Abstract
Methylosinus trichosporium OB3b produces an extracellular copper-binding ligand (CBL) with high affinity for copper. Wild-type cells and mutants that express soluble methane monooxygenase (sMMO) in the presence and absence of copper (sMMOc) were used to obtain cell exudates that were separated and analyzed by size exclusion high-performance liquid chromatography. A single chromatographic peak, when present, contained most of the aqueous-phase Cu(II) present in the culture medium. In mutant cultures that were unable to acquire copper, extracellular CBL accumulated to high levels both in the presence and in the absence of copper. Conversely, in wild-type cultures containing 5 μM Cu(II), extracellular CBL was maintained at a low, steady level during exponential growth, after which the external ligand was rapidly consumed. When Cu(II) was omitted from the growth medium, the wild-type organism produced the CBL at a rate that was proportional to cell density. After copper was added to this previously Cu-deprived culture, the CBL and copper concentrations in the medium decreased at approximately the same rate. Apparently, the extracellular CBL was produced throughout the period of cell growth, in the presence and absence of Cu(II), by both the mutant and wild-type cultures and was reinternalized or otherwise utilized by the wild-type cultures when it was bound to copper. CBL produced by the mutant strain facilitated copper uptake by wild-type cells, indicating that the extracellular CBLs produced by the mutant and wild-type organisms are functionally indistinguishable. CBL from the wild-type strain did not promote copper uptake by the mutant. The molecular weight of the CBL was estimated to be 500, and its association constant with copper was 1.4 × 1016 M−1. CBL exhibited a preference for copper, even in the presence of 20-fold higher concentrations of nickel. External complexation may play a role in normal copper acquisition by M. trichosporium OB3b. The sMMOc phenotype is probably related to the mutant’s inability to take up CBL-complexed copper, not to a defective CBL structure.
Methane monooxygenase (MMO) catalyzes the initial attack on methane by methanotrophic bacteria. This task is sufficiently difficult that nature tolerates an unusual lack of specificity in the substrates that MMO oxygenates; MMO can also initiate the aerobic (cometabolic) transformation of a variety of haloorganic compounds, including trichloroethylene, chloroform, dichloromethane, dichloroethane, trichloroethane, trifluoroethylene, and tribromoethylene among others (6, 9, 17).
Some species (type II species, some type X species, and recently a type I methanotroph) produce both a soluble MMO (sMMO) and a particulate (membrane-associated) MMO (pMMO) (1, 14, 24). Even a minuscule amount of available Cu(II) (0.85 to 1.0 μmol/g [dry weight] of cells) selects for pMMO at the expense of sMMO expression (13). Significantly, sMMO is better suited for catalyzing cometabolic transformations than pMMO is (5, 19, 24).
Copper is thought to be, along with iron, part of the catalytic site of pMMO from Methylococcus capsulatus (Bath) (10). Active pMMO complexes contain as many as 14.5 copper atoms per 99-kDa enzyme complex (25). Most of the copper, however, is loosely bound and may perform secondary functions, such as enzyme stabilization, copper storage, or maintenance of a specific redox state. At Cu concentrations ranging from ≥1 to ≤20 μM, the levels of membrane-associated Cu and Fe and the specific activity of pMMO are directly related to the external copper concentration (25).
Because copper is found in many polluted environments (8), capable organisms are frequently unable to produce sMMO when they are grown under conditions relevant to in situ bioremediation. A recent investigation by Bowman et al. (2) of a trichloroethylene-contaminated aquifer showed that methane injection stimulated growth of indigenous sMMO-producing methanotroph populations. However, the sMMO activity was 41 to 67% lower in the polluted groundwater (with aqueous-phase copper levels of less than 1 μM) than in copper-free, nitrate-salts media (NSM).
Copper has a central regulatory role in Methylosinus trichosporium OB3b, affecting the synthesis of sMMO and pMMO, membrane organization, and the growth rate (11, 15, 18, 23). To overcome the problem of sMMO suppression, Phelps et al. (21) developed mutant strains (sMMOc) that grew well and expressed sMMO in the presence of ≤12 μM copper. These mutants exhibited no pMMO activity and lacked characteristic cytoplasmic membrane structures that are normally present when cells are grown in copper-sufficient media. Fitch et al. (7) reported that sMMOc mutants solubilized extracellular copper but were not capable of copper assimilation.
Here we describe experiments designed to illuminate physiological aspects of the external, copper-binding ligands (CBLs) produced by Methylosinus trichosporium wild-type strain OB3b and by strain PP358, an sMMOc mutant. Our results provide information about the physiology of MMO selection in sMMO-producing methanotrophs and the nature of mutations that are responsible for stable sMMOc phenotypes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type Methylosinus trichosporium OB3b (= ATCC 35070), designated RTR, and sMMOC mutant PP358 (= ATCC 55315) were provided by G. Georgiou of the University of Texas at Austin. The mutant strain was described by Phelps et al. (21) and Fitch et al. (7).
NSM contained 5 mM Na2HPO4 and 5 mM KH2PO4 (adjusted to pH 7.0), as well as 10 mM NaNO3, 1.0 mM K2SO4, 1.0 mM MgSO4 · 7H2O, 0.1 mM CaCl2 · 2H2O, 10 μM NaMoO4 · 2H2O, 1 μM MnSO4 · H2O, 1 μM CoCl2 · 6H2O, 2 μM ZnSO4 · 7H2O, 1 μM KI, and 2 μM H3BO3. Copper and iron were provided from filter-sterilized stock solutions of CuSO4 and FeSO4 at final concentrations of 5 and 40 μM, respectively, after other medium components were autoclaved and cooled. To produce solid media, NSM was supplemented with 1.8% Noble agar (Difco) and 0.1 mg of cycloheximide (a fungal inhibitor) per ml. Stock cultures were maintained as described by Phelps et al. (21). Plates were used to inoculate overnight liquid cultures grown in 165-ml serum vials containing 13 ml of NSM. The serum vials were crimp sealed with Teflon-coated rubber septa. Methane was fed to the vials with sterile 20-ml syringes.
