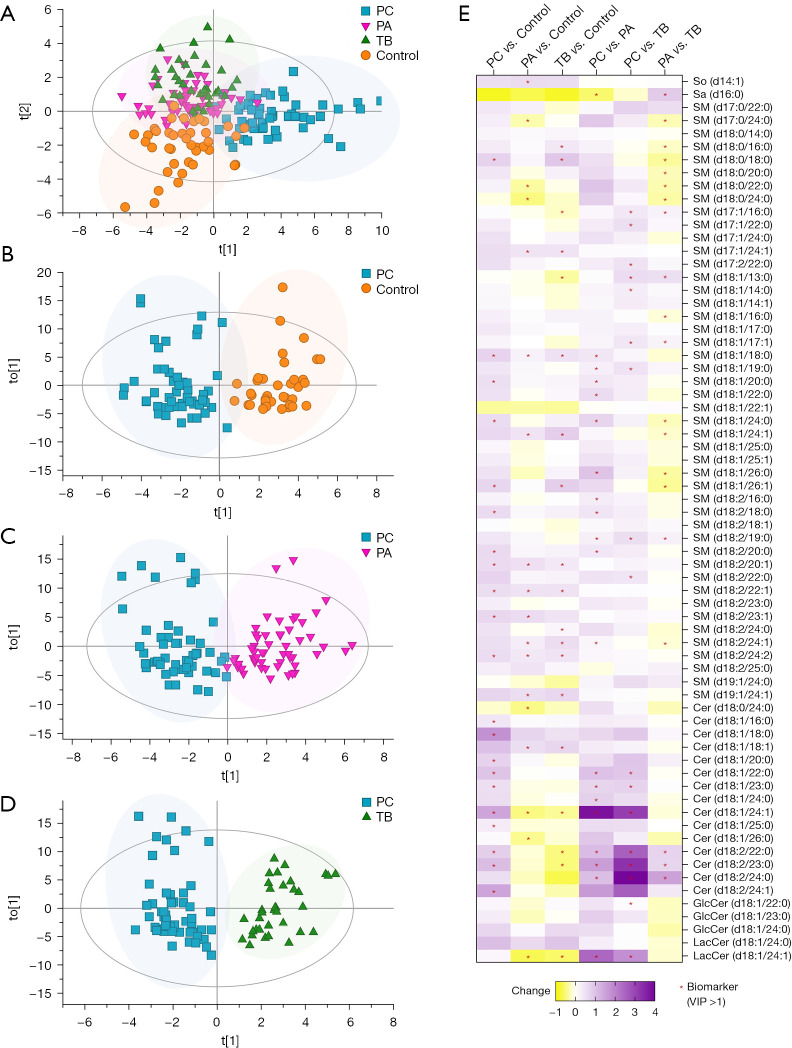
A total of 68 SPLs, including 46 sphingomyelins (SMs), 15 ceramides (Cers), 3 hexosylceramides (HexCers), 2 lactosylceramides (LacCers), 1 sphingosine (So), and 1 sphinganine (Sa), were quantified for each sample using the established UHPLC-QQQ-MS approach. Multivariate analysis was performed to explore the SPLs associated with the discrimination of different groups.
Figure 2A exhibits distinct separation between the control group and the patient groups, with the PC group showing clear discrimination from the control, PA and TB groups. However, some overlap was observed between the PA and TB groups. This indicated that the PC group exhibited unique serum SPL profiles compared to the control group and other pulmonary infection groups.
Figure 2.
