Abstract
Whole-cell assays of methane and trichloroethylene (TCE) consumption have been performed on Methylosinus trichosporium OB3b expressing particulate methane monooxygenase (pMMO). From these assays it is apparent that varying the growth concentration of copper causes a change in the kinetics of methane and TCE degradation. For M. trichosporium OB3b, increasing the copper growth concentration from 2.5 to 20 μM caused the maximal degradation rate of methane (Vmax) to decrease from 300 to 82 nmol of methane/min/mg of protein. The methane concentration at half the maximal degradation rate (Ks) also decreased from 62 to 8.3 μM. The pseudo-first-order rate constant for methane, Vmax/Ks, doubled from 4.9 × 10−3 to 9.9 × 10−3 liters/min/mg of protein, however, as the growth concentration of copper increased from 2.5 to 20 μM. TCE degradation by M. trichosporium OB3b was also examined with varying copper and formate concentrations. M. trichosporium OB3b grown with 2.5 μM copper was unable to degrade TCE in both the absence and presence of an exogenous source of reducing equivalents in the form of formate. Cells grown with 20 μM copper, however, were able to degrade TCE regardless of whether formate was provided. Without formate the Vmax for TCE was 2.5 nmol/min/mg of protein, while providing formate increased the Vmax to 4.1 nmol/min/mg of protein. The affinity for TCE also increased with increasing copper, as seen by a change in Ks from 36 to 7.9 μM. Vmax/Ks for TCE degradation by pMMO also increased from 6.9 × 10−5 to 5.2 × 10−4 liters/min/mg of protein with the addition of formate. From these whole-cell studies it is apparent that the amount of copper available is critical in determining the oxidation of substrates in methanotrophs that are expressing only pMMO.
Methanotrophs are a group of gram-negative bacteria that can utilize methane as a sole source of carbon and energy. Ecological studies have shown that methanotrophs are widespread in nature, and strains have been isolated from a variety of environments, typically at oxic-anoxic interfaces where both oxygen and methane are found (21, 46). Two general categories of methanotrophs have been identified (type I and type II) based on several characteristics, including the pattern of internal membranes, carbon assimilation pathway, and predominant fatty acid chain length. One strain, Methylococcus capsulatus Bath, has characteristics of both types and is classified as type X (15). These cells are an important sink of methane (28) and can also be used for the remediation of sites contaminated with halogenated hydrocarbons, such as trichloroethylene (TCE), congeners of dichloroethylene (DCE), and polychlorinated biphenyls, via cometabolism. The first enzyme in the pathway of methane oxidation, methane monooxygenase (MMO), is also responsible for the cometabolism of these hazardous substances. MMO, however, has been shown to exist in two different forms for a small subset of methanotrophs, most notably in some type II and type X strains, but also in one type I strain (19). At low copper-to-biomass ratios the MMO activity for these cells is located in the cytoplasmic fraction and is termed soluble methane monooxygenase (sMMO) (9, 32, 36). If the copper-to-biomass ratio increases, methane oxidation is carried out by a different form of MMO, which is associated with the membranes (particulate methane monooxygenase, or pMMO). In the environment, the typical concentration of copper in fresh groundwater is less than 10 μg/liter (0.16 μM), but it can be as high as 200 μg/liter (3 μM) for groundwater in mineralized zones, and in polluted groundwater it has been reported to be as high as 470 μg/liter (7.4 μM) (14). In some of these situations, any methanotrophs present that have the genes for sMMO may be expressing sMMO, but in others, it is possible that pMMO is the predominant form of MMO expressed, due to a high copper/biomass ratio. pMMO and sMMO have very different transformation rates and substrate ranges (6, 37), and therefore it is important to understand more clearly how copper affects the whole-cell kinetics of substrate oxidation by these strains both for the global carbon cycle and also for environmental remediation.
Furthermore, the majority of known methanotrophs do not have the genes encoding sMMO (15, 23) and therefore can only express pMMO. It is necessary to determine what environmental parameters, including copper concentrations, affect the activity of pMMO. Increased pMMO activity as measured by the oxidation of propylene in whole cells, cell extracts, and membrane fractions has been reported in M. capsulatus Bath with increased copper concentrations in the growth medium (8, 24, 25, 30, 34, 47). In recent studies, electron paramagnetic resonance spectroscopy, correlated with metal analysis and pMMO activity as measured by propylene oxidation, showed that increasing the concentration of copper in the growth medium caused an increase in pMMO activity and that copper binds to specific sites in the membrane preparations that may constitute the active site of pMMO (24, 25, 34). The phospholipid content of the membranes of M. capsulatus Bath has also been seen to change as the amount of copper increases (29). Another study has also shown that varying the amounts of several nutrients, including copper, iron, manganese, and ammonia, affects the ability of an uncharacterized mixed culture of methanotrophs to oxidize methane (4). Other physiological studies of the effect of copper on methanotrophs have shown that increasing copper in the growth medium increases cell yield and pMMO activity in Methylomicrobium albus BG8 (7). These researchers have suggested that this methanotroph may produce two forms of pMMO or one form whose activity may be regulated by the amount of bioavailable copper. Genetic analysis of chromosomal DNA from M. albus BG8 and M. capsulatus Bath show that multiple copies of the gene encoding one of the polypeptides of pMMO exist (33). It is possible that the two gene copies encode different forms of pMMO that have different amounts of copper and possibly different kinetic characteristics.