At the mid-log phase (A600, ≈0.4), the cultures described above were used as inocula for 1-liter sealed Erlenmeyer flasks in which the total liquid volume was 100 ml. Methane was added periodically by establishing a partial vacuum in the flask and backfilling with 99.0% pure methane (Union Carbide). The gas phase methane level was maintained at about 20% (vol/vol) by exchanging the headspace volume three times per day. Copper was added to a final concentration of 5 μM or was omitted from the medium. Copper impurities produced a constant background copper level of about 0.25 μM in the “copper-free” medium. Cultures were incubated at 30°C and agitated at 240 rpm on a rotary shaker. Periodically, 3-ml samples were removed and subsamples were used to measure optical density and sMMO activity (naphthalene assay) (see below). High-performance liquid chromatography (HPLC) separations were performed with 200-μl subsamples after passage through a 0.22-μm-pore-size type GS filter (Millipore). Each filtrate was stored at 4°C before analysis. Each of the four culture-growth combinations investigated (RTR with and without copper, PP358 with and without copper) was tested for external ligand production by using two or more independently grown cultures.
At times, experimental objectives required higher ligand concentrations than those that could be generated without concentrating the aqueous medium. On these occasions, the log-phase contents of the 165-ml serum vial reactors were used to inoculate 1-liter sealed Erlenmeyer flasks containing 200 ml of NSM, and the methane levels and incubation conditions were maintained as described above. At an A600 of ≈1.0 (1-cm light path) the contents were used to inoculate a baffled 2-liter bioreactor (Omni-Culture bench top fermentor; Virtis Co., Gardiner, N.Y.). Methane and ambient air were bubbled through the fermentor continuously to maintain the dissolved oxygen concentration at or above 80% of saturation throughout the growth period. Dissolved oxygen was measured with a galvanic electrode (New Brunswick Scientific Co.). Cultures were maintained at 30°C and stirred at 400 rpm during growth. Both wild-type strain RTR and mutant strain PP358 were grown to optical densities (A600) between 1.5 and 2.0 (1-cm light path) in the presence of 5 μM Cu(II) and in the absence of Cu(II) [0.25 μM Cu(II)]. Suspensions were centrifuged (10,000 × g, 10 min), after which the concentrate was filtered with a 0.22-μm-pore-size type GS filter. The filtrates were lyophilized (4.5-liter Labconco unit) and stored at −20°C as source materials for subsequent experiments.
sMMO activity.
Culture sMMO activity was examined by using the naphthalene colorimetric assay of Brusseau et al. (3). To obtain a quantitative estimate of sMMO activity, several 0.5-ml aliquots of a cell-formate suspension were mixed with 0.5 ml of the saturated naphthalene preparation and incubated at 30°C. These preparations were sacrificed at 10-min intervals and used to develop and measure color at 528 nm. A528 values were converted to concentrations of the naphthol-azo dye by using an assumed extinction coefficient of 38,000 M−1 cm−1 (3).
Analytical techniques.
In all experiments, growth was monitored by measuring light scattering at a wavelength of 600 nm (A600; 1-cm cuvette) with a Shimadzu model UV160U recording spectrophotometer.
Copper and nickel were analyzed by using a Perkin-Elmer model 303 atomic absorption spectrophotometer equipped with a model HGA-400 graphite furnace and single-element copper and nickel lamps (λCu = 324.8 nm; λNi = 222.0 nm; Photron). Copper standards were prepared from oven-dried Cu(NO3)2 dissolved in water with 0.1% HNO3. Nickel standards were purchased from Perkin-Elmer.
Paper chromatography.
Ascending paper chromatography was performed as described by Smith and Seakins (22) with a solvent mixture consisting of acetic acid, n-butanol, and water (3:12:5, by volume). Fractions of the size exclusion HPLC (SE-HPLC) peak eluting near 25 ml (see below) were pooled for analysis. A 5-μl aliquot was applied to the chromatographic paper (8 by 20 cm; type 3MM; Whatman) and dried with a hair drier at the lowest setting. In order to increase the amount of sample applied to the paper, the application procedure was repeated 10 times. Chamber equilibration with the solvent occurred overnight. The prepared paper was then introduced into the solvent chamber to initiate the separation. Runs were terminated after 4 h. After the chromatogram was removed from the chamber, it was dried at 100°C in an oven for 10 min and sprayed with a 5-mg/ml solution of ninhydrin (Sigma) in acetone for staining. Chromatographs were analyzed under visible and/or UV light.
Several other qualitative staining techniques were used to investigate possible functional moieties of the CBL without chromatographic separation. Tests were performed on Whatman 3MM paper after 10 depositions (5 μl each) of a 20-mg/ml solution of lyophilized filtrate from PP358 grown in copper-free media; 20 μl of the reagents was added to each sample spot, and the results were compared with positive and negative controls. The presence of sugars was determined with an aniline-based solution, the presence of histidines was determined with Pauly’s reagent, and the presence of phenols was determined with Folin-Ciocalteu reagent. Tryptophan-containing metabolites were assayed with Ehrlich reagent. Reagents were prepared as described by Smith and Seakins (22).
SE-HPLC.
