Abstract
Cervical cancer (CC) is the fourth most common cancer in women worldwide. It develops through precancerous lesions (cervical intraepithelial neoplasia (CIN), graded from low-grade (CIN1) to high-grade (CIN2-3)). It is well established that precancerous and cancerous cervical lesions are caused by a persistent infection with high-risk types of the human papilloma virus (hrHPV). To have a deeper understanding of the pathogenesis of CIN and CC, we systematically analyzed the landscape of genomic alterations and HPV integration profiles in high-grade CIN2/3. We performed deep whole genome sequencing on exfoliated cervical cells and matched peripheral blood samples from a cohort of 51 Chinese patients (of whom 35 were HPV+) with high-grade CIN from 3 ethnic groups and constructed strict integrated workflow of genomic analysis. In addition, the HPV types and integration breakpoints in the exfoliated cervical cells from these patients were examined. Genomic analysis identified 6 significantly mutated genes (SMGs), including CDKN2A, PIK3CB, FAM20A, RABEP1, TMPRSS2 and SS18L1, in 51 CIN2/3 samples. As none of them had previously been identified as SMGs in the Cancer Genome Atlas cervical squamous cell carcinoma and endocervical adenocarcinoma (TCGA-CESC) cohort, future studies with larger sample size of CINs may be needed to validate our findings. Mutational signature analysis showed that mutational signatures of CINs were dramatically different from CCs, highlighting their different mutational processes and etiologies. Moreover, non-silent somatic mutations were detected in all of the CIN2/3 samples, and 88% of these mutations occurred in genes that also mutated in CCs of TCGA cohort. CIN2 samples had significantly less non-silent mutations than CIN3 samples (P = .0006). Gene ontology and pathway level analysis revealed that functions of mutated genes were significantly associated with tumorigenesis, thus these genes may be involved in the development and progression of CC. HPV integration breakpoints occurred in 28.6% of the CIN2/3 samples with HPV infection. Integrations of common high risk HPV types in CCs, including HPV16, 52, 58 and 68, also occurred in the CIN samples. Our results lay the groundwork for a deeper understanding of the molecular mechanisms underlying the pathogenesis of CC and pave the way for new tools for screening, diagnosis and treatment of cervical precancerous and cancerous lesions.
Keywords: cervical cancer, cervical intraepithelial neoplasia, human papilloma virus, mutational signatures, significantly mutated genes, whole-genome sequencing
1. Introduction
Cervical cancer (CC) is the third most common cancer in women worldwide and has a high mortality rate among women. In 2008, CC was responsible for 275,000 deaths, thereby being the fourth leading cause of cancer death in females worldwide.[1,2] In China, CC is the second most common gynecological malignancy.[3,4] Over the last few years, the incidence of CC is rising in many countries and the prognosis of it remains very poor. A comprehensive description of the mutational landscape in CC may provide insights into a better selection of treatments and improve prognosis. Virtually all CCs result from a persistent infection with certain high-risk types of the human papilloma virus (hrHPV) family.[5] Following an hrHPV infection, CC develops through a series of subsequent steps: hrHPV persistence, hrHPV-mediated epithelial transformation, development of precancerous lesions (cervical intraepithelial neoplasia graded 1 to 3 (CIN1-3)) and, finally, progression to invasive CC.[6] CC development, in particular the step from precancerous lesion to invasive cancer, usually takes a long time in most patients. High-grade precancerous CIN2 and CIN3 lesions can develop within 3 to 5 years following an hrHPV infection,[7] whereas further progression to invasive cancer can take up to 20 to 30 years.[8,9] This long period offers many opportunities for intervention and has probably contributed to the success of Papanicolaou (Pap) screening to reduce the incidence and mortality of CC.[10]
HPV DNA test and liquid-based cytology (such as Thinprep Cytology Test) have been widely used in CC screening. However, both methods have their limitations.[11] The combined use of expressions of HPV E6/E7 mRNA and tumor suppressor protein p16 was reported to improve the diagnosis of cervical neoplasia.[12] Combined methylation marker analysis of 2 genes, MAL and miR-124-2, on HPV-positive self-collected cervicovaginal samples could distinguish CIN2 or worse and CIN3 or worse with minimum sensitivities of 71 · 3% and 77 · 0%, respectively, at specificity of 50%, thus exceeding the sensitivity of combined HPV16 and HPV18 genotyping.[13] Therefore, the discovery and validation of new molecular biomarkers will play an increasingly important role in the prevention of CC. Previous studies[14,15] have identified many significant mutated genes (SMGs) in CC, such as PIK3CA, KMT2C, KMT2D, FBXW7, EP300, HLA-B, PTEN, NFE2L2, ARID1A, KRAS, MAPK1, SHKBP1, ERBB3, CASP8, HLA-A and TGFBR2, and so on. However, the genomic profiles and virus integrations in precancerous cervical lesions, especially in CIN, are largely unknown.
In the current study, we systematically analyzed the genomic profiles of 51 CIN2/3 specimens with matched germline DNA. Somatic mutations, mutation signatures and HPV integration events were identified. The discoveries in the current study may facilitate the identification and validation of potential biomarkers for the screening and treatment of CIN and CC.
2. Methods
2.1. Patients and samples
This project and protocols were conducted in accordance with the Declaration of Helsinki (as revised in 2013). Exfoliated cervical cells and matched peripheral blood were collected from patients attending a sexually transmitted disease clinic at the First Affiliated Hospital of Dali University, from January 2019 to January 2020. The study was approved by Ethics Committee of the First Affiliated Hospital of Dali University. Sample collection, HPV typing, liquid-based cytology and biopsy were performed according to guidelines or at the patients’ request with appropriate informed consent. Cervical biopsy results were reviewed by an experienced pathologist to confirm the histopathological diagnosis. Genomic DNA isolated from peripheral blood was obtained from each participant as a control.
2.2. Whole genome-sequencing and data analysis
Genomic DNA was extracted from exfoliated cervical cells and matched peripheral blood samples using the DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer instructions. After DNA extraction, agarose gel electrophoresis was used to confirm that there was no obvious DNA degradation or RNA contamination. Next, Agilent 2100 Bioanalyzer was used to examine the distribution of the size of the DNA fragments (Agilent Technologies, Santa Clara, USA), and a NanoDrop (Thermo Scientific, MA) was used to determine the OD values.
A total amount of 3.0 μg genomic DNA per sample was used as input material for the sequencing library preparation. Briefly, DNA shearing was carried out by a Covaris S2 ultrasonicator (Covaris, Massachusetts) to generate 300-500bp fragments. The remaining overhangs were converted into blunt ends via 3’ to 5’ exonuclease and 5’ to 3’ polymerase activities of T4 DNA Polymerase. After adenylation of the 3’ ends of DNA fragments, adapter were ligated. DNA fragments with ligated adapter on both ends were selectively amplified in a PCR. The amplified libraries were then purified using an AMPure XP system (Beckman Coulter, Beverly, USA) and quantified using an Agilent high-sensitivity DNA assay on an Agilent 2100 Bioanalyzer. The prepared libraries were sequenced with paired-end, 150 base pair (bp)-long reads on an Illumina NovaSeq 6000 system.
