Abstract
Accurately predicting survival in patients with early hepatocellular carcinoma (HCC) is essential for making informed decisions about treatment and prognosis. Herein, we have developed a machine learning (ML) model that can predict patient survival and guide treatment decisions. We obtained patient demographic information, tumor characteristics, and treatment details from the SEER database. To analyze the data, we employed a Cox proportional hazards (CoxPH) model as well as 3 ML algorithms: neural network multitask logistic regression (N-MLTR), DeepSurv, and random survival forest (RSF). Our evaluation relied on the concordance index (C-index) and Integrated Brier Score (IBS). Additionally, we provided personalized treatment recommendations regarding surgery and chemotherapy choices and validated models’ efficacy. A total of 1136 patients with early-stage (I, II) hepatocellular carcinoma (HCC) who underwent liver resection or transplantation were randomly divided into training and validation cohorts at a ratio of 3:7. Feature selection was conducted using Cox regression analyses. The ML models (NMLTR: C-index = 0.6793; DeepSurv: C-index = 0.7028; RSF: C-index = 0.6890) showed better discrimination in predicting survival than the standard CoxPH model (C-index = 0.6696). Patients who received recommended treatments had higher survival rates than those who received unrecommended treatments. ML-based surgery treatment recommendations yielded higher hazard ratios (HRs): NMTLR HR = 0.36 (95% CI: 0.25–0.51, P < .001), DeepSurv HR = 0.34 (95% CI: 0.24–0.49, P < .001), and RSF HR = 0.37 (95% CI: 0.26–0.52, P = <.001). Chemotherapy treatment recommendations were associated with significantly improved survival for DeepSurv (HR: 0.57; 95% CI: 0.4–0.82, P = .002) and RSF (HR: 0.66; 95% CI: 0.46–0.94, P = .020). The ML survival model has the potential to benefit prognostic evaluation and treatment of HCC. This novel analytical approach could provide reliable information on individual survival and treatment recommendations.
Keywords: DeepSurv, hepatocellular carcinoma (HCC), liver resection, liver transplantation, machine learning, survival analysis
1. Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide, responsible for approximately 780,000 deaths in 2018.[1] Majority of HCC cases occur in individuals with coexisting cirrhosis, primarily caused by hepatitis B or C virus (HCV) infection.[2] The survival rates and clinical courses of HCC are dependent on the stage of the disease. Unfortunately, the prognosis for HCC patients is often poor, particularly for those at high risk of developing HCC, with a 5-year survival rate below 10%.[3]
Various therapeutic modalities, including liver transplantation (LT), liver resection (LR), local ablation therapies, and transarterial therapies have been utilized to treat HCC.[4] Among these treatments, LT and LR are still considered the most effective methods for achieving long-term survival, with a 5-year survival rate of 60% to 80%.[5] Both treatments can be performed for the subset of patients with early HCC within specific criteria.[5] Both procedures can be performed on patients with early HCC that meet specific criteria. LR is the preferred option for early-stage HCC patients without cirrhosis, as even major resections are associated with low likelihood of life-threatening complications and satisfactory outcomes.[5] Conversely, LT can cure cancer and its underlying causative diseases, which are the leading risk factors for developing new tumors. According to the Milan criteria, LT is highly recommended as the first-line option for HCC, while liver resection is considered unsuitable for tumors.[6] However, these criteria may be too restrictive, preventing many patients from receiving advanced treatment. Therefore, there have been proposals[7–10] to expand selection criteria in anticipation of further refinement of treatment regimens. The most appropriate surgical option for patients diagnosed with HCC, particularly those who fall just outside the criteria, remains a matter of controversy.[11] Although studies have reported that chemotherapy has a beneficial effect on the survival and recurrence rates of patients who have undergone LT and LR,[12] evidence for this remains insufficient.[6]
In discussing whether a treatment or criteria is suitable for a patient, the critical issue is how to accurately predict patients’ outcomes following treatment. While the criteria can be simplified by using parameters such as size and number of tumors, many possible prognostic factors have been explored, including inflammatory markers,[13] alpha-fetoprotein (AFP) score,[14] des-γ-carboxyprothrombin level,[15] and genomic features,[16] among others. The Cox proportional hazards (CoxPH) model has been commonly employed in survival analysis and building predictive models. The CoxPH calculates the effects of prognostic factors on the risk of death based on the assumption that log-risk of death is a linear combination of covariates of a patient. However, in the analysis of real-world practice, it may be too simplistic to assume that the log-risk function is linear.[17] Hence, it is necessary to develop novel approaches for survival analysis.
Machine learning (ML) is a fast-expanding branch of data analysis that can facilitate the handling of numerous variables and complex interactions in extensive data and can yield accurate predictions in healthcare contexts. Many ML algorithms have been established exclusively for the analysis of right-censored survival data. A pioneering method is the random survival forest (RSF),[18] which estimates a hazard function through the ensemble prediction of many decision trees. DeepSurv,[17] a recently updated Cox proportional hazards deep learning model, was proven to enhance personalized RSF model treatment recommendations. The multitask logistic regression model is a proportional hazards model that considers time-varying risks. A deep learning extension has been constructed that outperforms ordinary linear survival models. These 3 models improve upon standard survival analysis by enabling the prediction of an individual hazard ratio (HR) in order to determine a particular treatment for each patient based on their unique disease characteristics. While numerous studies have applied ML models to predict survival and guide treatment in various cancers, few have examined their utility in HCC, particularly for early-stage patients who qualify for curative resection or transplantation. Therefore, this study aims to compare the performances of the ML models and the CoxPH model in terms of their prediction of survival and their ability to provide personalized treatment recommendations for early-stage HCC patients.
2. Materials & methods
2.1. Reporting guidelines
This study follows the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines[19] for reporting observational studies. Additionally, we adhered to the Prediction model Risk of Bias Assessment Tool (PROBAST)[20] and Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) guidelines[21] which provide a framework for reporting details essential to assessing risk of bias and evaluating validity of prognostic models.