Normally, samples were obtained for SE-HPLC directly from cell cultures, as described previously. Alternatively, lyophilized filtrate was suspended in Tris buffer (0.02 M Tris [Sigma], 0.1 M NaCl; pH 7.5) at a concentration of 30 mg/ml for use in SE-HPLC. The solution was passed through a type SE-100 17-μm column (30 by 1 cm; Innovata, Sweden). A 200-μl sample loop was used for all SE-HPLC experiments. Cu(II) (5 μl of a 10 mM CuSO4 stock solution) was added to some samples to mark fractions with a high affinity for copper and to establish the binding capacity of the ligand present. All samples were eluted at a flow rate of 0.4 ml/min by using the same Tris buffer. Fractions (0.5 ml) were collected and assayed spectrophotometrically (A280). The copper was measured in each sample before and after spiking with CuSO4 and in eluent fractions.
Molecular weight determination.
The molecular weight standards used included bovine serum albumin (molecular weight, 67,000), ovalbumin (43,000), RNase A (13,700), horse heart cytochrome c (12,400), eledoisin-related peptide (700), and tryptophan (204) (Sigma). Urea (3 M) was added to the Tris buffer to eliminate potential hydrophobic interactions between solutes and the SE-100 HPLC column material. The elution volume was then correlated with the molecular weight of the standards by performing a linear regression analysis.
Characterization of CBL binding strength.
In competition experiments, the biochelator (CBL) was mixed with copper and a chelator of known affinity and binding strength for copper. The chelators used (which were obtained from Sigma) included triethylenetetraamine (TRIEN), EDTA, nitrilotriacetic acid (NTA), ethylenediamine (EN), diethylenetriamine (DEN), iminodiacetic acid (IDA), and tryptophan. In order to isolate CBL for competition experiments, lyophilized cell filtrate from PP358 cultures grown in medium containing 5 μM copper was prepared for SE-HPLC as described previously. Fractions of the peak eluting near 25 ml (designated the peak of interest [POI]) were pooled, and 120 μl of the combined fractions was mixed with 70 μl of SE-HPLC Tris buffer and 50 μl of a 50 μM stock solution of a second (competing) copper chelator prepared in SE-HPLC Tris buffer. The molar concentration of the competing ligand was approximately equal to the concentration of copper and, as such, approximately equal to the copper binding capacity of the POI added in these blends. The resulting mixture was stored at room temperature for 24 h before components were again separated by SE-HPLC. The copper in the eluted fractions was measured to establish the distribution of this element in the biochelator and the competing ligand. Additional analyses were performed by using DEN mixed with pooled fractions of the POI at three DEN/Cu molar ratios. The amount of POI was kept constant (130 μl from the pooled fractions), and the amount of DEN was varied to produce DEN/Cu molar ratios of 0.5, 1.0, and 2.0. SE-HPLC was again used to separate the mixtures, and the copper in the fractions was measured to determine its distribution in DEN and the CBL.
The constant for association between copper and the CBL was estimated by using the following stoichiometric expressions:
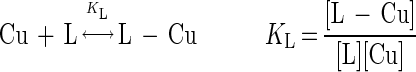 |
1 |
 |
2 |
where L is the competing ligand (in this case DEN) and n is the number of Cu atoms bound to a single CBL molecule.
The concentrations of both copper complexes (L-Cu and CBL-Cun) were determined experimentally by performing copper analyses of the respective peak fractions after SE-HPLC. The concentrations of free chelators (L and CBL) were estimated from mass balances, after the amount of the copper-complexed fraction (L-Cu and CBL-Cun) was subtracted from the original amount of chelator provided. KCBL was then calculated by assuming an integer value for n (n = 1, 2, or 3) and solving equations 1 and 2 simultaneously. The equilibrium constant for the copper-DEN complex was log KL = 16.0 (20).
CBL specificity for copper.
POI solutions were obtained by pooling fractions from the SE-HPLC 25-ml peak, after the original PP358 sample was spiked with copper as described above. Portions (300 μl) of the resultant solutions were mixed with copper and nickel stock solutions to give final liquid phase metal concentrations of 5 μM (Cu) and 100 μM (Ni). Citrate was added to each mixture to a final concentration of 200 μM to complex excess metal ions not bound to the CBL. We prepared a control solution in which 300 μl of HPLC Tris buffer replaced the POI solution. After storage at room temperature for 24 h (for attainment of equilibrium), the mixtures were separated by SE-HPLC. Copper and nickel analyses were performed to determine which metal ion was bound to the CBL.
CBL-Cu uptake experiments.
The CBL-Cu uptake experiments were designed to determine the manner in which Cu affects the production and utilization of the CBL and the nature of the sMMOC mutation in PP358. Both strain RTR and strain PP358 were grown without copper in 1-liter Erlenmeyer flasks to optical densities (A600) of 1.0 to 1.5. Then the cultures were divided, and copper (10 μM in the RTR culture, 5 μM in the PP358 culture) was added to one-half of the reactor contents. In each case the second half of the culture was left undisturbed. Incubation and growth were continued for both fractions. The periodic measurements obtained included measurements of A600, sMMO activity (quantitative assay), the POI area (as determined by SE-HPLC), and the aqueous- and particulate-phase Cu concentrations. For Cu measurements, 1.5-ml culture samples were centrifuged for 10 min at 6,600 × g (Eppendorf model 5415 Centrifuge). The concentrate was passed through a 0.22-μm-pore-size type GS filter (Millipore) before the soluble Cu in the filtrate was measured. The cell pellet was resuspended in 1.5 ml of a 10 mM EDTA solution (pH 4.8) and incubated for 10 min with constant agitation. The resultant solution was again centrifuged, and the Cu in the filtered concentrate was measured to determine the amount of EDTA-extractable, particulate Cu. A second pellet was resuspended in 1.5 ml of deionized water to measure the particulate, nonextractable copper. The entire procedure was carried out twice by using two sets of independently grown RTR and PP358 cultures.
Culture and medium transfer experiments.