The sequencing quality of raw reads was evaluated by FastQC software (v0.11.4). Adapters and low-quality bases were trimmed from raw sequencing reads using fastp (v0.19.6).[16] Clean reads were aligned to the human reference genome (hg19 build) using the BWA (v0.7.17).[17] Sorted BAM files were created using the samtools (v1.5-10). Duplicate reads were marked using the Picard (v 2.5.0). Single nucleotide variants (SNVs) and small insertions and deletions (indels) were called using the Mutect2 (v3.5).[18] The candidate mutations were checked manually with the IGV.[19]
We used ANNOVAR (2020-06-07 version) to annotate the detected mutations which was in variant call format.[20] Ensembl database were used to determine amino acid changes. The dbSNP (142 build), COSMIC (v94), 1000 Genomes Project, and Exome Aggregation Consortium databases were used to obtain the allele frequencies across different populations. MutSig2CV algorithms was used to identify significantly mutated genes (SMGs) in our CIN cohort.[21] MutSig2CV identifies genes that are mutated more frequently than expected by chance given background mutational processes and other covariates. Genes were classified as significantly mutated by MutSig2CV analyses if they had a false discovery rate of <0.1 after correction for multiple hypothesis testing.
2.3. Mutational signature analysis
The number of somatic SNP mutations falling into the 96 single base substitutions (SBS) contexts (https://cancer.sanger.ac.uk/signatures/signatures_v2/) was determined using the R package mutSigExtractor (https://github.com/UMCUGenetics/mutSigExtractor, v1.27). Then, SigProfilerExtractor (v1.1.3) was used (with default settings) to extract de novo mutational signatures.[22] Meanwhile, the extracted de novo mutational signatures with high cosine similarity (≥0.7) to any reference COSMIC mutational signatures with known cancer-type associations were labeled accordingly. Besides, we used deconstructSigs (v1.8.0) to estimate the contribution of each signature to each sample using COSMIC mutational signatures v2 (30 mutational signatures in total) as reference signatures. The deconstructSigs package determines the linear combination of pre-defined signatures (i.e., reference signatures) that most precisely reconstructs the mutational profile of a single tumor sample.[23] It uses a multiple linear regression model with the restriction that every coefficient must be >0, as negative contributions make no biological sense.
2.4. HPV typing and HPV integration detection
HPV typing and HPV integration detection were performed with FastViFi.[24] The dominant HPV type was determined as the type with the most read evidence. HPV integration sites were identified with chimeric reads mapping to both human and HPV genomes. Within each sample, integration sites were merged into a single integration event if they were <500 kb apart. For each integration event, we identified the genes that was interupted or nearby genes which fell within the event ± 10 kb.
2.5. Public dataset acquisition and preprocessing
The MAF files and clinical information of CC patients were obtained from the TCGA database (https://portal.gdc.cancer.gov), which contains the mutation results of 289 samples. Since TCGA uses whole-exome sequencing (WES), while this project used the whole-genome sequencing, we only included mutations within the WES coding regions (TCGA used the Agilent SureSelect whole-exome capture kit of size 35.8 MB).
2.6. Gene ontology and KEGG pathway enrichment analysis
Gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database were used to perform functional enrichment analysis. DAVID analysis tool (https://david.ncifcrf.gov/, The DAVID Knowledgebase version v2023q2) was used to conduct over-representation test.[25] We used FDR<=0.05 as criteria to select significantly enriched Geng ontology terms and KEGG pathways. FDR was obtained by using Benjamini and Hochberg (BH) false discovery rate (FDR) algorithm.
2.7. Statistical analysis
All statistical analyses were conducted using R software (v3.32). Continuous variables were compared using Student t test or Mann–Whitney U test as indicated, while categorical variables were compared using the chi-square test. Unless otherwise specified, all statistical tests were conducted using a 2-sided alpha level of 0.05.
3. Results
3.1. Patient samples and clinical data
A total of 51 patients with CIN2/3 cervical lesions from 3 ethnic groups in China, namely Han (17/51, 33%), Yi (17/51, 33%), Bai (17/51, 33%), were enrolled in the present study. HPV + patients (35/51, 69%) were 4 years older, on average, than HPV-negative (HPV–) patients (mean, 41.5 vs 37.8 years). The demographic characteristics of these patients are summarized in Table 1.
Table 1.
Patient characteristics.
| Case No. | HPV infection | Histopathological diagnosis | Age | Ethnic group |
|---|---|---|---|---|
| YYH01 | HPV+ | CIN2 | 54 | Han |
| XSW02 | HPV+ | CIN2 | 35 | Han |
| ZZZ03 | HPV+ | CIN3 | 54 | Han |
| LZD04 | HPV+ | CIN3 | 55 | Han |
| LZX05 | HPV+ | CIN2 | 47 | Han |
| ZLP06 | HPV+ | CIN2 | 38 | Han |
| CFQ07 | HPV+ | CIN2 | 39 | Bai |
| SGL08 | HPV+ | CIN3 | 56 | Bai |
| YTF10 | HPV+ | CIN3 | 47 | Bai |
| YWC11 | HPV+ | CIN3 | 49 | Bai |
| XHT12 | HPV+ | CIN3 | 40 | Bai |
| HZK09 | HPV+ | CIN2 | 38 | Bai |
| LLJ31 | HPV+ | CIN3 | 29 | Yi |
| YSY39 | HPV+ | CIN3 | 32 | Yi |
| YPZ42 | HPV+ | CIN3 | 46 | Yi |
| PCH46 | HPV+ | CIN3 | 40 | Yi |
| MBY47 | HPV+ | CIN3 | 42 | Yi |
| BGF48 | HPV+ | CIN3 | 45 | Yi |
| ZJY13 | HPV+ | CIN3 | 56 | Han |
| SCY14 | HPV+ | CIN3 | 40 | Han |
| YYY15 | HPV+ | CIN3 | 42 | Han |
| SXJ16 | HPV+ | CIN2 | 36 | Han |
| WYP17 | HPV+ | CIN3 | 27 | Han |
| HGL18 | HPV+ | CIN2 | 41 | Bai |
| HM19 | HPV+ | CIN2 | 43 | Bai |
| XQY20 | HPV+ | CIN2 | 34 | Bai |
| ZLF21 | HPV+ | CIN3 | 28 | Bai |
| WCL22 | HPV+ | CIN3 | 42 | Bai |
| HSL23 | HPV+ | CIN3 | 45 | Bai |
| GYX13 | HPV+ | CIN3 | 56 | Yi |
| SHH16 | HPV+ | CIN3 | 47 | Yi |
| ZY19 | HPV+ | CIN3 | 28 | Yi |
| CGJ28 | HPV+ | CIN3 | 26 | Yi |
| ZLM7 | HPV+ | CIN3 | 30 | Yi |
| ZZQ32 | HPV+ | CIN3 | 46 | Yi |
| LCX24 | HPV− | CIN2 | 34 | Han |
| SMJ25 | HPV− | CIN2 | 40 | Han |
| WXT26 | HPV− | CIN2 | 35 | Han |
| YJL27 | HPV− | CIN2 | 42 | Han |
| YSY28 | HPV− | CIN3 | 45 | Han |
| HYH29 | HPV− | CIN3 | 34 | Han |
| ZBF30 | HPV− | CIN2 | 40 | Bai |
| YYQ31 | HPV− | CIN2 | 40 | Bai |
| LJ32 | HPV− | CIN2 | 42 | Bai |
| ZXM33 | HPV− | CIN3 | 40 | Bai |
| YLH34 | HPV− | CIN3 | 43 | Bai |
| ZXF18 | HPV− | CIN2 | 27 | Yi |
| FJL28 | HPV− | CIN3 | 58 | Yi |
| LXR30 | HPV− | CIN3 | 20 | Yi |
| WSM40 | HPV− | CIN3 | 29 | Yi |
| LLH37 | HPV− | CIN3 | 36 | Yi |
CIN = cervical intraepithelial neoplasia, HPV = human papilloma virus.
