Abstract
Aim:
This work aimed to investigate the antiviral activity of two 1,4-disubstituted-1,2,3-triazole derivatives (1 and 2) against Chikungunya virus (CHIKV) replication.
Materials & methods:
Cytotoxicity was analyzed using colorimetric assays and the antiviral potential was evaluated using plaque assays and computational tools.
Results:
Compound 2 showed antiviral activity against CHIKV 181-25 in BHK-21 and Vero cells. Also, this compound presented a higher activity against CHIKV BRA/RJ/18 in Vero cells, like compound 1. Compound 2 exhibited virucidal activity and inhibited virus entry while compound 1 inhibited virus release. Molecular docking suggested that these derivatives inhibit nsP1 protein while compound 1 may also target capsid protein.
Conclusion:
Both compounds exhibit promising antiviral activity against CHIKV by blocking different steps of virus replication.
Keywords: alphavirus, antiviral, arbovirus, chikungunya, molecular modeling, triazole
Tweetable abstract
Two 1,2,3-triazole derivatives against #Chikungunya virus showed promising antiviral activity, acting at different steps of the virus replication cycle.
Chikungunya virus (CHIKV) is an arthritogenic old world Alphavirus belonging to the Togaviridae family. It is an enveloped virus with a single-stranded positive-sense RNA genome of nearly 11.8 Kb that has two open reading frames (ORFs) that encode for four non-structural proteins (nsP1 to nsP4) and six structural proteins or peptides, namely capsid protein (C), envelope proteins 1, 2 and 3 (E1, E2 and E3), 6K and transframe (TF) [1]. According to its genome diversity, this virus is classified into four distinct genotypes: West African, East/Central/South African (ECSA), Asian and Indian Ocean lineages [2].
CHIKV is the etiological agent of Chikungunya fever which exhibited a substantial burden in the last decade on several continents, mostly in America [3]. It is transmitted mainly by infected mosquitoes, especially Aedes aegypti and Aedes albopictus, but vertical transmission has also been confirmed [4]. It was first isolated in 1952 in Tanzania and since its re-emergence in Kenya in 2004, this virus has spread to many other countries [5]. In the last decade, many countries in South Asia and Southeast Asia have experienced the re-emergence of CHIKV, including India and Thailand [4,6]. Infections are most commonly caused by ECSA and Asian lineages [7]. Climate and social changes are key factors that have increased the spread of Aedes mosquitoes and, consequently, the risk area for this disease [8]; approximately 75% of the world's population are living in areas where CHIKV transmission has been reported and 3–5 million cases are reported every year [3,4]. In 2022, more than 100 countries from different continents have reported CHIKV transmission, with the highest number of cases reported in Brazil [7,9].
Typically, infected individuals present mild symptoms like fever, rash, arthralgia and myalgia. Although the disease is generally self-limited and deaths are rare, nearly 40% of patients progress to a state of chronic disease in which debilitating muscle and joint pain persist for months to years and endangers the quality of life [10,11]. Many patients can develop other long-term and/or more severe complications such as heart failure, hemorrhage, multi-organ failure, Guillain-Barre syndrome, encephalitis and cognitive delay [4,12].
Many efforts have been put into developing a CHIKV vaccine to control Chikungunya fever. Multiple different platforms have been explored such as RNA, subunit, virus-like particle and live-attenuated or inactivated vaccines [13]. Recently, a live-attenuated vaccine candidate (VLA1553) showed promising immunogenic and safety properties in a phase III trial, but has not been approved yet [14]. The lack of approved antiviral drugs to combat this disease makes the development of novel drugs an urgent need [15].
In the last decade, 1,2,3-triazole derivatives have received great attention due to novel synthetic routes developed and their biological applications [16]. The 1,2,3-triazole moiety can act as bioisosteres of different groups, as well as improve metabolic stability and solubility [17]. In addition, this scaffold has diverse pharmacological activities such as anticancer, neuroprotective, antiparasitic, antifungal and antiviral, including against herpes simplex type 1 and 2 (HSV-1 and HSV-2), Ebola and hepatitis C viruses [18,19]. Previously, our group has synthesized 1,4-disubstituted-1,2,3-triazole derivatives, in particular compounds 1 and 2 (Figure 1), harboring an interesting biological versatility with anticancer [20], antileishmanial [21] and anti-HSV-1 [22] activities. These results have prompted us to investigate the antiviral potential of these derivatives against CHIKV by combining in vitro and computational methods.
Figure 1. . Chemical structure of 1,4-disubstituted-1,2,3-triazole derivatives tested herein.
Materials & methods
Synthesis
The synthesis of 1,4-disubstituted-1,2,3-triazole derivatives (1 and 2) was carried out by copper (I) catalyzed-azide-alkyne cycloaddition (CuAAC) reaction, as described by da Silva et al. [20], and Viegas et al. [22]. Structural characterization of the compounds was performed by nuclear magnetic resonance (1H NMR, 13C NMR) (Supplementary Figures 1 & 2) and high-resolution mass spectrometry (HRMS) techniques. The purity of compounds was confirmed by HRMS according to the following data.
2,2′-(4,4′-((1,3-phenylenebis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1 diyl)) dibenzaldehyde (1)
1H NMR (400 MHz, CDCl3) δ 9.91 (s, 2H), 8.11 (dd, J = 7.8, 1.5 Hz, 2H), 8.05 (s, 2H, triazole-H), 7.77 (td, J = 7.7, 1.6 Hz, 2H), 7.68 (t, J = 7.5 Hz, 2H), 7.54 (dd, J = 7.9, 0.9 Hz, 2H), 7.25 (t, J = 8.2 Hz, 1H), 6.73 (t, J = 2.3 Hz, 1H), 6.69 (dd, J = 8.2, 2.4 Hz, 1H), 5.33 (s, 4H). 13C NMR (101 MHz, CDCl3) δ 189.17, 160.18, 145.69, 138.87, 135.30, 131.14, 131.08, 130.83, 130.34, 126.18, 125.51, 108.51, 102.95, 62.87. HRMS (ESI) m/z calculated for C26H20N6O4+Na [M + Na]+, 503.1438; found 503.1438.
(E)-4-methyl-N-(2-(4-(phenoxymethyl)-1H-1,2,3triazol1yl)benzylidene)benzenesulfonohydrazide (2)
1H NMR (400 MHz, CDCl3) δ 8.57 (s, 1H), 8.09 (dd, J = 7.3, 2.1 Hz, 1H), 7.82–7.79 (m, 3H), 7.54–7.48 (m, 3H), 7.35–7.27 (m, 4H), 7.05–6.95 (m, 3H), 5.26 (s, 2H), 2.39 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 157.97, 144.63, 144.14, 141.29, 135.47, 130.66, 130.21, 129.65, 129.12, 127.84, 127.32, 125.74, 125.29, 121.51, 114.82, 61.58, 21.58. HRMS (ESI) m/z calculated for C23H21N5O3S + Na [M + Na]+, 470.1265; found 470.1261.
Cells, viruses & compounds
Baby hamster kidney cells (BHK-21; ATCC CCL-10) were cultured in minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco). African green monkey kidney cells (Vero; ATCC CCL-81) were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. All experiments were performed using these conditions unless otherwise stated.
In the present study, a Brazilian clinical isolate of CHIKV named CHIKV BRA/RJ/18 (ECSA lineage) [23] and the CHIKV 181/25 strain were used. The Brazilian CHIKV isolate was propagated in Vero cells at a multiplicity of infection (MOI) of 0.1 for 48 hours post-infection (hpi), while the CHIKV 181/25 strain was propagated in BHK-21 cells at an MOI of 0.1 for 72 hpi. Then, infected cells were lysed by three cycles of freezing and thawing, the homogenates were clarified by centrifugation, and the supernatant was harvested and stored at -80°C until use. MAYV (ATCC VR-66, TR 4675 strain) was kindly provided by Dr. Davis Fernandes Ferreira from Universidade Federal do Rio de Janeiro. MAYV stocks were prepared in Vero cells at an MOI of 0.1 for 72 hpi, followed by the collection of clarified supernatant as described for CHIKV strains.
Chloroquine (CQ; >98% purity) was used as the positive control due to its in vitro anti-CHIKV activity reported elsewhere [24] and was purchased from Sigma-Aldrich (Brazil). CQ was dissolved in sterile water while the other compounds were dissolved in sterile dimethyl sulfoxide (DMSO). Compounds´ stocks were prepared at a final concentration of 50 mM and were stored at -20°C until use. The final working concentrations were obtained after dilution with the working medium.
Cytotoxicity studies
The cytotoxicity of the compounds in BHK-21 cells was evaluated using the WST-1 assay and Quick Cell Proliferation Colorimetric Assay kit (BioVision) according to the manufacturer's instructions. Briefly, BHK-21 (2 × 104 cells/well) cells were seeded in 96-well plates, and plates were incubated overnight at 37°C in a 5% CO2 atmosphere. Then, the medium was replaced by a fresh medium containing 10% FBS and compounds at different concentrations (62.5, 125, 250, 500 and 1000 μM), and cells were further incubated for 72 h at 37°C in a 5% CO2 atmosphere. After 72 h, the media containing compounds were replaced by 100 μl of fresh media supplemented with 10% FBS and 10 μl of the WST-1 dye, and cells were incubated for further 3 h at 37°C in a 5% CO2 atmosphere. Finally, the optical density at 450 nm and 650 nm was determined using the FlexStation 3 Multi-Mode microplate reader (Molecular Devices).
