Abstract
Triple-negative breast cancer (TNBC) is an aggressive form of breast cancer, and the majority of TNBC lacks targeted therapies. Previous studies have shown that TNBC cells are highly sensitive to TNF-related apoptosis-inducing ligand (TRAIL), making it a potentially viable treatment option for TNBC. However, the development of TRAIL resistance limits its potential for clinical use, and the underlying mechanisms are not fully understood. To better understand the mechanism of resistance to TRAIL, we performed RNA sequencing to identify the candidates that are responsible for resistance to TRAIL in two previously established TRAIL-resistant MDA231 and SUM159 cells. This approach led us to identify differentially expressed genes (DEGs) and pathways in TRAIL-resistant MDA231 and SUM159 cells compared to their TRAIL-sensitive counterparts. We showed that several DEGs and pathways were associated with inflammation in TRAIL-resistant cells, including IL-1α and IL6. By downregulating IL-1α and IL6 expression, we showed that TRAIL sensitivity can be significantly restored in TRAIL-resistant cells. Therefore, this study identifies a mechanism by which the inflammation pathway promotes TRAIL resistance, which could be targeted for enhancing TRAIL-based therapies in TNBC cells.
Keywords: TRAIL, resistance, inflammation, TNBC
Introduction
TRAIL (also known as the Apo2 ligand) is a tumor necrosis factor (TNF) superfamily cytokine [1,2], which plays an important role in apoptosis and tumor surveillance [3,4]. TRAIL selectively induces apoptosis in tumor cells with little to no toxicity toward normal cells, making it a promising agent for cancer therapy [5-7]. There are two membrane-bound death receptors for TRAIL, including TRAIL-R1 (DR4) and TRAIL-R2 (DR5) [8-14] and three decoy receptors, including DcR1, DcR2, and osteoprotegerin (OPG) [9,11,15-18]. The binding of TRAIL to DR4 or DR5 promotes death receptor trimerization, which brings together the Fas-associated death domain (FADD) adaptor protein and pro-caspase 8 to form the death-inducing signaling complex (DISC) [19,20]. The latter activates caspase 8, followed by caspases 3, 6, and 7 activation, to induce cell death [19,20]. In some cells, caspase 8 cleaves the Bcl-2 family member Bid, resulting in the generation of truncated Bid (tBID) protein [21,22]. The tBID protein then translocates to the mitochondria, where it activates the mitochondrial apoptosis pathway and thus enhances TRAIL-induced apoptosis [23]. However, binding of TRAIL to decoy receptors inhibits TRAIL-induced apoptosis because this binding competes for TRAIL binding to death receptors DR4 and DR5.
TRAIL-based treatment has been shown in phase I/II clinical trials to be safe and effective in some cancer patients [24,25]. However, the intrinsic and acquired resistance to TRAIL limits its utility in clinical settings. Previous studies have shown that resistance mechanisms to TRAIL can develop at any stage of the TRAIL apoptotic pathway [26]. For example, aberrant expression of the DR4/DR5 death receptors and the decoy receptors can undoubtedly impact the binding of TRAIL to death receptors DR4/DR5 on the surface of TRAIL-resistant tumors, resulting in decreased apoptosis [26]. Furthermore, c-FLIP overexpression can prevent TRAIL-induced apoptosis by inhibiting caspase 8 activation. In addition, anti-apoptotic Bcl-2 family members and Inhibitor of Apoptosis family proteins (IAPs) have been shown to be upregulated in TRAIL-resistant tumors [27-29]. Furthermore, a recent study found that the expression of the immune checkpoint protein programmed death ligand-1 (PD-L1) can confer resistance to TRAIL in tumor cells [30]. In addition, activation of oncogenic signaling pathways such as ERK1/2, AKT, and NF-kB has been shown to confer resistance to the TRAIL signaling pathway [31-33]. Therefore, further research is needed to understand better the mechanisms underlying TRAIL resistance to improve its use in clinical settings.
In this study, we performed RNA sequencing and gene expression analysis to identify the genes and associated pathways that confer resistance to TRAIL in TNBC cells. Among these pathways, we showed that the inflammatory pathway plays a critical role in TRAIL-resistant MDA231 and SUM159 TNBC cells. By down-regulating cytokines such as IL-1α and IL-6, we showed that TRAIL sensitivity is greatly enhanced in TRAIL-resistant cells. Therefore, this study identifies the contribution of the cytokine and inflammatory pathways to TRAIL resistance and suggests that targeting the cytokine and inflammatory pathways could improve TRAIL-based therapies in TNBC.
Materials and methods
Cell lines and culture
MDA-MB-231 (MDA231) and SUM159 TNBC cell lines were obtained from the American Type Culture Collection (ATCC). Dulbecco’s modified Eagle’s medium (DMEM) was used to culture MDA231 and SUM159 cells. All cells received 10% fetal bovine serum and 1% penicillin-streptomycin (PS) and kept at 37°C in a humidified environment of 5% CO2.
Reagents and antibodies
PVDF membranes (catalogue no. IPVH00010) and actin antibody (catalogue no. A1978) were purchased from Sigma-Aldrich. Recombinant Human sTRAIL/Apo2L (catalog no. 310-04) was purchased from PeproTech. Trypsin-EDTA (catalogue no. 25300-054), DMEM (catalogue no. 11995-065), bovine serum albumin (catalogue no. BP1605-100), FBS (catalogue no. 10437028), goat anti-mouse alexa fluor 680 IgG (catalog no. A21058), LipofectamineTM RNAiMAX transfection reagent (catalog no. 13778150), Opti-MEM (catalog no. 11058021), penicillin-streptomycin (catalog no. 15140122), Power SYBR Green PCR mix (catalog no. 4367659), SuperScript™ III First-Strand Synthesis System (catalog no. 18080051), goat anti-rabbit alexa fluor 680 IgG (catalogue no. A21109), SuperSignalTM West PICO PLUS chemiluminescent substrate (catalog no. 34580), and TRIzolTM reagent (catalog no. 15596026) were purchased from ThermoFisher Scientific. Anti-rabbit IgG HRP-linked (catalog no. 7074) and anti-mouse IgG HRP-linked (catalog no. 7076) were purchased from Cell Signaling. Protein assay dye (catalog no. 500-0006) was purchased from Bio-Rad. Human siRNA for IL-6 (sc-39627), IL-1α (sc-39613), and control (sc-37007) was purchased from Santa Cruz Biotechnology, INC.