2.2. Data source
All patients with HCC included in this research were selected from the SEER “18 Regs Research Plus Nov 2020 Sub (2000–2018 varying)” data set (http://seer.cancer.gov). The SEER database contains data on cancer patients from 18 regions of the United States and accounts for about 28% of the total population of cancer patients in the US.[22] This database contains a considerable amount of related information on patients, including tumor data, and information on causes of death and survival times and so on. By signing the SEER Research Data Agreement form and sending it via email, we were authorized to access the database.
2.3. Study population
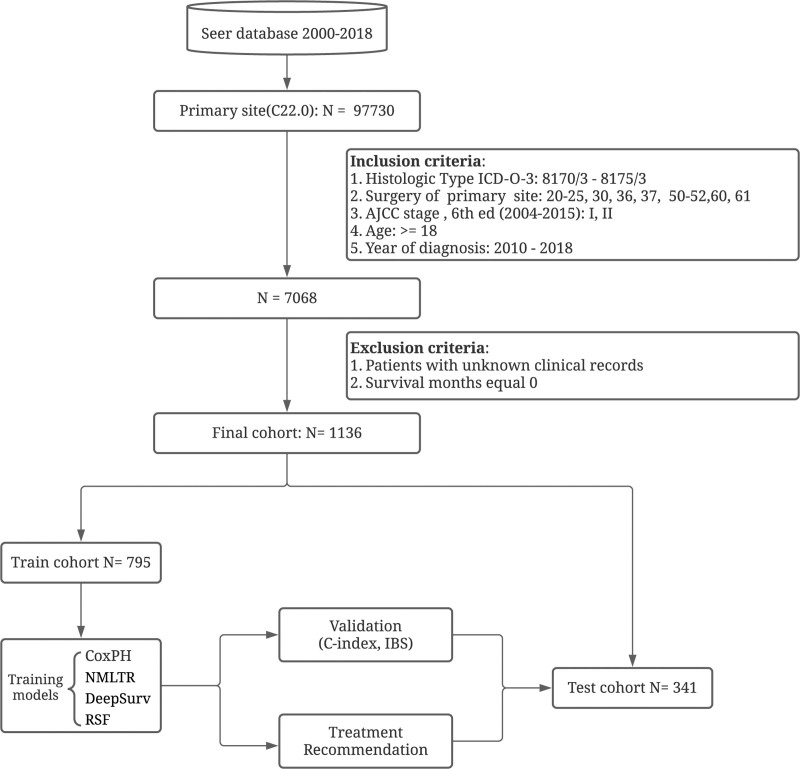
We extracted data from patients newly diagnosed with primary HCC between January 1, 2010 and December 31, 2017 using the International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) histology codes 8170/3 to 8175/3, with the liver site code C22.0. We collected the baseline information of cases (race, sex, age, ethnicity and marriage status), tumor characteristics (number, size, histologic grade, stage, and histologic type), AFP, Fibrosis Score, and treatment details (radiotherapy, surgical type, and chemotherapy). Patients with early-stage (stage I and II, AJCC, 7th) HCC who had undergone LR or LT were enrolled in our final analysis. Patients with unknown clinical records were excluded. As shown in Figure 1, the detailed selection process is represented by a flowchart.
Figure 1.
Study profile and analysis pipeline.
2.4. Model development
Feature selection was conducted using univariate and multivariate Cox regression. Only significant factors (P < .05) were included in the model development. Additionally, least absolute shrinkage and selection operator (LASSO) regularization was performed for variable selection. Variables identified by LASSO were chosen based on the commonly used 1 standard error (1SE) rule. The variables selected by both Cox regression and LASSO were then combined and included in model development. Three ML models were developed, namely DeepSurv, RSF, and neural network multitask logistic regression (NMLTR). Meanwhile, a multivariate CoxPH model was also constructed for comparison. The dataset was randomly divided into training and testing datasets at a ratio of 7:3. Hyperparameter tuning was obtained through random search with 10-fold cross-validation on the training dataset. The concordance index (C-index)[23] is used to evaluate the performance of models with different combinations of hyperparameters.
2.5. Model evaluation and validation
The accuracy of the Cox model was evaluated by calculating the Harrell C-index, which measures the correlation between predicted survival risks and actual survival times. A C-index of 0.5 describes a random prediction, while a C-index of 1.0 describes a perfectly predicting model. Based on Kang method,[24] we tested the difference between 2 models’ C-indexes. A Brier score—used to evaluate the accuracy of a predicted survival function at a given time—was also reported; it represents the average squared distances between the observed survival status and the predicted survival probability and is always a number between 0 and 1, with 0 being the best possible value. As a benchmark, a useful model will have a Brier score below 0.25. Additionally, the Integrated Brier Score (IBS) provided an overall calculation of the model performance for all available times.[25]
2.6. Feature importance
In order to study the relationship between individual features and model performance, we take the contribution of the feature to the model discrimination as the feature importance and measure the contribution by calculating the decrease in the C-index caused by sequentially replacing the value of each feature with a random value. The greater the decrease in the C-index caused by a feature after replacing it with a random value, the greater the contribution of this feature to the model.
2.7. Treatment recommendation
The CoxPH model computes a constant recommender function and recommends the same treatment option for all patients. In contrast, ML methods provide personalized treatment recommendations, predicting an individual treatment hazard by computing relevant interaction terms.[17] Our study recommends a treatment if predicted survival was found to be longer with this treatment than with others. After the ML models made personalized treatment recommendations for each patient surgical method and chemotherapy choice, we performed a log-rank test to validate the difference between patients who aligned with the model recommended treatment and those who did not experience the recommended treatment.
2.8. Statistical analysis
Throughout all clinical data, continuous variables are presented as the mean value ± standard deviation (SD). Categorical variables are described as frequencies and percentages. The chi-square test and unpaired 2-sided t test were used to compare the differences in variables between groups. Data preprocessing and plot were completed using the R programming language (version 4.1.2). ML models were built using the PySurvival package in the Python programming language (version 3.6.8).