3.2. Genomic alterations in CIN2/CIN3 samples
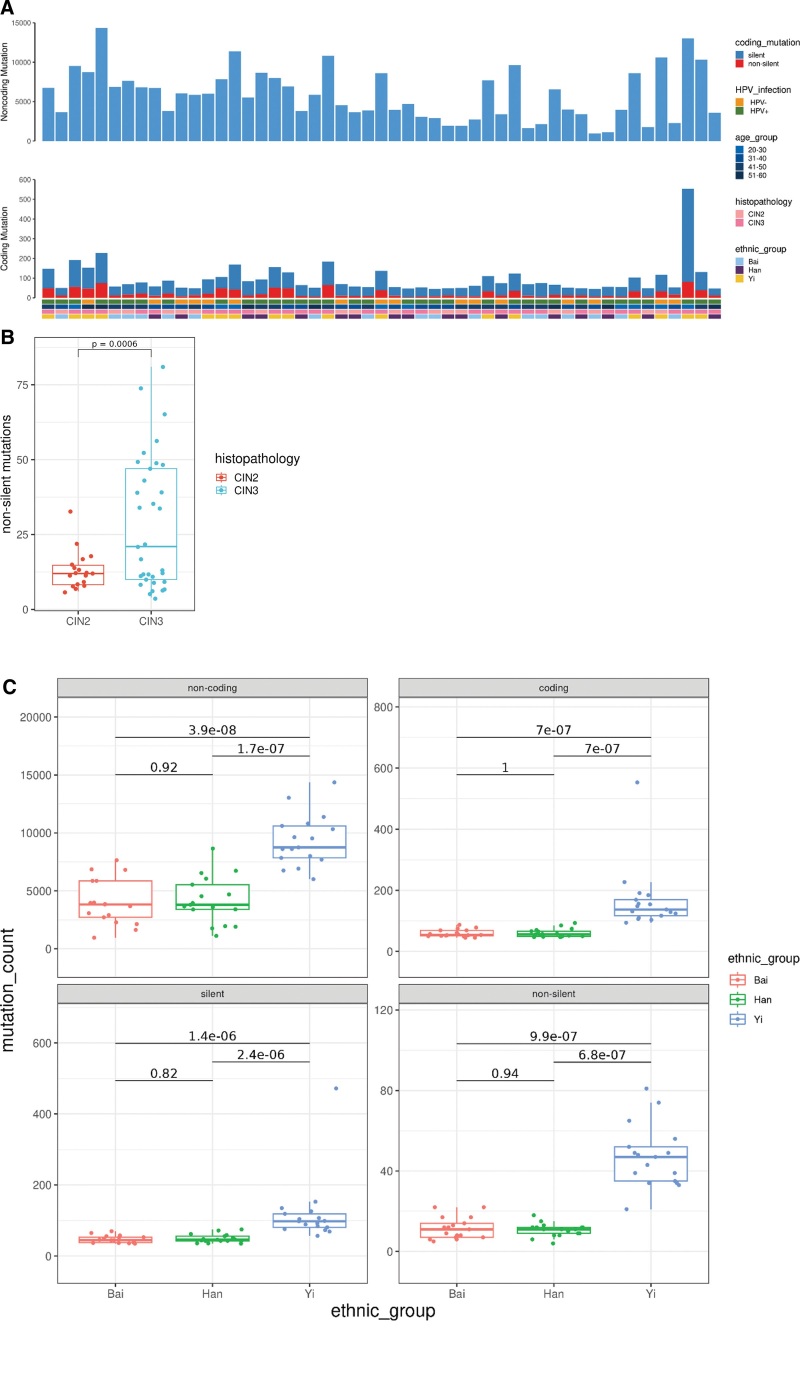
To explore the profiles of molecular variants in CIN samples, we selected 51 CIN2/CIN3 samples and matched peripheral blood. We then performed whole-genome sequencing with the NovaSeq 6000 system. The average depth of coverage for exfoliated cervical cells was 32 × (range, 25–39×) and the average depth of coverage for the matched peripheral blood cells was 20 × (range, 17–26×). The sequencing depth for each sample was listed in Supplementary Table 1, http://links.lww.com/MD/K588. Whole-genome sequencing of samples from our discovery cohort identified an average of 5942 somatic mutations (range, 1003-14,583) per sample, including 95 coding mutations (range, 45–553), of which an average of 23 (range, 4–80) is non-silent mutations that were predicted to alter protein-coding sequence (including missense, nonsense, readthrough, Splice Site SNVs and small coding insertions or deletions) (see Fig. 1A). The non-silent mutations detected in each CIN samples were listed in Supplementary Table 2, http://links.lww.com/MD/K589. Notably, we found CIN2 samples had significantly less non-silent mutations than CIN3 samples (P = .0006, Student t test, see Fig. 1B), suggesting that somatic mutations accumulate as the neoplasia progress to the higher grade. Additionally, we compared the genomic alterations in our CIN patients from 3 different ethnic groups. We found that CIN patients from Yi harbored significantly more non-coding, coding, silent and non-silent mutations than patients from Han and Bai, while we found no difference in patients from the latter 2 ethnic groups, Han and Bai (see Fig. 1C). This suggested that ethnicity may have an impact on the generation of the genomic variations. Considering the small sample size of 3 ethnic groups in this study, more ethnic groups including ethnic groups in other countries and larger sample size would be needed to reveal the impact of ethnicity on genomic variations.
Figure 1.