Given such information—which clearly shows that copper is important not only for regulating the expression of sMMO and pMMO by those strains that are known to express both forms but also for the activity of cells expressing only pMMO—it is important to determine more precisely how varying copper concentrations affect the abilities of cells expressing the pMMO to oxidize methane and cometabolites such as TCE. TCE is a solvent widely used for the degreasing of metal parts, scouring of textiles, and production of organic chemicals and pharmaceuticals (43). The widespread use of TCE, however, has caused extensive pollution of groundwater (45). It may be very difficult to remove TCE from contaminated soils and aquifers with conventional pump-and-treat methods because of adsorption onto the soil matrix, low soil permeability, and the presence of non-aqueous-phase liquids that may be difficult to locate. It has been suggested that it may take on the order of 100 years to remove pollutants such as TCE from contaminated areas by current pump-and-treat techniques (41). Furthermore, methods such as air stripping, the use of landfills, and incineration are undesirable, as these procedures may transfer pollution from one medium to another. A “holistic” approach to environmental remediation is being advocated whereby the polluted area is decontaminated without polluting another part of the environment (44). Biodegradation may achieve this goal, as the waste(s) can be completely mineralized to CO2 and H2O.
Much of the work carried out to study TCE degradation by methanotrophs has examined uncharacterized mixed cultures in which the predominant form of the MMO is unknown and the effect of varying nutrient concentrations on TCE degradation is difficult to ascertain (1, 2, 5, 13, 17, 39). Those studies that have examined well-characterized or pure cultures of methanotrophs have almost exclusively involved sMMO, due to the high rate of TCE turnover by cells expressing this enzyme (6, 12, 19, 26, 27, 40, 42). Studies by DiSpirito et al. and Zahn and DiSpirito have shown that pMMO can also degrade TCE, but at a much lower rate than sMMO (10, 47). Because the majority of known methanotrophs express only pMMO and, under certain environmental conditions, the copper/biomass ratio can be expected to be high, it is important to carefully measure the kinetics of both TCE and methane oxidation by pMMO. A recent paper has reported that increasing the concentration of copper in the methanotrophic growth medium can affect both methane and TCE degradation by a recently isolated marine methanotroph (35). In the present paper, the effects of available copper on the ability of Methylosinus trichosporium OB3b expressing pMMO to oxidize methane and TCE are examined. The results collected here generally agree with the results of Smith et al. (35), but in this paper, more data documenting the effect of copper are presented and we can better compare our results with those of other researchers to explain the role of copper in the activity of pMMO and how to optimize TCE degradation by pMMO.
MATERIALS AND METHODS
Chemicals and analytical techniques.
All chemicals used in the preparation of media were of reagent grade or better. The highest purity methane (>99.99%) was obtained from Matheson Gas Company. Spectrophotometric-grade TCE (>99.9% pure) was obtained from Fisher Scientific Company for the TCE degradation experiments. Distilled deionized water from a Corning Millipore D2 system was used for all experiments. All glassware was washed with detergent and then acid washed in 2 N HNO3 overnight to remove trace metals, including copper. The acid was subsequently removed by repeated rinses with distilled deionized water.
Protein measurements.
Biomass concentrations were measured in the form of protein by using the Bio-Rad protein assay kit with bovine serum albumin as a standard. The cells were digested at 90°C for 30 min in 5 N NaOH. Serial dilutions were prepared to achieve final protein concentrations within the linear range of the assay. The amount of protein is determined by measuring the absorbance at 595 nm after the Bio-Rad assay dye has been added.
Culture conditions.
M. trichosporium OB3b was grown on nitrate mineral salt medium (46) at 30°C in batch flasks shaken at 250 rpm in a methane-air atmosphere (1:2 ratio) at 1 atm of pressure. The culture medium was no more than 15% of the total flask volume to prevent mass transfer limitations of methane from affecting growth. Copper was added aseptically as Cu(NO3)2 · 2.5(H2O) after autoclaving to vary the copper concentration from 2.5 to 20 μM, and the concentration was equilibrated for at least 2 days before the media were inoculated. This strain can express both sMMO and pMMO; therefore, the lowest concentration of copper in the growth medium was purposely set at 2.5 μM to avoid expression of sMMO. For the growth of the cells, the method developed by Tsien et al. was used (42). In brief, the cells were grown to mid-exponential phase (i.e., an optical density at 600 nm [OD600] of 0.75 to 0.8). For the methane consumption experiments, the cells were then diluted to an OD600 of 0.30 with prewarmed fresh medium with the same copper concentration as the growth medium. The protein concentrations of these cell suspensions varied between 0.081 and 0.092 mg/ml. For the TCE degradation experiments, the cells were grown and harvested in a similar fashion but were diluted to an OD600 of 0.25 with prewarmed fresh medium with the same copper concentration as the growth medium. The protein concentrations in these cell suspensions varied from 0.057 to 0.059 mg/ml. To monitor expression of sMMO, the naphthalene assay specific for sMMO activity was done as described by Brusseau et al. (6) for all cell suspensions used for both TCE and methane consumption assays.
Methane and TCE degradation assays.
After cells in the exponential phase were harvested for the experiments, methane was removed from the growth flasks by evacuating the flasks five times and allowing air to reequilibrate after each evacuation. Three-milliliter aliquots were then aseptically transferred to 20-ml serum vials. The vials were capped with Teflon-coated rubber butyl stoppers (Wheaton) and sealed with aluminum crimp seals. For each concentration of methane and TCE examined in these assays, triplicate samples were created, with one control. The control was made by adding 50 μl of 5 N NaOH to lyse the cells and was used to assess abiotic disappearance of the substrate, which was less than 1.5% in 2 h for both methane and TCE.
To obtain a range of aqueous methane concentrations from 1 to 85 μM in solution, methane was added to the vials with Dynatech A-2 gastight syringes. Aqueous concentrations were calculated with a dimensionless Henry’s constant of 27.02 (22). The serum vials were then shaken at 270 rpm and 24°C. At several time points, headspace samples of 100 μl were removed from each vial, again with a Dynatech A-2 gastight syringe. The headspace samples were then injected into an HP 5890 series II gas chromatograph with a flame ionization detector (FID) and two DB-624 analytical columns (J&W Scientific Co.). The injector, oven, and detector temperatures were set at 160, 120, and 250°C, respectively.
