Abstract
Background:
To date, deep learning–based detection of optic disc abnormalities in color fundus photographs has mostly been limited to the field of glaucoma. However, many life-threatening systemic and neurological conditions can manifest as optic disc abnormalities. In this study, we aimed to extend the application of deep learning (DL) in optic disc analyses to detect a spectrum of nonglaucomatous optic neuropathies.
Methods:
Using transfer learning, we trained a ResNet-152 deep convolutional neural network (DCNN) to distinguish between normal and abnormal optic discs in color fundus photographs (CFPs). Our training data set included 944 deidentified CFPs (abnormal 364; normal 580). Our testing data set included 151 deidentified CFPs (abnormal 71; normal 80). Both the training and testing data sets contained a wide range of optic disc abnormalities, including but not limited to ischemic optic neuropathy, atrophy, compressive optic neuropathy, hereditary optic neuropathy, hypoplasia, papilledema, and toxic optic neuropathy. The standard measures of performance (sensitivity, specificity, and area under the curve of the receiver operating characteristic curve (AUC-ROC)) were used for evaluation.
Results:
During the 10-fold cross-validation test, our DCNN for distinguishing between normal and abnormal optic discs achieved the following mean performance: AUC-ROC 0.99 (95 CI: 0.98–0.99), sensitivity 94% (95 CI: 91%–97%), and specificity 96% (95 CI: 93%–99%). When evaluated against the external testing data set, our model achieved the following mean performance: AUC-ROC 0.87, sensitivity 90%, and specificity 69%.
Conclusion:
In summary, we have developed a deep learning algorithm that is capable of detecting a spectrum of optic disc abnormalities in color fundus photographs, with a focus on neuro-ophthalmological etiologies. As the next step, we plan to validate our algorithm prospectively as a focused screening tool in the emergency department, which if successful could be beneficial because current practice pattern and training predict a shortage of neuroophthalmologists and ophthalmologists in general in the near future.
In recent years, artificial intelligence (AI) in the form of deep learning (DL) has been used to classify medical images in a wide variety of medical disciplines, particularly in specialties where large, well-annotated data sets are readily available, such as dermatology (1), pathology (2–4), radiology (5–8), oncology (8–14), and ophthalmology. Within ophthalmology, deep learning systems (DLSs) have been developed to analyze various retinal conditions, such as age-related macular degeneration (15–17), diabetic retinopathy (18–20), and retinopathy of prematurity (21,22). DL techniques also have been used to detect optic nerve abnormalities in color fundus photographs. Most publications to date in this area have largely been limited to the field of glaucoma (20,23–25), enabled by the availability of a large number of color fundus photographs (CFPs) containing optic disc changes that are suspicious for or confirmed to be due to glaucomatous damage. However, other than glaucoma, numerous other processes can lead to pathologic optic disc changes. For example, many life-threatening neurological and systemic conditions, such as increased intra-cranial pressure, intracranial tumors, and malignant hypertension, may first manifest as optic disc swelling. As an extension of our previous work in applying DL to neuro-ophthalmology (26), in this study, we aimed to extend the application of DL in optic disc analyses and to test the hypothesis that small databases, when enhanced with machine learning techniques such as transfer learning and data augmentation, can be used to develop a robust DLS to detect a spectrum of nonglaucomatous optic neuropathies from two-dimensional color fundus photographs.
METHODS
Our training data set included 944 deidentified CFPs (abnormal 364; normal 580). Our testing data set included 151 deidentified CFPs (abnormal 71; normal 80). CFPs that were blurry, grossly out of focus, or did not display the optic disc in its entirety were excluded. A detailed breakdown of the data sets, including image specifications and patient demographics, is presented in Table 1. This research study adhered to the Declaration of Helsinki, was reviewed by our institutional review board (IRB), and deemed to be IRB-exempt.
TABLE 1.
Image sources, image specifications, and patient demographics of the training and testing datasets
| Training Data set | % | Image Specifications | Demographics |
|---|---|---|---|
| Abnormal (364 images) | |||
| NRM practice | 100 | Digital and digitized analog; 30° | Multiethnic (mostly Caucasians) |
| Normal (580 images) | |||
| NRM practice | 55 | Digital and digitized analog; 30° | Multiethnic (mostly Caucasians) |
| ACRIMA | 45 | Digital; 35° | Spain |
|
| |||
| Testing data set | % | Image Specifications | Demographics |
|
| |||
| Abnormal (71 images) | |||
| PSS practice | 86 | Digital; 30° | Multiethnic (mostly Caucasians) |
| Paxos | 11 | Digital; ~45° | Multiethnic |
| Normal (80 images) | |||
| PSS practice | 33 | Digital; 30° | Multiethnic (mostly Caucasians) |
| DRIONS | 63 | Digitized analog; ~45° | Caucasian |
| Paxos | 4 | Digital; ~45° | Multiethnic |
NRM, Neil R. Miller; PSS, Prem S. Subramanian.