Somatic mutations in 51 high-grade cervical intraepithelial neoplasia genomes by whole-genome sequencing. (A) Non-coding and coding somatic mutation counts by HPV infection status, age group, histopathology and ethnic groups. (B) Comparison of non-silent mutations between CIN2 and CIN3 samples. HPV, human papillomavirus; CIN, cervical intraepithelial neoplasia. (C) Comparison of non-coding, coding, silent and non-silent mutations among CIN patients from 3 ethnic groups, Han, Bai, Yi.
3.3. Mutational signatures in CIN2/CIN3 samples
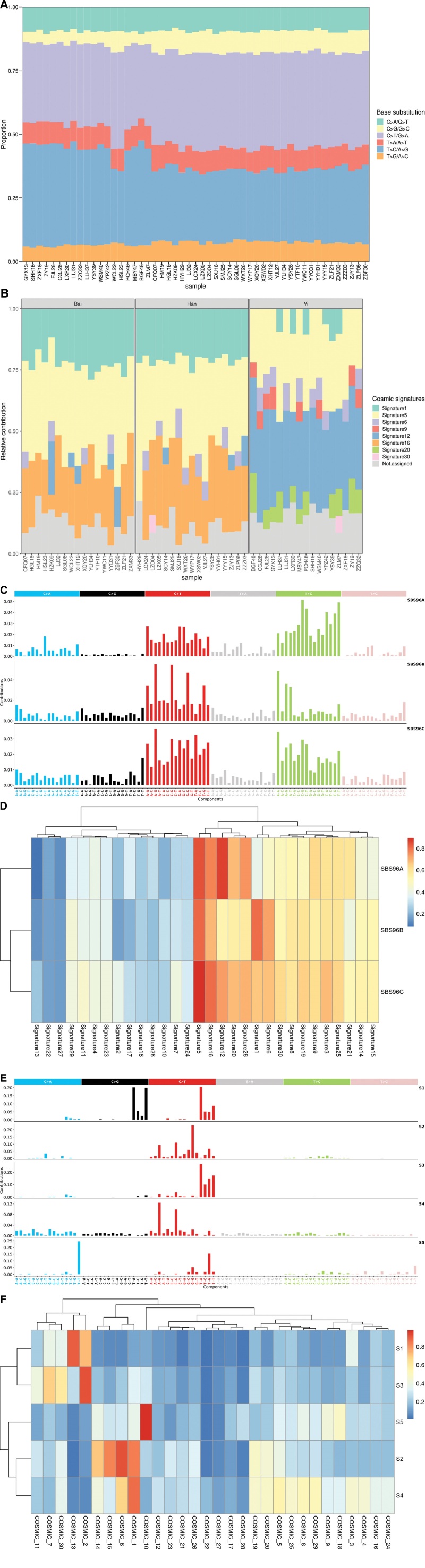
In this study, we found that C > T/G > A and T > C/A > G transitions dominated mutation spectrum in both CIN2 and CIN3 samples (Fig. 2A), which was consistent with a previous report.[26] Mutational signatures reveal the specific mutational processes. It is well known that a variety of mutational processes underlie the development of cancer, with potential implications for understanding of cancer etiology, prevention, and therapy. We thus tried to reveal the mutational signatures in CIN samples using 2 strategies. First, we employed the deconstructSigs approach which determines the linear combination of pre-defined signatures that most accurately reconstructs the mutational profile of a single tumor sample. The proportions of the COSMIC v2 reference mutational signatures in each CIN sample were shown in Figure 2B. We found that signature 5 was most common in the CIN samples, followed by signature 1, 12 and 16. The signature 6, 9, 20 and 30 were also present in a proportion of samples. The etiology of signature 5, 12, 16 and 30 remains unknown. Signature 1 is the result of an endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine. The signature 6 and 20 are associated with defective DNA mismatch repair and are found in microsatellite unstable tumors. The signature 9 is characterized by a pattern of mutations that has been attributed to polymerase η, which is implicated with the activity of AID during somatic hypermutation. When we compared the mutational signatures among CIN patients from 3 ethnic groups, we discovered that Han and Bai patients had similar mutational signature contributions, while Yi patients had clearly distinct mutational signature constitutions (Fig. 2B). Again, given the small sample size of 3 ethnic groups in this study, more ethnic groups including ethnic groups in other countries and larger sample size would be required to disclose the impact of ethnicity on mutational signatures. Next, we used a nonnegative matrix factorization (NMF) based tool SigProfilerExtractor to identify de novo mutation signatures in CIN samples. we identified 3 de novo mutational signatures (SBS96A, SBS96B, SBS96C; Fig. 2C) in our CIN cohort, which are related to signatures described in the COSMIC v2 reference mutational signatures, as reflected by cosine similarity scores (Fig. 2D). SBS96A which accounts for 40.0% somatic mutations in our CIN cohort is related to signature 5, 12, 16 and 26. The etiology of signature 26 is believed to be associated with defective DNA mismatch repair. The etiology of other signatures has been described above. SBS96B which accounts for 34.9% somatic mutations is related to signature 1, 5 and 6. The etiology of these signatures has also been described. SBS96C which accounts for 25.1% somatic mutations is related to signature 5, 12, 16 and 26. The etiology of these signatures has already been described. Although we used 2 orthogonal approaches to investigate the mutation signatures in our CIN cohort, we got similar results. In comparison with the mutational signatures of CC, we extracted the de novo mutational signatures using SigProfilerExtractor for the CC cohort in the TCGA dataset. We identified 5 de novo mutational signatures (S1-S5) that, respectively, account for 36.2%, 28.2%, 17.5%, 9.6% and 8.6% somatic mutations in the TCGA CC cohort (Fig. 2E). We then calculated their similarity with the COSMIC V2 reference mutational signatures (Fig. 2F). S1 is related to signature 13, whose proposed etiology has been attributed to activity of the AID/APOBEC family of cytidine deaminases converting cytosine to uracil. S2 is related to signature 6. S3 is related to signature 2, whose proposed etiology has also been attributed to activity of the AID/APOBEC family of cytidine deaminases. S4 is related to signature 1. S5 is related to signature 10, whose proposed etiology is the altered activity of the error-prone polymerase POLE. Overall, these findings suggested that the predominant mutation signatures in CCs were dramatically distinct from CINs, highlighting their different mutational processes and etiologies.
Figure 2.
The mutation spectrums and mutational signatures in CIN2/3. (A) Distribution of 6 base substitution types in 51 CIN2/3. The x-axis indicated the patients ID, and the y-axis depicted the proportion of each mutation categories. (B) Mutational signature profiles identified with deconstructSigs tool in 51 CIN2/3. The x-axis indicated the patients ID, and the y-axis showed the proportion of each mutational signatures. (C) De novo mutational signatures identified with SigProfilerExtractor tool in 51 CIN2/3. (D) Heatmap of cosine similarity between extracted de novo mutational signatures in CIN2/3 and COSMIC v2 reference mutational signatures. (E) De novo mutational signatures identified with SigProfilerExtractor tool in TCGA-CESC. (F) Heatmap of cosine similarity between extracted de novo mutational signatures in TCGA-CESC and COSMIC v2 reference mutational signatures. CIN, cervical intraepithelial neoplasia; TCGA-CESC, the Cancer Genome Atlas cervical squamous cell carcinoma and endocervical adenocarcinoma.
