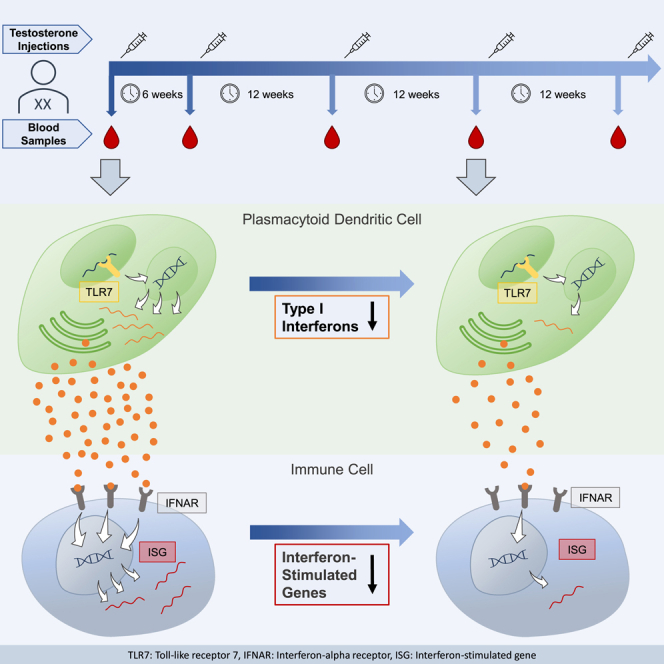
Summary
Type I interferons (IFN-I) are important mediators of antiviral immunity and autoimmune diseases. Female plasmacytoid dendritic cells (pDCs) exert an elevated capacity to produce IFN-I upon toll-like receptor 7 (TLR7) activation compared to male pDCs, and both sex hormones and X-encoded genes have been implicated in these sex-specific differences. Using longitudinal samples from a trans men cohort receiving gender-affirming hormone therapy (GAHT), the impact of testosterone injections on TLR7-mediated IFN-I production by pDCs was assessed. Single-cell RNA analyses of pDCs showed downregulation of IFN-I-related gene expression signatures but also revealed transcriptional inter-donor heterogeneity. Longitudinal quantification showed continuous reduction of IFN-I protein production by pDCs and reduced expression of IFN-I-stimulated genes in peripheral blood mononuclear cells (PBMCs). These studies in trans men demonstrate that testosterone administration reduces IFN-I production by pDCs over time and provide insights into the immune-modulatory role of testosterone in sex-specific IFN-I-mediated immune responses.
Subject areas: Health sciences, Patient social context, Gender
Graphical abstract
Highlights
-
•
Longitudinal study of IFN-I pDC responses in 10 trans men
-
•
IFN-I production by pDCs upon TLR7/8 ligation decreased on mRNA and protein level
-
•
Downstream induction of interferon-stimulated genes was reduced
-
•
Data demonstrate regulation of the TLR7/8 pathway by sex hormones
Health sciences; Patient social context; Gender
Introduction
Sex differences in immune responses are increasingly recognized to play an important role in the outcome of infectious and autoimmune diseases.1,2,3,4 In particular, a strong sex bias has been described in type I interferon (IFN-I) responses, with immune cells of females having the ability to produce higher quantities of IFN-I following toll-like receptor 7 (TLR7) stimulation.5 While more vigorous IFN-I responses can enable stronger antiviral immunity, IFN-I-driven autoimmune diseases such as systemic lupus erythematosus (SLE) are also more prevalent in females.2,6,7,8,9,10,11 IFN-I are produced by plasmacytoid dendritic cells (pDCs), a low-frequency innate immune cell with the capacity to produce up to 1,000 times more IFN-I than other immune cells.12 IFN-I production is triggered upon activation of endosomal TLR 7/8 and 9, sensing single-stranded RNA (ssRNA) and CpG-DNA, respectively.13,14,15,16,17 The TLR7 pathway in pDCs and B cells exhibits sex-specific differences, triggering more vigorous IFN-I responses in female pDCs.5,18,19,20,21 Both, genes encoded by the X chromosome and sex hormones, have been implicated in modulating TLR7-mediated responses. TLR7 is encoded by the X chromosome, and the TLR7 gene can escape X chromosomal inactivation (XCI) in females, resulting in quantitative transcriptional differences.22,23,24 Higher quantities of TLR7 can result in elevated production of IFN-I and thus in more vigorous IFN-I-mediated effects. Besides this gene-dose effect, sex hormones can also affect the TLR7 pathway. In a cohort of postmenopausal females, IFN-I production by pDCs was reduced compared to premenopausal females but was increased to levels observed in premenopausal females by hormone replacement treatment with estradiol.19 Furthermore, blocking estrogen receptor 1 and 2 during pDC differentiation in vitro reduced TLR7-mediated IFN-I production,22 while testosterone has been described to exert dampening effects on IFN-I release by pDCs.20,25 However, the relative contribution of genes encoded by sex chromosomes and hormonal factors to sex differences in IFN-I responses by human pDCs remains insufficiently understood, as sex chromosomes and sex hormones are interlinked.
Trans men are individuals with two X chromosomes who were assigned female at birth but whose gender identity is incongruent with that assignment. Some trans men decide to undergo gender-affirming hormone therapy (GAHT) to produce physical characteristics that are more congruent with their gender. Onset of physical masculinization ranges from one month after initiation of GAHT to one year, depending on the physical property.26 Little is known about the kinetics of immunological changes and their underlying mechanisms; however, it has been proposed that sex hormones exert their effects on immune cell progenitors in the hematopoietic compartment rather than on differentiated immune cells in the circulation or tissue.19,22 Some case studies of trans women under GAHT reported development of the female-biased autoimmune diseases systemic sclerosis and SLE after initiation of estrogen treatment,27,28,29 while one case report of a trans man described symptomatic improvement of subacute cutaneous lupus after initiation of testosterone regimen.30 These case reports suggest a potential role of GAHT on IFN-I-mediated immune responses involved in the pathogenesis of these autoimmune diseases. While longitudinal studies in trans persons have been performed focusing on T cells and autoantibodies,31 in-depth studies investigating longitudinal changes in IFN-I-related immune functions in transgender individuals under GAHT are missing. To elucidate the role of testosterone in TLR7-mediated IFN-I responses, we analyzed peripheral blood samples collected from a longitudinal cohort of trans men receiving GAHT. Here, we report that IFN-I production by pDCs is continuously reduced under testosterone injections in trans men and describe shifts in pDC transcriptomes upon TLR7/8 stimulation that are modulated by testosterone administration. Our findings contribute to a better understanding of the immunomodulatory role of testosterone for IFN-I responses and its contribution to sex differences in immunity.
Results
Testosterone injections in trans men led to shifts in the sex hormone profile and resulted in increased HGB and HCT levels
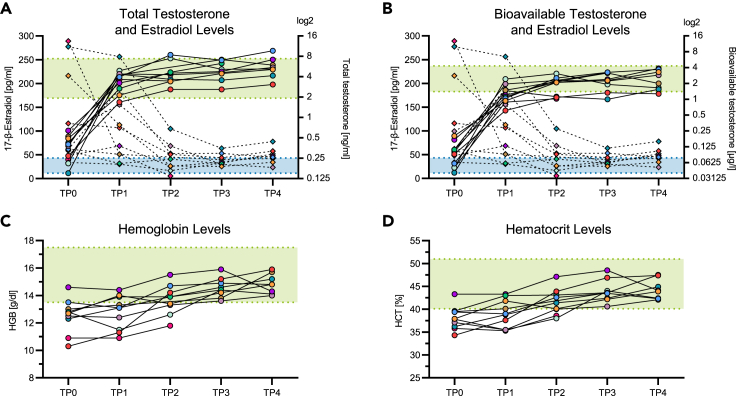
To determine the effects of testosterone injections on immune functions, we investigated peripheral blood samples collected from a cohort of 10 trans men who received GAHT consisting of 1,000 mg intramuscular testosterone undecanoate. Blood samples were collected longitudinally, with the first sample being collected prior to initiation of testosterone administrations. After the first injection, the second dose was scheduled six weeks later, followed by a steady rhythm of three months. Blood sample collection was scheduled at the same time points, with samples taken prior to testosterone injections. The mean age of cohort participants at initiation of GAHT was 28 years, with a range from 19 to 41 years. In blood samples, levels of testosterone and estradiol were quantified. Both total testosterone and bioavailable testosterone serum levels increased significantly over time (p < 0.0001, Figures 1A and 1B). 17-β-estradiol serum levels initially showed a wide distribution, most likely due to different phases of the menstrual cycle at the time of blood collection, but decreased significantly over the course of testosterone injections (p = 0.0003, Figures 1A and 1B). Testosterone and 17-β-estradiol serum concentrations in trans men subsequently stayed in or around male reference ranges over the median 43 weeks observation period (Figures 1A and 1B). Testosterone stimulates erythropoiesis in the bone marrow, resulting in elevated hemoglobin (HGB) and hematocrit (HCT) levels.32,33,34 In line with this, hemoglobin and hematocrit levels increased continuously and significantly over time in the study cohort (p < 0.0001, Figures 1C and 1D). Taken together, these data demonstrate that injected testosterone in trans men increased testosterone and reduced 17-β-estradiol plasma concentrations within the range of male reference levels and was biologically active in the hematopoietic compartment.
Figure 1.
