Abstract
Rapidly growing tumors often encounter energy stress, such as glutamine deficiency. However, how normal and tumor cells differentially respond to glutamine deficiency remains largely unclear. Here, we demonstrate that glutamine deprivation activates PERK, which phosphorylates FBP1 at S170 and induces nuclear accumulation of FBP1. Nuclear FBP1 inhibits PPARα-mediated β-oxidation gene transcription in normal lung epithelial cells. In contrast, highly expressed OGT in non-small cell lung cancer (NSCLC) cells promotes FBP1 O-GlcNAcylation, which abrogates FBP1 phosphorylation and enhances β-oxidation gene transcription to support cell proliferation under glutamine deficiency. In addition, FBP1 pS170 is negatively correlated with OGT expression in human NSCLC specimens, and low expression of FBP1 pS170 is associated with poor prognosis in NSCLC patients. These findings highlight the differential regulation of FBP1 in normal and NSCLC cells under glutamine deprivation and underscore the potential to target nuclear FBP1 for NSCLC treatment.
Keywords: FBP1, non-small cell lung cancer (NSCLC), glutamine deprivation, phosphorylation, O-GlcNAcylation, prognostic biomarker
Introduction
Lung cancer is the second most common cancer worldwide and has the highest mortality rate among all tumors [1]. Non-small cell lung cancer (NSCLC) is the most typical pathological type with a poor prognosis due to high mortality, metastasis rates, and resistance to chemotherapy. Rapidly growing tumors, such as NSCLC, often encounter energy stresses, including a shortage of glutamine, and respond to them by reprogramming their metabolism [2]. Glutamine is an essential amino acid for tumor cell proliferation and progression. However, the mechanism by which NSCLC cells adapt to glutamine deficiency remains largely unknown.
Cancer cells prefer to generate energy through aerobic glycolysis, also known as the Warburg effect [3,4]. As a reverse process of glycolysis, gluconeogenesis plays a pivotal role in cancer progression [5]. The gluconeogenesis enzyme fructose-1,6-bisphosphatase (FBP), which hydrolyzes fructose 1,6-bisphosphate (F1,6BP) to fructose 6-phosphate (F6P), is regarded as a tumor suppressor due to its inhibition of glycolysis. FBP1 is downregulated in many types of cancer [6]. Downregulated FBP1 has been reported to promote tumor progression in hepatocellular carcinoma (HCC) and clear cell renal cell carcinoma (ccRCC) [7-9]. Recently, we demonstrated that FBP1 can act as a histone phosphatase and is involved in glucose deprivation-induced metabolic reprogramming [10]. In normal hepatocytes, activated RNA-like endoplasmic reticulum kinase (PERK) was found to phosphorylate FBP1 at S170, resulting in conversion of the FBP1 tetramer to a monomer for nuclear translocation under glucose deprivation. Nuclear accumulated FBP1 interacts with PPARα, dephosphorylates histone H3T11 and suppresses PPARα-mediated β-oxidation gene transcription. In contrast, highly expressed O-linked N-acetylglucosamine transferase (OGT) in HCC cells induces FBP1 S124 O-GlcNAcylation. O-GlcNAcylation abrogates FBP1 S170 phosphorylation and promotes β-oxidation to support HCC cell survival. Although the differential regulation of FBP1 in normal hepatocytes and HCC cells was revealed, whether FBP1 can inhibit PPARα-mediated β-oxidation in NSCLC cells under other energy stresses remains unknown.
In this study, we showed that glutamine deprivation induced PERK-dependent FBP1 phosphorylation and nuclear translocation. Nuclear FBP1 interacted with PPARα and inhibited β-oxidation gene expression in normal lung epithelial cells. In contrast, NSCLC cells exhibited much greater OGT expression, leading to FBP1 O-GlcNAcylation, which blocked PERK-mediated FBP1 phosphorylation and its nuclear translocation. This inhibition abrogated the inhibitory effect of FBP1 on PPARα and resulted in strongly enhanced β-oxidation and tumor growth in mice.
Materials and methods
Cell culture
HEK 293T, BEAS-2B and A549 cells were obtained from ATCC. HEK 293T, BEAS-2B and A549 cells were grown in DMEM with 10% fetal bovine serum and 1% penicillin streptomycin. The cells were cultured in 5% CO2 at 37°C in a humidified incubator. Glutamine deprivation was performed by washing the cells twice with phosphate-buffered saline (PBS) and culturing the cells in glutamine-free DMEM (Gibco).
Lentivirus production and infection
Lentivirus production and infection were performed as described previously [11]. The following shRNA sequences were used: FBP1, 5’-CCTTGATGGATCTTCCAACAT-3’; OGT, 5’-TTTAGCACTCTGGCAATTAAA-3’; and PERK, 5’-GGAACGACCTGAAGCTATAAA-3’.
Immunoprecipitation and immunoblotting analysis
The extraction of proteins from cultured cells was performed with a modified buffer and was followed by immunoprecipitation and immunoblotting using corresponding antibodies [12].
Immunofluorescence analysis
Immunofluorescence analysis was performed as previously described [13]. Primary antibodies were diluted 1:100, and fluorescence dye-conjugated secondary antibodies were diluted 1:1000.
Subcellular fractionation
Nuclear fractions were isolated from the cells using a nuclei/cytosol fractionation kit (BioVision).
Cell proliferation assay
A total of 4 × 103 cells were seeded in 96-well plates in DMEM with 10% bovine calf serum with or without glutamine deprivation. Cell viability was detected by CCK-8 assays [14].
Real-time PCR
Total RNA extraction, reverse transcription and quantitative real-time PCR were performed as previously described [10]. TRIzol (Invitrogen), TaqMan reverse transcription reagent kit (Applied Biosystems) and SYBR Premix Ex Taq kit (TaKaRa) were used. The primers used for qRT-PCR were consistent with a previously published paper [10].
