Abstract
The dinucleotide cyclic di-AMP (c-di-AMP) is synthesized as a second messenger in the Gram-positive model bacterium Bacillus subtilis as well as in many bacteria and archaea. Bacillus subtilis possesses three diadenylate cyclases and two phosphodiesterases that synthesize and degrade the molecule, respectively. Among the second messengers, c-di-AMP is unique since it is essential for B. subtilis on the one hand but toxic upon accumulation on the other. This role as an “essential poison” is related to the function of c-di-AMP in the control of potassium homeostasis. C-di-AMP inhibits the expression and activity of potassium uptake systems by binding to riboswitches and transporters and activates the activity of potassium exporters. In this way, c-di-AMP allows the adjustment of uptake and export systems to achieve a balanced intracellular potassium concentration. C-di-AMP also binds to two dedicated signal transduction proteins, DarA and DarB. Both proteins seem to interact with other proteins in their apo state, i.e. in the absence of c-di-AMP. For DarB, the (p)ppGpp synthetase/hydrolase Rel and the pyruvate carboxylase PycA have been identified as targets. The interactions trigger the synthesis of the alarmone (p)ppGpp and of the acceptor molecule for the citric acid cycle, oxaloacetate, respectively. In the absence of c-di-AMP, many amino acids inhibit the growth of B. subtilis. This feature can be used to identify novel players in amino acid homeostasis. In this review, we discuss the different functions of c-di-AMP and their physiological relevance.
Keywords: potassium transport, osmoregulation, Bacillus subtilis, cyclic di-AMP, second messenger, third messenger, signal transduction
Cyclic di-AMP (c-di-AMP) signaling in Bacillus subtilis. The second messenger c-di-AMP is synthesized by the diadenylate cyclases DisA, CdaA, and CdaS and degraded by the phosphodiesterases GdpP and PgpH. C-di-AMP binds to signal transduction proteins that can be regarded as third messengers and can control the expression and activity of several transporters by binding to mRNA riboswitches and proteins, respectively. The Gram-positive bacterium uses c-di-AMP to control potassium and osmotic homeostasis and to adapt cellular activities to the potassium availability.
Introduction
Bacteria are exposed to constantly changing environments. Due to their small size, they experience this exposure much more severely than larger organisms such as complex multicellular eukaryotes do. Moreover, the rapid growth of bacteria makes very fast responses extremely important. However, in some cases, the original signals are not suited for direct signal transduction, or they have to reach out to a variety of different cellular targets. Indeed, to achieve coordinated regulation of a single biological process at multiple steps (such as gene expression and protein activity), which has been called sustained sensing (Orr et al. 2016), classical protein-based signal transduction systems are often not well suited. In such instances, bacteria use so-called second messengers; dedicated molecules that orchestrate a cellular response, often by interaction with a variety of target molecules. These target molecules may be protein or RNA molecules (Nelson and Breaker 2017, Hengge et al. 2019). The second messengers are typically specific mono- or dinucleotides that are not used as building blocks for cellular nucleic acids. The most intensively studied of these second messengers is cyclic AMP (cAMP), which governs the paradigmatic glucose-lactose diauxie in Escherichia coli (Görke and Stülke 2008). In addition to carbon catabolite repression, cAMP participates in other processes, including virulence or nitrogen utilization (Görke and Stülke 2008, Green et al. 2014).
The Gram-positive model bacterium Bacillus subtilis is unable to form the classical 3’,5’-cAMP; however, this bacterium synthesizes cyclic dinucleotide second messengers, i.e. cyclic di-AMP (c-di-AMP) and cyclic di-GMP (c-di-GMP), as well as the alarmones guanosine tetraphosphate and guanosine pentaphosphate (collectively referred to as (p)ppGpp) (Rhaese et al. 1975, Oppenheimer-Shaanan et al. 2011, Diethmaier et al. 2014). In addition, 5-amino-4-imidazole carboxamide riboside 5′-triphosphate (ZTP) and diadenosine tetraphosphate (Ap4A) were recently shown to be second messenger molecules in B. subtilis (Chandrangsu et al. 2019, Giammarinaro et al. 2022).
Even though the role of c-di-GMP is poorly explored in B. subtilis, it seems to contribute to the decision-making between two mutually exclusive lifestyles: motility and biofilm formation. C-di-GMP binds to the receptor protein MotI, and this complex interacts with the flagellar stator protein MotA to inhibit motility (Chen et al. 2012). In contrast, the synthesis of an extracellular polysaccharide by the YdaJLMN complex requires a functional interaction with the c-di-GMP target protein YdaK (Bedrunka and Graumann 2017). Thus, c-di-GMP-mediated signal transduction seems to control lifestyle choice in B. subtilis, as observed for many other bacteria (Hengge 2009, Jenal et al. 2017).
The alarmone (p)ppGpp is synthesized by the bifunctional (p)ppGpp-synthetase/hydrolase Rel in response to amino acid starvation as well as by the so-called small alarmone synthetases SasA and SasB (Ronneau and Hallez 2019, Bange et al. 2021). As the name alarmone suggests, the formation of (p)ppGpp as a result of amino acid starvation indicates to the cell the need for a global re-organization of cellular activities. Transcription, translation, and DNA replication are shut down, whereas pathways for amino acid syntheses are activated. This principal logic is conserved in different bacteria; however, the precise mechanisms of (p)ppGpp action are different. While the molecules directly bind to the RNA polymerase in E. coli, the control of transcription is indirect in Gram-positive bacteria, including B. subtilis. Here, (p)ppGpp specifically inhibits the synthesis of purine nucleotides and particularly of GTP. Since many mRNAs use a GTP as the initiation nucleotide, the transcription of these mRNAs is inhibited upon the accumulation of (p)ppGpp. In contrast, mRNAs that initiate with an ATP, are more strongly expressed during the stringent response. Similarly, (p)ppGpp negatively affects the accumulation of purine nucleotides in both E. coli and B. subtilis, but this is achieved in different ways. In E. coli, the stringent response triggers the degradation of the nucleotides, whereas (p)ppGpp binds to multiple enzymes of purine biosynthesis to inhibit their activities in B. subtilis (Liu et al. 2015, Anderson et al. 2019, Zhang et al. 2019).