For the measurement of TCE degradation, formate in the form of sodium formate was added in some experiments to examine what possible limitations due to lack of reducing equivalents existed in cells expressing the pMMO. After the removal of methane, TCE was added with Hamilton 1700 series gastight syringes from a bottle of TCE-saturated water solution prepared by the method of Alvarez-Cohen and McCarty (1) to obtain a range of aqueous concentrations from 1 to 65 μM. For the partitioning of TCE between the liquid space and the headspace, a dimensionless Henry’s constant of 0.42 was used (18). The serum vials were shaken at 270 rpm at 30°C. TCE remaining in the control and sample vials was measured at several time points by removing 100 μl of headspace with Dynatech A-2 gastight syringes and injecting the sample into an HP 6890 gas chromatography analyzer with an FID and a DB-5 capillary column (J&W Scientific Co.). The injector, oven, and detector temperatures were 250, 120, and 250°C, respectively. The FID was used to assay TCE in the headspace instead of the more sensitive electron capture detector because the electron capture detector gave a nonlinear response at the higher TCE concentrations needed in the experiments.
Methane and TCE degradation rates were determined by monitoring the decrease in the amounts of both substrates over an 8-h period based on the headspace analysis. The initial rate of degradation for each initial substrate concentration was calculated by using a 2-h period (from t = 0 to t = 2 h). These rates were then normalized to the initial cell concentration. The average initial degradation rate of the triplicate samples is reported here along with the standard deviation. The kinetic parameters of maximal degradation rate, Vmax (nanomoles per milligram of protein per minute) and substrate concentration at half the maximal degradation rate in whole cells, Ks (micromolar), were determined by applying nonlinear regression on the Michaelis-Menten formula with Systat V version 5.21 for the Macintosh.
RESULTS
Examination of mass transfer limitations.
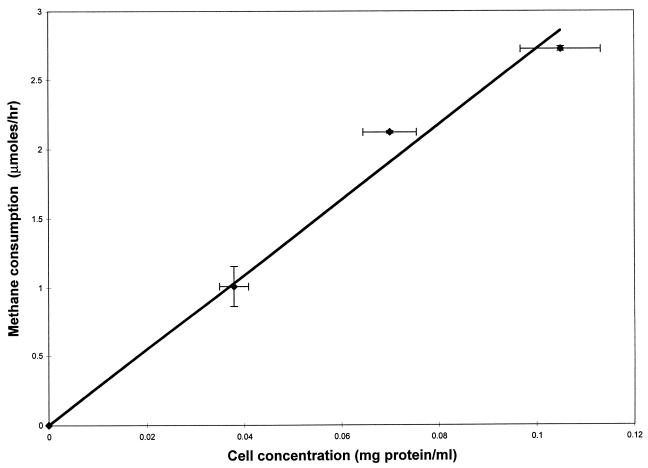
Substrate degradation in these experiments can be limited by mass transfer from the vial headspace to the liquid phase if the numbers of cells used in the experiments are large, causing the biological rate of degradation to be greater than the rate of dissolution of the substrates into the media. To obtain a cell concentration range in which the degradation experiments were performed without mass transfer limitation, methane degradation by different numbers of cells was measured. Three-milliliter aliquots of concentrations of cells ranging from 0.035 to 0.1 mg of protein/ml were transferred aseptically to 20-ml vials. The vials were then capped, and methane was added with a Dynatech A-2 gastight syringe as described earlier to obtain a methane concentration in solution of 32 μM. The methane degradation assay was then performed. As shown in Fig. 1, the rate of methane oxidation increased in proportion to the number of cells, indicating that, for the methane consumption experiments, the rate of mass transfer from the gas phase to the liquid phase was greater than the rate at which the cells oxidized methane. Similar experiments were performed for TCE. Because TCE was added as a saturated water solution, it was important to determine how long it took for TCE to partition between the gaseous and liquid phases. In abiotic control experiments, equilibrium was achieved in less than 3 min regardless of the TCE concentration (data not shown).
FIG. 1.
Rate of methane consumption by M. trichosporium OB3b as a function of cell concentration. The symbols represent the means of three samples, and the bars represent ±1 standard deviation.
Change of culture conditions over time.
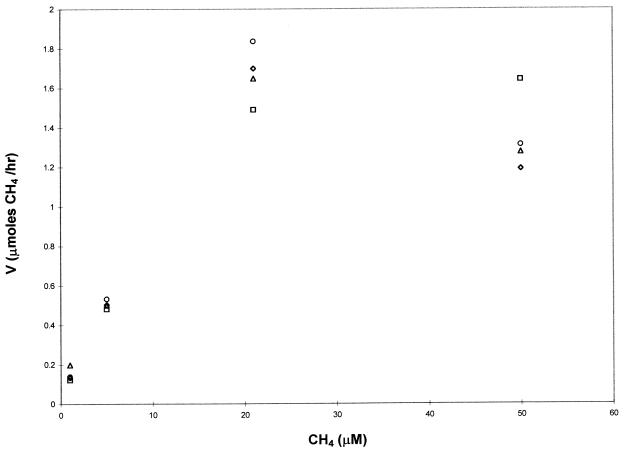
To ensure that the 2-h period utilized to calculate the initial rates of degradation was appropriate, one methane consumption experiment was run over 2 h, with samples taken every 30 min. As shown in Fig. 2, the rate of methane consumption plotted against the initial methane concentration was similar regardless of whether uptake was monitored over 30, 60, 90, or 120 min. The 2.5-min response time of the gas chromatograph, however, limited the number of samples that could be prepared and sampled within 30 min. For this experiment, only four concentrations in duplicate could be prepared and sampled as opposed to seven concentrations in triplicate with a 2-h reference frame. In order to have more data for a better statistical analysis, a 2-h period was used in the methane and TCE degradation experiments. To determine if the cells increased substantially in number over the 2-h period, cell measurements were taken before and after one methane consumption experiment. The protein concentration before the addition of methane was 0.084 ± 0.007 mg/ml, and after incubation for 2 h it was 0.089 ± 0.009 mg/ml. Such a small variation is not surprising due to the low growth rate of M. trichosporium OB3b (∼0.09 h−1). Therefore, the rates presented here are not affected by changes in biomass or the time used to calculate the Michaelis-Menten kinetic parameters.