IHC analysis and histological evaluation of human lung adenocarcinoma
Human lung adenocarcinoma and adjacent matched nontumor tissue samples were obtained from The First Affiliated Hospital, Zhejiang University School of Medicine. All tissue samples were collected in compliance with the informed consent policy. Table 1 summarizes the clinical features of the patients. IHC analysis was performed as described previously [15]. The quantification and scoring of the staining of tissue sections and its comparison with the corresponding overall survival duration was determined as described previously [16].
Table 1.
Patient characteristics (N=79)
| Clinicopathological Variables | Number of patients (%) |
|---|---|
| Gender | |
| Male | 40 (50.6%) |
| Female | 39 (49.4%) |
| Age, years | |
| ≤ 60 | 41 (51.8%) |
| > 60 | 38 (48.2%) |
| Tumor size, cm | |
| ≤ 2 | 39 (49.4%) |
| > 2 | 40 (50.6%) |
| Lymph node metastasis | |
| Negative | 33 (41.8%) |
| Positive | 46 (58.2%) |
| Tumor grade | |
| Well or moderately differentiated (I & II) | 39 (49.4%) |
| Poorly differentiated (III) | 40 (50.6%) |
| TNM stage | |
| Early (I & II) | 45 (57.0%) |
| Late (III & IV) | 34 (43.0%) |
Abbreviation: TNM stage, tumor-node-metastasis stage.
Animal studies
A total of 4 × 106 A549 cells with reconstituted expression of different FBP1 mutants were subcutaneously injected into 6-week-old male BALB/c athymic nude mice. The injections were performed as described previously [17]. Animals were sacrificed 28 days after injection. The tumors were dissected and weighed. The animal study was approved by the Institutional Review Board at The First Affiliated Hospital, Zhejiang University School of Medicine. No statistical method was used to predetermine the sample size.
Statistical analysis
We used SPSS 21.0 (IBM, Armonk, NY) to perform statistical analyses. The association between marker expression and clinical factors was analyzed by the chi-squared test. The survival analyses were performed by the Kaplan-Meier method (log-rank test). We used univariate Cox regression to calculate the risk factors for progress, and the risk factors were then included in a multivariate Cox regression model to identify the independent prognostic factors. P < 0.05 was considered statistically significant. Statistically significant differences between groups were analyzed by two-tailed t test using GraphPad Prism 6.
Results
Glutamine deprivation induces nuclear accumulation of FBP1 and inhibits PPARα-mediated β-oxidation gene transcription in normal lung epithelial cells but not in NSCLC cells
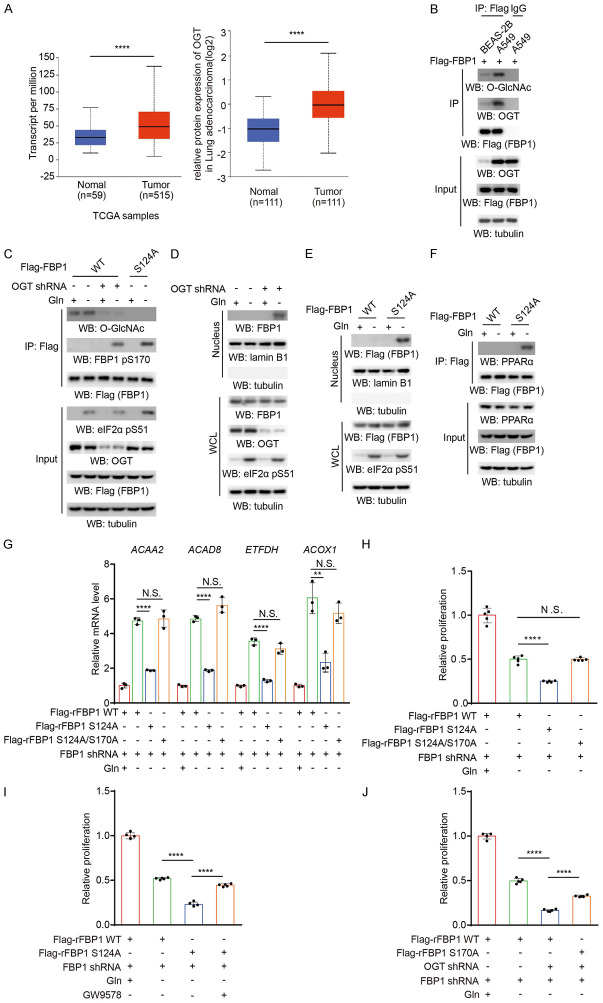
Glutamine deprivation is known to induce ER stress and PERK activation [18]. To determine whether glutamine deprivation differentially regulates FBP1 in normal lung epithelial cells and NSCLC cells, we exposed BEAS-2B normal lung epithelial cells and A549 NSCLC cells to glutamine deprivation. Immunofluorescence analyses revealed that FBP1 was translocated into the nucleus in BEAS-2B cells but not in A549 cells (Figure 1A). Cell fractionation analyses showed that a small portion of cytosolic FBP1 accumulated in the nucleus of BEAS-2B cells but not A549 cells (Figure 1B). In addition, glutamine deprivation induced FBP1 binding with PPARα in BEAS-2B cells but not in A549 cells (Figure 1C). To determine whether FBP1 regulates PPARα activity, we examined PPARα-mediated β-oxidation gene expression. We found that glutamine deprivation substantially increased the expression of acetyl-CoA acyltransferase 2 (ACAA2), acyl-CoA dehydrogenase family member 8 (ACAD8), electron transfer flavoprotein dehydrogenase (ETFDH), and acyl-CoA oxidase 1 (ACOX1) in A549 cells and to a much lesser extent in BEAS-2B cells (Figure 1D). These results suggested that glutamine deprivation promotes FBP1 nuclear accumulation and inhibits PPARα-mediated β-oxidation gene transcription in normal lung epithelial cells but not in NSCLC cells.
Figure 1.