Color Fundus Photographs with Abnormal Optic Discs
All of the CFPs with abnormal optic discs in the training data set were obtained from the neuro-ophthalmological practice of author Neil R. Miller (N.R.M.), captured by a 30° camera centered on the disc and deidentified over several decades. Various optic disc abnormalities were represented, including but not limited to papilledema, hypoplasia, hereditary optic neuropathy, arteritic and non-arteritic anterior ischemic optic neuropathy, and toxic optic neuropathy. The spectrum of optic disc abnormalities in the training data set is summarized in Table 2. Eighty-six percent of the CFPs with abnormal optic discs in the testing data set were obtained from the neuro-ophthalmological practice of author Prem S. Subramanian (P.S.S.) (~2056 ×·2048 pixels; 24 bit/pixel), captured by a 30° camera centered on the disc and deidentified between years 2012 and 2018. These photographs were selected to represent a wide range of optic disc abnormalities, such as, but not limited to hereditary optic neuropathy, atrophy, papilledema, optic neuritis, compressive optic neuropathy, ischemic optic neuropathy, hypoplasia, and optic nerve sheath meningioma. The spectrum of optic disc abnormalities in the testing data set is summarized in Table 3. Although the images were chosen randomly to include a wide range of abnormalities, each image represented a unique eye. The diagnosis of each optic disc abnormality was established by a combination of detailed clinical history, clinical examination, ophthalmic imaging, and/or neurological imaging.
TABLE 2.
Spectrum of optic disc abnormalities represented in our training data set
| Condition | % Distribution among images with an abnormal optic disc |
|---|---|
| AAION | 1.9 |
| Atrophy | 1.4 |
| Compressive optic neuropathy | 0.8 |
| Congenital anomaly | 5.5 |
| Hereditary optic neuropathy | 11.5 |
| Hypoplasia | 16.5 |
| Infiltration | 1.6 |
| Malignant hypertension | 0.5 |
| Miscellaneous | 21.9 |
| Morning glory disc | 1.6 |
| NAAION | 5.2 |
| Nonglaucomatous cupping | 1.6 |
| Optic nerve sheath meningioma | 0.8 |
| Papilledema | 22.8 |
| Papillorenal syndrome | 1.6 |
| Toxic optic neuropathy | 4.4 |
AAION, arteritic anterior ischemic optic neuropathy; NAAION, nonarteritic anterior ischemic optic neuropathy.
TABLE 3.
Spectrum of optic disc abnormalities represented in our external, independent testing data set
| Condition | % Distribution among images with an abnormal optic disc |
|---|---|
| AAION | 5.6 |
| Atrophy/Pallor | 11.3 |
| Compressive optic neuropathy | 1.4 |
| Hereditary optic neuropathy | 9.9 |
| Hypoplasia | 1.4 |
| Miscellaneous | 18.3 |
| NAAION | 9.9 |
| Nonglaucomatous cupping | 1.4 |
| Optic nerve sheath meningioma | 2.8 |
| Optic neuritis | 5.6 |
| Papilledema | 32.4 |
AAION, arteritic anterior ischemic optic neuropathy; NAAION, nonarteritic anterior ischemic optic neuropathy.
Other Databases
CFPs with normal optic discs were obtained from the publicly available database DRIONS (27). The DRIONS database contained images taken with a color analog fundus camera (roughly 45° field of view) and digitized at a resolution of 600 · 400 and 8 bits/pixel. This cohort contained 46.2% males and 100% Caucasians, with a mean age of 53.0 years. CFPs with normal optic discs were obtained from the publicly available database ACRIMA (28), which included 35-degree images from a Spanish cohort. Finally, CFPs with both normal and abnormal optic discs were captured separately from a different cohort of patients with a mobile, smartphone-based, ophthalmic imaging adapter (Paxos) (29).
Deep Learning System Development
Using transfer learning, we adopted a readily available ResNet-152 (30) deep convolutional neural network (DCNN) that was pretrained on ImageNet (31) (a database of 1.2 million color images of everyday objects sorted into 1,000 categories). We started with the ResNet-152 and redefined the last fully connected layer to have 2 outputs instead of the default 1,000 for our underlying binary classification problem. Additional details for the DCNN are available in Supplemental Digital Content 1 (see Appendix, http://links.lww.com/WNO/A495).