FIG. 2.
Rate of methane consumption over 30 (diamond), 60 (triangle), 90 (circle), and 120 (square) min by M. trichosporium OB3b. The symbols represent the means of two samples.
Activity of sMMO.
As noted in a recent review (15), sMMO expression is not seen in those strains that can express sMMO at copper/biomass ratios greater than 0.89 μmol of copper/g (dry weight) of cells. Assuming that protein is 50% of the dry cell mass (31), the minimum copper/biomass ratio was 13.6 μmol of copper/g (dry weight) of cells in the methane consumption experiments. For the TCE degradation experiments, the minimum copper/biomass ratio was 21.1 μmol of copper/g (dry weight) of cells. Both these ratios are well above the maximum value above which sMMO expression is not observed. Furthermore, the naphthalene assay specific for sMMO developed by Brusseau et al. was used to monitor the expression of sMMO (6). In all experiments, the assay did not detect any naphthol production, indicating that pMMO was the only MMO expressed by these cells under the provided culture conditions.
Effect of copper on methane degradation by M. trichosporium OB3b expressing pMMO.
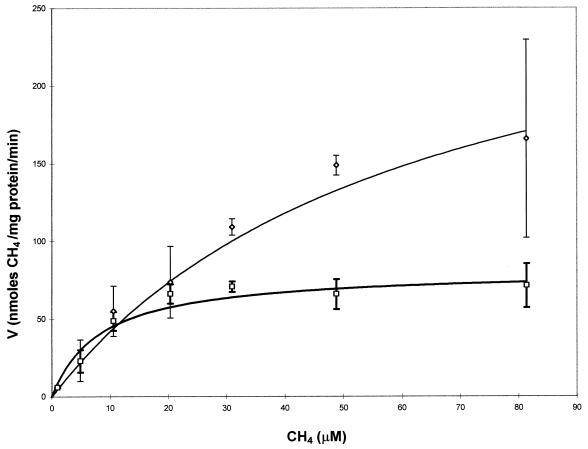
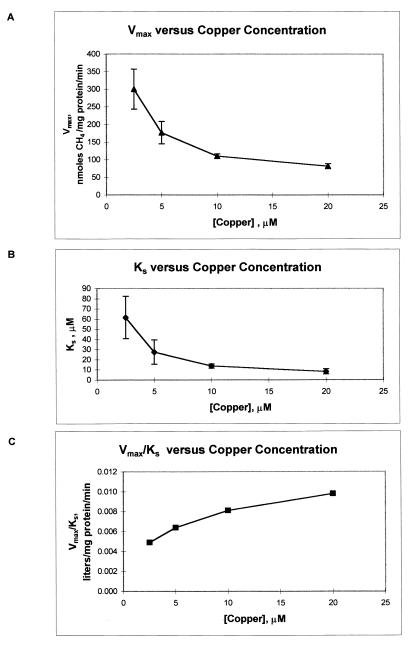
The results of the experiments to determine the effect of copper on methane degradation by M. trichosporium OB3b are shown in Fig. 3 and 4 and Table 1. In these experiments, the cells were grown with either 2.5, 5, 10, or 20 μM copper. In Fig. 3, the initial rate of methane degradation of the cells is plotted against the initial methane concentration for the lowest and highest concentrations of copper in the growth medium. From this figure it is apparent that as the concentration of copper increased, the kinetics of methane degradation changed significantly. Using nonlinear regression analysis, we fitted the methane consumption results for all the growth concentrations of copper with the Michaelis-Menten model, and the calculated parameters of Vmax and Ks are shown in Table 1. As can be seen in Fig. 3, the value of Vmax dropped from 300 ± 57 nmol of methane/mg of protein/min to 82 ± 6.7 nmol of methane/mg of protein/min as the concentration of copper in the growth medium increased from 2.5 to 20 μM. At methane concentrations below 20 μM, however, the rates of methane oxidation for all concentrations of copper were similar and were within experimental error. Also from Fig. 3, it is clear that the rate at which Vmax is approached increases appreciably as the concentration of copper in the growth medium increases. Therefore, not only does the whole-cell rate of methane oxidation by M. trichosporium OB3b expressing pMMO vary with respect to the concentration of copper, the affinity of the cells for methane as measured by the half-saturation constant, Ks, also changes. As shown in Table 1, the Ks value decreased from 62 ± 21 μM methane to 8.3 ± 2.5 μM, indicating that the cells had a higher affinity for methane as the concentration of copper increased. The decrease of the calculated values for both Ks and Vmax is shown in Fig. 4. For both kinetic parameters a noticeable decrease was seen over the range of copper concentrations examined. The pseudo-first-order rate constant, Vmax/Ks, is also shown in Table 1 and Fig. 4. Changing the concentration of copper in the growth medium caused this value to increase by 100%, clearly showing that varying concentrations of copper affect the ability of cells expressing pMMO to oxidize methane.
FIG. 3.
Michaelis-Menten plot of methane consumption by M. trichosporium OB3b grown with 2.5 (◊) or 20 (□) μM copper. The symbols represent the means of three samples, and the bars represent ±1 standard deviation.
FIG. 4.