3. Results
3.1. Basic characteristics
A total of 1136 patients with early-stage (I, II) HCC met our inclusion criteria. The baseline information of the patients at the time of enrollment is shown in Table 1. Their mean age was 61.48 ± 9.19 years, and 74.7% were male. The surgery classes comprised 718 (63.20%) patients for LR and 418 (36.80%) for LT. The mean overall survival (OS) was 49.25 ± 26.34 months in the LR group and 61.89 ± 27.30 in the LT group. Receipt of LT was significantly in connection with younger age, white ethnicity, better grading of the differentiated cells, earlier stage of development, having undergone chemotherapy, higher fibrosis score, smaller tumor size and greater number of tumors (Table 1).
Table 1.
Patient demographic, disease and treatment characteristics.
| Level | Overall | LR | LT | P value | |
|---|---|---|---|---|---|
| n | 1136 | 718 | 418 | ||
| Sex (%) | Female | 287 (25.3) | 188 (26.2) | 99 (23.7) | .387 |
| Male | 849 (74.7) | 530 (73.8) | 319 (76.3) | ||
| Age (mean (SD)) | 61.48 (9.19) | 63.18 (9.95) | 58.56 (6.79) | <.001 | |
| Race (%) | White | 717 (63.1) | 382 (53.2) | 335 (80.1) | <.001 |
| Black | 134 (11.8) | 92 (12.8) | 42 (10.0) | ||
| Other | 285 (25.1) | 244 (34.0) | 41 (9.8) | ||
| Marital status (%) | Not married | 380 (33.5) | 244 (34.0) | 136 (32.5) | .665 |
| Married | 756 (66.5) | 474 (66.0) | 282 (67.5) | ||
| Ethnicity (%) | Non-Hispanic | 977 (86.0) | 637 (88.7) | 340 (81.3) | .001 |
| Hispanic | 159 (14.0) | 81 (11.3) | 78 (18.7) | ||
| Grade (%) | Well differentiated | 297 (26.1) | 167 (23.3) | 130 (31.1) | <.001 |
| Moderately differentiated | 646 (56.9) | 396 (55.2) | 250 (59.8) | ||
| Poorly differentiated | 184 (16.2) | 147 (20.5) | 37 (8.9) | ||
| Undifferentiated | 9 (0.8) | 8 (1.1) | 1 (0.2) | ||
| SEER stage (%) | Localized | 996 (87.7) | 672 (93.6) | 324 (77.5) | <.001 |
| Regional | 140 (12.3) | 46 (6.4) | 94 (22.5) | ||
| AJCC stage (%) | I | 704 (62.0) | 498 (69.4) | 206 (49.3) | <.001 |
| II | 432 (38.0) | 220 (30.6) | 212 (50.7) | ||
| Radiotherapy (%) | No | 1111 (97.8) | 703 (97.9) | 408 (97.6) | .900 |
| Yes | 25 (2.2) | 15 (2.1) | 10 (2.4) | ||
| Chemotherapy (%) | No | 889 (78.3) | 642 (89.4) | 247 (59.1) | <.001 |
| Yes | 247 (21.7) | 76 (10.6) | 171 (40.9) | ||
| AFP (%) | Negative or normal | 459 (40.4) | 292 (40.7) | 167 (40.0) | .861 |
| Positive or elevated | 677 (59.6) | 426 (59.3) | 251 (60.0) | ||
| Fibrosis score (%) | Ishak 0–4 | 410 (36.1) | 373 (51.9) | 37 (8.9) | <.001 |
| Ishak 5–6 | 726 (63.9) | 345 (48.1) | 381 (91.1) | ||
| Tumor size (%) | <3cm | 473 (41.6) | 194 (27.0) | 279 (66.7) | <.001 |
| >=3 and < 5 cm | 389 (34.2) | 267 (37.2) | 122 (29.2) | ||
| >=5 cm | 274 (24.1) | 257 (35.8) | 17 (4.1) | ||
| Tumor number (mean (SD)) | 1.23 (0.53) | 1.25 (0.56) | 1.19 (0.48) | .107 | |
| Survival months (mean (SD)) | 53.90 (27.38) | 49.25 (26.34) | 61.89 (27.30) | <.001 | |
| Status (%) | Alive | 755 (66.5) | 426 (59.3) | 329 (78.7) | <.001 |
| Dead | 381 (33.5) | 292 (40.7) | 89 (21.3) |
For categorical values, the P value for a χ2 test comparing the LR and LT groups is provided; for numerical values, the P value for an unpaired 2-sided t test is provided.
LR = liver resection, LT = liver transplantation.
3.2. Feature selection
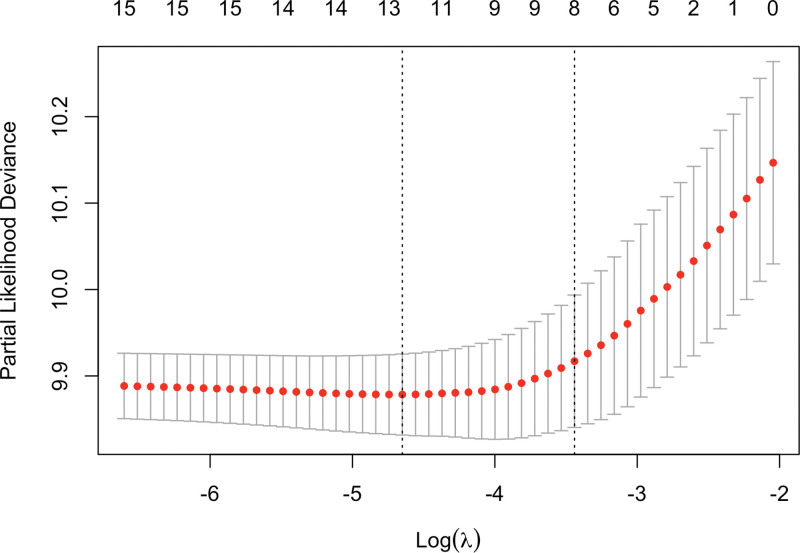
Univariate and multivariate Cox regression analyses were performed for all data. In addition, LASSO regularization was conducted, identifying 8 variables (Fig. 2): Race, Marital status, Grade, AJCC stage, Surgery, AFP, Fibrosis score, and Tumor size. As presented in Table 2, ten significant factors (race, age, marital status, AJCC stage, grade, surgery, AFP, fibrosis score, tumor size, and number of tumors) were initially selected by the Cox regression analysis. The 8 variables identified through LASSO were then combined with the variables selected by Cox regression. Chemotherapy was also included, due to the significant differences between treatment groups (Table 1). Ultimately, after combining the LASSO and Cox selected variables, a total of 11 features were included in the final model development.
