Summary
Intrauterine infection/inflammation (IUI) is a frequent complication of pregnancy leading to preterm labor and fetal inflammation. How inflammation is modulated at the maternal-fetal interface is unresolved. We compared transcriptomics of amnion (a fetal tissue in contact with amniotic fluid) in a preterm Rhesus macaque model of IUI induced by lipopolysaccharide with human cohorts of chorioamnionitis. Bulk RNA sequencing (RNA-seq) amnion transcriptomic profiles were remarkably similar in both Rhesus and human subjects and revealed that induction of key labor-mediating genes such as IL1 and IL6 was dependent on nuclear factor κB (NF-κB) signaling and reversed by the anti-tumor necrosis factor (TNF) antibody Adalimumab. Inhibition of collagen biosynthesis by IUI was partially restored by Adalimumab. Interestingly, single-cell transcriptomics, flow cytometry, and immunohistology demonstrated that a subset of amnion mesenchymal cells (AMCs) increase CD14 and other myeloid cell markers during IUI both in the human and Rhesus macaque. Our data suggest that CD14+ AMCs represent activated AMCs at the maternal-fetal interface.
Subject areas: Immunology, Bioinformatics, Omics
Graphical abstract
Highlights
-
•
Amnion participates in host-immune response to IUI via NF-κB activation
-
•
NF-κB activation in the amnion is TNF dependent
-
•
During IUI, AMCs get activated to express myeloid and innate immune genes
Immunology; Bioinformatics; Omics
Introduction
The amnion is a thin but tough, avascular membrane that straddles the maternal-fetal interface. The amnion tissue is of fetal origin comprising a layer of cuboidal epithelial cells with underlying mesenchymal cells firmly adherent to a thick basal membrane composed of extracellular matrix and collagen with interspersed fibroblast-like mesenchymal cells. The amnion is in contact with the amniotic fluid which harbors a high load of inflammatory products during intrauterine infection.1,2,3 Similar to other mucosal barrier niches, amnion epithelial cells express a set of Toll-like receptors (TLRs) suggesting that amnion can recognize a wide variety of pathogen-derived molecular patterns,4 but unlike other mucosal barriers it is external to the fetus but internal to the mother. The total surface area of the amniotic membrane in vivo at term is about 0.2 m2 (roughly the same area as the fetal skin).5 When stimulated or exposed to inflammatory signals (e.g., endotoxin), amnion can also secrete pro-inflammatory mediators including prostaglandins and cytokines/chemokines.4,6 In our previous studies focused on chorio-decidua immune response, we reported that the amnion secretes neutrophilic chemoattractants in a tumor necrosis factor (TNF)- and interleukin-1 (IL-1) signaling-dependent manner during experimental chorioamnionitis (inflammation in the amnion, chorion, decidua, and the amniotic fluid).7,8,9 However, the mechanisms and precise cell types orchestrating inflammation at the maternal-fetal interface are not known.
Understanding the pathogenesis of chorioamnionitis is important because ∼40% of preterm labor (PTL) cases are caused by chorioamnionitis.10,11 Biologically, unlike other mucosal barriers, chorioamnionitis presents the host a challenge in balancing immunotolerance of the antigenically mismatched fetus and host defense at the maternal-fetal interface. Inflammatory products during chorioamnionitis can rupture the normally tough amniotic membrane12 and cause preterm labor.10 Since collagen scaffolding is a primary source of tensile strength of the amnion,12,13 understanding the mechanisms of collagen biosynthesis during chorioamnionitis is also important.
We recently reported that injection of lipopolysaccharide (LPS) in the amniotic space in pregnant Rhesus macaque at about 80% term gestation closely mimics human pathology.7 In this model, LPS induces a massive neutrophilic infiltration and upregulation of pro-inflammatory cytokines in the chorio-decidua and amniotic fluid.7 These changes are similar to human chorioamnionitis.7 Both animal experiments and clinical studies implicate TNF signaling as an important mediator of intrauterine inflammatory response and preterm labor.8,14,15,16,17 We therefore used the clinically prescribed anti-TNF antibody Adalimumab (Humira) to inhibit TNF signaling in the present study.
To gain mechanistic insights and simultaneously maintain clinical relevance, we used carefully collected and archived isolated amnion tissue from preterm Rhesus macaques and human subjects with and without chorioamnionitis. We used the amnion tissue in contact with the extra-placental chorion and decidua called the reflected amnion,18 since we previously demonstrated this tissue to be highly responsive to intrauterine infection/inflammation (IUI) signals.8,19 To permit a comprehensive and unbiased assessment of the response of amnion, we used a transcriptomic profiling and analytical approach. To comprehensively understand the transcriptomic changes, we used both bulk and single-cell RNA sequencing (scRNA-seq) approach. These studies provide a new understanding of the response of amnion tissue during chorioamnionitis.
Results
Rhesus and human amnion transcriptomes change similarly upon IUI exposure
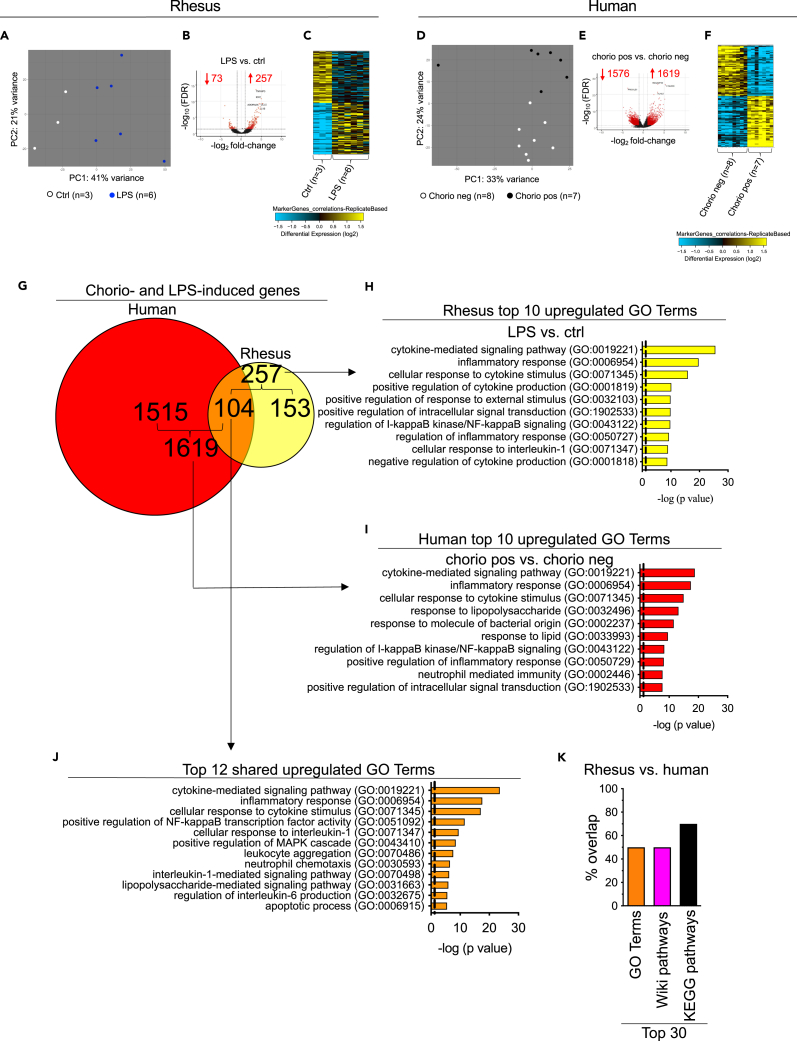
We previously reported that intra-amniotic injection of 1 mg LPS in the Rhesus macaque results in a predominantly neutrophilic infiltration in the chorio-decidua tissues of the placenta and upregulates pro-inflammatory cytokines in the amniotic fluid.7,8 Furthermore, the inflammatory response was higher at 16 h compared to a 48 h or 5 days exposure7,20 and therefore the time point chosen represented the height of inflammatory response. These previous publications give a detailed report identifying different immune cells during chorioamnionitis. We now explore response of the amnion tissue. We first compared transcriptomic profiles of the amnion in controls (Ctrls) vs. LPS-exposed Rhesus macaques. Unbiased principal-component analysis (PCA) of global amnion gene-expression profiles demonstrated distinct clustering based on prenatal inflammation exposures (Figure 1A). Compared to Ctrls, LPS-exposed amnion had 330 differentially expressed genes (DEGs) (257 upregulated and 73 downregulated; DEG>2-fold change; false discovery rate (FDR) adjusted p value<0.05) (Figure 1B). Heatmaps of DEGs showed distinct expression profiles and consistency in different samples within the same Rhesus subgroup (Figure 1C). Similar results were observed in human samples: PCA of global amnion gene-expression profiles demonstrated distinct clustering based on human chorioamnionitis cases (Figure 1D). 3,195 genes were differentially expressed in chorioamnionitis-positive samples compared to the negative ones (1,619 upregulated and 1,576 downregulated; DEG>2-fold change; FDR adjusted p value<0.05) (Figure 1E); heatmaps of DEGs showed distinct expression profiles and consistency in different samples within the same human subgroup (Figure 1F).
Figure 1.
Similar transcriptomic changes between Rhesus macaque and human subjects after exposure to chorioamnionitis (chorio)
Amnion tissue from the extra-placental fetal membranes was dissected free from the underlying chorion and decidua immediately after delivery in both Rhesus and human.
(A) Principal-component analysis (PCA) of RNA-seq data obtained from Rhesus amnion samples of Ctrl (n = 3) and LPS (n = 6) animals showing distinct clustering based on exposure to LPS.
(B) Volcano plots displaying differentially expressed genes (DEGs) in Rhesus LPS vs. Ctrl. Red dots and numbers indicate genes with an FDR adjusted p value <0.05 (DEG>2-fold change).
(C) Heatmap of the top 60 genes differentially expressed between Rhesus amnion samples upon exposure to inflammation.
(D) Similar to Rhesus data, PCA of RNA-seq data obtained from human amnion samples of chorio neg (n = 8) and chorio pos (n = 7) women showing distinct clustering based on exposure to chorio.
(E) Volcano plots displaying DEGs in human chorio pos vs. chorio neg. Red dots and numbers indicate genes with an FDR adjusted p value <0.05 (DEG>2-fold change).
(F) Heatmap of the top 60 genes differentially expressed between human amnion samples in upon exposure to inflammation.
(G) Venn diagrams displaying DEGs > 2-fold change (FDR adjusted p value <0.05) in both Rhesus and human upon chorio. Approximately 40% (i.e., 104/257 genes, p < 0.05 Bonferroni) of the genes induced by LPS exposure in Rhesus are shared with the genes induced by chorioamnionitis in human.
(H) Top 10 GO terms of 257 induced genes in Rhesus upon LPS exposure.
(I) Top 10 GO terms of 1,619 induced genes in human upon chorioamnionitis.
(J) Top 12 common upregulated GO terms between Rhesus and human.
(K) Histograms show a substantial overlap between the top 30 GO terms, Wiki Pathways, and KEGG pathways between Rhesus and human.
Interestingly, Rhesus amnion shared approximately 40% (i.e., 104/257 genes, p < 0.05) of human amnion IUI-induced genes (Figure 1G; Table S4). Functional annotation analyses revealed significant enrichment and similarity (FDR adjusted p value<0.05) of Gene Ontology (GO) terms involved in inflammation-related processes including the cytokine-mediated signaling pathway, inflammatory response, regulation of I-kappa B (IkB) kinase/nuclear factor κB (NF-κB) signaling, cellular response to IL-1, neutrophil mediated immunity, and response to LPS in both Rhesus (Figure 1H; Table S5) and human chorioamnionitis cases (Figure 1I; Table S6). Importantly, the functional annotation analysis of the common 104 genes induced by inflammation in both Rhesus and human (Figure 1J) were similar to the induced pathways in the Rhesus and human alone (compare Terms in 1J vs. 1H, 1I). Representative genes associated with these upregulated GO terms common to Rhesus and human were S100A9, CCL5, ADORA2A, ICAM1, CXCL3, BCL2A1, IL1B, TNF, and CSF3 (Figures S3A and S3B). These findings were further validated by quantitative PCR (qPCR) analysis of selected genes both in Rhesus (Figure S3C and STAR methods) and human (Figure S3D and STAR methods). Of note, shared pathways include 50% of the top 30 GO terms (Figure 1K and shaded rows in Tables S5 and S6), 50% of the top 30 Wiki pathways (Figure 1K and shaded rows in Tables S7 and S8), and 70% of the top 30 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Figure 1K and shaded rows in Tables S9 and S10).