The second messenger ZTP is involved in the mobilization of zinc under conditions of zinc limitation. This molecule binds to the metallochaperone ZagA, which in turn mobilizes zinc ions from dispensable ribosomal proteins to the essential and strictly zinc-dependent GTP cyclohydrolase FolE that is required for folate biosynthesis and tRNA modification (Chandrangsu et al. 2019).
Finally, the dinucleotide Ap4A was only very recently shown to be a bona-fide second messenger. This molecule binds to the IMP dehydrogenase GuaB, an enzyme of the GMP biosynthetic pathway, and inhibits its activity (Giammarinaro et al. 2022).
Thus, second messengers control a wide range of functions in B. subtilis (see http://www.subtiwiki.uni-goettingen.de/v4/category?id=SW.3.5; Pedreira et al. 2022). In this review, we will focus on the regulatory functions of the second messenger c-di-AMP in B. subtilis. Strikingly, c-di-AMP and (p)ppGpp-mediated signal transduction as well as their biosynthesis and degradation are tightly intertwined, highlighting the central role of both second messengers for the physiology of B. subtilis.
Among all second messengers, c-di-AMP is unique as it is on the one hand essential for many of the bacteria that use it, and on the other hand, it becomes toxic if the intracellular concentration gets too high. The molecule was therefore dubbed an essential poison (Gundlach et al. 2015b, Huynh and Woodward 2016). In addition, c-di-AMP is the only known second messenger that can control a biological process by binding both to a protein and the encoding mRNA molecule to prevent activity and expression of the protein. This was reported for two potassium transporters in B. subtilis, KtrAB and KimA, which are controlled at both levels by c-di-AMP (Gundlach et al. 2017b, Nelson et al. 2013, Gundlach et al. 2019).
Synthesis and degradation of c-di-AMP in B. subtilis
The cellular levels of c-di-AMP depend on the opposing activities of diadenylate cyclases and phosphodiesterases that synthesize and degrade the second messenger, respectively. For an overview on the enzymes involved in c-di-AMP synthesis and degradation, see Fig. 1. All known diadenylate cyclases consist of a conserved enzymatically active DAC domain that is responsible for the production of the second messenger from two molecules of ATP. This DAC domain is then fused to other domains that control the activity of the protein (see Commichau et al. 2019, Galperin 2023 for review). Diadenylate cyclases are present in bacteria of the classes Firmicutes, Actinobacteria, and Cyanobacteria, and they are also found in Chlamydia, Delta-proteobacteria, Spirochaetes, and in bacteria of the Cytophaga/Flavobacterium/Bacteroides group. Moreover, diadenylate cyclases have recently been discovered in the Euryarchaeota group of the Archaea (Braun et al. 2019, Galperin et al. 2021). The most widespread type of diadenylate cyclase, called CdaA, contains three N-terminal transmembrane helices fused to a cytoplasmatic DAC domain (Rosenberg et al. 2015, Heidemann et al. 2019). Most bacteria contain only one diadenylate cyclase, and this is usually of the CdaA type. Bacillus subtilis and its close relatives are peculiar in possessing three diadenylate cyclases. In addition to CdaA, they encode the DisA and CdaS proteins. DisA is an octameric DNA-binding diadenylate cyclase in which the DAC domain is fused to a linker and a DNA-binding helix-hairpin-helix domain (Witte et al. 2008). Moreover, B. subtilis and its close relatives possess a third enzyme, CdaS, which consists of a N-terminal autoinhibitory domain fused to the DAC domain. CdaS is only expressed during sporulation and is required for efficient germination of B. subtilis spores (Mehne et al. 2014).
Figure 1.
Components of c-di-AMP signaling in B. subtilis. The second messenger c-di-AMP is synthesized by the diadenylate cyclases DisA, CdaA, and CdaS through the condensation of two ATP molecules. Conversely, degradation of c-di-AMP to pApA is facilitated by the phosphodiesterases GdpP and PgpH. C-di-AMP binds to signal transduction proteins that can be regarded as third messengers and can control the expression and activity of several transporters by binding to mRNA riboswitches and proteins, respectively. Red arrows, inhibition; green arrows, activation; gray arrows, regulatory consequences unknown.
The degradation of c-di-AMP is catalyzed by dedicated phosphodiesterases. These enzymes typically contain a so-called DHH-DHHA1 (aspartate-histidine-histidine) domain or a HD (histidine-aspartate) domain. In B. subtilis, an enzyme of each of these classes is present. Both phosphodiesterases, GdpP and PgpH, are membrane proteins, and both are inhibited by (p)ppGpp (Rao et al. 2010, Huynh et al. 2015). Both enzymes cleave c-di-AMP to the linear dinucleotide pApA, which is finally cleaved to two molecules of ATP by the nano-RNase NrnA (Gall et al. 2022).
The mechanisms of c-di-AMP synthesis and degradation have been reviewed recently (Commichau et al. 2019, Stülke and Krüger 2020) and will therefore not be detailed here. However, recent research has shed some light on the regulation of the catalytic activity of the major diadenylate cyclase CdaA. This enzyme is encoded in a conserved operon with a regulatory protein, CdaR, and with the phosphoglucosamine mutase GlmM. CdaA interacts with both proteins, and these interactions control the activity of CdaA in B. subtilis as well as in Staphylococcus aureus and Listeria monocytogenes (Gundlach et al. 2015b, Mehne et al. 2013, Tosi et al. 2019, Gibhardt et al. 2020, Pathania et al. 2021). CdaR consists of a transmembrane domain and four so-called YbbR domains that are structurally similar to ribosomal protein L25 (Barb et al. 2011). L25 binds the 5S ribosomal RNA (Schmalisch et al. 2002,Korobeinikova et al. 2008), suggesting that CdaR might also interact with RNA. However, the YbbR domains of CdaR are faced to the extracellular side of the membrane. Depending on the conditions, stimulation, and inhibition of CdaA by CdaR were observed (Mehne et al. 2013, Rismondo et al. 2015). GlmM clearly inhibits the activity of CdaA, and it was shown that this inhibition occurs under conditions of osmotic stress in L. monocytogenes (Tosi et al. 2019, Gibhardt et al. 2020, Pathania et al. 2021). This latter observation provides an immediate explanation for how the bacteria can initiate potassium uptake to counteract osmotic stress (see below).