TRAIL-resistant cell lines
TRAIL-resistant MDA231 (MDA231-R) and SUM159 (SUM159-R) cells were generated over a 6-month period by gradually exposing parental MDA231 and SUM159 cells to increasing concentrations of TRAIL (5 to 120 ng/ml) as previously described [34]. TRAIL-resistant cells were maintained in TRAIL (120 ng/ml).
MTT assay
The 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay was performed as described previously [30]. In brief, a total of 2000 cells were seeded in 96-well plates. Following overnight attachment, cells were incubated with different concentrations of drugs in 200 µl media for 48 or 72 hours. Each well received 20 µl of MTT solution (5 mg/ml) and was incubated at 37°C for 2 hours. MTT-containing media were removed, and formazan crystals were dissolved in 100 µl DMSO. A SynergyTM-2 microplate reader (BioTek Instruments Inc., Winooski, VT, USA) was used to measure the optical density at 570 nm. The IC50 values were calculated using linear regression analysis with Microsoft Excel. Experiments were performed in triplicates.
Colony formation assay
A total of 250 cells were seeded in 24 or 6-well plates. Following overnight attachment, cells were incubated with different concentrations of drug in 2 ml for 48 or 72 hours. Cells were then maintained in a drug-free medium for an additional 14 days. The media was changed every two days. The resulting cells were washed with PBS and fixed for 10 min in cold 100% methanol. Cells were stained with 0.25% crystal violet for 30 min and washed in water. After air drying, colonies with more than 50 cells were counted using Oxford Optronix GELCOUNTTM.
Western blot analysis
Western blot analysis was performed, as previously described [30]. Cells were lysed with 1× NP40 lysis buffer that contained protease and phosphatase inhibitors. Total protein was collected after a 20 min centrifugation at 12,000 rpm at 4°C. The protein concentration was determined using a Bio-Rad protein assay. A total of 50-200 μg of protein was electrophoresed in a 10% sodium dodecyl sulphate-polyacrylamide gel (SDS-PAGE). Proteins were then transferred to 0.45 µM polyvinylidene difluoride (PVDF) membranes and blocked with 2% casein. The membranes were incubated with specific primary antibodies overnight at 4°C before being incubated for 1 hour with Alexa Fluor conjugated secondary antibodies. The Odyssey infrared imaging system (LICOR) at 700 nm (Li-Cor Biosciences, Lincoln, NE, USA) or enhanced chemiluminescence (ThermoFisher Scientific, catalogue no. 34580) was used to detect protein signals.
RNA extraction, sequencing, and bioinformatics
Total RNA was extracted from MDA231-P/R and SUM159-P/R cells preserved in TRIzol (Life Technologies) using the Direct-zol RNA kit (Zymo). Sequencing libraries were generated from 200 ng of RNA using the NEBNext Ultra II Directional RNA Library Prep Kit before sequencing on the NovaSeq (2 × 50 bp). The reads were aligned to the human genome (Build hg38) [35] and tabulated for each gene region [36]. Differential analyses were performed between conditions, utilizing three replicates of each cell line [37]. Significantly altered genes (log fold change ≥ 2; FDR ≤ 0.05) were used to identify affected pathways [38].
DEG, gene ontology (GO), and kyoto encyclopedia of genes and genomes (KEGG) analysis
The RNA expression levels were log2-transformed, and an unpaired t-test, followed by FDR correction, was used to compare between groups: MDA231-P/R and SUM159-P/R cells. Differentially expressed genes (DEGs) were then identified using a volcano plot with a false discovery rate (FDR) ≤ 0.05 and a fold-change (FC) of ≥ 2. The iPathwayGuide (Advaita Bio, Inc.) was used for gene ontology (GO) analysis, KEGG pathway analysis, and meta-analysis.
Real-time PCR
cDNA was synthesized using Invitrogen SuperScript III Reverse Transcriptase kit (ThermoFisher Scientific) from 1 µg-5 μg total RNA and random primers. The following primer sequences were used for semiquantitative and real-time PCR: GAPDH forward (5’-ATC AAG AAG GTG GTG AAG CAG-3’), GAPDH reverse (5’-TGT CGC TGT TGA AGT CAG AGG-3’), IL-6 forward (5’-AGA CAG CCA CTC ACC TCT TCA G-3’), IL-6 reverse (5’-TTC TGC CAG TGC CTC TTT GCT G-3’), CCL5 forward (5’-CCT GCT GCT TTG CCT ACA TTG C-3’), CCL5 reverse (5’-ACA CAC TTG GCG GTT CTT TCG G-3’), CD24 forward (5’-ATG GGC AGA GCA ATG GTG GCC A-3’), CD24 reverse (5’-AGA GTG AGA CCA CGA AGA GAC T-3’), CSF2 forward (5’-CTC AGA AAT GTT TGA CCT CCA G-3’), CSF2 reverse (5’-TGA CAA GCA GAA AGT CCT TCA G-3’), IL-1α forward (5’-AGT AGC AAC CAA CGG GAA GG-3’), and IL-1α reverse (5’-TGG TTG GTC TTC ATC TTG GG-3’). Primer sequences were obtained from Invitrogen. The Power SYBR Green PCR mix (ThermoFisher Scientific) was used for real-time RT-PCR on the Step One Plus Real-Time PCR System. The following were the thermal cycling conditions: 95°C for 10 min, then forty cycles of 95°C for 15 sec and 60°C for 1 min. The ΔΔCt method was used to calculate relative RNA levels, with GAPDH serving as an internal control. The results displayed are representative of at least three independent experiments.