To evaluate the cytotoxicity of the compounds in Vero cells, the MTT assay was employed with a similar methodology as described for the assays with BHK-21 cells. Vero cells (2 × 104 cells/well) were incubated overnight and treated with the compounds at the same concentrations under the same conditions described for BHK-21 cells. After 72 h, the culture medium was discarded, MTT solution (2 mg/ml) was added to each well and cells were further incubated for 4 h at 37°C and 5% CO2. Then, the MTT solution was removed, 100 μl of DMSO was added to each well to solubilize formazan crystals and cell plates were gently shaken for 20 min. The optical density values were then measured at 545 nm using the Thermo Plate microplate reader.
The optical densities of treated cells were compared with those obtained for untreated cells. DMSO (1%) was used as vehicle control. The concentration of the compounds required to decrease 50% cell viability (CC50) was estimated from dose-response curves in GraphPad Prism 8.
Antiviral activity evaluation
BHK-21 (1.75 × 105 cells/well) or Vero cells (2 × 105 cells/well) were seeded into 24-well plates and incubated at 37°C in a 5% CO2 atmosphere. One day later, cells were infected with CHIKV 181/25 or BRA/RJ/18 at an MOI of 1.0. After 2 h, the virus inoculum was discarded, and cells were washed with PBS (pH 7.4). Fresh media containing the compounds were added and cells were further incubated for 24 h at 37°C and 5% CO2. At 24 hpi, cells were lysed by three cycles of freezing and thawing to release internalized viruses to the supernatant which was collected for quantification of total virus load by plaque reduction assays.
The compounds were first screened by their antiviral activity at 50 μM and the ones with inhibition rates greater than 50% were further evaluated at other concentrations (3.125, 6.25, 12.5, 25 and 50 μM). CQ and DMSO (0.1%) were used as controls and were evaluated under the same conditions. Inhibition rates of the tested compounds were determined relative to the infected and untreated control. The compounds' concentration required to reduce virus yield by 50% (EC50) was determined by nonlinear regression from dose-response curves using GraphPad Prism 8.
For anti-MAYV assays, Vero cells (2 × 105 cells/well) were infected (MOI of 1.0) for 1 h at 37°C and 5% CO2. After virus attachment, virus inoculum was discarded, and cells were washed with PBS (pH 7.4) and treated with the compounds (50 μM) or not for 24 h at 37°C and 5% CO2. Finally, cells were lysed by three cycles of freezing and thawing, and virus titers of the collected supernatants were determined using plaque assays. DMSO (0.1%) was employed as the vehicle control.
Plaque reduction assay
The virus titer in the previously collected supernatants was determined by standard plaque assays. Confluent monolayers of BHK-21 or Vero cells were grown in 24-well plates and were infected with tenfold dilutions of the collected content. After 2 h, the virus inoculum was removed, and cells were covered with fresh medium supplemented with 5% FBS and 1.5% methylcellulose, followed by incubation at 37°C with a 5% CO2 atmosphere for 48 h (CHIKV BRA/RJ/18 infection) or 72 h (CHIKV 181/25 strain or MAYV infections). After incubation, the overlay medium was removed and cells were fixed and stained with formaldehyde 10% and crystal violet 0.2%. Viral plaques were counted and virus titers were determined as plaque-forming units per milliliter (PFU/ml).
Time-of-addition assay
Vero cells were seeded in 24-well plates at a density of 2 × 105 cells/well 24 h before infection. Cells were infected with CHIKV BR/RJ/18 isolate at an MOI of 1.0 for 2 h to allow virus attachment at 37°C under 5% CO2. After this time, virus inoculum was discarded, cells were washed with PBS (pH 7.4), fresh medium in the presence or absence of the compounds was added, and cells were incubated for further 24 h at 37°C and 5% CO2. At 24 hpi, cells were lysed by three cycles of freezing and thawing, and virus titers of the supernatants were determined using plaque assays. The triazole derivatives and CQ were added to the medium at 100 μM at different times along the virus replication cycle, as follows: cells were pre-treated with the compounds 2 h prior to virus infection (-2 hpi); cells were treated with the compounds during virus attachment (co-treatment); cells were treated with the compounds after virus attachment period at 0, 2, 4, 6, 8 and 10 hpi. Once added, compounds were kept in the medium throughout the remaining assay period. DMSO (0.2%) was used as vehicle control. Inhibition rates were calculated by comparing the number of plaques formed in the infected and untreated cells and infected and treated ones.
Attachment inhibition assay
The effects of the triazole derivatives on CHIKV attachment were evaluated according to Fox and coworkers [25] with few modifications. Vero cells (2 × 105 cells/well) grown in 24-well plates were pre-chilled at 4°C for 1 h, then, infected with CHIKV BRA/RJ/18 isolate (MOI = 1.0) for 1 h at 4°C. The triazole derivatives (100 μM) were added to the cells during the same period. CQ (100 μM) and DMSO (0.2%) were used as controls. After attachment, media containing virus inoculum and compounds were removed, cells were washed twice with cold PBS (pH 7.4) and fresh media were added. Cells were further incubated for 24 h at 37°C and 5% CO2. At 24 hpi, cells were lysed by three cycles of freezing and thawing, and virus titers of the supernatants were determined by plaque assays. Attachment inhibition rates were determined relative to infected and untreated control.
Entry inhibition assay
To evaluate whether the triazole derivatives could impair virus entry, we used the protocol described by Lani and coworkers [26] with modifications. Vero cells (2 × 105 cells/well) were grown in 24-well plates at 37°C and 5% CO2 on the day before the experiment. Cells were pre-chilled at 4°C for 1 h, followed by infection with BRA/RJ/18 isolate (MOI = 1.0) for 1 h under the same conditions. Then, the virus inoculum was discarded, cells were washed twice with cold PBS (pH 7.4), and media containing the triazole derivatives (100 μM) were added for 1 h at 37°C. CQ (100 μM) and DMSO (0.2%) were used as controls. After treatment, media were removed, cells were washed with PBS (pH 3.0) for 1 min and then washed three-times with PBS (pH 7.4). Posteriorly, fresh media without compounds were added to the cells which were further incubated for 23 h at 37°C and 5% CO2. At 24 hpi, cells were lysed by three cycles of freezing and thawing, and virus titration was carried out using plaque assays. The inhibition rates of CHIKV entry were determined in relation to the infected and untreated control group.
Egress inhibition assay
The evaluation of the effects of the triazole derivatives on virus egress was carried out as described by Fox and coworkers [25] with few modifications. Vero cells (2 × 105 cells/well) were infected with CHIKV BRA/RJ/18 (MOI = 1.0) for 2 h at 37°C and 5% CO2 atmosphere, after which the inoculum was removed. Then, cells were thoroughly washed with PBS (pH 7.4), fresh media containing the compounds (100 μM) were added, and cells were incubated for further 6 h. CQ (100 μM) and DMSO (0.2%) were also used as controls. At 6 hpi, supernatants were collected as the extracellular content (released virus particles). Directly after this, cells were washed twice with PBS (pH 7.4) and fresh media were added. Thereafter, cells were further submitted to three cycles of freezing and thawing to release intracellular viruses into the supernatants that were collected as the intracellular content (internalized virus particles). Virus titers in both extracellular and intracellular contents were determined by plaque assays. Inhibition rates were calculated relative to the infected and untreated cells and the intracellular/extracellular rates were calculated as the ratio of virus titers determined in the intracellular and extracellular contents.
Virus particle inactivation assay
The virucidal activity of the triazole derivatives was evaluated as described by Lani and coworkers [26] with few modifications. Viral suspensions containing 2 × 105 PFU of the CHIKV BRA/RJ/18 isolate were treated or not with 100 μM of the triazoles, CQ, or DMSO (0.2%) for 2 h at 37°C. Then, all suspensions were diluted (1:100) to prevent the potential effects of the compounds on virus replication steps in cells. To evaluate the remaining infectivity of the treated and untreated viral suspensions, the diluted suspensions were used to infect confluent monolayers of Vero cells for 2 h at 37°C and 5% CO2. After virus attachment, cells were washed twice with PBS (pH 7.4), and a fresh medium was added, followed by incubation for 24 h at 37°C and 5% CO2. Finally, cells were lysed by three cycles of freezing and thawing, and virus titers of the supernatants were quantified by plaque assays. The virucidal activity of the compounds was determined by their ability to reduce virus infectivity relative to the untreated control.
In silico target fishing strategy
Structural similarity analyses
Initially, a literature search was conducted to build a library of small molecules with experimentally confirmed binding or inhibitory activity on any CHIKV protein. By the time this study was done, this library was comprised of 50 compounds with binding affinity or inhibitory activity against the nsP1, nsP2, nsP3, nsP4 and C proteins of CHIKV. Structural comparison of the triazole derivatives with proven ligands of CHIKV`s proteins was performed using the MACCS fingerprints available within the OpenBabel 3.1.1 software [27] and 2D structural similarity was calculated by the Tanimoto coefficient (Tc). The molecular targets of the most similar compounds to each triazole derivative were selected for further molecular docking studies.