Statistical Analysis
We performed a 10-fold cross-validation test (training 75%; validation 15%; and testing 10%) using images from our training data set for internal validation and model development. After the 10-fold cross-validation was completed, we then tested the cross-validation model with the best performance against the testing data set. Using the prefixed operating threshold value determined on the training data set (F-1 score), we calculated the diagnostic performance on the testing data set using receiver operating characteristic (ROC) curve, sensitivity, and specificity.
Heat Map Generation
We created heatmaps through class activation mapping (CAM) (29) to identify regions in the CFPs used by the DCNN to detect optic disc abnormalities. This technique visually highlights areas of activation during decision making within an image (the “warmer” the color, e.g., red, the more highly activated a particular region is). We performed CAM analyses on all 151 photographs in the testing data set, and these CAM images were reviewed and interpreted by one of the authors (T.Y.A.L.).
RESULTS
During the 10-fold cross-validation test, our DCNN for distinguishing between a normal and an abnormal optic disc achieved the following mean performance: area under the curve (AUC) of ROCs 0.99 (95 CI: 0.98–0.99, range 0.96–1), sensitivity 94% (95 CI: 91%–97%, range 86%–100%), and specificity 96% (95 CI: 93%–99%, range 83%–100%) (Figs. 1A–J). When evaluated against the external testing data set, our most robust (by AUC) cross-validation model achieved the following overall performance: AUC 0.87, sensitivity 90%, specificity 69%, positive predictive value (positive defined as abnormal) 72%, and negative predictive value 89%. Specifically, when tested against images taken with a smartphone-based, ophthalmic imaging adapter (a small subset within the testing data set), our model showed a sensitivity of 100% and a specificity of 50%. Of the 151 images in the testing data set, there were 7 false negatives, which included the following diagnoses: dominant optic atrophy (n = 1), Leber hereditary optic neuropathy (n = 3), optic disc drusen (n = 1), optic neuritis (n = 1), and nonglaucomatous cupping (n = 1).
FIG. 1.
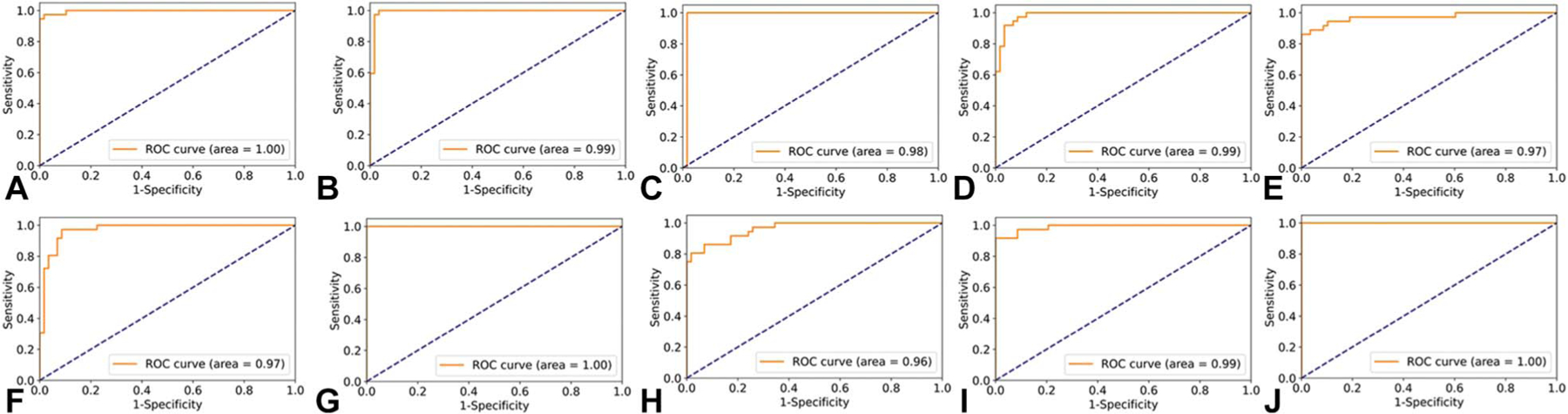
Receiver operating characteristic (ROC) curves for our 10-fold cross-validation (CV) models. A. CV1, (B) CV2, (C) CV3, (D) CV4, (E) CV5, (F) CV6, (G) CV7, (H) CV8, (I) CV9, and (J) CV10. CV model 7 was validated by an external independent testing data set.
We performed CAM analyses for all 151 images to gain insights into the decision-making process of our DCNN. Sample CAM analyses for incorrectly classified images are shown in Figure 2. Among images that were incorrectly classified, strong activation outside of the disc was seen in 32% of the eyes. Sample CAM analyses for correctly classified images are shown in Figure 3. By contrast, among the correctly classified images, strong activation at the disc was seen in 90% of the eyes.
FIG. 2.