The role of c-di-AMP in potassium and osmotic homeostasis
The initial identification of targets of c-di-AMP in S. aureus revealed binding to proteins involved in potassium homeostasis (Corrigan et al. 2013, see Stülke and Krüger 2020 and He et al. 2020 for a list of c-di-AMP targets in different bacteria). In fact, many bacteria that use c-di-AMP have multiple potassium-related targets. The overview on c-di-AMP targets in B. subtilis (Table 1, see Fig. 1) already suggests that the control of potassium homeostasis is a major function of the second messenger in this bacterium.
Table 1.
Targets of c-di-AMP in B. subtilis.
| Target | Nature of the target | Function | Effect of c-di-AMP binding | Reference(s) |
|---|---|---|---|---|
| Targets related to potassium homeostasis | ||||
| KimA | Protein | Potassium uptake | Inhibition of activity | Gundlach et al. 2019 |
| c-di-AMP riboswitch (kimA) | RNA | Control of kimA expression | Transcription termination | Nelson et al. 2013 |
| KtrA | Protein | Potassium uptake | Inhibition of activity | Gundlach et al. (2019) |
| c-di-AMP riboswitch (ktrAB) | RNA | Control of ktrAB expression | Transcription termination | Nelson et al. (2013) |
| KtrC | Protein | Potassium uptake | Inhibition of activity | Gundlach et al. (2019) |
| CpaA | Protein | Potassium export | Stimulation of activity | Gundlach et al. (2019) |
| KhtT | Protein | Potassium export | Stimulation of activity | Gundlach et al. (2019) |
| Other targets | ||||
| OpuCA | Protein | Uptake of compatible solutes | Inhibition of activity | Gundlach et al. (2019) |
| MgtE | Protein | Magnesium uptake | ? | Gundlach et al. (2019) |
| Third messenger targets | ||||
| DarA | Protein | Signal transduction | Not known | Gundlach et al. (2015b) |
| DarB | Protein | Signal transduction, control of Rel and PycA activities | No interaction with Rel and PycA | Gundlach et al. (2019) |
Potassium is the most abundant metal ion in any living cell. This essential ion is important for the activity of many enzymes and proteins complexes such as the ribosome. On the other hand, the accumulation of too large amounts of the ion is toxic for the cells (Gundlach et al. 2017a, Danchin and Nikel 2019). Potassium is closely linked to glutamate, the by far most abundant metabolite in any living cell, which, in addition to many other functions, also serves as a counterion to potassium. Thus, the concentrations of potassium and glutamate must be carefully balanced. In B. subtilis and other Gram-positive bacteria, the intracellular levels of c-di-AMP are controlled by potassium and the nitrogen source. The c-di-AMP concentrations increase with increasing potassium concentrations. Moreover, the presence of glutamate stimulates c-di-AMP accumulation in B. subtilis (Gundlach et al. 2015b). It is important to note that potassium uptake is facilitated in the presence of glutamate and vice versa (Krüger et al. 2020, Krüger et al. 2021a). The direct response of c-di-AMP to potassium as well as the fact that most of the c-di-AMP targets are involved in potassium homeostasis suggest that c-di-AMP serves as a second messenger that reports on the potassium availability.
As mentioned above, c-di-AMP can control KtrAB and KimA, two potassium transporters, by binding to the corresponding mRNA, thus preventing their expression at a riboswitch as well as by binding to the proteins to inhibit their activity (Gundlach et al. 2019). Thus, the nucleotide binds completely different classes of molecules—protein and RNA—to achieve a unified regulation, i.e. to reduce potassium uptake when the ion is already present at high concentrations in the cell. The potassium transporters KtrAB and KimA are not related to each other and belong to different protein families. Thus, c-di-AMP can also bind completely different proteins to control their activity.
Bacillus subtilis encodes three potassium importers, KtrAB, KtrCD, and KimA that are all inhibited by c-di-AMP. Of these, KtrAB and KtrCD are potassium channels that are driven by the proton motive force. The KtrB and KtrD subunits are integral membrane proteins that can transport potassium. The KtrA and KtrC subunits consist of two different regulator of potassium conductance (RCK) domains that bind to the membrane subunits to modulate their activity (Schrecker et al. 2019, Stautz et al. 2021). Of the two paralogous potassium channels, KtrAB has a very high affinity for potassium, whereas KtrCD has a low affinity (Holtmann et al. 2003, Gundlach et al. 2017a). However, KtrCD also becomes a high-affinity channel in the presence of glutamate (Krüger et al. 2020). The third potassium importer, KimA, is a potassium/proton symporter. This protein has a medium affinity for potassium. Under standard conditions, potassium is imported via KtrCD and KimA, whereas KtrAB is only expressed under conditions of extreme potassium starvation (Gundlach et al. 2017a).
The ktrC and ktrD genes are constitutively expressed in B. subtilis (Krüger et al. 2021a). This is a typical feature of the major low-affinity transporters. In contrast, the kimA gene and the ktrAB operon are strongly induced at low potassium concentrations. This regulation is achieved by a c-di-AMP-responsive riboswitch in the 5’ leader regions of the corresponding mRNAs (Nelson et al. 2013, Gundlach et al. 2017a,b). Binding of c-di-AMP to the riboswitch region of the nascent transcript results in a structural rearrangement of the mRNA and in the formation of a transcription terminator. Therefore, these transcripts are no longer elongated (Gao and Serganov 2014, Ren and Patel 2014). In this way, the expression of the high-affinity uptake systems can be reduced if sufficient potassium is available for the bacteria.
Many bacteria also possess the high-affinity ATP-driven potassium importer KdpFABC. In L. monocytogenes and S. aureus, the expression of this transporter is controlled by the KdpDE two-component system. In these bacteria, c-di-AMP binds to the KdpD sensor kinase via its cytoplasmic N-terminal universal stress protein domain and inhibits its activity (Corrigan et al. 2013, Moscoso et al. 2015, Gibhardt et al. 2019). Interestingly, the close pathogenic relatives of B. subtilis, B. anthracis, B. cereus, and B. thuringiensis also have the KdpFABC potassium transporter. However, these bacteria lack the KdpDE two-component system. Instead, their kdpFABC operon is controlled by the c-di-AMP-sensitive riboswitch (Wang et al. 2019). This regulation is remarkable for two reasons: (i) the c-di-AMP-controlled sensor kinase KdpD represents yet another class of independently evolved c-di-AMP target that is involved in the regulation of potassium homeostasis. (ii) The replacement of KdpDE by the c-di-AMP riboswitch in the pathogenic Bacilli shows that the components of c-di-AMP signaling in the control of potassium homeostasis can be plugged together in multiple ways.