We also investigated which genes and pathways were downregulated in both Rhesus and human upon inflammation. Rhesus amnion shared ∼10% (i.e., 11/73 genes, p = 0.7) of human amnion IUI-downregulated genes (Figure S4A). Functional annotation analysis revealed significant enrichment (FDR adjusted p value<0.05) of GO terms involved in a wide range of biological processes including cell development in both rhesus (Figure S4B) and human (Figure S4C). Functional annotation analysis of the 11 common genes downregulated by inflammation in both Rhesus and human revealed GO terms involved in cell junction assembly (Figure S4D).
LPS exposure and response to TNF inhibition identify unique functional clusters of genes in the amnion
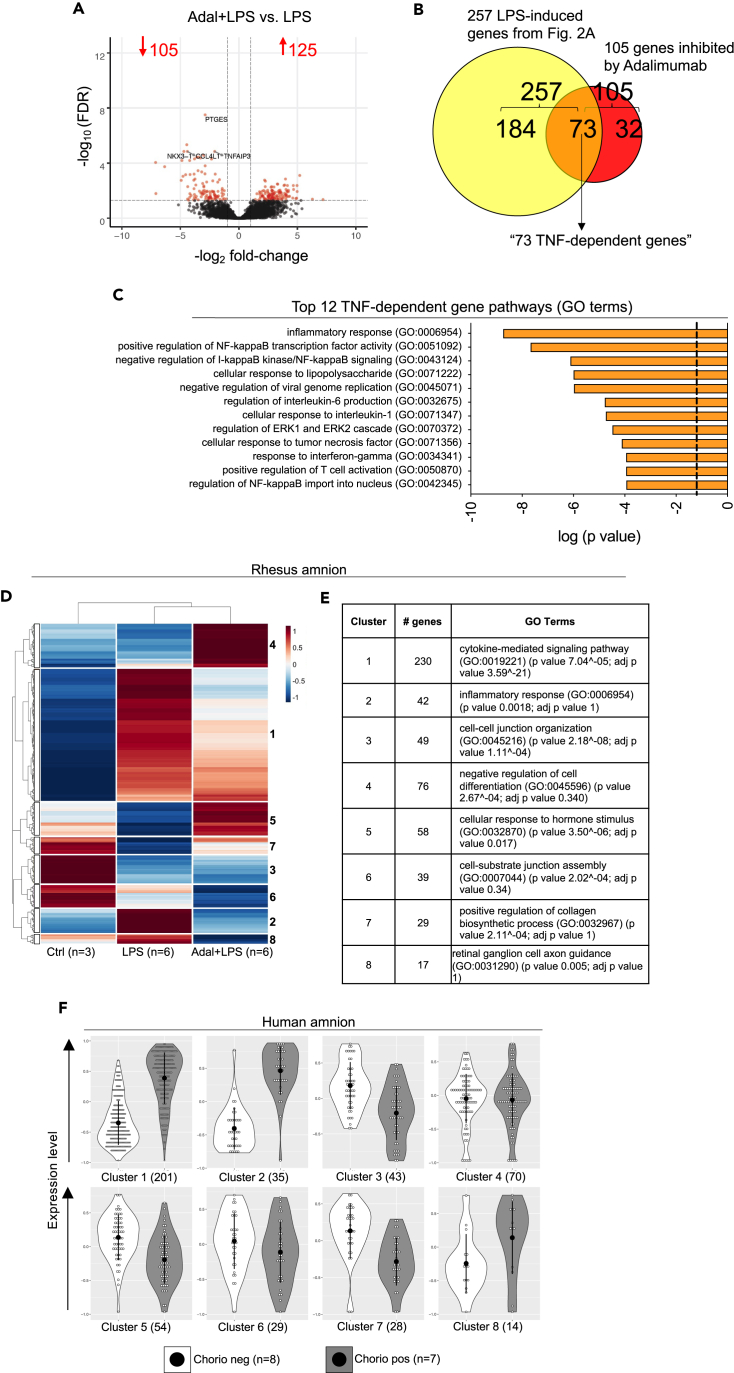
We previously demonstrated that TNF blockade by the anti-TNF antibody Adalimumab blocked chorio-decidua neutrophil recruitment and activation during Rhesus IUI and reduced LPS-driven IUI during chorioamnionitis.8 To investigate the effect of TNF blockade on the amnion upon IUI, we compared the transcriptomic landscape between Rhesus LPS-exposed vs. Adalimumab+LPS (Adal+LPS) groups. There were 230 DEGs (125 upregulated and 105 downregulated; DEG>2-fold change; FDR adjusted p value<0.05) (Figure 2A). Of the 257 LPS-induced genes compared to Ctrl animals shown in Figure 1B, 73 genes were inhibited by Adalimumab (designated as “TNF-dependent genes”) (Figure 2B; Table S11). Functional annotations of TNF-dependent genes identified alterations in the inflammatory response, cellular response to LPS, and regulation of IL-6 production (Figure 2C). On the other hand, functional annotations of 125 upregulated genes by Adal+LPS vs. LPS predicted alterations including cellular response to hormone stimulus and axonogenesis (Figure S5).
Figure 2.
TNF signaling is an important mediator of LPS-induced inflammatory pathways in the Rhesus amnion
TNF signaling was inhibited by anti-TNF antibody Adalimumab (Adal).
(A) Volcano plots displaying DEGs in Adal+LPS vs. LPS Rhesus groups. Red dots and numbers indicate genes with an FDR adjusted p value <0.05 and DEG > 2-fold change vs. LPS.
(B) Venn diagram showing Adalimumab decreased 73 out of 257 LPS-induced genes (40%) (downregulated “TNF-dependent” genes).
(C) Top 12 GO terms of 73 TNF-dependent genes. Clusters of genes based on patterns of differential expression in the three Rhesus groups (Ctrl, LPS, Adal+LPS). The functional annotation of each cluster was done using Enrichr.
(D) Heatmap of differentially expressed genes for each cluster is shown. Dendrogram of unsupervised hierarchical clustering for both the gene clusters and exposure groups are shown.
(E) Functional annotation of the 8 clusters identified in (D) with the number of genes and GO terms is shown for the Rhesus.
(F) The gene clusters identified in the Rhesus were compared in the two human groups (chorio neg and chorio pos) using violin plots. Open circles within the violin represent the mean expression value for each gene within the defined cluster. The black dot represents the mean of the violin, and the bar represents one standard deviation. The numbers in parenthesis define the number of genes in each cluster. Note the extensive similarities of differential expression patterns in each functional cluster in the human samples compared to Rhesus.
To further identify similarities and dissimilarities of gene expression patterns among the three different groups of animals (i.e., Ctrl, LPS, and Adal+LPS), we performed differential expression analysis and clustering. We identified 8 clusters based on differential biological process annotations between the exposure groups (Figures 2D and 2E). Gene expression patterns showed that compared to Ctrl animals, LPS exposure alone had a marked induction of genes in clusters 1 and 2 (Figure 2D), which were associated with cytokine-mediated signaling pathway and inflammatory response, respectively (Figure 2E). The treatment with Adalimumab showed an intermediate profile between Ctrl and LPS-exposed animals for cluster 1 and a similar profile with Ctrl animals in cluster 2 (Figure 2D). Cluster 3 genes, which were involved in cell-cell junction organization, and cluster 6 genes, which were involved in cell-substrate junction assembly, were higher in the Ctrl samples and decreased in both the LPS and Adal+LPS groups (Figures 2D and 2E). On the other hand, cluster 4 genes, involved in negative regulation of cell differentiation, were higher in the Adal+LPS compared to both Ctrl and LPS (Figures 2D and 2E). While LPS alone suppressed cluster 5 genes associated with cellular response to hormone stimulus, the combination of Adal+LPS significantly induced these genes in this cluster (Figures 2D and 2E).
Rupture of membranes due to weakening collagen scaffolding is an important determinant of preterm labor in the setting of IUI.12 Interestingly, genes associated with positive regulation of collagen biosynthesis (cluster 7) were suppressed by LPS compared with Ctrl, while the treatment with Adalimumab showed a profile more similar to Ctrl (Figures 2D and 2E). Next, we asked if the genes identified in Rhesus amnion had a similar profile of expression in the human samples. Indeed, the expression patterns in the human chorio samples were concordant, indicating the high degree of similarity between the Rhesus model and human pathology (Figure 2F).
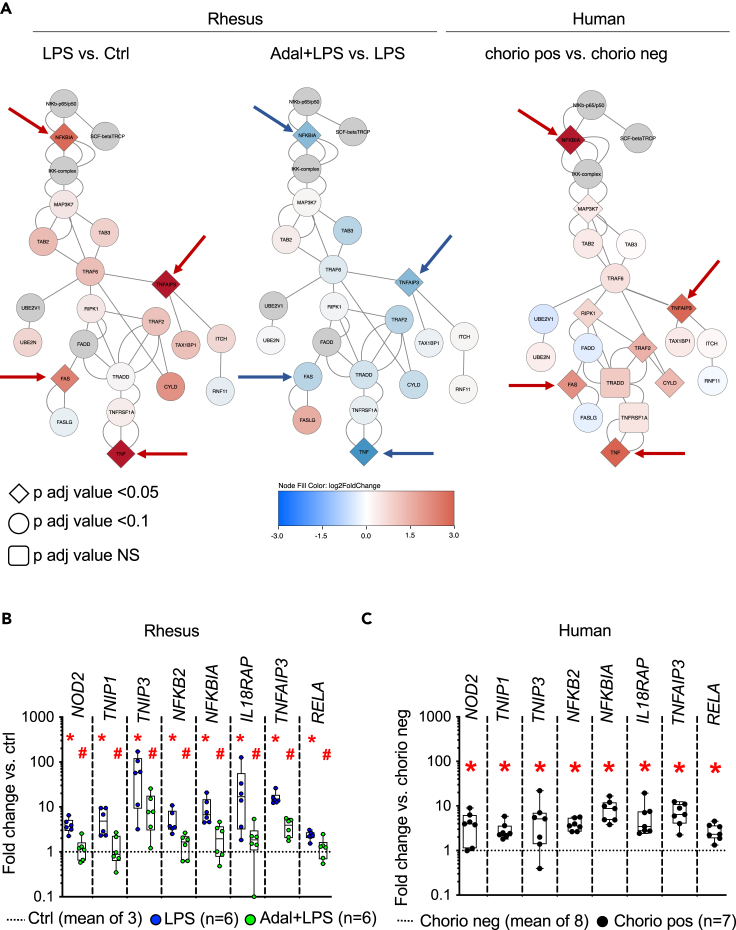
TNF-signaling blockade decreases inflammatory response in amnion by inhibiting NF-κB pathway
Based on our data and the key roles played by NF-κB in the regulation of inflammation including during the perinatal period,21,22 we examined the NF-κB pathway more closely. Interestingly, our bulk RNA sequencing (RNA-seq) data show that TNF-dependent GO terms were enriched in the regulation of NF-κB pathway (Figure 2C). Furthermore, we overlaid our entire dataset of Rhesus and human genes on the NF-κB canonical pathway23 and found that the most upregulated genes were NFKBIA, TNFAIP3, FAS, and TNF in both LPS-exposed amnion and human chorioamnionitis-positive amnion samples (Figure 3A). Other genes associated with regulation of NF-κB signaling GO terms were similarly upregulated in both LPS-exposed amnion (Figure 3B) and human chorioamnionitis-positive amnion samples (Figure 3C). Of note, Adalimumab treatment significantly decreased the expression of those genes (Figures 3A and 3B). These data were further validated by qPCR analysis of key selected genes in both Rhesus and human (Figure S6 and STAR methods).
Figure 3.
TNF blockade reverses activation of NF-κB pathway in the amnion during chorio
(A) RNA-seq data were superimposed on the canonical NF-κB pathway.23 Signaling molecules are color intensity and shape coded based on the expression level and FDR adjusted p values. Note the similarities between the key signaling molecules between the Rhesus and human intrauterine inflammation (arrow), and the reversal in the Adal+LPS group.