Testosterone and estradiol blood levels in the trans men cohort and corresponding HGB and HCT values
(A) Total testosterone and (B) bioavailable testosterone as well as 17-β-estradiol serum levels in the longitudinal cohort of n = 10 trans men. Testosterone values are represented by circles and straight lines, and estradiol values are represented by rhombuses and dotted lines. Each symbol color represents an individual donor. The male serum testosterone reference range is indicated as a green area (total testosterone: 1.93–7.4 ng/mL, bioavailable testosterone: 1.4–4.3 μg/L), and the male serum estradiol reference range is indicated as a blue area (11.3–43.2 pg/mL). The increase of testosterone levels and the decrease of estradiol levels reached statistical significance (total testosterone: R2 = 0.73, p < 0.0001; bioavailable testosterone: R2 = 0.72, p < 0.0001; estradiol: R2 = 0.51, p = 0.0003), as assessed by extra sum-of-squares F test comparing a linear regression model with a shared slope and individual intercepts to a respective model with a hypothetical slope of 0.
(C and D) Hemoglobin and hematocrit levels in the cohort increased significantly over time (HBG: R2 = 0.75, p < 0.0001; HCT: R2 = 0.78, p < 0.0001), as assessed by an equivalent statistical model used in (A), (B). Male reference ranges are indicated as green areas (HGB: 13.5–17.5 g/dL, HCT: 40.1–51%). n = 10.
Immune cell frequencies in the trans men cohort were unaffected by testosterone injections
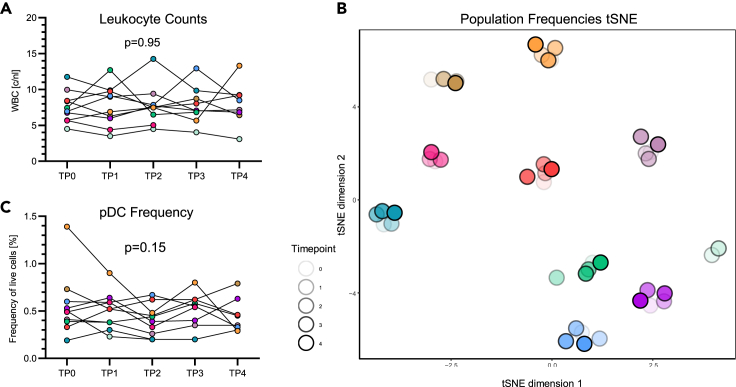
Given the observed impact of intramuscular testosterone injections on hemoglobin and hematocrit levels, immune cell population frequencies were assessed for testosterone-mediated changes in the trans men cohort. As overall leukocyte frequencies did not change over time (Figure 2A), a more specific longitudinal analysis of immune cell frequencies within peripheral blood mononuclear cells (PBMCs) was performed using multiparameter flow cytometry and antibody panels designed to identify 106 different immune cell subpopulations. T-distributed stochastic neighbor embedding (t-SNE) analyses showed that samples from each donor formed distinct clusters, indicating intrinsic inter-individual differences in immune cell compositions and stability of the frequencies of immune cell subpopulations during GAHT (Figure 2B). In line with the global observation of subpopulation frequency stability, frequencies of pDCs did not change significantly over the course of testosterone administration (Figure 2C), nor did frequencies of other common immune cell populations (Figure S1). Altogether, these data indicate that testosterone injections did not significantly impact immune cell population composition over the study period of up to 15 months in the trans men cohort.
Figure 2.
Immune cell frequencies in the trans men cohort were unaffected by testosterone injections
(A) Automated leukocyte counts in the hemogram of longitudinal trans men cohort samples. Total counts per nL remained stable during the study period (R2 = 0.58, p = 0.95).
(B) t-distributed stochastic neighbor embedding (t-SNE) plot of 106 immune cell population frequencies for all five time points in the trans men cohort (n = 10), as identified by flow cytometry. Samples from each donor form a separate cluster, showing stability between the different time points. Each symbol color represents an individual donor, and different shades indicate the different time points.
(C) Frequencies of pDCs, depicted from the identical dataset used in (B), did not change significantly over time (p = 0.15). Statistical significance in (A), (C) was as assessed by extra sum-of-squares F test comparing a linear regression model with a shared slope and individual intercepts to a respective model with a hypothetical slope of 0. n = 10.
TLR7/8 stimulation of PBMCs induced distinct transcriptomic changes in pDCs
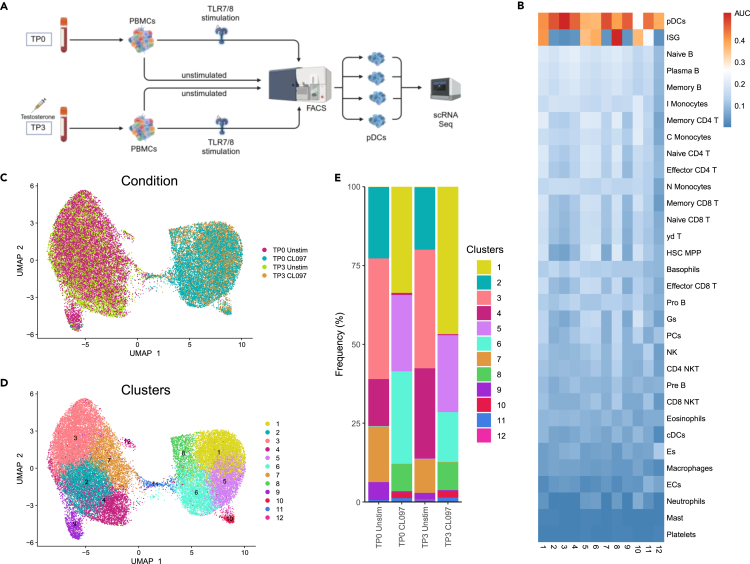
Previous studies by our group and others have reported significant differences in IFN-I responses of pDCs following TLR7 stimulation between females and males5,18,20,35 and have also suggested that sex hormones modulate TLR7 responses of pDCs.19,22,25,36 However, to date, no longitudinal study has been performed assessing intra-individual effects of testosterone administration on IFN-I production by pDC subsets. We therefore investigated longitudinal changes in pDC subset distributions and functions in three trans men using single-cell RNA sequencing (scRNA-seq) of sorted pDCs from TLR7/8-stimulated (2 h) and unstimulated PBMC samples from TP0 (baseline) and TP3 (28–32 weeks of testosterone administration). pDCs from the four different conditions per study participant were labeled with barcoded hashtag oligonucleotide antibodies directed against ubiquitously expressed surface proteins, fluorescence-activated cell sorting (FACS)-isolated (gating strategy see Figure S2), and scRNA-seq was performed using the 10× Chromium platform (Figure 3A). Following removal of low-quality cells, the purity of FACS-isolated pDCs was initially validated by assessing mRNA expression of surface markers used for pDC gating. Consistent with the specificity of antibodies used for pDC sorting (Figure S2), mRNA expression levels of the pDC markers HLA-DR (Human Leukocyte Antigen - DR isotype) and CD123 (IL3RA) were high, whereas mRNA expression of markers for other immune cell types, such as CD3, CD19, CD56 (NCAM1), CD14, CD11c (ITGAX), was low or absent (Figure S3A). To further validate pDC identity, pDC mRNA expression profiles were compared to a public immune cell marker dataset37 (Table S1). Expression of 17 pDC genes was enriched in sorted cells from the three study subjects, while enrichment of non-pDC immune cells markers was low or missing (Figure 3B). A subset of CX3CR1+ CD2+ CD56+ AXL+ dendritic cells, which has previously been described to represent a classical dendritic cell (cDC) population phenotypically and functionally distinct from pDCs,38,39 was excluded from further analysis of pDCs. After exclusion of these cDCs, a total of 32,731 cells with a median expression of 2,028 genes per cell were used for analysis.
Figure 3.
TLR7/8 stimulation of PBMCs induced distinct transcriptomic changes in pDCs
(A) Workflow of the scRNA-seq experiment, shown for one donor. Peripheral blood mononuclear cells (PBMCs) from time points before (TP0) and under (TP3) testosterone injections were isolated and stimulated with the TLR7/8 ligand CL097 for 2 h or left unstimulated before FACS-isolation of pDCs and subsequent scRNA-seq analysis (n = 3). Created with BioRender.com.
(B) Heatmap depicting enrichment (AUC values) of immune cell subset marker gene sets (ref.37) across all pDC clusters. Both unstimulated and stimulated pDC clusters displayed high AUC values for pDC marker genes, confirming pDC identity. Stimulated pDC clusters exhibited high AUC values for the set of ISGs, indicating effects of CL097 stimulation.
(C) Uniform manifold approximation and projection (UMAP) colored by condition. Unstimulated and stimulated pDCs from both assessed time points each form a separate group.
(D) UMAP colored by unsupervised clusters determined using Louvain method after constructing a shared nearest neighbor (SNN) graph based on k-nearest neighbors. Five clusters were associated with unstimulated pDCs and six clusters with stimulated pDCs; cluster 11 was equally distributed between both groups.
(E) Bar plots showing the percentage of cells in each pDC cluster at different stimulation conditions and time points. Cluster frequencies did not change significantly after testosterone administration (Wilcoxon signed-rank tests).
Uniform manifold approximation and projection (UMAP) of pDC single-cell transcriptomic data from all conditions produced two clearly distinguishable groups diverging by stimulation condition (Figure 3C). Hence, CL097 stimulation induced considerable shifts in pDC transcriptomes. Unsupervised clustering of transcriptomic data from both stimulation conditions and both time points combined resulted in identification of 12 pDC clusters that expressed varying numbers of marker genes comparing cells from each cluster to all other clusters (Figure 3D; Table S2). Clusters 2, 3, 4, 7, 9, and 12 were predominantly associated with unstimulated pDCs, whereas clusters 1, 5, 6, 8, and 10 were associated with CL097-stimulated pDCs (Figures 3C and 3D). Accordingly, CL097-stimulation-associated pDC clusters showed high expression of a set of 11 interferon-stimulated genes (ISGs) derived from the public dataset,37 while unstimulated pDC clusters presented with low ISG expression levels (Figure 3B; Table S1). pDC cluster 11 was equally distributed in both unstimulated and CL097-stimulated pDCs and showed intermediate ISG expression (Figures 3B–3E), possibly indicating that these pDCs were in the process of initiating the TLR7/8 response. In line with CL097-stimulated pDCs showing high ISG expression, the top gene ontology (GO) terms “IFN-I signaling pathway”, “defense response to virus”, “defense response to symbiont” and “cellular response to IFN-I″ were exclusively associated with these clusters (Figure S3B). To assess whether testosterone injections affected pDC cluster composition, cluster frequencies were compared between TP0 and TP3 (Figure 3E). Although shifts in frequencies between pDC clusters were observed between time points, cluster frequencies were not significantly changed after testosterone administration. Together, these results using scRNA-seq analyses of pDCs in three trans men showed that CL097 stimulation changed pDC transcriptomes; however, the composition of identified pDC clusters was not significantly affected by testosterone administration.