siRNA transfection for IL-6 or IL-1α knockdown
siRNA knockdown was performed, as described previously [30]. Specifically, cells were seeded in 60 mm plates overnight. The following day, cells were transfected with IL-6 and IL-1α siRNA or control siRNA (Santa Cruz Biotechnology, Inc) using LipofectamineTM RNAiMAX transfection reagent (ThermoFisher Scientific) as directed by the manufacturer’s instruction. IL-6 (sc-39627A) siRNA is a pool of 3 different siRNA duplexes: sc-39627A (sense: 5’-CAGAACGAAUUGACAAACAtt-3’, antisense: 5’-UGUUUGUCAAUUCGUUCUGtt-3’), sc-39627B (sense: 5’-GCACAGAACUUAUGUUGUUtt-3’, antisense: 5’-AACAACAUAAGUUCUGUGCtt-3’), sc-39627C (sense: 5’-GUGUAGGCUUACCUCAAAUtt-3’, antisense: 5’-AUUUGAGGUAAGCCUACACtt-3’). IL-1α (sc-39613) is a pool of 3 different siRNA duplexes: sc-39613A (sense: 5’-CAUCCAAGCUUACCUUCAAtt-3’, antisense: 5’-UUGAAGGUAAGCUUGGAUGtt-3’), sc-39613B (sense: 5’-CCACAGACCUAGGAUUUCAtt-3’, antisense: 5’-UGAAAUCCUAGGUCUGUGGtt-3’), sc-39613C (sense: 5’-GGAGACCUGUAAUCAUAUAtt-3’, antisense: 5’-UAUAUGAUUACAGGUCUCCtt-3’). After 72 hours, the cells were treated or left untreated at the indicated time points. After treatment, cells were harvested to confirm IL-1α or IL-6 expression by Western blot analysis. Cell viability was then determined using MTT or colony formation assays in both treated and untreated cells.
Statistical analysis
All data were processed and analyzed using Microsoft Excel. Densitometry analysis was performed using ImageJ. To compare between two groups, the Student’s unpaired t-test was used. No correction was made for multiplicity.
Results
Characterization of gene expressions that alter as TRAIL-sensitive TNBC cells acquire TRAIL resistance
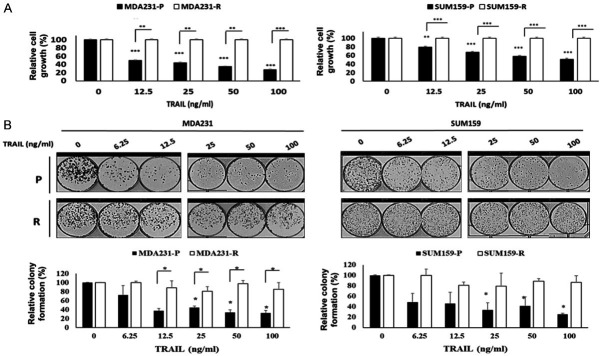
We previously established two TRAIL-resistant TNBC cell lines, MDA231-R and SUM159-R, by exposing TRAIL-sensitive MDA231 (MDA231-P) and SUM159 (SUM159-P) cells to increasing concentrations of TRAIL for more than six months, as previously described [34]. We confirmed that MDA231-R and SUM159-R cells had IC50s greater than 100 ng/ml compared to MDA231-P (IC50 = 39.6 ng/ml) and SUM159-P cells (IC50 = 87.5 ng/ml) cells (Figure 1A). In addition, colony formation assays also verified that MDA231-R and SUM159-R cells are resistant to TRAIL compared to their corresponding TRAIL sensitive counterparts, MDA231-P and SUM159-P cells (Figure 1B). Therefore, these cells are good models for studying resistance to TRAIL.
Figure 1.
TRAIL sensitivity of TNBC cells. A. The MTT assay was performed to determine relative cell growth of MDA231 (left) and SUM159 (right) cells treated with the indicated doses of TRAIL for 48 h. B. Colony formation assays and quantification of MDA231 (left) and SUM159 (right) cells treated with the indicated doses of TRAIL for 48 h. Top: Colony formation assay. Bottom: Densitometry bar graph depicting relative colony formation. P, parental; R, resistant to TRAIL. In the bar graphs, data are represented as mean ± SD, where the error bars denote the standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test.
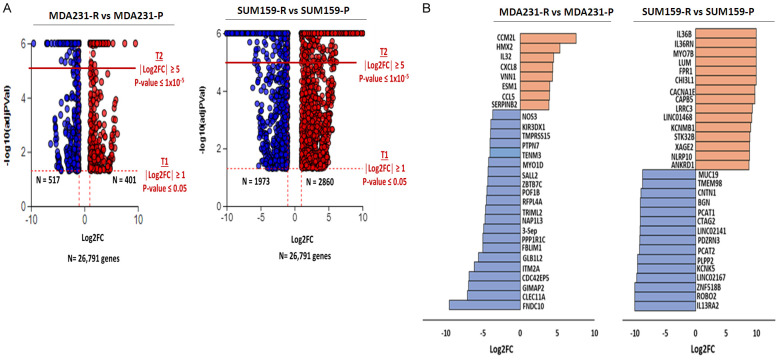
To gain insight into the genes and pathways that confer resistance to TRAIL in these TNBC, we performed RNA sequencing to identify differentially expressed genes (DEGs) between MDA231 and SUM159 cells sensitive to and resistant to TRAIL. A total of 26,791 genes in each cell line were included in the RNA-seq analysis (Figure 2A). DEGs were defined by an absolute value log2FC ≥ 1 with FDR ≤ 0.05. Based on this criterion, MDA231-R cells had 918 DEGs with 401 upregulated and 517 downregulated, while SUM159-R cells had 4,833 DEGs with 2,860 upregulated and 1,973 downregulated. Next, we examined the data to identify the 30 most significant DEGs in MDA231-R and SUM159-R cells. Accordingly, we narrowed down the top 30 DEGs using additional criteria (log2FC ≥ 1 and FDR ≤ 1 × 10-5) and identified that in MDA231-R cells, 8 DEGs were upregulated and 22 were downregulated, whereas 15 in SUM159-R cells were both up- and downregulated (Figure 2B). In particular, 4 of the upregulated DEGs (IL32, CXCL8, VNN1, CCL5) in MDA231-R cells were associated with an immune response, as were 6 (IL36B, IL36RN, CHI3L1, FPR1, NLRP10, ANKRD1) in SUM159-R cells. These results suggest that the immune response may play a critical role in TRAIL resistance in TNBC cells.