(B and C) Normalized counts from RNA-seq data of representative genes associated with the GO terms in the NF-κB pathways are shown as fold change compared to the no inflammation group (dashed line) in (B) Rhesus, and (C) human amnion. Note the similar increases in expression of NF-κB pathway genes in the Rhesus amnion exposed to LPS and human amnion exposed to chorio. Data are represented as mean ± SE (∗p < 0.05 compared to no inflammation group by Mann Whitney U-test). Adalimumab decreased LPS-induced increases in NF-κB pathway genes in B (#p < 0.05 compared to LPS group by Mann Whitney U-test).
Myeloid cell markers in a subset of amnion mesenchymal cells (AMCs)
We were intrigued by a large number of immune cell-associated genes in our bulk RNA-seq data. Therefore, we hypothesized that amnion cells may sense the inflammatory insult in the amniotic fluid and in turn trigger the immune response. To more exhaustively assess our data, we selected the genes belonging to the GO term related to antigen-presenting cells (APCs).24 A large number of genes associated with APCs were significantly upregulated in both the Rhesus and human amnion exposed to inflammation (Figure S7). These genes could be expressed by resident amnion cells or by the infiltrating immune cells. Hematoxylin and eosin (H&E) staining of sections of chorioamnion-decidua (CAD) tissue revealed that no Rhesus sample Ctrls (0/3) or LPS-exposed (0/6) had any immune cell infiltration in the amnion (Figure S8). However, in the human cases 0/8 no chorio cases and 5/7 chorio-positive cases had neutrophil infiltration in the amnion, a hallmark of severe chorioamnionitis25 (Figure S8).
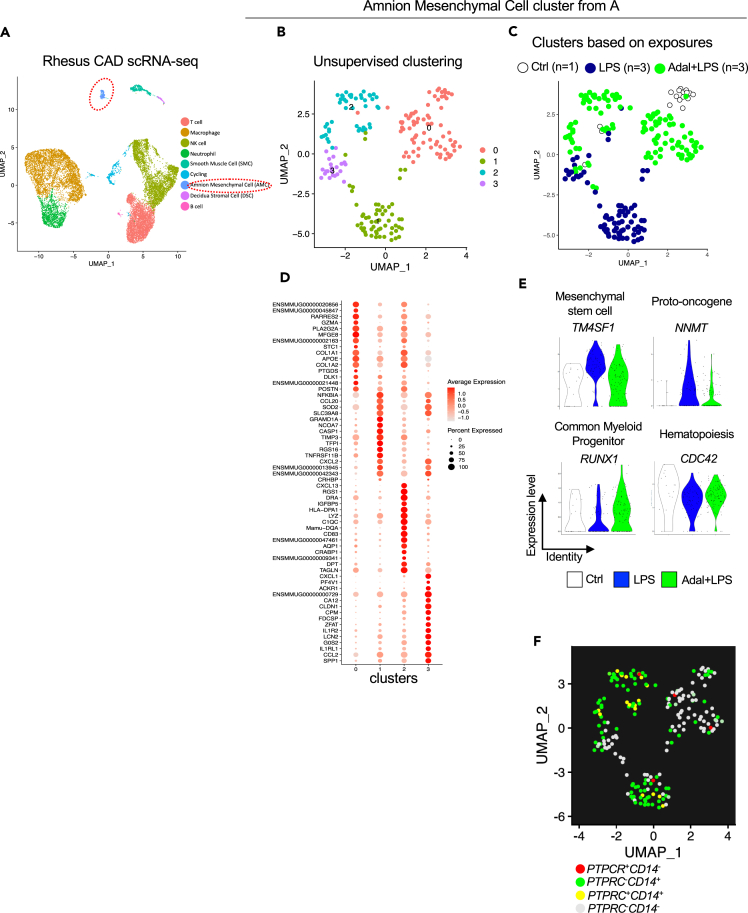
To more definitively assess cell of origin, we performed scRNA-seq analyses. We chose Rhesus rather than human samples since the Rhesus amnion did not have histologic evidence of immune cell infiltration. Furthermore, the role of TNF signaling could be assessed in the Rhesus tissues. To capture all cells more comprehensively, we used CAD tissue for scRNA-seq rather than the dissociated amnion that was used for bulk RNA-seq. Notably six out of eight samples for the scRNA-seq were also common to bulk RNA-seq (Table S1).
For cell type annotations, we pooled scRNA-seq data from all 8 samples. Consistent with our previous flow cytometry data showing a majority of dissociated CAD cells being CD45,26 clusters could be annotated into various hematopoietic and non-hematopoietic chorio-decidua cells based on established marker genes (Figure 4A). However, to our knowledge, there are no reliable markers for the amnion. We therefore used multiple methods to annotate amnion cells. First, we used detailed comparative bulk RNA-seq data from dissected amnion, chorion, decidua, and 16 other non-placenta human tissues.27 We calculated a module score using the top 100 genes with the highest proportional median found by Kim et al., in physically peeled amnion.27 The amnion module from Kim et al. overlaid nicely with our putative amnion cluster along with a few other scattered cells (Figure S9A). Second, we took advantage of the fact that amnion tissue is of fetal origin, while decidua and most of the immune cells in the decidua are of maternal origin, and that 50% of our fetuses were male (See Table S1). We observed a clear overlap with our amnion cluster with two Y chromosome gene markers (RPS4Y1 and RPS4Y2) (Figure S9B). Since amnion is known to contain epithelium and mesenchymal cells but no blood vessel cells, we interrogated our data against known epithelial and mesenchymal markers. We did not find any epithelial/trophoblast cell marker genes represented in the putative amnion cells (Figure S9C). By contrast, mesenchymal cell marker genes were readily expressed by these amnion cells (Figure S9D). We therefore designated the cluster as AMCs. One of the 8 samples did not have any identifiable amnion cells and hence subsequent analyses were conducted on 7 samples (Ctrl n = 1; LPS n = 3; and Adal+LPS n = 3).
Figure 4.
scRNA-seq reveals increased expression of CD14 and innate immune activation genes in a subset of amniotic mesenchymal cells after exposure to LPS in the Rhesus
Single-cell (sc) RNA-seq was done on dissociated chorioamnion-decidua (CAD) cells in the Rhesus.
(A) UMAP plot of genes in the CAD cells combined from the three Rhesus groups (Ctrl, LPS, Adal+LPS). Cells were annotated based on marker genes. Amniotic mesenchymal cell (AMC) cluster is circled.
(B and C) UMAP plot of the AMCs is shown by (B) unsupervised clustering and (C) clusters based on exposures.
(D) Dot plot showing the top representative marker genes for each cluster. The color scale represents the scaled expression average of each gene. The size of the dot represents the percentage of cells expressing each gene of interest.
(E) Violin plots showing the expression of marker genes for stem cell/proliferation, common myeloid progenitor, and hematopoiesis in the AMCs.
(F) UMAP plot of AMCs showing expression of PTPRC (CD45) and CD14. Cluster 1 (AMCs exposed to LPS) had the highest number of CD14 positive PTPRC (CD45) negative AMCs.
Unsupervised clustering of AMCs revealed four different subclusters (Figure 4B). Similar to the bulk RNA-seq data, scRNA-seq also showed distinct clustering of these cells dependent on prenatal exposures (Figure 4C). Cluster 0 comprised AMCs from Ctrl and Adal+LPS groups (Figure 4C). Cluster 1 and 3 AMCs were almost entirely from the LPS group (Figure 4C). Cluster 2 AMCs were predominantly from the Adal+LPS group (Figure 4C). Next, we analyzed DEGs characterizing each of the AMC clusters. Cluster 0 AMCs were represented by collagen synthesis, retinoid signaling, immune homeostasis, and prostaglandin synthesis genes (Figures 4D and S10A). Cluster 1 and 3 AMCs were enriched in inflammation mediators and NF-κB signaling (Figures 4D and S10A). Interestingly, cluster 2 AMCs were enriched in antigen presentation and innate immune response (Figures 4D and S10A). Similar to bulk RNA-seq, scRNA-seq data of the AMCs showed LPS induction and TNF inhibition of S100A9, ADORA2A, and BCL2A1 (Figure S10B) and genes involved in the NF-κB pathway (Figure S10C). LPS decreased the AMC expression of genes involved in collagen biosynthesis (Figure S10D), consistent with the results from bulk RNA-seq analysis (see Figures 2D‒2F, 3, and S3 , respectively).
Next, we analyzed different stem cell and hematopoietic cell markers in the AMCs noting that conventional hematopoietic cells occupy different clusters in Figure 4A. We detected expression of mesenchymal stem cell (MSC) factor TM4SF1 and the proto-oncogene NNMT (Figure 4E). However, classic MSC markers like OCT4, Sox-2, and Nanog were not detected, suggesting that these cells are not MSCs (data not shown). The common myeloid progenitor RUNX1 and hematopoietic stem cell quiescence marker CDC42 were also expressed in the AMCs (Figure 4E). However, common lymphoid progenitor markers e.g., EBF1, FLT3, GATA3, IKZF1, MYB, and TCF3 were not expressed (data not shown). LPS downregulated both RUNX1 and CDC42, while increasing TM4SF1 and NNMT expression predicting increased proliferation and differentiation along the myeloid lineage (Figure 4E). Consistently, we observed expression of the monocyte/macrophage marker CD14 in Ctrls AMCs that appeared to increase after LPS (Figures 4B and 4F: compare expression in cluster 0 vs. clusters 1 and 3). Interestingly, most of the CD14 expressing AMCs were negative for PTPRC (CD45) (Figure 4F).
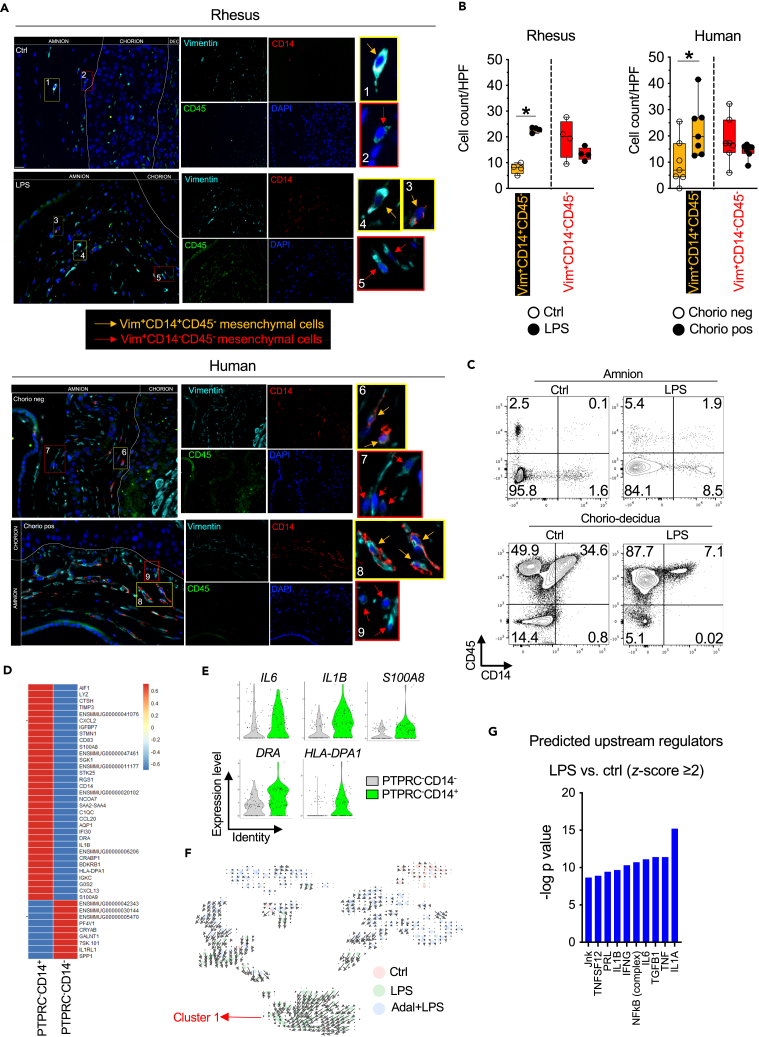
To further confirm these results, we performed immune co-localization of CD45, CD14, and Vimentin (a type III intermediate filament protein expressed in mesenchymal cells) on both Rhesus Ctrl and LPS-exposed animals and human chorio-negative and chorio-positive fetal membrane samples. Consistent with our scRNA data, the number of amnion mesenchymal Vim+CD14+CD45− cells significantly increased in both Rhesus and human subjects upon exposure to inflammation (Figures 5A and 5B). These CD14-expressing AMCs (Vim+CD14+CD45−) persisted 2 days and 5 days after LPS exposure (Figure S11A and STAR methods). Furthermore, we performed flow cytometry on dissociated amnion cells. Consistent with our scRNA and immune co-localization, CD326(EpCAM)−CD14+CD45− AMCs could be detected in about 1.5% Ctrl amnion cells that increased to almost 9% after exposure to LPS (Figure 5C and STAR methods). Approximately 2% CD14+CD45+ double-positive AMCs were detected mostly after LPS exposure. CD14+CD45− cells were restricted to the amnion since the adjacent chorio-decidua in the same animals did not have single positive CD14 cells and all the CD14-expressing cells in the chorio-decidua also expressed CD45 denoting a monocyte/macrophage cell type (Figure 5C and STAR methods). Interestingly, few CD14+CD45− were present in the uterus upon LPS exposure (Figure S11B and STAR methods). Compared to classic amnion mesenchymal PTPRC-CD14− cells, PTPRC-CD14+ cells expressed higher level of several molecules associated with myeloid cells such as AIF1 (IBA1), CD14, CD83, S100A8, IL6, IL1B, DRA, and HLA-DPA1 and innate immune activation markers such as C1QC, LYZ, CTSH, SAA2, and CXCL2 (Figures 5D and 5E). RNA velocity analysis demonstrated that the length of the arrows was the highest for AMCs exposed to LPS suggesting a recent increase in unspliced transcripts in this population (Figure 5F). The direction of arrows converged toward cluster 1 (Figure 5F), inferring a trajectory of differentiation toward cells expressing inflammatory markers (Figure 4D) and those serving inflammatory response to external stimuli (Figure S10A). Notably, cluster 1 cells had the highest numbers of PTPRC-CD14+ cells (Figure 4F). For these reasons, we designated these cluster 1 AMCs as activated AMCs. Notably, RNA velocity of AMCs from Ctrls or Adal+LPS group appeared to have either shorter arrows or direction pointing away from cluster 1 (Figure 5F), suggesting TNF signaling playing a role in AMC activation. Consistently, predicted upstream regulator analyses using the Ingenuity Pathway Analysis revealed that the genes induced by LPS exposure in the AMCs were IL1, TNF, and NF-κB signaling similar to the inference in bulk RNA-seq (Figures 5G and 3A).