Biochemical studies on the regulation of the potassium channels revealed that KtrA has a 100-fold lower affinity for c-di-AMP as compared to KtrC. Thus, c-di-AMP-mediated control of KtrAB is exerted mainly at the level of the c-di-AMP-sensitive riboswitch (Rocha et al. 2019), whereas the constitutively expressed KtrC is the main target for c-di-AMP-mediated control at the protein level (Rocha et al. 2023). Thus, by controlling the expression and activities of potassium transporters to different extents, c-di-AMP can ensure that the cell can transport the required amounts of potassium under each condition.
As mentioned above, potassium becomes toxic for the cell upon accumulation. This requires a shutdown of potassium uptake by c-di-AMP-mediated switch-off of the transporters. In addition, c-di-AMP can bind to potassium exporters to stimulate their activity, thus resulting in a reduction of the intracellular potassium concentration. In B. subtilis, c-di-AMP binds to the potassium exporter CpaA and to the KhtT subunit of the KhtUT potassium/proton antiporter (Gundlach et al. 2019). In both cases, c-di-AMP binds to the RCK_C domains of these proteins, as in the potassium importers (Chin et al. 2015, Cereija et al. 2021).
In conclusion, the antagonistic control of potassium importers and exporters provides B. subtilis an efficient way to achieve the optimal intracellular potassium concentration under any condition (Fig. 1).
In B. subtilis and many other bacteria, a sudden increase of the intracellular potassium concentration is the first response to osmotic stress (Bremer and Krämer 2019). Thus, the control of potassium homeostasis by c-di-AMP might also be part of a larger picture of the regulation of osmoadaptation. Indeed, c-di-AMP also binds and inhibits transporters of compatible compounds that are toxic in the absence of an osmotic upshift. In B. subtilis and several other bacteria, c-di-AMP binds and inhibits the ATPase subunit of the Opu-compatible compound ABC transporter (Huynh et al. 2016, Schuster et al. 2016, Devaux et al. 2018, Gundlach et al. 2019, Sikkema et al. 2020). In lactic acid bacteria, even a transcription factor that controls the expression of compatible solute uptake systems is controlled by c-di-AMP (Devaux et al. 2018, Pham et al. 2018, Bandera et al. 2021). For B. subtilis, the direct control of potassium homeostasis has been shown to be the major essential function of c-di-AMP (Gundlach et al. 2017b, Stülke and Krüger 2020). However, this is not the case for L. monocytogenes (Gibhardt et al. 2019), and the more general control of osmotic homeostasis has been suggested to be the main function of c-di-AMP in this organism (Wang et al. 2022, Schwedt et al. 2023). Thus, the precise functions of c-di-AMP may differ even between closely related organisms.
DarA and DarB: c-di-AMP-binding third messengers?
The search for c-di-AMP-binding proteins in S. aureus, L. monocytogenes, and B. subtilis also identified proteins without an obvious enzymatic or transport functions. Two of these proteins, DarA and DarB, are present in B. subtilis. DarA is a member of the PII superfamily of signal transduction proteins. This protein is also present in S. aureus and L. monocytogenes but is interestingly absent from the pathogenic Bacilli (Gundlach et al. 2015a, Forchhammer and Lüddecke 2016). The DarB protein consists of two nucleotide-binding CBS domains that together form the so-called Bateman domain (Gundlach et al. 2019, Heidemann et al. 2022, Biemans-Oldehinkel et al. 2006). DarB is also present in L. monocytogenes but not in S. aureus. DarA and DarB can be present in their apo form or in the c-di-AMP-bound form, depending on the intracellular potassium and c-di-AMP concentrations (Krüger et al. 2021b). It is tempting to speculate that both proteins act as a kind of third messenger that transduces signals to other proteins and thus controls their activity.
The proteins of the large PII superfamily are signal transduction proteins that bind small-molecule effectors and control the activity of enzymes, transporters, and transcription factors. The classical PII proteins are best known for their role in the control of nitrogen assimilation (Forchhammer and Lüddecke 2016). It therefore seems likely that the c-di-AMP-binding PII-like protein DarA also binds to one or more other proteins to regulate them in response to potassium availability. Unfortunately, and despite extensive efforts, the potential interaction partner of DarA has escaped its identification until today, in B. subtilis, L. monocytogenes, and S. aureus. Under the standard growth conditions, the inactivation or overexpression of B. subtilis DarA does not result in phenotypic effects (Gundlach et al. 2015a). Thus, the role of DarA remains enigmatic and a task for future research.
For DarB, functions have been identified both in B. subtilis and in L. monocytogenes as well as in Streptococcus agalactiae. In these bacteria, DarB binds the (p)ppGpp synthetase/hydrolase Rel (Peterson et al. 2020, Krüger et al. 2021b, Covaleda-Cortés et al. 2023). Under conditions of potassium starvation, apo-DarB binds to the Rel protein to stimulate and inhibit its alarmone synthetase and hydrolase activities, respectively. The result is an intracellular accumulation of (p)ppGpp under conditions of potassium starvation (Krüger et al. 2021b). The second alarmone (p)ppGpp in turn binds to several proteins of nucleotide biosynthesis and the central genetic processes, resulting in a shutdown of central cellular processes, among them translation (Hauryliuk et al. 2015, Bange et al. 2021). As the ribosome depends on potassium ions for activity, alarmone production and the resulting stop of translation help the cell to adapt to conditions of potassium starvation (Krüger et al. 2021b). Recent structural studies have suggested that the binding of c-di-AMP to DarB results in protrusion of the nucleotide at the site of interaction with Rel, thus preventing binding of the two proteins (Heidemann et al. 2022). Binding of Apo-DarB to Rel supports a conformation of the synthetase domain that increases the affinity of Rel for ATP and switches off hydrolase activity (Ainelo et al. 2023).