Another set of experiments was designed to see if the PP358 phenotype was related to a defect in the CBL produced by that strain. Our objectives were pursued by transferring (i) wild-type (RTR) cells to cell-free media containing CBL produced by the mutant strain and (ii) PP358 cells to cell-free media including CBL that had previously supported growth of strain RTR.
Both the RTR and PP358 cultures were grown to the mid-log phase (A600, ≈1.0-1.5) in 1-liter Erlenmeyer flasks containing copper-free NSM (liquid volume, 150 ml). Then, cells were separated from the liquid phase by a combination of centrifugation and filtration (0.22-μm-pore-size type GS filter [Millipore]), which yielded cell-free medium and a washed cell suspension for each original culture. These preparations were recombined by resuspending RTR cells in the cell-free PP358 growth medium and the mutant cells in the cell-free RTR medium (initial A600, ≈1.0). Copper was added to each culture-medium combination to a final concentration of 5 μM. Periodic samples obtained from each recombined culture were used to measure growth (A600), the sMMO activity, the CBL concentration, and the soluble, extractable, and particulate copper concentrations.
RESULTS
sMMO activity.
PP358 cultures expressed sMMO during growth on NSM containing methane with and without 5 μM Cu, as shown by naphthol production from naphthalene. As expected, sMMO activity was absent in RTR cultures containing 5 μM Cu, although sMMO was expressed in RTR cultures without copper. In copper-amended cultures, the ratio of total copper to cell dry weight at the maximum cell density was about 5.8 μmol/g of cell dry weight (CDW), a value that should have prevented sMMO expression in the RTR culture (13).
SE-HPLC.
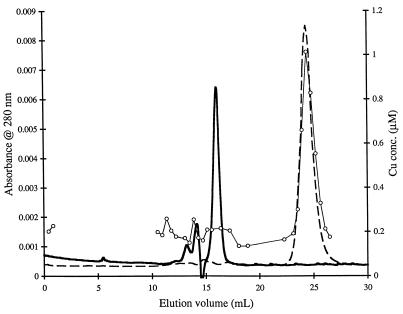
Representative chromatograms derived from the four cultures tested (PP358 and RTR, each with and without 5 μM Cu) are shown in Fig. 1 and 2. Each of the curves was developed without concentrating aqueous-phase constituents by lyophilization. Although the results of replicate experiments are not presented, the chromatograms were reproducible. Copper-complexing agents were detectable at 280 nm, as shown by the coelution of high copper levels and chromatogram peaks. The general features of the chromatograms derived from the mutant cultures grown with and without copper were similar (Fig. 1). In each case, copper was associated primarily with a single peak (the POI) at an elution volume of about 25 ml. The medium blank produced no such peak. Thus, in the presence or absence of external copper, the mutant strain excreted one or more (coeluting) ligands with affinity for copper. The POI was also present in the wild-type cultures with and without copper, but accumulated to only modest levels when 5 μM Cu was provided (Fig. 2). There was evidence of copper accumulation in a second peak at an elution volume of about 12 ml in chromatograms derived from the copper-free RTR culture.
FIG. 1.
Typical SE-HPLC chromatograms and corresponding copper analysis results for filtrates derived from batch PP358 cultures. The copper concentrations in the original growth media and the culture optical densities at the points of analysis were as follows: 0.25 μM Cu (copper free) and A600 = 1.69 (A); and 5 μM Cu and A600 = 1.06 (B). ○——○, copper concentration; ——, A280. A dashed line indicates an absence of data in a region. Abs., absorbance.
FIG. 2.
Typical SE-HPLC chromatograms and corresponding copper analysis results for filtrates derived from batch RTR cultures. The copper concentrations in the original growth media and the culture optical densities at the points of analysis were as follows: 0.25 μM Cu (copper free) and A600 = 1.15 (A); and 5 μM Cu and A600 = 1.12 (B). ○——○, copper concentration; ——, A280. A dashed line indicates an absence of data in a region. Abs., absorbance.
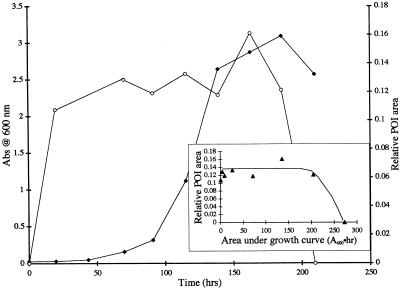
The accumulation of compounds that contributed to the POI, as indicated by the time-dependent peak area at 25 ml (on SE-HPLC chromatograms), differed significantly in the mutant and wild-type cultures (Fig. 3 and 4). Until PP358 reached the stationary growth phase, the POI area increased at a rate that was about proportional to culture optical density (Fig. 3). Upon entry into the stationary phase, however, no further increase in POI area was evident. The patterns of growth and POI development were similar for mutant cultures grown in the presence and absence of copper, and a similar pattern was obtained with the copper-free, wild-type culture (data not shown). In the RTR culture supplemented with 5 μM Cu(II), however, the POI area stabilized quickly at a low level and remained steady throughout the period of cell growth (Fig. 4). At the onset of the stationary growth phase the POI area decreased to undetectable levels in the culture medium. In replicate experiments (at least two replicates for each of the four culture-copper level combinations), the results were qualitatively identical. Only illustrative cases are included here for the sake of brevity.
FIG. 3.
Growth (A600) and relative POI area (obtained from HPLC chromatograms) as a function of time in a PP358 batch culture grown without copper. Symbols: ⧫, culture A600; ○, relative POI area. (Inset) Relative POI area (▴) as a function of the first moment of bacterial growth (area under bacterial growth curve). The relative POI area is the area of the SE-HPLC peak at a 25-ml elution volume divided by a representative peak area derived from the PP358 cultures (A600 = 1.06; 5 μM Cu). The line is a manual fit to the data. The experiment was repeated, and the same qualitative results were obtained. Abs, absorbance.