Comparison of serum SPLs between different groups of participants. OPLS-DA score plots show discrimination between (A) all participant groups, including control (test cohort, n=20), PC (test cohort, n=27), PA (n=27), and TB (n=18) (R2X =0.806, R2Y =0.543, Q2 =0.356) groups; (B) control (test cohort) and PC (test cohort) (R2X =0.796, R2Y =0.823, Q2 =0.544) groups; (C) PC (test cohort) and PA (R2X =0.724, R2Y =0.784, Q2 =0.645) groups; and (D) PC (test cohort) and TB (R2X =0.767, R2Y =0.854, Q2 =0.714) groups. (E) A heatmap plot displays the change in each SPL between groups, including PC vs. control, PA vs. control, TB vs. control, PC vs. PA, PC vs. TB, and PA vs. TB; biomarkers (VIP >1) between the groups are labelled with a red asterisk (*). Change (PC vs. control) = (level in PC − level in control)/level in control. PC, patients with pulmonary cryptococcosis; PA, patients with pulmonary aspergillosis; TB, patients with pulmonary tuberculosis; control, healthy controls; So, sphingosine; Sa, sphinganine; SM, sphingomyelin; Cer, ceramide; GlcCer, glucosylceramide; LacCer, lactosylceramide; VIP, variable importance in the projection; SPLs, sphingolipids; OPLS-DA, orthogonal partial least squares discriminate analysis.
The OPLS-DA score plot (shown in Figure 2B) further confirmed the clear separation between the PC group and the control group, with R2X =0.796, R2Y =0.823, Q2 =0.544. Subsequently, the PC group was compared with the PA and TB groups. As shown in Figure 2C,2D, the PC group demonstrated obvious discrimination from the PA group (R2X =0.724, R2Y =0.784, Q2 =0.645) and the TB group (R2X =0.767, R2Y =0.854, Q2 =0.714). Twenty biomarkers, including 11 SMs, 7 Cers, 1 LacCer and 1 Sa, distinguished the PC group from the PA group, while 17 biomarkers, comprising 9 SMs, 6 Cers, 1 HexCer and 1 LacCer, distinguished the PC group from the TB group. The changes in these SPLs are visually presented in a heatmap plot (Figure 2E).
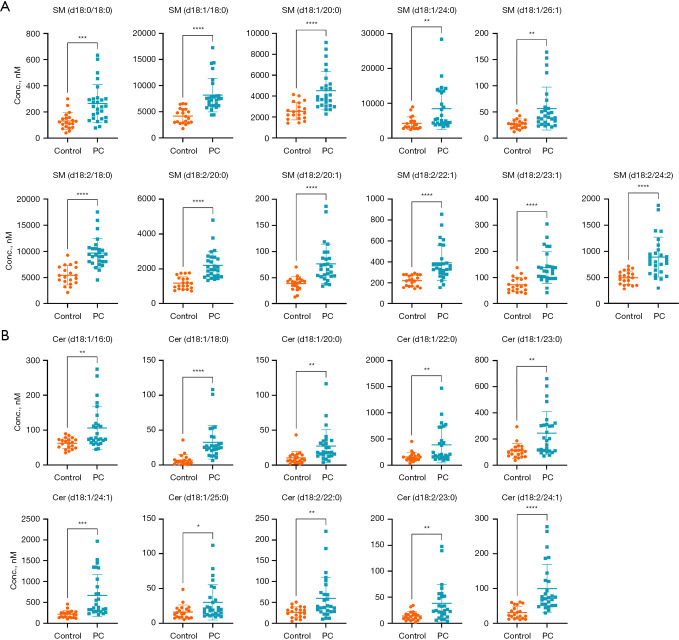
Notably, 21 SPLs, consisting of 11 SMs (Figure 3A) and 10 Cers (Figure 3B), were upregulated in the PC group.
Figure 3.
Potential biomarkers for differentiating PC patients from controls. The (A) 11 SMs and (B) 10 Cers differentially expressed in PC patients. Each dot represents a participant sample. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. SM, sphingomyelin; control, healthy controls; PC, pulmonary cryptococcosis; Cer, ceramide.