Figure 5.
Both Rhesus macaques and human subjects exposed to chorioamnionitis increase numbers of CD14-expressing AMCs
(A) Rhesus and human chorioamnion-decidua section from paraffin-embedded blocks were co-stained with Vimentin (cyan), CD14 (red), CD45 (green), and DAPI (blue). Orange arrows in the magnified yellow insets (#1, 3, 6, and 8) indicate Vimentin(Vim)+CD14+CD45− cells, while red arrows in red insets (#2, 5, 7, and 9) indicate Vim+CD14−CD45− cells. Dotted white lines denote the different layers (amnion, chorion, and decidua (dec)). Of note, Vim+CD14+CD45− were not present in the chorion layer. Representative sections (Rhesus: n = 4/group; Human n = 7/group) are shown.
(B) Quantification of Vim+CD14+CD45− amniotic mesenchymal-myeloid cells and Vim+CD14−CD45− amniotic mesenchymal cells. Average of 5 randomly selected HPF fields were plotted as the representative value for the animal or human sample. Counts were performed in a blinded manner. Data are mean ± SEM (∗p < 0.05 compared to no inflammation group by Mann Whitney U-test).
(C) In a different set of Rhesus macaques, amnion was physically separated from chorio-decidua. Both tissues were proteolytically processed and cell suspension was analyzed by flow cytometry. Representative (n = 2/treatment) contour plots showing the expression of CD45 and CD14 in amnion and chorio-decidua CD326 (EpCAM)- cells. Right lower quadrant cells represent CD45−CD14+, while right upper quadrant CD45+CD14+ cells. Note that CD45−CD14+ cells can only be seen in the amnion but not chorio-decidua. Also note the similarities between the scRNA-seq data (Figure 4F), immune co-localization (A and 5B), and amnion flow cytometry for CD45 and CD14 (C and STAR methods).
(D) Heatmap showing differentially expressed genes (p < 0.05 and abs(log2FC) > 0.5) between PTPRC-CD14+ cells and PTPRC-CD14− AMC. Red: upregulated genes; blue: downregulated genes.
(E) Violin plots showing the expression of IL6, IL1β, S100A8, DRA, and HLA-DPA1 genes for PTPRC-CD14+ cells and PTPRC-CD14− cells.
(F) RNA velocity shows increased length of arrows denoting a recent increase in unspliced transcripts in AMCs exposed to LPS and a direction pointing toward cluster 1 inferring a trajectory toward AMCs expressing innate immune activation markers.
(G) Bar graph showing the top 10 predicted upstream regulators of genes in cluster 1 in panel B (LPS exposure) ranked by p value based on Ingenuity Pathway Analysis.
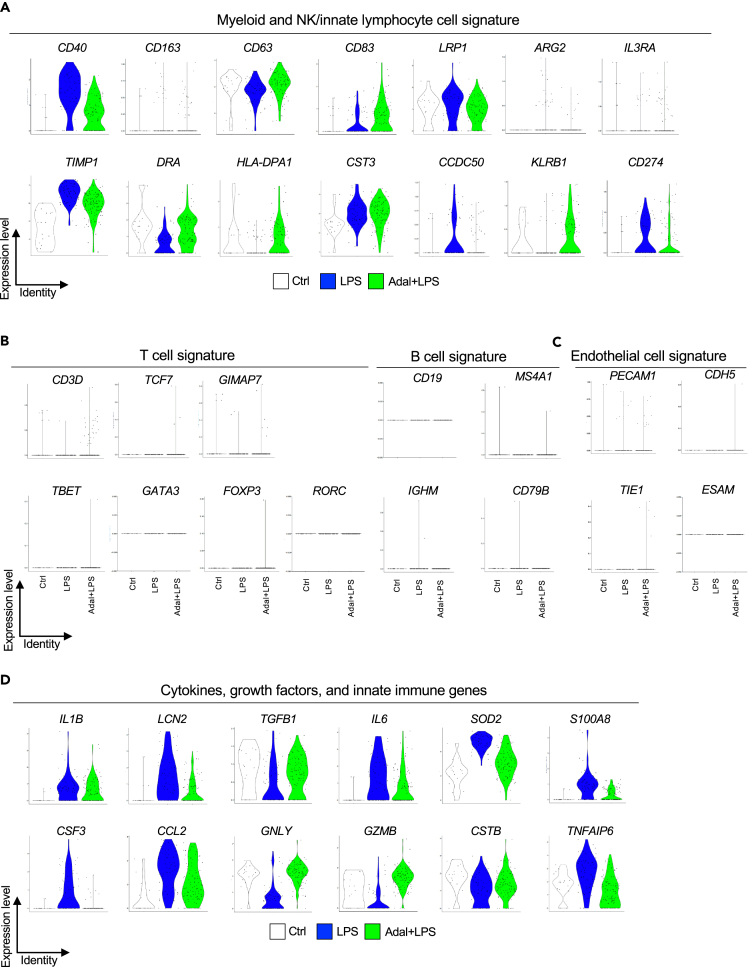
A survey of other hematopoietic markers revealed AMC expression of other myeloid cell markers (Figure 6A). AMC expression of CD40 (co-stimulatory protein on APCs), LRP1 (low-density lipoprotein related protein 1, CD91), TIMP1 (Tissue inhibitor of metalloproteinase 1), CST3 (Cystatin C), CD274 (PDL1), and CCDC50 (Coiled-coiled domain containing 50) increased after LPS exposure. LRP1, TIMP1, and CST3 are involved in would healing and repair. PDL1 and CCDC50 are markers associated with plasmacytoid dendritic cells, and PDL1 can suppress adaptive immunity. On the other hand, DRA and HLA-DPA1, markers associated with myeloid dendritic cells, were downregulated by LPS (Figure 6A). KLRB1 (CD161) expressed by natural killer (NK) and innate lymphoid cells (ILCs) was also represented (Figure 6A). Few AMCs expressed NCAM1 (CD56) (data not shown). Among lymphocyte lineage markers, a few AMCs expressed CD3D, TCF7, and GIMAP7, but other markers for T and B cell including subsets were absent (Figure 6B). Similarly, endothelial cell markers were absent (Figure 6C). Consistent with a predominant myeloid and NK cell marker gene signature in the AMCs, cytokine and innate immune genes including IL1B, LCN2, TGFB1, IL6, SOD2, S100A8, CSF3, CCL2 (MCP1), GNLY (Granulysin), GZMB (Granzyme B), CSTB (Cathepsin B), and TNFAIP6 were represented (Figure 6D).
Figure 6.
Myeloid and NK/innate lymphocyte cell gene signature in the AMC cluster of chorioamnion-decidua scRNA-seq
Violin plots for different exposure groups in the AMC clusters is shown.
(A) Expression of myeloid cell markers CD40, LRP1 (CD91), CD63, CD163, CD83, ARG2, CD274 (PDL-1), and IL3RA (CD123) and expression of NK cell/innate lymphocyte gene KLRB1 (CD161).
(B) Low-level expression of CD3D, but absence of key T subset transcription factors and B lymphocyte marker genes.
(C) Absence of endothelial cell markers.
(D) Expression of genes for cytokine, growth factors, and innate immune genes associated with myeloid and NK/innate lymphoid cells.
To further investigate the variations and functional modules of AMC types after IUIs in our scRNA-seq data, we used high dimensional weighted gene co-expression network analysis (hdWGCNA), a comprehensive framework for constructing and analyzing co-expression network data.28 We identified 21 functional modules of genes whose expression profiles are tightly interconnected. Out of these 21 modules, five modules (M1, M3, M5, M11, and M21) were more represented based on prenatal exposure (Figure S12A). In line with the biological processes found in the bulk RNA-seq analysis, GO term enrichment analysis associated those five modules with response to cytokines, inflammatory response, regulation of neutrophil activation and migration, regulation of NF-κB signaling, collagen fibril organization, extracellular matrix organization, and response to TNF (Figure S12B). Thus, an unsupervised co-expression analysis further confirmed functional changes in AMCs exposed to IUI.
Taken together, these results suggest that the AMCs are remarkably plastic, that a subset expresses myeloid and NK/innate lymphocyte cell markers, associated cytokines, and activated innate immune cell markers, and that these increase after exposure to IUI.