FIG. 4.
Growth (A600) and relative POI area (obtained from HPLC chromatograms) as a function of time in an RTR batch culture grown with 5 μM Cu. Symbols: ⧫, culture A600; ○, relative POI area. (Inset) Relative POI area (▴) as a function of the first moment of bacterial growth (area under bacterial growth curve). The relative POI area is the area of the SE-HPLC peak at a 25-ml elution volume divided by a representative peak area derived from the PP358 cultures (A600 = 1.06; 5 μM Cu). The line is a manual fit to the data. The experiment was repeated, and the same qualitative results were obtained. Abs, absorbance.
Notice that the relative POI area (Fig. 3 and 4, insets) represents the area of the SE-HPLC peak at a 25-ml elution volume normalized by using the peak area from the Fig. 1B chromatogram (PP358 culture; A600 = 1.06; culture grown in the presence of 5 μM Cu). Clearly, much less of the CBL accumulated in the copper-amended RTR culture than in the PP358 cultures.
When chromatograms were derived from samples containing excess copper, the peak area at 25 ml was assumed to be proportional to the concentration of the CBL in the sample. The constant of proportionality (r) was obtained from the copper present in pooled HPLC fractions for the same peak by assuming that the copper-CBL stoichiometry was 1:1. Based on the results of 10 such measurements of peak area and copper, the r was estimated to be 0.137 ± 0.043 μM CBL per unit of POI area. The mean value was at times used to calculate CBL concentrations based on the measured peak areas at an elution volume of 25 ml. During mid-log-phase growth, the apparent CBL masses (peak areas) for both mutant cultures (with and without copper) and for the wild type grown with no copper were 1 order of magnitude greater than the apparent CBL mass of the RTR culture grown in the presence of 5 μM Cu (Table 1).
TABLE 1.
Copper-complexing capacities of accumulated CBL in growth media derived from Methylosinus trichosporium OB3b (=RTR) and PP358
| Culture | Cu concn (μM) | A600 | Cu-complexing capacity (μmol of copper · g of CDW−1)a |
|---|---|---|---|
| RTR | 0.25 | 2.87 | 1.85 |
| 5 | 2.90 | 0.20 | |
| PP358 | 0.25 | 2.76 | 2.07 |
| 5 | 1.69 | 2.55 |
One A600 unit corresponds to 0.43 g of CDW/liter (7). POI mass was obtained by measuring the POI area and converting it by using the following conversion factor: 0.137 μM CBL per HPLC POI area unit (see text).
To determine whether the POI contained a single compound or multiple compounds, eluent fractions including the 25-ml peak (PP358 culture grown without Cu) were subjected to a second separation step by using SE-HPLC. Again, only one major peak was observed in SE-HPLC preparations (Fig. 5). The peaks of absorbance and copper concentration again coincided. In the absence of the POI, copper eluted from the column at an elution volume of approximately 15 ml (data not shown). The 15-ml peak was also examined by paper chromatography. Only one (ninhydrin-UV) spot was detected in the POI lane (data not shown), again suggesting that the POI contained only one primary compound. The spot was pink in the visible spectrum and fluoresced under UV light. There was no spot prior to the ninhydrin treatment. Primary amines typically yield a purple color when they are treated with ninhydrin (22).
FIG. 5.
SE-HPLC chromatograms developed from fractions that comprised the POI at 25 ml (PP358 culture grown with 5 μM Cu). – – –, SE-HPLC obtained with Tris as the elution buffer; ——, SE-HPLC obtained with Tris–3 M urea as the elution buffer; ○——○, copper concentration in fractions derived from Tris eluent containing no urea.
Qualitative analysis of CBL functional groups.
POI compounds, including the CBL, failed to produce color with chromogenic reagents designed to react with sugars, histidine, tryptophan, and phenols. In the UV range, pooled fractions from the SE-HPLC peak at 25 ml showed a peak at about 270 nm. There were no other distinguishing spectral features.
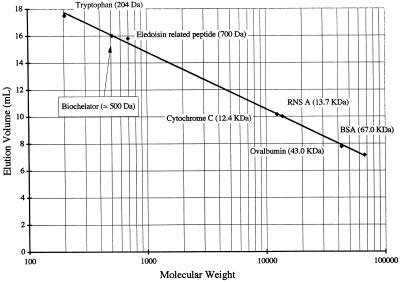
Molecular mass determination.
The POI elution time in Tris eluent containing 3 M urea suggested that molecules contributing to the POI have a molecular mass of about 500 Da (Fig. 5 and 6). The peaks at elution volumes of 13 to 14 ml (Fig. 5) corresponded approximately to the molecular masses of POI multimers.
FIG. 6.
SE-HPLC volumes for elution of standards having known molecular weights. The eluent consisted of Tris buffer plus 3 M urea. The calibration curve was used to estimate the molecular weight of the CBL, as shown. RNS A, RNase A; BSA, bovine serum albumin.
Characterization of CBL binding strength.
TRIEN and EDTA had greater affinity for the free cupric ion than the external chelator produced by Methylosinus trichosporium OB3b did (Table 2). Conversely, NTA, EN, tryptophan, and IDA bound copper less effectively than the CBL. The association constant for the CBL-Cu complex apparently is between the association constants for NTA (log K1 = 13.1) (20) and EDTA (log K1 = 18.8) and close to the association constant for DEN (log K1 = 16.0). The residual copper complexed with NTA, tryptophan, and IDA represents a stoichiometric excess (above the capacity of the biochelator). Similarly, the residual copper associated with the POI in experiments involving TRIEN, EDTA, and DEN suggested that the capacity of the competing ligands to bind copper was exceeded slightly in these experiments or that equilibrium was not completely attained over the 24-h incubation period that preceded the separation.