Evaluation of SPL biomarkers for the diagnosis of PC
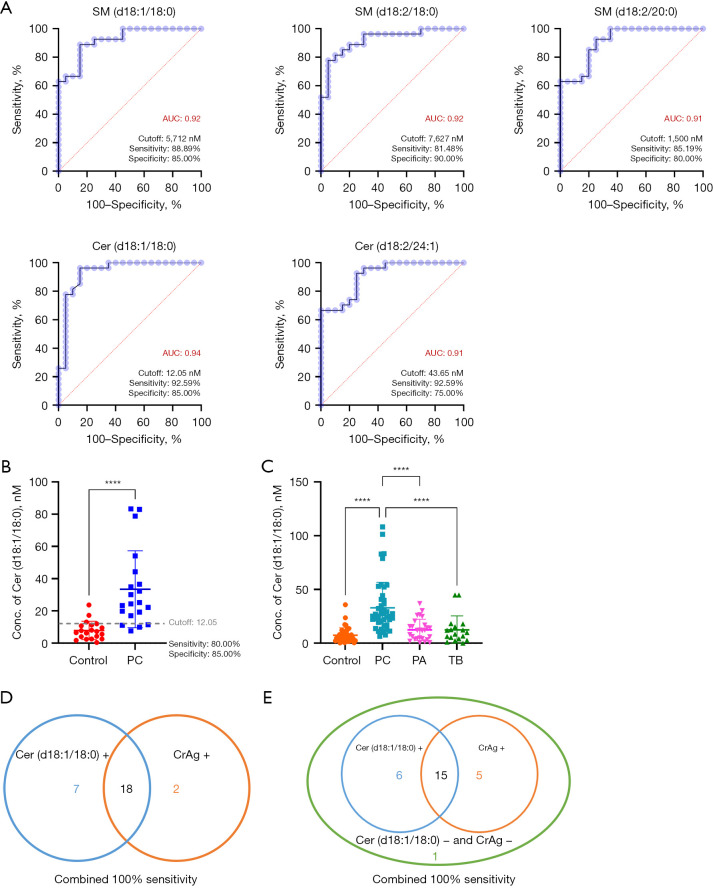
The evaluation of specific diagnostic biomarkers for PC encompassed two key aspects: (I) verification of biomarker candidates in the validation cohort, comprising 20 controls and 20 PC patients; and (II) assessment of the sensitivity and specificity of the potential biomarkers using other pulmonary infection groups (Figure 1B).
As mentioned earlier, multivariate analysis revealed 21 differentially expressed SPLs in the PC group (Figure 3A,3B). SPLs with an AUC ≥0.9 were considered potential diagnostic biomarkers for PC. Among them, 5 SPLs, namely, SM (d18:1/18:0), SM (d18:2/18:0), SM (d18:2/20:0), Cer (d18:1/18:0) and Cer (d18:2/24:1), displayed an AUC ≥0.9 (Figure 4A). Subsequently, these 5 SPLs were validated using a separate cohort, and the optimal cut-off value (12.05 nM) determined from the test cohort were applied. Among these SPLs, Cer (d18:1/18:0) presented a sensitivity of 80.00% and specificity of 85.00% in the validation cohort (Figure 4B), making it a promising diagnostic biomarker,
Figure 4.
Diagnostic performance of the selected biomarker. (A) The ROC curves of 5 SPLs with an AUC ≥0.9, namely, SM (d18:1/18:0), SM (d18:2/18:0), SM (d18:2/20:0), Cer (d18:1/18:0) and Cer (d18:2/24:1). (B) Scatter plot showing the serum level of Cer (d18:1/18:0) in the validation cohort (control: n=20; PC: n=20). (C) Scatter plot showing the serum level of Cer (d18:1/18:0) in different participant groups (control: n=40; PC: n=47; PA: n=27; TB: n=18). (D) The sensitivity compared between Cer (d18:1/18:0) (12.05 nM as the cut-off value) and the CrAg assay. (E) The sensitivity compared between Cer (d18:1/18:0) (18.00 nM as the cut-off value) and the CrAg assay. ****, P<0.0001. SM, sphingomyelin; AUC, area under curve; Cer, ceramide; control, healthy controls; PC, pulmonary cryptococcosis; PA, pulmonary aspergillosis; TB, pulmonary tuberculosis; CrAg, cryptococcal antigen; ROC, receiver operating characteristic; SPL, sphingolipid.