Molecular docking studies
Initially, the protonation state of the triazole derivatives at pH 7.4 was predicted using the MarvinSketch v. 16.2.29.0, 2016, Chemaxon (www.chemaxon.com) program and their 3D structure was constructed and optimized using the Spartan'10 software (Wavefunction Inc., CA, USA). First, a conformational analysis was carried out in vacuum using the Merck molecular force field (MMFF). Then, the lowest-energy conformation was subjected to a geometry optimization step using the semi-empirical RM1 method which was further submitted to an energy calculation using the DFT method with the B3LYP/6-31* basis set. The 3D structure of the known inhibitors with the highest similarity to the triazole derivatives was obtained using the same method and they were used for comparative purposes in docking studies.
According to the similarity comparison, the binding mode of both triazole derivatives with CHIKV nsP1 and nsP2 protease was evaluated while the interaction of compound 1 with the CHIKV C protein was also investigated. The 3D structures of CHIKV nsP1 (PDB 7DOP), nsP2 protease (PDB 3TRK) and C protein (PDB 5H23) were retrieved from the PDB. Solvent molecules and ions were removed.
Docking studies were performed using the Autodock Tools 1.5.7 (ADT) and Autodock 4.2.6 software. ADT was used to prepare proteins by adding hydrogen atoms and Gasteiger charges while ligands' torsions were automatically set. The docking protocol for nsP2 protease was validated previously and is described elsewhere [28]. Due to the lack of experimental 3D structure of the other proteins in complex with ligands, the search spaces were defined according to putative binding sites described by experimental and theoretical methods previously [29–31]. For nsP1 studies, the grid box was centered on the CD atom of Q151 and had dimensions of 60 × 60 × 70 points. For C protein, the grid box with dimensions of 44 × 44 × 44 points was centered on the CE1 atom of H139. In both cases, a 0.375 Å grid spacing was used. The Lamarckian genetic algorithm was used as the search engine and 50 binding poses were calculated using the default searching parameters. The binding pose with the lowest energy was selected for analysis using Discovery Studio Visualizer 2019 and Pymol 2.5.0a0 (The PyMOL Molecular Graphics System, Schrödinger, LLC).
Statistical analyses
In vitro assays were carried out in triplicate in three independent experiments and data were expressed as mean ± standard deviation (SD). Statistical analyses were performed by using one-way ANOVA followed by the Tukey test within the GraphPad Prism v. 8 software. Statistical significance was considered when p-value < 0.05.
Results
Cytotoxicity & antiviral activity of the 1,4-disubstituted-1,2,3-triazole derivatives in different cell lines
Initially, we evaluated the toxicity of the 1,4-disubstituted-1,2,3-triazole derivatives on BHK-21 cells using different concentrations. The marketed drug CQ was also evaluated as it was used as a positive control in the antiviral assays. Both triazole derivatives, 1 and 2, exhibited CC50 values higher than 1000 μM which indicated low cytotoxicity while CQ showed higher toxicity with a CC50 of 591.1 μM (Table 1). No cytotoxic effects were observed after the treatment of cells with DMSO (1.0%).
Table 1. . Cytotoxicity and anti-CHIKV activity of the 1,4-disubstituted-1,2,3-triazole derivatives evaluated in this work and the control drug chloroquine (CQ).
| Compound | CHIKV 181/25 | CHIKV BRA/RJ/18 | ||||||
|---|---|---|---|---|---|---|---|---|
| BHK-21 | Vero | Vero | ||||||
| CC50 (μM) | EC50 (μM) | SI† | CC50 (μM) | EC50 (μM) | SI† | EC50 (μM) | SI† | |
| 1 | >1000 | >50 | ND | 466.6 ± 74.4 | >50 | ND | 19.9 ± 4.0 | 23.4 |
| 2 | 1053.2 ± 87.0 | 28.6 ± 3.6 | 36.9 | 211.1 ± 49.6 | 30.0 ± 1.3 | 7.1 | 19.7 ± 5.5 | 10.7 |
| CQ | 591.1 ± 62.4 | 18.8 ± 2.2 | 32.5 | 469.1 ± 84.4 | ND | ND | 47.5 ± 10.3 | 9,88 |
The selectivity index (SI) was calculated as the ratio between the CC50 and EC50 values.
CHIKV: Chikungunya virus; CQ: Chloroquine; ND: Not determined.
Next, we screened these compounds against the replication of CHIKV 181/25 in BHK-21 cells at 50 μM. Compound 1 decreased virus titer by 39.9% (±7.49) while compound 2 inhibited 77.2% (±1.3) of virus replication, which is comparable to the inhibitory activity of CQ (88.5% ± 1.1) (Supplementary Figure 3).
Consequently, compound 2 and CQ were tested at different concentrations to determine their EC50 (Table 1). Although the CQ exhibited a nearly twofold higher antiviral activity (EC50 = 18.8 μM; SI = 32.5) than compound 2, the triazole derivative presented an interesting antiviral activity with EC50 of 28.6 μM. Yet, this compound showed a slightly increased selectivity (SI = 36.9) than CQ (SI = 32.5) which indicates greater safety and efficacy in comparison to the control drug.
To verify whether the triazoles had a cell-dependent antiviral activity, we assessed their inhibitory activity on the CHIKV 181/25 replication in Vero cells. As observed in BHK-21 cells, compound 1 exhibited moderate antiviral activity (32.79% ±13.09) whereas compound 2 (90.4% ± 2.5) and CQ (98.2% ± 0.2) strongly inhibited CHIKV replication at 50 μM (Supplementary Figure 3). The EC50 of compound 2 was 30.0 μM, pointing to its cell-independent antiviral activity. However, both triazoles presented higher cytotoxic activity toward Vero cells, which resulted in a lower SI value for compound 2 (Table 1).
Additionally, the inhibitory activity of these derivatives against the replication of a Brazilian clinical isolate (BRA/RJ/18) in Vero cells was analyzed. Interestingly, both derivatives 1 and 2 significantly reduced viral yield by 67.8% (±1.6) and 79.2% (±5.1), respectively, at 50 μM (Supplementary Figure 3). Both triazoles exhibited a stronger inhibitory potency than CQ with EC50 values of 19.9 μM for compound 1, 19.7 μM for compound 2, and 47.5 μM for CQ (Table 1). They also showed an improved selectivity (SI of 23.4 and 10.7 for compounds 1 and 2) in comparison to the control drug CQ (SI = 9.9).
Moreover, the antiviral activity of compounds 1 and 2 was evaluated against MAYV, which belongs to the same genus and family as CHIKV. No marked reduction in virus titer was observed after treatment with any compounds at 50 μM (Supplementary Figure 4), suggesting that they inhibit CHIKV replication specifically.
Evaluation of the mechanism of action of the 1,4-disubstituted-1,2,3-triazole derivatives against CHIKV
Having confirmed the antiviral potential of compounds 1 and 2, we investigated their mechanism of action against the Brazilian CHIKV isolate in Vero cells at a highly effective and non-cytotoxic concentration (100 μM).
Time-course evaluation of the anti-CHIKV activity of the 1,4-disubstituted-1,2,3-triazoles
Initially, we carried out a time-of-addition assay to investigate at which step of viral replication these compounds might inhibit. Both derivatives showed a different antiviral profile than the one observed for CQ which mainly acts at viral entry (Figure 2). Compound 2 showed a more prominent inhibitory activity (>60%) when added up to 4 hpi, followed by a significant decrease in its antiviral activity. These results suggest that this compound may act at early steps of viral replication including post-entry stages. On the other hand, compound 1 maintained a prominent activity when added up to 8 hpi which suggests that this compound might act at early and late steps of viral replication.
Figure 2. . Antiviral activity of the 1,4-disubstituted-1,2,3-triazole derivatives and chloroquine after treatment at different times during Chikungunya virus infection (MOI 1.0) in Vero cells according to the superior scheme.