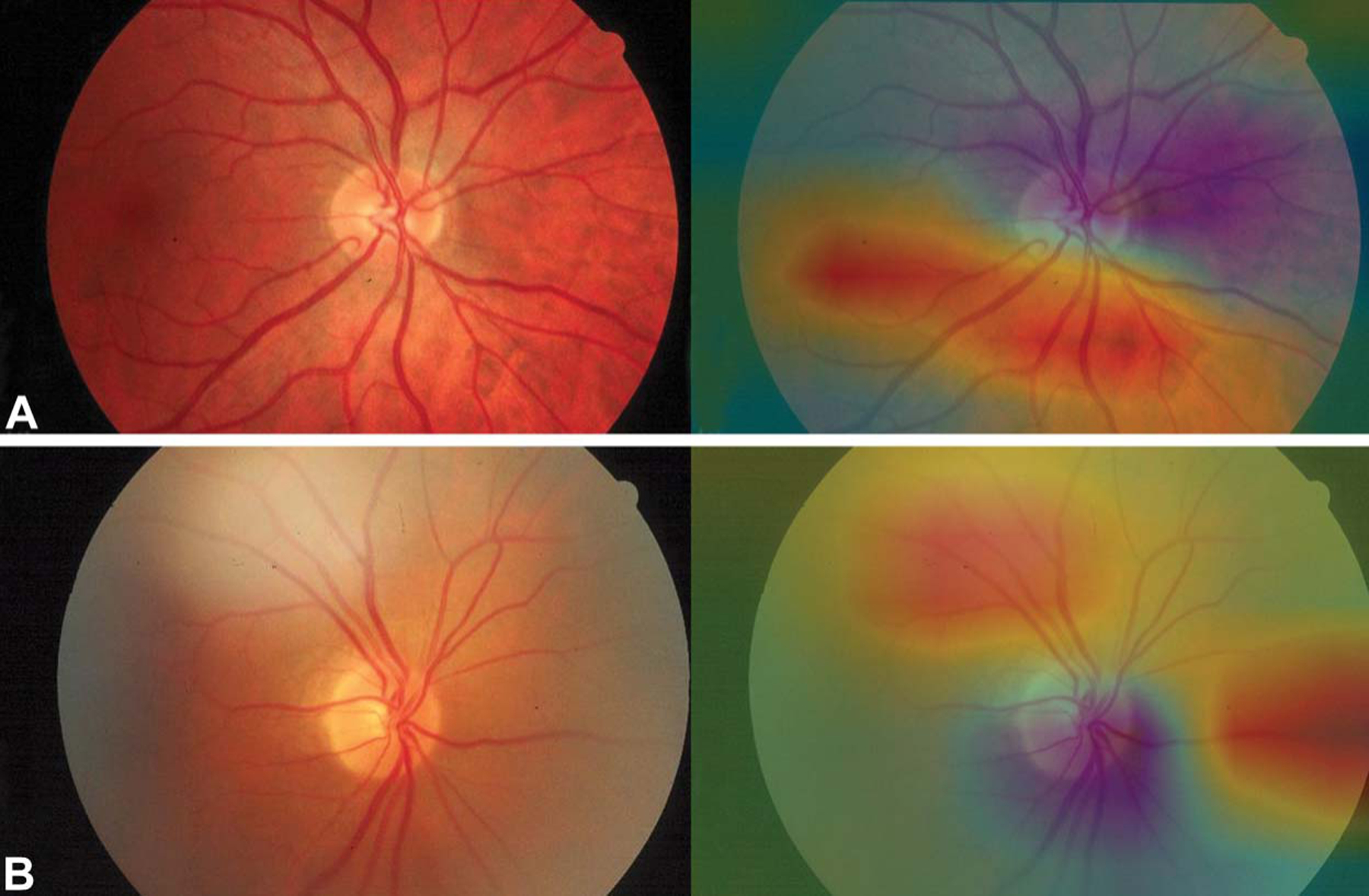
Sample heatmaps generated by class activation mapping analyses, showing strong activation outside of the optic disc. Both (A and B) contain a normal optic disc but were erroneously identified as “abnormal” by the algorithm. Note the areas of highlighted image artifacts in (B).
FIG. 3.