Heterogeneity of IFN-I-related transcriptomic responses upon testosterone injections in pDCs of trans men
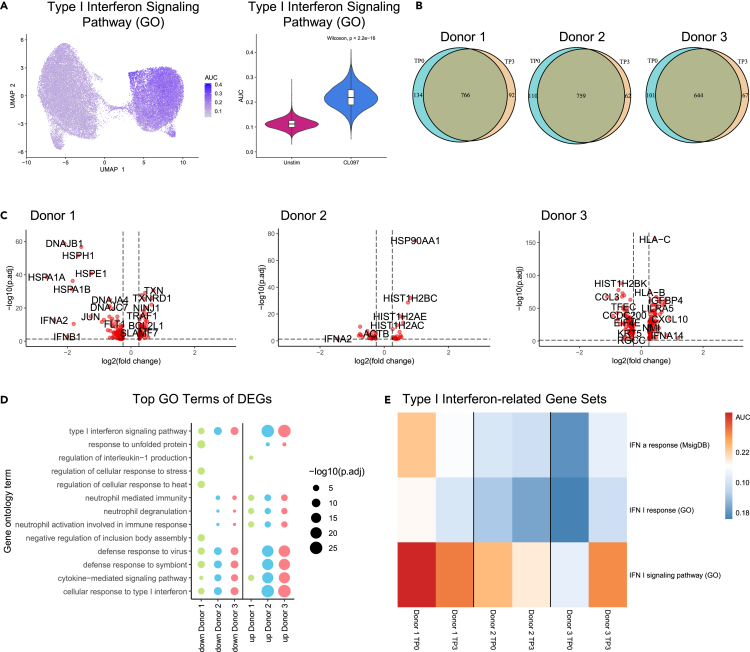
We next assessed changes in IFN-I responses of pDCs over time more specifically, focusing on genes in the GO terms “IFN-I signaling pathway” in CL097-stimulated pDC clusters. CL097-stimulated pDCs exhibited a significant increase (p = 2.2e-16) in IFN-I signaling gene enrichment, as compared to unstimulated pDCs, demonstrating a profound IFN-I response following TLR7/8 stimulation (Figure 4A), with largely overlapping upregulation of gene counts at both time points (Figure 4B). To further investigate the potential role of testosterone injections in modulation of IFN-I responses upon CL097 stimulation, changes in transcriptional responses to CL097 stimulation in pDCs comparing TP0 and TP3 were assessed using differential expression analysis (Table S3). In the three trans men investigated, the number of differentially expressed genes between TP0 and TP3 differed (donor 1: 152 genes, donor 2: 196 genes, donor 3: 44 genes). In all three trans men, the genes belonging to the IFN-I family ranked among those with strongest differential expression. In donors 1 and 2, genes of the IFN-I family were downregulated at TP3 (IFNA14, IFNB1, and IFNA2 mRNA in donor 1; IFNA2 mRNA in donor 2), while donor 3 exhibited upregulation of IFNA14 mRNA at TP3 (Figure 4C), reflecting inter-individual heterogeneity in the three donors. Changes in receptor expression can account for differences in IFN-I responses; however, in our dataset, TLR7 was not differentially expressed between TP0 and TP3, and TLR8 mRNA transcripts were not detectable in pDCs (Table S3).
Figure 4.
Heterogeneity of IFN-I-related transcriptomic responses upon testosterone injections in pDCs of trans men
(A) Enrichment of genes associated with the GO term “IFN-I signaling pathway” visualized as AUC values on UMAP and violin plot. Upregulation of respective genes upon CL097 stimulation was significant (p = 2.2e-16, Wilcoxon signed-rank test).
(B) Venn diagrams of upregulated genes upon CL097 stimulation at TP0 and TP3 in the three donors used for scRNA-seq analysis. Gene counts were largely overlapping between the two time points.
(C) Volcano plots of differentially expressed genes (DEGs) for the three individual donors comparing TP3 to TP0 in TLR7/8-stimulated pDCs. IFN-I genes belonged to genes with the strongest differential expression, exhibiting downregulation in donor 1 and 2, and upregulation in donor 3 comparing TP0 to TP3. p value threshold: <0.05; Log2FC threshold: >0.25.
(D) Top 5 GO terms associated with DEGs (TP3 CL097 vs. TP0 CL097) from each donor and enrichment of these GO terms across all donors, reflecting differential expression of IFN-I-related genes and inter-donor heterogeneity.
(E) Heatmap displaying enrichment of gene sets (AUC values) associated with IFN-I responses, displayed for each donor.
In line with the decreased mRNA expression of individual IFN-I genes after testosterone administration observed before, several GO terms related to IFN-I response were associated with genes downregulated at TP3 upon CL097 stimulation in all three donors, namely “IFN-I signaling pathway”, “defense response to virus”, “cytokine-mediated signaling pathway”, and “cellular response to IFN-I” (Figure 4D). Enrichment of IFN-I-related GO terms was however also found in genes upregulated in TP3 compared to TP0 after CL097 stimulation in donor 3, again indicating inter-individual heterogeneity in IFN responses (Figure 4D; Table S4). This was further confirmed by specific assessment of the gene sets “IFN-I response”, “IFN-I signaling pathway”, and “Interferon-α response”, which decreased in donors 1 and 2 under testosterone injections but increased in donor 3 (Figure 4E). Next, mRNA transcripts of ESR1, ESR2, and XIST were assessed, since their gene products have been suggested to affect IFN-I production.19,24,40,41 While changes in ESR1, ESR2, and XIST transcript levels were observed between TP0 and TP3 (Figure S3C), differential expression of these genes did not meet the thresholds used for analysis of overall scRNA-seq data (p < 0.05; log2FC > 0.25; minimum percent of cells expressing the respective gene: 0.1). Taken together, transcriptomic assessment of three trans men revealed overall downregulation, but also heterogeneity, in IFN-I-related transcriptomic profiles of sorted TLR7/8-stimulated pDCs in response to testosterone administration.
IFN-I protein production by pDCs decreased under testosterone injections
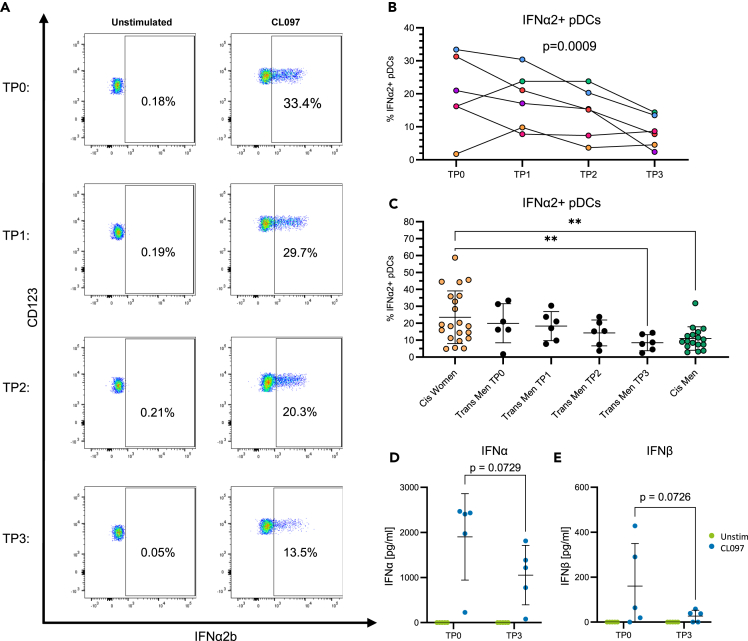
To further assess the impact of testosterone injections on IFN-I protein production of pDCs following TLR7/8 stimulation in a larger set of trans men, PBMCs isolated longitudinally from six trans men were stimulated with CL097 for 17 h and intracellular production of IFNα2, IFNβ, TNFα, IL6, IL12, and CXCL10 was quantified by flow cytometry.21 Percentages of IFNα2-producing pDCs decreased continuously over time under testosterone injections, as exemplified for one trans man (Figure 5A), and the decrease in the percentage of IFNα2-producing pDCs was significant over the study period in the six trans men (p = 0.0009, Figure 5B). Similarly, the percentage of IFNβ-producing pDCs also declined significantly over time (p = 0.03, Figure S4A). Of note, the frequency of IFNα2 protein-producing pDCs following 17 h CL097 stimulation in donor 3 also decreased from 33.4% to 13.5% between TP0 and TP3 (primary flow cytometry data for donor 3 shown in Figure 5A), while IFNA14 mRNA levels of stimulated pDCs measured after 2 h increased (Figure 4C), highlighting the impact of stimulation kinetics on IFN-I production by pDCs. In contrast to IFNα and IFNβ, frequencies of pDCs producing TNFα, IL6, IL12, and CXCL10 remained unchanged (Figures S4B–S4E), suggesting that pDC cytokine production is not generally suppressed under testosterone administration.