In addition to the control of the stringent response, B. subtilis apo-DarB also binds to the pyruvate carboxylase, an enzyme that replenishes the citric acid cycle by providing oxaloacetate. Binding of apo-DarB under conditions of potassium starvation results in a stimulation of pyruvate carboxylase activity and, thus, in increased oxaloacetate production (Krüger et al. 2022). This meets the increased demand for citric acid cycle intermediates during potassium limitation (Krüger et al. 2022). Interestingly, c-di-AMP-mediated control of pyruvate carboxylase activity was also found in L. monocytogenes. In this bacterium, it is not DarB but c-di-AMP directly that binds the pyruvate carboxylase resulting in enzyme inhibition (Sureka et al. 2014). While the mechanisms are different, the logic of c-di-AMP-dependent regulation of pyruvate carboxylase is similar in both bacteria: at high potassium concentration, c-di-AMP is present and causes inhibition or lack of DarB-dependent activation of the enzyme in L. monocytogenes and B. subtilis, respectively. In contrast, the enzyme is not inhibited or even activated by apo-DarB at low potassium concentrations. As a result, c-di-AMP-mediated control results in low or high pyruvate carboxylase activity in the presence or absence of potassium, respectively, in both bacteria.
It is interesting to note that it is apo-DarB that binds to and controls the Rel and pyruvate carboxylase enzymes. No activity has so far been assigned to DarB in complex with c-di-AMP. Importantly, in the absence of c-di-AMP, inactivation of either DarA or DarB allows the adaptation to growth on otherwise toxic complex medium. This has been observed for both B. subtilis and L. monocytogenes (Whiteley et al. 2015, Krüger et al. 2021a) and suggests that the apo-proteins have some activity that is detrimental to the growth of the bacteria. This indirect control by c-di-AMP may help to reach out to proteins that are unable to submit to direct control by the second messenger.
The lack of c-di-AMP makes the cells susceptible to growth inhibition by amino acids
A B. subtilis strain lacking c-di-AMP (the Δdac mutant) is unable to grow in the presence of otherwise well-tolerated potassium concentrations, and growth is also severely inhibited in the presence of glutamate (Krüger et al. 2021a). Similarly, c-di-AMP is essential for growth on complex medium in several bacteria. In addition to glutamate, other amino acids inhibit the growth of B. subtilis strains lacking c-di-AMP. These include histidine, alanine, asparagine, glutamine, arginine, and proline (Meißner et al. 2022, unpublished results). One particular reason for the growth inhibition of the Δdac mutant is the generation of glutamate as a result of amino acid degradation. Glutamate converts the low-affinity potassium channel KtrCD to a high-affinity channel, thus even the low concentrations of potassium that are tolerated by the strain lacking all diadenylate cyclases become toxic for the cells (Krüger et al. 2020, Krüger et al. 2021a).
While the physiology of B. subtilis is in general very well understood, there are still severe gaps in our knowledge on amino acid homeostasis (Wicke et al. 2023). The growth inhibition of the Δdac mutant by amino acids is an excellent tool to identify novel components of amino acid homeostasis. Suppressor analyses with the Δdac mutant grown in the presence of glutamate revealed that many of mutants had mutations affecting one of the potassium channel proteins. A similar result was obtained at high histidine concentration (Krüger et al. 2021a, Meißner et al. 2022) (Fig. 2). These observations support the idea that the stimulation of potassium uptake is a major problem for the strain in the presence of amino acids.
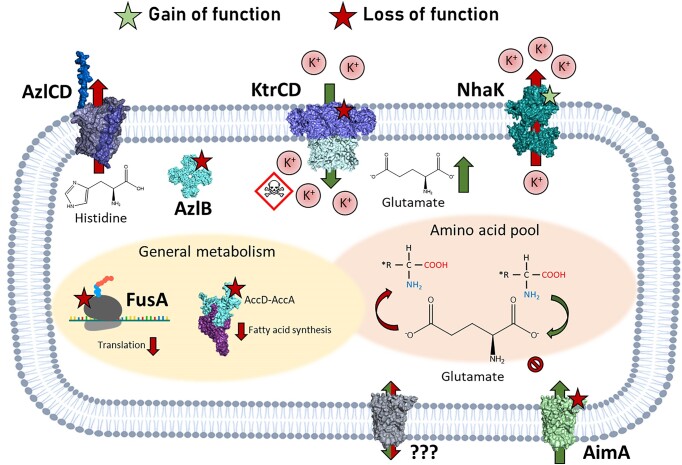
Figure 2.
Effects of amino acids on a c-di-AMP-free strain of B. subtilis. Amino acids, primarily glutamate, make up by far the largest fraction of the metabolite pool of B. subtilis cells. Without c-di-AMP, however, glutamate becomes toxic as it causes an affinity change of the potassium-channel KtrCD, leading to an increased influx of potassium ions, which are harmful to the cell in the absence of c-di-AMP. Bacillus subtilis responds to glutamate stress via mutations in AimA, the major glutamate transporter, and KtrCD itself (Krüger et al. 2021). Higher concentrations of glutamate cause downregulations in general metabolism. Here, the essential genes fusA (involved in translation) and accA (involved in fatty acid biosynthesis) are affected. Furthermore, nhaK also acquires a gain-of-function mutation resulting in enhanced potassium export under glutamate stress. Other amino acids feed into glutamate production and therefore might also be harmful in a c-di-AMP-free setting. Stressing the cells with histidine leads to mutations in the transcriptional repressor AzlB, which causes an overexpression of the bipartite exporter AzlCD. This causes histidine to be exported out of the cell with higher efficiency (Meißner et al. 2022).
However, all glutamate-resistant mutants also had mutations that more directly address the amino acid used in the suppressor screen. In the case of glutamate, most suppressor mutants had the aimA gene inactivated. AimA was only recently identified as the main serine transporter of B. subtilis (Klewing et al. 2020) and is also the major low-affinity transporter for glutamate. Moreover, in several mutants, glutamate degradation to 2-oxoglutarate was activated by a mutation that decryptifies the normally inactive glutamate dehydrogenase GudB. Finally, some of the mutants were able to export glutamate by a combination of mutations affecting lipid biosynthesis and the mechanosensitive channel YfkC (Krüger et al. 2021a). Thus, the use of the Δdac mutant allowed the identification of the so far unknown major importer and an export mechanism for glutamate.
