Abstract
Claudins are four-transmembrane proteins, which were found in tight junctions. They maintain cell barriers and regulate cell differentiation and proliferation. They are involved in maintaining cellular polarity and normal functions. Different claudins show different expression patterns. The expression level and localization of claudins are altered in various cancers. They promote or inhibit proliferation, invasion, and migration of cancer cells through multiple signaling pathways. Therefore, claudins may serve as diagnostic markers, novel therapeutic targets, and prognostic risk factors. The important roles of claudins in cancer aroused our great interest. In the present review, we provide a summary of insights into expression patterns of claudins in cancer, which is more comprehensive and provides new ideas for further research.
Keywords: Claudins, Cancer, Biomarker, Pathological diagnosis, Expression
1. Introduction
Tight junctions (TJs) are a type of intercellular junctions. They are widely distributed at the apical regions of epithelial cells and endothelial cells. TJs act as mechanical junctions and selective barriers. As a result, TJs are essential for the maintenance of cellular polarity [1]. TJs are formed of the intricate combinations of multiple proteins. In 1998, Furuse et al. successfully identified and designated the claudin (CLDN) protein within the complex structure of TJs [2]. The CLDN family is a major component of TJs (Fig. 1). At least 24 members have been identified. CLDNs are important to cell barriers, cell differentiation, and proliferation [1]. The expression profile of CLDNs is tissue-specific. For example, CLDN18.1 was mainly expressed in the lung, whereas CLDN18.2 was mainly found in the stomach [3,4].
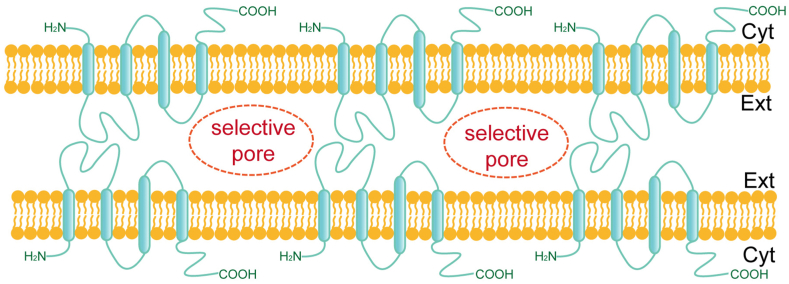
Fig. 1.
Claudins are essential components of intercellular tight junctions. Claudins are four-transmembrane proteins that act as mechanical barriers by interconnecting and are involved in forming intercellular selective pores.
Previous studies have shown apparent changes in the expression level and localization of CLDNs in several pathological conditions [[5], [6], [7], [8], [9], [10]]. Depending on the different types of cancer, CLDNs can exert either tumor suppressor or tumor promoting effects through different pathways. For example, CLDN1 could promote cancer cell proliferation or invasion in colorectal cancer [11], thyroid cancer [12], estrogen receptor (ER)-negative breast cancer [13], gastric cancer [14], pancreatic cancer [15]. In contrast, it could inhibit cancer cell proliferation in prostate cancer [16], lung adenocarcinoma [17], and ER-positive breast cancer [13]. Therefore, CLDNs may provide new insights into pathological diagnosis and perhaps new targets for therapy. Compared with previous reviews, this paper summarized CLDNs-related studies and progress more comprehensively, which would provide new ideas for subsequent intensive research.
1.1. Classification, expression and function of claudins
Mammalian CLDNs are divided into classic and non-classic CLDNs. Classic CLDNs include 1–10, 14, 15, 17, and 19. Non-classic CLDNs comprise 11–13, 16, 18, and 20–24 (Fig. 2). The C-terminus of classic CLDNs is short. Nonclassic CLDNs have a longer C-terminus, which interacts with cytoplasmic scaffolding proteins [18]. They can also be divided into two types according to the main functions: pore-forming and sealing CLDNs. The pore-forming CLDNs could form paracellular channels, such as CLDN2, 10, and 15. The sealing CLDNs could limit paracellular permeability, like CLDN1, 3, 4, 5, 7, and 11 [18]. CLDNs are expressed in a variety of normal human tissues. We summarized the expression patterns (Table 1) and functions (Table 2) of CLDNs in normal tissues.
Fig. 2.
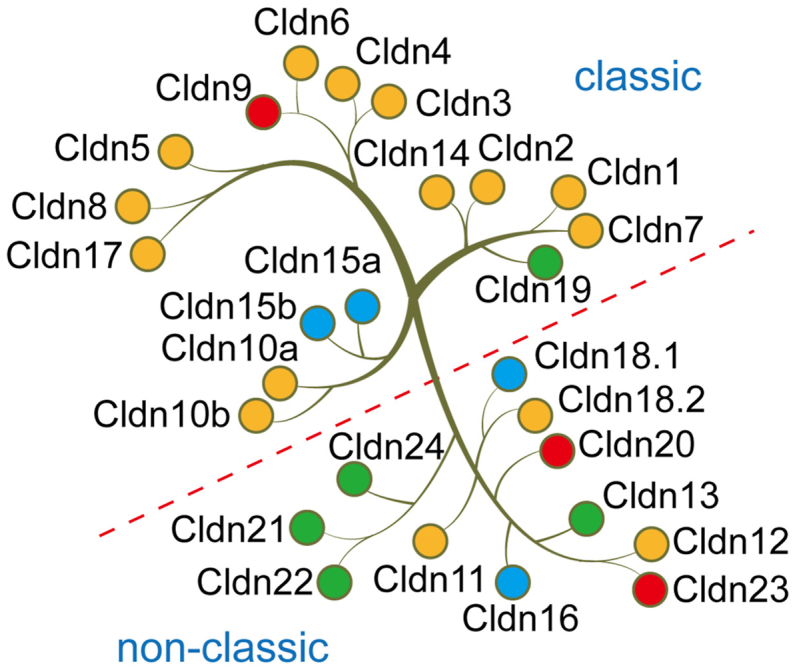
Mammalian claudins are mainly divided into two categories. One is classic, and the other is non-classic. The blue dots indicate that the claudin may mostly play a tumor suppressor role in tumors. The red dots indicate that the claudin may mainly play a role in promoting tumors. Yellow dots mean that the claudin plays a tumor-promoting or tumor-suppressing role in different tumors. Green dots indicate that the specific role of claudin in tumors has not been reported yet.
Table 1.
Expression patterns of claudins in normal tissues.
| Claudins | Expression patterns | Reference |
|---|---|---|
| CLDN1 | Intestine, brain, kidney, liver, testis, fetal lung alveolar epithelial (HFL) cells, bronchiolar epitheliums, and pancreatic ducts and exocrine glands | [[19], [20], [21]] |
| CLDN2 | Pancreatic ducts and exocrine glands | [21] |
| CLDN3 | Colorectal, thyroid, salivary gland, pancreas, prostate cancer, liver, kidney, HFL cells, bronchiolar epitheliums, pancreatic ducts and exocrine glands, and pancreatic islet endocrine glands | [[19], [20], [21]] |
| CLDN4 | Breast, ovary, prostate, bladder, gastrointestinal mucosa, bile duct, HFL cells, type II alveolar epitheliums, bronchiolar epitheliums, and pancreatic ducts and exocrine glands | [[19], [20], [21]] |
| CLDN5 | HFL cells, type II alveolar epitheliums, and vascular endothelial cells | [20,22,23] |
| CLDN6 | Normal tissues during embryonic development | [24,25] |
| CLDN7 | Bronchiolar epitheliums, pancreatic ducts and exocrine glands, and pancreatic islet endocrine glands | [20,21] |
| CLDN10a | Kidney | [26] |
| CLDN14 | Thick ascending limb of Henle | [27,28] |
| CLDN18.1 | HFL cells and type II alveolar epitheliums | [19] |
| CLDN18.2 | Gastric mucosal epithelial cells | [4] |
Table 2.
Function of claudins in normal tissues.
| Claudins | Function | Reference |
|---|---|---|
| CLDN2 | Formation of cation channels | [29] |
| Promotes the permeability of Na+ and water through solvent dragging | [30] | |
| CLDN5 | Formation of blood-brain barrier | [22] |
| CLDN10a | Formation of anion channels | [29,31] |
| CLDN10b | Formation of cation channels | [29,31] |
| CLDN14 | Regulated the permeability of Ca2+ in the thick ascending limb of Henle (TAL) | [27,28] |
| CLDN15 | Formation of cation channels | [29] |
| Decoupled the solvent dragging effect of CLDN2 | [32] | |
| CLDN16 | Regulated the permeability of Mg2+ in the TAL | [33,34] |
| CLDN17 | Formation of anion channels | [29] |
| CLDN18.1 | Maturation and differentiation of the alveolar epithelium | [3,[35], [36], [37]] |
| CLDN18.2 | Regulated the permeability of H+ between gastric mucosal epithelial cells | [38] |
| Formation of gastric mucosal barrier | [39] | |
| CLDN19 | Regulated the permeability of Mg2+ in the TAL | [34] |
1.2. Claudins in cancer
Alterations of the expression or localization of CLDNs have been observed in various tumors. Their expression profiles can be regulated by different mechanisms. Furthermore, these changes could result in alterations of cancer cell functions through various pathways.
Epithelial-mesenchymal transition (EMT) is the process by which epithelial cells transform into cells with a mesenchymal phenotype, allowing epithelial cells to acquire mesenchymal characteristics such as increased migration rate [40]. EMT is therefore one of the keys to cancer cell metastasis. High expression of CLDN1 in colorectal cancer [41], hepatocellular carcinoma [42], and lung cancer [43] might enhance cancer cell aggressiveness through promotion of EMT. CLDN3 could inhibit EMT in hepatocellular carcinoma [44] and lung cancer [45]. Whether CLDN6 promotes EMT depends on the tumor type. CLDN6 promoted EMT in gastric cancer [46], but suppressed EMT in breast cancer [47].
Cell proliferation and apoptosis can also be affected by changes of CLDNs expression. In colorectal cancer, high expression of CLDN1 could promote proliferation and resist apoptosis [48]. However, in pancreatic cancer, increased CLDN1 might suppress cancer cell proliferation and promote apoptosis [15]. In lung cancer, CLDN18.1 expression was down-regulated, which promoted cancer cell proliferation and inhibited apoptosis [49].
Furthermore, CLDNs are associated with cancer chemoresistance. In pancreatic cancer cells, CLDN7 promoted cell resistance to cisplatin [50]. High expression of CLDN1 and CLDN2 could also confer resistance to cisplatin in lung cancer cells [51,52].
1.3. Claudins in gastric cancer
CLDNs play vital roles in gastric cancer (Table 3). Different CLDNs can affect the occurrence and development of gastric cancer through multiple pathways.
Table 3.