Figure 2.
RNA sequence analysis of TRAIL-resistant TNBC cells. A. Volcano plots showing DEGs in each cell line pair. Left: MDA231 cells. Right: SUM159 cells. B. Clustered bar graph of the top 30 DEGs in each cell line pair. Left: MDA231 cells. Right: SUM159 cells. Two thresholds were used. T1: FDR ≤ 0.05 and |Log2FC| ≥ 1. T2: FDR ≤ 1 × 10-5 and |Log2FC| ≥ 1. Red, upregulated; Blue, downregulated; DEG, differentially expressed gene; FC, fold change; adjPVal, FDR; P, parental; R, resistant to TRAIL; T, threshold.
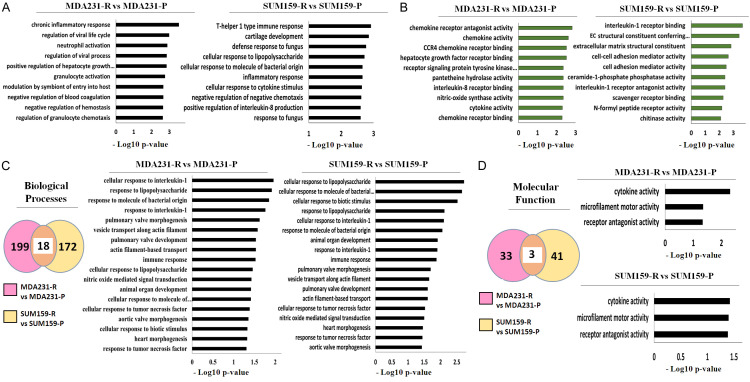
DEGs in TRAIL resistance are enriched in several immune-dependent biological processes
Because our findings suggest that the immune pathways are altered in TRAIL resistance, we wondered what functional annotations these DEGs in MDA231 and SUM159 cells involved and whether they were also associated with immune response. To this end, we used the iPathwayGuide software to perform a Gene Ontology (GO) analysis on the top 30 DEGs of each TNBC cell line. We divided them into two categories: biological processes and molecular functions. Cellular components were omitted since they were primarily linked with cellular structure. As shown in Figure 3A and 3B, the top 30 DEGs in MDA231-R cells were enriched in 217 biological processes and 36 molecular functions, whereas SUM159-R cells’ top 30 DEGs were enriched in 190 biological processes and 44 molecular functions. We then used the criteria (-log10 p-value ≥ 2.0, i.e., p-value ≤ 0.01) to narrow the data further down so that each category contained the top 10 statistically significant biological processes or molecular functions. The analysis revealed that many biological processes and molecular functions in MDA231-R and SUM159-R cells were associated with immunity. This led us to investigate whether MDA231-R and SUM159-R cells shared any biological processes or molecular functions that could be related to immune response. Consequently, we performed a meta-analysis comparing the biological processes and molecular functions associated with the top 30 DEGs of each TNBC cell line. We showed that MDA231-R and SUM159-R cells share 18 biological processes and 3 molecular functions. Figure 3C shows that MDA231-R and SUM159-R cells shared 7 biological processes associated with the immune response, including cellular responses and interleukin-1 responses (GO:0071347; GO:0070555), bacterial molecules (GO:0002237; GO:0071219), tumor necrosis factor (GO:0071356; GO:0034612), and immune response (GO:0006955). We also showed that MDA231-R and SUM159-R cells shared a molecular function that was related to the immune response (cytokine activity [GO:0005125]) (Figure 3D). These data suggest that DEGs in TRAIL-resistant TNBC cells are enriched in several immune-dependent biological processes and molecular functions.
Figure 3.
Gene ontology analysis shows that DEGs have immune-related functional annotations and that TRAIL-resistant TNBC cells have similar immune-related gene ontologies. (A, B) Gene ontology analysis of the top 30 DEGs in each pair of cell lines for (A) biological processes and (B) molecular functions. Left: MDA231 cells. Right: SUM159 cells. (C, D) Meta-analysis of all DEGs in each cell line pair for (C) biological processes and (D) molecular functions. Left: Venn diagram of shared gene ontologies (C, D). Middle: Clustered bar graph for MDA231 cells (C). Right: Clustered bar graph for SUM159 cells (C). Upper Right: Clustered bar graph for MDA231 cells (D). Lower Right: Clustered bar graph for SUM159 cells (D). DEG, differentially expressed gene; P, Parental; R, resistant to TRAIL.