The Δdac mutant also adapted to the presence of histidine by the acquisition of mutations. In this case, of a large collection of suppressor mutants, each contained a mutation that inactivates the azlB gene. This gene encodes the repressor of the azlBCD-brnQ-yrdK operon (Meißner et al. 2022). The AzlCD complex is a bipartite amino acid exporter that has already been shown to export toxic analogues of branched-chain amino acids, such as 4-azaleucine (Belitsky et al. 1997). The identification of histidine as an additional substrate of AzlCD suggests that this exporter has a broad substrate specificity.
The identification of novel importers and exporters for glutamate and histidine suggests that the Δdac mutant is an excellent tool to study novel players in amino acid homeostasis.
A diadenylate cyclase involved in monitoring and repairing DNA
c-di-AMP was originally identified as a so-far unknown metabolite bound to the DNA integrity-scanning protein DisA and as the product of the diadenylate cyclase activity of DisA (Witte et al. 2008). The DisA protein consists of a N-terminal enzymatically active DAC domain, a linker, and a C-terminal DNA-binding helix-hinge-helix domain. DisA scans the chromosome and causes a delay in sporulation if it detects chromosome damage (Bejerano-Sagie et al. 2006, Oppenheimer-Shaanan et al. 2011). By interactions with DNA repair and recombination proteins such as RadA, RecA, and RuvB DisA contribute to the maintenance of DNA integrity (Torres et al. 2019, Torres et al. 2019a, Gándara et al. 2021). It has also been proposed that DisA might actually monitor the potassium concentration in the nucleoid since potassium neutralizes the negative charge of DNA (Gundlach et al. 2018). The precise link between DNA damage, c-di-AMP synthesis, and the contribution of DisA to DNA repair remains to be elucidated.
Future directions of research
Even though the very existence of c-di-AMP was discovered only 15 years ago, an impressive amount of knowledge on the diverse functions of this unique essential second messenger has accumulated. However, there are still open questions that should be addressed in future research:
Which signals and mechanisms control the expression and activity of the enzymes that make and break c-di-AMP?
What is the function of the third messenger protein DarA?
How does DisA contribute to DNA integrity?
Another exciting prospect is the possible use of the diadenylate cyclase CdaA as a novel target for antibiotic compounds. CdaA is the only diadenylate cyclase in many pathogens, and it is essential for these bacteria. A recent report on such an inhibitor that was identified based on computer-based design (Neumann et al. 2023) shows that this is a promising line of research.
Acknowledgement
We are grateful to Jan Gundlach, Larissa Krüger, and Fabian Commichau as well as all lab members who have made valuable contributions to the investigation of c-di-AMP signaling in B. subtilis. Ralf Ficner, Volkhard Kaever, and Rolf Daniel are acknowledged for longstanding close cooperation. Research on c-di-AMP signaling in our lab is supported by the Deutsche Forschungsgemeinschaft (DFG) via Priority Program SPP1879 to JS.
Contributor Information
Christina Herzberg, Department of General Microbiology, GZMB, Georg-August-University Göttingen, Grisebachstr. 8, 37077 Göttingen, Germany.
Janek Meißner, Department of General Microbiology, GZMB, Georg-August-University Göttingen, Grisebachstr. 8, 37077 Göttingen, Germany.
Robert Warneke, Department of General Microbiology, GZMB, Georg-August-University Göttingen, Grisebachstr. 8, 37077 Göttingen, Germany.
Jörg Stülke, Department of General Microbiology, GZMB, Georg-August-University Göttingen, Grisebachstr. 8, 37077 Göttingen, Germany.
Conflict of interest
None declared.
References
- Ainelo A, Caballero-Montes J, Bulvas Oet al. The structure of DarB on complex with RelNTD reveals nonribosomal activation of Rel stringent factors. Sci Adv. 2023;9:eade4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BW, Liu K, Wolak Cet al. Evolution of (p)ppGpp-HPRT regulaton through diversification of an allosteric oligomeric interaction. eLife. 2019;8:e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandera AM, Bartho J, Lammens Ket al. BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture. Nucleic Acids Res. 2021;49:10166–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange G, Brodersen DE, Liuzzi Aet al. Two P or not two P: understanding regulation by bacterial second messengers (p)ppGpp. Annu Rev Microbiol. 2021;75:383–406. [DOI] [PubMed] [Google Scholar]
- Barb AW, Cort JR, Seetharaman Jet al. Structures of domains I and IV from YbbR are representative of a widely distrubuted protein family. Protein Sci. 2011;20:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrunka P, Graumann PL. New functions and subcellular localization patterns of c-di-GMP components (GGDEF domain proteins) in B. subtilis.Front Microbiol. 2017;8:794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky Iet al. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–90. [DOI] [PubMed] [Google Scholar]
- Belitsky BR, Gustafsson MC, Sonenshein ALet al. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J Bacteriol. 1997;179:5448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E, Mahmood NABN, Poolman B. A sensor for intracellular ionic strength. Proc Natl Acad Sci USA. 2006;103:10624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F, Thomalla L, van der Does Cet al. Cyclic nucleotides in archaea: cyclic di-AMP in the archaeon Haloferax volcanii and its putative role. Front Microbiol. 2019;8:00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer E, Krämer R. Responses of microorganisms to osmotic stress. Annu Rev Microbiol. 2019;73:313–34. [DOI] [PubMed] [Google Scholar]
- Cereija TB, Guerra JPL, Jorge JMPet al. c-di-AMP, a likely master regulator of bacterial K(+) homeostasis machinery, activates a K(+) exporter. Proc Natl Acad Sci USA. 2021;118:e2020653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Huang X, Gaballa Aet al. Bacillus subtilis FolE is sustained by the ZagA metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol Microbiol. 2019;112:751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chai Y, Guo JHet al. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol. 2012;194:5080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KH, Liang JM, Yang JGet al. Structural insights into the distinct binding mode of cyclic di-AMP with SaCpaA-RCK. Biochemistry. 2015;54:4936–51. [DOI] [PubMed] [Google Scholar]
- Commichau FM, Heidemann JL, Ficner Ret al. Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J Bacteriol. 2019;201:e00462–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan Tet al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110:9084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covaleda-Cortés G, Mechaly A, Brissac Tet al. The c-di-AMP-binding protein CbpB modulates the level of the ppGpp alarmone in Streptococcus agalactiae. FEBS J. 2023;290:2968–92. [DOI] [PubMed] [Google Scholar]