Figure 2.
The cross-validated deviance plot for the Lasso regression model. The Lasso regression model was fitted with Cox family and evaluated using 5-fold cross-validation. The plot provides insights into the performance of the model by displaying the deviance values across different regularization parameter values.
Table 2.
Univariate and multivariate Cox regression results.
| Characteristic | Univariate Cox | Multivariate Cox | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Sex | .77 | .30 | ||||
| Female | — | — | — | — | ||
| Male | 1.04 | 0.82, 1.31 | 1.14 | 0.89, 1.45 | ||
| Age | 1.02 | 1.01, 1.03 | <.001 | 1.01 | 0.99, 1.02 | .34 |
| Race | .12 | .024 | ||||
| White | — | — | — | — | ||
| Black | 1.33 | 0.99, 1.79 | 1.21 | 0.89, 1.64 | ||
| Other | 0.94 | 0.74, 1.20 | 0.75 | 0.58, 0.98 | ||
| Marital status | <.001 | .008 | ||||
| Not married | — | — | — | — | ||
| Married | 0.70 | 0.57, 0.86 | 0.75 | 0.60, 0.92 | ||
| Ethnicity | .84 | .87 | ||||
| Non-Hispanic | — | — | — | — | ||
| Hispanic | 0.97 | 0.72, 1.30 | 0.97 | 0.72, 1.33 | ||
| Grade | <.001 | <.001 | ||||
| Well differentiated | — | — | — | — | ||
| Moderately differentiated | 1.66 | 1.27, 2.18 | 1.41 | 1.07, 1.86 | ||
| Poorly differentiated | 2.63 | 1.91, 3.62 | 2.10 | 1.50, 2.93 | ||
| Undifferentiated | 2.61 | 1.05, 6.46 | 1.52 | 0.60, 3.84 | ||
| SEER stage | .12 | .36 | ||||
| Localized | — | — | — | — | ||
| Regional | 0.77 | 0.56, 1.08 | 0.85 | 0.60, 1.21 | ||
| AJCC stage | .007 | <.001 | ||||
| I | — | — | — | — | ||
| II | 1.32 | 1.08, 1.62 | 1.57 | 1.26, 1.96 | ||
| Surgery | <.001 | <.001 | ||||
| Hepatic resection/lobectomy | — | — | — | — | ||
| Hepatectomy with transplant | 0.42 | 0.33, 0.54 | 0.42 | 0.31, 0.57 | ||
| Radiotherapy | .78 | .82 | ||||
| No | — | — | — | — | ||
| Yes | 1.11 | 0.55, 2.24 | 0.92 | 0.45, 1.87 | ||
| Chemotherapy | .082 | .78 | ||||
| No | — | — | — | — | ||
| Yes | 0.80 | 0.62, 1.04 | 1.04 | 0.78, 1.38 | ||
| AFP | .002 | .10 | ||||
| Negative or normal | — | — | — | — | ||
| Positive or elevated | 1.39 | 1.13, 1.72 | 1.21 | 0.96, 1.51 | ||
| Fibrosis score | .56 | .004 | ||||
| Ishak 0–4 | — | — | — | — | ||
| Ishak 5–6 | 0.94 | 0.76, 1.16 | 1.42 | 1.12, 1.79 | ||
| Tumor size | <.001 | .001 | ||||
| <3cm | — | — | — | — | ||
| >=3 and < 5 cm | 1.65 | 1.30, 2.11 | 1.35 | 1.04, 1.74 | ||
| >=5 cm | 2.16 | 1.67, 2.79 | 1.71 | 1.28, 2.28 | ||
| Tumor number | 1.23 | 1.05, 1.45 | .017 | 1.34 | 1.13, 1.59 | .002 |
AFP = alpha-fetoprotein, CI = confidence interval, HR = hazard ratio.
3.3. Model comparisons
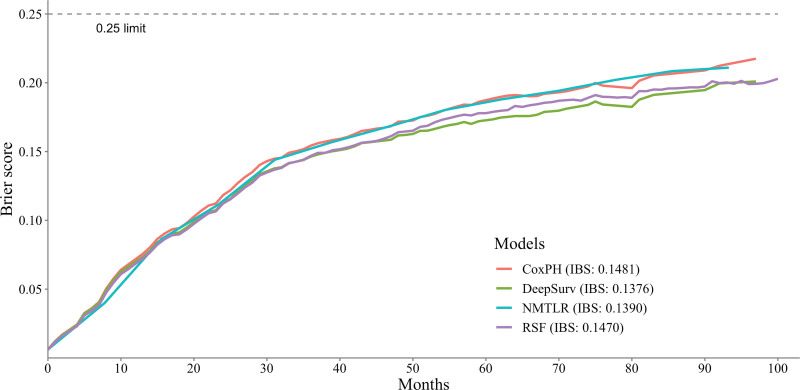
Table 3 shows the predictive performance of the ML and CoxPH models. In the test dataset, the 3 ML models demonstrated significantly better discrimination (P < .01) than the standard CoxPH model with C-index values of NMLTR: 0.6793, DeepSurv: 0.7028, and RSF: 0.6890, while the CoxPH model had a C-index of 0.6696. Among the 3 ML models, DeepSurv achieved the highest C-index of 0.7028. Figure 3 displays the IBS of the 4 models: 0.1481 (CoxPH), 0.1390 (NMLTR), 0.1376 (DeepSurv), and 0.6890 (RSF). The C-index values obtained from the training data set and the test set were similar, indicating that the models did not suffer from overfitting, with values of CoxPH: 0.6705, NMLTR: 0.6993, DeepSurv: 0.7387, and RSF: 0.7302.