Figure 5.
IFN-I production by pDCs decreased under testosterone injections
(A) Frequency of IFNα2+ pDCs over time under testosterone injections of one donor from the trans men cohort.
(B) Upon TLR7/8 stimulation of PBMCs, frequencies of IFNα2-producing pDCs in n = 6 trans men decreased significantly over time under testosterone administration (R2 = 0.76, p = 0.0009). Statistical significance was assessed by extra sum-of-squares F test comparing a linear regression model with a shared slope and individual intercepts to a respective model with a hypothetical slope of 0.
(C) IFNα+ pDC frequencies upon TLR7/8 stimulation of PBMCs in an age-matched control cohort of cis women (n = 21) and cis men (n = 17) and in the trans men cohort, displayed for each time point. Cis women controls vs. cis men controls: p = 0.003; cis women controls vs. trans men TP3: p = 0.007; Mann-Whitney test, mean with SD displayed as error bars. ∗p < 0.05, ∗∗p < 0.01.
(D) IFNα and (E) IFNβ levels in supernatant of TLR7/8-stimulated PBMCs showed a decreasing trend over time under testosterone injections (IFNα and IFNβ: p = 0.073, two-way ANOVA followed by Fisher’s least significant difference test, n = 5). Blue dots represent CL097 stimulation, and green dots represent the unstimulated condition. Mean with SD indicated as error bars.
Next, IFNα2+ pDC frequencies were compared to a cross-sectional control cohort of cis-gender women (n = 21) and men (n = 17) to account for different constellations of sex hormones and sex chromosomes. As described previously,5,18,19,20,21 cis women exhibited significantly higher frequencies of IFNα2+ pDCs compared to cis men (p = 0.003, Figure 5C). Furthermore, cis women presented with a broader inter-individual range, which was mirrored in trans men at TP0. After continuous reduction of IFNα2+ pDC frequencies over time in the trans men cohort, levels were significantly lower compared to cis women controls (p = 0.007, Figure 5C) and stabilized in a range comparable to cis men controls. To also determine total IFN-I production by PBMCs, the amount of secreted IFNα and IFNβ protein was quantified by ELISA in the supernatant of PBMCs from TP0 and TP3 stimulated with CL097 for 17 h in five of the six trans men for which sufficient cell numbers were available. In line with the intracellular cytokine data, a reduction of secreted IFNα and IFNβ under testosterone administration was observed (IFNα and IFNβ: p = 0.07, Figures 5D and 5E). Taken together, these data demonstrate that longitudinal testosterone injections reduce TLR7/8-induced IFN-I protein production by pDCs and total IFN-I secretion by PBMCs in trans men.
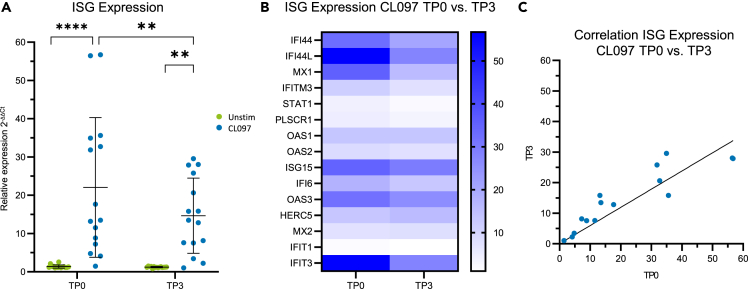
IFN-I exert some of their effector functions by inducing the expression of ISGs.42 Therefore, mRNA expression of ISGs (IFI44, IFI44L, MX1, MX2, IFITM3, STAT1, PLSCR1, IFIT1, IFIT3, HERC5, IFI6, ISG15, OAS1, OAS2, OAS3) in five of the six trans men studied before with available samples was assessed in PBMCs following 17 h CL097 stimulation. CL097 stimulation induced significant upregulation of relative ISG mRNA expression (TP0: p < 0.0001, TP3: p = 0.0027, Figure 6A). In line with the above data demonstrating reduced IFN-I production by pDCs under testosterone injections, the mean upregulation of ISG expression in PBMCs upon CL097 stimulation was significantly reduced at TP3 compared to TP0 (p = 0.001, Figures 6A and 6B). This reduction of ISG expression under testosterone administration was mainly driven by a strong reduction in expression levels of four ISGs (IFI44, IFI44L, MX1, IFITM3; Figure S5). Furthermore, linear regression analysis of relative expression values upon CL097 stimulation from TP0 and TP3 resulted in a slope of 0.6 of the regression line, demonstrating an average 40% reduction of ISG expression upon CL097 stimulation of PBMCs under testosterone injections (Figure 6C). Together, these data show that the observed reduction of CL097-induced IFN-I production of pDCs under testosterone administration in trans men also resulted in decreased downstream induction of ISGs.
Figure 6.
Expression of interferon-stimulated genes in PBMCs is reduced under testosterone administration
(A) Relative interferon-stimulated gene (ISG) expression levels of PBMCs upon TLR7/8 stimulation (blue) and without stimulation (green). CL097 stimulation induced significant upregulation of ISG expression (TP0: p < 0.0001, TP3: p = 0.0027, two-way ANOVA followed by Šidák multiple comparison test, n = 5). TLR7/8-mediated ISG expression was significantly reduced at TP3 compared to TP0 (p = 0.001, two-way ANOVA followed by Šidák multiple comparison test). Mean with SD indicated as error bars. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(B) Heatmap of mean relative expression values of 15 ISGs from TP0 and TP3, depicted from the same dataset used in (A). ISG expression was generally reduced under testosterone injections.
(C) Linear regression of relative ISG expression values upon CL097 stimulation of PBMCs from TP0 and TP3 (R2 = 0.75, Slope = 0.6).
Discussion
Sex represents an important variable in immunity, and sex-specific differences in immune responses can be mediated by sex hormones and genes encoded by allosomes. In particular, IFN-I production by pDCs in response to TLR7 stimulation has been shown in multiple studies to be more elevated in females compared to males.5,21,35 Different factors have been implicated in these sex differences in IFN-I production, including modulation of the TLR7 pathway by sex hormones and gene-dosage effects resulting from escape of the TLR7 gene encoded on the X chromosome from inactivation of the second X chromosome in females.19,20,22,23,24,25,36,40 However, the relative contributions of sex hormones to these sex differences in IFN-I production by pDCs in humans are not well understood, and longitudinal data on the effects of testosterone on TLR7-responsiveness of pDCs in humans are lacking. Here, we demonstrate in a longitudinal trans men cohort that testosterone administration negatively modulates TLR7/8-induced IFN-I production by pDCs and consequently reduces downstream ISG expression.
While sex differences in TLR7-mediated IFN-I responses of pDCs have been well established in mice and humans,5,18,21,22,35 the underlying mechanisms in humans are less well understood, since allosomal and sex hormonal effects cannot be distinguished in human studies comparing women and men. Furthermore, previous cross-sectional studies have been hampered by inter-individual variability of IFN-I-producing pDC frequencies, which occurs especially in individuals carrying two X chromosomes,18,19 and are further impacted by variability resulting from genetic variation,43 environmental factors, lifestyle, and behavior. The longitudinal design of the current study in trans men receiving GAHT enabled us to overcome some of these limitations.44 Over the median 43 weeks observation period, testosterone levels increased, while estradiol levels decreased; both stabilizing in respective male reference ranges. In line with the described stimulatory effects of testosterone on erythropoiesis,32,33,34 hemoglobin and hematocrit levels in trans men increased over time, indicating biological activity of testosterone. In contrast to this longitudinal increase of hemoglobin and hematocrit levels, no changes in leukocyte numbers were observed, and principal intra-individual immune subpopulation frequencies were stable over the course of GAHT. In particular, frequencies of pDCs that represent the main IFN-I-producing immune cells12 remained stable in the trans men cohort. These data suggest that testosterone administration modulates intrinsic pDC functioning rather than cell type frequencies, providing a unique opportunity to assess the longitudinal effects of testosterone on IFN-I production by human pDCs.
Single-cell RNA sequencing analyses enable the quantification of transcriptional changes in individual immune cells over time. pDCs reacted to 2 h of TLR7/8 stimulation with major changes within the transcriptome, demonstrating the prominent role of this pathway in pDCs. Assessment of longitudinal transcriptomic differences under testosterone injections in three trans men demonstrated that IFN-I-related genes belonged to the most differentially expressed genes. Specific assessment of IFN-I-related GO terms revealed that IFN-I-related gene signatures were downregulated in two and elevated in one individual. However, transcriptomic analyses do not always allow for sufficient prediction of protein presence.45,46,47 Importantly, on protein level, frequencies of CL097-stimulated IFNα- and IFNβ-producing pDCs decreased over time in the six trans men investigated in this study, regardless of donor age and despite heterogeneity in inter-individual responses. In line with that, total IFNα- and IFNβ secretion by PBMCs was reduced. These findings demonstrate the capacity of testosterone administration to reduce IFN-I production by pDCs over time. Considering the rather short lifespan of pDCs between several days and a few weeks,13,48 changes in sex hormone levels likely had a progressive effect on pDC development and epigenetic imprinting, resulting in altered TLR7 signaling and IFN-I production. For one donor, findings on protein level diverged from findings on mRNA level, with a clear decrease of IFNα- and IFNβ-producing pDC frequencies and lower total secretion by PBMCs, while IFN-I profiles on mRNA level increased. This divergence underlines the biological importance of TLR7 stimulation kinetics (2 h for mRNA vs. 17 h for protein) and also indicates the potential relevance of IFN-I-mediated positive feedback loops in IFNα- and IFNβ-production of pDCs.49 Furthermore, as some genes involved in regulation and modulation of IFN-I responses are encoded on the X chromosome and can escape XCI, including TLR7 or TASL,19,24,50,51 it is possible that testosterone administration impacted expression levels of these genes, as well as other known regulators of IFN-I responses like ESR1 (and ESR2),40 contributing to the observed heterogeneity between donors. However, changes in gene expression levels in the small sample size investigated here were inconclusive, and further studies using larger sample sizes are needed to specifically evaluate the impact of sex hormones on IFN-I modulators. Our study furthermore revealed that in line with the dampened IFN-I response under testosterone injections, ISG expression following TLR7/8 stimulation was also reduced by 40% over time in trans men. This reduction in ISG expression might, however, not be exclusively ascribed to the effects of lower IFN-I levels but potentially also be mediated through direct regulation of ISG expression by sex hormones.