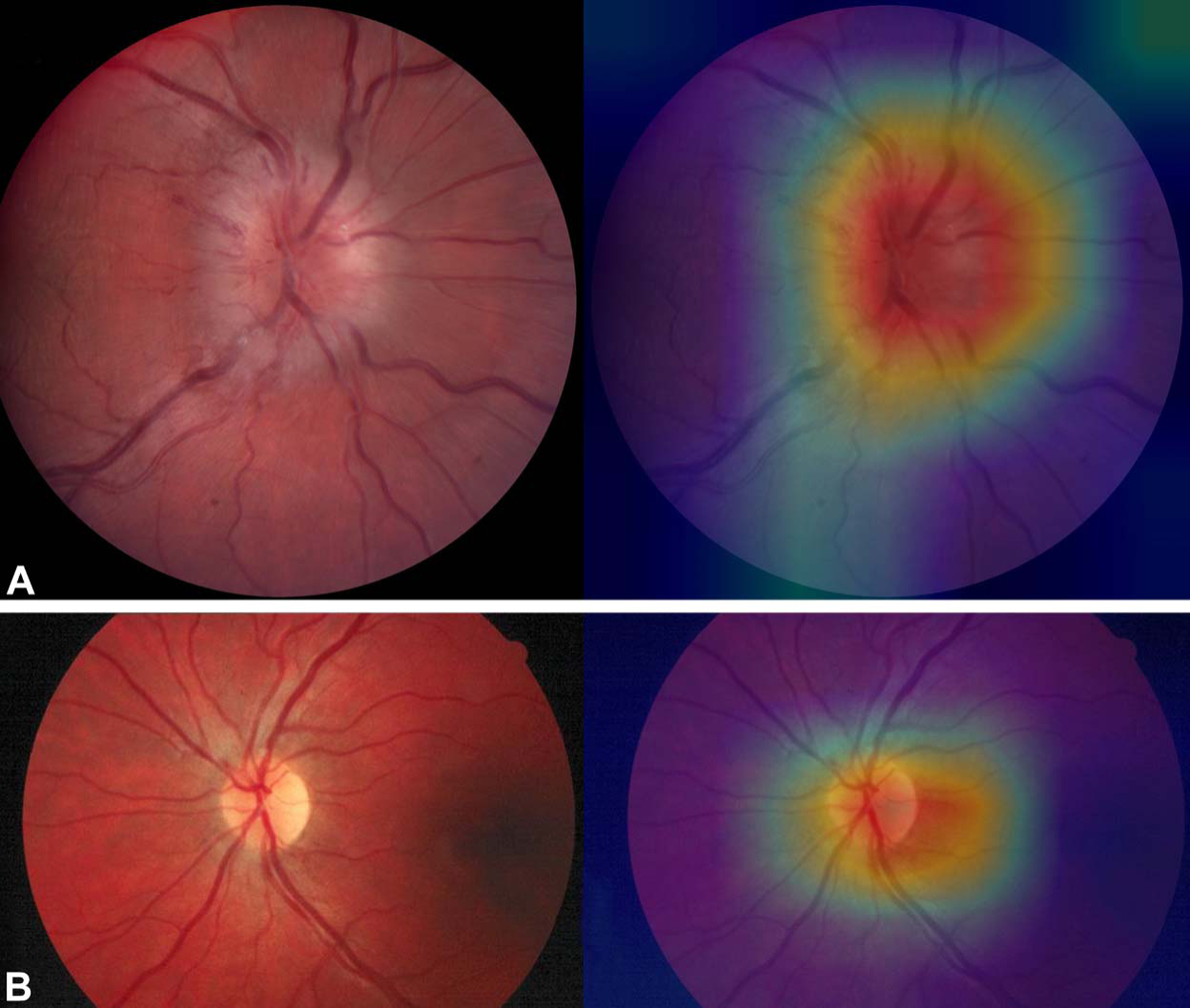
Sample heatmaps generated by class activation mapping analyses, showing strong activation at the optic disc: optic disc swelling with hemorrhages (A) and normal (B).
DISCUSSION
Previously, classical machine learning techniques, such as random forest classification and support vector machine, have been applied to optic disc analyses, including papilledema severity grading (32,33), distinguishing between papilledema and normal discs (34), and detection of optic disc pallor (35). DL is currently the cutting-edge machine learning technique for medical image analysis and is superior to classical machine learning techniques in that DL does not require labor-intensive feature engineering and can be directly trained with raw data such as CFPs. However, DL requires a large amount of data for training, which limit its applicability in fields, such as neuro-ophthalmology, where large image databases are typically not available. Previously, our group demonstrated the feasibility of applying DL techniques to a small neuro-ophthalmological data set for the basic task of determining disc laterality, using transfer learning and data augmentation (26). The current study sought to build on our previous work and to develop a DLS that can detect a spectrum of nonglaucomatous optic neuropathies in CFPs.