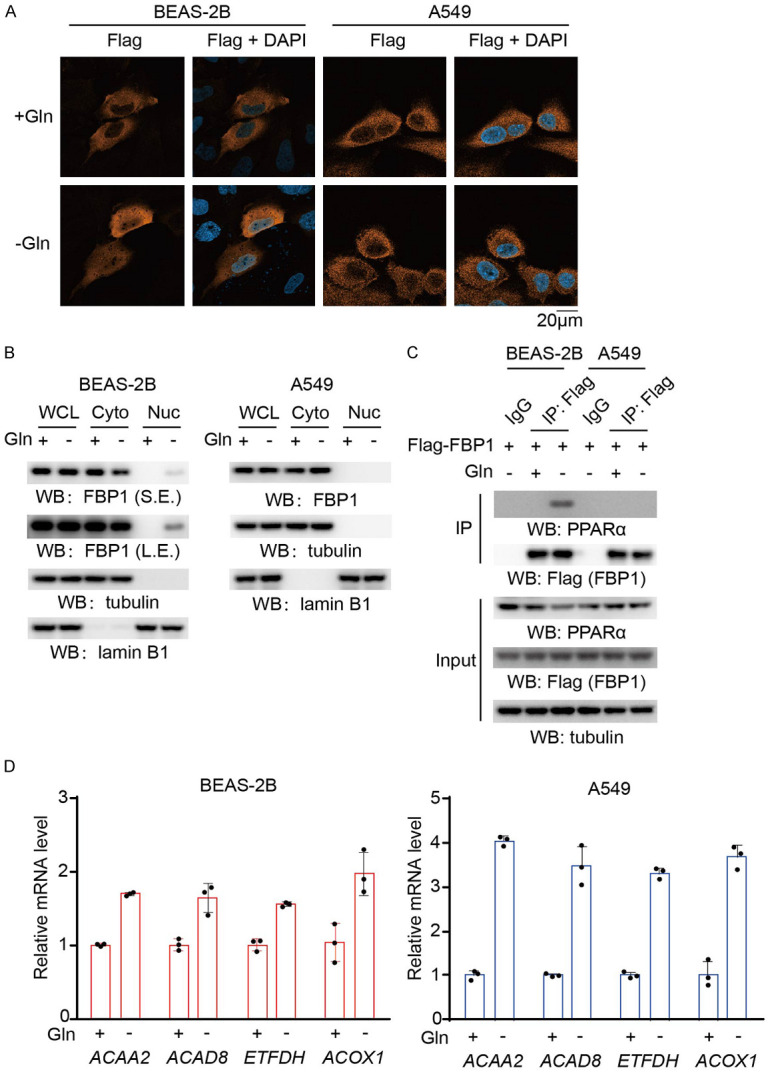
Glutamine deprivation induces nuclear accumulation of FBP1 and inhibits PPARα-mediated β-oxidation gene transcription in normal lung epithelial cells but not in NSCLC cells. A. Flag-FBP1 proteins were expressed in BEAS-2B and A549 cells, which were then treated with or without glutamine deprivation for 6 h. Representative images of immunofluorescence and DAPI staining are shown. B. BEAS-2B and A549 cells were treated with or without glutamine deprivation for 6 h. Cytosolic and nuclear fractions were prepared, and the expression levels of the indicated proteins were detected. WCL, whole cell lysate; Cyto, cytosol; Nuc, nucleus; S.E., short exposure; L.E., long exposure. C. BEAS-2B and A549 cells expressing Flag-FBP1 were treated with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. D. BEAS-2B and A549 cells were treated with or without glutamine deprivation for 12 h. The relative mRNA levels of the indicated genes were determined. The data represent the mean ± SD of 3 independent experiments.
Glutamine deprivation-activated PERK phosphorylates FBP1 and mediates FBP1 nuclear accumulation in normal lung epithelial cells
Glutamine deprivation was reported to activate PERK [18], a kinase that phosphorylates FBP1 at serine 170 [10]. To determine whether PERK is involved in glutamine deprivation-induced FBP1 nuclear accumulation, we performed a coimmunoprecipitation assay and showed that glutamine deprivation induced the interaction of PERK with FBP1 in BEAS-2B cells (Figure 2A). In addition, glutamine deprivation induced FBP1 S170 phosphorylation, and this phosphorylation was abrogated by expression of the FBP1 S170A mutant or treatment with the PERK inhibitor GSK2656157 (Figure 2B), which also inhibited the eukaryotic initiation factor 2 (eIF2) α subunit pS51, a known PERK substrate [19]. In addition, both GSK2656157 (Figure 2C) and PERK depletion via PERK shRNA (Figure 2D) blocked the FBP1 nuclear accumulation induced by glutamine deprivation. Consistently, FBP1 S170A or FBP1 nuclear localization signal mutant (NLS) (R23A/K24A) mutations blocked FBP1 nuclear accumulation (Figure 2E) and the subsequently interaction with PPARα (Figure 2F) under glutamine deprivation. In addition, the expression of PPARα-mediated β-oxidation genes induced by glutamine deprivation was substantially enhanced by reconstituted expression of RNA interference-resistant (r) FBP1 S170A or the rFBP1 NLS mutant in BEAS-2B cells (Figure 2G). Notably, cell proliferation was decreased upon glutamine deprivation, but this decrease was alleviated by the expression of rFBP1 S170A or the rFBP1 NLS mutant in BEAS-2B cells (Figures 2H, S1A). These results indicated that glutamine deprivation-induced and PERK-mediated FBP1 S170 phosphorylation promotes FBP1 nuclear accumulation and subsequently suppresses PPARα-dependent β-oxidation gene transcription and proliferation of BEAS-2B cells.
Figure 2.