3.4. Significantly mutated genes in CIN2/CIN3 samples
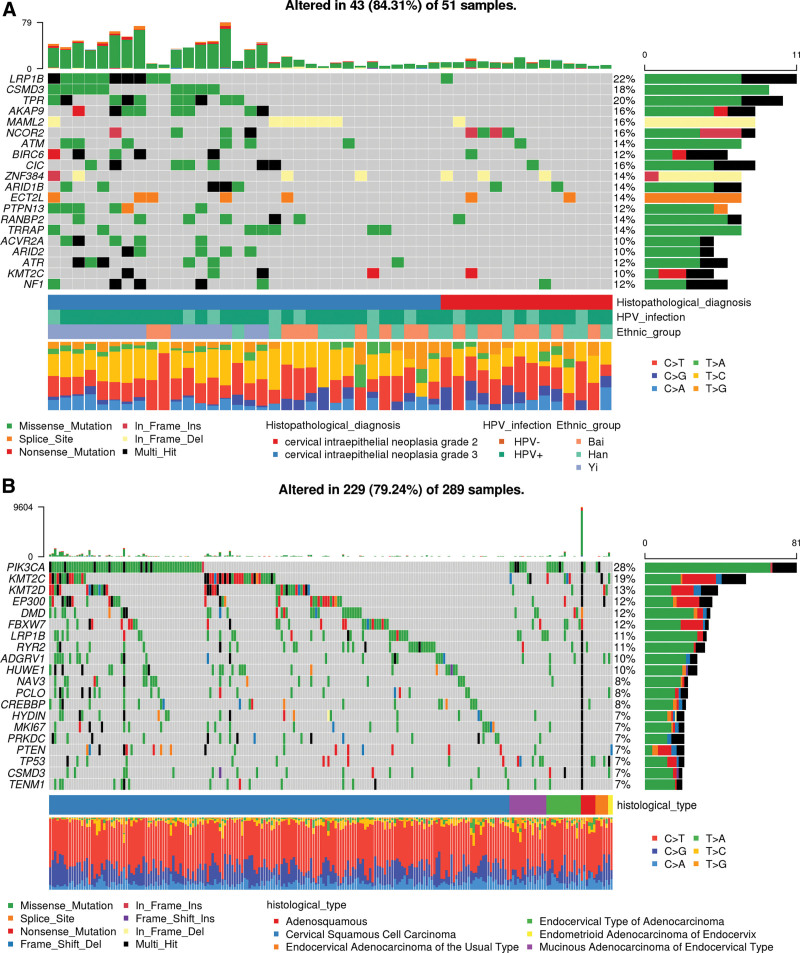
The top 20 most frequently mutated genes are shown in Figure 3A. As a comparison, the top 20 most frequently mutated genes in the Cancer Genome Atlas cervical squamous cell carcinoma and endocervical adenocarcinoma (TCGA-CESC) cohort are shown in Figure 3B. Three genes, LRP1B, CSMD3, KMT2C, were found to be shared by the top 20 most frequently mutated genes in our CINs cohort and TCGA-CESC cohort. In the TCGA-CESC cohort, fourteen genes were identified as significantly mutated genes (SMGs) with false discovery rates (FDR) < 0.1 using the MutSig2CV algorithm. These genes are PIK3CA, EP300, FBXW7, HLA-B, PTEN, NFE2L2, ARID1A, KRAS, MAPK1, SHKBP1, ERBB3, CASP8, HLA-A and TGFBR2.[15] Using MutSig2CV algorithms, we identified 6 significantly mutated genes (SMGs), CDKN2A, PIK3CB, FAM20A, RABEP1, TMPRSS2 and SS18L1. The detailed MutSig2CV analysis result was listed in Supplementary Table 3, http://links.lww.com/MD/K590. Of the 6 SMGs identified in our cohort, none of them were reported as SMGs in CCs from the TCGA cohort. Although these SMGs were novel in CIN or CC, one of them, CDKN2A, is a well-known tumor suppressor gene. One study reported CDKN2A can inhibit cell proliferation and invasion in CC through LDHA-mediated AKT/mTOR pathway.[27] Somatic mutations in another novel SMG, PIK3CB, was identified in a small patient cohort of metastatic castrate-resistant prostate cancer (mCRPC).[28] In addition, to investigate the potential impact of ethnicity on SMGs, we tried to identify SMGs using somatic mutations in CIN patients from each ethnic groups. We identified PIK3CB, CDKN2A, TMPRSS2 as SMGs that passed FDR < 0.1 criterion in Yi patients. However, we didn’t identify any SMGs that passed FDR < 0.1 criterion in Han and Bai, possibly because their sample sizes were too small and we detected much less genomic variations in them than in Yi. The detailed MutSig2CV analysis result for each ethnic group was listed in Supplementary Table 4&5&6, http://links.lww.com/MD/K591, http://links.lww.com/MD/K592, http://links.lww.com/MD/K593. Overall, we had identified 6 novel SMGs in our CIN cohort and some of them were found to play important roles in the development and progression of various cancer types. However, considering that the sample size in our study may not be large enough, future studies with a larger sample size of CINs may be needed to validate our findings.
Figure 3.
Landscapes of frequently mutated genes in our CIN2/3 cohort and in cervical cancer from TCGA. (A) Landscapes of frequently mutated genes in our CIN2/3 cohort. (B) Landscapes of frequently mutated genes in cervical cancer from TCGA. CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus.