Effect of increasing the concentration of copper in the growth medium on the maximal consumption rate (Vmax) (A), affinity (Ks) (B), and pseudo-first-order rate constant (Vmax/Ks) (C) of methane by M. trichosporium OB3b expressing pMMO. The symbols represent the means, and the bars represent ±1 standard deviation.
TABLE 1.
Calculated kinetic parameters for methane consumption by M. trichosporium OB3b expressing pMMO with varying concentrations of coppera
| Copper concn (μM) | Vmax (nmol/min/mg of protein)b | Ks (μM)b | Vmax/Ks (liters/min/mg of protein) |
|---|---|---|---|
| 2.5 | 300 (57) | 62 (21) | 4.9 × 10−3 |
| 5 | 177 (32) | 28 (12) | 6.3 × 10−3 |
| 10 | 110 (6.7) | 14 (2.1) | 7.9 × 10−3 |
| 20 | 82 (6.7) | 8.3 (2.5) | 9.9 × 10−3 |
Calculated by nonlinear least-squares regression analysis. Cell densities in these experiments ranged from 0.081 to 0.092 mg of protein/ml.
Numbers in parentheses indicate the standard deviation of three replicates.
Effect of copper on TCE cometabolism by M. trichosporium OB3b expressing pMMO.
TCE oxidation by methanotrophs can be difficult to model accurately due to limitations caused by substrate toxicity, product toxicity, and/or lack of endogenous reducing equivalents (1, 2, 16, 17). To avoid substrate toxicity, TCE concentrations were kept at levels below that found to be toxic (38). Since the TCE degradation rate generally increased with increasing TCE concentrations up to a calculated Vmax and never decreased with increasing TCE concentrations, substrate toxicity was not encountered under these conditions (up to 65 μM). To avoid problems associated with product toxicity, the time course of the experiments was designed to be sufficient to obtain initial degradation rates but not long enough to have a buildup of potentially toxic products. The effects of copper and exogenous reducing equivalents on TCE oxidation by cells expressing pMMO could then be directly assessed.
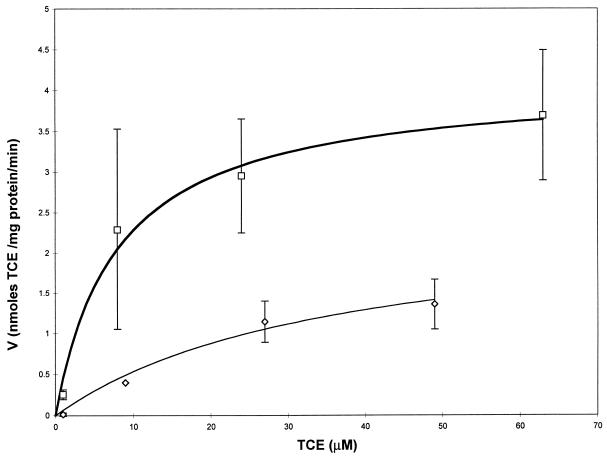
The kinetic parameters for TCE degradation by M. trichosporium OB3b with varying copper and formate concentrations are compiled in Table 2. From the TCE degradation assays, it is apparent that copper not only affects the degradation of methane by cells expressing pMMO, it also affects the degradation of TCE. TCE was not degraded to any noticeable extent by the cells grown at 2.5 μM copper even after 8 h in the absence and presence of exogenous reducing equivalents in the form of formate. When the cells were grown in the presence of 20 μM copper, TCE degradation was observed within 2 h in both the presence and absence of formate, although formate addition did have a major impact on TCE degradation, as shown in Fig. 5. In the figure, the initial rate of TCE degradation is plotted against the initial TCE concentration. The data in the figure have been fitted with a Michaelis-Menten model, and the calculated initial maximal degradation rate for TCE was seen to increase from 2.5 ± 0.7 to 4.1 ± 0.4 nmol of TCE/mg of protein/min with the addition of formate. The affinity of the cells for TCE also increased with the addition of formate, as measured by the fourfold drop in Ks (36 ± 18 μM without formate to 8 ± 2.5 μM with formate). Furthermore, the pseudo-first-order rate constant, Vmax/Ks (indicative of the rate of degradation of TCE by whole cells at concentrations much less than Ks), increased over sevenfold, from 6.9 × 10−5 to 5.2 × 10−4 liters/min/mg of protein.
TABLE 2.
Calculated kinetic parameters for TCE degradation by M. trichosporium OB3b expressing pMMO under varying concentrations of copper and formatea
| Copper concn (μM) | Formate concn (mM) | Vmax (nmol/min/mg of protein)b | Ks (μM)b | Vmax/Ks (liters/min/mg of protein) |
|---|---|---|---|---|
| 2.5 | 0 | NMc | NM | NM |
| 2.5 | 20 | NM | NM | NM |
| 20 | 0 | 2.5 (0.7) | 36 (18) | 6.9 × 10−5 |
| 20 | 20 | 4.1 (0.4) | 7.9 (2.5) | 5.2 × 10−4 |
Determined by nonlinear least-squares regression analysis. Cell densities in these experiments ranged from 0.057 to 0.059 mg of protein/ml.
Numbers in parentheses indicate the standard deviation of three replicates.
NM, no measurable TCE degradation.
FIG. 5.
Michaelis-Menten plot of TCE consumption by M. trichosporium OB3b grown with 20 μM copper and either with or without an exogenous source of reducing equivalents in the form of formate. ◊, no formate added; □, 20 μM formate added. The symbols represent the means of three samples, and the bars represent ±1 standard deviation.