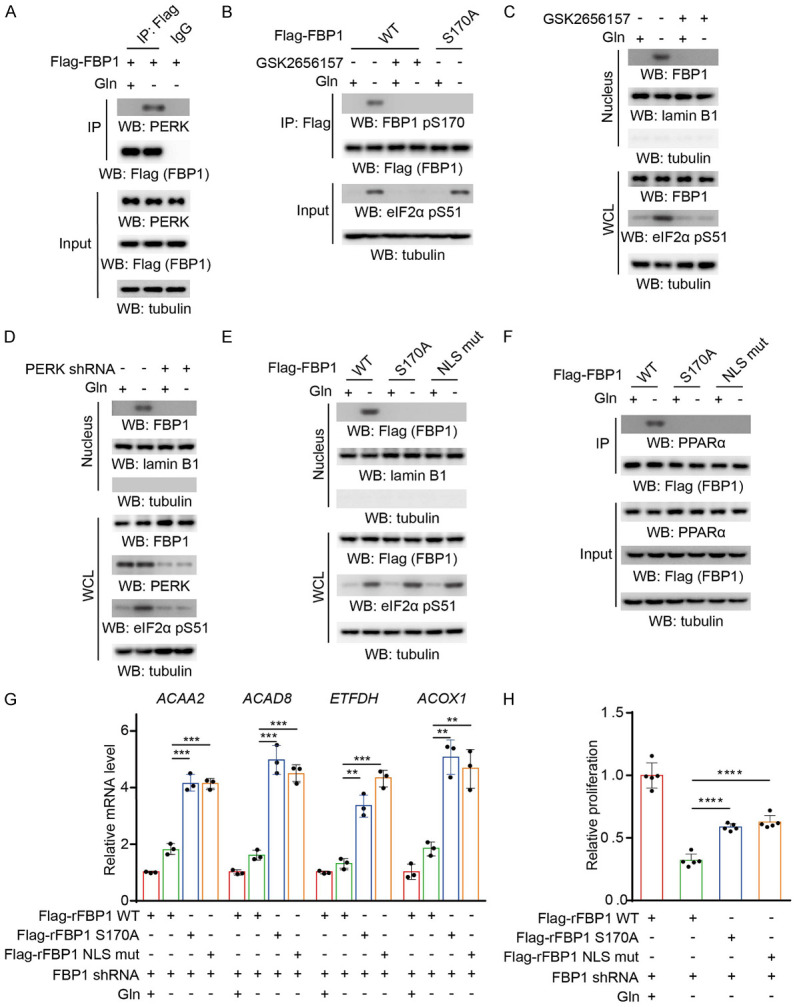
Glutamine deprivation-induced and PERK-mediated FBP1 S170 phosphorylation promotes the nuclear accumulation of FBP1 in normal lung epithelial cells. A. BEAS-2B cells expressing Flag-FBP1 were treated with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. B. The indicated Flag-FBP1 proteins were expressed in BEAS-2B cells, which were pretreated with or without GSK2656157 (10 μM) for 30 min before treatment with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. C. BEAS-2B cells were pretreated with or without GSK2656157 (10 μM) for 30 min before treatment with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. D. BEAS-2B cells with or without PERK shRNA expression were treated with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. E. The indicated Flag-FBP1 proteins were expressed in BEAS-2B cells, which were treated with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. F. The indicated Flag-FBP1 proteins were expressed in BEAS-2B cells treated with or without glutamine deprivation for 6 h. IP analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. G. BEAS-2B cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation for 12 h. The relative mRNA levels of the indicated genes were determined. The data represent the mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. H. BEAS-2B cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test.
O-GlcNAcylation inhibits FBP1 nuclear translocation and promotes β-oxidation gene expression and proliferation of NSCLC cells
In HCC cells, FBP1 S124 is O-GlcNAcylated by OGT, leading to inhibition of its nuclear translocation and promotion of tumor growth [10]. Both O-GlcNAcylation and OGT expression were reported to be increased in lung squamous cell carcinoma tissue compared with normal lung tissue [20,21]. Analyses of The Cancer Genome Atlas (TCGA) data also revealed that OGT is highly expressed in lung adenocarcinoma (LUAD) compared to normal lung tissue (Figure 3A). Consistently, analyses of the Clinical Proteomic Tumor Analysis Consortium (CPTAC) datasets revealed elevated OGT protein level in LUAD (Figure 3A). To determine whether FBP1 is O-GlcNAcylated in NSCLC, we performed an immunoprecipitation assay and showed that OGT expression, FBP1 O-GlcNAcylation, and the interaction between OGT and FBP1 were notably higher in A549 cells than in BEAS-2B cells (Figure 3B). In addition, depletion of OGT or ectopic expression of the FBP1 S124A mutant reduced FBP1 O-GlcNAcylation and increased FBP1 S170 phosphorylation (Figure 3C) with subsequently increased FBP1 nuclear accumulation (Figure 3D) upon glutamine deprivation, suggesting that FBP1 S124 O-GlcNAcylation blocks FBP1 S170 phosphorylation and nuclear translocation in NSCLC cells. Consistently, in response to glutamine deprivation, FBP1 S124A increased its nuclear accumulation and its association with PPARα in A549 cells (Figure 3E, 3F). In addition, PPARα-regulated β-oxidation gene transcription (Figure 3G) and cell proliferation (Figures 3H, S1B) were inhibited by the expression of rFBP1 S124A but not rFBP1 S124A/S170A in A549 cells upon glutamine deprivation. Treatment of A549 cells expressing FBP1 S124A with PPARα agonist GW9578 alleviated FBP1-S124A-induced inhibition of the cell proliferation (Figures 3I, S1C). Of note, depletion of OGT in A549 cells decreased cell proliferation upon glutamine deprivation, and this decrease was partially rescued by the expression of rFBP1 S170A (Figures 3J, S1D). These results indicated that FBP1 S124 O-GlcNAcylation inhibits FBP1 S170 phosphorylation and its subsequent nuclear translocation, thereby promoting PPARα-regulated β-oxidation gene expression and cell proliferation in NSCLC cells.