Table 3.
Performance of each of the 4 models.
| C-index | IBS | ||
|---|---|---|---|
| Train | Test | Test | |
| CoxPH | 0.6705 | 0.6696 | 0.1481 |
| NMLTR | 0.6993 | 0.6793 | 0.1390 |
| DeepSurv | 0.7387 | 0.7028 | 0.1376 |
| RSF | 0.7302 | 0.6890 | 0.1470 |
Metrics in train and test dataset are calculated separately.
CoxPH = Cox proportional hazards, IBS = Integrated Brier Score, NMLTR = neural multi-task logistic regression, RSF = random survival forest.
Figure 3.
Prediction error curve. As a benchmark, a useful model will have a Brier score below 0.25.
3.4. Feature importance
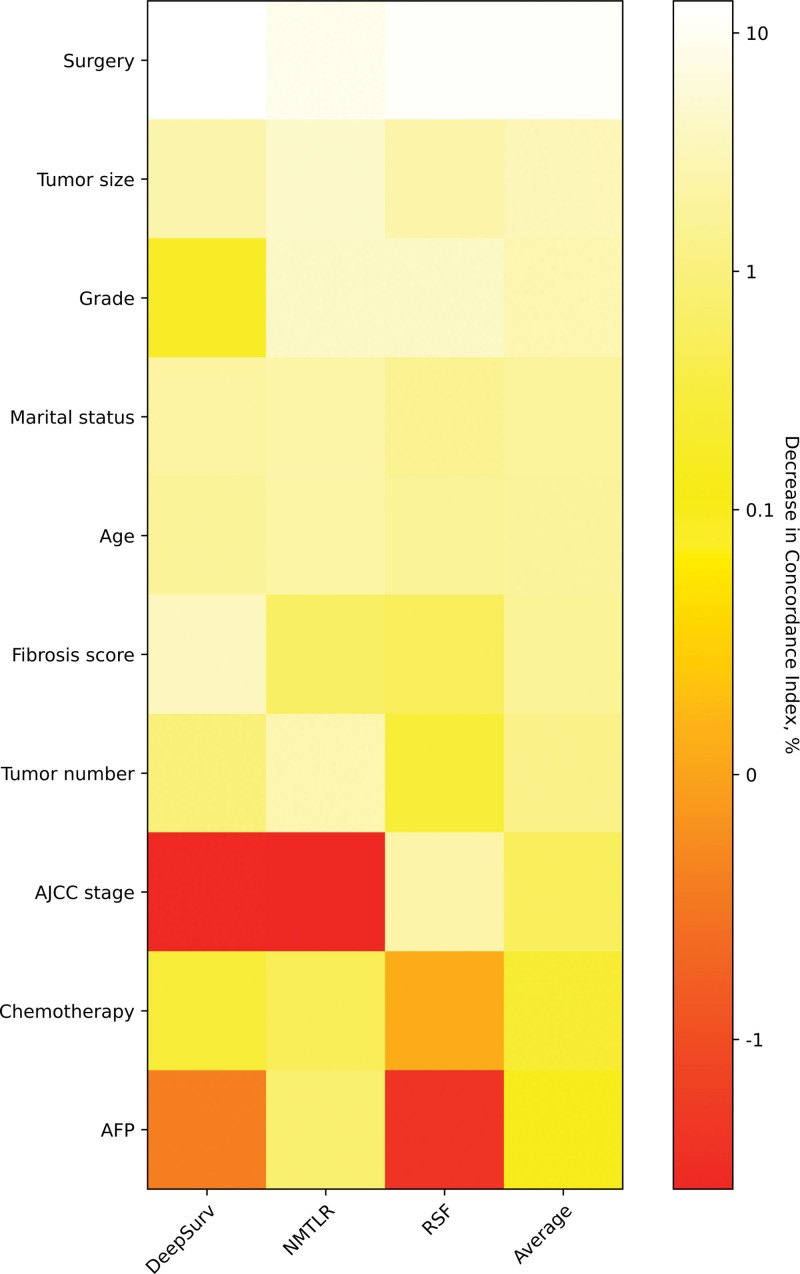
The reduction degree of the C-index caused by feature replacement is expressed as a percentage and is reflected in the figure by color. The whiter the color, the more important the feature is. For feature importance to each feature, with a more than 0.1% loss in C-index when surgery, tumor size, grade, marital status, age, fibrosis score, tumor number, and AJCC stage are replaced (Fig. 4).
Figure 4.
Heatmap of feature importance for DeepSurv, neural network multitask logistic regression (N-MLTR) and random survival forest (RSF) models. The values are expressed as a percentage reduction in the C-index after the value of a feature has been replaced by random numbers. Higher values suggest that a feature is more important in influencing the predictive accuracy of the corresponding deep learning model. C-index = concordance index.