DISCUSSION
From the data, it is evident that copper plays an important role in the ability of whole cells expressing pMMO to degrade both methane and TCE. As shown in Tables 1 and 2, the pseudo-first-order rate constant (Vmax/Ks) for TCE and methane degradation increased with increasing copper concentrations, as did the affinity for the two substrates (Ks decreased). The changes in whole-cell rates of methane and TCE oxidation agree with results of experiments to determine the activity of pMMO in cell extracts and membrane preparations. Recent studies have shown that increasing the concentration of copper in the growth medium causes an increase in pMMO activity as measured by propylene oxidation, that copper binds to specific sites in the membranes, and that copper is involved in electron transfer (24, 25, 34). Evidence collected by Zahn and DiSpirito also suggests that copper is involved in electron transfer in pMMO, although they postulate that iron is also part of the active site (47). These researchers found that the majority of copper associated with pMMO is bound to a small (1,200-Da) polypeptide that may be involved in maintaining favorable redox conditions. The role of copper in transferring electrons in pMMO is supported by the whole-cell TCE degradation experiments performed here. The pseudo-first-order rate constant for TCE degradation by M. trichosporium OB3b grown in the presence of 20 μM copper was seven times greater with the addition of formate, an exogenous source of reducing equivalents, than in its absence. Furthermore, as the cells were not seen to degrade TCE when they were grown with low (2.5 μM) copper concentrations in both the presence and absence of formate, it is possible that pMMO was deficient in copper, causing inefficient electron transfer from the in vivo electron donor.
These results are interesting when compared to those of Brusseau et al. (6) and DiSpirito et al. (10). Brusseau et al. examined the ability of M. trichosporium OB3b at an OD600 of 0.2 in the presence of either 0 or 1 μM copper to degrade TCE. As shown in Table 3, TCE degradation by sMMO was easily measured at a copper concentration of 0 μM. TCE degradation, however, was absent at a growth concentration of 1 μM copper that repressed sMMO synthesis, but the cells still oxidized methane due to expression of pMMO. These results are in agreement with the lack of measurable TCE degradation by M. trichosporium OB3b grown with 2.5 μM copper in our laboratory. DiSpirito et al., however, did see low rates of TCE degradation by M. trichosporium OB3b and other methanotrophs expressing pMMO when the cells were grown in the presence of 2.5 μM copper. These results may appear to contradict the data collected here and by Brusseau et al., but in the experiments performed by DiSpirito et al., TCE degradation was monitored for up to 12 h with a reaction mixture of 20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer with 20 μM copper after the cells were harvested (11). In the experiments reported here and by Brusseau et al., the copper concentration was the same in the growth medium and in the TCE degradation assays. Based on the data collected in our laboratory, it is likely that Brusseau et al. did not observe any TCE degradation by pMMO, due to the low concentration of copper, while DiSpirito et al. did, due to the high copper concentration in the assay mixture that stimulated pMMO. Therefore, pMMO can oxidize TCE, but only when the cells are exposed to high concentrations of copper.
TABLE 3.
Reported kinetic values for degradation of TCE by methanotrophs known to be expressing either sMMO or pMMO
| Straina | Enzyme | Copper concn (μM) | Vmax (nmol/min/mg of protein)b | Ks (μM)b | Vmax/Ks (liters/min/mg of protein) | Reference |
|---|---|---|---|---|---|---|
| BB5.1 | pMMO | 28 | 0.10–0.15c | 8–10c | 1.2 × 10−5–1.5 × 10−5c | 35 |
| BB5.1 | pMMO | 3 | NMd | NM | NM | 35 |
| OB3b | pMMO | 20 | 2.5–4.1 | 7.9–36 | 6.9 × 10−5–5.2 × 10−4 | This study |
| OB3b | pMMO | 2.5 | NM | NM | NM | This study |
| OB3b | pMMO | 1.0 | NM | NM | NM | 6 |
| OB3b | pMMO | 4.8 | <0.20e | NDf | ND | 26 |
| OB3b | pMMO | 2.5g | 0.296 | ND | ND | 10 |
| A45 | pMMO | 2.5g | 0.233 | ND | ND | 10 |
| MN | pMMO | 2.5g | 0.677 | ND | ND | 10 |
| OBBP | pMMO | 2.5g | 0.202 | ND | ND | 10 |
| MC | pMMO | 2.5g | 0.58 | ND | ND | 10 |
| OB3b | sMMO | 0h | 54–300e | ND | ND | 26 |
| OB3b | sMMO | 0h | 456 | 138 | 3.3 × 10−3 | 6 |
| OB3b | sMMO | 0h | 995 (160) | 126 (8) | 7.9 × 10−3 | 19 |
| OB3b | sMMO | 0h | 580 (200)e | 145 (61) | 4 × 10−3 | 27 |
| 68-1 | sMMO | 0h | 2,235 (260) | 225 (13) | 9.9 × 10−3 | 19 |
A45, Methylobacter marinus A45; BB5.1, Methylobacter sp. strain BB5.1; MN, Methylomonas strain MN; OBBP, Methylocystis parvus OBBP; MC, Methylococcus capsulatus Bath; OB3b, Methylosinus trichosporium OB3b; 68-1, Methylomonas methanica 68-1.
Numbers in parentheses indicate the standard deviation of the reported value.
It should be noted that the units reported in reference 35 are incorrect but are shown here in the corrected form (20).
NM, no measurable TCE degradation.
Converted from reported units of nanomoles per minute per milligram of cells, assuming cell dry weight is 50% protein.
ND, not determined.
Cells grown with 2.5 μM copper, but TCE degradation assays performed with 20 μM copper in the reaction mixture.
No copper added to growth medium.
Other researchers have shown that increasing copper in the growth medium can stimulate TCE oxidation by an estuarine methanotroph (35). It should be noted that the units for the rates of methane and TCE oxidation in the article by Smith et al. (35) are incorrectly reported as micromoles per hour per microgram of protein. The correct units are nanomoles per hour per milligram of protein for TCE consumption and micromoles per hour per milligrams of protein for methane consumption (20). The corrected values for TCE consumption are shown in Table 3. In their study, Smith et al. saw a similar effect of copper on the ability of a methanotroph expressing pMMO to oxidize TCE, i.e., no TCE oxidation was apparent at low concentrations of copper although TCE oxidation was evident at high concentrations. These rates of TCE oxidation are less than the values presented here, possibly due to the use of copper/biomass ratios in those experiments different from those used in the experiments reported here for M. trichosporium OB3b expressing pMMO.