TABLE 2.
Fractions of copper associated with different chelators when they were mixed with a Cu-saturated POI solutiona
The estimated values for the association constant between the CBL and copper, based on partitioning of copper between DEN and the CBL (Table 2) and application of equations 1 and 2, are presented in Table 3. If it is assumed, in the absence of stoichiometric data, that n = 1 (1:1 stoichiometry for the Cu-CBL complex), the Cu-CBL association constant can be estimated to be 1.4 × 1016 M−1. Estimates obtained with other integer values for n are also provided in Table 3.
TABLE 3.
Copper association constants for CBL assuming different n values for the Cun-CBL stoichiometrya
| DEN/Cu ratio in POI |
KCBL (M−1)
|
||
|---|---|---|---|
| n = 1 | n = 2 | n = 3 | |
| 0.5:1 | 1.6 × 1016 | 5.1 × 1032 | 1.7 × 1049 |
| 1:1 | 2.6 × 1016 | 9.0 × 1032 | 3.4 × 1049 |
| 2:1 | 3.1 × 1015 | 4.2 × 1031 | 5.7 × 1047 |
| Avg | 1.4 × 1016 | 1.4 × 1032 | 1.7 × 1049 |
See equations 1 and 2 in the text.
CBL specificity for copper.
Analyses of HPLC fractions from the POI-Cu-Ni mixture revealed that copper remained bound to the CBL even when the total nickel concentration was 20 times the total copper concentration (data not shown); that is, the complexation of copper with a ligand(s) that comprised the POI was not affected by excess nickel. Furthermore, under the conditions used, the distribution of Ni in chromatograms was not affected by the presence of the biochelator.
CBL disappearance and copper uptake: copper mass balance.
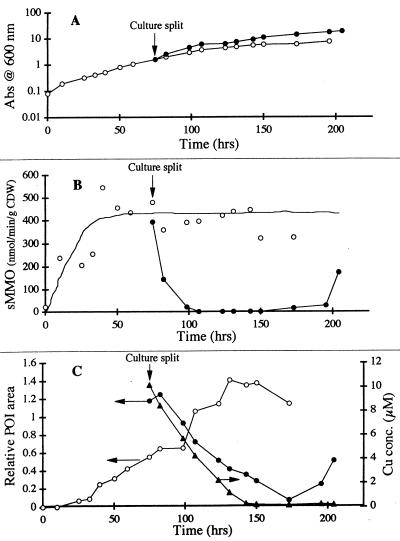
The effect of adding copper to an RTR culture that was grown under copper-free conditions is illustrated by the results of a comparison of copper-amended and unamended fractions (Fig. 7). In the unamended (copper-free) culture, CBL accumulated in the medium to levels that were similar to those observed previously (Table 1). As expected, the sMMO activity in the unamended fraction was not affected by splitting the culture. Conversely, the sMMO activity in the Cu-amended, wild-type culture decreased dramatically after copper was added, and the POI concentration decreased until the POI was virtually undetectable. Growth was slightly faster in the Cu-amended fraction.
FIG. 7.
Response of RTR batch cultures grown without copper to the addition of 5 μM copper. (A) Growth of cultures with and without copper. (B) sMMO activity (as determined by the naphthalene assay). (C) Copper and CBL accumulation and uptake. The original culture was split, and copper was added to one aliquot at the point shown. Symbols: •, culture to which copper was added; ○, copper-free culture. The lines represent manual fits to the data. The entire experiment was carried out two times, and the same qualitative results were obtained. The relative POI area is the area of the SE-HPLC peak at a 25-ml elution volume divided by a representative peak area derived from the PP358 cultures (A600 = 1.06; 5 μM Cu). Abs, absorbance.
It is also noteworthy that in the copper-amended fraction, the level of aqueous-phase copper decreased parallel to the level of POI until it reached background levels. When the added copper was exhausted near the end of the experiment, the wild-type cells again produced sMMO, and the extracellular levels of CBL also increased. Periodic measurements of particulate copper contents indicated that copper lost from the liquid phase was, in fact, assimilated into cell mass (data not shown). A reasonable mass balance (±20%) was maintained for total copper throughout the experiment. Relatively little (<10%) of the particulate copper was solubilized during EDTA extraction. In the mutant culture, about 80% of the 5 μM copper added remained soluble throughout the 5-day (post-copper-addition) incubation period. Essentially no copper was assimilated, and sMMO remained active throughout the experiment (data not shown). As expected, the extracellular levels of CBL also remained high. The results of a replicate experiment (data not shown) displayed all of the same general features.
Culture-medium transfer experiments.
RTR cells grown in copper-free NSM were washed and resuspended in cell-free media derived from the PP358 culture containing 5 μM Cu. Under these conditions, extracellular copper was taken up by the wild-type organism, and the extracellular CBL concentration declined proportionally (Table 4). Furthermore, the sMMO activity in copper-starved wild-type cells decreased immediately with no interruption of growth.
TABLE 4.
CBL-Cu uptake experiments: summary of resultsa
| Exptl conditions
|
Results obtained during CBL-Cu uptake phase
|
||||
|---|---|---|---|---|---|
| Strain used for uptake exptb | Origin of CBL used for uptake exptc | Initial Cu concn during uptake phase (μM) | sMMO activityd | CBL uptake (+/−)e | Cu uptake |
| RTR | RTR | 0.0 | + | − | NAf |
| RTR | RTR | 5.0 | − | + | + |
| RTR | PP358 | 0.0 | + | − | NA |
| RTR | PP358 | 5.0 | − | + | + |
| PP358 | RTR | 5.0 | + | − | − |
RTR and PP358 cultures were grown in NSM without copper. Washed RTR cells were then added to media containing CBL from the PP358 mutant; PP358 cells were added to media containing CBL from the wild-type strain. Copper was provided as indicated. The results (sMMO expression, CBL uptake, and copper uptake) suggest that (i) RTR cells take up copper that is complexed with CBL from the mutant strain and (ii) PP358 cannot acquire copper, even when it is bound to CBL from the wild-type organism.