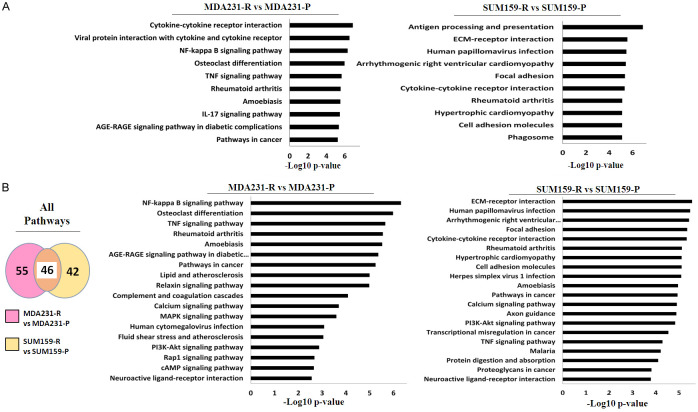
Inflammatory pathways in TRAIL-resistant TNBC cells are impacted
Because several biological processes and molecular functions are associated with the immune response in MDA231-R and SUM159-R cells, we then asked which pathways were affected in these TNBC cells. To address this, we used the top 30 DEGs of MDA231 and SUM159 cells in a Kyoto and Encyclopedia of Genes and Genomes (KEGG) pathway analysis to determine which pathways had been significantly impacted. The pathways were then classified into the 10 most statistically significant pathways for each TNBC cell line. As shown in Figure 4A, 6 of the top 10 most significantly impacted pathways in the KEGG analysis were associated with immune response in MDA231-R cells, while 5 were for SUM159-R cells. This further led us to ask if any pathways were impacted and shared by MDA231-R and SUM159-R cells. Consequently, we performed a meta-analysis of all genes in MDA231-R and SUM159-R cells. We identified 46 pathways that were affected and shared by both TNBC cells (Figure 4B). Among the 20 most statistically significant pathways, 2 had an association with immune response, such as cytokine-cytokine interaction and tumor necrosis factor (TNF) signaling pathways shared by both MDA231 and SUM159 cells. These pathways are functionally associated with inflammation as part of the immune response. Thus, these results indicate that inflammatory pathways are impacted in TRAIL-resistant TNBC cells.
Figure 4.
KEGG analysis shows that DEG alter immune-related canonical pathways and that TRAIL-resistant TNBC cells share altered inflammatory pathways. A. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of altered canonical pathways by the top 30 DEGs in each cell line pair. Left: Clustered bar graph MDA231 cells. Right: Clustered bar graph SUM159 cells. B. Meta-analysis of shared altered canonical pathways using KEGG of all DEGs in each pair of cell lines. Left: Venn diagram. Middle: Clustered bar graph MDA231 cells. Right: Clustered bar graph SUM159 cells. DEG, differentially expressed gene; FC, fold change; P, Parental; R, resistant to TRAIL.
Validation of some gene candidates in TRAIL resistance
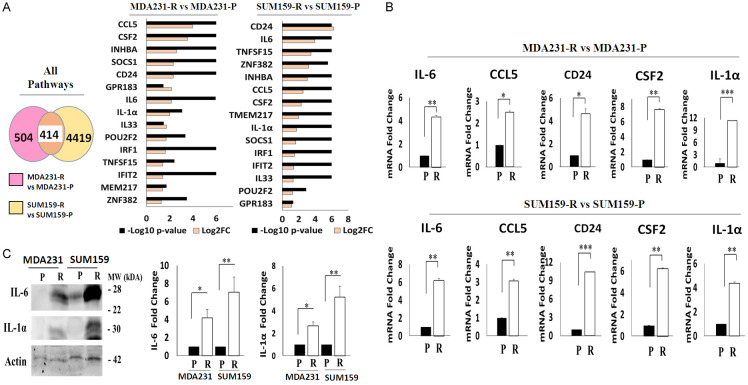
Because immune pathways are up-regulated in TRAIL-resistant TNBC cells, we wondered if there were DEGs shared by MDA231-R and SUM159-R cells that were associated with immune response or inflammation. To this end, we performed a meta-analysis of all DEGs that were upregulated in these TNBC cells, using the absolute value log2FC ≥ 1 and FDR ≤ 0.05 criterion. 15 of the 414 identified shared DEGS were related to immunity (Figure 5A). Those DEGs include CCL5, CD24, CSF2, INHBA, SOCS1, GPR183, IL33, IL-6, IL-1α, POUF2, TNFSF15, IFIT2, IRF1, TMEM217, and ZNF382. We then used RT-PCR to confirm the expression of some of these DEGs. Due to their association with inflammation, we validated CCL5, CD24, IL-6, CSF2, and IL-1α expression. Furthermore, CCL5, IL-6, IL-1α, and CSF2 were chosen for their roles in the signaling pathways of TNF and the cytokine-cytokine receptor interaction. IL-1α was selected because of its role in the latter. Furthermore, CD24 was chosen for validation due to its ability to evade immune surveillance. As shown in Figure 5B, RT-PCR showed that all gene candidates were upregulated in MDA231-R and SUM159-R cells compared to their corresponding TRAIL-sensitive counterparts. Furthermore, we tested the basal levels of these cytokines in MDA231 and SUM159 cells by Western blot analysis. Figure 5C shows that MDA231-R and SUM159-R cells had higher basal levels of IL-6 and IL-1α than TRAIL-sensitive cells. These results indicate that TRAIL resistance in TNBC is mediated in part by inflammation.
Figure 5.
TRAIL-resistant TNBC cells have similar inflammatory genes. A. Meta-analysis of shared DEGs in each cell line pair. Left: Venn diagram. Middle: Clustered bar graph for MDA231 cells. Right: Clustered bar graph for SUM159 cells. B. RT-PCR of IL-6, CCL5, CD24, CSF2, IL-1α. Upper: MDA231 cells. Lower: SUM159 cells. C. Western blot of IL-6 and IL-1α in MDA231 and SUM159 cells. Actin was used as a loading control. All experiments are representative of three independent experiments. Individual mRNA expression levels were normalized to GAPDH. DEG, differentially expressed gene; P, parental; R, resistant to TRAIL. In the bar graphs, data are represented as mean ± SD, where the error bars denote the standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test.