Figure 3.
O-GlcNAcylation inhibits the nuclear translocation of FBP1 and promotes β-oxidation gene expression in NSCLC cells. A. Left: A TCGA dataset was used to analyze OGT expression levels in normal lung tissues and LUAD tissues. Normal samples: minimum, 9.876; maximum, 76.777; and median, 33.21; LUAD samples: minimum, 5.37; maximum, 138.023; and median, 48.894. The horizontal lines mark the median. ****P < 0.0001. Right: The CPTAC dataset was used to analyze OGT protein levels in normal lung tissues and LUAD tissues (https://ualcan.path.uab.edu/analysis-prot.html). Normal samples: minimum, -2.722; maximum, 0.317; and median, -1.026; LUAD samples: minimum, -2.014; maximum, 2.103; and median, -0.032. The horizontal lines mark the median. ****P < 0.0001. B. BEAS-2B and A549 cells expressing Flag-FBP1 were harvested for immunoprecipitation analysis with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. C. The indicated Flag-FBP1 proteins were expressed in A549 cells with or without OGT shRNA expression, which were treated with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. D. A549 cells with or without OGT shRNA expression were treated with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. E. The indicated Flag-FBP1 proteins were expressed in A549 cells, which were treated with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. F. The indicated Flag-FBP1 proteins were expressed in A549 cells treated with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. G. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation for 12 h. The relative mRNA levels of the indicated genes were determined. The data represent the mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. H. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. I. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation and GW9578 (500 nM) for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. J. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins with or without OGT shRNA expression were treated with or without glutamine deprivation for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test.
FBP1 S170 phosphorylation inhibits tumor growth and is negatively correlated with poor prognosis in NSCLC patients
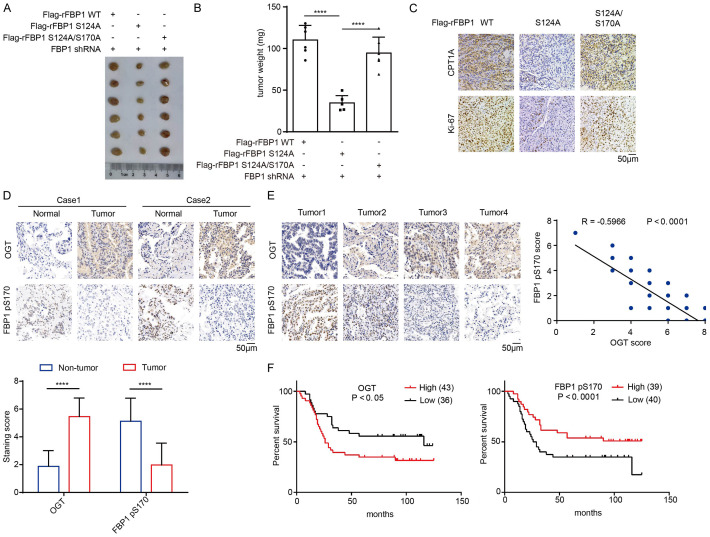
To determine the role of FBP1 in tumor development, we subcutaneously injected A549 cells with reconstituted expression of rFBP1, rFBP1 S124A, or rFBP1 S124A/S170A into athymic nude mice. We showed that expression of rFBP1 S124A reduced tumor growth and tumor weight (Figure 4A, 4B). The inhibitory effects of the S124A mutation were abrogated by the expression of the rFBP1 S124A/S170A mutant with the combined mutation of S124A and S170A (Figure 4A, 4B). Immunohistochemical (IHC) analyses showed that expression of rFBP1 S124A, but not rFBP1 S124A/S170A, decreased CPT1A and Ki-67 expression in tumor tissues (Figure 4C). These results suggested that FBP1 S124 O-GlcNAcylation abrogates the S170 phosphorylation-dependent nuclear accumulation of FBP1 and promotes tumor growth in mice.
Figure 4.
FBP1 S170 phosphorylation inhibits tumor growth and is negatively associated with poor prognosis in NSCLC patients. A. A549 cells (4 × 106) with reconstituted expression of different FBP1 mutants were subcutaneously injected into the flank region of athymic nude mice (n=6 per group). The resulting tumors were resected 28 days after injection. A representative image of tumor sizes is shown. B. The tumors in the mice were weighed. The data represent the mean ± SD of 6 mice. ****P < 0.0001. C. IHC analyses of the tumor tissues were performed. Representative images are shown. D. Representative IHC staining of OGT and FBP1 pS170 in NSCLC and adjacent nontumor tissues is shown. The expression levels of OGT and FBP1 pS170 in NSCLC and adjacent nontumor tissues were quantified and compared between NSCLC and adjacent nontumor tissues. ****P < 0.0001. E. Representative IHC staining of OGT and FBP1 pS170 in NSCLC tissues is shown. The correlations between the expression levels of OGT and FBP1 pS170 were analyzed. F. Analysis of the expression levels of OGT and FBP1 pS170 with the prognosis of NSCLC patients was performed.
Analyses of OGT protein expression in tumor tissues from a previously published dataset [22] showed that OGT protein expression was positively associated with poor prognosis of patients with LUAD (Figure S1E). In addition, we analyzed the TCGA dataset and showed that FBP1 mRNA levels were negatively associated with poor prognosis (Figure S1F). We next performed IHC analyses of FBP1 S170 phosphorylation and O-GlcNAcylation in LUAD (N=79) and adjacent nontumor tissue specimens. Compared with adjacent nontumor tissues, LUAD tissues exhibited reduced FBP1 pS170 levels and increased OGT expression (Figure 4D). In NSCLC specimens, the levels of FBP1 pS170 were negatively correlated with OGT expression (Figure 4E). In addition, low levels of FBP1 S170 phosphorylation or high levels of OGT expression were positively correlated with worse prognosis (Figure 4F), lymph node metastasis, higher tumor grade and advanced TNM stage (Table 2, P < 0.05) in patients with NSCLC.