Roles of claudins in gastric cancer.
| Claudins | Activity | Findings | Reference |
|---|---|---|---|
| CLDN1 | Tumor promoter | Associated with lymph node metastasis, higher TNM stage, worse overall survival (OS), and postoperative survival | [14,53] |
| Tumor suppressor | Reduced the tumorigenic effect of GC cells | [54] | |
| CLDN2 | Tumor promoter | Promoted the invasion and growth of GC cells | [55] |
| CLDN4 | Tumor suppressor | Inhibited the invasiveness of gastric cancer cells | [56] |
| CLDN6 | Tumor promoter | Promoted EMT through the LAST1/2-YAP1 pathway | [46] |
| Tumor suppressor | Promoted GC cell differentiation by inhibiting the JNK pathway | [57] | |
| CLDN7 | Tumor promoter | Promoted the invasiveness of GC | [55] |
| CLDN9 | Tumor promoter | Promoted the migration of GC | [55] |
| CLDN10 | Tumor promoter | Low expression of CLDN10 correlated with better OS | [58] |
| CLDN11 | Tumor suppressor | Low expression of CLDN11 was associated with increased cancer cell invasion, migration, and proliferation | [59] |
| CLDN18.2 | Tumor promoter | CLDN18-ARHGAP26 gene fusion reduced the adhesion of cancer cells and makes cancer cells more invasive | [60] |
| Tumor suppressor | Deletion of Cldn18.2 could cause gastric inflammation and create conditions for GC | [61] | |
| CLDN23 | Tumor promoter | Patients expressing CLDN23 had a worse prognosis and were associated with vessel cancer embolus | [62] |
CLDN1's role in gastric cancer is debated. Recent research suggested that CLDN1 was the direct target of the tumor suppressor RUNX3. RUNX3 could up-regulate CLDN1 expression and reduce tumorigenicity by binding to its promoter region. Meanwhile, transforming growth factor beta (TGF-β) might promote the combination of the two [54]. Besides, some research showed that β-catenin which was increased in gastric cancer could up-regulate CLDN1. High CLDN1 expression was associated with lymph node involvement, higher TNM stage, poor overall survival (OS) [14], and poor postoperative survival in gastric cancer patients [53], suggesting that CLDN1 may promote gastric cancer development.
Researches on CLDN6 in gastric cancer are controversial. Several studies showed that CLDN6 expression was increased in gastric cancer which might promote cell migration [55]. By interacting with large tumor suppressor 1/2 (LATS1/2), CLDN6 reduced the phosphorylation level of Yes-associated protein 1 (YAP1) and promoted YAP1 entry into the nucleus, thereby increasing the transcriptional activity of the EMT-associated transcription factor Snail1, which promoted EMT and increased cancer cell invasion [46]. Other works showed that CLDN6 might be a tumor suppressor in gastric cancer. Overexpression of CLDN6 in gastric cancer cells could inhibit the c-Jun N-terminal kinase (JNK) pathway, promote the differentiation of tumor cells which would make them more similar to normal gastric epithelial cells, and reduce the degree of malignancy of gastric cancer cells [57].
Furthermore, the clinicopathological analysis showed that low expression of CLDN6 in gastric cancer was associated with lymph node metastasis, late stage, distant metastasis and poor prognosis [63]. Although its function is disputed, it has been developed as a therapeutic target. The potential of CLDN6 as a tumor immunotherapy target was demonstrated by a virus-like particle (VLP) vaccine against CLDN6, which induced a complement-dependent cytotoxic (CDC) response in mice [64]. In addition, a chimeric antigen receptor (CAR) T-cell therapy targeting CLDN6 in combination with RNA lipoplex (LPX) is in phase I/II clinical trials for solid tumors expressing CLDN6 (NCT04503278), which has shown high efficacy [65].
CLDN11 expression was decreased in gastric cancer. Decreased lncRNA PCAT18 in gastric adenocarcinoma would increase the miR-135b, thereby inhibiting the transcription of CLDN11 and promoting the invasion, migration, and proliferation of cancer cells [59]. Meanwhile, miR-421 could also inhibit CLDN11 and arrest the G1/S cell cycle [66]. Furthermore, CLDN11 expression was regulated by epigenetic mechanisms as well. Hypermethylation of the CLDN11 promoter leads to its silencing [67,68]. However, CLDN11 was increased in gastric cancer metastases [69], suggesting that CLDN11 has different functions at different stages in the occurrence and development of gastric cancer.
Infection with Helicobacter pylori (H. pylori) causes persistent inflammation and damage to the gastric mucosa, eventually resulting in gastric cancer. However, Cldn18.2-KO mice could develop refractory gastritis in the absence of H. pylori infection, leading to spasmolytic polypeptide expressing metaplasia (SPEM) and ultimately gastric cancer. When incubated with H. pylori, deleting CLDN18.2 could cause permeability defects [70,61]. These studies show that CLDN18.2 deletion can cause gastritis and increased gastric cancer susceptibility.
The clinicopathological analysis of CLDN18.2 in gastric cancer is disputed. Some studies showed that CLDN18.2 had no significant relationship with gender [71,72], age [73], primary tumor location [73], TNM stage [72], Lauren classification, and prognosis [5,71,[74], [75], [76], [77], [78]]. However, other studies showed that high CLDN18.2 expression was correlated with age (<70 years old) [77], higher initial stage [77], lymph node and distant metastasis, diffuse gastric cancer in Lauren classification, and better prognosis [72,[79], [80], [81]].
In normal gastric mucosa, CLDN18.2 is located in the cell membrane. In contrast, gastric cancer cells showed diffuse CLDN18.2 positivity in the membrane, cytoplasm, and even nucleus [10]. Meanwhile, CLDN18.2 expression in gastric cancer cells was correlated with mucin-5AC (MUC5AC) expression and mucin-2 (MUC2) deletion [79,82]. In the invasive front of gastric cancer, higher CLDN18.2 expression was associated with lower Ki67 expression [83]. CLDN18.2 was also linked to the human epidermal growth factor receptor 2 (HER2) expression [71,78] and Epstein-Barr virus (EBV) infection [5,73,77].
It is interesting to note that CLDN18.2 has become an emerging gastric cancer therapeutic target. A phase I clinical trial of CAR T-cell therapy targeting CLDN18.2 is currently underway (NCT03874897) and has shown promising efficacy in patients with advanced or refractory gastric cancer [84]. IMAB362 is a newly developed chimeric monoclonal antibody capable of killing CLDN18.2-positive tumor cells by antibody-dependent cell-mediated cytotoxicity (ADCC) and CDC [85]. In addition, in some gastric cancers, mainly diffuse gastric cancer and signet ring cell carcinoma, the CLDN18-ARHGAP26 gene fusion can be observed. It was associated with patient age, stage, metastasis, prognosis, and genomically stable (GS) gastric cancer subtype and exclusively with RHOA and cadherin 1 (CDH1) mutations. CLDN18.2 is diffusely positive in gastric cancer cells with the gene fusion. The fusion protein could disrupt the post-synaptic density-95, disks-large, and zonula occludens-1 (PDZ)-binding domain of wild-type CLDN18.2, reduce cancer cell adhesion, and promote cancer cell invasion [[60], [86], [87]].
CagA is a virulence-related factor of H. pylori. After H. pylori infection, cagA could up-regulate the expression of caudal-related homeobox transcription factor 2 (CDX2) and promote the binding of CDX2 to the promoter region of CLDN2, which increased the CLDN2 expression. Upregulated CLDN2 may promote the invasion of gastric cancer cells [88]. These studies indicate CLDN2 primarily plays a promoting role in gastric cancer progression. Instead, CLDN4 mainly acted as a tumor suppressor in gastric cancer by inhibiting the invasion and migration of gastric cancer cells. CLDN4 could be regulated by epigenetic mechanisms of methylation and histone modifications [56]. Furthermore, CLDN7 promoted cancer cell invasion, and CLDN9 enhanced cell migration [55]. Reduced CLDN10 was associated with better OS and had a negative correlation with immune cell infiltration [58]. CLDN23 was decreased in gastric cancer, but patients with higher CLDN23 expression had worse prognosis and more vascular tumor thrombus [62].
1.4. Claudins in colorectal cancer
CDX2 is up-regulated in most colorectal cancer cases. By cooperating with CDX1 and GATA binding protein 4 (GATA4), it increased the expression of β-catenin, which then raised the transcriptional activity of T-cell factor (Tcf) and initiated the transcription of CLDN1 [89,90]. As a tumor suppressor, SMAD family member 4 (SMAD4) was down-regulated in colorectal cancer and lost its inhibitory effect on CLDN1 expression [91]. Tumor necrosis factor-α (TNFα), one of the critical cytokines in colitis-associated colorectal cancer (CAC) development, could also up-regulate CLDN1 expression [41]. CLDN1 up-regulated the EMT-related transcription factors Snail and Slug by the activation of the c-Abl-Ras-Raf-1-ERK1/2 pathway, thereby promoting EMT and increasing cell invasion and migration [41]. Another EMT-related transcription factor is zinc finger E-box binding homeobox 1 (ZEB1). CLDN1 could up-regulate the activity of ZEB1 through the Wnt and PI3K/AKT pathways, which down-regulated E-cadherin and promoted EMT. In a feedback loop, downregulation of E-cadherin activated the Wnt pathway. Simultaneously, the PI3K/AKT pathway could activate Snail and Slug transcriptional activity [92]. In addition, CLDN1 could also activate the Notch pathway by upregulating ERK and matrix metalloproteinase 9 (MMP9), which increased inflammation and cancer cell proliferation [93].
Anoikis is a type of programmed cell death induced by separating cells from the extracellular matrix, which is one of the essential mechanisms of tumor metastasis. In colorectal cancer, CLDN1 bound to Src protein, activated Akt, and up-regulated Bcl-2, allowing cancer cells to acquire the anti-anoikis ability and enhanced metastasis [48].
High expression of CLDN1 in colorectal cancer was associated with poor prognosis [92]. In conclusion, CLDN1 plays a promoting role in the progression of colorectal cancer by enhancing the invasion, migration, and proliferation of cancer cells. However, some studies indicated that high CLDN1 expression was associated with a better prognosis [94]. Differences in the length of follow-up may explain the discrepancy with previous studies. The former was followed for over 10 years, while the latter was followed for 3 and 5 years. This suggests that high CLDN1 expression might have a negative correlation with long-term prognosis. Moreover, CLDN1 was a novel target for colorectal cancer [95].
CLDN2 is up-regulated in colorectal cancer and mainly plays a promoting role. The microenvironment of colorectal cancer activated the ERK1/2 and PI3K pathways in an epidermal growth factor receptor (EGFR)-dependent manner and up-regulated CLDN2 [96]. Inhibition of CLDN2 expression could dissociate the CLDN2/zonula occludens 1 (ZO1)/ZO-1-associated Y-box factor (ZONAB) complex. ZONAB translocated to the nucleus which promoted the transcription of n-myc downstream-regulated gene 1 (NDRG1). NDRG1 could increase cyclin-dependent kinase inhibitor activity, thereby arresting the cell cycle. At the same time, NDRG1 could ubiquitinate caveolin-1 and inhibit EMT. In addition, CLDN2 stabilized the CLDN2/ZO1/ZONAB complex and prevented ZONAB from entering the nucleus, thereby inhibiting NDRG1 expression and promoting cancer cell proliferation, invasion, and migration [97,98]. Highly expressed CLDN2 could also promote liver and lung metastasis, which was linked to its PDZ-binding motif [99].