It is also clear in Table 3 that the reported pseudo-first-order rate constant and Vmax values of TCE oxidation by pMMO are 1 to 2 orders of magnitude lower than those reported for sMMO. The lower rates of TCE oxidation by pMMO may be beneficial, however, in obtaining more complete removal of TCE over longer periods rather than achieving fast initial degradation that does not remove TCE to mandated levels. For example, Anderson and McCarty discovered that 1,1-DCE was less toxic to a mixed culture of methanotrophs believed to be expressing pMMO than to the same mixed culture grown in conditions under which sMMO would be expressed (3). The researchers postulated that sMMO rapidly oxidized 1,1-DCE, producing a toxic intermediate. In conditions in which pMMO could be expressed, toxicity was much less, probably due to a reduction in the rate of toxic product formation that led to a reduction of inhibition. This supports the use of pMMO in select situations, particularly for the degradation of mixed chlorinated solvents over extended times.
Conclusions.
Whole-cell studies of methane and TCE degradation by M. trichosporium OB3b expressing pMMO indicate that the kinetics of both methane and TCE consumption change in response to changing copper concentrations. The data indicate that copper is not only a key parameter in regulating the relative expression of sMMO and pMMO in those strains that have both forms but is also an important factor in the activity and specificity of pMMO itself. The Vmax for methane decreased with increasing concentrations of copper in the growth medium, but both the affinity for methane and the pseudo-first-order rate constant increased as the concentration of copper increased from 2.5 to 20 μM. With low concentrations of copper that preclude expression of sMMO, no measurable TCE degradation was seen, but increasing the concentration of copper in the growth medium enabled M. trichosporium OB3b to degrade TCE. This suggests that as M. trichosporium OB3b is grown in higher copper concentrations, pMMO may more effectively bind and oxidize substrates. Furthermore, the ability of this cell to degrade TCE was enhanced with the addition of formate. The changes reported here in pMMO activity and substrate specificity in whole cells expressing pMMO support other studies that show that copper plays a major role in the activity and structure of pMMO. This information can be useful for in situ bioremediation, as many methanotrophs can express only pMMO and copper concentrations can vary significantly in environmental systems.
ACKNOWLEDGMENT
Research support from the National Science Foundation (grant MCB-9708557) is gratefully acknowledged.
REFERENCES
- 1.Alvarez-Cohen L, McCarty P L. Effects of toxicity, aeration, and reductant supply on trichloroethylene transformation by a mixed methanotrophic culture. Appl Environ Microbiol. 1991;57:228–235. doi: 10.1128/aem.57.1.228-235.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Cohen L, McCarty P L. Product toxicity and cometabolic competitive inhibition modeling of chloroform and trichloroethylene transformation by methanotrophic resting cells. Appl Environ Microbiol. 1991;57:1031–1037. doi: 10.1128/aem.57.4.1031-1037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson J E, McCarty P L. Effect of three chlorinated ethenes on growth rates for a methanotrophic mixed culture. Environ Sci Technol. 1996;30:3517–3524. [Google Scholar]
- 4.Boisen A, Arvin E, Broholm K. Effect of mineral nutrients on the kinetics of methane utilization by methanotrophs. Biodegradation. 1993;4:163–170. [Google Scholar]
- 5.Broholm K, Christensen T H, Jensen B K. Different abilities of eight mixed cultures of methane-oxidizing bacteria to degrade TCE. Water Res. 1993;27:215–224. [Google Scholar]
- 6.Brusseau G A, Tsien H-C, Hanson R S, Wackett L P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1:19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- 7.Collins M L P, Buchholz L A, Remsen C C. Effect of copper on Methylomonas albus BG8. Appl Environ Microbiol. 1991;57:1261–1264. doi: 10.1128/aem.57.4.1261-1264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook S A, Shiemke A K. Evidence that copper is a required cofactor for the membrane-bound form of the methane monooxygenase. J Inorg Biochem. 1996;63:273–284. [Google Scholar]
- 9.Dalton H, Prior S D, Stanley S H. Regulation and control of methane monooxygenase. In: Crawford C R L, Hanson R S, editors. Microbial growth on C1 compounds. Washington, D.C: American Society for Microbiology; 1984. pp. 75–82. [Google Scholar]
- 10.DiSpirito A A, Gulledge J, Shiemke A K, Murrell J C, Lidstrom M E, Krema C L. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II, and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 11.DiSpirito, A. A. Personal communication.
- 12.Eng W, Palumbo A V, Sriharan S, Strandberg G W. Methanol suppression of trichloroethylene degradation by Methylosinus trichosporium (OB3b) and methane-oxidizing mixed cultures. Appl Biochem Biotechnol. 1991;28:887–899. doi: 10.1007/BF02922658. [DOI] [PubMed] [Google Scholar]
- 13.Fliermans C B, Phelps T J, Ringelberg D, Mikell A T, White D C. Mineralization of trichloroethylene by heterotrophic enrichment cultures. Appl Environ Microbiol. 1988;54:1709–1714. doi: 10.2172/666263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forstner U, Wittmann G T W. Metal pollution in the aquatic environment. Berlin, Germany: Springer-Verlag; 1981. p. 356. [Google Scholar]
- 15.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry S M, Grbic-Galic D. Influence of endogenous and exogenous electron donors and trichloroethylene oxidation toxicity on trichloroethylene oxidation by methanotrophic cultures from groundwater aquifers. Appl Environ Microbiol. 1991;57:236–244. doi: 10.1128/aem.57.1.236-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrysson T, McCarty P L. Influence of the endogenous storage lipid poly-β-hydroxybutyrate on the reducing power availability during cometabolism of trichloroethylene and naphthalene by resting methanotrophic mixed cultures. Appl Environ Microbiol. 1993;59:1602–1606. doi: 10.1128/aem.59.5.1602-1606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard P H, Sage G W, Jarvis W F, Gray D A. Handbook of environmental fate and exposure data for organic chemicals. II. Solvents. Chelsea, Mich: Lewis Publishers; 1991. [Google Scholar]
- 19.Koh S-C, Bowman J P, Sayler G S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lidstrom, M. E. 1997. Personal communication.