Table 2.
Correlation analyses of OGT and FBP1 pS170 expressions with patient characteristics
| Clinicopathological Variables | N | OGT Expression | chi-square value | P Value | FBP1 pS170 Expression | chi-square value | P Value | ||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Low (n=36) | High (n=43) | Low (n=40) | High (n=39) | ||||||
| Gender | |||||||||
| Male | 40 | 17 | 23 | 0.308 | 0.579 | 24 | 16 | 2.844 | 0.092 |
| Female | 39 | 19 | 20 | 16 | 23 | ||||
| Age, years | |||||||||
| < 60 | 41 | 17 | 24 | 0.579 | 0.447 | 21 | 20 | 0.012 | 0.914 |
| ≥ 60 | 38 | 19 | 19 | 19 | 19 | ||||
| Tumor size, cm | |||||||||
| ≤ 2 | 39 | 16 | 23 | 0.641 | 0.423 | 17 | 22 | 1.529 | 0.216 |
| > 2 | 40 | 20 | 20 | 23 | 17 | ||||
| Lymph node metastasis | |||||||||
| Negative | 33 | 20 | 13 | 5.166 | 0.023 | 12 | 21 | 4.617 | 0.032 |
| Positive | 46 | 16 | 30 | 28 | 18 | ||||
| Tumor grade | |||||||||
| I & II | 39 | 23 | 16 | 5.58 | 0.018 | 14 | 25 | 6.691 | 0.010 |
| III | 40 | 13 | 27 | 26 | 14 | ||||
| TNM stage | |||||||||
| Early (I & II) | 45 | 26 | 19 | 6.283 | 0.012 | 17 | 28 | 6.913 | 0.009 |
| Late (III & IV) | 34 | 10 | 24 | 23 | 11 | ||||
Analysis by using the univariate Cox regression model revealed that higher tumor grade, TNM stage, OGT expression, and lower FBP1 S170 phosphorylation were significantly associated with inferior overall survival (OS) (Table 3, all P < 0.05). In addition, after adjusting a series of confounding variables, including age, gender, tumor grade and TNM stage, higher OGT expression (HR: 1.285, 95% CI: 1.021-1.616, P=0.0323) and lower FBP1 S170 phosphorylation (HR: 0.751, 95% CI: 0.593-0.952, P=0.0178) remained statistically significant for inferior OS in the multivariate Cox regression model (Table 3). These results suggested that the levels of OGT expression and FBP1 pS170 are negatively correlated in clinical specimens and serve as prognostic factors for NSCLC patients.
Table 3.
Univariate and multivariate Cox regression analyses of risk factors associated with overall survival
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P Value | HR | 95% CI | P Value | |
| OGT | ||||||
| Gender (Male vs. Female) | 1.423 | 0.794-2.553 | 0.2364 | 0.840 | 0.460-1.533 | 0.5697 |
| Age (≥ 59 vs. < 59) | 1.587 | 0.864-2.913 | 0.1362 | 1.764 | 0.941-3.306 | 0.0766 |
| Grade (High vs. Low) | 8.624 | 2.083-35.710 | 0.0030 | 6.273 | 1.452-27.111 | 0.0139 |
| TNM stage (Late vs. Early) | 5.231 | 2.810-9.737 | < 0.0001 | 3.248 | 1.599-6.597 | 0.0011 |
| OGT expression | 1.436 | 1.149-1.794 | 0.0015 | 1.285 | 1.021-1.616 | 0.0323 |
| FBP1 pS170 | ||||||
| Gender (Female vs. Male) | 1.423 | 0.794-2.553 | 0.2364 | 0.808 | 0.439-1.488 | 0.4942 |
| Age (≥ 59 vs. < 59) | 1.587 | 0.864-2.913 | 0.1362 | 1.748 | 0.925-3.302 | 0.0853 |
| Grade (High vs. Low) | 8.624 | 2.083-35.710 | 0.0030 | 6.048 | 1.395-26.221 | 0.0162 |
| TNM stage (Late vs. Early) | 5.231 | 2.810-9.737 | < 0.0001 | 3.400 | 1.734-6.668 | 0.0004 |
| FBP1 pS170 expression | 0.708 | 0.569-0.882 | 0.0021 | 0.751 | 0.593-0.952 | 0.0178 |
Discussion and conclusions
Rapidly growing tumors, including NSCLC, generally have a greater ability than their normal counterparts to adapt to energy stress by altering many pathways in which some metabolic enzymes function in a noncanonical way [23-26]. Importantly, metabolic enzymes and metabolites can directly or indirectly regulate histone modification, promoter accessibility and transcription [27]. Here, we report that glutamine deprivation induces PERK-dependent FBP1 S170 phosphorylation in normal lung epithelial cells. This phosphorylation promotes FBP1 nuclear translocation and subsequent inhibition of PPARα-mediated β-oxidation gene transcription. However, the OGT expression level was much higher in human NSCLC than in adjacent normal tissues. Elevated OGT expression abrogates FBP1 S170 phosphorylation by O-GlcNAcylating FBP1 at S124, thus inhibiting its subsequent regulation of β-oxidation gene expression to ultimately support tumor cell survival and proliferation. Moreover, OGT expression and FBP1 S170 phosphorylation were negatively correlated with each other. Highly expressed OGT was associated with poor prognosis of NSCLC patients, while the levels of FBP1 S170 phosphorylation showed the opposite tendency.