To ascertain the specificity of Cer (d18:1/18:0) for the diagnosis of PC, its levels were compared among PC patients and other groups with potential confounding conditions. As shown in Figure 4C, the level of Cer (d18:1/18:0) was significantly elevated in PC patients compared to controls, as well as PA and TB patients. However, when a cut-off value of 12.05 nM for Cer (d18:1/18:0) was employed, only 51.85% of PA cases and 61.11% of TB cases were correctly identified as negative, indicating a high probability of misdiagnosis. Adopting a cut-off value of 18.00 nM could significantly decrease the false-positive rate for PA and TB, although the sensitivity for PC slightly decreased. Considering the overall classification performance for PC, PA and TB, a serum concentration of Cer (d18:1/18:0) higher than 18.00 nM can be deemed as the optimal cut-off value for PC diagnosis. Consequently, Cer (d18:1/18:0) exhibited a sensitivity of 76.60%, true negative rate of 95.00% for controls, true negative rate of 77.78% for PA, and true negative of 83.33% for TB (Table 3).
Table 3. Sensitivity and specificity of different Cer (d18:1/18:0) cut-offs.
| Sensitivity or specificity | Cut-off value (nM) | |
|---|---|---|
| 12.05 | 18.00 | |
| Sensitivity (%) (true positive for PC) | 87.23 | 76.60 |
| Specificity (%) (true negative for control) | 85.00 | 95.00 |
| Specificity (%) (true negative for PA) | 51.85 | 77.78 |
| Specificity (%) (true negative for TB) | 61.11 | 83.33 |
Cer, ceramide; PC, pulmonary cryptococcosis; PA, pulmonary aspergillosis; TB, pulmonary tuberculosis.
Furthermore, the diagnostic performance of Cer (d18:1/18:0) was compared with that of the serum CrAg assay. The CrAg assay indicated that 20 out of 27 PC patients in the test cohort as positive for cryptococcosis, resulting in a sensitivity of 74.07%. In contrast, the sensitivity of Cer (d18:1/18:0) for PC in the test cohort was 92.59% (using a cut-off value of 12.05 nM) and 77.78% (using a cut-off value of 18.00 nM). Therefore, Cer (d18:1/18:0) exhibited a relatively higher diagnostic performance compared to the CrAg assay. Combining Cer (d18:1/18:0) with the serum CrAg assay achieved a diagnostic sensitivity of 100% for PC patients (Figure 4D,4E). This suggested that Cer (d18:1/18:0) can be used in combination with the CrAg assay in clinical settings to achieve high diagnostic accuracy.
Changes in Cer (d18:1/18:0) levels upon treatment
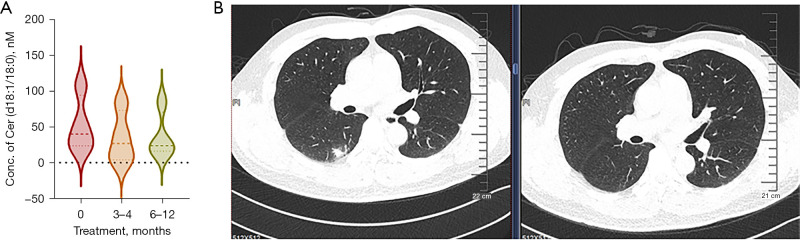
To gain further insights into the role of Cer (d18:1/18:0) in the pathogenesis of PC, serum samples were collected from 4 PC patients who underwent fluconazole treatment for duration of 3–4 and 6–12 months. The objective was to monitor the alterations in Cer (d18:1/18:0) levels over the course of treatment. As shown in Figure 5A, the levels of Cer (d18:1/18:0) showed a declining trend during the initial 3–4 months of treatment; however, statistical significance was not observed. Moreover, no substantial further reduction in the levels of Cer (d18:1/18:0) was observed beyond the 6–12 months treatment period. Notably, the levels of Cer (d18:1/18:0) appeared to be associated with improvements in lung imaging. For example, a patient with an initial Cer (d18:1/18:0) level of 22.04 nM displayed exudative lesions in the middle lobe as revealed by lung imaging. Following 3 months of treatment, the Cer (d18:1/18:0) level decreased to 0.99 nM, accompanied by a significant improvement in lung imaging (Figure 5B).