Our DLS showed robust performance during the 10-fold cross-validation test, with a mean performance of AUC 0.99, sensitivity 94%, and specificity 96%. As expected, when our model was tested against an independent external testing data set, there was a drop in performance; as by definition, the external testing data set contained a different data distribution (for image field of view, image quality, range of pathologies, and artifacts, etc.) that our algorithm did not encounter during training. During external validation, our DLS achieved the following performance: AUC 0.87, sensitivity 90%, and specificity 69%. The drop in performance was driven mainly by the decrease in specificity or the increase in false positives. CAM analyses of the false-positive images suggested that strong activation outside of the disc was likely contributory. For example, our algorithm likely erroneously identified image artifacts, such as those seen in Figure 2B, as “abnormal.”
The strength of our study lies in the wide range of neuro-ophthalmological optic disc abnormalities that were included in both our training and testing data sets. These abnormalities included arteritic and nonarteritic anterior ischemic optic neuropathy, primary optic atrophy, compressive optic neuropathy, congenital anomalies (including hypoplasia, morning glory disc, and papillorenal syndrome disc), hereditary optic neuropathy, infiltration, optic disc swelling from malignant hypertension, optic nerve sheath meningioma, papilledema, and toxic optic neuropathy. Most of the images in the external testing data set were obtained as part of routine, standard care from the clinical practice of author PSS, a neuro-ophthalmologist. Thus, we believe our experimental results reflected real-world outcomes. Our testing data set was also diverse, including images taken by traditional desktop fundus cameras and mobile smartphone-based cameras. Specifically, although our algorithm was developed exclusively with images taken by traditional desktop fundus cameras, it achieved a reasonable performance when tested against images taken by smartphone cameras, with an accuracy of 83%, sensitivity of 100%, specificity of 50%, and negative predictive value of 100%. This suggests that an updated version of our algorithm, further trained and fine-tuned with smartphone-based photographs, could be used as a focused screening tool in the emergency department setting if validated prospectively. Many life-threatening neurological and systemic conditions, such as increased intracranial pressure, present with changes in the appearance of the optic disc and are a frequent reason for ophthalmology consults in the emergency department. A DLS that can reliably detect these changes in photographs taken by smartphones could be a beneficial screening tool in the emergency department setting.
Our study also supports a small but growing body of literature that DL techniques can be applied to useful neuro-ophthalmological tasks. For example, Ahn et al (36) constructed their own DL model, which achieved an AUC of 0.992 in distinguishing between normal discs, pseudopapilledema, and optic neuropathies. However, in contrast to our study, Ahn et al did not evaluate their DLS against an external testing data set obtained from a different source. Using data from 19 sites in 11 countries, Milea et al (37) trained and tested a DLS that was able to detect papilledema with a sensitivity of 96.4% and specificity of 84.7%. Their DLS was subsequently compared in a new data set with 2 expert neuro-ophthalmologists and was found to have comparable performances (38).
Our study has several weaknesses. First, it is unclear whether our algorithm could identify a normal-appearing optic disc with an active retrobulbar process, such as idiopathic or multiple sclerosis-related optic neuritis. Second, most images in both our training and testing data sets are derived from Caucasian patients. In the future, we plan to test our algorithm against a data set with a more diverse or different ethnic makeup. Third, our training data set is not large enough for us to train our DLS to differentiate among specific pathologies, for example, differentiating optic disc swelling due to anterior ischemic optic neuropathy from papilledema due to increased intracranial pressure.
In summary, we have developed a DLS that is capable of detecting a spectrum of optic disc abnormalities in CFPs with robust performance. Although our algorithm demonstrated high sensitivity in detecting optic disc abnormalities in photographs taken by smartphone-based cameras, its specificity for this type of photograph was low, highlighting the need for additional training and refinement of our algorithm with smartphone-based photographs. As the next step, we plan to validate our algorithm prospectively as a focused screening tool in the emergency department, which if successful could be beneficial because current practice pattern and training predict a shortage of neuro-ophthalmologists (39) and ophthalmologists in general (40) in the near future.
Footnotes
D. Myung is a co-inventor of a patent on the Paxos portable camera that was used in the study. The remaining authors report no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.jneuro-ophthalmology.com).
Contributor Information
T. Y. Alvin Liu, Department of Ophthalmology, Wilmer Eye Institute, Johns Hopkins University, Baltimore, Maryland.
Jinchi Wei, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, Maryland.
Hongxi Zhu, Malone Center for Engineering in Healthcare, Johns Hopkins University, Baltimore, Maryland.
Prem S. Subramanian, Department of Ophthalmology, University of Colorado School of Medicine, Aurora, Colorado.
David Myung, Department of Ophthalmology, Byers Eye Institute, Stanford University, Palo Alto, California.
Paul H. Yi, Department of Radiology, Johns Hopkins University, Baltimore, Maryland.
Ferdinand K. Hui, Department of Radiology, Johns Hopkins University, Baltimore, Maryland.
Mathias Unberath, Malone Center for Engineering in Healthcare, Johns Hopkins University, Baltimore, Maryland.