Discussion
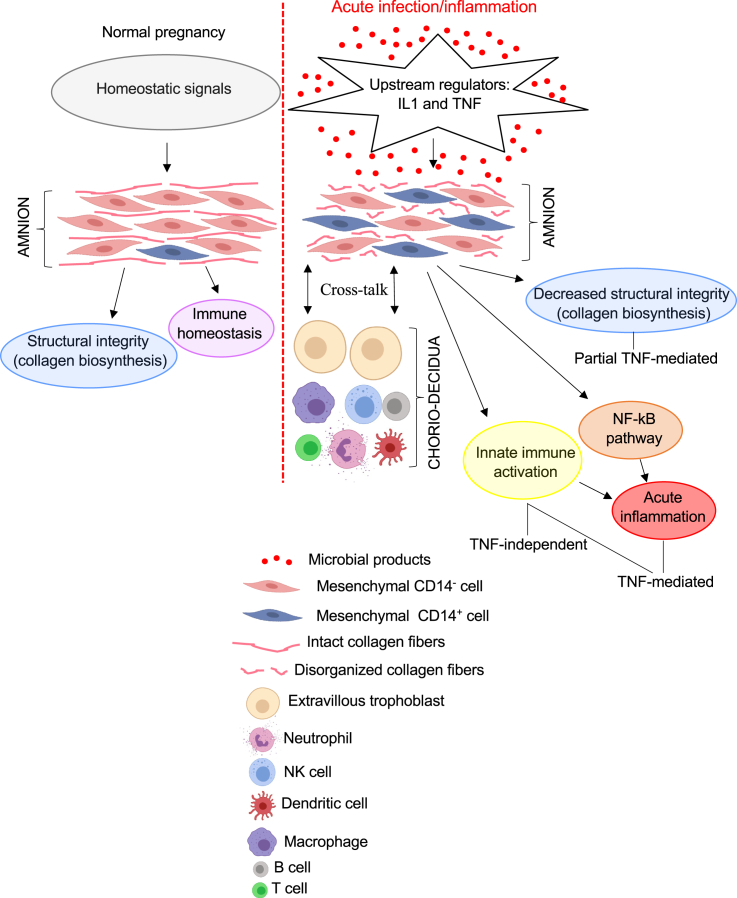
Using tissues from both Rhesus macaques and human subjects, we demonstrate that AMCs increased expression of myeloid cell markers and innate immune activation genes during IUI. AMC expression of CD14, the LPS co-receptor classically expressed on monocytes/macrophages, was confirmed by scRNA-seq, immunohistology, and flow cytometry. Prior studies have reported expression of CD14 in bone marrow and placenta mesenchymal cells.29,30 CRISPR-mediated over-expression of CD14 caused a shift of bone marrow mesenchymal stromal cell to a more activated pro-inflammatory phenotype.29 Consistently, CD14-expressing AMCs had a higher expression of activation and myeloid cell markers compared to CD14-negative AMC (Figures 5D and 5E). In addition to CD14 expression, our study also shows a previously unreported extensive repertoire of myeloid cell and activation markers in the AMCs that increase during IUI. We propose that during IUI, upstream regulators like IL-1 and TNF (possibly from amnion epithelial cells sensing microbial products) increase the proportion of AMCs expressing myeloid cell markers, leading to acute inflammation in a largely TNF-dependent fashion (Figure 7). Microbial sensing in the amnion epithelium and mesenchyme may be accomplished via pattern recognition receptors such as TLRs and NOD2. During IUI, the collagen biosynthetic pathway is suppressed, predicting the weakening of membranes, an important predisposing factor to preterm labor (Figure 7).
Figure 7.
Model for amnion mesenchymal cell immunomodulatory function
During normal pregnancy, homeostatic signals drive AMCs to synthesize proteins important for structural integrity, e.g., collagen and subserve immune homeostasis. During IUI, microbial products induce upstream regulators like IL-1 and TNF possibly in amnion epithelial cells. These upstream signals induce innate immune activation, and NF-κB pathway activation. Under baseline conditions, the mesenchymal cells are largely CD14− (classical mesenchymal cells). IUI increases amnion mesenchymal CD14+ cells and innate immune activation genes (representing activated AMCs) that modulate the host response to intrauterine infection. Acute inflammation driven by NF-κB pathway is TNF signaling mediated. Collagen biosynthesis, responsible for the structural integrity of the membranes, is decreased upon inflammation and partially dependent on TNF signaling.
Amnion cell epithelial-to-mesenchymal transition (EMT) and a “mesenchymal state” of amnion were previously shown to be associated with chorioamnionitis, predisposition to rupture of membranes, and induction of labor.31,32,33,34 RNA velocity inferred a trajectory toward a more pro-inflammatory and activated AMC during IUI (Figure 5F). TNF signaling appeared to be an important mediator of activation of amnion tissue in general and AMC in particular during IUI (Figures 2C, 3, 4C–D, 5F, and 6). We previously reported that activation of IRAK1 and secretion of neutrophil chemoattractants by the amnion during IUI are dependent on IL-1 signaling.7 Taken together, the studies suggest that EMT and mesenchymal activation in the amnion may be important mechanisms of innate host defense during IUI, and TNF and IL-1 signaling mediate many of the processes (Figure 7).
Amnion is thought to be an immune-privileged tissue of fetal origin due to the expression of non-classical major histocompatibility complex (MHC) such as human leukocyte antigen (HLA)-G to evade graft vs. host response during pregnancy.35 Thus, the amnion is being investigated for cell-based therapies of inflammatory disorders.36,37 Magatti et al. reported that amnion cells can express classic HLA-DR antigens38 and further that the DR− amnion cells suppressed blood mononuclear cell-induced allogenic T cell proliferation, while the DR+ amnion cells increased antiCD3-primed allogenic T cell proliferation.38 Our scRNA-seq revealed that AMCs from Ctrl animals expressed classical MHC antigens DRA and HLA-DPA1 that appeared to decrease during IUI (Figure 6A). By contrast, the expression of co-stimulatory molecules usually present on APCs (CD40, CD80, and CD83) were induced during IUI (Figures 6A and S7). Whether the AMCs can function as opportunistic atypical APCs as suggested by the expression of activated APC markers and upregulation of the interferon (IFN) γ and IL-12/IL-23 axis during IUI (Figures S13A‒S13C and STAR methods) similar to intestinal epithelial cells39 is not known. Our data suggest that AMCs could be the amnion cells that were shown previously to possess both stimulatory or suppressive immunomodulatory function40,41 and further suggest that these phenotypes could be altered based on exposure to IUI. Attention to IUI, a relatively common pregnancy complication, needs to be factored before deriving the amnion for novel cell-based therapies.
In addition to the APC markers, we could detect expression of several other myeloid cell markers such as CD14, LRP1 (CD91), CD163, ARG2, CD274 (PDL1), S100A8, and IL3RA (CD123) and expression of KLRB1 (CD161) expressed by NK and innate lymphocyte cells42 (Figures 6A and 6D). The expression of these myeloid markers during IUI was largely independent of TNF signaling. Of note, a majority of these AMCs expressing myeloid or NK markers were CD45 negative. These CD14+CD45− AMC seems to be unique to the amnion and to a lesser extent to the uterus, since the chorio-decidua did not have these single positive cells (Figures 5A–5C and S11B; STAR methods). Of note, the AMCs expressed a variety of cytokines/growth factors including IL1B, IL6, CCL2, LCN2, TGFB1, and cytotoxic granules such as granulysin and granzyme B (Figure 6D). These CD45−CD14+-expressing AMCs were readily found both in Rhesus macaques upon acute (16 h) and chronic (48 h and 5 days) inflammation induced by LPS exposure, and in human subjects with chorioamnionitis (Figures 5A, 5B, and S11A and STAR methods), with a higher expression of antimicrobial peptide and other innate immune activation genes (Figure 5D). We suggest a nomenclature of activated amnion mesenchymal cells for these unusual cells.
The canonical NF-κB pathway has been co-opted by different epithelia, e.g., skin and intestine, to perform critical functions relating to maintenance of epithelial integrity and immune homeostasis.43 In the skin and intestinal epithelia, a tight control of NF-κB constitutive low-level activity seems to be important for immune homeostasis.43 In human amnion, constitutive activity of NF-κB resulting in expression of several pro-labor genes appears to be important in mediating functional progesterone withdrawal and triggering normal term labor,44,45 and exosomal delivery of an NF-κB inhibitor decreased inflammation and preterm birth in a mouse model.46 We previously reported that NF-κB-driven IL6 and PTGS2 expression in the CAD tissue is upregulated during inflammation-induced preterm labor in the Rhesus.47 Although the net activity of NF-κB is increased, an important negative regulator A20 (also called TNFAIP3) was also induced in a TNF-dependent fashion. A20 is an essential component of the ubiquitin-editing protein complex and potently decreases NF-κB activation.48 Contrary to our results in both the Rhesus and human samples, a previous study reported decreased expression of A20 in the amnion during chorioamnionitis.49 Importantly, similar to epithelia of the skin and intestines,48 our data demonstrate that TNF signaling in the amnion is an important regulator of NF-κB activity and may be a key regulator of IUI-induced preterm labor.
A cluster of 29 genes involved in the positive regulation of collagen synthesis (cluster 7 in Figure 2) was downregulated in the amnion during both Rhesus and human chorioamnionitis. This collagen synthesis downregulation was partially reversed by Adalimumab treatment in the Rhesus macaque. Collagen of different types imparts tensile strength to the amnion and is known to be in lower concentrations in women with preterm rupture of membranes, an important cause of preterm labor.50 In a sheep model of LPS-induced chorioamnionitis, we reported that collagen architecture and content are decreased after exposure to LPS.51 Inflammation and oxidative stress are known to be important in the pathogenesis of rupture of fetal membranes.52 In vitro studies of human fetal membranes demonstrate that TNF and granulocyte-macrophage colony-stimulating factor (GMCSF) can decrease the tensile strength of the membranes.53 Whether inhibition of TNF signaling can prevent the collagen breakdown during IUI and prevent preterm rupture of membranes needs further study. A retrospective study of pregnant women with inflammatory disorders revealed a higher rate of prematurity with those treated with anti-TNF vs. non-biologic therapy.54 While the effect could be due to underlying medical conditions rather than the therapy, caution is warranted before using potent anti-inflammatory agents during pregnancy.
A few scRNA-seq studies of placenta and fetal membranes have been published using human,55,56,57,58 or mouse placenta.59 Amnion cells including the AMCs were not specifically identified in these publications, which focused on immune cells and cells of villous placenta. Two marker genes, the stem cell marker TM4SF160 and the proto-oncogene NNMT,61 in our AMC data were represented as marker genes in 2 populations of epithelial glandular cells previously reported in the first-trimester human placenta.56 Our scRNA-seq did not capture amnion epithelial cells. We suspect that the relative rarity of amnion cells (∼5% of CAD cells), exclusion due to doublet formation (inherent tendency of epithelial cells to reaggregate after dissociation), and the large size of epithelial cells may have led to exclusion in the microfluidic capture during scRNA-seq, library preparation, and downstream data cleaning. One strength of our annotation is that it was based both on rigorous marker gene assignment and corroboration by bulk RNA-seq of isolated amnion.
In conclusion, we demonstrate that during chorioamnionitis the amnion transcriptome is dominated by immune- and inflammation-related genes, largely driven by the NF-κB pathway. The AMCs expressing CD14 and other myeloid cell markers increase in number with increased expression of innate immune activation markers during IUIs. TNF signaling is an important regulator of acute inflammation. Our study alters the notion of amnion being a “simple physical barrier.”
Limitations of the study
Functional characterization of AMCs and amnion epithelial cells in a variety of pregnancy conditions should be investigated using both in vitro and in vivo approaches in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Live/Dead | ThermoFisher Scientific | Cat# L34957 |
| Anti-CD45 | BD Biosciences | Cat# 562394; RRID: AB_11152948 |
| Anti-CD326 (EpCam) | Biolegend | Cat# 324204: RRID: AB_756078 |
| Anti-CD14 | Biolegend | Cat# 301828; RRID: AB_2275670 |
| Anti-CD45 | Abcam | Cat# ab10558; RRID: AB_442810 |
| Sytox Green | ThermoFisher Scientific | Cat# S-34860 |
| Anti-Vimentin | Invitrogen | Cat# MA5-11883; RRID: AB_10985392 |
| CD14 | LSBio | Cat# LS-B3012-50; RRID: AB_1965166 |
| donkey anti-goat IgG | Invitrogen | Cat# A-11057; RRID: AB_2534104 |
| donkey anti-rabbit IgG | Invitrogen | Cat# A-21206; RRID: AB_2535792 |
| donkey anti-mouse IgG | Invitrogen | Cat# A-31571; RRID: AB_162542 |
| DAPI | Molecular Probe | Cat# D1306; RRID: AB_2629482 |
| rabbit isotype IgG control | Invitrogen | Cat# 08-6199; RRID: AB_2532942 |
| mouse isotype IgG control | Invitrogen | Cat# 08-6599; RRID: AB_2532952 |
| normal goat IgG | Vector | Cat# I-5000; RRID: AB_2336353 |
| Biological samples | ||
| Human amnion | University of Cincinnati | |
| Rhesus amnion | University of California Davis | |
| Rhesus fetal membranes (i.e., chorioamnion-decidua) | University of California Davis | |
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase I | Roche | Cat# 11088793001 |
| DNAseI | Roche | Cat# 10104159001 |
| Dispase II | Life technologies | Cat# 11320-033 |
| LPS | Sigma | Cat# L2880 |
| DMEM/F-12 | Gibco | Cat#11320-033 |
| Trizol | Sigma | Cat# T8424 |
| Adalimumab | AbbVie | |
| Critical commercial assays | ||
| Non-Human Primate Cytokine | Millipore | Cat# PRCYTOMAG-40K-13 |
| NEBNext rRNA Depletion kit | New England BioLabs | Cat# E6350 |
| NEBNext Ultra II RNA kit | New England BioLabs | Cat# E7765 |
| Illumina TruSeq Exome | Illumina | Cat# #20020490 |
| Illumina Exome Panel | Illumina | Cat# 20020183 |
| 10X Single Cell 3′ v2 Reagent Kits | 10X Genomics | Cat# PN-120237 |
| Chromium Single-Cell 3′ Library Kit | 10X Genomics | Cat# PN-1000121 |
| Deposited data | ||
| Bulk RNA-seq, human amnion | This paper | GSE243830 |
| Bulk RNA-seq, Rhesus amnion | This paper | GSE243830 |
| scRNA-seq, Rhesus chorioamnion-decidua | This paper | GSE243830 |
| Experimental models: Organisms/strains | ||
| Macaca mulatta | University of California Davis | |
| Oligonucleotides | ||
| S100A9 | Thermofisher | Cat# Rh02801277_m1 |
| CCL5 | Thermofisher | Cat# Rh02621811_m1 |
| ADORA2A | Thermofisher | Cat# Rh02902837_m1 |
| ICAM1 | Thermofisher | Cat# Rh02621706_m1 |
| NOD2 | Thermofisher | Cat# Rh02879855_m1/Cat# Hs00223394_m1 |
| TNFAIP3 | Thermofisher | Cat# Rh02860236_m1/Cat# Rh01568117 |
| CXCL3 | Thermofisher | Cat# Rh02788128_gH/Cat# Hs00171061_m1 |
| TNF | Thermofisher | Cat# Hs99999043_m1 |
| NFKB2 | Thermofisher | Cat# Rh02621752_m1/Cat# Rh01028900_m1 |
| RELA | Thermofisher | Cat# Rh02802962/Cat# Rh01042017_m1 |
| Software and algorithms | ||
| FlowJo, Version 10 | FlowJo | https://www.flowjo.com |
| GraphPad Prism, Version 9 | Prism | https://www.graphpad.com |
| 10X Cell Ranger software, Version 2.1.1 | 10X Genomics | https://www.10xgenomics.com/ |
| R package Seurat, Version 3.1.2 | CRAN | https://cran.r-project.org/web/packages/Seurat/index.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Suhas Kallapur, skallapur@mednenet.ucla.edu.