- Danchin A, Nikel PI. Why nature chose potassium. J Mol Evol. 2019;87:271–88. [DOI] [PubMed] [Google Scholar]
- Devaux L, Sleiman D, Mazzuoli MVet al. Cyclic di-AMP regulation of osmotic homeostasis is essential in group B Streptococcus. PLoS Genet. 2018;14:e1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier C, Newman JA, Kovács ATet al. The YmdB phosphodiesterase is a global regulator of late adaptive responses in Bacillus subtilis. J Bacteriol. 2014;196:265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Lüddecke J. Sensory properties of the PII signaling family. FEBS J. 2016;283:425–37. [DOI] [PubMed] [Google Scholar]
- Gall AR, Hsueh BY, Siletti Cet al. NrnA is a linear dinucleotide phosphodiesterase with limited function in cyclic dinucleotide metabolism in Listeria monocytogenes. J Bacteriol. 2022;204:e00206–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. All DACs in a row: domain architecture of bacterial and archaeal diadenylate cyclases. J Bacteriol. 2023;205:e00023–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Wolf YI, Makarova KSet al. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021;49:D274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gándara C, Torres R, Carrasco Bet al. DisA restrains the processing and cleavage of reversed replication forks by the RuvAB-RecU resolvasome. Int J Mol Sci. 2021;22:11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A, Serganov A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol. 2014;10:787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammarinaro PI, Young MKM, Steinchen Wet al. Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis. Nat Microbiol. 2022;7:1442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibhardt J, Heidemann JL, Bremenkamp Ret al. An extracytoplasmic protein and a moonlighting enzyme modulate synthesis of c-di-AMP in Listeria monocytogenes. Environ Microbiol. 2020;22:2771–91. [DOI] [PubMed] [Google Scholar]
- Gibhardt J, Hoffmann G, Turdiev Aet al. C-di-AMP assists osmoadaptation by regulating the Listeria monocytogenes potassium transporters KimA and KtrCD. J Biol Chem. 2019;294:16020–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Micro. 2008;6:613–24. [DOI] [PubMed] [Google Scholar]
- Green J, Stapleton MR, Smith LJet al. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr Opin Microbiol. 2014;18:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach J, Commichau FM, Stülke J. Perspective of ions and messengers: an intricate link between potassium, glutamate, and cyclic di-AMP. Curr Genet. 2018;64:191–5. [DOI] [PubMed] [Google Scholar]
- Gundlach J, Dickmanns A, Schröder-Tittmann Ket al. Identification, characterization, and structure analysis of the cyclic di-AMP-binding PII-like signal transduction protein DarA. J Biol Chem. 2015a;290:3069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach J, Herzberg C, Hertel Det al. Adaptation of Bacillus subtilis to life at extreme potassium limitation. mBio. 2017a;8:e00861–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach J, Herzberg C, Kaever Vet al. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci Signal. 2017b;10:eaal3011. [DOI] [PubMed] [Google Scholar]
- Gundlach J, Krüger L, Herzberg Cet al. Sustained sensing in potassium homeostasis: cyclic di-AMP controls potassium uptake by KimA at the levels of expression and activity. J Biol Chem. 2019;294:9605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach J, Mehne FM, Herzberg Cet al. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol. 2015b;197:3265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KSet al. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Micro. 2015;13:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yin W, Galperin MYet al. Cyclic-di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 2020;48:2807–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann JL, Neumann P, Dickmanns Aet al. Crystal structures of the c-di-AMP synthesizing enzyme CdaA. J Biol Chem. 2019;294:10463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann JL, Neumann P, Krüger Let al. Structural basis for c-di-AMP-dependent regulation of the bacterial stringent response by receptor protein DarB. J Biol Chem. 2022;298:102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Micro. 2009;7:263–73. [DOI] [PubMed] [Google Scholar]
- Hengge R, Häussler S, Pruteanu Met al. Recent advances and current trends in nucleotide second messenger signaling in bacteria. J Mol Biol. 2019;431:908–27. [DOI] [PubMed] [Google Scholar]
- Holtmann G, Bakker EP, Uozumi Net al. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol. 2003;185:1289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Choi PH, Sureka Ket al. Cyclic di-AMP targets the cystathione beta-synthase domain of the osmolyte transporter OpuC. Mol Microbiol. 2016;102:233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Luo S, al PD. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci USA. 2015;112:E747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Woodward JJ. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol. 2016;30:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Micro. 2017;15:271–84. [DOI] [PubMed] [Google Scholar]
- Klewing A, Koo BM, Krüger Let al. Resistance to serine in Bacillus subtilis: identification of the serine transporter YbeC and of a metabolic network that links serine and threonine metabolism. Environ Microbiol. 2020;22:3937–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobeinikova AV, Gongadze GM, Korepanov APet al. 5S rRNA-recognition module of CTC family proteins and its evolution. Biochemistry. 2008;73:156–73. [DOI] [PubMed] [Google Scholar]
- Krüger L, Herzberg C, Rath Het al. Essentiality of c-di-AMP in Bacillus subtilis: bypassing mutations converge in potassium and glutamate homeostasis. PLoS Genet. 2021a;17:e1009092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger L, Herzberg C, Warneke Ret al. Two ways to convert a low-affinity potassium channel to high affinity: control of Bacillus subtilis KtrCD by glutamate. J Bacteriol. 2020;202:e00138–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger L, Herzberg C, Wicke Det al. A meet-up of two second messengers: the c-di-AMP receptor DarB controls (p)ppGpp synthesis in Bacillus subtilis. Nat Commun. 2021b;12:1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger L, Herzberg C, Wicke Det al. Sustained control of pyruvate carboxylase by the essential second messenger cyclic di-AMP in Bacillus subtilis. mBio. 2022;13:e0360221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Myers AR, Pisithkul Tet al. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell. 2015;57:735–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Micro. 