Down-regulation of IL-1α and IL-6 increases TRAIL sensitivity in TRAIL-resistant cells
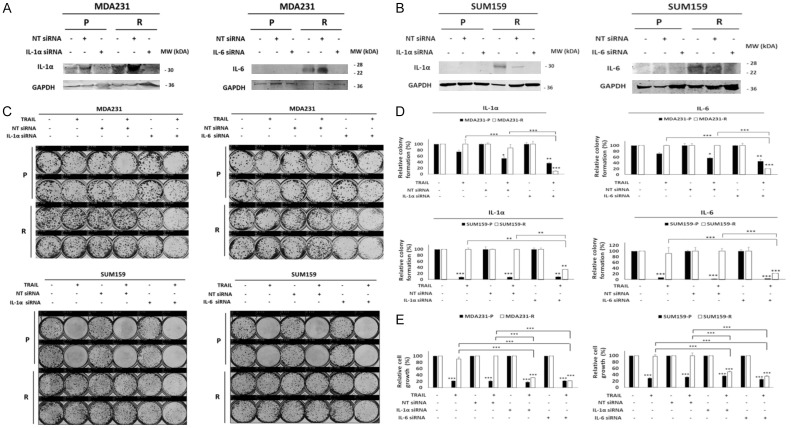
Once the upregulation of these cytokines was confirmed, we wanted to know if they play a role in TRAIL resistance in these TNBC cells. To address this, we used siRNA to knock down the expression of IL-1α or IL-6 expression to assess the effect of their knockdown on the sensitivity of TRAIL. We chose IL-6 and IL-1α as a proof of principle to test these cytokines in resistance to TRAIL. Figure 6A and 6B show that siRNA effectively knocked down IL-1α and IL-6 in MDA231 and SUM159 cells. Importantly, IL-6 and IL-1α knockdown increased the cytotoxicity of TRAIL in MDA231-R and SUM159-R cells, as determined by colony formation and MTT assays (Figure 6C-E). Taken together, these data strongly suggest that the expression of IL-1α and IL-6 expression have a negative effect on the sensitivity of TNBC cells to TRAIL.
Figure 6.
Knockdown of IL-6 and IL-1α increases the sensitivity of TRAIL. MDA231 and SUM159 cells were transfected with IL-6, IL-1α, or nontarget siRNAs for 72 H. (A, B) Western blot of IL-6 and IL-1α in MDA231 (A) and SUM159 (B) cells. (C) Colony formation assay of the resulting MDA231 and SUM159 cells transfected with IL-6, IL-1α, or nontarget siRNA and treated with TRAIL (50 ng/ml) for 72 h. Upper: MDA231 cells. Lower: SUM159 cells. (D) Densitometry bar graph depicting relative colony formation. Upper: MDA231 cells. Lower: SUM159 cells. Left: IL-1α. Right: IL-6. (E) MTT assay was performed to determine relative cell growth in MDA231 (left), and SUM159 (right) cells transfected with IL-1α, IL-6, or nontarget siRNA and treated with TRAIL (50 ng/ml) for 72 h. GAPDH was used as a loading control. IL, interleukin; NT, nontarget; P, parental; R, resistant to TRAIL. In the bar graphs, data are represented as mean ± SD, where the error bars denote the standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test.
Discussion
The TRAIL pathway remains a promising target for cancer therapy because of its ability to selectively reduce cell growth in tumor cells without harming normal cells. However, the development of resistance to TRAIL in tumor cells is a significant problem that needs to be addressed. In this study, we profiled gene expression changes between TRAIL-sensitive and TRAIL-resistant cells and identified a number of genes that are differentially expressed. By performing pathway analysis, we identified the immune and inflammatory pathways in TRAIL resistance that are altered as TRAIL-sensitive cells acquire TRAIL resistance. Identifying a link of the immune/inflammatory pathways to resistance to TRAIL may provide a strategy to improve the efficacy of TRAIL-based treatment in cancer cells.
The mechanisms of TRAIL resistance have been extensively studied but are not completely understood. Previous studies have implicated several pathways in resistance to TRAIL, but these studies have focused primarily on intrinsic and extrinsic apoptosis pathways, and many tumor cells remain resistant to TRAIL [7]. By performing an RNA-seq analysis in TNBC cells, we identified 918 DEGs in MDA231-R cells (401 up-regulated and 517 downregulated) and 4,833 DEGs in SUM159-R cells (2,860 up-regulated and 1,973 downregulated). We identified that some of the genes that were up-regulated in TRAIL-resistant cells were associated with immune response and inflammation. This is consistent with the findings of a recent study of TRAIL-resistant glioblastoma cell expression profiles, which revealed that immune activation and inflammatory pathways were upregulated [39]. The latter has also been shown in TRAIL-resistant MCF10A breast cancer cells [40].
Although previous studies indicated that most TNBC cells are non-inflamed [41], our study found that TRAIL can increase the expression of genes associated with an inflammatory phenotype in TRAIL-resistant cells. We showed through GO analysis that the top 30 DEGs targeted biological processes and molecular functions related to immune and inflammatory responses. We also showed that many of these GOs are shared by TRAIL-resistant cells. Notably, responses to interleukins and TNFs were identified as shared biological processes, whereas cytokine activity was identified as a shared molecular function. These results support previous studies showing that interleukins can reduce the sensitivity of transformed and cancer cells to TRAIL [42,43] and that IL-1 can protect keratinocytes from the cytotoxicity of TRAIL by activating NF-κB [42].
The underlying mechanism by which inflammation confers resistance to TRAIL remains to be defined. Several lines of evidence suggest that TRAIL can activate several signaling pathways that stimulate the expression of NF-κB, AKT, and ERK, resulting in the production of pro-inflammatory cytokines [43-45]. In our previous studies, we showed that the preceding pathways were associated with TRAIL resistance in TNBC cells [30,32]. Furthermore, studies have shown that TRAIL can induce cytokine release in TRAIL-resistant cancer cells in a FADD-dependent manner, which is mediated by caspase 8 [46,47]. In this study, we showed that pro-inflammatory cytokines were increasingly expressed in TRAIL-resistant cells. An analysis of the KEGG pathway in these cells revealed that the top 30 DEGs impacted inflammatory pathways. Furthermore, a KEGG analysis of all DEGs revealed that TNF and cytokine-cytokine receptor signaling pathways were the most altered and shared among TRAIL-resistant cells. These cells also shared 414 DEGs, 15 of which were associated with immunity. An association with TNF or cytokine-cytokine receptor signaling is consistent with a previous study in which TRAIL resistant cells had higher levels of programmed death ligand-1 (PD-L1) expression [30].