One limitation of our study is the small sample size, especially given the diverging results on mRNA expression level of pDCs in one donor. Furthermore, PBMCs were used for TLR7/8 stimulation, instead of either unseparated peripheral blood or isolated pDCs, and this needs to be considered in the interpretation of the results. Nonetheless, the unique longitudinal design of this transgender cohort study provided important new insights into the role of testosterone administration in immune regulation. While the results from these studies clearly demonstrate a suppressive effect of testosterone injections on IFN-I production by pDCs, the observed effects can be mediated by elevated testosterone levels or resulting decreased estradiol levels in trans men- or a combination of both. Further research will be required to identify the precise regulators by which sex hormones modulate gene expression and posttranscriptional processing of molecules involved in the TLR7/8 pathway to identify novel immunotherapeutic targets for IFN-I-mediated diseases. In conclusion, our study demonstrates that testosterone administration in trans men reduces the frequency of IFN-I-producing pDCs, the total secreted amount of IFN-I by PBMCs, and resulting ISG expression levels, providing a better understanding of sex hormonal modulation of immunity in humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TotalSeq™-C0251 anti-human Hashtag 1 Antibody | Biolegend | Cat#: 394661 / RRID:AB_2801031 |
| TotalSeq™-C0252 anti-human Hashtag 2 Antibody | Biolegend | Cat#: 394663 / RRID:AB_2801032 |
| TotalSeq™-C0253 anti-human Hashtag 3 Antibody | Biolegend | Cat#: 394665 / RRID:AB_2801033 |
| TotalSeq™-C0254 anti-human Hashtag 4 Antibody | Biolegend | Cat#: 394667 / RRID:AB_2801034 |
| BDCA2 (CD303) / PE-Cy7 / 201A | Biolegend | Cat#: 354214 / RRID: AB_2563141 |
| CCR2 (CD192) / BV421 / K036C2 | Biolegend | Cat#: 357210 / RRID:AB_2563463 |
| CCR4 (CD194) / BV605 / L291H4 | Biolegend | Cat#: 359418 / RRID:AB_2562483 |
| CCR5 (CD195) / BV421 / J418F1 | Biolegend | Cat#: 359118 / RRID:AB_2563577 |
| CCR6 (CD196) / BV711 / G034E3 | Biolegend | Cat#: 353436 / RRID:AB_2629608 |
| CCR7 (CD197) / APC / G043H7 | Biolegend | Cat#: 353213 / RRID:AB_10915474 |
| CD11c / BUV737 / B-ly6 | BD Biosciences | Cat#: 741827 / RRID:AB_2871162 |
| CD11c / PE-Cy7 / Bu15 | Biolegend | Cat#: 337216 / RRID:AB_2129790 |
| CD123 / BV711 / 6H6 | Biolegend | Cat#: 306030 / RRID:AB_2566354 |
| CD123 / PE-Cy7 / 6H6 | Biolegend | Cat#: 306010 / RRID:AB_493576 |
| CD123 / BV711 / 9F5 | Biolegend | Cat#: 563161 / RRID:AB_2738038 |
| CD127 / PE/Dazzle 594 / A019D5 | Biolegend | Cat#: 351336 / RRID:AB_2563637 |
| CD14 / BV510 / M5E2 | Biolegend | Cat#: 301841 / RRID:AB_2561379 |
| CD14 / PE-Cy7 / M5E2 | Biolegend | Cat#: 301814 / RRID:AB_389353 |
| CD14 / APC-Cy7 / HCD14 | Biolegend | Cat#: 325620 / RRID:AB_830693 |
| CD14 / PerCP-Cy5.5 / HCD14 | Biolegend | Cat#: 325622 / RRID:AB_893250 |
| CD141 / PE / M80 | Biolegend | Cat#: 344104 / RRID:AB_2255842 |
| CD16 / BV785 / 3G8 | Biolegend | Cat#: 302045 / RRID:AB_2561367 |
| CD16 / PE-Cy7 / 3G8 | BD Biosciences | Cat#: 302016 / RRID:AB_314216 |
| CD16 / BUV737 / 3G8 | BD Biosciences | Cat#: 612786 / RRID:AB_2833077 |
| CD161 / BV421 / HP-3G10 | Biolegend | Cat#: 339914 / RRID:AB_2561421 |
| CD19 / PE/Dazzle 594 / HIB19 | Biolegend | Cat#: 302252 / RRID:AB_2563560 |
| CD19 / PE-Cy7 / SJ25C1 | Biolegend | Cat#: 363012 / RRID:AB_2564203 |
| CD19 / BUV395 / SJ25C1 | Biolegend | Cat#: 563549 / RRID:AB_2738272 |
| CD19 / FITC / HIB19 | Biolegend | Cat#: 302206 / RRID:AB_314236 |
| CD1a / PE-Cy7 / HI149 | Biolegend | Cat#: 300122 / RRID:AB_2073128 |
| CD1c / APC / L161 | Biolegend | Cat#: 331524 / RRID:AB_10719956 |
| CD24 / BUV737 / ML5 | Biolegend | Cat#: 741831 / RRID:AB_2871166 |
| CD25 / BUV737 / 2A3 | Biolegend | Cat#: 612807 / RRID:AB_2916878 |
| CD27 / FITC / O323 | BD Biosciences | Cat#: 302806 / RRID:AB_314298 |
| CD27 / APC / O323 | Biolegend | Cat#: 302810 / RRID:AB_314302 |
| CD3 / AF700 / SK7 | Biolegend | Cat#: 300424 / RRID:AB_493741 |
| CD3 / BUV737 / UCHT1 | Biolegend | Cat#: 564307 / RRID:AB_2744390 |
| CD3 / BUV395 / UCHT1 | BD Biosciences | Cat#: 563546 / RRID:AB_2744387 |
| CD34 / PE-Cy7 / 581 | Biolegend | Cat#: 343516 / RRID:AB_1877251 |
| CD38 / PE-Cy7 / HIT2 | Biolegend | Cat#: 303516 / RRID:AB_2072782 |
| CD4 / BV785 / RPA-T4 | Biolegend | Cat#: 300554 / RRID:AB_2564382 |
| CD4 / PerCP-Cy5.5 / RPA-T4 | Biolegend | Cat#: 300530 / RRID:AB_893322 |
| CD4 / APC / RPA-T4 | Biolegend | Cat#: 300514 / RRID:AB_314082 |
| CD40 / BV421 / 5C3 | BD Biosciences | Cat#: 334332 / RRID:AB_2564211 |
| CD40L (CD154) / PE / 24-31 | Biolegend | Cat#: 310805 / RRID:AB_314828 |
| CD45 / BUV395 / HI30 | Biolegend | Cat#: 563792 / RRID:AB_2869519 |
| CD45RA / PerCP-Cy5.5 / HI100 | Biolegend | Cat#: 304122 / RRID:AB_893357 |
| CD56 / PE-Cy7 / 5.1H11 | Biolegend | Cat#: 362510 / RRID:AB_2563927 |
| CD56 / BUV395 / NCAM16.2 | Biolegend | Cat#: 563554 / RRID:AB_2687886 |
| CD56 / BV421 / HCD56 | Biolegend | Cat#: 318328 / RRID:AB_11218798 |
| CD8 / BV711 / SK1 | Biolegend | Cat#: 344734 / RRID:AB_2565243 |
| CD8 / BUV737 / SK1 | Biolegend | Cat#: 612754 / RRID:AB_2870085 |
| CD80 / BV605 / 2D10 | Biolegend | Cat#: 305225 / RRID:AB_11123909 |
| CD86 / FITC / BU63 | BD Biosciences | Cat#: 374204 / RRID:AB_2721574 |
| CD95 / BV510 / DX2 | Biolegend | Cat#: 305640 / RRID:AB_2629738 |
| c-kit (CD117) / BV510 / 104D2 | Biolegend | Cat#: 313220 / RRID:AB_2563804 |
| CRTH2 (CD294) / FITC / BM16 | Biolegend | Cat#: 350108 / RRID:AB_11204086 |
| CX3CR1 / BV605 / 2A9-1 | BD Biosciences | Cat#: 744488 / RRID:AB_2742268 |
| CXCL10 / PE / 4NY8UN | Biolegend | Cat#: 12-9744-42 / RRID:AB_2572728 |
| CXCR3 (CD183) / PE / 1C6/CXCR3 | BD Biosciences | Cat#: 353705 / RRID:AB_10959652 |
| CXCR5 (CD185) / BV711 / J252D4 | BD Biosciences | Cat#: 356934 / RRID:AB_2629526 |
| FoxP3 / PE / 206D | BD Biosciences | Cat#: 320108 / RRID:AB_492986 |
| HLA-DR / PerCP-Cy5.5 / L243 | Biolegend | Cat#: 307630 / RRID:AB_893567 |
| HLA-DR / BV605 / L243 | BD Biosciences | Cat#: 307640 / RRID:AB_2561913 |
| IFNα2b / BV421 / 7N4-1 | PBL Assay Science | Cat#: 561382 / RRID:AB_10716058 |
| IFNβ / FITC / MMHB-3 | Biolegend | Cat#: PBL-21400-3 / RRID:AB_387831 |
| IgD / BV510 / IA6-2 | BD Biosciences | Cat#: 348220 / RRID:AB_2561945 |
| IgG / PerCP-Cy5.5 / M1310G05 | Biolegend | Cat#: 410709 / RRID:AB_2565787 |
| IgM / BV605 / MHM-88 | Biolegend | Cat#: 314523 / RRID:AB_2562373 |
| IL-12 / APC / C11.5 | BD Biosciences | Cat#: 501818 / RRID:AB_2124518 |
| IL-6 / PE-CF594 / MQ2-13A5 | Biolegend | Cat#: 563543 / RRID:AB_2714015 |
| KLRG1 / PE-Cy7 / 14C2A07 | BD Biosciences | Cat#: 368614 / RRID:AB_2728371 |
| PD-1 (CD279) / FITC / NAT105 | BD Biosciences | Cat#: 367412 / RRID:AB_2572163 |
| TCRγδ / BV786 / 11F2 | BD Biosciences | Cat#: 744743 / RRID:AB_2742451 |
| TNFα / PerCP-Cy5.5 / MAb11 | Biolegend | Cat#: 502926 / RRID:AB_2204081 |
| Vα7.2 / APC / 3C10 | Biolegend | Cat#: 351708 / RRID:AB_10933246 |
| Vδ2 / APC / B6 | Biolegend | Cat#: 331418 / RRID:AB_2687324 |
| Biological samples | ||
| Human peripheral blood, trans men cohort | amedes Medizinisches Versorgungszentrum Hamburg | N/A |