3.5. Treatment recommendation
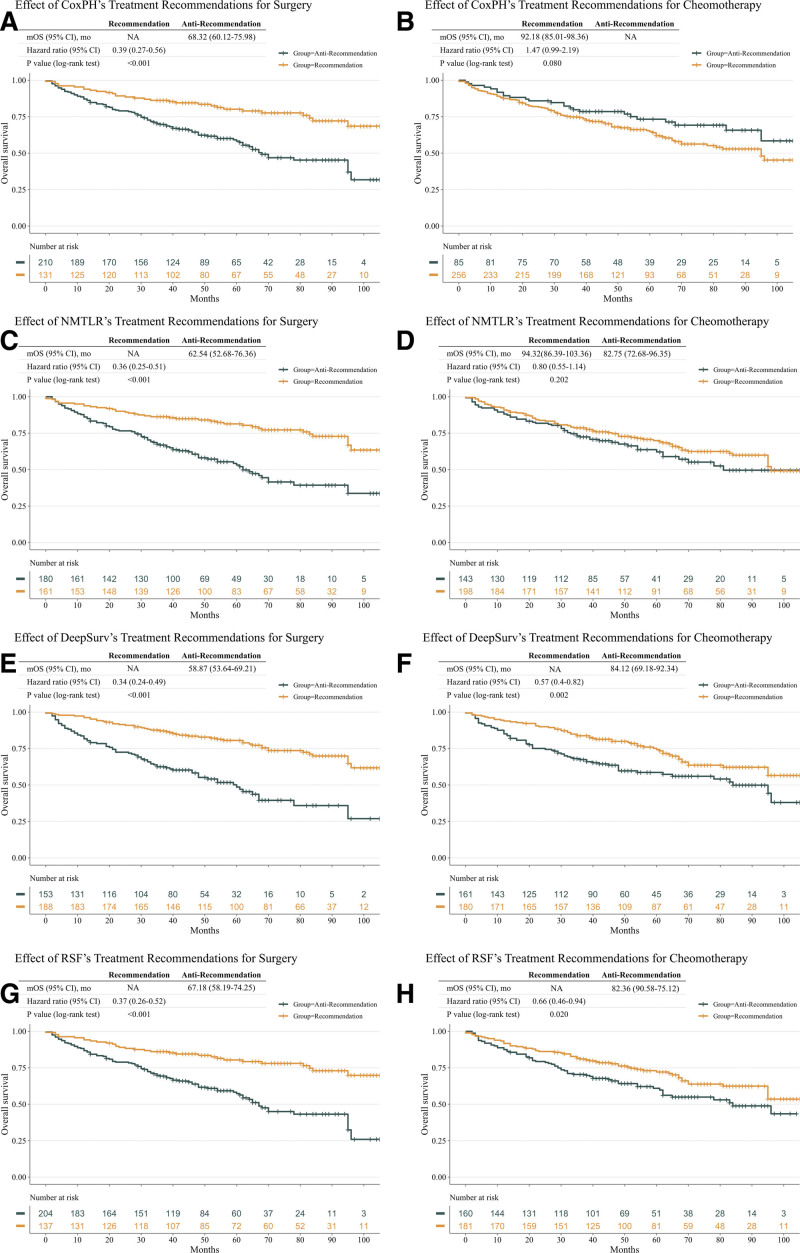
Since the CoxPH model makes a constant treatment recommendation for all patients, the anti-recommendation group in CoxPH refers to patients who received a treatment option with a higher risk of death (Fig. 5). Although fixed CoxPH model recommendations can also benefit patients (HR: 0.39; 95% CI: 0.27–0.56; P < .001), personalized treatment recommendations based on the ML models achieved higher HR values, with an HR of 0.36 (95% CI, 0.25–0.51; P < .001) for NMTLR, 0.34 (95% CI, 0.24–0.49; P < .001) for DeepSurv, and 0.37 (95% CI, 0.26–0.52; P = < 0.001) for RSF. Treatment recommendations for chemotherapy according to the ML models was associated with significantly improved survival for DeepSurv (HR: 0.57; 95% CI: 0.4–0.82; P = .002) and RSF (HR: 0.66; 95% CI: 0.46–0.94; P = .020). No improved survival was seen with recommending chemotherapy in CoxPH (HR: 1.47; 95% CI: 0.99–2.19; P = .08) and NMTLR (HR: 0.80; 95% CI: 0.55–1.14; P = .202).
Figure 5.
Survival outcomes of the treatment recommendations of surgery and chemoradiotherapy in the test dataset. The results are presented for Cox proportional hazards (CoxPH) (A, B), neural network multitask logistic regression (N-MLTR) (C, D), DeepSurv (E, F), and random survival forest (RSF) (G, H) models. The panels on the left show the effect of surgery treatment recommended by each of the 4 models, with a higher HR value being achieved by the 3 machine learning models. A recommendation benefit is seen for patients receiving the chemotherapy treatment recommended by the DeepSurv and RSF models (F, H) on the right panel. CoxPH = Cox proportional hazards.
4. Discussion
ML algorithms have increasingly conspicuous applications within health care, with applications in HCC including the prediction of tumor characteristics by biochemical and clinical indicators,[26,27] prediction of postoperative adverse events by preoperative features,[28,29] and diagnosis by imaging,[30–32] among others. Since standard survival analysis is limited by the assumption of a linear combination of covariates, ML is proposed as a novel method for survival analysis. It has been verified with several real-world sets of data.[17,28,33,34] In this study, we trained and internally validated 3 ML models, and demonstrated the advantages of ML in predicting survival and making personalized treatment recommendations in patients with early-stage HCC.
Accurate prediction of survival of HCC is not only one of the cornerstones of establishing criteria for treatment selection but also a necessary condition for personalized treatment recommendations. We firstly conducted a Cox proportional-hazards regression analysis of 1136 patients with HCC for feature selection. The selected features comprised race, age, marital status, tumor grade, surgery status, chemotherapy status, AJCC stage, AFP, fibrosis score, tumor size, and the number of tumors (Table 2). We then developed 3 ML-based models for predicting the survival of HCC patients. The evaluation result of the models is summarized in Table 3. All 3 ML methods outperformed the standard method in predicting survival. In addition, we carried out 10-fold cross-validation for comparison of models, as well as independent internal validation of the optimal model, which maximized the generalization and reliability of models. Nowadays, with the increasing application of imaging[35] and genetic data[36,37] in the survival prediction of HCC, ML methods—combined with the increased performance of computers—are able to fully explore and analyze high-dimensional and large-scale data.
Applying deep learning based on survival analysis to clinical treatment recommendations was first proposed by Katzman[17] in 2018. Its performance advantages over standard methods have been demonstrated by several subsequent studies, including studies on the treatment of lung cancer[38] and head and neck cancer.[39] According to our results, compared with the CoxPH model of recommending LT for all patients, the personalized surgical recommendations of the ML models resulted in a higher HR value, which means that patients may survive for longer if they receive a recommended treatment. The chemotherapy factor was not shown to be significant in the CoxPH model, but the treatment recommendations of DeepSurv and RSF showed significant differences between patients who aligned with the recommendation and those who did not. Although LT is a highly effective treatment for HCC, physicians must select those patients who they believe to have a significant survival advantage following transplantation in order to efficiently utilize a limited supply of liver grafts.[6] After so many years of carefully revising selection criteria for LR and LT, no conclusions can be made in this regard;[6] applying ML to integrate patients’ biochemical, imaging, and genetic information in order to make personalized treatment recommendations may be a possible solution.