Daniel S. W. Ting, Singapore Eye Research Institute, Singapore National Eye Center, Duke-NUS Medical School, National University of Singapore, Singapore.
Neil R. Miller, Department of Ophthalmology, Wilmer Eye Institute, Johns Hopkins University, Baltimore, Maryland.
REFERENCES
- 1.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Arnam J; Cancer Genome Atlas Research Network; Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep 2018;23:181–193 e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirinukunwattana K, Ahmed Raza SE, Yee-Wah T, Snead DR, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging 2016;35:1196–1206. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 2016;191:214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung SW, Han SS, Lee JW, Oh KS, Kim NR, Yoon JP, Kim JY, Moon SH, Kwon J, Lee HJ, Noh YM, Kim Y. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop 2018;89:468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017;284:574–582. [DOI] [PubMed] [Google Scholar]
- 7.Lakhani P Deep convolutional neural networks for endotracheal tube position and X-ray image classification: challenges and opportunities. J Digit Imaging 2017;30:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, Su M-Y, Cha S, Filippi CG, Bota D, Baldi P, Poisson LM, Jain R, Chow D. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 2018;39:1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehteshami Bejnordi B, Veta M, Diest J, Ginneken B, Karssemeijer N, Litjens G, van der Laak JAM; the CAMELYON16 Consortium, Hermsen E, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Haß C, Bruni E, Wong Q, Halici U, Ümit Öner M, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Phoulady HA, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venâncio R. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318:2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couture HD, Williams LA, Geradts J, Nyante SJ, Butler EN, Marron JS, Perou CM, Troester MA, Niethammer M. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 2018;4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc 2015;2015:1899–1908. [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Roa AA, Arevalo Ovalle JE, Madabhushi A, Gonzalez Osorio FA. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Med Image Comput Comput Assist Interv 2013;16:403–410. [DOI] [PubMed] [Google Scholar]
- 13.Mishra R, Daescu O, Leavey P, Rakheja D, Sengupta A. Convolutional neural network for histopathological analysis of osteosarcoma. J Comput Biol 2018;25:313–325. [DOI] [PubMed] [Google Scholar]
- 14.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridge J, Harding S, Zheng Y. Development and validation of a novel prognostic model for predicting AMD progression using longitudinal fundus images. BMJ Open Ophthalmol 2020;5:e000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y, Keenan TD, Chen Q, Agrón E, Allot A, Wong WT, Chew EY, Lu Z. Predicting risk of late age-related macular degeneration using deep learning. NPJ Digit Med 2020;3:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhuiyan A, Wong TY, Ting DSW, Govindaiah A, Souied EH, Smith RT. Artificial intelligence to stratify severity of age-related macular degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol 2020;9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig CA, Perera C, Myung D, Greven MA, Smith SJ, Chang RT, Leng T. Automatic identification of referral-warranted diabetic retinopathy using deep learning on mobile phone images. Transl Vis Sci Technol 2020;9:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burlina P, Paul W, Mathew P, Joshi N, Pacheco KD, Bressler NM. Low-shot deep learning of diabetic retinopathy with potential applications to address artificial intelligence bias in retinal diagnostics and rare ophthalmic diseases. JAMA Ophthalmol 2020;138:1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, Hamzah H, Garcia-Franco R, Yeo IYS, Lee SY, Wong EYM, Sabanayagam C, Baskaran M, Ibrahim F, Tan NC, Finkelstein EA, Lamoureux EL, Wong IY, Bressler NM, Sivaprasad S, Varma R, Jonas JB, Guang He M, Cheng CY, Cheung GCM, Aung T, Hsu W, Lee ML, Wong TY. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017;318:2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Paul Chan RV, Dy J, Erdogmus D, Ioannidis S, Kalpathy-Cramer J, Chiang MF. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol 2018;136:803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell JP, Kim SJ, Brown JM, Ostmo S, Paul Chan RV, Kalpathy-Cramer J, Chiang MF, of the Imaging and Informatics in Retinopathy of Prematurity Consortium. Evaluation of a deep learning-derived quantitative retinopathy of prematurity severity scale. Ophthalmology 2020. [DOI] [PMC free article] [PubMed]
- 23.Shibata N, Tanito M, Mitsuhashi K, Fujino Y, Matsuura M, Murata H, Asaoka R. Development of a deep residual learning algorithm to screen for glaucoma from fundus photography. Sci Rep 2018;8:14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masumoto H, Tabuchi H, Nakakura S, Ishitobi N, Miki M, Enno H. Deep-learning classifier with an ultrawide-field scanning laser ophthalmoscope detects glaucoma visual field severity. J Glaucoma 2018;27:647–652. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Li L, Wormstone IM, Qiao C, Zhang C, Liu P, Li S, Wang H, Mou D, Pang R, Yang D, Zangwill LM, Moghimi S, Hou H, Bowd C, Jiang L, Chen Y, Hu M, Xu Y, Kang H, Ji X, Chang R, Tham C, Cheung C, Ting DSW, Wong TY, Wang Z, Weinreb RN, Xu M, Wang N. Development and validation of a deep learning system to detect glaucomatous optic neuropathy using fundus photographs. JAMA Ophthalmol 2019. [DOI] [PMC free article] [PubMed]
- 26.Liu TYA, Ting DSW, Yi PH, Wei J, Zhu H, Subramanian PS, Li T, Hui FK, Hager GD, Miller N. Deep learning and transfer learning for optic disc laterality detection: implications for machine learning in neuro-ophthalmology. J Neuroophthalmol 2020;40:178–184. [DOI] [PubMed] [Google Scholar]
- 27.Carmona EJ, Rincon M, Garcia-Feijoo J, Martinez-de-la-Casa JM. Identification of the optic nerve head with genetic algorithms. Artif Intell Med 2008;43:243–259. [DOI] [PubMed] [Google Scholar]
- 28.Available at: https://figshare.com/s/c2d31f850af14c5b5232. Accessed April 1, 2021.