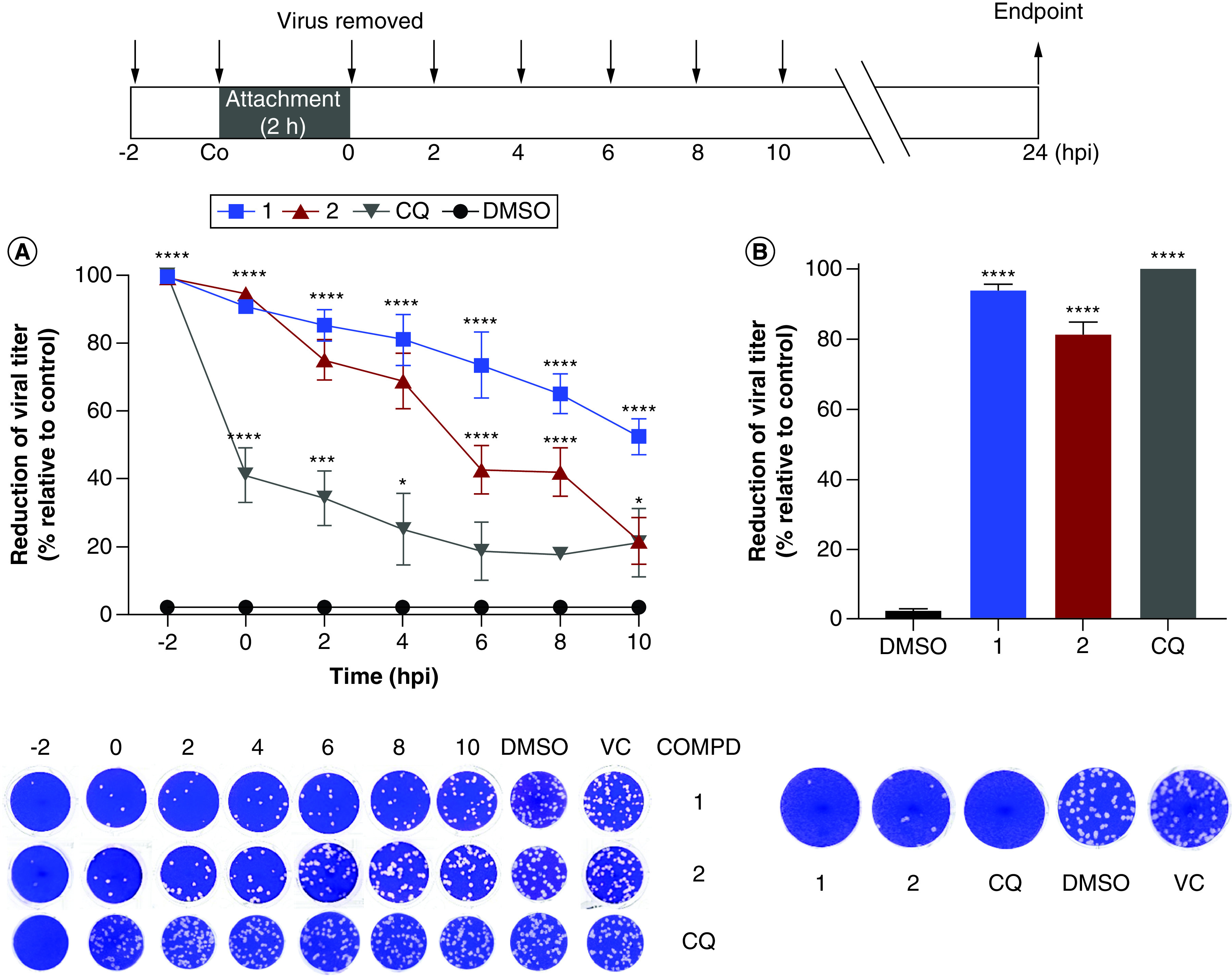
Compounds (100 μM) were added (A) at different times pre- or post-infection, or (B) during the virus attachment period (co-treatment). At 24 hpi, cells were lysed and supernatants were collected. Inhibitory rates were determined by comparison to the infected and untreated cell control (VC) using plaque assays. Representative viral plaques of Vero cells are shown. Data are expressed as mean ± standard deviation (n = 3). One-way ANOVA and Tukey post-test were employed to evaluate statistical significance relative to DMSO-treated (0.2%) control (*p < 0.05; ***p < 0.001; ****p < 0.0001).
CHIKV: Chikungunya virus; COMPD: Compound; CQ: Chloroquine; DMSO: Dimethyl sulfoxide; VC: Virus control.
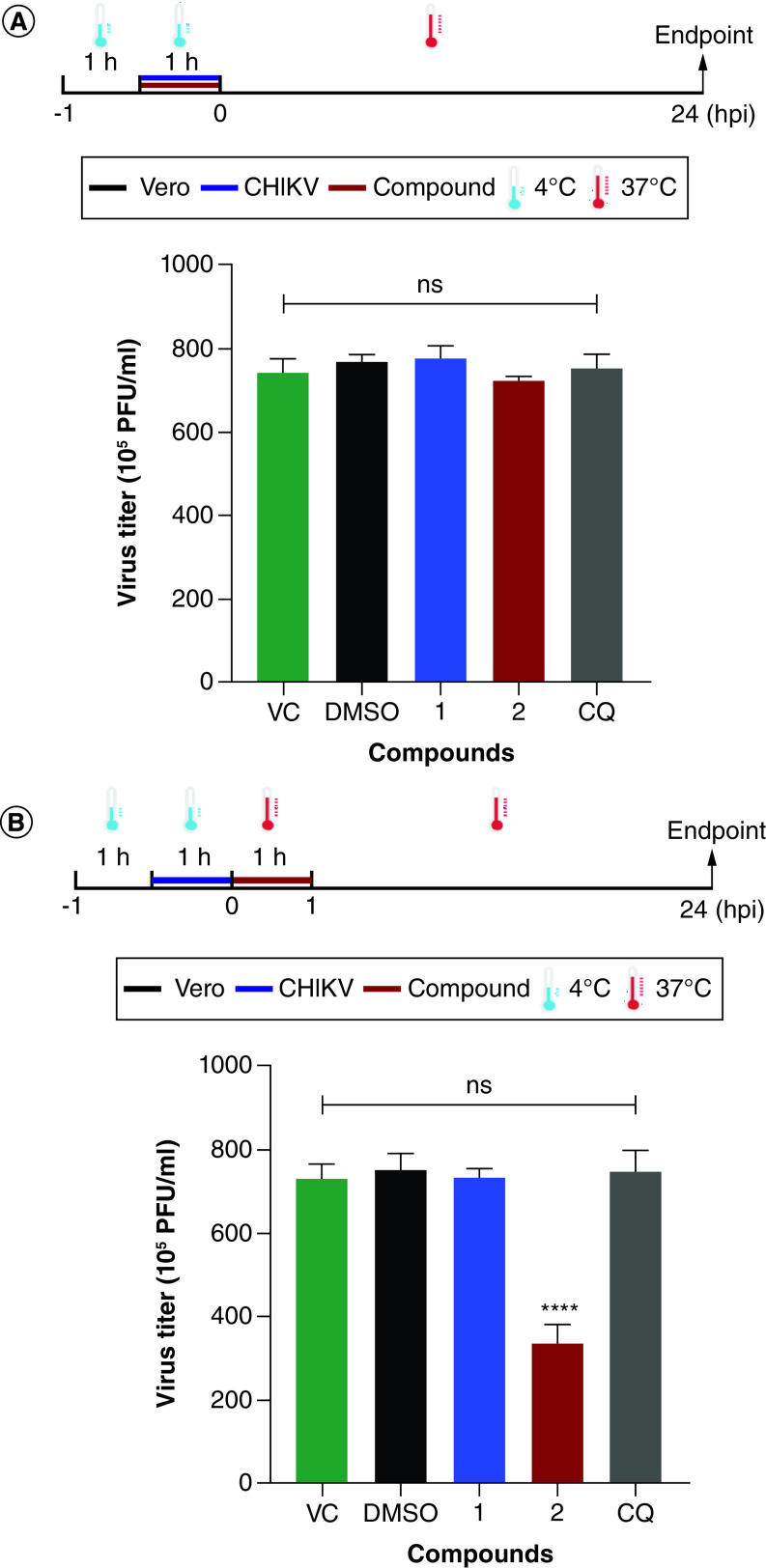
Effects of the triazole derivatives on CHIKV attachment & entry
Since these compounds may act at early steps of viral replication, we evaluated whether they inhibit virus attachment and entry. None of the derivatives blocked CHIKV attachment to Vero cells since no differences were observed in the virus titers of the different experimental groups (Figure 3A). However, compound 2 showed significant inhibition against virus entry in Vero cells and reduced 54.4% (±5.2) of virus titer in comparison to the untreated control (Figure 3B).
Figure 3. . Effects of the 1,4-disubstituted-1,2,3-triazole derivatives, 1 and 2, on the early steps of Chikungunya virus replication.
(A) Attachment and (B) entry. Vero cells were infected with CHIKV BRA/RJ/18 isolate at an MOI of 1.0 and compounds (100 μM) were added according to the superior scheme. Infected and untreated cells (VC) or infected cells treated with DMSO (0.2%, vehicle control) or chloroquine (CQ, 100 μM) were used as controls. At 24 hpi, virus titers were determined by plaque assays. Data are expressed as mean ± standard deviation (n = 3) and statistical analysis was carried out using ANOVA followed by the Tukey test. Statistical significance was assessed relative to the VC group (****p < 0.0001).
CHIKV: Chikungunya virus; COMPD: Compound; CQ: Chloroquine; DMSO: Dimethyl sulfoxide; VC: Virus control.