Because TRAIL is a TNF superfamily cytokine, it is worth investigating whether it plays a noncanonical role in inducing inflammation and whether this can be exacerbated by pro-inflammatory cytokines in TRAIL-resistant cells. This prompted us to ask if inhibiting the expression of pro-inflammatory cytokines IL-6 and IL-1α renders TRAIL-resistant cells more sensitive to TRAIL. As shown in this study, the knockdown of IL-6 and IL-1α expression significantly impacted TRAIL sensitivity in TRAIL-resistant cells (Figure 6). Consistent with this observation, it has been shown that IL-6 contributes to TRAIL resistance and that its interruption sensitizes cells to TRAIL-induced cytotoxicity [48]. In addition, it has been demonstrated that IL-1α can render cells TRAIL-resistant [42]. Therefore, these results validate the role of inflammation in resistance to TRAIL in TNBC cells.
In summary, inflammation is correlated with TRAIL resistance in TNBC cells. Several DEGs and pathways were identified in TRAIL-resistant cells that are associated with inflammation. The cytokine and cytokine receptor interaction pathways were among the most affected, with IL-6 and IL-1α being two of the most upregulated DEGs. More importantly, TRAIL sensitivity in TRAIL resistant cells was affected by downregulation of IL-6 and IL-1α. Based on these findings, additional research is needed to determine the efficacy of combining IL-6 and IL-1α inhibition with TRAIL in TRAIL-resistant TNBC cells in vivo. Furthermore, more research is required to better understand the roles of IL-6 and IL-1α in resistance to TRAIL and how TRAIL up-regulates these pro-inflammatory cytokines. We believe that these findings may also apply to other TRAIL-resistant tumors and warrant additional investigation. Therefore, our results suggest that targeting inflammation may be a viable strategy for overcoming TRAIL resistance in TNBC cells.
Acknowledgements
This work was, in part, supported by Grants R01CA174949 (GS Wu), R21CA249376 (GS Wu), T32-CA009531 (JM Pimentel), R21GM140352 (S Kim), and P30 CA022453 (S Kim) from the National Institute of Health, National Cancer Institute, the Michigan Ovarian Cancer Alliance (GS Wu) and Dean’s Diversity Fellowship of Wayne State University (JM Pimentel).
Disclosure of conflict of interest
None.
Abbreviations
- DEG
differentially expressed gene
- DR
death receptor
- FADD
fas-associated death domain
- FDR
false discovery rate
- FC
fold change
- GO
geneontology
- IAP
Inhibitor of Apoptosis family proteins
- IL
interleukin
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OPG
osteoprotegerin
- PD-L1
programmed death-ligand 1
- tBID
truncated bid
- TNF
tumor necrosis factor
- TNBC
triple-negative breast cancer
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
References
- 1.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 2.Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 4.Wajant H. CD95L/FasL and TRAIL in tumour surveillance and cancer therapy. Cancer Treat Res. 2006;130:141–165. doi: 10.1007/0-387-26283-0_7. [DOI] [PubMed] [Google Scholar]
- 5.von Karstedt S, Montinaro A, Walczak H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat Rev Cancer. 2017;17:352–366. doi: 10.1038/nrc.2017.28. [DOI] [PubMed] [Google Scholar]
- 6.Wu GS. TRAIL as a target in anti-cancer therapy. Cancer Lett. 2009;285:1–5. doi: 10.1016/j.canlet.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Yuan X, Gajan A, Chu Q, Xiong H, Wu K, Wu GS. Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 2018;37:733–748. doi: 10.1007/s10555-018-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 9.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 10.Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA, Goodwin RG, Rauch CT. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI, Goddard AD, Godowski P, Ashkenazi A. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 12.Wu GS, Burns TF, McDonald ER 3rd, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, el-Deiry WS. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity. 1997;7:821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 14.Wu GS, Burns TF, Zhan Y, Alnemri ES, El-Deiry WS. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res. 1999;59:2770–2775. [PubMed] [Google Scholar]
- 15.Degli-Esposti MA, Smolak PJ, Walczak H, Waugh J, Huang CP, DuBose RF, Goodwin RG, Smith CA. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J Exp Med. 1997;186:1165–1170. doi: 10.1084/jem.186.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsters SA, Sheridan JP, Pitti RM, Huang A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard AD, Godowski P, Ashkenazi A. A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol. 1997;7:1003–1006. doi: 10.1016/s0960-9822(06)00422-2. [DOI] [PubMed] [Google Scholar]
- 17.Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity. 1997;7:813–820. doi: 10.1016/s1074-7613(00)80399-4. [DOI] [PubMed] [Google Scholar]
- 18.Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, Dul E, Appelbaum ER, Eichman C, DiPrinzio R, Dodds RA, James IE, Rosenberg M, Lee JC, Young PR. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–14367. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 19.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 20.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 22.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 23.Yamada H, Tada-Oikawa S, Uchida A, Kawanishi S. TRAIL causes cleavage of bid by caspase-8 and loss of mitochondrial membrane potential resulting in apoptosis in BJAB cells. Biochem Biophys Res Commun. 1999;265:130–133. doi: 10.1006/bbrc.1999.1641. [DOI] [PubMed] [Google Scholar]