Materials availability
This study did not generate new unique reagents.
Experimental model and study partecipant details
Animals
Normally cycling, adult female Rhesus macaques (Macaca mulatta) (n = 33) were time mated. At ∼130 days of gestation (∼80% of term gestation), the pregnant Rhesus received either a 1 mL saline solution (controls) (n = 12, by the intraamniotic (n = 11) or the intramuscular (n = 1) route) or 1 mg LPS (derived from E. coli O55:B5, Sigma-Aldrich, St. Louis, MO, n = 15) in 1 mL saline solution by ultrasound-guided intraamniotic (IA) injection. The gestational age of study was based on chorioamnionitis being more frequent at preterm gestation. The dose of LPS was based on our previous dose response experiments in the sheep.20 We previously reported that this 1 mg LPS dose in the Rhesus macaque results in an intrauterine inflammatory response that is comparable to humans.7 Tumor necrosis factor (TNF) signaling was inhibited in the amniotic and systemic compartments by the TNF blocker Adalimumab (Humira, AbbVie Inc. North Chicago, IL) given IA (40 mg) + maternal subcutaneous (SC) (40 mg) 1 and 3 h before LPS respectively (n = 6) (Figure S1 and STAR methods). The dose and route of administration of TNF inhibitor were reported to have total specific TNF inhibitory activity in the amniotic fluid and significantly reduced intrauterine inflammation induced by LPS.8 Fetuses were surgically delivered 16 h after LPS-exposure. We previously reported that the inflammatory response was higher at 16 h compared to a 48 h or 5days exposure.7,62 Thus, the time-point chosen represented the height of inflammatory response. All the animals treated with saline and Adalimumab were used in previous studies focused on chorio-decidua,7,8 while some LPS-exposed animals were new to this study. For immunohistology, two extra groups of animals with a delayed IA LPS exposure of 48 h (1 mg) and 5 days (1 mg) were used. The animals belonging to these groups were used in previous studies.47,62 The clinical characteristics of the animals were similar among the treatments (Table S1 and STAR methods). There were no spontaneous deaths or preterm labor in the animals. The numbers of animals for each experiment are shown in the Table S1 as well as in the corresponding figure.
Human samples
Fifteen pregnant women at pregnancies from 26° to 366 weeks were recruited. Cohorts were developed based on a detailed placenta histo-pathologic diagnosis of chorioamnionitis based on Redline’s criteria,25 and divided in two groups: preterm chorioamnionitis (chorio) negative (n = 8) and preterm chorio positive amnion samples (n = 7). Clinical characteristics of the recruited cohorts are shown in Table S2 and STAR methods. All the samples from human subjects were used in a previous study.7
Study approval and ethics statement
All animal procedures were approved by the IACUC (protocol # 22121) at the University of California Davis and endorsed by the University of California, Los Angeles. The committee that reviewed and approved is the UC Davis Institutional Animal Care and Use Committee. Care and housing of animals met all Institutional Animal Care and Use Committee, US Department of Agriculture, and US NIH guidelines for humane macaque husbandry, including the presence of enrichment objects, daily foraging enrichment, and auditory and olfactory access to conspecifics in the same room.
Pregnant women provided a written informed consent from 2014 to 2017 under a protocol approved by the Institutional Review Boards (IRBs) of Cincinnati Children’s Hospital and University of Cincinnati (#2013–2243) for use of their placenta samples.
Method details
Histologic evaluation of fetal membranes for chorioamnionitis
H&E staining was performed for Rhesus and human fetal membrane sections, and staining was photographed. H&E-stained sections of human fetal membranes were scored in a blinded manner (by S.G. Kallapur) for chorioamnionitis using criteria outlined by Redline et al. based on numbers and depth of neutrophil infiltration of the tissue.25
Amnion isolation for bulk RNA-seq analysis
Extra-placental membranes were dissected away from the placenta within 30 min after delivery for both human and Rhesus, as previously described.7 For bulk RNA-seq analysis restricted to amnion, chorio-decidua cells were scraped from the amnion. Amnion was then surgically separated and flash-frozen for RNA studies.
Cell suspension preparation
For amnion cell, chorio-decidua cell, and uterine cell flow cytometry experiments, separated amnion tissues, chorio-decidua tissues, and uterus tissues were finely minced and digested with Dispase II (Life Technologies, Grand Island, NY) plus collagenase A (Roche, Indianapolis, IN) followed by DNase I (Roche) treatment, as previously described.7,26 Cell suspensions were filtered, and the red blood cells lysed. Viability was >90% by trypan blue exclusion test. For scRNA-seq analysis and some FACS-sorting we used intact Rhesus chorioamnion-decidua (CAD) without surgical separation using the same cell dissociation protocol as isolated amnion and uterus.
FACS-sorting and flow cytometry
FACS-sorting was done to identify live CAD cells for single-cell RNA-seq experiments. CAD cell suspension was stained with Sytox Green according to the manufacturer instructions (Thermofisher) and the viability was >90%. Figure S2 (and STAR methods) shows a representative gating strategy used to sort live CAD cells. For flow cytometry experiments, amnion cells, chorio-decidua cells, and uterine cells were treated with 20 μg/mL human immunoglobulin G (IgG) to block Fc receptors, stained for surface markers CD326 (EpCAM clone 9C4; Biolegend), CD45 (clone D058-1283; BD Bioscience) and CD14 (clone ME52; Biolegend) for 30 min at 4°C in PBS, washed, and fixed in fixative stabilizing buffer (BD Bioscience). Samples were acquired with LSR Fortessa 2 (BD Bioscience) within 30 min after the staining. All antibodies were titrated for optimal detection of positive populations and mean fluorescence intensity. At least 500,000 events were recorded for each sample. Doublets were excluded based on forward scatter properties, and dead cells were excluded using LIVE/DEAD Fixable Aqua dead cell stain (Life Technologies). Unstained and negative biological population were used to determine positive staining for each marker. Data were analyzed using FlowJo version 10 software (TreeStar Inc., Ashland, OR).
RNA extraction and qPCR
Total RNA was extracted from snap-frozen amnion, after homogenizing in TRIzol (Invitrogen, Carlsbad, CA). RNA concentration and quality were measured by Nanodrop spectrophotometer (ThermoScientific). Reverse transcription of the RNA was performed using Verso cDNA synthesis kit (ThermoScientific). Quantitative RT-PCR (qPCR) was carried out in a StepOnePlus real-time PCR system (Life Technologies) following standard cycling conditions. qPCR assays were performed with Rhesus and human specific TaqMan gene expression primers (Life Technologies). A list of probes is provided in Table S3. Eukaryotic 18S rRNA (Life Technologies) was the endogenous control for normalization of the target RNAs, and a sample from an IA saline injected Rhesus animal or human chorio neg sample was used to calibrate. The values were expressed relative to the average value of the control group.
Bulk RNA-seq: Library construction, sequencing, and analysis
For bulk RNA-seq analysis, fifteen Rhesus amnion samples (ctrl n = 3; LPS n = 6; and Adal+LPS n = 6) and fifteen human amnion samples (preterm chorio negative n = 8 and preterm chorio positive n = 7) were used. The integrity of purified total RNA from amnion was assessed using the RNA High-Sensitivity Assay on the TapeStation 2200 (Agilent Technologies). 200–300 ng of starting material were used as input material for the NEBNext rRNA Depletion kit (cat# E6350). RNA libraries were then prepared using the NEBNext Ultra II RNA kit (cat# E7765). Two hundred ng of each final library was subject to capture hybridization using Illumina TruSeq Exome, (cat #20020490) using Illumina Exome Panel (cat #20020183) according to the manufacturer’s instructions. Quality control for the final libraries was performed using the DNA D1000 Assay (TapeStation 2200 - Agilent Technologies) and quantified using a Qubit dsDNA BR Assay (Life Technologies). Diluted libraries were pooled and sequenced 50 single-end on a HiSeq3000 (Illumina). The reads were mapped with STAR 2.5.3a to the Macaca mulatta genome (Mmul 8.0.1) or to the human genome (GRCh38). The counts for each gene were obtained by using the options --quantMode GeneCounts. Differential expression analyses were carried out using DESeq2. The normalized counts were obtained from the DESeq2 analysis. Principal Component Analysis (PCA) was performed with the plotPCA function in DESeq2 after regularized log transformation. Heatmaps were plotted on the log2 value of the normalized counts. Inference of GO terms, Wiki pathways, and KEGG pathways were generated using Enrichr.63
Single-cell (sc)RNA-seq: Library construction, sequencing, and analysis
For scRNA-seq analysis, eight unseparated chorioamnion-decidua (CAD) samples were used (Ctrl n = 2; LPS n = 3; and Adal+LPS n = 3. Note that 6/8 tissues were also used for bulk RNA-seq analysis (Table S1). Live-sorted CAD cells were microfluidically partitioned with single cell capture, barcoding, and library construction (10X genomics chromium platform, Pleasanton, California).
Single-cell RNA-seq libraries were prepared using the 10X Single Cell 3′ v2 Reagent Kits. Specifically, single cell suspensions were loaded on a Chromium Controller instrument to generate single-cell Gel Bead-In- EMulsions (GEMs). GEM-RT were performed in a Veriti 96-well thermal cycler (Thermo Fisher Scientific, Waltham, MA), following which, GEMs were harvested and the cDNAs were amplified and cleaned up with SPRIselect Reagent Kit. Indexed sequencing libraries were constructed using Chromium Single-Cell 3′ Library Kit for enzymatic fragmentation, end-repair, A-tailing, adapter ligation, ligation cleanup, sample index PCR, and PCR cleanup. The barcoded sequencing libraries were quantified using the KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, MA). Sequencing libraries were loaded on a NovaSeq2 (Illumina, San Diego, CA) with a custom sequencing setting (26 bp for Read 1 and 91 bp for Read 2). Data processing including quality control, read alignment (to the reference genome Mmul 8.0.1), and gene quantification was conducted using the 10X Cell Ranger software (version 2.1.1). The samples were then merged into a single expression matrix using the cellranger aggr pipeline.