In colorectal cancer cells, EGF bound to EGFR and activated the ERK1/2 and PI3K/AKT pathways, thereby upregulating CLDN3 expression [100]. However, another study showed that CLDN3 expression was down-regulated by epigenetic mechanisms in colorectal cancer. Low expression of CLDN3 promoted EMT through activation of the Wnt/β-catenin pathway and enhanced cell migration ability. Low CLDN3 expression could also activate the IL-6/IL-23/STAT3 pathway and further activate the Wnt pathway. Accordingly, patients with higher CLDN3 expression showed improved outcomes [101].
The role of CLDN7 in colorectal cancer is still controversial. The Wnt/Tcf4 pathway inhibited CLDN7 expression via SRY box transcription factor 9 (SOX9) in colorectal epithelial cells. In colorectal cancer, this inhibitory effect was relieved, and high CLDN7 expression disrupted cell polarity and promoted proliferation [102]. However, another study suggested that low expression of CLDN7 increased Snail1 activity and promoted EMT [103]. Meantime, low expression of CLDN7 was associated with poor differentiation, vein invasion, and liver metastasis [103,104]. CLDN8 mainly plays a promoting role in colorectal cancer progression. The LncRNA LINC00662 was up-regulated in colorectal cancer and competed with miR-340-5p for binding, thereby increasing the co-expression of CLDN8/IL22. CLDN8 then activated the MAPK/ERK pathway and up-regulated the expression of MMP-9, promoting cell proliferation, invasion, and migration [105,106]. CLDN11 expression was under epigenetic regulation, which was reduced by promoter hypermethylation. Low level of CLDN11 was associated with metastasis and poor prognosis [107]. In vitro, CLDN12 induced cell proliferation which could be inhibited by anti-CLDN12 antibody [108]. This suggests that anti-CLDN12 may be one of the targeted therapies for colorectal cancer. The expression of CLDN14 was increased, which promoted cancer cell proliferation, invasion, and migration by activating the PI3K/AKT/mTOR pathway [109]. CLDN23 could also enhance the proliferation and migration of colorectal cancer cells [110].
1.5. Claudins in pancreatic cancer
CLDNs play key roles in the pathogenesis of pancreatic cancer (Table 4). Pancreatic cancer usually originates from precancerous lesions, including pancreatic intraepithelial neoplasia (PanIN), mucinous cystic neoplasia (MCN), and intraductal papillary mucinous neoplasia (IPMN), of which PanIN is the most common [111]. CLDN4 was highly expressed in pre-cancerous pancreatic lesions, and its expression was increased with the histological grade of MCN and IPMN, especially in pancreaticobiliary IPMN [112]. K-ras mutations were found in pancreatic cancer, and activating K-ras could up-regulate CLDN4 [113]. TGFβ could promote cancer cell invasion by inhibiting CLDN4 expression. Meanwhile, patients with higher CLDN4 expression had a better prognosis [113]. CLDN3 and CLDN4 were specific receptors for Clostridium perfringens enterotoxin (CPE). CPE could reduce cell permeability and induce cell apoptosis by binding to CLDN3 and CLDN4 [114]. As a result, CPE can be used for the treatment of CLDN4-positive pancreatic cancer.
Table 4.
Roles of claudins in pancreatic cancer.
| Claudins | Activity | Findings | Reference |
|---|---|---|---|
| CLDN1 | Tumor suppressor | Promoted apoptosis and inhibited proliferation | [15] |
| CLDN4 | Tumor suppressor | Inhibited the invasiveness of cancer cells | [113] |
| CLDN7 | Tumor promoter | Improved the invasion and metastasis, and promoted the resistance to cisplatin | [50] |
| CLDN12 | Tumor promoter | Promoted EMT | [115] |
| CLDN18.2 | Tumor suppressor | High expression of CLDN18.2 correlated with a better prognosis | [116] |
CLDN18.2 was also highly expressed in high-grade pancreatic intraepithelial neoplasia (PanIN-2, 3), IPMN, MCN, well-differentiated pancreatic cancer, and pancreatic cancer metastases [112,117,118]. The expression level of CLDN18.2 increased with the degree of malignancy of pancreatic cancer. It could be used as a diagnostic marker for early pancreatic cancer, and patients with higher CLDN18.2 expression had a better outcome [119,116]. CLDN18.2 was mainly regulated by the PKC pathway and could be epigenetically regulated by DNA methylation [120,121]. Previous studies showed that IMAB362 for CLDN18.2-positive gastric and pancreatic cancer patients could achieve significant curative effects with fewer side effects [122]. Furthermore, the chemotherapeutic drugs gemcitabine or 5-fluorouracil were able to increase CLDN18.2 expression in vitro [123,124].
CLDN1 was expressed in the membranes of normal pancreatic ducts and well-differentiated pancreatic cancer cells. CLDN1 has the potential to be used as a diagnostic marker for pancreatic cancer. TNFα could promote cell apoptosis and inhibit proliferation by up-regulating CLDN1 [15]. Therefore, CLDN1 mainly plays a tumor suppressor role. CLDN2 was mainly expressed on benign pancreatic lesions and decreased in MCN and IPMN [112]. CLDN7 decreased with pancreatic ductal adenocarcinoma differentiation but was not associated with patients' prognosis [119]. Besides, CLDN7 could form a complex with EpCAM, improving cell invasion, metastasis, and resistance to cisplatin [50]. Zinc finger protein 460 (ZNF460) promoted the expression of LINC00857 which competed with miR-150-5p to promote CLDN12 expression. LINC00857 could also enhance the binding of SRSF1 to pre-CLDN12, increase alternative splicing and promote the expression of CLDN12. High CLDN12 expression could promote EMT, cell invasion, and migration [115].
1.6. Claudins in hepatocellular carcinoma
In hepatocellular carcinoma (HCC), miR-29a expression was decreased and the inhibitory effect on CLDN1 was lost [125]. CLDN1 activated the Ras-Raf-1-ERK pathway through c-Abl, up-regulating the transcriptional activities of Slug and ZEB1 to promote EMT [42]. CLDN1 could also activate PKCδ through c-Abl and up-regulate MMP-2 to promote cancer cell invasion [126]. Hypermethylation of its promoter led to the downregulation of CLDN3 in HCC and the loss of its inhibitory effect on the Wnt/β-catenin pathway, which promoted EMT and enhanced cell metastasis and proliferation [44]. CLDN6 was up-regulated in HCC, promoting cell proliferation, invasion, and migration by activating EGFR/AKT/mTOR pathway [127]. CLDN9 was also increased and promoted cell migration through the Tyk2/STAT3 pathway [128].
The expression of miR-486 was decreased in HCC, while CLDN10 was increased. CLDN10 was able to promote EMT, migration, and angiogenesis [129]. High level of CLDN10 expression were associated with disease recurrence after radical hepatectomy [130,131]. miR-99b was up-regulated in HCC which inhibited the transcription of CLDN11, thereby promoting cancer cell migration [132]. EZH2 was an enhancer of transcriptional regulators. In HCC, EZH2 could silence CLDN14 via H3K27me3. Low CLDN14 expression activated the Wnt/β-catenin pathway, which promoted EMT and cancer cell invasion [133]. CLDN17 was also increased, which activated the STAT3 pathway via Tyk2 and enhanced cell migration capacity. Simultaneously, patients with higher CLDN17 expression had a poorer prognosis [134].
1.7. Claudins in lung cancer
Lung cancer can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC comprises adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. CLDNs exert vital funtions both in SCLC and NSCLC (Table 5).
Table 5.
Roles of claudins in lung cancer.
| Claudins | Activity | Findings | Reference |
|---|---|---|---|
| CLDN1 | Tumor promoter | Promoted EMT in NSCLC | [43] |
| Initiated autophagy and induced resistance to CDDP in NSCLC | [51] | ||
| High expression of CLDN1 in lung adenocarcinoma was associated with poor prognosis | [135] | ||
| Tumor suppressor | Loss of CLDN1 expression in lung adenocarcinoma correlated with disease progression and cancer cell invasiveness | [17] | |
| High expression of CLDN1 was associated with better prognosis in lung SCC | [136] | ||
| CLDN2 | Tumor promoter | Up-regulated MMP9 and promoted the migration of lung adenocarcinoma cells | [137] |
| Increased drug resistance to CDDP | [52] | ||
| CLDN3 | Tumor promoter | High expression of CLDN3 was associated with poor prognosis in lung SCC | [136] |
| Tumor suppressor | Decreased expression in lung SCC promoted EMT through the Wnt/β-catenin signaling pathway | [45] | |
| CLDN6 | Tumor suppressor | Low expression of CLDN6 was associated with poor prognosis in NSCLC | [138] |
| CLDN7 | Tumor suppressor | Improved the adhesion of lung cancer cells, inhibited cell proliferation, invasion, and migration | [139,140] |
| CLDN9 | Tumor promoter | Promoted the movement of lung cancer cells | [141] |
| CLDN12 | Tumor promoter | Promoted EMT in SCC cells via Tyk2/Stat1 pathway | [142] |
| CLDN18.1 | Tumor suppressor | Inhibited proliferation and invasion by inhibiting YAP1/PI3K/PDK1/AKT pathway | [49] |
In NSCLC cells, TNFα could promote EMT by up-regulating CLDN1 [43]. Increased CLDN1 initiated autophagy by increasing the phosphorylation level of unc-51 like autophagy activating kinase 1 (ULK1), resulting in the resistance of NSCLC to cisplatin [51]. High expression of CLDN1 in lung adenocarcinoma was correlated with poor prognosis (Sun et al., 2016). In summary, CLDN1 may have an oncogenic role in NSCLC. However, another study suggests that loss of CLDN1 was linked to disease progression and cancer cell invasion [17]. Higher CLDN1 expression was associated with a better prognosis in lung squamous cell carcinoma (SCC) [136]. In invasive lepidic predominant adenocarcinoma (LPA), CLDN1 expression was lower than adenocarcinoma in situ (AIS), and the survival rate was better in patients with higher CLDN1 expression [143]. In the process from AIS to LPA, CLDN1 is thought to play a key role. These studies suggest that CLDN1 may be involved in suppressing tumor growth in lung SCC.
MMP9 cleaved EGF to activate the EGFR/MEK/ERK pathway and up-regulated c-Fos expression in lung adenocarcinoma cells. C-Fos interacted with c-Jun to form the AP-1 dimer complex, which bound to the AP-1 site in CLDN2 promoter region to promote CLDN2 expression [144]. CLDN2 could increase the nuclear distribution of Sp1 to up-regulate MMP9 expression which would promote cancer cell migration [137]. These works suggest a possible crosstalk between CLDN2 and MMP9. ATP-binding cassette transporters were able to transport substrates across the cell membrane. In lung cancer, CLDN2 stimulated the expression and function of ATP-binding cassette subfamily C member 2 (ABCC2) by upregulating Sp1. ABCC2 increased chemotherapy resistance by reducing chemotherapy drug accumulation in cancer cells [52]. Chemotherapy resistance may therefore be more common in patients with high CLDN2 expression. Kaempferol and luteolin are natural flavonoids found in various plants. They could reduce the interaction between STAT3 and CLDN2 promoter to down-regulate the expression of CLDN2 which would inhibit the proliferation of lung adenocarcinoma cells [145]. Therefore, CLDN2 could be developed as a therapeutic target for lung cancer, and flavonoids exert an anti-tumor effect by inhibiting CLDN2.