Figure 5.
Change in the Cer (d18:1/18:0) level upon treatment. (A) Violin plot showing the alteration in the serum level of Cer (d18:1/18:0) in PC patients after treatment; (B) chest HRCT of a confirmed PC patient, image comparison before and after 3–6 months of treatment. Cer, ceramide; PC, pulmonary cryptococcosis; HRCT, high resolution computed tomography.
Discussion
The present study established a cohort of hospitalized patients with pulmonary infection, including PC, PA, and TB, as well as healthy controls, to explore the potential use of SPLs in aiding the diagnosis of PC. Several key findings emerged from this study. Firstly, the composition of SPLs demonstrated clear distinctions among different groups, such as PC versus PA, PC versus TB, and PC versus healthy control. Secondly, the serum marker Cer (d18:1/18:0) was identified as a diagnostic biomarker for PC with a sensitivity and specificity exceeding 80%, thus serving as a specific diagnostic biomarker for PC. Thirdly, the diagnostic performance of Cer (d18:1/18:0) comparable or even surpassed that of the traditional serum CrAg assay. Lastly, changes in the level of Cer (d18:1/18:0) exhibited a positive correlation with the prognosis of PC.
Previous studies have extensively demonstrated the value of SPLs as diagnostic markers for various diseases. These SPLs have been associated with conditions such as heart disease, diabetes, neurodegenerative diseases, tumours, chronic obstructive pulmonary disease, and cystic fibrosis (18,19). Additionally, SPLs have been closely related to infectious diseases. For example, increased plasma levels of SM (d18:0/16:0), SM (d18:1/16:0) and certain glycosphingolipids have been reported observed in patients with community-acquired pneumonia (25). In cases of dengue fever, the level of SM (d18:1/16:0) decreased significantly by 5-fold during the early fever stage (26). The combined assessment of SM (d18:1/22:3) and glycerophospholipid showed excellent diagnostic sensitivity for sepsis (27). Even in coronavirus disease 2019 (COVID-19) patients, SPLs were identified as differentiating metabolites from healthy controls, with S1P (d18:1) significantly decreased in COVID-19 patients (28). These prior studies provide compelling evidence supporting the use of SPLs as diagnostic markers for infectious diseases. In our study, we successfully demonstrated that the SPL composition of PC patients could be clearly distinguished from that of other clinically confounding groups. Of note, serum Cer (d18:1/18:0) exhibited relatively high sensitivity and specificity for the diagnosis of PC, effectively discriminating PC from PA and TB. Thus, Cer (d18:1/18:0) can be served as an auxiliary diagnostic marker for PC. Previous studies have suggested that neutrophils can counteract Cryptococcus through sphingomyelinase activity (29). However, the underlying mechanism governing the generation of Cer (d18:1/18:0) in PC patients remains unclear and will be a focal point of future research.
To further validate the utility of Cer (d18:1/18:0) as an auxiliary diagnostic marker, we compared its diagnostic performance with that of the serum CrAg assay, a confirmed diagnostic marker for PC (20). The sensitivity of the serum CrAg assay was reported to be lower in HIV-negative patients compared to HIV-positive patients, and the sensitivity of serum CrAg in patients with only PC was lower than that in patients with cryptococcal meningitis (30). In addition, infected patients, such as those with TB, may exhibit false-positive reactions in the CrAg assay (7). In our study, the sensitivity of the serum CrAg assay in HIV-negative PC patients was 74.07%, while Cer (d18:1/18:0) exhibited a relatively higher sensitivity in the cohort, reaching 92.59%. Additionally, the relationship between the CrAg assay and treatment efficacy and prognosis is controversial (31,32). Conversely, our study revealed that the level of Cer (d18:1/18:0) gradually returned to normal following treatment and exhibited a positive correlation with the prognosis of PC. These findings suggest that SPLs could serve as biomarkers for monitoring treatment effectiveness.