| Human peripheral blood, control cohort | University Medical Center Hamburg-Eppendorf | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ack Lysing Buffer | Lonza Bioscience | Cat#: BP10-548E |
| Brefeldin A | Sigma Aldrich | Cat#: B7651-5MG |
| CL097 | InvivoGen | Cat#: tlrl-c97 |
| Dimethyl sulfoxide (DMSO) | Sigma Aldrich | Cat#: D85897 |
| Fetal Bovine Serum | Sigma Aldrich | Cat#: S0615 |
| Lymphocyte Separation Media | Capricorn | Cat#: LSM-A |
| Medium A (Fixation Medium) | Life Technologies | Cat#: GAS001S100 |
| Medium B (Permeabilization Medium) | Life Technologies | Cat#: GAS002S100 |
| Trizol | Life Technologies | Cat#: 15596018 |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell 5' Kit v2, 4 rxns | 10x Genomics | Cat#: PN-1000265 |
| Human IFN Alpha All Subtype ELISA Kit, High Sensitivity (Serum, Plasma, TCM) | PBL Assay Science | Cat#: 41115-1 |
| Human IFN Beta ELISA Kit, High Sensitivity (Serum, Plasma, TCM) | PBL Assay Science | Cat#: 41415-1 |
| qScriber™ cDNA Synthesis Kit | highQU GmbH | Cat#: RTK0101 |
| Oligonucleotides | ||
| See Table S5 for a comprehensive list of primers. | ||
| Deposited data | ||
| scRNA seq data | This paper | GEO: GSE231370 |
| Code used for analysis of scRNA seq data | This paper | https://github.com/malteborggrewe/IFN_pDC_transmencohort |
| Software and algorithms | ||
| AUCell v. 1.16.0 | Aibar et al.54 | https://scenic.aertslab.org/ |
| Cellranger v. 7.0.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| enrichR v. 3.0 | Kuleshov et al.56 | https://github.com/wjawaid/enrichR |
| FlowJo v. 10.8.1 | BD Biosciences | https://www.flowjo.com/ |
| GraphPad Prism v. 9.4.0 | GraphPad Software | https://www.graphpad.com/ |
| MAST algorithm v. 1.20.0 | Finak et al.53 | https://github.com/RGLab/MAST |
| R v. 4.2.2 | The R Foundation | https://www.r-project.org/ |
| Rtsne v. 0.15 | Krijthe et al.55 | https://github.com/jkrijthe/Rtsne |
| Seurat v. 4.0.4 | Hao et al.52 | https://satijalab.org/seurat/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marcus Altfeld (marcus.altfeld@leibniz-liv.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Peripheral blood samples from trans men enrolled in this study (n=10) were collected under protocols approved by the ethics committee of the medical association Hamburg (Ethik-Kommission der Ärztekammer Hamburg, permission number PV5245). Individuals in the cisgender control cohort were recruited at the University Medical Center Hamburg-Eppendorf (permission number PV4780). Written informed consent was obtained from all individuals prior to enrolment in the study. Participants in the trans men cohort were recruited and received 1,000 mg testosterone undecanoate i.m. (Nebido, Jenapharm, Jena, Germany) injections at the medical practice amedes Medizinisches Versorgungszentrum Hamburg GmbH (Hamburg, Germany), where blood draws were also performed. Whole blood was collected from participants in 9 mL EDTA tubes (catalog no. 02.1066.001, Sarstedt, Nümbrecht, Germany). After the initial testosterone injection, the second injection was scheduled 6 weeks later, followed by a steady rhythm of 3 months, corresponding to TP1: 42 days, TP2: 126 days, TP3: 210 days, TP4: 294 days. Median injection dates of the cohort fit the scheduled rhythm exactly, ranging from TP1: 39-118 days, TP2: 96-203 days, TP3: 197-287 days, TP4: 268-488 days. Participants were observed over a median of 43 weeks (246-468 days), with median TP1: 42 (39-217) days, TP2: 130 (112-313) days, TP3: 219 (197-452) days, TP4: 303 (246-468) days. In six donors, the baseline blood sample was taken immediately prior to the initial testosterone injection, while in the other donors the time between first collected blood sample and initial testosterone injection was 6 / 9 / 23 / 161 days, respectively. Participants' ages at initiation of GAHT ranged from 19-41 years, with four participants in the age group 18-25 years, three in the age group 26-30 years, two in the group 31-40 years and one participant was 41 years of age. Individuals in the cis women control cohort (n=21) had a mean age of 33 years (range 29-40 years) and in the cis men control cohort (n=17) of 32 years (range 26-39 years). In the trans men cohort, five individuals received leuprorelin acetate (Trenantone, Takeda, Tokyo, Japan) in addition to testosterone undecanoate injections. None of the participants took anovulants prior to the first testosterone injection. Before initiation of testosterone administration, individuals were screened for chronic infections, autoimmune and metabolic diseases, and cancers by medical history and blood tests, and were only included in the study if negative.
Method details
Sex hormone analysis & hemogram
Sex hormone levels in serum were quantified from all respective timepoints at the laboratory facility amedes MVZ für Laboratoriumsmedizin, Mikrobiologie und Genetik (Hamburg, Germany) using electrochemiluminescence immunoassays for total testosterone, bioavailable testosterone, and 17-β-estradiol. Leukocyte counts and hemogram analyses from EDTA blood of all respective timepoints were performed at the same facility, using a Sysmex XN3100 device (Sysmex, Norderstedt, Germany).
PBMC isolation and cryopreservation
After venepuncture, processing of transgender cohort blood samples was started after an average of 2.5 hours. PBMC isolation was performed by density centrifugation (500 g, 30 min, 21°C) using Lymphocyte Separation Media (catalog no. LSM-A, Capricorn, Ebsdorfergrund, Germany). After collection of PBMCs at the interphase of the layers and washes using Hank's Balanced Salt (catalog no. H6648, Sigma Aldrich, St. Louis, MO, USA), remaining erythrocytes were lysed with ACK Lysing Buffer (catalog no. BP10-548E, Lonza, Basel, Switzerland) for 3 min. Cell counts were determined using the Trypan Blue (catalog no. T8154, Sigma Aldrich) exclusion method on a TC20 Automated Cell Counter (Bio-Rad, Hercules, CA, USA) and aliquots of different proportions were prepared. For cytokine production experiments by flow cytometry, PBMCs were used immediately. For population frequency and scRNA seq experiments, PBMCs were frozen in heat inactivated fetal bovine serum (FBS, catalog. no S0615, Sigma Aldrich) + 10% (v/v) dimethyl sulfoxide (catalog no. D85897, Sigma Aldrich) using a freezing container special for cryopreservation at -80°C and transferred into liquid nitrogen storage the next day until further processing.
TLR7/8 stimulation
Freshly isolated PBMCs (2.5 x 106 for cytokine production experiments by flow cytometry, 5 x 105 for cytokine quantification by ELISA and ISG qPCR) were stimulated with 1 μg/mL of the synthetic TLR7/8 agonist CL097 (catalog no. tlrl-c97, InvivoGen, San Diego, CA, USA) for 17 hours at 37°C, 5% CO2 in RPMI Medium (catalog no. 21875-034, Thermo Fisher, Waltham, MA, USA) + 10% (v/v) FBS (Sigma Aldrich) + Penicillin-Streptomycin (catalog no. P4333-100ML, Sigma Aldrich). For intracellular cytokine staining, 5 μg/mL Brefeldin A (catalog no. B7651-5MG, Sigma Aldrich) was added immediately after addition of CL097. For scRNA seq experiments, CL097 stimulation of thawed PBMCs was performed for 2 hours under the same conditions in the absence of Brefeldin A.