The R package Seurat (v3.1.2) was used to cluster the cells in the merged matrix. Cells with less than 500 transcripts or 100 genes, or more than 5% of mitochondrial expression were first filtered out as low-quality cells. The Seurat function NormalizeData was used to normalize the raw counts. Variable genes were identified using the FindVariableGenes function. The ScaleData function was used to scale and center expression values in the dataset, the number of unique molecular identifiers (UMI) was regressed against each gene. Principal component analysis (PCA), and uniform manifold approximation and projection (UMAP) were used to reduce the dimensions of the data, and the first two dimensions were used in the plots. The FindClusters function was used to cluster the cells. Marker genes were found using the FindAllMarkers function for each cluster, which employs the Wilcoxon Rank-Sum Test to determine the significance and the Benjamini-Hochberg Procedure to correct for multiple comparisons. Cell types were annotated based on the marker genes and their match to canonical markers. The module scores were calculated using the AddModuleScore function. Sub-clustering on the amnion cells was performed. The same functions described above were used to obtain the sub-clusters. We obtained eight high-quality scRNA-seq profiles from CAD samples. Analysis of cellular trajectory by RNA velocity was performed using the Python package scVelo.64 In one Ctrl sample we did not detect any amnion cells and therefore we excluded this sample from the analysis. Thus, final analyses were conducted on 7 samples (ctrl n = 1; LPS n = 3; and Adalimumab+LPS n = 3).
In order to illustrate the variations of cell types in amnion after intrauterine infection we used hdWGCNA package28 to perform co-expression network analysis on genes expressed in at least 5% of the cells. MetacellsByGroups function was used to construct metacells in each treatment group. The resulting metacells expression matrix was normalized by calling NormalizeMetacells function and then used for network analysis. In order to ensure scale-free topology of the co-expression network TestSoftPowers function was used to perform parameter sweep. The sweep identified 10 as the optimum soft power threshold to use when constructing the co-expression network by calling ConstructNetwork function with otherwise default parameters. WGCNA dendrogram was visualized with PlotDendrogram function whereas ModuleFeaturePlot was used to map harmonized module eigengenes, computed with ModuleEigengene function, onto the dimensionality reduction plot. The modules identified by hdWGCNA analysis were functionally characterized by performing enrichment tests with enrichR package. For each of the modules, a set of 100 most connected genes (ModuleConnectivity function) was tested for enrichment. Top 10 GO Biological Process terms for each module with adjusted p value <0.05 were shown along with the enrichment score.
Immunohistology
Immunofluorescence was performed as previously described with some modifications.65 Briefly, paraffin-embedded Rhesus and human fetal membrane blocks were sectioned and subjected to an antigen retrieval step consisting of incubation in AR10 (Biogenex) for 2 min at 125°C in the Digital Decloaking Chamber (Biocare) which was followed by cooling to 90°C for 10 min. All sections were incubated with 5% bovine serum and 10% normal donkey serum for 30 min before the incubation with goat polyclonal CD14 (1:125 dilution; LSBio), rabbit polyclonal CD45 (1:40 dilution; Abcam), and mouse monoclonal Vimentin (V9; 1:125 dilution, Invitrogen) in antibody diluent (DAKO) at 4°C overnight. Binding of the antibodies was detected simultaneously using donkey anti-goat IgG Alexa Fluor 568 (1:400 dilution, Invitrogen), donkey anti-rabbit IgG Alexa Fluor 488 (1:400 dilution, Invitrogen), and donkey anti-mouse IgG Alexa Fluor 647 (1:300 dilution, Invitrogen) for 1 h at room temperature. Nuclear counterstain was achieved using DAPI (Vector Laboratories). All slides were coverslipped using ProLong Gold Antifade Mountant (Invitrogen). Primary antibodies were replaced by rabbit isotype IgG control (Invitrogen), mouse isotype IgG control (Invitrogen), and normal goat IgG (4 μg/mL, Santa Cruz Biotechnology) with the stain as the negative control. Stained slides were imaged by a Zeiss Axio Imager Z1 (Carl Zeiss Inc., Thornwood, NY).
Cytokines
IFNγ and IL12/IL23p40 concentrations in amniotic fluid were determined by Luminex using non-human primate multiplex kits (Millipore).
Quantification and statistical analysis
Statistical analyses
Prism version 9 software (GraphPad, La Jolla, CA) was used to analyze data. Values were expressed as means ± SE or mean ± SD. Two-tailed Mann-Whitney U tests for non-normally distributed continuous variables and Student’s t test for Gaussian distributed data points were used. Fisher’s exact test was used for categorical variables were used to determine differences between groups. Bonferroni correction was used for the overlap between Rhesus and human gene sets.66 Results were considered significant for p values of ≤0.05. Details of statistical tests are reported in each figure legend.
Acknowledgments
This study was supported by R01 HD98389 (SK) and by U01 ES029234 (CC).
We thank Sarah Davis, Jennifer Kendrick, Sarah Lockwood, Anne Gibbons, Paul-Michael Sosa, Ma Zhong-Min, and Marie Jose-Lemoy research personnel at the CNPRC, UC Davis, for help with the animals. We also thank the Technology Center for Genomics & Bioinformatics (TCGB) at UCLA for scRNA library construction and sequencing; the Broad Stem Cell Research Center (BSCRC) at UCLA for bulk RNA-seq; the Institute for Quantitative & Computational Biosciences – Collaboratory for RNA-seq analysis; and the Immune Assessment Core (IAC) for ELISA multiplex. The Graphical Abstract was created with BioRender.com (https://BioRender.com).
Author contributions
P.P., M.C., M.M., and P.S. participated in data generation. P.P., M.C., M.M., B.B.N., F.M., G.P., L.A., M.E., N.P., L.S., M.P., E.D.F., A.H.J., C.A.C., and S.G.K. participated in analysis and interpretation of data. A.H.J., C.A.C., and S.G.K. participated in the conception and design of the study and obtained the funding. P.P. and S.G.K. wrote the manuscript. All authors have reviewed the manuscript and approve the final version.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 6, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108118.
Supplemental information
Data and code availability
-
•
All original bulk RNA-seq and scRNA-seq data have been deposited at GEO and is publicly available (GEO: GSE243830).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Romero R., Miranda J., Chaiworapongsa T., Korzeniewski S.J., Chaemsaithong P., Gotsch F., Dong Z., Ahmed A.I., Yoon B.H., Hassan S.S., et al. Prevalence and Clinical Significance of Sterile Intra-amniotic Inflammation in Patients with Preterm Labor and Intact Membranes. Am. J. Reprod. Immunol. 2014;72:458–474. doi: 10.1111/aji.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buhimschi C.S., Dulay A.T., Abdel-Razeq S., Zhao G., Lee S., Hodgson E.J., Bhandari V., Buhimschi I.A. Fetal inflammatory response in women with proteomic biomarkers characteristic of intra-amniotic inflammation and preterm birth. BJOG An Int. J. Obstet. Gynaecol. 2009;116:257–267. doi: 10.1111/j.1471-0528.2008.01925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bredeson S., Papaconstantinou J., Deford J.H., Kechichian T., Syed T.A., Saade G.R., Menon R. HMGB1 promotes a p38MAPK associated non-infectious inflammatory response pathway in human fetal membranes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillaux C., Méhats C., Vaiman D., Cabrol D., Breuiller-Fouché M. Functional screening of TLRs in human amniotic epithelial cells. J. Immunol. 2011;187:2766–2774. doi: 10.4049/jimmunol.1100217. [DOI] [PubMed] [Google Scholar]
- 5.Joyce E.M., Diaz P., Tamarkin S., Moore R., Strohl A., Stetzer B., Kumar D., Sacks M.S., Moore J.J. In-vivo stretch of term human fetal membranes. Placenta. 2016;38:57–66. doi: 10.1016/j.placenta.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell C.M., Hirst J.J., Mitchell M.D., Murray H.G., Zakar T. Genes upregulated in the amnion at labour are bivalently marked by activating and repressive histone modifications. Mol. Hum. Reprod. 2019;25:228–240. doi: 10.1093/molehr/gaz007. [DOI] [PubMed] [Google Scholar]
- 7.Presicce P., Park C.W., Senthamaraikannan P., Bhattacharyya S., Jackson C., Kong F., Rueda C.M., DeFranco E., Miller L.A., Hildeman D.A., et al. IL-1 signaling mediates intrauterine inflammation and chorio-decidua neutrophil recruitment and activation. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Presicce P., Cappelletti M., Senthamaraikannan P., Ma F., Morselli M., Jackson C.M., Mukherjee S., Miller L.A., Pellegrini M., Jobe A.H., et al. TNF-Signaling Modulates Neutrophil-Mediated Immunity at the Feto-Maternal Interface During LPS-Induced Intrauterine Inflammation. Front. Immunol. 2020;11:558. doi: 10.3389/fimmu.2020.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain V.G., Kong F., Kallapur S.G., Presicce P., Senthamaraikannnan P., Cappelletti M., Chougnet C.A., Bhattacharyya S., Pasare C., Muglia L.J. IRAK1 Is a Critical Mediator of Inflammation-Induced Preterm Birth. J. Immunol. 2020;204:2651–2660. doi: 10.4049/jimmunol.1901368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldenberg R.L., Culhane J.F., Iams J.D., Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero R., Dey S.K., Fisher S.J. Preterm labor: one syndrome, many causes. Science. 2014;345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar D., Moore R.M., Mercer B.M., Mansour J.M., Redline R.W., Moore J.J. The physiology of fetal membrane weakening and rupture: Insights gained from the determination of physical properties revisited. Placenta. 2016;42:59–73. doi: 10.1016/j.placenta.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Hampson V., Liu D., Billett E., Kirk S. Amniotic membrane collagen content and type distribution in women with preterm premature rupture of the membranes in pregnancy. Br. J. Obstet. Gynaecol. 1997;104:1087–1091. doi: 10.1111/j.1471-0528.1997.tb12073.x. [DOI] [PubMed] [Google Scholar]
- 14.Arntzen K.J., Kjøllesdal A.M., Halgunset J., Vatten L., Austgulen R. TNF, IL-1, IL-6, IL-8 and soluble TNF receptors in relation to chorioamnionitis and premature labor. J. Perinat. Med. 1998;26:17–26. doi: 10.1515/jpme.1998.26.1.17. [DOI] [PubMed] [Google Scholar]
- 15.Romero R., Grivel J.C., Tarca A.L., Chaemsaithong P., Xu Z., Fitzgerald W., Hassan S.S., Chaiworapongsa T., Margolis L. Evidence of perturbations of the cytokine network in preterm labor. Am. J. Obstet. Gynecol. 2015;213:836.e1–836.e18. doi: 10.1016/j.ajog.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christiaens I., Zaragoza D.B., Guilbert L., Robertson S.A., Mitchell B.F., Olson D.M. Inflammatory processes in preterm and term parturition. J. Reprod. Immunol. 2008;79:50–57. doi: 10.1016/j.jri.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Sadowsky D.W., Adams K.M., Gravett M.G., Witkin S.S., Novy M.J. Preterm labor is induced by intraamniotic infusions of interleukin-1beta and tumor necrosis factor-alpha but not by interleukin-6 or interleukin-8 in a nonhuman primate model. Am. J. Obstet. Gynecol. 2006;195:1578–1589. doi: 10.1016/j.ajog.2006.06.072. [DOI] [PubMed] [Google Scholar]
- 18.Han Y.M., Romero R., Kim J.S., Tarca A.L., Kim S.K., Draghici S., Kusanovic J.P., Gotsch F., Mittal P., Hassan S.S., Kim C.J. Region-specific gene expression profiling: novel evidence for biological heterogeneity of the human amnion. Biol. Reprod. 2008;79:954–961. doi: 10.1095/biolreprod.108.069260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toothaker J.M., Presicce P., Cappelletti M., Stras S.F., McCourt C.C., Chougnet C.A., Kallapur S.G., Konnikova L. Immune Cells in the Placental Villi Contribute to Intra-amniotic Inflammation. Front. Immunol. 2020;11:866. doi: 10.3389/fimmu.2020.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer B.W., Moss T.J., Willet K.E., Newnham J.P., Sly P.D., Kallapur S.G., Ikegami M., Jobe A.H. Dose and time response for inflammation and lung maturation after intra-amniotic endotoxin in preterm lambs. Am. J. Respir. Crit. Care Med. 2001;164:982–988. doi: 10.1164/ajrccm.164.6.2103061. [DOI] [PubMed] [Google Scholar]