- 24.Montinaro A, Walczak H. Harnessing TRAIL-induced cell death for cancer therapy: a long walk with thrilling discoveries. Cell Death Differ. 2023;30:237–249. doi: 10.1038/s41418-022-01059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Cristofano F, George A, Tajiknia V, Ghandali M, Wu L, Zhang Y, Srinivasan P, Strandberg J, Hahn M, Sanchez Sevilla Uruchurtu A, Seyhan AA, Carneiro BA, Zhou L, Huntington KE, El-Deiry WS. Therapeutic targeting of TRAIL death receptors. Biochem Soc Trans. 2023;51:57–70. doi: 10.1042/BST20220098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralff MD, El-Deiry WS. TRAIL pathway targeting therapeutics. Expert Rev Precis Med Drug Dev. 2018;3:197–204. doi: 10.1080/23808993.2018.1476062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun SY, Yue P, Zhou JY, Wang Y, Choi Kim HR, Lotan R, Wu GS. Overexpression of bcl2 blocks TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in human lung cancer cells. Biochem Biophys Res Commun. 2001;280:788–797. doi: 10.1006/bbrc.2000.4218. [DOI] [PubMed] [Google Scholar]
- 28.Chawla-Sarkar M, Bae SI, Reu FJ, Jacobs BS, Lindner DJ, Borden EC. Downregulation of Bcl-2, FLIP or IAPs (XIAP and survivin) by siRNAs sensitizes resistant melanoma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2004;11:915–923. doi: 10.1038/sj.cdd.4401416. [DOI] [PubMed] [Google Scholar]
- 29.Burns TF, El-Deiry WS. Identification of inhibitors of TRAIL-induced death (ITIDs) in the TRAIL-sensitive colon carcinoma cell line SW480 using a genetic approach. J Biol Chem. 2001;276:37879–37886. doi: 10.1074/jbc.M103516200. [DOI] [PubMed] [Google Scholar]
- 30.Pimentel JM, Zhou JY, Wu GS. Regulation of programmed death ligand 1 (PD-L1) expression by TNF-related apoptosis-inducing ligand (TRAIL) in triple-negative breast cancer cells. Mol Carcinog. 2023;62:135–144. doi: 10.1002/mc.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harper N, Farrow SN, Kaptein A, Cohen GM, MacFarlane M. Modulation of tumor necrosis factor apoptosis-inducing ligand- induced NF-kappa B activation by inhibition of apical caspases. J Biol Chem. 2001;276:34743–34752. doi: 10.1074/jbc.M105693200. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Zhou JY, Wei WZ, Wu GS. Activation of the Akt survival pathway contributes to TRAIL resistance in cancer cells. PLoS One. 2010;5:e10226. doi: 10.1371/journal.pone.0010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nesterov A, Lu X, Johnson M, Miller GJ, Ivashchenko Y, Kraft AS. Elevated AKT activity protects the prostate cancer cell line LNCaP from TRAIL-induced apoptosis. J Biol Chem. 2001;276:10767–10774. doi: 10.1074/jbc.M005196200. [DOI] [PubMed] [Google Scholar]
- 34.Uddin MH, Pimentel JM, Chatterjee M, Allen JE, Zhuang Z, Wu GS. Targeting PP2A inhibits the growth of triple-negative breast cancer cells. Cell Cycle. 2020;19:592–600. doi: 10.1080/15384101.2020.1723195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fresno C, Fernandez EA. RDAVIDWebService: a versatile R interface to DAVID. Bioinformatics. 2013;29:2810–2811. doi: 10.1093/bioinformatics/btt487. [DOI] [PubMed] [Google Scholar]
- 39.Cingoz A, Ozyerli-Goknar E, Morova T, Seker-Polat F, Esai Selvan M, Gumus ZH, Bhere D, Shah K, Solaroglu I, Bagci-Onder T. Generation of TRAIL-resistant cell line models reveals distinct adaptive mechanisms for acquired resistance and re-sensitization. Oncogene. 2021;40:3201–3216. doi: 10.1038/s41388-021-01697-6. [DOI] [PubMed] [Google Scholar]
- 40.Flusberg DA, Roux J, Spencer SL, Sorger PK. Cells surviving fractional killing by TRAIL exhibit transient but sustainable resistance and inflammatory phenotypes. Mol Biol Cell. 2013;24:2186–2200. doi: 10.1091/mbc.E12-10-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y, Ma D, Zhao S, Suo C, Shi J, Xue MZ, Ruan M, Wang H, Zhao J, Li Q, Wang P, Shi L, Yang WT, Huang W, Hu X, Yu KD, Huang S, Bertucci F, Jiang YZ, Shao ZM AME Breast Cancer Collaborative Group. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res. 2019;25:5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 42.Kothny-Wilkes G, Kulms D, Poppelmann B, Luger TA, Kubin M, Schwarz T. Interleukin-1 protects transformed keratinocytes from tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem. 1998;273:29247–29253. doi: 10.1074/jbc.273.44.29247. [DOI] [PubMed] [Google Scholar]
- 43.Trauzold A, Siegmund D, Schniewind B, Sipos B, Egberts J, Zorenkov D, Emme D, Roder C, Kalthoff H, Wajant H. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene. 2006;25:7434–7439. doi: 10.1038/sj.onc.1209719. [DOI] [PubMed] [Google Scholar]
- 44.Gao J, Wang D, Liu D, Liu M, Ge Y, Jiang M, Liu Y, Zheng D. Tumor necrosis factor-related apoptosis-inducing ligand induces the expression of proinflammatory cytokines in macrophages and re-educates tumor-associated macrophages to an antitumor phenotype. Mol Biol Cell. 2015;26:3178–3189. doi: 10.1091/mbc.E15-04-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoller V, Funcke JB, Roos J, Dahlhaus M, Abd El Hay M, Holzmann K, Marienfeld R, Kietzmann T, Debatin KM, Wabitsch M, Fischer-Posovszky P. Trail (TNF-related apoptosis-inducing ligand) induces an inflammatory response in human adipocytes. Sci Rep. 2017;7:5691. doi: 10.1038/s41598-017-05932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartwig T, Montinaro A, von Karstedt S, Sevko A, Surinova S, Chakravarthy A, Taraborrelli L, Draber P, Lafont E, Arce Vargas F, El-Bahrawy MA, Quezada SA, Walczak H. The TRAIL-induced cancer secretome promotes a tumor-supportive immune microenvironment via CCR2. Mol Cell. 2017;65:730–742. e735. doi: 10.1016/j.molcel.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henry CM, Martin SJ. Caspase-8 Acts in a non-enzymatic role as a scaffold for assembly of a pro-inflammatory “FADDosome” complex upon TRAIL stimulation. Mol Cell. 2017;65:715–729. e715. doi: 10.1016/j.molcel.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054–2065. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]