To our knowledge, this is the first study harnessing advanced ML techniques to provide individualized treatment suggestions for early-stage HCC based on predictive modeling of survival outcomes. By capturing complex variable relationships, our approach overcomes the limitations of traditional statistical methods that rely on linearity assumptions. The framework presented allows generating nuanced, patient-specific clinical recommendations by integrating various disease features. This demonstrates the translational potential of ML for supporting personalized treatment decisions in early HCC. The novelty lies in the application of modern ML methodology to address an important clinical challenge in HCC management.
There are several limitations to our current study. Prediction accuracy relies heavily on the quality and completeness of the input data. The SEER database may be subject to measurement error or missing data, which could introduce bias and affect model performance. We did not explicitly account for or impute missing values in this analysis. In addition, we have not directly evaluated the measurement properties or reliability of variables in the SEER dataset. Model accuracy statements pertain specifically to prediction on the available SEER data, which may differ from accuracy in unseen real-world clinical populations due to data limitations. In addition, the model requires external validation in other populations before clinical application. Predictor variables were limited to those available in the registry data. Incorporating omics and imaging data could potentially improve model performance. The model does not provide individual risk thresholds to guide decision making. Additional work is needed to determine how best to integrate model predictions into clinical practice.
5. Conclusions
In conclusion, this study demonstrates the potential of ML models to predict survival and guide personalized treatment decisions in early-stage HCC. Our models outperformed traditional Cox regression analysis. By capturing nonlinear effects, they provide nuanced, individualized treatment suggestions. Further validation and incorporation of emerging data modalities may enhance model performance. This illustrates the promise of advanced analytics to aid clinical decision-making in early HCC through personalized predictive modeling.
Author contributions
Conceptualization: Xianguo Li, Haijun Bao, Jinhuang Chen, Xiaogang Shu.
Data curation: Xianguo Li, Yongping Shi, Wenzhong Zhu, Zuojie Peng, Lizhao Yan.
Formal analysis: Xianguo Li, Haijun Bao, Wenzhong Zhu.
Funding acquisition: Jinhuang Chen, Xiaogang Shu.
Methodology: Haijun Bao, Yongping Shi.
Resources: Xianguo Li, Lizhao Yan.
Software: Yongping Shi, Zuojie Peng, Lizhao Yan.
Supervision: Jinhuang Chen, Xiaogang Shu.
Validation: Jinhuang Chen, Xiaogang Shu.
Writing – original draft: Xianguo Li, Yongping Shi.
Writing – review & editing: Xianguo Li, Haijun Bao, Jinhuang Chen, Xiaogang Shu.
Abbreviations:
- AFP
- alpha-fetoprotein
- CI
- confidence interval
- C-index
- concordance index
- CoxPH
- Cox proportional hazards
- HCC
- hepatocellular carcinoma
- HR
- hazard ratio
- HRs
- higher hazard ratios
- IBS
- Integrated Brier Score
- LR
- liver resection
- LT
- liver transplantation
- ML
- machine learning
- N-MLTR
- neural network multitask logistic regression
- OS
- overall survival
- RSF
- random survival forest
- SD
- standard deviation
- SEER
- Surveillance, Epidemiology, and End Results
XL, HB, and YS contributed equally to this work.
The datasets generated during and/or analyzed during the current study are publicly available.
The authors have no conflicts of interest to disclose.
This study was supported by the National Natural Science Foundation of China (NSFC) (Grant number: 82072744,82204874).
Ethical approval for this study was not necessary, since all patients with HCC included in this research were selected from the SEER “18 Regs Research Plus Nov 2020 Sub (2000–2018 varying)” data set. We were authorized to access the database by signing the SEER Research Data Agreement form and sending it via email.
How to cite this article: Li X, Bao H, Shi Y, Zhu W, Peng Z, Yan L, Chen J, Shu X. Machine learning methods for accurately predicting survival and guiding treatment in stage I and II hepatocellular carcinoma. Medicine 2023;102:45(e35892).
Contributor Information
Xianguo Li, Email: lxg555hust@126.com.
Haijun Bao, Email: 1353644739@qq.com.
Yongping Shi, Email: d202282047@hust.edu.cn.
Wenzhong Zhu, Email: d202281886@hust.edu.com.
Zuojie Peng, Email: peng1240454342@qq.com.
Lizhao Yan, Email: yanlizhaozzz@gmail.com.