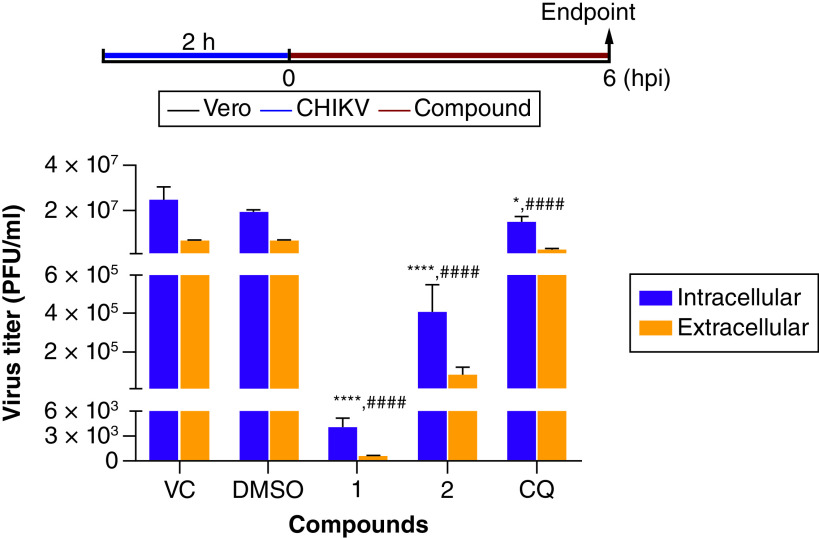
Evaluation of the inhibitory activity on CHIKV egress
As compound 1 was suggested to inhibit late steps of CHIKV replication, we also investigated whether the triazole derivatives could affect virus egress from Vero cells. All compounds tested reduced significantly the virus titer in the extracellular content when compared with untreated control (VC) (Figure 4). Taking the intracellular/extracellular titers ratio into account, we observed a nearly twofold increased viral yield in the intracellular content after compound 1 treatment (7.3) in comparison to the untreated (3.7) or DMSO (2.8) controls while a more modest effect was observed for compound 2 (5.2) and CQ (6.2). This result demonstrated that compound 1 blocks CHIKV release or related events.
Figure 4. . Analysis of the inhibitory activity of the 1,4-disubstituted-1,2,3-triazole derivatives, 1 and 2, on Chikungunya virus egress.
Vero cells were infected with CHIKV BRA/RJ/18 (MOI = 1.0) and then treated with different compounds (100 μM) according to the top scheme. Infected and untreated cells (VC) or infected cells treated with DMSO (0.2%, vehicle control) or chloroquine (CQ, 100 μM) were used as controls. At 6 hpi, extracellular and intracellular contents were collected and titrated by plaque assays. Data are expressed as mean ± standard deviation (n = 3). Statistical significance was determined by ANOVA and Tukey post-test relative to VC group (for intracellular content, *p < 0.05; ****p < 0.0001; for the extracellular content, ####p < 0.0001).
CHIKV: Chikungunya virus; COMPD: Compound; CQ: Chloroquine; DMSO: Dimethyl sulfoxide; VC: Virus control.
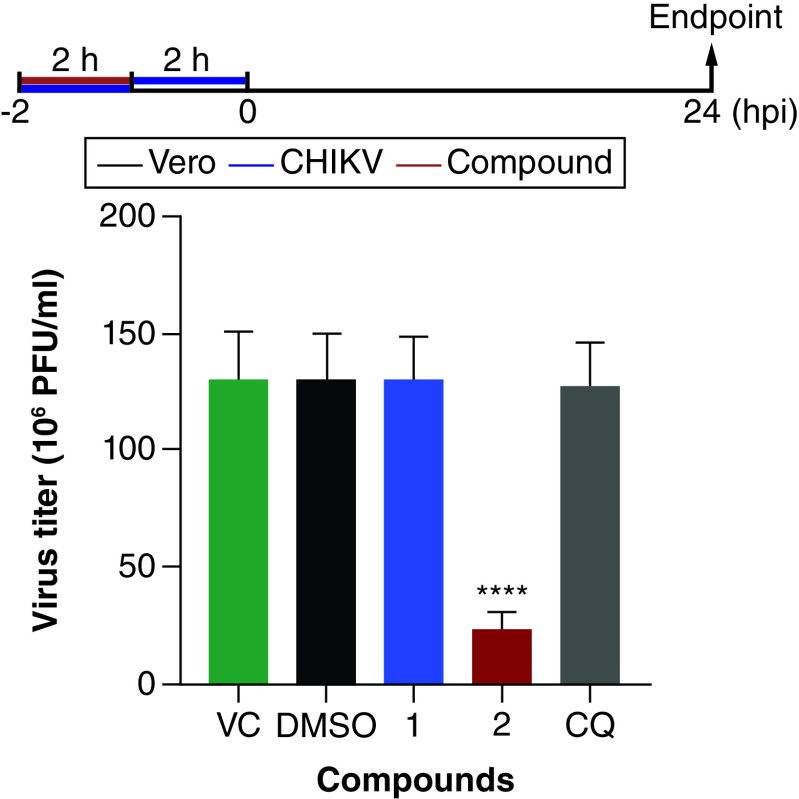
Analysis of the virucidal activity of the triazoles on CHIKV particles
To investigate the direct effect of the compounds on extracellular virus particles, CHIKV suspensions were treated and their remaining infectivity was evaluated in Vero cells. Interestingly, compound 2 exhibited a strong virucidal activity and virus titer was reduced by 81.8% (±4.0) in comparison to the untreated control (Figure 5). By contrast, compound 1 and CQ did not show any virucidal activity against CHIKV particles.
Figure 5. . Inactivation effects of the 1,4-disubstituted-triazoles and chloroquine on Chikungunya virus particles.
Viral suspensions of CHIKV BRA/RJ/18 isolate (2 × 105 PFU) were incubated in the presence (100 μM) or absence of the compounds for 2 h at 37°C. Dimethyl sulfoxide (0.2%) was used as vehicle control. Then, all suspensions were diluted (1:100) and Vero cells monolayers were infected and incubated for further 24 h at 37°C and 5% CO2. Cells were lysed and virus titers were quantified by plaque assays. Results are shown as mean ± standard deviation (n = 3) and statistical significance was measured by one-way ANOVA and Tukey's tests (****p < 0.0001).
CHIKV: Chikungunya virus; COMPD: Compound; CQ: Chloroquine; DMSO: Dimethyl sulfoxide; VC: Virus control.
Prediction of the antiviral targets of the 1,4-disubstituted-1,2,3-triazole derivatives
We employed ligand- and structure-based computational methods to gain further insights of the molecular targets of the triazole derivatives. Based on the principle of similarity, similar ligands likely exert similar biological activity by interacting with similar targets. So, we built a library of experimentally proven ligands of CHIKV proteins and performed a structural similarity analysis with the triazole derivatives. Compound 1 presented the highest similarity with the following ligands: P1-1 (Tc = 0.64), an nsP1 inhibitor [32]; C-5 (Tc = 0.57), an inhibitor of the proteolytic activity of C protein [31]; and P2–19 (Tc = 0.57), an nsP2 protease inhibitor [33] (Figure 6A). Similarly, compound 2 showed the highest similarity with P1-1 (Tc = 0.53) and P2–19 (Tc = 0.53). Further, molecular docking studies were carried out to investigate the interaction pattern of the triazole derivatives with the targets of the most similar compounds.
Figure 6. . Prediction of molecular targets of the 1,4-disubstituted-1,2,3-triazole derivatives, 1, and 2.
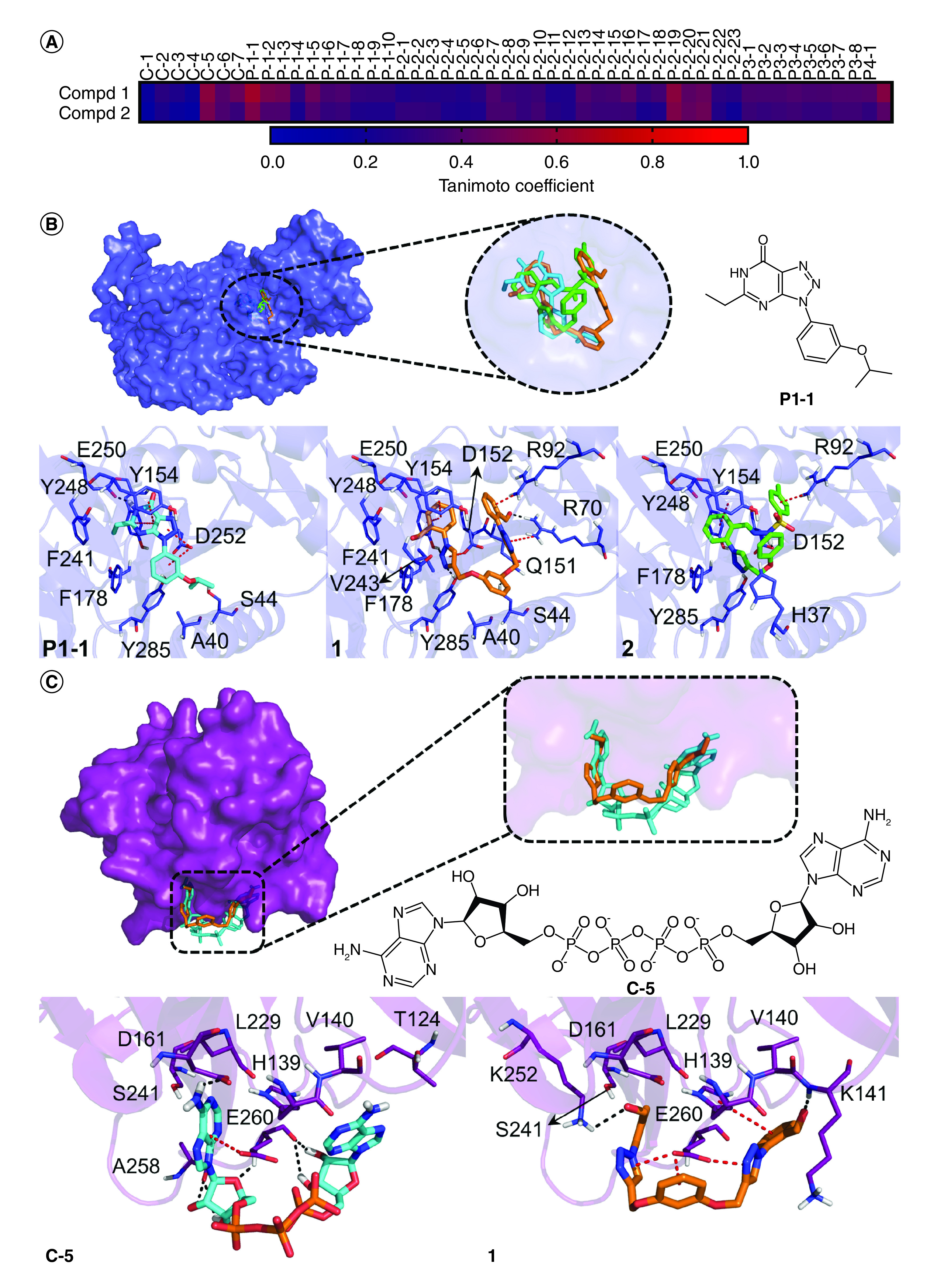
(A) Structural similarity comparison between the triazole derivatives and proven ligands of Chikungunya virus proteins reported in the literature. The similarity was measured by the Tanimoto coefficient (Tc); Predicted binding mode and interactions of the compounds and similar known inhibitors with Chikungunya virus proteins: (B) nsP1 and (C) C protein. Hydrogen bonds and π-stacking interactions are represented as black and red dashed lines, respectively.