The expression of CLDN7 was reduced in lung cancer. Low expression of CLDN7 decreased the expression of integrin beta1, resulting in poor adhesion between tumor cells and the stroma [139]. It could also promote cancer cell proliferation. In NSCLC, hepatocyte growth factor (HGF) could bind to its receptor c-Met to activate the ERK/MAPK pathway which would promote cell invasion and migration. CLDN7 could be an inhibitor of HGF activation of the ERK/MAPK pathway. However, decreased expression of CLDN7 led to the loss of this inhibitory ability, resulting in increased invasion and migration [140]. In addition, low expression of CLDN7 was found to be related to poor prognosis in lung SCC [146]. CLDN7 has been shown to be an important tumor suppressor for lung cancer.
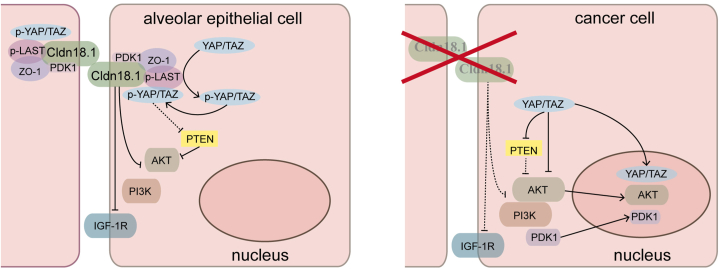
CLDN18.1 was essential for alveolar epithelial maturation and differentiation. Immunohistochemistry showed that CLDN18.1 expression was down-regulated in lung cancer. Lung adenocarcinoma could spontaneously develop in Cldn18.1-KO mice. In vitro, assays indicated that CLDN18.1 was able to inhibit cell proliferation and invasion and promote cell death. CLDN18.1 inhibited the translocation of Yes-associated protein (YAP) to the nucleus, thereby inhibiting the PI3K/PDK1/AKT pathway (Fig. 3). Taken together, these results suggest that CLDN18.1 functions primarily as a tumor suppressor in lung cancer [49].
Fig. 3.
Claudin 18.1 (CLDN18.1) is a tumor suppressor in alveolar epithelial cells. In normal alveolar epithelial cells, CLDN18.1 binds to Zonula Occludens-1 (ZO-1) and Hippo kinases (p-LATS) in normal alveolar epithelial cells to form a complex. Yes-associated protein (YAP) is phosphorylated by p-LAST and anchored at this complex, inhibiting YAP translocation to the nucleus. Meanwhile, inhibition of YAP can preserve the activity of phosphatase and tensin homolog (PTEN), thereby inhibiting phosphatidylinositol 3-kinase/phosphoinositide-dependent kinase 1 (PI3K/PDK1)/AKT signaling pathway. CLDN18.1 can also directly inhibit PDK1, AKT, and insulin-like growth factor-1 receptor (IGF-1R). In lung cancer cells, deletion of Cldn18.1 causes YAP activation and translocation to the nucleus to promote cancer cell proliferation. Meanwhile, activated YAP can inhibit the activity of PTEN and activate the PI3K/PDK1/AKT signaling pathway, which further promotes cancer cell proliferation.
Reduced expression of CLDN3 in lung SCC activated the Wnt/β-catenin pathway to promote EMT which would increase the ability of cancer cells to invade and migrate [45]. However, there was literature suggesting that high CLDN3 expression in pulmonary SCC was associated with poor prognosis [136]. In lung SCC, CLDN5, 7, and 18 could reduce the phosphorylation level of AKT by inhibiting the interaction between AKT and PDK1, which could reduce cyclin D1 to suppress G1-S cell cycle progression [147]. CLDN6 was down-regulated in NSCLC. Low CLDN6 expression was associated with poor prognosis and was an independent risk factor for predicting NSCLC prognosis [138]. CLDN18.2 could also be expressed in NSCLC and can be used as one of the therapeutic targets [148]. CLDN9 was overexpressed in lung cancer, promoted tumor cell motility, and might be associated with metastasis [141]. Similar to CLDN1, the expression of CLDN10 was decreased in LPA compared to AIS, and the survival rate of CLDN10-positive patients was higher, suggesting that CLDN10 may also play a role in the progression from AIS to LPA [143]. By activating the Tyk2/Stat1 pathway, increased CLDN12 in lung SCC reduced E-cadherin expression and promoted EMT. Meanwhile, high CLDN12 expression was associated with lymph node metastasis and poor prognosis [142]. As in the case of colorectal cancer, an anti-CLDN12 antibody also inhibited the proliferation of lung cancer cells in vitro [108]. This makes CLDN12 a promising target for cancer therapy.
Sulforaphane (SFN) is one of the best anti-cancer substances from plants as it inhibits the carcinogenesis of cells and induces the apoptosis of cells. SFN-Cys is an intermediate metabolite with a longer retention time in the body and higher concentrations in lung. SFN-Cys could down-regulate CLDN5 and up-regulate CLDN7 through ERK1/2 in lung cancer cells. The type of lung cancer determined the effect of SFN-Cys on CLDN1. SFN-Cys increased the expression of CLDN1 in lung adenocarcinoma but decreased the expression of CLDN1 in SCC. At the same time, SFN-Cys reduced the interaction of CLDN1, 5, and 7 with α-tubulin to disrupt microtubule dynamics and induce cancer cell apoptosis [149]. Chrysin is also a type of flavonoid. CLDN1 and 11 were up-regulated in SCC. Chrysin could down-regulate the expression of CLDN1 and 11 by reducing the phosphorylation level of AKT and inhibiting the interaction between AKT and PDK1. Ultimately, this increased the toxicity of doxorubicin (DXR) to lung SCC [150].
1.8. Claudins in breast cancer
CLDN6 was reduced in breast cancer and functioned primarily as a tumor suppressor. Epigenetic modifications might affect the expression of CLDN6. Abnormal hypermethylation of CpG islands in the promoter region led to a decrease in the expression of CLDN6 [151]. Meanwhile, MeCP2 was capable of binding to CpG islands in the aberrantly methylated promoter region of CLDN6. It recruited histone deacetylase (HDAC) to deacetylate H3Ac and H4Ac, thereby inhibiting transcription and leading to a reduction in CLDN6 expression [152]. SMAD2 also down-regulated CLDN6 by DNA methyltransferase 1 (DNMT1)-mediated promoter methylation [47].
The low levels of CLDN6 could trigger the activation of the p38 MAPK pathway, leading to an increase in the expression of MMP2. This, in turn, enhanced cell proliferation and migration capabilities [153]. Conversely, CLDN6 downregulation could reduce the transcriptional activity of apoptosis signal-regulating kinase 1 (ASK1) and inhibited JNK and p38 pathway activation. This led to an increased Bcl2/Bax ratio, a decreased caspase3 expression, and an inhibition of the mitochondrial apoptosis pathway [154,155]. Additionally, in breast cancer cells, the activation of ERβ with estradiol could boost the transcription of CLDN6. High levels of CLDN6 promoted autophagy through the autophagy regulator beclin1 while inhibiting cell invasion and migration [156]. To summarize, CLDN6 acts as a tumor suppressor in breast cancer by reducing cell invasion and migration, promoting cell apoptosis, and inducing autophagy.
However, the expression of CLDN6 was linked to resistance to chemotherapy. Over-expressed CLDN6 interacted with p53 and caused the translocation of p53 from the nucleus to the cytoplasm. This led to increased expression of glutathione S-transferase-p1 (GSTP1), which rendered cancer cells resistant to drugs such as adriamycin, 5-fluorouracil, and cisplatin [157]. In triple-negative breast cancer, overexpression of CLDN6 resulted in increased expression of afadin (AF-6) and decreased phosphorylation of ERK, which in turn increased cancer cell resistance to adriamycin (ADM) [158].
Snail and Slug could bind to the E-box in the promoter region of CLDN1 and decrease its expression [159]. In ER-negative breast cancer, CLDN1 had a pro-cancer effect with anti-apoptotic properties and promoted the migration of cancer cells [13]. However, in ER-positive breast cancer, CLDN1 promoted apoptosis of cancer cells and acted as a tumor suppressor [13].
The level of CLDN2 expression was decreased in breast cancer, and this reduction had been found to be associated with advanced stage and lymph node metastasis [160]. This suggests that CLDN2 may play a role as a tumor suppressor. However, in cases of liver metastases from breast cancer, there was an increase in CLDN2 expression. The elevated CLDN2 in this context promoted the metastasis of cancer cells by facilitating the formation of complexes between integrin receptors α2β1 and α5β1, which led to increased adhesion between cancer cells and the extracellular matrix [161].
The level of CLDN3 expression was positively correlated with the occurrence of lymph node metastasis in triple-negative breast cancer [162]. CLDN4 expression was negatively correlated with tumor size and positively correlated with disease-free survival (DFS) [162]. Elevated expression of CLDN5 was linked to a poor prognosis. This increased expression of CLDN5 could facilitate cell movement and migration through the neural Wiskott-Aldrich syndrome protein (N-WASP) and Rho-associated coiled-coil containing protein kinase (ROCK) pathways [163]. CLDN8 expression was down-regulated and correlated with androgen receptor (AR) expression levels. Co-expression of CLDN8 and AR in breast cancer was correlated with a better prognosis [164]. CLDN10 was decreased in breast cancer, and the CLDN10 rs1325774 polymorphism was related to an increased breast cancer risk [165]. Reduced miR-205 resulted in the loss of its inhibitory effect on CLDN11, leading to increased CLDN11 expression. CLDN11 promoted cancer cell proliferation and inhibited apoptosis [166]. A higher level of CLDN11 in invasive breast ductal carcinoma was associated with a worse overall survival rate [167].
In breast cancer, IL-18 was elevated, leading to the inhibition of CLDN12. Low CLDN12 expression enhanced cell migration by activating the p38 MAPK pathway [168]. However, in ER-negative breast cancer, high expression of CLDN12 was associated with a poor overall survival rate [169]. In triple-negative breast cancer, twist1 inhibited CLDN15 expression by binding to its promoter region, resulting in enhanced cell invasion, migration, and angiogenic mimicry formation [170]. CLDN16 could reduce cancer cell invasiveness and proliferation, and patients with low CLDN16 expression had a poor prognosis [171]. On the other hand, overexpression of CLDN20 in breast cancer cells promoted cell invasiveness, and high expression of CLDN20 was associated with a poor prognosis [172].
1.9. Claudins in tumors of genital organs and urinary system
The lipolysis stimulated receptor (LSR) is a critical component of the tricellular junction. Downregulation of LSR could enhance nscriptional activity of Sp1, which would lead to increased expression of CLDN1 in endometrial cancer. High level of CLDN1 could then increase the expression of MMP2, 9, 10, and MT1-MMP, which ultimately increased the invasiveness of cancer cells [173]. CLDN2, 3, and 4 were also elevated, and the heightened CLDN3 and 4 were correlated with myometrial invasion [174,175]. Additionally, CLDN6 expression was increased and linked to poor prognosis, serving as an independent prognostic marker for this type of cancer [176]. On the other hand, CLDN7 was decreased in endometrial cancer. Low level of CLDN7 could promote cell proliferation and invasion [177].