1012;10:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehne FM, Gunka K, Eilers Het al. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehne FMP, Schröder-Tittmann K, Eijlander RTet al. Control of the diadenylate cyclase CdaS in Bacillus subtilis: an autoinhibitory domain limits c-di-AMP production. J Biol Chem. 2014;289:21098–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meißner J, Schramm T, Hoßbach Bet al. How to deal with toxic amino acids: the bipartite AzlCD complex exports histidine in Bacillus subtilis. J Bacteriol. 2022;204:e0035322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso JA, Schramke H, Zhang Yet al. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the Universal Stress Protein domain and downregulates the expression of the Kdp potassium transporter. J Bacteriol. 2015;198:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Breaker RR. The lost language of the RNA world. Sci Signal. 2017;10:eaam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa Ket al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Kloskowski P, Ficner R. Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor. Microlife. 2023;4:uqad021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler Jet al. C-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Galperin MY, Lee VT. Sustained sensing as an emerging principle in second messenger signaling systems. Curr Opin Microbiol. 2016;34:119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M, Tosi T, Millership Cet al. Structural basis for the inhibition of the Bacillus subtilis c-di-AMP cyclase CdaA by the phosphoglucomutase GlmM. J Biol Chem. 2021;297:101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedreira T, Elfmann C, Stülke J. The current state of SubtiWiki, the database for the model organism Bacillus subtilis. Nucleic Acids Res. 2022;50:D875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BN, Young MKM, Luo Set al. (p)ppGpp and c-di-AMP homeostasis is controlled by CbpB in Listeria monocytogenes. mBio. 2020;11:e1625–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham HT, Nhiep NTH, Vu TNMet al. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet. 2018;14:e1007574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, See RY, Zhang Det al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Patel DJ. C-di-AMP binds the ydaO riboswitch in two pseudo-related pockets. Nat Chem Biol. 2014;10:780–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhaese HJ, Dichtelmüller H, Grade R. Studies on the control of development. Accumulation of guanosine tetraphosphate and pentaphosphate in response to inhibition of protein synthesis in Bacillus subtilis. Eur J Biochem. 1975;56:385–92. [DOI] [PubMed] [Google Scholar]
- Rismondo J, Gibhardt J, Rosenberg Jet al. Phenotypes associated with the essential diadenylate cyclase CdaA and its potential regulator CdaR in the human pathogen Listeria monocytogenes. J Bacteriol. 2015;198:416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha R, Jorge JMP, Teixeira-Duarte CMet al. c-di-AMP determines the hierarchical organization of bacterial RCK proteins. Biorxiv. 2023. 10.1101/2023.04.24.538052. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha R, Teixeira-Duarte CM, Jorge JMPet al. Characterization of the molecular properties of KtrC, a second RCK domain that regulates a Ktr channel in Bacillus subtilis. J Struct Biol. 2019;205:34–43. [DOI] [PubMed] [Google Scholar]
- Ronneau S, Hallez R. Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol Rev. 2019;43:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J, Dickmanns A, Neumann Pet al. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J Biol Chem. 2015;290:6596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalisch M, Langbein I, Stülke J. The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J Mol Microbiol Biotechnol. 2002;4:495–501. [PubMed] [Google Scholar]
- Schrecker M, Wunnicke D, Hänelt I. How RCK domains regaulate gating of K+ channels. Biol Chem. 2019;400:1303–22. [DOI] [PubMed] [Google Scholar]
- Schuster CF, Bellows LE, Tosi Tet al. The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci Signal. 2016;9:ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedt I, Wang M, Gibhardt Jet al. Cyclic di-AMP, a multifaceted regulator of central metabolism and osmolyte homeostasis in Listeria monocytogenes. Microlife. 2023;4:uqad005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema HR, van den Noort M, Rheinberger Jet al. Gating by ionic strength and safety check by cyclic-di-AMP in the ABC Transporter OpuA. Sci Adv. 2020;6:eabd7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stautz J, Hellmich Y, Fuss MFet al. Molecular mechanisms for bacterial potassium homeostasis. J Mol Biol. 2021;433:166968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J, Krüger L. Cyclic di-AMP signaling in bacteria. Annu Rev Microbiol. 2020;74:159–79. [DOI] [PubMed] [Google Scholar]
- Sureka K, Choi PH, Precit Met al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014;158:1389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Carrasco B, Gándara Cet al. Bacillus subtilis DisA regulates RecA-mediated DNA strand exchange. Nucleic Acids Res. 2019a;47:5141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Serrano E, Tramm Ket al. Bacillus subtilis RadA/Sms contributes to chromosomal transformation and DNA repair in concert with RecA and circumvents replicative stress in concert with DisA. DNA Repair (Amst). 2019;77:45–57. [DOI] [PubMed] [Google Scholar]
- Tosi T, Hoshiga F, Millership Cet al. Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM. PLoS Pathog. 2019;15:e1007537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wamp S, Gibhardt Jet al. Adaptation of Listeria monocytogenes to perturbations of c-di-AMP metabolism underpins its role in osmoadaptation and identifies a fosfomycin uptake system. Environ Microbiol. 2022;24:4466–88. [DOI] [PubMed] [Google Scholar]
- Wang X, Cai X, Ma Het al. A c-di-AMP riboswitch controlling kdpFABC operon transcription regulates the potassium transporter system in Bacillus thuringiensis. Commun Biol. 2019;2:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Pollock AJ, Portnoy DA. The PAMP c-di-AMP is essential for Listeria monocytogenes growth in rich but not in minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe. 2015;17:788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke D, Meißner J, Warneke Ret al. Understudied proteins and understudied functions in the model organism Bacillus subtilis—a major challenge in current research. Mol Microbiol. 2023;120: 8–19. [DOI] [PubMed] [Google Scholar]
- Witte G, Hartung S, Büttner Ket al. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–78. [DOI] [PubMed] [Google Scholar]
- Zhang YE, Baerentsen RL, Führer Tet al. (p)ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Mol Cell. 2019;74:1239–49. [DOI] [PubMed] [Google Scholar]