Compd: Compound.
We first docked these compounds with CHIKV nsP1 and compared them with the inhibitor P1-1. Compounds 1 and 2 showed binding energy values of -6.7 and -5.9 kcal/mol, respectively, exhibiting a greater theoretical affinity with the enzyme in comparison to the inhibitor P1-1 (-5.4 kcal/mol). Both derivatives bound within the GTP binding site and were superimposed with P1-1 (Figure 6B). One of the benzaldehyde groups of compound 1 was positioned at the same region as the purine base of GTP and was sandwiched by Y154 and Y248 by π-π-stacking interactions whereas the neighbor triazole nucleus was hydrogen bonded to Y154 and Y285 and established an anion-π interaction with D152. The other benzaldehyde group bound at the phosphates binding region and was involved in hydrogen bond and cation-π interactions with R70 and R92, respectively. The neighbor triazole ring was also stacked to R70. The central linking group was hydrogen-bonded to S44 and this compound interacted with other residues such as A40, Q151, F178, F241, V243 and E250 by van der Waals contacts.
Compound 2 exhibited a similar interaction network (Figure 6B). For instance, the phenyl ring attached to the triazole moiety was anchored at the binding region of the purine ring of GTP and interacted with Y154, F178, Y248, E250 and Y285. The tosylhydrazone group of the compound was positioned at the binding region of the phosphate groups of GTP and established hydrogen bond and cation-π interactions with D152 and R92. In addition, the phenol ring interacted with the catalytic residue H37. Since most of these interactions were also observed for the inhibitor P1-1 and are important to GTP binding, nsP1 is likely a target of these compounds.
Following this, compounds were docked to CHIKV nsP2 protease and their binding mode and interactions were compared with the ones observed for the inhibitor P2–19. Although compounds 1 and 2 presented a similar binding manner with the known inhibitor, we noticed a slight change in their binding pose, probably due to their higher volume which, in turn, compromised their fit within the tunnel-shaped active site (Supplementary Figure 5). Consequently, this change prevented the interaction of the derivatives with S1 subsite residues (e.g., C1013) that are important for enzyme function and ligand recognition, suggesting that these compounds do not inhibit CHIKV nsP2 protease.
Since compound 1 showed high structural similarity with the C protein inhibitor C-5 as well, docking studies with CHIKV C protein and these compounds were performed (Figure 6C). Although both compounds 1 and C-5 shared a comparable binding manner, the triazole derivative exhibited significantly lower binding energy (-5.8 kcal/mol) than the known ligand (-0.9 kcal/mol). The benzaldehyde and triazole groups of 1 occupied the same regions as the purine moiety of C-5 while the central linking group of the triazole derivative was positioned at the same region as the phosphates of the proven inhibitor. As a result of this, one of the benzaldehyde was hydrogen-bonded to K242, and its neighbor triazole ring was involved in an anion-π interaction with E260 as well as the central phenyl and the other triazole ring. By contrast, the other benzaldehyde group established hydrogen bond and π-π-stacking interactions with K141 and the catalytic residue H139, respectively. Additionally, van der Waals contacts were observed with the catalytic D161 and other residues like V140, L229, and S241. Interestingly, this compound resembled several contacts formed by the known inhibitor C-5 which suggests that compound 1 could bind to the C protein and block its proteolytic activity.
Discussion
Chikungunya fever is a serious threat to world public health and yet there are no antiviral agents available to combat this disease. In this work, we evaluated the anti-CHIKV activity of two 1,4-disubstituted-1,2,3-triazole derivatives, 1 and 2, synthesized previously by our group and endowed with different pharmacological properties and good in vitro safety profiles [20–22].
We showed that compound 2, but not 1, efficiently inhibited the replication of CHIKV 181/25 strain in both BHK-21 and Vero cells with EC50 values of 28.6 μM and 30.0 μM, respectively. Although the control drug CQ had a nearly twofold higher potency in BHK-21 cells, compound 2 exhibited slightly greater selectivity. Interestingly, this compound was able to inhibit the replication of a Brazilian clinical isolate (CHIKV BRA/RJ/18) belonging to another lineage (ECSA) in Vero cells with higher activity and selectivity than CQ (EC50 = 19.7 μM and SI = 10.72). Surprisingly, compound 1 also had significant antiviral activity against the clinical isolate with similar potency but improved selectivity (EC50 = 19.9 μM and SI = 23.4). Despite having moderate toxicity toward this cell line, effective concentrations of these compounds (EC50) are, at least, tenfold lower than the concentrations required to induce toxicity (CC50) which, in turn, may allow the establishment of effective and safe dosage regimens [34], unlike CQ. These derivatives also showed interesting safety in human cells such as fibroblasts and astrocytes [20,22].
Although CQ exhibited more potent activity against the CHIKV 181/25 replication, this drug failed to treat or protect humans and/or non-human primates. Instead, it exacerbated CHIKV infection in animal models [35,36]. On the other hand, ribavirin is a broad-spectrum antiviral drug bearing a 1,2,4-triazole ring and has a promising anti-CHIKV potential. In fact, ribavirin significantly inhibited CHIKV BRA/RJ/18 strain in Vero cells with an EC50 of 2.4 μM as reported by our group [37]. However, this drug showed weaker activity when evaluated against CHIKV 181/25 replication in different cell lines as Vero (EC50 = 407.69 μM and SI = 0.65), Huh-7 (EC50 = 10.54 μM and SI = 4.64) and A549 (EC50 = 479.52 μM and SI = 0.43) [38], unlike compound 2 which showed similar activity in both Vero and BHK-21 cells. Although ribavirin showed higher antiviral activity against an ECSA clinical isolate in Vero cells (EC50 = 15.51 μM and SI > 6.45) [39], the selectivity was comparable to the one observed for both compounds 1 and 2. Further, ribavirin therapy apparently improved the symptoms of patients such as joint pain and swelling [40]. Nonetheless, the use of this drug is limited due to its side effects and contraindications [41], so alternative drugs should still be pursued.
Besides ribavirin, there are few reports of triazole derivatives with anti-CHIKV activity [42–44]. Among these compounds, a triazolopyrimidine derivative containing a piperidine ring functionalized as urea (namely 15e) showed the highest antiviral activity with EC50 values ranging from 1.0 μM to 8.9 μM against replication of different CHIKV strains (IOL 899, Venturini, Congo 95 and St. Martin), and CC50 value of 201 μM in Vero cells [44]. Despite not having a higher antiviral activity in comparison to this derivative, compound 2 showed a wide activity against two CHIKV genotypes as well as a cell-independent activity that has not been reported yet for any triazole derivative while compound 1 presented similar selectivity.
Considering that Chikungunya chronic disease is possibly more associated with the ECSA genotype [10], we investigated the mechanism of action of both derivatives against the Brazilian CHIKV isolate replication in Vero cells. According to the time-of-addition assay, both derivatives probably act at early steps of CHIKV replication, but compound 1 may also inhibit late steps. In fact, compound 2 inhibited virus entry but not attachment whereas compound 1 did not block these processes. Since the effects of lysosomotropic agents, such as CQ, are reversible when removed within 90 min after infection by alphaviruses [45], which is confirmed by our experiment, we believe that compound 2 does not affect endosomal pH to block virus entry. In addition, compounds that block early steps such as virus attachment, entry, or fusion (e.g., arbidol, baicalin and suramin) usually exhibit more prominent activity when added up to 2 hpi [46–48], which suggests that the triazole derivatives may also act at other post-entry steps. In agreement, lycorine and favipiravir are inhibitors of CHIKV RNA translation and replication, respectively, and have significant antiviral action when added up to 4 hpi [49,50], like compound 2.