CLDN6 expression was reduced in cervical cancer. As in breast cancer, reduced CLDN6 expression inhibited the activity of ASK1, which in turn suppressed the programmed cell death of cancer cells [178,179]. Cervical intraepithelial neoplasia (CIN) is a precursor of cervical cancer. The expression of CLDN12 was increased in CIN and cervical cancer. However, low CLDN12 expression in cervical cancer was correlated with poor prognosis [180].
The expression of miR-155 was reduced in ovarian cancer, resulting in a loss of its ability to inhibit CLDN1. CLDN1 could then increase the expression of MMP2 and 9 while reducing the expression of E-cadherin and β-catenin. These changes promoted cancer cell invasion and migration [181]. CLDN3 and 4 were elevated in ovarian cancer [182], which might increase cancer cell aggressiveness, possibly by increasing MMP2 activity [183]. However, in contrast to colorectal cancer, EGF actually reduced the expression of CLDN3 and 4. This was due to the activation of the MEK/ERK and PI3K/AKT pathways [184]. The low expression of them led to activation of the PI3K/AKT pathway and increased transcriptional activity of twist, which promoted EMT [185]. Low CLDN3 and 4 in ovarian cancer were linked to chemotherapy resistance. The decreased expression could lead to decreased mRNA levels of Copper Transporter 1 (CTR1), which would reduce cellular copper absorption and disrupt copper homeostasis. This, in turn, increased the resistance of cancer cells to cisplatin, a commonly used chemotherapy drug [186]. However, it is still unclear why CLDN3 and 4 are highly expressed in ovarian cancer, despite the ability of EGF to down-regulate these CLDNs and the presence of high EGFR expression in these tumors [187]. In addition, some studies suggested that chemotherapy-resistant or recurrent ovarian cancers actually exhibited high levels of CLDN3 and 4 [188,189]. There was also research suggesting that CLDN4 expression did not have a significant correlation with cisplatin resistance [190].
CLDN6 had been found to be up-egulated in ovarian cancer and was inversely associated with the infiltration of immune cells [191]. In addition, CLDN6 was one of the receptors for CPE, which induced cytotoxicity and increases the sensitivity of ovarian cancer cells to CPE [192]. A phase I clinical trial (NCT02054351) was conducted with IMAB027, an immune-activating monoclonal antibody targeting CLDN6. Furthermore, BNT142, a drug that specifically targeted CLDN6 was currently in Phase I/II clinical trials (NCT05262530). These studies highlight the potential of CLDN6 as a promising therapeutic target for ovarian cancer. Elevated CLDN7 expression in epithelial ovarian cancer (EOC) was linked to shorter progression-free survival. CLDN7 could also decrease the sensitivity of EOC cells to cisplatin, but treatment with CLDN7 siRNA could improve the situation [193]. Therefore, combination therapy targeting CLDN7 can effectively overcome chemoresistance to cisplatin. CLDN10 expression was elevated and positively correlated with immune cell infiltration. Higher CLDN10 expression was related to improved overall survival [191]. The role of CLDN10 might be associated with the TGFβ and Wnt/β-catenin pathways in ovarian cancer [194].
CLDN1 had been implicated as a tumor suppressor in prostate cancer. Loss of CLDN1 was linked to disease progression and cancer cell invasion [16]. CLDN4 was associated with a poor prognosis [195]. Another important factor in prostate cancer was AR, which could increase the expression of CLDN8. CLDN8 in turn activated the AKT/MAPK pathway, leading to an increase in cell proliferation and migration [196].
The expression of CLDN1 was increased in papillary renal cell carcinoma and could be used as a diagnostic marker. However, loss of CLDN1 was associated with tumor dedifferentiation and aggressiveness. In clear cell renal cell carcinoma (ccRCC), CLDN1 was associated with the occurrence of cancer and metastasis [197]. CLDN8 expression was reduced in ccRCC and patients with low expression had a poor prognosis. CLDN8 might be an independent prognostic factor for ccRCC. Besides, CLDN8 could inhibit cell invasion, migration, and proliferation through the AKT and EMT pathways [198]. Promoter hypermethylation in renal cell carcinoma led to reduced expression of CLDN10 which would reduce expression and acetylation of ATP synthase subunit O (ATP5O) [199]. This led to reduced expression of NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 (NDUFS2), which in turn affected reactive oxygen species (ROS) production, succinate dehydrogenase subunit B (SDHB) expression, and caspase3 expression. Ultimately, this inhibited the mitochondrial apoptosis pathway and promoted cell proliferation, migration, and invasion [200]. As for CLDN14 and 16, although they were associated with renal ion transport, their role in renal cancer needs to be further investigated.
1.10. Claudins in other tumors
In oral SCC, the expression of CLDN1 was increased. Increased CLDN1 led to cleavage of the laminin-5 gamma 2 chain by promoting the expression of MMP2, MMP9, and MT1-MMP. Lysate promoted the invasion of cancer cells by binding with EGFR [201]. Besides, high expression of CLDN1 was associated with high grade, late stage, nerve and vascular invasion, and lymph node invasion [202]. On the other hand, lacking CLDN7 in the center of oral SCC could be used as a marker to predict regional recurrence [203]. The expression of CLDN8 was reduced, but a higher expression of CLDN8 was linked to a lower overall survival rate [204]. In addition, the expression of CLDN17 was also down-regulated in oral cancer, which in turn increased the transcriptional activity of Snail and twist to promote EMT and enhance cell invasion and migration [205].
The expression of CLDN1 was increased in hypopharyngeal SCC. Elevated CLDN1 might stimulate the growth of new lymphatic vessels within the tumor, leading to an increased likelihood of lymph node metastasis [206,207]. In laryngeal carcinoma, CLDN1 and 7 were reduced, while CLDN3 and 8 were increased. These changes in expression levels can be used as molecular markers for the diagnosis [208].
In esophageal squamous cell carcinoma (ESCC), the transcription factor twist could prevent CLDN4 expression by binding to E-box site in the promoter region of CLDN4 [209]. Hypermethylation of its promoter region could also reduce the expression. Low expression of CLDN4, an independent risk factor for overall survival and recurrence, was linked to lymph node metastasis, the depth of invasion, the degree of differentiate, and the T stage of cancer [210,211]. CLDN6 could also be silenced by methylation in ESCC [212]. Loss of CLDN7 in ESCC would promote EMT to increase the invasion of cancer cells [213].
In melanoma, PKC increased CLDN1 expression and translocated it from the cell membrane to the cytoplasm. The high level of CLDN1 could then increase the expression of MMP2 and facilitate the invasion of tumor cells [214]. B-1 lymphocytes could secrete IL-10, which promoted CLDN10 expression. CLDN10 then activated the ERK pathway to facilitate the tumor cells metastasis [215]. Dysplastic nevus is an intermediate stage between common nevus and melanoma. Melanoma showed methylation in the promoter region of CLDN11 compared to dysplastic nevus. This suggests that loss of CLDN11 in melanoma is accompanied by an increased ability of tumor cells to invade surrounding tissues [216].
CLDN1 was increased in papillary thyroid disease (PTC) and its lymph node metastases [217]. In metastatic follicular thyroid carcinoma (FTC), PKC increased the nuclear expression of CLDN1. Elevated CLDN1 might facilitate cancer cell invasion and migration [12]. In thyroid cancer, CLDN10 was highly expressed in PTC but not in FTC, making it a potential discriminator between the two types of cancer [218]. Increased CLDN10 could promote proliferation, invasion, and migration of thyroid cells. Furthermore, the expression of CLDN10 was also related to lymph node metastasis in thyroid carcinoma [219].
In nasopharyngeal carcinoma (NPC), hypermethylation of the CLDN11 promoter prevents GATA1 from binding to the transcription start site. This led to CLDN11 being down-regulated which would reduce the interaction of CLDN11 with tubulin alpha-1b and beta-3. As a result, cell invasion and migration were enhanced. Using tubulin polymerization inhibitors could block the invasion and migration of NPC cells with low CLDN11 expression, suggesting their potential as therapeutic drugs [220].
In retinoblastoma (RB), miR-361-5p was reduced, preventing it from binding to the 3′ untranslated region of CLDN8. This resulted in increased expression of CLDN8, which would promote cell proliferation and inhibit cell apoptosis [221].
In pituitary oncocytoma, CLDN9 was increased. Furthermore, aggressive oncocytomas have higher expression of CLDN9 compared to non-invasive ones [222]. This suggests that CLDN9 may be involved in enhancing the invasiveness of cancer cells.
In glioblastoma, polycomb repressor complex 2-mediated H3K27me3 silenced the expression of miRNA-1275, which would up-regulate CLDN11. High expression of CLDN11 could inhibit cell proliferation [223].
In osteosarcoma, CLDN2 was reduced. This reduction resulted in the silencing of afadin. This activated the Ras/Raf/MEK/ERK pathway to promote the migration of cancer cells [224]. In addition, silencing CLDN8 could promote apoptosis. It could also reduce the level of CDK2, induce cell cycle arrest in G0/G1 phase, and inhibit cell proliferation [225]. Furthermore, CLDN12 could increase cancer cell invasion through PI3K/AKT pathway. High CLDN12 expression was associated with TNM stage, lung metastasis, and shorter survival time [226].
CLDN15 was less commonly altered in tumors. However, it could be expressed with high sensitivity and specificity in malignant pleural mesothelioma. This makes it a potential diagnostic marker for this particular cancer [227].
2. Discussion
CLDNs are the major constituents of tight junctions. CLDNs maintain cell polarity and regulate paracellular permeability under normal physiological conditions. However, when tissues are damaged, changes in the cellular microenvironment can affect the expression of CLDNs through various pathways. These changes in CLDNs can disrupt paracellular permeability, cell polarity, and overall cell function in different ways. Different tissues will be affected by different types of CLDN alterations. Some changes may help to repair tissues, while others may exacerbate tissue damage. Therefore, investigating the role of CLDNs in both normal and pathological conditions is crucial for understanding the development and occurrence of cancer. These findings may provide new insights into cancer diagnosis and therapy.
The potential of CLDNs as prognostic factors has been suggested by extensive analysis of clinicopathological data. By examing the expression of CLDNs, patients can be divided into positive and negative groups, which facilitates prognosis prediction and the formulation of appropriate treatment and follow-up plans. However, it is important to note that there may be conflicting results when analyzing certain clinicopathological data due to possible racial and regional differences in CLDNs expression. In addition, the final results may be significantly affected by the fact that some anti-CLDN antibodies may react with other subtypes of CLDNs. Therefore, in order to make the experimental results more accurate, it is necessary to validate the effectiveness and cross-reactivity of the antibodies based on specific circumstances before conducting the experiment.