- 21.Lidstrom M E, Somers L. Seasonal study of methane oxidation in Lake Washington. Appl Environ Microbiol. 1984;47:1255–1260. doi: 10.1128/aem.47.6.1255-1260.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morel F M, Hering J G. Principles and applications of aquatic chemistry. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 23.Murrell J C. Genetics and molecular biology of methanotrophs. FEMS Microbiol Rev. 1992;88:233–248. doi: 10.1111/j.1574-6968.1992.tb04990.x. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen H-H T, Nakagawa K H, Hedman B, Elliot S J, Lidstrom M E, Hodgson K O, Chan S I. X-ray absorption and epr studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath): Cu(I) ions and their implications. J Am Chem Soc. 1996;118:12766–12776. [Google Scholar]
- 25.Nguyen H-H, Shiemke A K, Jacobs S J, Hales B J, Lidstrom M E, Chan S I. The nature of copper ions in the membranes containing the particulate methane monooxygenase from Methylococcus capsulatus Bath. J Biol Chem. 1994;269:14995–15005. [PubMed] [Google Scholar]
- 26.Oldenhuis R, Vink R L J M, Janssen D B, Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989;55:2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldenhuis R, Oedzes J Y, van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oremland R S, Culbertson C W. Importance of methane-oxidizing bacteria in the methane budget as revealed by the use of a specific inhibitor. Science. 1992;356:421–423. [Google Scholar]
- 29.Peltola P, Priha P, Laakso S. Effect of copper on membrane lipids and on methane monooxygenase activity of Methylococcus capsulatus (Bath) Arch Microbiol. 1993;159:521–525. [Google Scholar]
- 30.Prior S D, Dalton H. The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath) J Gen Microbiol. 1985;131:155–163. [Google Scholar]
- 31.Schlegel H G. General microbiology. 7th ed. Cambridge, United Kingdom: Cambridge University Press; 1993. [Google Scholar]
- 32.Scott D, Brannan J, Higgins I J. The effect of growth conditions on intracytoplasmic membranes and methane monooxygenase activities in Methylosinus trichosporium OB3b. J Gen Microbiol. 1981;125:63–72. [Google Scholar]
- 33.Semrau J D, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes A J, Finch R, Murrell J C, Lidstrom M E. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semrau J D, Zolandz D, Lidstrom M E, Chan S I. The role for copper in the pMMO of Methylococcus capsulatus Bath: a structural vs. catalytic function. J Inorg Biochem. 1995;58:235–244. doi: 10.1016/0162-0134(94)00056-g. [DOI] [PubMed] [Google Scholar]
- 35.Smith K S, Costello A A, Lidstrom M E. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl Environ Microbiol. 1997;63:4617–4620. doi: 10.1128/aem.63.11.4617-4620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanley S H, Prior S D, Leak D J, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane monooxygenase methane-oxidizing organisms: studies in batch and continuous culture. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 37.Stirling D I, Colby J, Dalton H. A comparison of the substrate and electron-donor specificities of the methane monooxygenase from three strains of methane-oxidizing bacteria. Biochem J. 1979;177:361–364. doi: 10.1042/bj1770361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strand S E, Bjelland M D, Stensel H D. Kinetics of chlorinated hydrocarbon degradation by suspended cultures of methane-oxidizing bacteria. J Water Pollut Control Fed. 1990;62:124–129. [Google Scholar]
- 39.Strandberg G W, Donaldson T L, Farr L L. Degradation of trichloroethylene and trans-1,2-dichloroethylene by a methanotrophic consortium in a fixed-film, packed-bed bioreactor. Environ Sci Technol. 1989;23:1422–1425. [Google Scholar]
- 40.Taylor R T, Hanna M L, Shah N N, Shonnard D R, Duba A G, Durham W B, Jackson K J, Knapp R B, Wijesinghe A M, Knezoich J P, Jovanivich M C. In situ bioremediation of trichloroethylene-contaminated water by a resting-cell methanotrophic microbial filter. Hydrol Sci J. 1993;38:323–342. [Google Scholar]
- 41.Travis C C, Doty C B. Can contaminated aquifers at Superfund sites be remediated? Environ Sci Technol. 1990;24:1464–1466. [Google Scholar]
- 42.Tsien H-C, Brusseau G A, Hanson R S, Wackett L P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989;55:3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.U.S. Department of Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry. Toxicological profile for trichloroethylene. U.S. Atlanta, Ga: Department of Health and Human Services; 1993. [Google Scholar]
- 44.Verstraete W, Top E. Holistic environmental biotechnology. In: Fry J C, Gadd G M, Herbert R A, Jones C W, Watson-Carik I A, editors. Microbial control of pollution. Cambridge, United Kingdom: Cambridge University Press; 1992. pp. 1–17. [Google Scholar]
- 45.Westrick J J, Mello J W, Thomas R F. The groundwater supply survey. J Am Water Works Assoc. 1984;5:52–59. [Google Scholar]
- 46.Whittenbury R K, Philips K D, Wilkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 47.Zahn J A, DiSpirito A A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]