- 29.Ludwig CA, Murthy SI, Pappuru RR, Jais A, Myung DJ, Chang RT. A novel smartphone ophthalmic imaging adapter: user feasibility studies in Hyderabad, India. Indian J Ophthalmol 2016;64:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. The IEEE Conference on Computer Vision and Pattern Recognition: CVPR, 2016:770–778.
- 31.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. Imagenet: a large-scale hierarchical image database IEEE Conf Comp Vis Pattern Recognition (Cvpr). 2009:248–255. [Google Scholar]
- 32.Frisén L. Swelling of the optic nerve head: a staging scheme. J Neurol Neurosurg Psychiatry 1982;45:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Echegaray S, Zamora G, Yu H, Luo W, Soliz P, Kardon R. Automated analysis of optic nerve images for detection and staging of papilledema. Invest Ophthalmol Vis Sci 2011;52:7470–7478. [DOI] [PubMed] [Google Scholar]
- 34.Akbar S, Akram MU, Sharif M, Tariq A, Yasin UU. Decision support system for detection of papilledema through fundus retinal images. J Med Syst 2017;41:66. [DOI] [PubMed] [Google Scholar]
- 35.Yang HK, Oh JE, Han SB, Kim KG, Hwang JM. Automatic computer-aided analysis of optic disc pallor in fundus photographs. Acta Ophthalmol (Copenh) 2019;97:e519–e525. [DOI] [PubMed] [Google Scholar]
- 36.Ahn JM, Kim S, Ahn KS, Cho SH, Kim US. Accuracy of machine learning for differentiation between optic neuropathies and pseudopapilledema. BMC Ophthalmol 2019;19:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milea D, Najjar RP, Zhubo J, Ting D, Vasseneix C, Xu X, Fard MA, Fonseca P, Vanikieti K, Lagrèze WA, Morgia CL, Cheung CY, Hamann S, Chiquet C, Sanda N, Yang H, Mejico L, Rougier MB, Kho R, Chau TTH, Singhal S, Gohier P, Clermont-Vignal C, Cheng CY, Jonas JB, Man PYW, Fraser CL, Chen JJ, Ambika S, Miller NR, Liu Y, Newman NJ, Wong TY, Biousse V, BONSAI Group. Artificial intelligence to detect papilledema from ocular fundus photographs. N Engl J Med 2020;382:1687–1695. [DOI] [PubMed] [Google Scholar]
- 38.Biousse V, Newman NJ, Najjar RP, Vasseneix C, Xu X, Ting DS, Milea LB, Hwang JM, Kim DH, Yang HK, Hamann S, Chen JJ, Liu Y, Wong TY, Milea D; BONSAI (Brain and Optic Nerve Study with Artificial Intelligence) Study Group. Optic disc classification by deep learning versus expert neuro-ophthalmologists. Ann Neurol 2020;88:785–795. [DOI] [PubMed] [Google Scholar]
- 39.Frohman LP. How can we assure that neuro-ophthalmology will survive? Ophthalmology 2005;112:741–743. [DOI] [PubMed] [Google Scholar]
- 40.National and Regional Projections of Supply and Demand for Surgical Specialty Practitioners:2013–2025: U.S. Department of Health and Human Services; Health Resources and Services Administration; Bureau of Health Workforce Washington, DC: National Center for Health Workforce Analysis; 2016. [Google Scholar]