CLDNs are essential in the regulation of cellular functions through multiple signaling pathways. Some of them are particularly involved in promoting cancer development. For example, they contribute to the induction of EMT through the Wnt/β-catenin pathway, as well as cancer cell proliferation, migration, and invasion through the MAPK and PI3K/AKT pathways. Targeted therapy can inhibit these tumor-promoting pathways and demonstrate anti-tumor efficacy. However, it is important to be cautious about potential side effects, as CLDNs are also expressed in normal cells.
Chimeric monoclonal antibodies specifically recognize highly expressed CLDNs on the surface of tumor cells, leading to the activation of ADCC and CDC mechanisms that effectively eliminate tumor cells. Although CLDNs are also present in normal tissues, they are predominantly localized at cellular junctions, where they are tightly connected and less accessible to binding by antibodies. This characteristic of CLDNs in normal cells contributes to the significant therapeutic effects and reduced side effects of chimeric monoclonal antibodies against CLDNs, making them highly promising in the field of tumor-targeted therapy.
Cisplatin is a commonly used anti-cancer drug, but its effectiveness is limited by the development of drug resistance. CLDNs have been found to be linked to cisplatin resistance. Combining anti-CLDN therapy may help reduce drug resistance in cancer cells. Therefore, further research into the relationship between CLDNs and drug resistance may potentially improve the effectiveness of chemotherapy for cancer patients.
Tumor immunotherapy has brought about a significant revolution in the field of tumor treatment, and the infiltration of immune cells is closely tied to prognosis and response to immunotherapy. A potential role for CLDNs in tumor immunotherapy has been indicated by their association with immune cell infiltration in the tumor stroma.
Extracting anti-tumor compounds from natural plants offers the benefits of reduced side effects and readily available resources. CLDNs have the potential to be targeted by a range of naturally derived compounds from plants, making them useful in the development of anti-tumor drugs.
However, the specific roles of various CLDN subtypes in tumorigenesis and development are not well understood and there is a lack of clinicopathological data. Nevertheless, CLDNs hold great promise for various applications in cancer research. Further investigation of CLDNs may lead to new approaches to cancer diagnosis, treatment, and prevention.
Data availability statement
No data was used for the research described in the article.
Funding
This research was supported by the grant (NO. 81802646 To Hui Li) from the National Natural Science Foundation of China, China.
CRediT authorship contribution statement
Daoyu Tao: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Writing – original draft. Bingxin Guan: Conceptualization, Data curation, Formal analysis, Methodology. Hui Li: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. Chengjun Zhou: Conceptualization, Formal analysis, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Hui Li, Email: 201990000019@sdu.edu.cn.
Chengjun Zhou, Email: chengjunzhou@sdu.edu.cn.
References
- 1.Angelow S., Ahlstrom R., Yu A.S. Biology of claudins. Am. J. Physiol. Ren. Physiol. 2008;295(4):F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse M., et al. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998;141(7):1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlingmann B., Molina S.A., Koval M. Claudins: gatekeepers of lung epithelial function. Semin. Cell Dev. Biol. 2015;42:47–57. doi: 10.1016/j.semcdb.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin U., et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin. Cancer Res. 2008;14(23):7624–7634. doi: 10.1158/1078-0432.CCR-08-1547. [DOI] [PubMed] [Google Scholar]
- 5.Dottermusch M., et al. Expression of the potential therapeutic target claudin-18.2 is frequently decreased in gastric cancer: results from a large Caucasian cohort study. Virchows Arch. 2019;475(5):563–571. doi: 10.1007/s00428-019-02624-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halimi S.A., et al. Comprehensive immunohistochemical analysis of the gastrointestinal and Müllerian phenotypes of 139 ovarian mucinous cystadenomas. Hum. Pathol. 2021;109:21–30. doi: 10.1016/j.humpath.2020.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Zou J., et al. Idiopathic pulmonary fibrosis is associated with tight junction protein alterations. Biochim. Biophys. Acta Biomembr. 2020;1862(5) doi: 10.1016/j.bbamem.2020.183205. [DOI] [PubMed] [Google Scholar]
- 8.Xu S., et al. Caveolin-1 regulates the expression of tight junction proteins during hyperoxia-induced pulmonary epithelial barrier breakdown. Respir. Res. 2016;17(1):50. doi: 10.1186/s12931-016-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen C.H., et al. SPAK-p38 MAPK signal pathway modulates claudin-18 and barrier function of alveolar epithelium after hyperoxic exposure. BMC Pulm. Med. 2021;21(1):58. doi: 10.1186/s12890-021-01408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y., et al. Nuclear staining of claudin-18 is a new immunohistochemical marker for diagnosing intramucosal well-differentiated gastric adenocarcinoma. Pathol. Int. 2020;70(9):644–652. doi: 10.1111/pin.12978. [DOI] [PubMed] [Google Scholar]
- 11.Dhawan P., et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Invest. 2005;115(7):1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwanziger D., et al. The impact of CLAUDIN-1 on follicular thyroid carcinoma aggressiveness. Endocr. Relat. Cancer. 2015;22(5):819–830. doi: 10.1530/ERC-14-0502. [DOI] [PubMed] [Google Scholar]
- 13.Blanchard A.A., et al. Claudins 1, 3, and 4 protein expression in ER negative breast cancer correlates with markers of the basal phenotype. Virchows Arch. 2009;454(6):647–656. doi: 10.1007/s00428-009-0770-6. [DOI] [PubMed] [Google Scholar]
- 14.Huang J., et al. The expression of claudin 1 correlates with β-catenin and is a prognostic factor of poor outcome in gastric cancer. Int. J. Oncol. 2014;44(4):1293–1301. doi: 10.3892/ijo.2014.2298. [DOI] [PubMed] [Google Scholar]
- 15.Kondo J., et al. Claudin-1 expression is induced by tumor necrosis factor-alpha in human pancreatic cancer cells. Int. J. Mol. Med. 2008;22(5):645–649. [PubMed] [Google Scholar]
- 16.Seo K.W., et al. Correlation between claudins expression and prognostic factors in prostate cancer. Korean J Urol. 2010;51(4):239–244. doi: 10.4111/kju.2010.51.4.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao Y.C., et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 2009;179(2):123–133. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- 18.Krause G., et al. Structure and function of claudins. Biochim. Biophys. Acta. 2008;1778(3):631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Hewitt K.J., Agarwal R., Morin P.J. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaarteenaho-Wiik R., Soini Y. Claudin-1, -2, -3, -4, -5, and -7 in usual interstitial pneumonia and sarcoidosis. J. Histochem. Cytochem. 2009;57(3):187–195. doi: 10.1369/jhc.2008.951566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyuno D., et al. Targeting tight junctions during epithelial to mesenchymal transition in human pancreatic cancer. World J. Gastroenterol. 2014;20(31):10813–10824. doi: 10.3748/wjg.v20.i31.10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene C., Hanley N., Campbell M. Claudin-5: gatekeeper of neurological function. Fluids Barriers CNS. 2019;16(1):3. doi: 10.1186/s12987-019-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiba H., et al. The region-selective regulation of endothelial claudin-5 expression and signaling in brain health and disorders. J. Cell. Physiol. 2021;236(10):7134–7143. doi: 10.1002/jcp.30357. [DOI] [PubMed] [Google Scholar]
- 24.Turksen K., Troy T.C. Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Dev Dyn. 2001;222(2):292–300. doi: 10.1002/dvdy.1174. [DOI] [PubMed] [Google Scholar]
- 25.Ushiku T., et al. Distinct expression pattern of claudin-6, a primitive phenotypic tight junction molecule, in germ cell tumours and visceral carcinomas. Histopathology. 2012;61(6):1043–1056. doi: 10.1111/j.1365-2559.2012.04314.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Itallie C.M., Anderson J.M. Claudins and epithelial paracellular transport. Annu. Rev. Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 27.Gong Y., et al. Claudin-14 regulates renal Ca⁺⁺ transport in response to CaSR signalling via a novel microRNA pathway. Embo j. 2012;31(8):1999–2012. doi: 10.1038/emboj.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T., et al. Parathyroid hormone controls paracellular Ca(2+) transport in the thick ascending limb by regulating the tight-junction protein Claudin14. Proc Natl Acad Sci U S A. 2017;114(16):E3344–e3353. doi: 10.1073/pnas.1616733114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Itallie C.M., Fanning A.S., Anderson J.M. Reversal of charge selectivity in cation or anion-selective epithelial lines by expression of different claudins. Am. J. Physiol. Ren. Physiol. 2003;285(6):F1078–F1084. doi: 10.1152/ajprenal.00116.2003. [DOI] [PubMed] [Google Scholar]
- 30.Krug S.M., Schulzke J.D., Fromm M. Tight junction, selective permeability, and related diseases. Semin. Cell Dev. Biol. 2014;36:166–176. doi: 10.1016/j.semcdb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Van Itallie C.M., et al. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am. J. Physiol. Ren. Physiol. 2006;291(6):F1288–F1299. doi: 10.1152/ajprenal.00138.2006. [DOI] [PubMed] [Google Scholar]
- 32.Alexander R.T. Claudin-15 is not a drag! Acta Physiol. 2020;228(1) doi: 10.1111/apha.13397. [DOI] [PubMed] [Google Scholar]
- 33.Hou J., et al. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J. Biol. Chem. 2007;282(23):17114–17122. doi: 10.1074/jbc.M700632200. [DOI] [PubMed] [Google Scholar]
- 34.Hou J., Goodenough D.A. Claudin-16 and claudin-19 function in the thick ascending limb. Curr. Opin. Nephrol. Hypertens. 2010;19(5):483–488. doi: 10.1097/MNH.0b013e32833b7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato K., et al. Deficiency of lung-specific claudin-18 leads to aggravated infection with Cryptococcus deneoformans through dysregulation of the microenvironment in lungs. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-00708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaFemina M.J., et al. Claudin-18 deficiency results in alveolar barrier dysfunction and impaired alveologenesis in mice. Am. J. Respir. Cell Mol. Biol. 2014;51(4):550–558. doi: 10.1165/rcmb.2013-0456OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G., et al. Knockout mice reveal key roles for claudin 18 in alveolar barrier properties and fluid homeostasis. Am. J. Respir. Cell Mol. Biol. 2014;51(2):210–222. doi: 10.1165/rcmb.2013-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi D., et al. Deficiency of claudin-18 causes paracellular H+ leakage, up-regulation of interleukin-1β, and atrophic gastritis in mice. Gastroenterology. 2012;142(2):292–304. doi: 10.1053/j.gastro.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 39.Tamura A., et al. Claudin-based paracellular proton barrier in the stomach. Ann. N. Y. Acad. Sci. 2012;1258:108–114. doi: 10.1111/j.1749-6632.2012.06570.x. [DOI] [PubMed] [Google Scholar]
- 40.Hay E.D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154(1):8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 41.Bhat A.A., et al. Claudin-1 promotes TNF-α-induced epithelial-mesenchymal transition and migration in colorectal adenocarcinoma cells. Exp. Cell Res. 2016;349(1):119–127. doi: 10.1016/j.yexcr.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh Y., et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2013;32(41):4873–4882. doi: 10.1038/onc.2012.505. [DOI] [PubMed] [Google Scholar]