The combination of various diagnostic methods has emerged as a prominent trend and has proven advantageous in augmenting the diagnostic accuracy for different diseases. For example, previous study has shown that the combination of polymerase chain reaction (PCR), fluorescence in situ hybridization, and immunohistochemical staining can significantly enhance the diagnostic accuracy for mucormycosis (33). Moreover, PCR in combination with galactomannan (GM) exhibits superior diagnostic performance compared to GM alone in identifying Aspergillus infections (34). We therefore conducted an assessment of the combined diagnostic capabilities of the Cer (d18:1/18:0) and CrAg assays. The findings unequivocally indicated that the integration of Cer (d18:1/18:0) and the serum CrAg assay obviously improves the diagnostic accuracy of PC, with even a 100% diagnosis rate. These two methods exhibit strong complementarity and hold promise as effective diagnostic method for future advancements in this field.
However, our study has several limitations. Firstly, lung cancer patients, who might be susceptible to misdiagnosis as PC patients, were not included in the study cohort due to age disparities between the originally enrolled patients and those in other cohorts. Given that age can significantly influence SPL levels, we opted to exclude lung cancer patients from our analysis. In future follow-up studies, we plan to include a substantial number of young lung cancer patients, expand the sample size, and validate the diagnostic efficacy of Cer (d18:1/18:0) in patients with PC and lung cancer. In addition, although adopting a cut-off value of 18.00 nM for Cer (d18:1/18:0) significantly reduces false positives for PA and TB patients, future studies should collect a new cohort of PA and TB patients to validate this finding. Moreover, a larger cohort comparison between the sensitivity of the serum CrAg assay and Cer (d18:1/18:0) should be conducted to enable better comparison. Furthermore, due to the limited availability of samples, only a small sample size was accessible, which could potentially limit the reliability of the identified biomarker’s diagnostic power. Therefore, in future studies, enrolling a larger cohort of PC patients from other medical institutions, including those who have undergone treatment and lung imaging, would be necessary to further evaluate the actual sensitivity and specificity of using Cer (d18:1/18:0) as a specific diagnostic biomarker for PC. Additionally, a larger cohort would allow for a better assessment of changes in Cer (d18:1/18:0) levels during treatment and its implication for prognosis.
Conclusions
This is the first study to explore biomarkers of PC based on sphingolipidomic analysis. We propose Cer (d18:1/18:0) as an alternative diagnostic method specific for PC in HIV-negative patients, and 18.00 nM is the optimal cut-off value with a diagnostic sensitivity of 76.60% and specificity of 95.00%. Of note, this SPL biomarker shows potential for the prediction of therapeutic efficacy, indicating its involvement in the pathology of PC. As a new biomarker for PC, Cer (d18:1/18:0) also demonstrates good complementarity with current biomarkers, as evidenced by its high diagnostic sensitivity when combined with the CrAg assay.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank the patients, along with the nurses and clinical staff who provided patient care, the staff at the hospital respiratory medicine departments, the staff at the hospital clinical laboratories, and the technical staff of the State Key Laboratory of Respiratory Disease for their excellent assistance. Furthermore, we would also like to thank the AJE team for polishing the English language of this manuscript.
Funding: This research was funded by the Guangdong Basic and Applied Basic Research Foundation (No. 2022A1515010089); the National Key Research and Development Project of China (No. 2022YFC0867500); the Science and Technology Program of Guangzhou (No. 202201020537); the Macao Science and Technology Development Fund, Macau Special Administrative Region (No. 082/2017/A2 to JRW); the Open Project of State Key Laboratory of Respiratory Diseases (Nos. SKLRD-OP-202210 and SKLRD-OP-202102); and the Independent Fund of the State Key Laboratory of Respiratory Diseases (No. SKLRD-Z-202019).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. 2019-26). All participants agreed with and signed the consent form before enrolment.
Footnotes
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-125/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-125/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-125/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-125/coif). The authors have no conflicts of interest to declare.