3.5. Significant enriched gene ontology terms and pathways in CIN2/CIN3 genomes
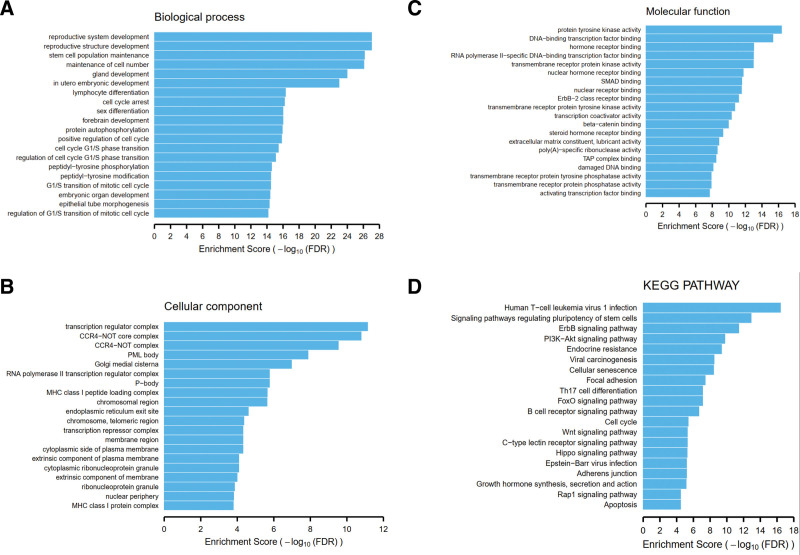
In order to investigate pathway level relationship of the mutated genes in CINs, we performed the functional enrichment analysis using the DAVID tool. As for gene ontology, we found that those genes mutated in the CINs were significantly associated with many tumorigenesis-related gene functions, including: stem cell population maintenance, cell cycle arrest, positive regulation of cell cycle, cell cycle G1/S phase transition, peptidyl-tyrosine phosphorylation (see Fig. 4A); transcription regulator complex, RNA polymerase II transcription regulator complex, chromosome and telomeric region, transcription repressor complex (see Fig. 4B); protein tyrosine kinase activity, DNA-binding transcription factor binding, transmembrane receptor protein kinase activity, ErbB-2 class receptor binding, beta-catenin binding (see Fig. 4C). As for KEGG pathway, consistent with gene ontology analysis, we noted that mutated genes were enriched for many tumorigenesis-related pathways: ErbB signaling pathway, PI3K-Akt signaling pathway, Viral carcinogenesis, Cellular senescence, Cell cylcle, Wnt signaling pathway (see Fig. 4D). Therefore, functions of mutated genes were associated with tumorigenesis, thus these genes may play important roles in the development and progression of CC.
Figure 4.
Functional enrichment analysis of mutated genes with DAVID tool. (A) Enrichment analysis using biological process gene ontology terms. (B) Enrichment analysis using cellular component gene ontology terms. (C) Enrichment analysis using molecular function gene ontology terms. (D) Enrichment analysis using KEGG pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.6. HPV DNA integrations in CIN2/CIN3 samples
To study the relationship between HPV integration in the human genome and the occurrence and development of CC, we analyzed HPV integration events in 35 HPV + CIN samples. A total of 25 HPV integration breakpoints were identified in 28.6% (10/35) of the HPV + CIN samples. HPV integration events seems to be more frequently detected in CIN3 (7 out of 18) than CIN2 (3 out of 17) samples (P = .164, Chi-squared test). In addition, we found that HPV integration breakpoints had more supported reads in CIN3 samples than those in CIN2 samples (P = .017, Mann–Whitney U test). Interestingly, we found that the integrations of certain HPV strains which were common in the CCs, including HPV16, 52, 58, and 68, had also occurred in the CIN2/CIN3 samples. Furthermore, of the 25 integration events we detected, HPV integration sites were found to locate within the exons or introns of many human genes, including ERBB4, LINC00486, LOC105377621, RPS6KB1, RASSF6, COL9A1, LINC00486, PLEKHM2, MGAT4C, ZNF586, FAM160A1, ARMC10. The detailed information about the detected integration events was listed in Table 2. Taken together, these data suggest that oncogenic HPV integration events may occur early in precancerous lesions and this could be the driving force to the development and progression of CC.
Table 2.
Human papilloma virus types and integration events in 35 human papilloma virus positive grade 2/3 cervical intraepithelial neoplasia samples.
| Case No. | Histopathology | HPV types | HPV integration sites and interrupted or flanking genes |
|---|---|---|---|
| YYH01 | CIN2 | HPV16 (1181) | NA |
| XSW02 | CIN2 | HPV68 (312); HPV16 (299) HPV52 (177) | NA |
| LZX05 | CIN2 | HPV16 (12373) | chr2:211459533-211459833, intronic, ERBB4; chr2:211489692-211489992, intronic, ERBB4 |
| ZLP06 | CIN2 | HPV51 (6317); HPV42 (1689) HPV16 (705) | chr12:66057591-66057891, intergenic, flanking genes (LINC02425;LLPH) |
| CFQ07 | CIN2 | HPV16 (116) | NA |
| HZK09 | CIN2 | HPV16 (842) | chr12:66057592-66057892, intergenic, flanking genes (LINC02425;LLPH); chr2:145665788-145666088, intergenic, flanking genes (TEX41;PABPC1P2); chr2:145935669-145935669, intergenic, flanking genes (TEX41;PABPC1P2) |
| SXJ16 | CIN2 | HPV39 (480) | NA |
| HGL18 | CIN2 | HPV111 (260) | NA |
| HM19 | CIN2 | HPV62 (8938) | NA |
| XQY20 | CIN2 | HPV39 (1144) | NA |
| ZZZ03 | CIN3 | HPV58 (4208) | NA |
| LZD04 | CIN3 | HPV81 (253); HPV62 (240) HPV16 (132) | NA |
| SGL08 | CIN3 | HPV16 (653) | NA |
| YTF10 | CIN3 | HPV59 (410) | NA |
| YWC11 | CIN3 | HPV62 (26605); HPV42 (15451) HPV89 (9480) | chr2:32916328-32916628, ncRNA_intronic, LINC00486; chr4:102485149-102485449, ncRNA_intronic, LOC105377621 |
| XHT12 | CIN3 | HPV52 (1299) | chr17:59930144-59930444, exonic, RPS6KB1; chr4:73618433-73618733, intronic, RASSF6; chr4:73799658-73799958, intergenic, flanking genes (CXCL8;CXCL6) |
| LLJ31 | CIN3 | HPV52 (33629) | NA |
| YSY39 | CIN3 | HPV102 (260) | NA |
| YPZ42 | CIN3 | HPV54 (990); HPV52 (120) | NA |
| PCH46 | CIN3 | HPV98 (610) | NA |
| MBY47 | CIN3 | HPV84 (278) | NA |
| BGF48 | CIN3 | HPV28 (814); HPV16 (411) | NA |
| ZJY13 | CIN3 | HPV58 (399) | NA |
| SCY14 | CIN3 | HPV58 (53106) | chr6:70255733-70256033, intronic, COL9A1 |
| YYY15 | CIN3 | HPV16 (4879) | NA |
| WYP17 | CIN3 | HPV68 (62927); HPV62 (44075) HPV87 (9292) | chr12:66057590-66057890, intergenic, flanking genes (LINC02425;LLPH); chr2:32916285-32916585, ncRNA_intronic, LINC00486 |
| ZLF21 | CIN3 | HPV52 (34916) | NA |
| WCL22 | CIN3 | HPV39 (15371); HPV16 (85) | chr1:66791073-66791373, intergenic, flanking genes (TCTEX1D1;INSL5) |
| HSL23 | CIN3 | HPV53 (91985) | NA |
| GYX13 | CIN3 | HPV31 (1148) | NA |
| SHH16 | CIN3 | HPV51 (260) | NA |
| ZY19 | CIN3 | HPV52 (557) | NA |
| CGJ28 | CIN3 | HPV39 (165899) | chr1:10831515-10831815, intergenic, flanking genes (CASZ1;C1orf127); chr1:15700754-15701054, intronic, PLEKHM2; chr12:30372396-30372696, intergenic, flanking genes (TMTC1;IPO8); chr12:86214572-86214872, intronic, MGAT4C; chr19:57769481-57769781, UTR5, ZNF586; chr6:126907436-126907736, intergenic, flanking genes (MIR588;RSPO3); chr9:111836839-111837139, intergenic, flanking genes (SHOC1;UGCG) |
| ZLM7 | CIN3 | HPV16 (11901) | NA |
| ZZQ32 | CIN3 | HPV52 (50790) | chr3:66859023-66859323, intergenic, flanking genes (LOC105377143;KBTBD8); chr3:104552877-104553177, intergenic, flanking genes (MIR548AB;ALCAM); chr4:151425628-151425928, intronic, FAM160A1; chr7:103090623-103090923, intronic, ARMC10 |
CIN = cervical intraepithelial neoplasia, HPV = human papilloma virus, NA = not available. Numbers in brackets represent sequencing reads assigned to HPV strains.