- 43.Shiozaki A., et al. Claudin 1 mediates TNFα-induced gene expression and cell migration in human lung carcinoma cells. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0038049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L., et al. CLDN3 inhibits cancer aggressiveness via Wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5(17):7663–7676. doi: 10.18632/oncotarget.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Che J., et al. Claudin-3 inhibits lung squamous cell carcinoma cell epithelial-mesenchymal transition and invasion via suppression of the wnt/β-catenin signaling pathway. Int. J. Med. Sci. 2018;15(4):339–351. doi: 10.7150/ijms.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu S., et al. CLDN6 promotes tumor progression through the YAP1-snail1 axis in gastric cancer. Cell Death Dis. 2019;10(12):949. doi: 10.1038/s41419-019-2168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y., et al. SMAD2 inactivation inhibits CLDN6 methylation to suppress migration and invasion of breast cancer cells. Int. J. Mol. Sci. 2017;18(9) doi: 10.3390/ijms18091863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A.B., Sharma A., Dhawan P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis. 2012;33(12):2538–2547. doi: 10.1093/carcin/bgs275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimobaba S., et al. Claudin-18 inhibits cell proliferation and motility mediated by inhibition of phosphorylation of PDK1 and Akt in human lung adenocarcinoma A549 cells. Biochim. Biophys. Acta. 2016;1863(6 Pt A):1170–1178. doi: 10.1016/j.bbamcr.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Nübel T., et al. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol. Cancer Res. 2009;7(3):285–299. doi: 10.1158/1541-7786.MCR-08-0200. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Z., et al. CLDN1 increases drug resistance of non-small cell lung cancer by activating autophagy via up-regulation of ULK1 phosphorylation. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2017;23:2906–2916. doi: 10.12659/MSM.904177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruhashi R., et al. Elevation of sensitivity to anticancer agents of human lung adenocarcinoma A549 cells by knockdown of claudin-2 expression in monolayer and spheroid culture models. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865(3):470–479. doi: 10.1016/j.bbamcr.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Eftang L.L., et al. Up-regulation of CLDN1 in gastric cancer is correlated with reduced survival. BMC Cancer. 2013;13:586. doi: 10.1186/1471-2407-13-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang T.L., et al. Claudin-1 has tumor suppressive activity and is a direct target of RUNX3 in gastric epithelial cells. Gastroenterology. 2010;138(1):255–265. doi: 10.1053/j.gastro.2009.08.044. e1-3. [DOI] [PubMed] [Google Scholar]
- 55.Zavala-Zendejas V.E., et al. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest. 2011;29(1):1–11. doi: 10.3109/07357907.2010.512594. [DOI] [PubMed] [Google Scholar]
- 56.Kwon M.J., et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab. Invest. 2011;91(11):1652–1667. doi: 10.1038/labinvest.2011.117. [DOI] [PubMed] [Google Scholar]
- 57.Lu Y.Z., et al. Claudin-6 is down-regulated in gastric cancer and its potential pathway. Cancer Biomarkers. 2020;28(3):329–340. doi: 10.3233/CBM-201554. [DOI] [PubMed] [Google Scholar]
- 58.Rao X., et al. Down-regulated CLDN10 predicts favorable prognosis and correlates with immune infiltration in gastric cancer. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.747581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X.Z., et al. lncRNA PCAT18 inhibits proliferation, migration and invasion of gastric cancer cells through miR-135b suppression to promote CLDN11 expression. Life Sci. 2020;249 doi: 10.1016/j.lfs.2020.117478. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W.H., et al. The significance of the CLDN18-ARHGAP fusion gene in gastric cancer: a systematic review and meta-analysis. Front. Oncol. 2020;10:1214. doi: 10.3389/fonc.2020.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki K., et al. Deficiency of stomach-type claudin-18 in mice induces gastric tumor formation independent of H pylori infection. Cell Mol Gastroenterol Hepatol. 2019;8(1):119–142. doi: 10.1016/j.jcmgh.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y., et al. Expression of claudin-11, -23 in different gastric tissues and its relationship with the risk and prognosis of gastric cancer. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0174476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao F., et al. Expression of CLDN6 in tissues of gastric cancer patients: association with clinical pathology and prognosis. Oncol. Lett. 2019;17(5):4621–4625. doi: 10.3892/ol.2019.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider I.C., et al. Displaying tetra-membrane spanning claudins on enveloped virus-like particles for cancer immunotherapy. Biotechnol. J. 2018;13(3) doi: 10.1002/biot.201700345. [DOI] [PubMed] [Google Scholar]
- 65.Reinhard K., et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science. 2020;367(6476):446–453. doi: 10.1126/science.aay5967. [DOI] [PubMed] [Google Scholar]
- 66.Yang P., et al. MicroRNA-421 promotes the proliferation and metastasis of gastric cancer cells by targeting claudin-11. Exp. Ther. Med. 2017;14(3):2625–2632. doi: 10.3892/etm.2017.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agarwal R., et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS One. 2009;4(11):e8002. doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X., Zhang Q., Chekouo T. Filtering high-dimensional methylation marks with extremely small sample size: an application to gastric cancer data. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.705708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang B., et al. Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq. Sci. Rep. 2021;11(1):1141. doi: 10.1038/s41598-020-80881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hagen S.J., et al. Loss of tight junction protein claudin 18 promotes progressive neoplasia development in mouse stomach. Gastroenterology. 2018;155(6):1852–1867. doi: 10.1053/j.gastro.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baek J.H., et al. Clinical implications of Claudin18.2 expression in patients with gastric cancer. Anticancer Res. 2019;39(12):6973–6979. doi: 10.21873/anticanres.13919. [DOI] [PubMed] [Google Scholar]
- 72.Lu Y., et al. Correlation between Claudin-18 expression and clinicopathological features and prognosis in patients with gastric cancer. J. Gastrointest. Oncol. 2020;11(6):1253–1260. doi: 10.21037/jgo-20-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coati I., et al. Claudin-18 expression in oesophagogastric adenocarcinomas: a tissue microarray study of 523 molecularly profiled cases. Br. J. Cancer. 2019;121(3):257–263. doi: 10.1038/s41416-019-0508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawada T., et al. New molecular staging with G-factor supplements TNM classification in gastric cancer: a multicenter collaborative research by the Japan Society for Gastroenterological Carcinogenesis G-Project committee. Gastric Cancer. 2015;18(1):119–128. doi: 10.1007/s10120-014-0338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S.R., et al. Clinical significance of CLDN18.2 expression in metastatic diffuse-type gastric cancer. J Gastric Cancer. 2020;20(4):408–420. doi: 10.5230/jgc.2020.20.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu B., et al. Highly expressed Claudin18.2 as a potential therapeutic target in advanced gastric signet-ring cell carcinoma (SRCC) J. Gastrointest. Oncol. 2020;11(6):1431–1439. doi: 10.21037/jgo-20-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pellino A., et al. Association of CLDN18 protein expression with clinicopathological features and prognosis in advanced gastric and gastroesophageal junction adenocarcinomas. J. Personalized Med. 2021;11(11) doi: 10.3390/jpm11111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira M.A., et al. RhoA, claudin 18, and c-MET in gastric cancer: clinicopathological characteristics and prognostic significance in curative resected patients. Med. Sci. 2021;10(1) doi: 10.3390/medsci10010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanada Y., et al. Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J. Pathol. 2006;208(5):633–642. doi: 10.1002/path.1922. [DOI] [PubMed] [Google Scholar]
- 80.Yasui W., et al. Transcriptome dissection of gastric cancer: identification of novel diagnostic and therapeutic targets from pathology specimens. Pathol. Int. 2009;59(3):121–136. doi: 10.1111/j.1440-1827.2009.02329.x. [DOI] [PubMed] [Google Scholar]
- 81.Jun K.H., et al. Expression of claudin-7 and loss of claudin-18 correlate with poor prognosis in gastric cancer. Int. J. Surg. 2014;12(2):156–162. doi: 10.1016/j.ijsu.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 82.Semba S., et al. Prognostic significance of intestinal claudins in high-risk synchronous and metachronous multiple gastric epithelial neoplasias after initial endoscopic submucosal dissection. Pathol. Int. 2008;58(6):371–377. doi: 10.1111/j.1440-1827.2008.02238.x. [DOI] [PubMed] [Google Scholar]
- 83.Oshima T., et al. Down-regulation of claudin-18 is associated with the proliferative and invasive potential of gastric cancer at the invasive front. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qi C., et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results. Nat Med. 2022;28(6):1189–1198. doi: 10.1038/s41591-022-01800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh P., Toom S., Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J. Hematol. Oncol. 2017;10(1):105. doi: 10.1186/s13045-017-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shu Y., et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat. Commun. 2018;9(1):2447. doi: 10.1038/s41467-018-04907-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanaka A., et al. Frequent CLDN18-ARHGAP fusion in highly metastatic diffuse-type gastric cancer with relatively early onset. Oncotarget. 2018;9(50):29336–29350. doi: 10.18632/oncotarget.25464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song X., et al. H. pylori-encoded CagA disrupts tight junctions and induces invasiveness of AGS gastric carcinoma cells via Cdx2-dependent targeting of Claudin-2. Cell. Immunol. 2013;286(1–2):22–30. doi: 10.1016/j.cellimm.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 89.Bhat A.A., et al. Caudal homeobox protein Cdx-2 cooperates with Wnt pathway to regulate claudin-1 expression in colon cancer cells. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0037174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miwa N., et al. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol. Res. 2001;12(11–12):469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 91.Shiou S.R., et al. Smad4 regulates claudin-1 expression in a transforming growth factor-beta-independent manner in colon cancer cells. Cancer Res. 2007;67(4):1571–1579. doi: 10.1158/0008-5472.CAN-06-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh A.B., et al. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. 2011;141(6):2140–2153. doi: 10.1053/j.gastro.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pope J.L., et al. Claudin-1 regulates intestinal epithelial homeostasis through the modulation of Notch-signalling. Gut. 2014;63(4):622–634. doi: 10.1136/gutjnl-2012-304241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang L., et al. Prognostic and clinical significance of claudin-1 in colorectal cancer: a systemic review and meta-analysis. Int. J. Surg. 2017;39:214–220. doi: 10.1016/j.ijsu.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 95.Nakagawa S., et al. Expression of CLDN1 in colorectal cancer: a novel marker for prognosis. Int. J. Oncol. 2011;39(4):791–796. doi: 10.3892/ijo.2011.1102. [DOI] [PubMed] [Google Scholar]