- 21.Lindström T.M., Bennett P.R. 15-Deoxy-{delta}12,14-prostaglandin j2 inhibits interleukin-1{beta}-induced nuclear factor-{kappa}b in human amnion and myometrial cells: mechanisms and implications. J. Clin. Endocrinol. Metab. 2005;90:3534–3543. doi: 10.1210/jc.2005-0055. [DOI] [PubMed] [Google Scholar]
- 22.Lappas M., Odumetse T.L., Riley C., Reti N.G., Holdsworth-Carson S.J., Rice G.E., Permezel M. Pre-labour fetal membranes overlying the cervix display alterations in inflammation and NF-kappaB signalling pathways. Placenta. 2008;29:995–1002. doi: 10.1016/j.placenta.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Lo Surdo P., Calderone A., Cesareni G., Perfetto L. SIGNOR: A Database of Causal Relationships Between Biological Entities-A Short Guide to Searching and Browsing. Curr. Protoc. Bioinformatics. 2017;58:8.23.1–8.23.16. doi: 10.1002/cpbi.28. [DOI] [PubMed] [Google Scholar]
- 24.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO Hub, and Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redline R.W., Faye-Petersen O., Heller D., Qureshi F., Savell V., Vogler C., Society for Pediatric Pathology Perinatal Section Amniotic Fluid Infection Nosology CommitteeP.S.A.F.I.N.C. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr. Dev. Pathol. 2003;6:435–448. doi: 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
- 26.Presicce P., Senthamaraikannan P., Alvarez M., Rueda C.M., Cappelletti M., Miller L.A., Jobe A.H., Chougnet C.A., Kallapur S.G. Neutrophil recruitment and activation in decidua with intra-amniotic IL-1beta in the preterm rhesus macaque. Biol. Reprod. 2015;92:56. doi: 10.1095/biolreprod.114.124420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., Zhao K., Jiang P., Lu Z.X., Wang J., Murray J.C., Xing Y. Transcriptome landscape of the human placenta. BMC Genom. 2012;13:115. doi: 10.1186/1471-2164-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morabito S., Reese F., Rahimzadeh N., Miyoshi E., Swarup V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods. 2023;3 doi: 10.1016/j.crmeth.2023.100498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirakawa M.P., Tjahjono N., Light Y.K., Celebi A.N., Celebi N.N., Chintalapudi P., Butler K.S., Branda S.S., Krishnakumar R. Upregulation of CD14 in mesenchymal stromal cells accelerates lipopolysaccharide-induced response and enhances antibacterial properties. iScience. 2022;25 doi: 10.1016/j.isci.2022.103759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilz G.A., Braun J., Ulrich C., Felka T., Warstat K., Ruh M., Schewe B., Abele H., Larbi A., Aicher W.K. Human mesenchymal stromal cells express CD14 cross-reactive epitopes. Cytometry A. 2011;79:635–645. doi: 10.1002/cyto.a.21073. [DOI] [PubMed] [Google Scholar]
- 31.Richardson L.S., Taylor R.N., Menon R. Reversible EMT and MET mediate amnion remodeling during pregnancy and labor. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aay1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janzen C., Sen S., Lei M.Y.Y., Gagliardi de Assumpcao M., Challis J., Chaudhuri G. The Role of Epithelial to Mesenchymal Transition in Human Amniotic Membrane Rupture. J. Clin. Endocrinol. Metab. 2017;102:1261–1269. doi: 10.1210/jc.2016-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson L., Menon R. Proliferative, Migratory, and Transition Properties Reveal Metastate of Human Amnion Cells. Am. J. Pathol. 2018;188:2004–2015. doi: 10.1016/j.ajpath.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weed S., Armistead B., Coleman M., Liggit H.D., Johnson B., Tsai J., Beyer R.P., Bammler T.K., Kretzer N.M., Parker E., et al. MicroRNA Signature of Epithelial-Mesenchymal Transition in Group B Streptococcal Infection of the Placental Chorioamniotic Membranes. J. Infect. Dis. 2020;222:1713–1722. doi: 10.1093/infdis/jiaa280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strom S.C., Gramignoli R. Human amnion epithelial cells expressing HLA-G as novel cell-based treatment for liver disease. Hum. Immunol. 2016;77:734–739. doi: 10.1016/j.humimm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Lim R., Malhotra A., Tan J., Chan S.T., Lau S., Zhu D., Mockler J.C., Wallace E.M. First-In-Human Administration of Allogeneic Amnion Cells in Premature Infants With Bronchopulmonary Dysplasia: A Safety Study. Stem Cells Transl. Med. 2018;7:628–635. doi: 10.1002/sctm.18-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q.W., Huang Q.M., Wu H.Y., Zuo G.S.L., Gu H.C., Deng K.Y., Xin H.B. Characteristics and Therapeutic Potential of Human Amnion-Derived Stem Cells. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22020970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magatti M., De Munari S., Vertua E., Gibelli L., Wengler G.S., Parolini O. Human amnion mesenchyme harbors cells with allogeneic T-cell suppression and stimulation capabilities. Stem Cell. 2008;26:182–192. doi: 10.1634/stemcells.2007-0491. [DOI] [PubMed] [Google Scholar]
- 39.Koyama M., Mukhopadhyay P., Schuster I.S., Henden A.S., Hülsdünker J., Varelias A., Vetizou M., Kuns R.D., Robb R.J., Zhang P., et al. MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity. 2019;51:885–898.e7. doi: 10.1016/j.immuni.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magatti M., Vertua E., Cargnoni A., Silini A., Parolini O. The Immunomodulatory Properties of Amniotic Cells: The Two Sides of the Coin. Cell Transplant. 2018;27:31–44. doi: 10.1177/0963689717742819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kronsteiner B., Wolbank S., Peterbauer A., Hackl C., Redl H., van Griensven M., Gabriel C. Human mesenchymal stem cells from adipose tissue and amnion influence T-cells depending on stimulation method and presence of other immune cells. Stem Cells Dev. 2011;20:2115–2126. doi: 10.1089/scd.2011.0031. [DOI] [PubMed] [Google Scholar]
- 42.Jackson C.M., Demmert M., Mukherjee S., Isaacs T., Thompson R., Chastain C., Gray J., Senthamaraikannan P., Presicce P., Chetal K., et al. A potent myeloid response is rapidly activated in the lungs of premature Rhesus macaques exposed to intra-uterine inflammation. Mucosal Immunol. 2022;15:730–744. doi: 10.1038/s41385-022-00495-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasparakis M. Role of NF-kappaB in epithelial biology. Immunol. Rev. 2012;246:346–358. doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
- 44.Allport V.C., Pieber D., Slater D.M., Newton R., White J.O., Bennett P.R. Human labour is associated with nuclear factor-kappaB activity which mediates cyclo-oxygenase-2 expression and is involved with the 'functional progesterone withdrawal. Mol. Hum. Reprod. 2001;7:581–586. doi: 10.1093/molehr/7.6.581. [DOI] [PubMed] [Google Scholar]
- 45.Lim S., MacIntyre D.A., Lee Y.S., Khanjani S., Terzidou V., Teoh T.G., Bennett P.R. Nuclear factor kappa B activation occurs in the amnion prior to labour onset and modulates the expression of numerous labour associated genes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheller-Miller S., Radnaa E., Yoo J.K., Kim E., Choi K., Kim Y., Kim Y.N., Richardson L., Choi C., Menon R. Exosomal delivery of NF-kappaB inhibitor delays LPS-induced preterm birth and modulates fetal immune cell profile in mouse models. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abd3865. [DOI] [PubMed] [Google Scholar]
- 47.Cappelletti M., Presicce P., Feiyang M., Senthamaraikannan P., Miller L.A., Pellegrini M., Sim M.S., Jobe A.H., Divanovic S., Way S.S., et al. The induction of preterm labor in rhesus macaques is determined by the strength of immune response to intrauterine infection. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner D., Blaser H., Mak T.W. Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 2015;15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 49.Lappas M. A20, an essential component of the ubiquitin-editing protein complex, is a negative regulator of inflammation in human myometrium and foetal membranes. Mol. Hum. Reprod. 2017;23:628–645. doi: 10.1093/molehr/gax041. [DOI] [PubMed] [Google Scholar]
- 50.Kanayama N., Terao T., Kawashima Y., Horiuchi K., Fujimoto D. Collagen types in normal and prematurely ruptured amniotic membranes. Am. J. Obstet. Gynecol. 1985;153:899–903. doi: 10.1016/0002-9378(85)90703-3. [DOI] [PubMed] [Google Scholar]
- 51.Regan J.K., Kannan P.S., Kemp M.W., Kramer B.W., Newnham J.P., Jobe A.H., Kallapur S.G. Damage-Associated Molecular Pattern and Fetal Membrane Vascular Injury and Collagen Disorganization in Lipopolysaccharide-Induced Intra-amniotic Inflammation in Fetal Sheep. Reprod. Sci. 2016;23:69–80. doi: 10.1177/1933719115594014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menon R., Richardson L.S. Preterm prelabor rupture of the membranes: A disease of the fetal membranes. Semin. Perinatol. 2017;41:409–419. doi: 10.1053/j.semperi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar D., Moore R.M., Sharma A., Mercer B.M., Mansour J.M., Moore J.J. In an in-vitro model using human fetal membranes, alpha-lipoic acid inhibits inflammation induced fetal membrane weakening. Placenta. 2018;68:9–14. doi: 10.1016/j.placenta.2018.06.305. [DOI] [PubMed] [Google Scholar]
- 54.Bröms G., Kieler H., Ekbom A., Gissler M., Hellgren K., Lahesmaa-Korpinen A.M., Pedersen L., Schmitt-Egenolf M., Sørensen H.T., Granath F. Anti-TNF treatment during pregnancy and birth outcomes: A population-based study from Denmark, Finland, and Sweden. Pharmacoepidemiol. Drug Saf. 2020;29:316–327. doi: 10.1002/pds.4930. [DOI] [PubMed] [Google Scholar]
- 55.Pique-Regi R., Romero R., Tarca A.L., Sendler E.D., Xu Y., Garcia-Flores V., Leng Y., Luca F., Hassan S.S., Gomez-Lopez N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife. 2019;8 doi: 10.7554/eLife.52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pavličev M., Wagner G.P., Chavan A.R., Owens K., Maziarz J., Dunn-Fletcher C., Kallapur S.G., Muglia L., Jones H. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 2017;27:349–361. doi: 10.1101/gr.207597.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsang J.C.H., Vong J.S.L., Ji L., Poon L.C.Y., Jiang P., Lui K.O., Ni Y.B., To K.F., Cheng Y.K.Y., Chiu R.W.K., Lo Y.M.D. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl. Acad. Sci. USA. 2017;114:E7786–E7795. doi: 10.1073/pnas.1710470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson A.C., Mould A.W., Bikoff E.K., Robertson E.J. Single-cell RNA-seq reveals cell type-specific transcriptional signatures at the maternal-foetal interface during pregnancy. Nat. Commun. 2016;7 doi: 10.1038/ncomms11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lv F.J., Tuan R.S., Cheung K.M.C., Leung V.Y.L. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cell. 2014;32:1408–1419. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 61.Eckert M.A., Coscia F., Chryplewicz A., Chang J.W., Hernandez K.M., Pan S., Tienda S.M., Nahotko D.A., Li G., Blaženović I., et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569:723–728. doi: 10.1038/s41586-019-1173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt A.F., Kannan P.S., Bridges J., Presicce P., Jackson C.M., Miller L.A., Kallapur S.G., Chougnet C.A., Jobe A.H. Prenatal inflammation enhances antenatal corticosteroid-induced fetal lung maturation. JCI Insight. 2020;5 doi: 10.1172/jci.insight.139452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 65.Vaccari M., Boasso A., Ma Z.M., Cecchinato V., Venzon D., Doster M.N., Tsai W.P., Shearer G.M., Fuchs D., Felber B.K., et al. CD4+ T-cell loss and delayed expression of modulators of immune responses at mucosal sites of vaccinated macaques following SIV(mac251) infection. Mucosal Immunol. 2008;1:497–507. doi: 10.1038/mi.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amand J., Fehlmann T., Backes C., Keller A. DynaVenn: web-based computation of the most significant overlap between ordered sets. BMC Bioinf. 2019;20:743. doi: 10.1186/s12859-019-3320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All original bulk RNA-seq and scRNA-seq data have been deposited at GEO and is publicly available (GEO: GSE243830).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.