Metabolic enzymes have been reported to possess noncanonical functions in tumor progression [4,27-31]. Pyruvate kinase muscle isozyme M2 (PKM2), phosphoglycerate kinase 1 (PGK1), ketohexokinase (KHK)-A, phosphoenolpyruvate carboxykinase 1 (PCK1), choline kinase (CHK) α, hexokinase 2 (HK2), and creatine kinase B (CKB) can act as protein kinase and phosphorylate various protein substrates. Consequently, these metabolism enzymes directly regulate many instrumental cellular activities, such as gene expression, mitochondrial metabolism, glycolysis, autophagy, mitosis, nucleotide synthesis, lipogenesis, consumption of lipid droplets, tumor cell immune evasion, and ferroptosis [3,13,17,24,26,31-38]. Interestingly, FBP1 was found to be a tumor suppressor that is lost in many cancers and was recently reported to act as a protein phosphatase to play a vital role in inhibiting cancer progression [10,39,40]. Our findings highlight the differential regulation of FBP1 in NSCLC cells and normal cells by manipulating its post-translational modification. Our results underscore the potential to target the moonlighting function of FBP1 as a strategy for lung cancer treatment.
Acknowledgements
This study was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (2020YFA0803300, Z.L.), the National Natural Science Foundation of China (82188102, 82030074, Z.L.), and the Zhejiang Natural Science Foundation Key Project (LD21H160003, Z.L.). Z.L. is the Kuancheng Wang Distinguished Chair.
Disclosure of conflict of interest
Z.L. owns shares in Signalway Biotechnology (Pearland, TX), which supplied rabbit antibodies that recognize FBP1 pS170.
Supporting Information
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Qian X, Lu Z. Local histone acetylation by ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Autophagy. 2017;13:1790–1791. doi: 10.1080/15548627.2017.1349581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Zheng Y, Lu Z. PGK1 is a new member of the protein kinome. Cell Cycle. 2016;15:1803–1804. doi: 10.1080/15384101.2016.1179037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Dong C. Gluconeogenesis in cancer: function and regulation of PEPCK, FBPase, and G6Pase. Trends Cancer. 2019;5:30–45. doi: 10.1016/j.trecan.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Grasmann G, Smolle E, Olschewski H, Leithner K. Gluconeogenesis in cancer cells - repurposing of a starvation-induced metabolic pathway? Biochim Biophys Acta Rev Cancer. 2019;1872:24–36. doi: 10.1016/j.bbcan.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huangyang P, Li F, Lee P, Nissim I, Weljie AM, Mancuso A, Li B, Keith B, Yoon SS, Simon MC. Fructose-1,6-bisphosphatase 2 inhibits sarcoma progression by restraining mitochondrial biogenesis. Cell Metab. 2020;31:174–188. e177. doi: 10.1016/j.cmet.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, Simon MC. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao K, Deng S, Xu L, Pan W, Yang S, Zheng F, Wu X, Hu H, Liu Z, Luo J, Zhang R, Kuang DM, Dong J, Wu Y, Zhang H, Zhou P, Bei JX, Xu Y, Ji Y, Wang P, Ju HQ, Xu RH, Li B. A feedback circuitry between polycomb signaling and fructose-1,6-bisphosphatase enables hepatic and renal tumorigenesis. Cancer Res. 2020;80:675–688. doi: 10.1158/0008-5472.CAN-19-2060. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Li M, Jiang H, Luo S, Shao F, Xia Y, Yang M, Ren X, Liu T, Yan M, Qian X, He H, Guo D, Duan Y, Wu K, Wang L, Ji G, Shen Y, Li L, Zheng P, Dong B, Fang J, Zheng M, Liang T, Li H, Yu R, Xu D, Lu Z. Fructose-1,6-bisphosphatase 1 functions as a protein phosphatase to dephosphorylate histone H3 and suppresses PPARα-regulated gene transcription and tumour growth. Nat Cell Biol. 2022;24:1655–1665. doi: 10.1038/s41556-022-01009-4. [DOI] [PubMed] [Google Scholar]
- 11.Zhu R, Yang Y, Shao F, Wang J, Gao Y, He J, Lu Z. Choline kinase alpha2 promotes lipid droplet lipolysis in non-small-cell lung carcinoma. Front Oncol. 2022;12:848483. doi: 10.3389/fonc.2022.848483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Z, Liang J, Li J, Lu Y, Ariyaratna V, Lu Z, Davies MA, Westwick JK, Mills GB. Physical association of PDK1 with AKT1 is sufficient for pathway activation independent of membrane localization and phosphatidylinositol 3 kinase. PLoS One. 2010;5:e9910. doi: 10.1371/journal.pone.0009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu D, Wang Z, Xia Y, Shao F, Xia W, Wei Y, Li X, Qian X, Lee JH, Du L, Zheng Y, Lv G, Leu JS, Wang H, Xing D, Liang T, Hung MC, Lu Z. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature. 2020;580:530–535. doi: 10.1038/s41586-020-2183-2. [DOI] [PubMed] [Google Scholar]