Jinhuang Chen, Email: huangxiong0603@sina.com.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Behary J, Amorim N, Jiang X-T, et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat Commun. 2021;12:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].He L, Fan X, Li Y, et al. Overexpression of zinc finger protein 384 (ZNF 384), a poor prognostic predictor, promotes cell growth by upregulating the expression of Cyclin D1 in Hepatocellular carcinoma. Cell Death Dis. 2019;10:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fitzmorris P, Shoreibah M, Anand BS, et al. Management of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2015;141:861–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. [DOI] [PubMed] [Google Scholar]
- [6].Sapisochin G, Bruix J. Liver transplantation for hepatocellular carcinoma: outcomes and novel surgical approaches. Nat Rev Gastroenterol Hepatol. 2017;14:203–17. [DOI] [PubMed] [Google Scholar]
- [7].Donadon M, Costa G, Cimino M, et al. Safe hepatectomy selection criteria for hepatocellular carcinoma patients: a validation of 336 consecutive hepatectomies The BILCHE score. World J Surg. 2015;39:237–43. [DOI] [PubMed] [Google Scholar]
- [8].Yin L, Li H, Li A-J, et al. Partial hepatectomy vs transcatheter arterial chemoembolization for resectable multiple hepatocellular carcinoma beyond Milan Criteria: a RCT. J Hepatol. 2014;61:82–8. [DOI] [PubMed] [Google Scholar]
- [9].Zheng S-S, Xu X, Wu J, et al. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation. 2008;85:1726–32. [DOI] [PubMed] [Google Scholar]
- [10].Yang SH, Suh K-S, Lee HW, et al. A revised scoring system utilizing serum alphafetoprotein levels to expand candidates for living donor transplantation in hepatocellular carcinoma. Surgery. 2007;141:598–609. [DOI] [PubMed] [Google Scholar]
- [11].Zhang K, Chen R, Gong X, et al. Survival outcomes of liver transplantation versus liver resection among patients with hepatocellular carcinoma: a SEER-based longitudinal study. J Formos Med Assoc. 2019;118:790–6. [DOI] [PubMed] [Google Scholar]
- [12].Bruix J, Takayama T, Mazzaferro V, et al. Adjuvant sorafenib for hepatocellular carcinoma after resection or ablation (STORM): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2015;16:1344–54. [DOI] [PubMed] [Google Scholar]
- [13].Halazun KJ, Najjar M, Abdelmessih RM, et al. Recurrence after liver transplantation for hepatocellular carcinoma: a new MORAL to the story. Ann Surg. 2017;265:557–64. [DOI] [PubMed] [Google Scholar]
- [14].Toso C, Meeberg G, Hernandez-Alejandro R, et al. Total tumor volume and alpha-fetoprotein for selection of transplant candidates with hepatocellular carcinoma: a prospective validation. Hepatology. 2015;62:158–65. [DOI] [PubMed] [Google Scholar]
- [15].Kaido T, Ogawa K, Mori A, et al. Usefulness of the Kyoto criteria as expanded selection criteria for liver transplantation for hepatocellular carcinoma. Surgery. 2013;154:1053–60. [DOI] [PubMed] [Google Scholar]
- [16].Miltiadous O, Sia D, Hoshida Y, et al. Progenitor cell markers predict outcome of patients with hepatocellular carcinoma beyond Milan criteria undergoing liver transplantation. J Hepatol. 2015;63:1368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Katzman JL, Shaham U, Cloninger A, et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].O’Brien RC, Ishwaran H, Szczotka-Flynn LB, et al. Random survival forests analysis of intraoperative complications as predictors of descemet stripping automated endothelial keratoplasty graft failure in the cornea preservation time study. JAMA Ophthalmol. 2021;139:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].von Elm E, Altman DG, Egger M, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61:344–9. [DOI] [PubMed] [Google Scholar]
- [20].Wolff RF, Moons KGM, Riley RD, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019;170:51–8. [DOI] [PubMed] [Google Scholar]
- [21].Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD). Circulation. 2015;131:211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hankey BF, Ries LA, Edwards BK. The Surveillance, Epidemiology, and End Results program: a national resource. Cancer Epidemiol Biomarkers Prev. 1999;8:1117–21. [PubMed] [Google Scholar]
- [23].Harrell FE, Jr., Lee KL, Califf RM, et al. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143–52. [DOI] [PubMed] [Google Scholar]
- [24].Kang L, Chen W, Petrick NA, et al. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34:685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Brier GW. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;78:1–3. [Google Scholar]
- [26].Liu W, Zhang L, Xin Z, et al. A promising preoperative prediction model for microvascular invasion in hepatocellular carcinoma based on an extreme gradient boosting algorithm. Front Oncol. 2022;12:852736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang J, Huang S, Xu Y, et al. Diagnostic accuracy of artificial intelligence based on imaging data for preoperative prediction of microvascular invasion in hepatocellular carcinoma: a systematic review and meta-analysis. Front Oncol. 2022;12:763842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ivanics T, Nelson W, Patel MS, et al. The toronto postliver transplantation hepatocellular carcinoma recurrence calculator: a machine learning approach. Liver Transpl. 2022;28:593–602. [DOI] [PubMed] [Google Scholar]
- [29].Liu S, Nalesnik MA, Singhi A, et al. Transcriptome and exome analyses of hepatocellular carcinoma reveal patterns to predict cancer recurrence in liver transplant patients. Hepatol Commun. 2022;6:710–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ren S, Li Q, Liu S, et al. Clinical value of machine learning-based ultrasomics in preoperative differentiation between hepatocellular carcinoma and intrahepatic cholangiocarcinoma: a multicenter study. Front Oncol. 2021;11:749137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen W, Zhang T, Xu L, et al. Radiomics analysis of contrast-enhanced CT for hepatocellular carcinoma grading. Front Oncol. 2021;11:660509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun K, Shi L, Qiu J, et al. Multi-phase contrast-enhanced magnetic resonance image-based radiomics-combined machine learning reveals microscopic ultra-early hepatocellular carcinoma lesions. Eur J Nucl Med Mol Imaging. 2022;49:2917–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Adeoye J, Koohi-Moghadam M, Lo AWI, et al. Deep learning predicts the malignant-transformation-free survival of oral potentially malignant disorders. Cancers (Basel). 2021;13:6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Oei RW, Lyu Y, Ye L, et al. Progression-Free survival prediction in patients with nasopharyngeal carcinoma after intensity-modulated radiotherapy: machine learning vs traditional statistics. J Pers Med. 2021;11:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Duncan JK, Ma N, Vreugdenburg TD, et al. Gadoxetic acid-enhanced MRI for the characterization of hepatocellular carcinoma: a systematic review and meta-analysis. J Magn Reson Imaging. 2017;45:281–90. [DOI] [PubMed] [Google Scholar]
- [36].Schulze K, Nault J-C, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65:1031–42. [DOI] [PubMed] [Google Scholar]
- [37].Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–24. [DOI] [PubMed] [Google Scholar]
- [38].She Y, Jin Z, Wu J, et al. Development and validation of a deep learning model for non-small cell lung cancer survival. JAMA Netw Open. 2020;3:e205842–42-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Howard FM, Kochanny S, Koshy M, et al. Machine learning-guided adjuvant treatment of head and neck cancer. JAMA Netw Open. 2020;3:e2025881. [DOI] [PMC free article] [PubMed] [Google Scholar]