References
- 1.Samarasinghe H, Xu J. Hybrids and hybridization in the Cryptococcus neoformans and Cryptococcus gattii species complexes. Infect Genet Evol 2018;66:245-55. 10.1016/j.meegid.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 2.Kwon-Chung KJ, Fraser JA, Doering TL, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med 2014;4:a019760. 10.1101/cshperspect.a019760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang W, Fa Z, Liao W. Epidemiology of Cryptococcus and cryptococcosis in China. Fungal Genet Biol 2015;78:7-15. 10.1016/j.fgb.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 4.Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 2009;23:525-30. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Li Y, Chen Y, et al. Trends of pulmonary fungal infections from 2013 to 2019: an AI-based real-world observational study in Guangzhou, China. Emerg Microbes Infect 2021;10:450-60. 10.1080/22221751.2021.1894902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gushiken AC, Saharia KK, Baddley JW. Cryptococcosis. Infect Dis Clin North Am 2021;35:493-514. 10.1016/j.idc.2021.03.012 [DOI] [PubMed] [Google Scholar]
- 7.Limper AH, Adenis A, Le T, et al. Fungal infections in HIV/AIDS. Lancet Infect Dis 2017;17:e334-43. 10.1016/S1473-3099(17)30303-1 [DOI] [PubMed] [Google Scholar]
- 8.Yamamura D, Xu J. Update on Pulmonary Cryptococcosis. Mycopathologia 2021;186:717-28. 10.1007/s11046-021-00575-9 [DOI] [PubMed] [Google Scholar]
- 9.Marr KA, Sun Y, Spec A, et al. A Multicenter, Longitudinal Cohort Study of Cryptococcosis in Human Immunodeficiency Virus-negative People in the United States. Clin Infect Dis 2020;70:252-61. 10.1093/cid/ciz193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escandón P, Lizarazo J, Agudelo CI, et al. Cryptococcosis in Colombia: Compilation and Analysis of Data from Laboratory-Based Surveillance. J Fungi (Basel) 2018;4:32. 10.3390/jof4010032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang D, Yu L, Luo J, et al. Characterization of Clinical and CT Manifestations of Pulmonary Cryptococcosis with Consolidation. Arch Iran Med 2021;24:508-11. 10.34172/aim.2021.73 [DOI] [PubMed] [Google Scholar]
- 12.Qiu S, Chen C, Li Y, et al. Pulmonary cryptococcosis misdiagnosed as lung cancer in a man with normal immune function: A case report. Radiol Case Rep 2022;17:1185-9. 10.1016/j.radcr.2022.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis JN, Wainwright H, Harrison TS, et al. Pulmonary cryptococcosis misdiagnosed as smear-negative pulmonary tuberculosis with fatal consequences. Int J Infect Dis 2010;14 Suppl 3:e310-2. 10.1016/j.ijid.2010.02.2255 [DOI] [PubMed] [Google Scholar]
- 14.Maziarz EK, Perfect JR. Cryptococcosis. Infect Dis Clin North Am 2016;30:179-206. 10.1016/j.idc.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hevey MA, George IA, Rauseo AM, et al. Performance of the Lateral Flow Assay and the Latex Agglutination Serum Cryptococcal Antigen Test in Cryptococcal Disease in Patients with and without HIV. J Clin Microbiol 2020;58:e01563-20. 10.1128/JCM.01563-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liaw YS, Yang PC, Yu CJ, et al. Direct determination of cryptococcal antigen in transthoracic needle aspirate for diagnosis of pulmonary cryptococcosis. J Clin Microbiol 1995;33:1588-91. 10.1128/jcm.33.6.1588-1591.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senghor Y, Guitard J, Angoulvant A, et al. Cryptococcal antigen detection in broncho-alveolar lavage fluid. Med Mycol 2018;56:774-7. 10.1093/mmy/myx092 [DOI] [PubMed] [Google Scholar]