We also observed that compound 1 significantly reduced CHIKV egress from Vero cells or any related process. For instance, berberine inhibits CHIKV replication when added after 4 hpi by interfering with C protein interactions with other protein units or newly-synthesized RNA strands and, consequently, affects viral assembly [51]. Also, the late residual antiviral effects of CQ have been associated with the blockade of post-translational modifications, especially the ones taking place at the endoplasmic reticulum or trans face of the Golgi apparatus [52]. Since these steps are required for CHIKV maturation, the triazole derivatives may inhibit virus release by targeting them. In addition, compound 2 displayed a significant virucidal activity that contributes to its antiviral profile and may be investigated as a prophylactic agent later. Baicalin also possesses virucidal potential which probably yields from changes in virion or virus envelope properties [47]. Overall, our results suggest that these compounds may act at different steps and further experiments remain to be performed to confirm the exact processes that are affected.
To shed light on their molecular targets, we applied combined ligand and structure-based computational strategies to improve the prediction accuracy [53]. Both derivatives exhibited the highest similarity with nsP1 and nsP2 protease inhibitors and further docking studies were carried out. For nsP1, both compounds exhibited a similar binding manner with the known inhibitor P1-1 at the GTP binding site. They also established interactions with important residues for the enzyme function and natural ligand binding, such as H37, Q151, D152, Y154, Y248 and Y285 [29], suggesting that they can inhibit this protein. Reinforcing this hypothesis, known inhibitors of CHIKV nsP1 exhibit a significant antiviral activity at early moments of infection (up to 4 hpi) as observed for the triazole derivatives [32,54–56]. Regarding nsP2 protease, none of the triazole derivatives established key interactions with the enzyme as observed for other proven inhibitors [28]. Consequently, these findings suggested that this protein is not a putative target of the studied derivatives. Since compound 1 also showed high similarity with C-5, a C protease inhibitor, we also performed docking studies with this enzyme. Interestingly, derivative 1 was superimposed to the proven inhibitor and resembled similar interactions, including the ones with the catalytic residues H139 and D161. Inhibitors of the proteolytic function of the C protein exert their antiviral activity at late stages of CHIKV replication [31], which supports our results and indicates that this compound can inhibit this protein to block virus release. However, since there are no experimentally proven ligands of E1 and E2 proteins of CHIKV to date, these proteins were not considered in the ligand-based strategy and, consequently, we cannot exclude that these compounds could interact with envelope proteins. Additionally, it is worth noting that further experimental assays should be carried out to confirm these proteins as antiviral targets of the studied triazole derivatives.
Due to the emergence and reemergence of viruses, the search for novel compounds with a broad antiviral spectrum is highly desired [57]. We also evaluated the antiviral activity of compounds 1 and 2 against MAYV, another arthritogenic alphavirus of relevance in Latin America. None of the derivatives inhibited MAYV replication, indicating that they are specific inhibitors of CHIKV. Indeed, finding compounds with a broad spectrum against CHIKV and other alphaviruses is still a challenge [58]. However, both compounds are known inhibitors of HSV-1 replication, which is a DNA virus [22]. Despite inhibiting both viruses, their antiviral activities are triggered by different mechanisms of action which may contribute to the inhibition of other viruses yet not investigated. In this context, compound 2 is more promising due to its antiviral properties against different CHIKV strains in a cell-independent way, but both compounds should still be evaluated against a panel of CHIKV strains of other lineages and other viruses to further accurately assess their broad-spectrum antiviral potential.
Conclusion
In this work, the antiviral activity of two triazole derivatives, 1 and 2, against CHIKV replication was uncovered. Compound 2 demonstrated a wider and cell-independent activity while compound 1 showed more selective activity against a Brazilian clinical isolate. Derivative 1 strongly inhibited virus release while compound 2 significantly inhibited virus entry and exhibited virucidal activity. Besides, they likely act at multiple steps of viral replication including post-entry steps. Computational target prediction suggested that both compounds block nsP1 protein while compound 1 could also target the C protease function which agrees with the experimental data obtained, but further experimental confirmation is still required. Additionally, we observed that they are specific inhibitors of CHIKV and did not inhibit MAYV replication. Thus, both compounds showed promising antiviral profiles and warrant further in-depth studies regarding their mechanism of action and in vivo activity as well as the design of new derivatives with higher potency and safety to combat CHIKV infection.
Summary points.
Chikungunya virus (CHIKV) is a reemerging arthropod-borne virus that has spread to over 100 countries worldwide and, yet there are no vaccines or antiviral drugs available to fight this disease yet.
In this study, we evaluated the antiviral potential of two 1,4-dissubstituted-1,2,3-triazole derivatives (1 and 2) against CHIKV using in vitro and in silico methods.
Both compounds did not present significant cytotoxicity effects on the cell lines studied.
Compound 2 exhibited similar activity against the replication of CHIKV 181/25 strain in both BHK-21 and Vero cells, with EC50 values of 28.6 and 30.0 μM, respectively. This compound also inhibited the replication of a Brazilian isolate (CHIKV BRA/RJ/18) in Vero cells with improved efficacy (EC50 = 19.7 μM; SI = 10.7).
Compound 1 was able to inhibit the CHIKV BRA/RJ/18 replication with similar potency and higher selectivity (EC50 = 19.9 μM; SI = 23.4), but not CHIKV 181/25.
Both compounds are likely to act at multiple steps of virus replication, including early post-entry stages.
Compound 2 inhibited virus entry (54.4%), but not attachment and release, besides having a strong virucidal activity (81.8%). By contrast, compound 1 showed high inhibitory effects against CHIKV egress, but not toward viral attachment and entry, nor direct effect in virus particles.
Computational studies suggested that both derivatives inhibit CHIKV nsP1 protein while compound 1 may also target capsid protein.
These triazole derivatives did not exhibit antiviral activity against Mayaro virus, another alphavirus.
Our study paves the way for further in-depth studies regarding their mechanism of action and in vivo activity evaluation as well as the design of new derivatives with higher potency and safety to combat CHIKV infections.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/fvl-2023-0142
Author contributions
VW Rabelo: methodology, validation, formal analysis, investigation, writing – original draft, writing – review and editing. VD Silva: methodology, validation, formal analysis, investigation, writing – original draft. ML Sanchez-Nuñez and LS Corrêa-Amorim: validation, investigation. CD Buarque and RJ Kuhn: funding acquisition, supervision, resources, writing – review and editing. PA Abreu: conceptualization, funding acquisition, supervision, writing – review and editing. ICNP Paixão: conceptualization, funding acquisition, supervision, resources, writing – review and editing, project administration.
Financial disclosure
This study was funded in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – finance code 001 as well as the Brazilian National Council for Scientific and Technological Development (CNPQ) and Research Support Foundation of the State of Rio de Janeiro (FAPERJ). RJ Kuhn also acknowledges the support of NIH National Institute of Allergy and Infectious Diseases award no. AI095366. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Competing interests disclosure
The authors have no competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Writing disclosure
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Kril V, Aiqui-Reboul-Paviet O, Briant L, Amara A. New insights into chikungunya virus infection and pathogenesis. Annu. Rev. Virol. 8(1), 327–347 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLOS Negl. Trop. Dis. 8(6), e2921 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puntasecca CJ, King CH, Labeaud AD. Measuring the global burden of chikungunya and Zika viruses: a systematic review. PLOS Negl. Trop. Dis. 15(3), e0009055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharif N, Sarkar MK, Ferdous RN et al. Molecular epidemiology, evolution and reemergence of chikungunya virus in South Asia. Front. Microbiol. 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manzoor KN, Javed F, Ejaz M et al. The global emergence of chikungunya infection: an integrated view. Rev. Med. Virol. 32(3), e2287 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Wimalasiri-Yapa BMCR, Stassen L, Huang X et al. Chikungunya virus in Asia - Pacific: a systematic review. Emerg. Microbes Infect. 8(1), 70–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lima Cavalcanti TYV, Pereira MR, Paula SO, Franca RFO. A review on chikungunya virus epidemiology, pathogenesis and current vaccine development. Viruses 14(5), 969 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chala B, Hamde F. Emerging and re-emerging vector-borne infectious diseases and the challenges for control: a review. Front. Public Heal. 9, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ECDC. Chikungunya worldwide overview (2023). www.ecdc.europa.eu/en/chikungunya-monthly
- 10.Paixão ES, Rodrigues LC, Costa MDCN et al. Chikungunya chronic disease: a systematic review and meta-analysis. Trans. R. Soc. Trop. Med. Hyg. 112(7), 301–316 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Ganesan V, Duan B, Reid S. Chikungunya virus: pathophysiology, mechanism, and modeling. Viruses 9(12), 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Ewijk R, Huibers MHW, Manshande ME et al. Neurologic sequelae of severe chikungunya infection in the first 6 months of life: a prospective cohort study 24-months post-infection. BMC Infect. Dis. 21(1), 179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S, Song S, Zhang L. Recent progress in vaccine development against chikungunya virus. Front. Microbiol. 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider M, Narciso-Abraham M, Hadl S et al. Safety and immunogenicity of a single-shot live-attenuated chikungunya vaccine: a double-blind, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 401(10394), 2138–2147 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battisti V, Urban E, Langer T. Antivirals against the Chikungunya Virus. Viruses 13(7), 1307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study reinforce that the search for novel specific antivirals against CHIKV is highly needed.