Both RTR and PP358 were initially grown without copper. Each strain expressed sMMO at the conclusion of the growth period.
Obtained by removing cells from (copper-free) media after cell growth.
+, continuation of sMMO activity, as indicated by naphthalene conversion; −, rapid decline in sMMO activity.
−, further accumulation of CBL in growth medium during the uptake phase (no CBL uptake); +, rapid loss of CBL following cell resuspension (CBL uptake).
NA, not applicable.
A control of sorts was prepared by resuspending RTR cells grown without copper in cell-free media (no Cu) derived from a PP358 culture that was grown without copper. As expected, sMMO expression and growth continued following resuspension of the RTR cells, and the extracellular concentration of CBL increased monotonically as long as cell growth was observed.
The inverse experimental procedure involved resuspension of mutant cells in cell-free media derived from an RTR culture that was grown without copper. Approximately 5 μM copper was added before growth was reinitiated with the resuspended cells. Neither growth nor sMMO activity was interrupted in the resuspended culture. Following the transfer, there was no copper assimilation by mutant cells, even in the presence of CBL produced by wild-type organisms, and the extracellular levels of the CBL increased throughout the subsequent period of growth.
DISCUSSION
Our observations are consistent with the following speculative picture. Both strain RTR and mutant derivatives of Methylosinus trichosporium OB3b excrete an extracellular CBL in the presence and absence of copper. Wild-type cells recover CBL and internalize copper only when copper is bound by the CBL. Cells with the sMMOc mutant phenotype cannot reacquire the CBL or copper. Our results suggest that the CBL plays a role in copper assimilation by the wild-type organism.
Several lines of evidence (data from sequential SE-HPLC separations and urea addition experiments, changes in ionic strength, data from ligand competition experiments and paper chromatography) suggest that a single primary compound was responsible for the SE-HPLC peak at an elution volume of 25 ml. There is reason to suspect that ligands produced by strains RTR and PP358 are functionally, if not structurally, identical. CBLs from the mutant and wild-type organisms both elute at 25 ml during SE-HPLC separations. Both readily bind copper, both promote copper uptake by wild-type cells, and neither can supply copper to the mutant.
In general, our findings support hypotheses concerning the origin of the sMMOc phenotype that were proposed by Fitch et al. (7) (i.e., that the mutant phenotype resulted from a defect in the mechanism of copper acquisition by Methylosinus trichosporium OB3b). Unlike wild-type cells, PP358 was Cu starved in the presence or absence of an external source of copper. It is evident that the mutant phenotype arises from the cells’ inability to take up complexed copper, perhaps due to the absence of or a defect in a membrane-bound receptor protein, but certainly not due to a defect in the CBL.
During growth of the wild-type culture without copper and in cultures of PP358 under all copper conditions investigated, the rate of CBL accumulation was about proportional to cell density (Fig. 3). Our results suggest that the ligand was produced at a rate that was proportional to cell number and that reinternalization of CBL was negligible under the growth conditions used. The early stabilization of the CBL concentration in the wild-type culture supplemented with copper (Fig. 4) suggests that CBL production was matched by uptake during exponential growth. Assuming that the intracellular copper concentration is about 1 μmol of Cu per g of CDW in Methylosinus trichosporium OB3b (under copper-sufficient conditions), the calculated rate of CBL production and copper acquisition is about 500 molecules · (cell · min)−1. This value is based on an apparent doubling time of 20 h and a conversion factor of 1012 cells per g of CDW and is of the same order of magnitude as the calculated specific rate of CBL accumulation in the mutant culture (Fig. 3). The calculation and comparison suggest that CBL is produced at a rate that nearly balances the cells’ need for copper under maximum-growth conditions and accounts for the lack of CBL accumulation in wild-type cultures grown with copper.
The presence of a peak near 280 nm in the POI UV spectrum suggests that the biochelator has at least one aromatic functional group. The results of ninhydrin chromogenic reactions are not conclusive with respect to the peptidic nature of the biochelator. The other qualitative tests performed indicated that histidine, tryptophan, and phenol derivatives do not contribute to the CBL structure.
Unlike other UV-absorbing peaks in Fig. 1 and 2, elution of the CBL was delayed significantly by hydrophobic interactions with the size exclusion gel. Higher eluent ionic strength (concentration range, 0.05 to 0.25 M) increased the elution volume of the POI but did not significantly modify the elution pattern of other components in the mixture (data not shown). Addition of 3 M urea to the elution buffer accelerated the appearance of the POI (Fig. 5), suggesting that hydrophobic interactions significantly retarded transport of the Cu-ligand complex. Urea is commonly added to elution buffers to eliminate hydrophobic interactions between the gel matrix and the analytes to be separated.
Although the effectiveness of CBL for copper detoxification has not been tested, the strength of copper complexation by the isolated ligand suggests that copper acquisition is its primary function. A weaker ligand might suffice for metal detoxification. Moffett and Brand (16) reported that the binding constant for complexes involving Cu and a biochelator produced by a marine bacterium in response to copper stress is between 1012 and 1013 M−1. Our results indicate that the binding constant for the Cu-CBL complex is greater by 3 to 4 orders of magnitude.