4. Discussion
Here we report a comprehensive analysis of genomic alterations and HPV integrations in CINs of Chinese patients. The aim of this study is to investigate both genomic alterations and viral integration profiles in CINs to document the potential mechanisms underlying the development of CC and provide potential biomarkers for screening, diagnosis and treatment of CINs and CCs.
Although it is known that CIN is a true neoplasm and often progresses to CC, the genetic alterations of CIN are largely unknown. There have been few studies that have attempted to investigate the somatic mutations that can drive normal cervical epithelial cells into neoplastic cells. But the sample sizes of these studies were relatively small. Meanwhile, it is largely unknown whether HPV infection status would affect CIN somatic mutation profiles. These facts prompted us to dissect CIN genome in this study. The aim of this study was twofold. First, we sought to uncover somatic mutations in the whole genome of CIN. Second, we attempted to reveal genomic differences between CIN and CC that might drive CIN to CC. We found that CIN harbored fewer somatic mutations than CC. More importantly, CIN harbored much fewer driver mutations than CC. Our data indicate that CINs have quantitatively and qualitatively naive genomes, which may require additional molecular events to evolve into CC genomes.
We report comprehensive mutation profile of CINs and compare the mutation profiles of CINs from our study with those of CCs in the TCGA cohort. It is worth noting that significantly mutated genes identified in TCGA-CESC cohort, such as PIK3CA, EP300, FBXW7, HLA-B, PTEN, NFE2L2, ARID1A, KRAS, MAPK1, SHKBP1, ERBB3, CASP8, HLA-A and TGFBR2, were not identified as significantly mutated genes (SMGs) in our CIN cohort. Moreover, TP53 mutation, which is very common in various cancers, was rarely identified in both CC and CINs. These observations were consistent with previous study that low mutation frequency of the TP53 and PIK3CA genes were found in CIN3 samples.[29] Interestingly, in our CIN cohort, we found somatic mutations in the most frequently mutated genes in TCGA-CESC cohort, such as PIK3CA, KMT2C, KMT2D, FBXW7, EP300, though their frequencies were much lower than in TCGA-CESC cohort. There are a large number of studies to investigate genomic profiles in CC patients, but very few studies to investigate genomic alterations in CIN patients. In a study conducted by Nikolai V. Litviakov et al, they evaluated 65 key driver mutations of the main oncogenes and tumor suppressor genes in 5 high grade squamous intraepithelial lesion (HSIL) patients who were later diagnosed with squamous-cell carcinoma and detected 18 different somatic driver mutations in the NRAS, EGFR, BRAF, KRAS, IDH2 oncogenes and TP53 suppressor genes in 4/5 HSIL cases.[30] We also found a total of 6 somatic mutations in EGFR, BRAF and IDH2 genes in 6/51 patients from our CIN cohort, but none of the mutations were shared between our cohort and Nikolai V. Litviakov cohort, suggesting ethnicity may have an impact on the heterogeneity of genomic variations.
We identified HPV integration in CIN2/3 samples with HPV infection, which was consistent with previous reports.[31] The different types of virus integration spectra were observed in different stages of cervical samples. HPV16 and HPV18 are the dominant HPV types integrated into CC samples. Integration of multiple HPV types, including HPV16, HPV52, HPV58 and HPV68, has been identified in CIN2/3 samples in our study. Large-scale screening of CC showed that HPV 16, 33 and 58 were the most common HPV types in high-grade squamous intraepithelial lesions in Chinese women,[32] which supported our finding in a relative small CIN2/3 cohort that HPV 18 type may not be the dominant type in precancerous lesions in patients from China. Recently, there were a few studies investigating genome-wide profiles of HPV integration in CCs and CINs. In the study conducted by Zheng Hu et al, they used high-throughput viral integration detection (HIVID) strategy to perform genome-wide profiling of HPV integration in CCs, CINs and cell line samples.[33] As reported in their manuscript, HIVID detected 10 times more integration breakpoints than whole-genome sequencing (WGS) did (145 vs 11) in the HPV-positive cell lines SiHa and HeLa, as well as 2 cervical carcinomas. This finding suggests that, compared with WGS, which may miss low-abundance HPV integrations, HIVID is a more sensitive method for genome-wide survey of HPV integration breakpoints. They also reported that during the progression of cervical lesions, the rate of HPV integration rose markedly from 53.8% (14 of 26) of CINs to 81.7% (85 of 104) of cervical carcinomas. And the number of integrations increased from a median of 1 integration per case in CINs (range 0–49) to a median of 8 integrations per case in cervical carcinomas (range 0–599). These 2 facts may explain why we detected significantly less integration events in our CIN patients. In line with our findings, they also reported lower HPV integration rate in CIN2 (4/9, 44.4%) than in CIN3 (5/7, 71.4%). In another study conducted by Jian Huang et al, they performed HPV probe-capture sequencing (HPCS) in 25 normal cervical samples, 44 CIN samples (including 24 CIN1 and 20 CIN2-3 samples) and 45 CC samples.[34] They identified a total of 2466 HPV integration breakpoints in 84.3% (75/89) of the abnormal samples, including 97.8% (44/45) of the CC samples and 70.5% (31/44) of the CIN samples. Since they used HPV probe-capture sequencing technology which is more sensitive for integration profiling as reported by Zheng Hu study, they reported higher integration prevalence than our study (70.5% vs 28.6%). In line with our findings, they reported much lower HPV16 and HPV18 integration events in CINs than in CCs. In their study, they also identified HPV51, HPV52, HPV58, HPV68 integration events in CINs as reported in our study. In addition, they found that HPV integration breakpoints had more supported reads in CC samples than those in the normal samples, CIN1 samples and CIN2-3 samples, which was consistent with our findings that HPV integration breakpoints in CIN3 had significantly more supported reads than in CIN2. It is worth noting that different integrated HPV types may indicate different outcomes of CIN samples, which require further follow-up and validation for these patients. Novel disease-specific biomarkers, such as gene mutations and HPV integrations, may serve as secondary markers after positive HPV DNA tests to identify women with prevalent precancers who require immediate colposcopy or treatment.