- 18.Blachnio-Zabielska A, Hajduch E, Le Stunff H. Editorial: The Role of Sphingolipid Metabolism in the Development of Type 2 Diabetes and Obesity. Front Endocrinol (Lausanne) 2021;12:835751. 10.3389/fendo.2021.835751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Zhang H, Si Y, et al. High-coverage lipidomics analysis reveals biomarkers for diagnosis of acute exacerbation of chronic obstructive pulmonary disease. J Chromatogr B Analyt Technol Biomed Life Sci 2022;1201-1202:123278. 10.1016/j.jchromb.2022.123278 [DOI] [PubMed] [Google Scholar]
- 20.Li ZT, Yau LF, Qiu Y, et al. Serum Sphingolipids Aiding the Diagnosis of Adult HIV-Negative Patients with Talaromyces marneffei Infection. Front Cell Infect Microbiol 2021;11:701913. 10.3389/fcimb.2021.701913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donnelly JP, Chen SC, Kauffman CA, et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium . Clin Infect Dis 2020;71:1367-76. 10.1093/cid/ciz1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO consolidated guidelines on tuberculosis: Module 3: Diagnosis – Tests for tuberculosis infection. Geneva: World Health Organization; 2022. [PubMed] [Google Scholar]
- 23.Li J, Xie LM, Song JL, et al. Alterations of Sphingolipid Metabolism in Different Types of Polycystic Ovary Syndrome. Sci Rep 2019;9:3204. 10.1038/s41598-019-38944-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JR, Zhang H, Yau LF, et al. Improved sphingolipidomic approach based on ultra-high performance liquid chromatography and multiple mass spectrometries with application to cellular neurotoxicity. Anal Chem 2014;86:5688-96. 10.1021/ac5009964 [DOI] [PubMed] [Google Scholar]
- 25.To KK, Lee KC, Wong SS, et al. Lipid metabolites as potential diagnostic and prognostic biomarkers for acute community acquired pneumonia. Diagn Microbiol Infect Dis 2016;85:249-54. 10.1016/j.diagmicrobio.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui L, Lee YH, Kumar Y, et al. Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl Trop Dis 2013;7:e2373. 10.1371/journal.pntd.0002373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neugebauer S, Giamarellos-Bourboulis EJ, Pelekanou A, et al. Metabolite Profiles in Sepsis: Developing Prognostic Tools Based on the Type of Infection. Crit Care Med 2016;44:1649-62. 10.1097/CCM.0000000000001740 [DOI] [PubMed] [Google Scholar]
- 28.Song JW, Lam SM, Fan X, et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab 2020;32:188-202.e5. 10.1016/j.cmet.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qureshi A, Subathra M, Grey A, et al. Role of sphingomyelin synthase in controlling the antimicrobial activity of neutrophils against Cryptococcus neoformans. PLoS One 2010;5:e15587. 10.1371/journal.pone.0015587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pappas PG, Perfect JR, Cloud GA, et al. Cryptococcosis in human immunodeficiency virus-negative patients in the era of effective azole therapy. Clin Infect Dis 2001;33:690-9. 10.1086/322597 [DOI] [PubMed] [Google Scholar]
- 31.Fisher JF, Valencia-Rey PA, Davis WB. Pulmonary Cryptococcosis in the Immunocompetent Patient-Many Questions, Some Answers. Open Forum Infect Dis 2016;3:ofw167. 10.1093/ofid/ofw167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohno S, Kakeya H, Izumikawa K, et al. Clinical features of pulmonary cryptococcosis in non-HIV patients in Japan. J Infect Chemother 2015;21:23-30. 10.1016/j.jiac.2014.08.025 [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Song Y, Li R. The use of combined PCR, fluorescence in situ hybridisation and immunohistochemical staining to diagnose mucormycosis from formalin-fixed paraffin-embedded tissues. Mycoses 2021;64:1460-70. 10.1111/myc.13382 [DOI] [PubMed] [Google Scholar]
- 34.Avni T, Levy I, Sprecher H, et al. Diagnostic accuracy of PCR alone compared to galactomannan in bronchoalveolar lavage fluid for diagnosis of invasive pulmonary aspergillosis: a systematic review. J Clin Microbiol 2012;50:3652-8. 10.1128/JCM.00942-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as