A detailed mechanistic explanation for the role of the CBL in copper acquisition remains to be established. Our observations could be explained, for example, by uptake of the copper-CBL complex, by degradation of CBL (accompanied by copper uptake), or by sudden changes in the balance of uptake and excretion. Hypotheses in this area remain speculation. When complete, the emerging physiological picture of copper acquisition by Methylosinus trichosporium OB3b may enable us to promote bioremediation efforts that are now inhibited by copper-mediated pMMO selection. Experiments designed to better establish biochelator function in Methylosinus trichosporium OB3b are in progress.
In summary, we identified and partially characterized a biochelator that accumulates in mutant cultures (sMMOc, PP358) of Methylosinus trichosporium OB3b in the presence and absence of external copper. The ligand is produced constitutively during periods of growth by both mutant and wild-type cultures. The wild-type cells are able to utilize the CBL when external copper is present. However, under no circumstances does the mutant reacquire the biochelator. The mutation in PP358 seems to be unrelated to possible defects in the CBL since CBL produced by PP358 was readily utilized by a growing RTR culture. Furthermore, CBL produced by strain RTR did not support copper uptake by the mutant. The biochelator is apparently a low-molecular-weight, hydrophobic molecule with high affinity and selectivity for copper. It seems to have some aromatic character. We hypothesize that the biochelator is part of the normal mechanism of copper uptake by Methylosinus trichosporium OB3b. Metal detoxification remains an alternative, although much less likely, physiological explanation for ligand production.
ACKNOWLEDGMENTS
This research was supported in part by grant ES04940 from the National Institute of Environmental Health Sciences. D.W.G. was funded at Kansas University by grant BES 9504383 from the National Science Foundation.
We thank A. Aguilar for help with operation of the bench top bioreactor.
REFERENCES
- 1.Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;27:113–209. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- 2.Bowman J P, Jimenez L, Rosario I, Hazen T C, Sayler G S. Characterization of the methanotrophic bacterial community present in a trichloroethylene-contaminated groundwater site. Appl Environ Microbiol. 1993;59:2380–2387. doi: 10.1128/aem.59.8.2380-2387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brusseau G A, Tsien H C, Hanson R S, Wackett L P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1:19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- 4.Chabarek S, Martell A E. Stability of metal chelates. I. Iminodiacetic and iminopropionic acids. J Am Chem Soc. 1952;74:5052–5056. [Google Scholar]
- 5.Colby J, Stirling D I, Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers and alicyclic, aromatic and heterocyclic compounds. Biochem J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ensley B D. Biochemical diversity of trichloroethylene metabolism. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- 7.Fitch M W, Graham D W, Arnold R G, Agarwal S K, Phelps P, Speitel G E, Georgiou G. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1993;59:2771–2776. doi: 10.1128/aem.59.9.2771-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forstner U, Wittman G T W. Metal pollution in the aquatic environment. New York, N.Y: Springer-Verlag; 1979. p. 356. [Google Scholar]
- 9.Fox B G, Froland W A, Dege J E, Lipscomb J D. Methane monooxygenase from Methylosinus trichosporium OB3b. J Biol Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 10.Green J, Dalton H. Protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath) J Biol Chem. 1985;20:15795–15801. [PubMed] [Google Scholar]
- 11.Green J, Prior S D, Dalton H. Copper ions as inhibitors of protein C of soluble methane monooxygenase of Methylococcus capsulatus (Bath) Eur J Biochem. 1985;153:137–144. doi: 10.1111/j.1432-1033.1985.tb09279.x. [DOI] [PubMed] [Google Scholar]
- 12.Hancock R D, Martell A E. Ligand design for selective complexation of metal ions in aqueous solution. Chem Rev. 1989;89:1875–1914. [Google Scholar]
- 13.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh S-C, Bowman J P, Sayler G S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leak D J, Dalton H. Growth yields of methanotrophs. I. Effect of copper on the energetics of methane oxidation. Appl Microbiol Biotechnol. 1986;23:470–476. [Google Scholar]
- 16.Moffett J W, Brand L E. Production of strong, extracellular Cu chelators by marine cyanobacteria in response to Cu stress. Limnol Oceanogr. 1996;41:388–395. [Google Scholar]
- 17.Oldenhuis R R L, Vink J M, Jannsen D B, Wiltholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989;55:2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S, Hanna M L, Taylor R T, Droege M W. Batch cultivation of Methylosinus trichosporium OB3b. I. Production of soluble methane monooxygenase. Biotechnol Bioeng. 1991;38:423–433. doi: 10.1002/bit.260380412. [DOI] [PubMed] [Google Scholar]
- 19.Patel R N, Hou C T, Laskin A I, Felix A. Microbial oxidation of hydrocarbons: properties of a soluble methane monooxygenase from a facultative methane-utilizing organism, Methylobacterium sp. strain CRL-26. Appl Environ Microbiol. 1982;44:1130–1137. doi: 10.1128/aem.44.5.1130-1137.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrin D D. Stability constants of metal-ion complexes, part B. Organic ligands. IUPAC Chemical Data Series no. 22. Oxford, United Kingdom: Pergamon Press; 1979. [Google Scholar]
- 21.Phelps P A, Agarwal S K, Speitel G E, Jr, Georgiou G. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl Environ Microbiol. 1992;58:3701–3708. doi: 10.1128/aem.58.11.3701-3708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith I, Seakins J W T. Chromatographic and electrophoretic techniques. Chicago, Ill: Year Book Medical Publishers; 1975. [Google Scholar]
- 23.Stanley S H, Prior S D, Leak D J, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 24.Tsien H-C, Hanson R S. Soluble methane monooxygenase component B gene probe for identification of methanotrophs that rapidly degrade trichloroethylene. Appl Environ Microbiol. 1992;58:953–960. doi: 10.1128/aem.58.3.953-960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahn J A, Dispirito A A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]