- 14.Ma Q, Jiang H, Ma L, Zhao G, Xu Q, Guo D, He N, Liu H, Meng Z, Liu J, Zhu L, Lin Q, Wu X, Li M, Luo S, Fang J, Lu Z. The moonlighting function of glycolytic enzyme enolase-1 promotes choline phospholipid metabolism and tumor cell proliferation. Proc Natl Acad Sci U S A. 2023;120:e2209435120. doi: 10.1073/pnas.2209435120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Yang W, Aldape K, He J, Lu Z. Epidermal growth factor (EGF)-enhanced vascular cell adhesion molecule-1 (VCAM-1) expression promotes macrophage and glioblastoma cell interaction and tumor cell invasion. J Biol Chem. 2013;288:31488–31495. doi: 10.1074/jbc.M113.499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Shao F, Shi S, Feng X, Wang W, Wang Y, Guo W, Wang J, Gao S, Gao Y, Lu Z, He J. Prognostic impact of metabolism reprogramming markers acetyl-CoA synthetase 2 phosphorylation and ketohexokinase-A expression in non-small-cell lung carcinoma. Front Oncol. 2019;9:1123. doi: 10.3389/fonc.2019.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian X, Li X, Shi Z, Xia Y, Cai Q, Xu D, Tan L, Du L, Zheng Y, Zhao D, Zhang C, Lorenzi PL, You Y, Jiang BH, Jiang T, Li H, Lu Z. PTEN suppresses glycolysis by dephosphorylating and inhibiting autophosphorylated PGK1. Mol Cell. 2019;76:516–527. e517. doi: 10.1016/j.molcel.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Shanware NP, Bray K, Eng CH, Wang F, Follettie M, Myers J, Fantin VR, Abraham RT. Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat Commun. 2014;5:4900. doi: 10.1038/ncomms5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Li M, Jiang H, Luo S, Shao F, Xia Y, Yang M, Ren X, Liu T, Yan M, Qian X, He H, Guo D, Duan Y, Wu K, Wang L, Ji G, Shen Y, Li L, Zheng P, Dong B, Fang J, Zheng M, Liang T, Li H, Yu R, Xu D, Lu Z. Fructose-1,6-bisphosphatase 1 functions as a protein phosphatase to dephosphorylate histone H3 and suppresses PPARalpha-regulated gene transcription and tumour growth. Nat Cell Biol. 2022;24:1655–1665. doi: 10.1038/s41556-022-01009-4. [DOI] [PubMed] [Google Scholar]
- 20.Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, Cong Q, Yu W. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011;1812:514–519. doi: 10.1016/j.bbadis.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu JY, Zhang C, Wang X, Zhai L, Ma Y, Mao Y, Qian K, Sun C, Liu Z, Jiang S, Wang M, Feng L, Zhao L, Liu P, Wang B, Zhao X, Xie H, Yang X, Zhao L, Chang Y, Jia J, Wang X, Zhang Y, Wang Y, Yang Y, Wu Z, Yang L, Liu B, Zhao T, Ren S, Sun A, Zhao Y, Ying W, Wang F, Wang G, Zhang Y, Cheng S, Qin J, Qian X, Wang Y, Li J, He F, Xiao T, Tan M. Integrative proteomic characterization of human lung adenocarcinoma. Cell. 2020;182:245–261. e217. doi: 10.1016/j.cell.2020.05.043. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee JH, Li W, Lyu J, Rao G, Zhang X, Qian CN, Rozen SG, Jiang T, Lu Z. Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol Cell. 2017;66:684–697. e689. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian X, Li X, Cai Q, Zhang C, Yu Q, Jiang Y, Lee JH, Hawke D, Wang Y, Xia Y, Zheng Y, Jiang BH, Liu DX, Jiang T, Lu Z. Phosphoglycerate kinase 1 phosphorylates beclin1 to induce autophagy. Mol Cell. 2017;65:917–931. e916. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, Li X, Shao F, Lv G, Lv H, Lee JH, Qian X, Wang Z, Xia Y, Du L, Zheng Y, Wang H, Lyu J, Lu Z. The protein kinase activity of fructokinase A specifies the antioxidant responses of tumor cells by phosphorylating p62. Sci Adv. 2019;5:eaav4570. doi: 10.1126/sciadv.aav4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, Lee JH, Li J, Yu R, Tan L, Xia Y, Zheng Y, Bian XL, Lorenzi PL, Chen Q, Lu Z. Choline kinase alpha 2 acts as a protein kinase to promote lipolysis of lipid droplets. Mol Cell. 2021;81:2722–2735. e2729. doi: 10.1016/j.molcel.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol. 2018;19:563–578. doi: 10.1038/s41580-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci. 2018;43:301–310. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian X, Jiang H, Meng Y, Li YP, Fang J, Lu Z. Regulation of gene expression by glycolytic and gluconeogenic enzymes. Trends Cell Biol. 2022;32:786–799. doi: 10.1016/j.tcb.2022.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metab. 2021;33:33–50. doi: 10.1016/j.cmet.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Qian X, Jiang H, Xia Y, Zheng Y, Li J, Huang BJ, Fang J, Qian CN, Jiang T, Zeng YX, Lu Z. Nuclear PGK1 alleviates ADP-dependent inhibition of CDC7 to promote DNA replication. Mol Cell. 2018;72:650–660. e658. doi: 10.1016/j.molcel.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, Wang L, Lu Z. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell. 2016;61:705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Qian X, Peng LX, Jiang Y, Hawke DH, Zheng Y, Xia Y, Lee JH, Cote G, Wang H, Wang L, Qian CN, Lu Z. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol. 2016;18:561–571. doi: 10.1038/ncb3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, Wei C, Guo F, Chen Y, Lu Z. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo D, Tong Y, Jiang X, Meng Y, Jiang H, Du L, Wu Q, Li S, Luo S, Li M, Xiao L, He H, He X, Yu Q, Fang J, Lu Z. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab. 2022;34:1312–1324. e1316. doi: 10.1016/j.cmet.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Wu K, Yan M, Liu T, Wang Z, Duan Y, Xia Y, Ji G, Shen Y, Wang L, Li L, Zheng P, Dong B, Wu Q, Xiao L, Yang X, Shen H, Wen T, Zhang J, Yi J, Deng Y, Qian X, Ma L, Fang J, Zhou Q, Lu Z, Xu D. Creatine kinase B suppresses ferroptosis by phosphorylating GPX4 through a moonlighting function. Nat Cell Biol. 2023;25:714–725. doi: 10.1038/s41556-023-01133-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhu W, Chu H, Zhang Y, Luo T, Yu H, Zhu H, Liu Y, Gao H, Zhao Y, Li Q, Wang X, Li G, Yang W. Fructose-1,6-bisphosphatase 1 dephosphorylates IκBα and suppresses colorectal tumorigenesis. Cell Res. 2023;33:245–257. doi: 10.1038/s41422-022-00773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, Tao J, Lin L, Qiu W, Lu Z. Repurposing FBP1: dephosphorylating IkappaBalpha to suppress NFkappaB. Cell Res. 2023;33:419–420. doi: 10.1038/s41422-023-00785-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.