PBMC staining, flow cytometry and FACS
For cytokine production experiments, freshly isolated PBMCs were stained immediately after CL097 stimulation. For population frequency and scRNA seq experiments, staining was performed after thawing of cryopreserved PBMCs. PBMCs were stained with the indicated surface antibodies and a dead stain (NEAR-IR for population frequency experiments and scRNA seq experiments, catalog no. L34976, Thermo Fisher; Zombie Aqua for cytokine production experiments, catalog no. 423101, Biolegend, San Diego, CA, USA) for 20 minutes at room temperature in the dark. For flow cytometry, PBMCs were fixed with Medium A (catalog no. GAS001S100, Life Technologies/Thermo Fisher), or left unfixed for FACS-isolation of pDCs. For intracellular cytokine staining, PBMCs were additionally permeabilized with Medium B (catalog no. GAS002S100, Life Technologies/Thermo Fisher) during incubation with the respective antibodies for 20 minutes at room temperature in the dark. Flow cytometry was performed using a LSRFortessa Flow Cytometer (Beckton Dickinson, Franklin Lakes, NJ, USA) in the core facility Fluorescence Cytometry at the Leibniz Institute of Virology, Hamburg, Germany. Generated FCS files were analyzed using the FlowJo software (version 10.8.1, Beckton Dickinson). Gating strategy is shown in Figure S2. A comprehensive list of all fluorescence antibodies is provided in the key resources table.
Cytokine quantification in supernatant
After stimulation of PBMCs with CL097 for 17 hours, supernatant was collected and stored at -80°C until further processing, keeping unstimulated supernatant as control. IFNα and IFNβ concentrations were quantified by ELISA using Human IFN Alpha All Subtype ELISA Kit, High Sensitivity (catalog no. 41115-1, PBL, Piscataway, NJ, USA) and Human IFN Beta ELISA Kit, High Sensitivity (catalog no. 41415-1, PBL) following the manufacturer's protocol. Samples were diluted 1:18 in Bio-Plex Sample Diluent (catalog no. 10041561, Bio-Rad) prior to incubation on the plates. Absorbance was measured at 450 nm at a Safire2 microplate reader (Tecan, Männedorf, Switzerland) using the Magellan software version 6.1. Samples were measured in duplicates and mean values were used for calculations. Values below the limit of detection were considered zero.
Reverse transcription and qPCR
1 x 106 unstimulated PBMCs and 5 x 105 CL097-stimulated PBMCs were lysed in TRIzol (catalog no. 15596018, Life Technologies/Thermo Fisher) following manufacturer’s recommendations and stored at -80°C until further processing. RNA was extracted with ethanol precipitation and treated with DNase I (catalog no. AM2222, Invitrogen/Thermo Fisher) as well as with a recombinant ribonuclease inhibitor (RNaseOUT, catalog no. 100000840, Invitrogen/Thermo Fisher) before reverse transcription into cDNA on a FlexCycler Block assembly T48 device (Analytik Jena, Jena, Germany), using the qScriber cDNA Synthesis Kit (catalog no. RTK0101, highQU, Kraichtal, Germany) following manufacturer’s recommendations. qPCR was performed with SybrGreen (catalog no. QPD0450, highQU) at a LightCycler 480 II-96 device (Roche, Basel, Switzerland) using the LightCycler480 software version 1.5.0. Primers for the following ISGs were utilized: HERC5, IFI6, IFI44, IFI44L, IFITM3, IFIT1, IFIT3, ISG15, MX1, MX2, OAS1, OAS2, OAS3, PLSCR1 and STAT1. A comprehensive list of all primer sequences is provided in Table S5. Normalized ISG expression was calculated using the 2-ΔΔCt method with B2M as housekeeping gene.
Cell sorting, scRNA seq, library generation and next generation sequencing
pDCs were FACS-isolated from CL097-stimulated and unstimulated PBMCs of 3 donors from the trans men cohort according to the gating strategy presented in Figure S2, using a FACSAria Fusion (Beckton Dickinson) device in the core facility Fluorescence Cytometry at the Leibniz Institute of Virology, Hamburg, Germany. Experiment conditions were labelled with respective TotalSeq™-C0251 anti-human Hashtag antibodies (Biolegend). After sorting, cells were kept on ice in autoMACS Running Buffer (Miltenyi Biotec, Bergisch Gladbach, Germany). Subsequently, cells were loaded onto the 10X Chromium Controller (10X Genomics, Pleasanton, CA, USA). Single-cell libraries were generated using the Chromium Next GEM Single Cell 5' Kit v2 (catalog no. PN-100026510, X Genomics) according to the manufacturer’s instructions. Libraries were sequenced on an Illumina NovaSeq 6000 system (S4 flow cell) with 150 base pairs and paired-end configurations.
scRNA seq analysis
Reads were aligned to the human genome (GRCh38) using cellranger (v7.0.0) (10X Genomics) before demultiplexing the conditions based on TotalSeq™-C0251 anti-human Hashtag antibodies (Biolegend) using the HTODemux function of Seurat (v4.0.4).52 After demultiplexing, alignment, and cell filtering (>5% mitochondrial reads/cell, median+/- 3∗MAD log10(reads)/cell), a total of 33,438 cells with a median expression of 2,028 genes/cell remained. Contaminating cells that expressed T cell marker CD3, monocyte marker CD14 or cDC markers AXL, CX3CR1, CD5 were excluded from further analysis, resulting in a final total number of 32,731 cells (Donor 1: 9,206; Donor 2: 3,524; Donor 3: 20,001). Data were normalized and integrated based on donors using Seurat and subsequently regressed for % mitochondrial reads/cell and number of genes/cell with Seurat ScaleData function. After determining the highly variable genes, jackstraw analysis and elbow plots were employed to find a suitable number of principal components used for UMAP dimensionality reduction, following a Louvain clustering based on shared nearest neighbor graph (FindClusters function, Seurat). Cluster markers and differentially expressed genes were determined using FindAllMarkers or FindMarkers function based on MAST algorithm (v1.20.0).53 Genes were regarded differentially expressed with an adjusted P value < 0.05 and a log2(Fold Change) > 0.25. Gene ontology enrichment for biological processes (2021) was performed using enrichR (v3.0). Enrichment of gene sets (GO terms and immune cell gene sets) was done using area under curve analysis (AUCell v1.16.0).54
Quantification and statistical analysis
Statistical analyses were performed using the GraphPad Prism software version 9.4.0 (GraphPad Software, San Diego, CA, USA) and R version 4.2.2 (The R Foundation, Vienna, Austria). The statistical tests applied for each analysis and number of subjects per group (n) are indicated in figure legends. A p value less than 0.05 was considered significant.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG) Research Unit 5068 - Sex Differences in Immunity. Open Access funding was enabled and organized by the Leibniz-Gemeinschaft. We thank the core facility Fluorescence Cytometry at the Leibniz Institute of Virology, Hamburg. We express our gratitude to all individuals who agreed to participate in this study.
Author contributions
B.G., S.H.H., S.M.Z., F.H., and B.P. planned experiments, and C.F.K., M.J.B., and M.A. gave important intellectual and practical input and guidance. B.G., S.H.H., F.H., R.J.T., M.P., V.S., and S.M.Z. conducted experiments. S.H.H., S.M.Z., M.A., and C.D. established the cohort, and S.H.H. and F.H. processed cohort blood samples. A.D. and J.H. provided essential support for flow cytometry. B.G. and F.H. analyzed flow cytometry data, and B.G. performed respective statistical tests. B.G. analyzed hormone, hemogram, qPCR, and ELISA data and conducted statistical tests. J.M.C. reviewed clinical cohort data. L.G. performed tSNE of population frequency data, and E.T. gave valued intellectual input. C.D. recruited participants for the cohort and provided clinical information. B.G. and M.A. had unrestricted access to all data. B.G., M.B., and M.A. conceptualized the publication. B.G. and M.A. wrote the manuscript, and M.B., S.B., and M.J.B. gave critical input in the writing process. The manuscript was reviewed by all authors and approved for publication.
Declaration of interests
S.H.H. is an employee at AdaptVac since 08/2021, a company commercializing virus-like particle display technology and vaccines. The other authors have declared that no competing interests exist.
Published: October 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108209.
Supplemental information
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at NCBI GEO and are publicly available as of the date of publication under accession number GSE231370.