- 16.Bozorov K, Zhao J, Aisa HA. 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: a recent overview. Bioorg. Med. Chem. 27(16), 3511–3531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonandi E, Christodoulou MS, Fumagalli G, Perdicchia D, Rastelli G, Passarella D. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov. Today 22(10), 1572–1581 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Kharb R, Yar MS, Sharma PC. Recent advances and future perspectives of triazole analogs as promising antiviral agents. Mini-Reviews Med. Chem. 11(1), 84–96 (2011). [DOI] [PubMed] [Google Scholar]
- 19.da S M Forezi L, Lima CGS, Amaral AAP et al. Bioactive 1,2,3-triazoles: an account on their synthesis, structural diversity and biological applications. Chem. Rec. 21(10), 2782–2807 (2021). [DOI] [PubMed] [Google Scholar]
- 20.da Silva VD, de Faria BM, Colombo E et al. Design, synthesis, structural characterization and in vitro evaluation of new 1,4-disubstituted-1,2,3-triazole derivatives against glioblastoma cells. Bioorg. Chem. 83, 87–97 (2019). [DOI] [PubMed] [Google Scholar]; •• This is the first report of the studied triazole derivatives.
- 21.Almeida-Souza F, da Silva VD, Silva GX et al. 1,4-disubstituted-1,2,3-triazole compounds induce ultrastructural alterations in Leishmania amazonensis promastigote: an in vitro antileishmanial and in silico pharmacokinetic study. Int. J. Mol. Sci. 21(18), 1–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viegas DJ, da Silva VD, Buarque CD, Bloom DC, Abreu PA. Antiviral activity of 1,4-disubstituted-1,2,3-triazoles against HSV-1 in vitro. Antivir. Ther. 25(8), 399–410 (2020). [DOI] [PubMed] [Google Scholar]; •• Our group uncovered the antiviral activity of the studied compounds against Herpes simplex virus type 1.
- 23.Cirne-Santos CC, Barros CS, Nogueira CCR et al. Inhibition by marine algae of chikungunya virus isolated from patients in a recent disease outbreak in rio de janeiro. Front. Microbiol. 10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan M, Santhosh SR, Tiwari M, Lakshmana Rao PV, Parida M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against chikungunya virus in vero cells. J. Med. Virol. 82(5), 817–824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox JM, Long F, Edeling MA et al. Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell 163(5), 1095–1107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lani R, Hassandarvish P, Shu MH et al. Antiviral activity of selected flavonoids against Chikungunya virus. Antiviral Res. 133, 50–61 (2016). [DOI] [PubMed] [Google Scholar]
- 27.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J. Cheminform. 3(1), 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabelo VWH, de Palmer Paixão ICN, Abreu PA. Structural insights into the inhibition of the nsP2 protease from Chikungunya virus by molecular modeling approaches. J. Mol. Model. 28(10), 311 (2022). [DOI] [PubMed] [Google Scholar]; • Computational tools can support the development of novel anti-CHIKV compounds and aid the investigation of their mechanism of action.
- 29.Jones R, Bragagnolo G, Arranz R, Reguera J. Capping pores of alphavirus nsP1 gate membranous viral replication factories. Nature 589(7843), 615–619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K, Law YS, Law MCY, Tan YB, Wirawan M, Luo D. Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1. Cell Host Microbe 29(5), 757–764.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Fatma B, Kumar R, Singh VA et al. Alphavirus capsid protease inhibitors as potential antiviral agents for Chikungunya infection. Antiviral Res. 179, (2020). [DOI] [PubMed] [Google Scholar]
- 32.Delang L, Li C, Tas A et al. The viral capping enzyme nsP1: a novel target for the inhibition of chikungunya virus infection. Sci. Rep. 6(1), 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanova L, Rausalu K, Žusinaite E, Tammiku-Taul J, Merits A, Karelson M. 1,3-thiazolbenzamide derivatives as chikungunya virus nsP2 protease inhibitors. ACS Omega 6(8), 5786–5794 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferraz AC, Moraes TFS, Nizer WSDC et al. Virucidal activity of proanthocyanidin against Mayaro virus. Antiviral Res. 168, 76–81 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Roques P, Thiberville SD, Dupuis-Maguiraga L et al. Paradoxical effect of chloroquine treatment in enhancing chikungunya virus infection. Viruses. 10(5), 268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study reported that chloroquine fails to combat CHIKV infection in humans and non-human primates.
- 36.De Lamballerie X, Boisson V, Reynier JC et al. On chikungunya acute infection and chloroquine treatment. Vector-Borne Zoonotic Dis. 8(6), 837–839 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Cirne-Santos CC, de Souza Barros C, de Oliveira MC et al. In vitro studies on the inhibition of replication of Zika and chikungunya viruses by dolastane isolated from seaweed Canistrocarpus cervicornis. Sci. Rep. 10(1), 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franco EJ, Rodriquez JL, Pomeroy JJ, Hanrahan KC, Brown AN. The effectiveness of antiviral agents with broad-spectrum activity against chikungunya virus varies between host cell lines. Antivir. Chem. Chemother. 26, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothan HA, Bahrani H, Mohamed Z et al. A combination of doxycycline and ribavirin alleviated chikungunya infection. PLOS ONE 10(5), 1–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravichandran R, Manian M. Ribavirin therapy for chikungunya arthritis. J. Infect. Dev. Ctries. 2(2), 140–142 (2008). [PubMed] [Google Scholar]
- 41.Sinclair SM, Jones JK, Miller RK, Greene MF, Kwo PY, Maddrey WC. The ribavirin pregnancy registry: an interim analysis of potential teratogenicity at the mid-point of enrollment. Drug Saf. 40(12), 1205–1218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study report that the clinical use of ribavirin has some limitations, which supports the search for novel alternatives to treat CHIKV infections.
- 42.Gigante A, Canela MD, Delang L et al. Identification of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones as novel inhibitors of chikungunya virus replication. J. Med. Chem. 57(10), 4000–4008 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Gigante A, Gómez-SanJuan A, Delang L et al. Antiviral activity of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones against chikungunya virus targeting the viral capping nsP1. Antiviral Res. 144, 216–222 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Gómez-Sanjuan A, Gamo AM, Delang L et al. Inhibition of the replication of different strains of chikungunya virus by 3-Aryl-[1,2,3]triazolo[4,5-d] pyrimidin-7(6 H)-ones. ACS Infect. Dis. 4(4), 605–619 (2018). [DOI] [PubMed] [Google Scholar]; • This study reports a triazole derivative with antiviral potential against the replication of different CHIKV strains in Vero cells.
- 45.Cassell S, Edwards J, Brown DT. Effects of lysosomotropic weak bases on infection of BHK-21 cells by Sindbis virus. J. Virol. 52(3), 857–864 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delogu I, Pastorino B, Baronti C, Nougairède A, Bonnet E, de Lamballerie X. In vitro antiviral activity of arbidol against Chikungunya virus and characteristics of a selected resistant mutant. Antiviral Res. 90(3), 99–107 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Oo A, Rausalu K, Merits A et al. Deciphering the potential of baicalin as an antiviral agent for Chikungunya virus infection. Antiviral Res. 150, 101–111 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Ho YJ, Wang YM, Lu JW et al. Suramin inhibits chikungunya virus entry and transmission. PLOS ONE 10(7), 1–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delang L, Guerrero NS, Tas A et al. Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J. Antimicrob. Chemother. 69(10), 2770–2784 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Li N, Wang Z, Wang R et al. In vitro inhibition of alphaviruses by lycorine. Virol. Sin. 36(6), 1465–1474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan JJ, Brown RS, Kielian M. Berberine chloride is an alphavirus inhibitor that targets nucleocapsid assembly. MBio 11(3), 1–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents 55(5), (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galati S, Di Stefano M, Martinelli E, Poli G, Tuccinardi T. Recent advances in in silico target fishing. Molecules 26(17), 5124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdelnabi R, Kovacikova K, Moesslacher J et al. Novel class of chikungunya virus small molecule inhibitors that targets the viral capping machinery. Antimicrob. Agents Chemother. 64(7), e00649–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mudgal R, Mahajan S, Tomar S. Inhibition of Chikungunya virus by an adenosine analog targeting the SAM-dependent nsP1 methyltransferase. FEBS Lett. 594(4), 678–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mudgal R, Bharadwaj C, Dubey A et al. Selective estrogen receptor modulators limit alphavirus infection by targeting the viral capping enzyme nsP1. Antimicrob. Agents Chemother. 66(3), e0194321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adalja A, Inglesby T. Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev. Anti. Infect. Ther. 17(7), 467–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez-Pérez MJ, Delang L, Ng LFP, Priego EM. Chikungunya virus drug discovery: still a long way to go? Expert Opin. Drug Discov. 14(9), 855–866 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.