In summary, we systematically analyzed the somatic mutation profiles and HPV integration profiles of CINs from Chinese patients. Our data suggest that CIN genomes contain quantitatively and qualitatively less aggressive alterations than CC genomes. The distinct genomic features of CIN from CC provide a useful resource for understanding this latent progressive disease and identifying molecular clues for better diagnosis and treatment options of CIN and CC. Overall, our findings provide the basis for developing new tools for screening, diagnosis and treatment of CIN and CC.
Author contributions
Conceptualization: Fuhui Duan, Guangming Wang.
Data curation: Jingjing Wang, Menghuan Li, Lixian Zhao.
Formal analysis: Jingjing Wang, Menghuan Li, Lixian Zhao.
Funding acquisition: Guangming Wang.
Methodology: Jingjing Wang, Menghuan Li, Lixian Zhao.
Software: Jingjing Wang, Bingjie Zhou, Huaqiu Chen.
Supervision: Guangming Wang.
Writing – original draft: Jingjing Wang, Menghuan Li, Lixian Zhao.
Writing – review & editing: Fuhui Duan, Guangming Wang.
Supplementary Material
Abbreviations:
- CC
- cervical cancer
- CIN
- cervical intraepithelial neoplasia
- HPV
- human papilloma virus
- SBS
- single base substitutions
- SMGs
- significant mutated genes
- TCGA-CESC
- the Cancer Genome Atlas cervical squamous cell carcinoma and endocervical adenocarcinoma
JW, ML, & LZ contributed equally to this work.
This study was supported by the Yunnan Provincial Natural Science Foundation project (grant number 202001BA070001-133), the Medical discipline leader of Yunnan Provincial Commission of Health and Family Planning (grant number D-2017057), the Yunnan Provincial Key Laboratory of Reproductive Health Research of Department of Education, and the key construction disciplines of The First Affiliated Hospital of Dali University.
Supplemental Digital Content is available for this article.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
How to cite this article: Wang J, Li M, Zhao L, Zhou B, Chen H, Duan F, Wang G. Whole genome sequencing in high-grade cervical intraepithelial neoplasia patients from different ethnic groups in China. Medicine 2023;102:45(e35953).
Contributor Information
Jingjing Wang, Email: wgm1991@dali.edu.cn.
Menghuan Li, Email: 1391904878@qq.com.
Lixian Zhao, Email: 269228896@qq.com.
Bingjie Zhou, Email: 985378815@qq.com.
Huaqiu Chen, Email: 275958206@qq.com.
Fuhui Duan, Email: 1427354417@qq.com.
References
- [1].Arbyn M, Castellsagué X, de Sanjosé S, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–86. [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J, Shin H, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. [DOI] [PubMed] [Google Scholar]
- [3].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [4].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [5].Walboomers JM, Jacobs VM, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. [DOI] [PubMed] [Google Scholar]
- [6].Steenbergen RDM, Snijders PJF, Heideman DAM, et al. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405. [DOI] [PubMed] [Google Scholar]
- [7].Snijders PJ, Steenbergen RD, Heideman DA, et al. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–64. [DOI] [PubMed] [Google Scholar]
- [8].McCredie M, Sharples K, Paul C, et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9:425–34. [DOI] [PubMed] [Google Scholar]
- [9].Vink MA, Bogaards JA, Kemenade FJ, et al. Clinical progression of high-grade cervical intraepithelial neoplasia: estimating the time to preclinical cervical cancer from doubly censored national registry data. Am J Epidemiol. 2013;178:1161–9. [DOI] [PubMed] [Google Scholar]
- [10].Peto J, Gilham C, Fletcher O, et al. The cervical cancer epidemic that screening has prevented in the UK. Lancet. 2004;364:249–56. [DOI] [PubMed] [Google Scholar]
- [11].Szarewski A, Ambroisine L, Cadman L, et al. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;17:3033–42. [DOI] [PubMed] [Google Scholar]
- [12].Cuschieri K, Wentzensen N. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2008;17:2536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Verhoef VM, Bosgraaf RP, Kemenade FJ, et al. A randomised controlled trial on triage of HPV positive women on self-collected cervicovaginal specimens: direct methylation marker testing versus indirect cytology. Lancet Oncol. 2014;15:315–22. [DOI] [PubMed] [Google Scholar]
- [14].Ojesina AI, Lichtenstein L, Freeman SS, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2013;506:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Islam SMA, Díaz-Gay M, Wu Y, et al. Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor. Cell Genom. 2022;2:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Javadzadeh S, Rajkumar U, Nguyen N, et al. FastViFi: fast and accurate detection of (Hybrid) viral DNA and RNA. NAR Genom Bioinform. 2022;4:lqac032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bao C, An N, Xie H, et al. Identifying potential neoantigens for cervical cancer immunotherapy using comprehensive genomic variation profiling of cervical intraepithelial neoplasia and cervical cancer. Front Oncol. 2021;11:672386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Luan Y, Zhang W, Xie J, et al. CDKN2A inhibits cell proliferation and invasion in cervical cancer through LDHA-mediated AKT/mTOR pathway. Clin Transl Oncol. 2021;23:222–8. [DOI] [PubMed] [Google Scholar]
- [28].Robinson D, Allen EMV, Wu Y, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tornesello ML, Annunziata C, Buonaguro L, et al. TP53 and PiK3CA gene mutations in adenocarcinoma, squamous cell carcinoma and high-grade intraepithelial neoplasia of the cervix. J Transl Med. 2014;12:255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Litviakov NV, Ibragimova MK, Tsyganov MM, et al. Changes in the genetic landscape during the malignization of high grade squamous intraepithelial lesion into cervical cancer. Curr Probl Cancer. 2020;44:100567. [DOI] [PubMed] [Google Scholar]
- [31].Hu Z, Zhu D, Wang W, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–63. [DOI] [PubMed] [Google Scholar]
- [32].Jing L, Zhong X, Huang W, et al. HPV genotypes and associated cervical cytological abnormalities in women from the pearl river delta region of Guangdong province, China: a cross-sectional study. BMC Infect Dis. 2014;14:388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zheng H, Zhu D, Wei W, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–63. [DOI] [PubMed] [Google Scholar]
- [34].Huang J, Qian Z, Gong Y, et al. Comprehensive genomic variation profiling of cervical intraepithelial neoplasia and cervical cancer identifies potential targets for cervical cancer early warning. J Med Genet. 2019;56:186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.