-
•
Code used for the analysis of scRNA seq data can be accessed through GitHub (https://github.com/malteborggrewe/IFN_pDC_transmencohort) and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Markle J.G., Fish E.N. SeXX matters in immunity. Trends Immunol. 2014;35:97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen H., Klein S.L. Sex Differences in Immunity to Viral Infections. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.720952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson N.M., Chen H.-C., Lechner M.G., Su M.A. Sex Differences in Immunity. Annu. Rev. Immunol. 2022;40:75–94. doi: 10.1146/annurev-immunol-101320-125133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berghöfer B., Frommer T., Haley G., Fink L., Bein G., Hackstein H. TLR7 ligands induce higher IFN-alpha production in females. J. Immunol. 2006;177:2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- 6.Whitacre C.C. Sex differences in autoimmune disease. Nat. Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 7.Rönnblom L., Alm G.V. Systemic lupus erythematosus and the type I interferon system. Arthritis Res. Ther. 2003;5:68–75. doi: 10.1186/ar625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockshin M.D. Sex differences in autoimmune disease. Lupus. 2006;15:753–756. doi: 10.1177/0961203306069353. [DOI] [PubMed] [Google Scholar]
- 9.Moulton V.R. Sex Hormones in Acquired Immunity and Autoimmune Disease. Front. Immunol. 2018;9:2279. doi: 10.3389/fimmu.2018.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angum F., Khan T., Kaler J., Siddiqui L., Hussain A. The Prevalence of Autoimmune Disorders in Women: A Narrative Review. Cureus. 2020;12:e8094. doi: 10.7759/cureus.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugrue J.A., Bourke N.M., O'Farrelly C. Type I Interferon and the Spectrum of Susceptibility to Viral Infection and Autoimmune Disease: A Shared Genomic Signature. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.757249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., Liu Y.J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 13.Colonna M., Trinchieri G., Liu Y.-J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 14.Collin M., Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154:3–20. doi: 10.1111/imm.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell D., Chintala S., Dey M. Plasmacytoid dendritic cell in immunity and cancer. J. Neuroimmunol. 2018;322:63–73. doi: 10.1016/j.jneuroim.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Reizis B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity. 2019;50:37–50. doi: 10.1016/j.immuni.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balan S., Saxena M., Bhardwaj N. Dendritic cell subsets and locations. Int. Rev. Cell Mol. Biol. 2019;348:1–68. doi: 10.1016/bs.ircmb.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Meier A., Chang J.J., Chan E.S., Pollard R.B., Sidhu H.K., Kulkarni S., Wen T.F., Lindsay R.J., Orellana L., Mildvan D., et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat. Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seillet C., Laffont S., Trémollières F., Rouquié N., Ribot C., Arnal J.-F., Douin-Echinard V., Gourdy P., Guéry J.C. The TLR-mediated response of plasmacytoid dendritic cells is positively regulated by estradiol in vivo through cell-intrinsic estrogen receptor α signaling. Blood. 2012;119:454–464. doi: 10.1182/blood-2011-08-371831. [DOI] [PubMed] [Google Scholar]
- 20.Wang J.P., Zhang L., Madera R.F., Woda M., Libraty D.H. Plasmacytoid dendritic cell interferon-α production to R-848 stimulation is decreased in male infants. BMC Immunol. 2012;13:35. doi: 10.1186/1471-2172-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler S.M., Beisel C., Sutter K., Griesbeck M., Hildebrandt H., Hagen S.H., Dittmer U., Altfeld M. Human pDCs display sex-specific differences in type I interferon subtypes and interferon α/β receptor expression. Eur. J. Immunol. 2017;47:251–256. doi: 10.1002/eji.201646725. [DOI] [PubMed] [Google Scholar]
- 22.Laffont S., Rouquié N., Azar P., Seillet C., Plumas J., Aspord C., Guéry J.C. X-Chromosome complement and estrogen receptor signaling independently contribute to the enhanced TLR7-mediated IFN-α production of plasmacytoid dendritic cells from women. J. Immunol. 2014;193:5444–5452. doi: 10.4049/jimmunol.1303400. [DOI] [PubMed] [Google Scholar]
- 23.Souyris M., Cenac C., Azar P., Daviaud D., Canivet A., Grunenwald S., Pienkowski C., Chaumeil J., Mejía J.E., Guéry J.C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- 24.Hagen S.H., Henseling F., Hennesen J., Savel H., Delahaye S., Richert L., Ziegler S.M., Altfeld M. Heterogeneous Escape from X Chromosome Inactivation Results in Sex Differences in Type I IFN Responses at the Single Human pDC Level. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb K., Peckham H., Radziszewska A., Menon M., Oliveri P., Simpson F., Deakin C.T., Lee S., Ciurtin C., Butler G., et al. Sex and Pubertal Differences in the Type 1 Interferon Pathway Associate With Both X Chromosome Number and Serum Sex Hormone Concentration. Front. Immunol. 2018;9:3167. doi: 10.3389/fimmu.2018.03167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hembree W.C., Cohen-Kettenis P.T., Gooren L., Hannema S.E., Meyer W.J., Murad M.H., Rosenthal S.M., Safer J.D., Tangpricha V., T'Sjoen G.G. Endocrine Treatment of Gender-Dysphoric/Gender-Incongruent Persons: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2017;102:3869–3903. doi: 10.1210/jc.2017-01658. [DOI] [PubMed] [Google Scholar]
- 27.Campochiaro C., Host L.V., Ong V.H., Denton C.P. Development of systemic sclerosis in transgender females: a case series and review of the literature. Clin. Exp. Rheumatol. 2018;36:50–52. [PubMed] [Google Scholar]
- 28.Chan K.L., Mok C.C. Development of systemic lupus erythematosus in a male-to-female transsexual: the role of sex hormones revisited. Lupus. 2013;22:1399–1402. doi: 10.1177/0961203313500550. [DOI] [PubMed] [Google Scholar]
- 29.Santos-Ocampo A.S. New onset systemic lupus erythematosus in a transgender man: possible role of feminizing sex hormones. J. Clin. Rheumatol. 2007;13:29–30. doi: 10.1097/01.rhu.0000256169.05087.ad. [DOI] [PubMed] [Google Scholar]
- 30.Ocon A., Peredo-Wende R., Kremer J.M., Bhatt B.D. Significant symptomatic improvement of subacute cutaneous lupus after testosterone therapy in a female-to-male transgender subject. Lupus. 2018;27:347–348. doi: 10.1177/0961203317734921. [DOI] [PubMed] [Google Scholar]
- 31.Giltay E.J., Fonk J.C., von Blomberg B.M., Drexhage H.A., Schalkwijk C., Gooren L.J. In vivo effects of sex steroids on lymphocyte responsiveness and immunoglobulin levels in humans. J. Clin. Endocrinol. Metab. 2000;85:1648–1657. doi: 10.1210/jcem.85.4.6562. [DOI] [PubMed] [Google Scholar]
- 32.Velho I., Fighera T.M., Ziegelmann P.K., Spritzer P.M. Effects of testosterone therapy on BMI, blood pressure, and laboratory profile of transgender men: a systematic review. Andrology. 2017;5:881–888. doi: 10.1111/andr.12382. [DOI] [PubMed] [Google Scholar]
- 33.Antun A., Zhang Q., Bhasin S., Bradlyn A., Flanders W.D., Getahun D., Lash T.L., Nash R., Roblin D., Silverberg M.J., et al. Longitudinal Changes in Hematologic Parameters Among Transgender People Receiving Hormone Therapy. J. Endocr. Soc. 2020;4:bvaa119. doi: 10.1210/jendso/bvaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahidi N.T. Androgens and erythropoiesis. N. Engl. J. Med. 1973;289:72–80. doi: 10.1056/NEJM197307122890205. [DOI] [PubMed] [Google Scholar]
- 35.Griesbeck M., Ziegler S., Laffont S., Smith N., Chauveau L., Tomezsko P., Sharei A., Kourjian G., Porichis F., Hart M., et al. Sex Differences in Plasmacytoid Dendritic Cell Levels of IRF5 Drive Higher IFN-α Production in Women. J. Immunol. 2015;195:5327–5336. doi: 10.4049/jimmunol.1501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seillet C., Rouquié N., Foulon E., Douin-Echinard V., Krust A., Chambon P., Arnal J.-F., Guéry J.C., Laffont S. Estradiol promotes functional responses in inflammatory and steady-state dendritic cells through differential requirement for activation function-1 of estrogen receptor α. J. Immunol. 2013;190:5459–5470. doi: 10.4049/jimmunol.1203312. [DOI] [PubMed] [Google Scholar]
- 37.Ianevski A., Giri A.K., Aittokallio T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat. Commun. 2022;13:1246. doi: 10.1038/s41467-022-28803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villani A.-C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes and progenitors. Science. 2017;356 doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.See P., Dutertre C.-A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K., et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356 doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laffont S., Seillet C., Guéry J.C. Estrogen Receptor-Dependent Regulation of Dendritic Cell Development and Function. Front. Immunol. 2017;8:108. doi: 10.3389/fimmu.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bost C., Arleevskaya M.I., Brooks W.H., Plaza S., Guery J.-C., Renaudineau Y. Long non-coding RNA Xist contribution in systemic lupus erythematosus and rheumatoid arthritis. Clin. Immunol. 2022;236 doi: 10.1016/j.clim.2022.108937. [DOI] [PubMed] [Google Scholar]
- 42.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azar P., Mejía J.E., Cenac C., Shaiykova A., Youness A., Laffont S., Essat A., Izopet J., Passaes C., Müller-Trutwin M., et al. TLR7 dosage polymorphism shapes interferogenesis and HIV-1 acute viremia in women. JCI Insight. 2020;5 doi: 10.1172/jci.insight.136047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shepherd R., Bretherton I., Pang K., Mansell T., Czajko A., Kim B., Vlahos A., Zajac J.D., Saffery R., Cheung A., Novakovic B. Gender-affirming hormone therapy induces specific DNA methylation changes in blood. Clin. Epigenetics. 2022;14:24. doi: 10.1186/s13148-022-01236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y., Beyer A., Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Sonneveld S., Verhagen B.M.P., Tanenbaum M.E. Heterogeneity in mRNA Translation. Trends Cell Biol. 2020;30:606–618. doi: 10.1016/j.tcb.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Zhan Y., Chow K.V., Soo P., Xu Z., Brady J.L., Lawlor K.E., Masters S.L., O'Keeffe M., Shortman K., Zhang J.-G., Lew A.M. Plasmacytoid dendritic cells are short-lived: reappraising the influence of migration, genetic factors and activation on estimation of lifespan. Sci. Rep. 2016;6 doi: 10.1038/srep25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michalska A., Blaszczyk K., Wesoly J., Bluyssen H.A.R. A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses. Front. Immunol. 2018;9:1135. doi: 10.3389/fimmu.2018.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz L.X., Lee J., Kapoor U., Kartnig F., Sedlyarov V., Papakostas K., César-Razquin A., Essletzbichler P., Goldmann U., Stefanovic A., et al. TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7-9. Nature. 2020;581:316–322. doi: 10.1038/s41586-020-2282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youness A., Miquel C.-H., Guéry J.C. Escape from X Chromosome Inactivation and the Female Predominance in Autoimmune Diseases. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.-C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krijthe J.H. Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation. 2015. https://github.com/jkrijthe/Rtsne
- 56.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at NCBI GEO and are publicly available as of the date of publication under accession number GSE231370.
-
•
Code used for the analysis of scRNA seq data can be accessed through GitHub (https://github.com/malteborggrewe/IFN_pDC_transmencohort) and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.