- 96.Dhawan P., et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene. 2011;30(29):3234–3247. doi: 10.1038/onc.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mi L., et al. The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene. 2017;36(30):4323–4335. doi: 10.1038/onc.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wei M., et al. Claudin-2 promotes colorectal cancer growth and metastasis by suppressing NDRG1 transcription. Clin. Transl. Med. 2021;11(12):e667. doi: 10.1002/ctm2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tabariès S., et al. Claudin-2 promotes colorectal cancer liver metastasis and is a biomarker of the replacement type growth pattern. Commun. Biol. 2021;4(1):657. doi: 10.1038/s42003-021-02189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Souza W.F., et al. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ahmad R., et al. Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer malignancy by hyperactivating Wnt/β-catenin signaling. Oncogene. 2017;36(47):6592–6604. doi: 10.1038/onc.2017.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Darido C., et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008;68(11):4258–4268. doi: 10.1158/0008-5472.CAN-07-5805. [DOI] [PubMed] [Google Scholar]
- 103.Wang K., et al. Claudin-7 downregulation induces metastasis and invasion in colorectal cancer via the promotion of epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2019;508(3):797–804. doi: 10.1016/j.bbrc.2018.10.049. [DOI] [PubMed] [Google Scholar]
- 104.Oshima T., et al. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol. Rep. 2008;19(4):953–959. [PubMed] [Google Scholar]
- 105.Cheng B., et al. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J. Exp. Clin. Cancer Res. 2020;39(1):5. doi: 10.1186/s13046-019-1510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng B., et al. CLDN8 promotes colorectal cancer cell proliferation, migration, and invasion by activating MAPK/ERK signaling. Cancer Manag. Res. 2019;11:3741–3751. doi: 10.2147/CMAR.S189558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li J., et al. Methylated claudin-11 associated with metastasis and poor survival of colorectal cancer. Oncotarget. 2017;8(56):96249–96262. doi: 10.18632/oncotarget.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kolchakova D., et al. Tight junction protein claudin-12 is involved in cell migration during metastasis. Biomolecules. 2021;11(5) doi: 10.3390/biom11050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qiao T.Y., et al. Claudin14 promotes colorectal cancer progression via the PI3K/AKT/mTOR pathway. Neoplasma. 2021;68(5):947–954. doi: 10.4149/neo_2021_210210N203. [DOI] [PubMed] [Google Scholar]
- 110.Chen W., et al. [The expression and biological function of claudin-23 in colorectal cancer] Sichuan Da Xue Xue Bao Yi Xue Ban. 2018;49(3):331–336. [PubMed] [Google Scholar]
- 111.Distler M., et al. Precursor lesions for sporadic pancreatic cancer: PanIN, IPMN, and MCN. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/474905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee J.H., et al. Immunohistochemical analysis of claudin expression in pancreatic cystic tumors. Oncol. Rep. 2011;25(4):971–978. doi: 10.3892/or.2011.1132. [DOI] [PubMed] [Google Scholar]
- 113.Michl P., et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63(19):6265–6271. [PubMed] [Google Scholar]
- 114.English D.P., Santin A.D. Claudins overexpression in ovarian cancer: potential targets for Clostridium Perfringens Enterotoxin (CPE) based diagnosis and therapy. Int. J. Mol. Sci. 2013;14(5):10412–10437. doi: 10.3390/ijms140510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y., et al. LINC00857 regulated by ZNF460 enhances the expression of CLDN12 by sponging miR-150-5p and recruiting SRSF1 for alternative splicing to promote epithelial-mesenchymal transformation of pancreatic adenocarcinoma cells. RNA Biol. 2022;19(1):548–559. doi: 10.1080/15476286.2021.1992995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tanaka M., et al. Claudin-18 is an early-stage marker of pancreatic carcinogenesis. J. Histochem. Cytochem. 2011;59(10):942–952. doi: 10.1369/0022155411420569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanada Y., et al. Immunohistochemical study of claudin 18 involvement in intestinal differentiation during the progression of intraductal papillary mucinous neoplasm. Anticancer Res. 2010;30(7):2995–3003. [PubMed] [Google Scholar]
- 118.Karanjawala Z.E., et al. New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am. J. Surg. Pathol. 2008;32(2):188–196. doi: 10.1097/PAS.0b013e31815701f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soini Y., et al. Expression of claudins 7 and 18 in pancreatic ductal adenocarcinoma: association with features of differentiation. J. Clin. Pathol. 2012;65(5):431–436. doi: 10.1136/jclinpath-2011-200400. [DOI] [PubMed] [Google Scholar]
- 120.Kojima T., Sawada N. Regulation of tight junctions in human normal pancreatic duct epithelial cells and cancer cells. Ann. N. Y. Acad. Sci. 2012;1257:85–92. doi: 10.1111/j.1749-6632.2012.06579.x. [DOI] [PubMed] [Google Scholar]
- 121.Ito T., et al. Transcriptional regulation of claudin-18 via specific protein kinase C signaling pathways and modification of DNA methylation in human pancreatic cancer cells. J. Cell. Biochem. 2011;112(7):1761–1772. doi: 10.1002/jcb.23095. [DOI] [PubMed] [Google Scholar]
- 122.Türeci Ӧ., et al. Characterization of zolbetuximab in pancreatic cancer models. OncoImmunology. 2019;8(1) doi: 10.1080/2162402X.2018.1523096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wöll S., et al. Claudin 18.2 is a target for IMAB362 antibody in pancreatic neoplasms. Int. J. Cancer. 2014;134(3):731–739. doi: 10.1002/ijc.28400. [DOI] [PubMed] [Google Scholar]
- 124.Kyuno D., et al. Claudin-18.2 as a therapeutic target in cancers: cumulative findings from basic research and clinical trials. Tissue Barriers. 2022;10(1) doi: 10.1080/21688370.2021.1967080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mahati S., et al. miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem. Biophys. Res. Commun. 2017;486(3):732–737. doi: 10.1016/j.bbrc.2017.03.110. [DOI] [PubMed] [Google Scholar]
- 126.Yoon C.H., et al. Claudin-1 acts through c-Abl-protein kinase Cdelta (PKCdelta) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J. Biol. Chem. 2010;285(1):226–233. doi: 10.1074/jbc.M109.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang L., et al. Downregulation of CLDN6 inhibits cell proliferation, migration, and invasion via regulating EGFR/AKT/mTOR signalling pathway in hepatocellular carcinoma. Cell Biochem. Funct. 2020;38(5):541–548. doi: 10.1002/cbf.3489. [DOI] [PubMed] [Google Scholar]
- 128.Liu H., et al. Claudin-9 enhances the metastatic potential of hepatocytes via Tyk2/Stat3 signaling. Turk. J. Gastroenterol. 2019;30(8):722–731. doi: 10.5152/tjg.2019.18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun H., et al. miR-486 regulates metastasis and chemosensitivity in hepatocellular carcinoma by targeting CLDN10 and CITRON. Hepatol. Res. 2015;45(13):1312–1322. doi: 10.1111/hepr.12500. [DOI] [PubMed] [Google Scholar]
- 130.Huang G.W., et al. Expression of claudin 10 protein in hepatocellular carcinoma: impact on survival. J. Cancer Res. Clin. Oncol. 2011;137(8):1213–1218. doi: 10.1007/s00432-011-0987-z. [DOI] [PubMed] [Google Scholar]
- 131.Cheung S.T., et al. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin. Cancer Res. 2005;11(2 Pt 1):551–556. [PubMed] [Google Scholar]
- 132.Yang J., et al. miR-99b promotes metastasis of hepatocellular carcinoma through inhibition of claudin 11 expression and may serve as a prognostic marker. Oncol. Rep. 2015;34(3):1415–1423. doi: 10.3892/or.2015.4104. [DOI] [PubMed] [Google Scholar]
- 133.Li C.P., et al. CLDN14 is epigenetically silenced by EZH2-mediated H3K27ME3 and is a novel prognostic biomarker in hepatocellular carcinoma. Carcinogenesis. 2016;37(6):557–566. doi: 10.1093/carcin/bgw036. [DOI] [PubMed] [Google Scholar]
- 134.Sun L., Feng L., Cui J. Increased expression of claudin-17 promotes a malignant phenotype in hepatocyte via Tyk2/Stat3 signaling and is associated with poor prognosis in patients with hepatocellular carcinoma. Diagn. Pathol. 2018;13(1):72. doi: 10.1186/s13000-018-0749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun B.S., et al. Claudin-1 correlates with poor prognosis in lung adenocarcinoma. Thorac Cancer. 2016;7(5):556–563. doi: 10.1111/1759-7714.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moldvay J., et al. Claudin-1 protein expression is a good prognostic factor in non-small cell lung cancer, but only in squamous cell carcinoma cases. Pathol. Oncol. Res. 2017;23(1):151–156. doi: 10.1007/s12253-016-0115-0. [DOI] [PubMed] [Google Scholar]
- 137.Ikari A., et al. Claudin-2 knockdown decreases matrix metalloproteinase-9 activity and cell migration via suppression of nuclear Sp1 in A549 cells. Life Sci. 2011;88(13–14):628–633. doi: 10.1016/j.lfs.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 138.Wang Q., et al. Low claudin-6 expression correlates with poor prognosis in patients with non-small cell lung cancer. OncoTargets Ther. 2015;8:1971–1977. doi: 10.2147/OTT.S85478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu Z., et al. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol. Cancer. 2015;14:120. doi: 10.1186/s12943-015-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lu Z., et al. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp. Cell Res. 2011;317(13):1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sharma R.K., et al. A spontaneous metastasis model reveals the significance of claudin-9 overexpression in lung cancer metastasis. Clin. Exp. Metastasis. 2016;33(3):263–275. doi: 10.1007/s10585-015-9776-4. [DOI] [PubMed] [Google Scholar]
- 142.Sun L., Feng L., Cui J. Increased expression of claudin-12 promotes the metastatic phenotype of human bronchial epithelial cells and is associated with poor prognosis in lung squamous cell carcinoma. Exp. Ther. Med. 2019;17(1):165–174. doi: 10.3892/etm.2018.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Z., et al. Expression of CLDN1 and CLDN10 in lung adenocarcinoma in situ and invasive lepidic predominant adenocarcinoma. J. Cardiothorac. Surg. 2013;8:95. doi: 10.1186/1749-8090-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ikari A., et al. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim. Biophys. Acta. 2012;1823(6):1110–1118. doi: 10.1016/j.bbamcr.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 145.Sonoki H., et al. Kaempherol and luteolin decrease claudin-2 expression mediated by inhibition of STAT3 in lung adenocarcinoma A549 cells. Nutrients. 2017;9(6) doi: 10.3390/nu9060597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yamamoto T., et al. Reduced expression of claudin-7 is associated with poor outcome in non-small cell lung cancer. Oncol. Lett. 2010;1(3):501–505. doi: 10.3892/ol_00000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Akizuki R., et al. Claudin-5, -7, and -18 suppress proliferation mediated by inhibition of phosphorylation of Akt in human lung squamous cell carcinoma. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864(2):293–302. doi: 10.1016/j.bbamcr.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 148.Micke P., et al. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int. J. Cancer. 2014;135(9):2206–2214. doi: 10.1002/ijc.28857. [DOI] [PubMed] [Google Scholar]
- 149.Zheng Z., et al. Sulforaphane metabolites inhibit migration and invasion via microtubule-mediated Claudins dysfunction or inhibition of autolysosome formation in human non-small cell lung cancer cells. Cell Death Dis. 2019;10(4):259. doi: 10.1038/s41419-019-1489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maruhashi R., et al. Chrysin enhances anticancer drug-induced toxicity mediated by the reduction of claudin-1 and 11 expression in a spheroid culture model of lung squamous cell carcinoma cells. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-50276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Osanai M., et al. Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci. 2007;98(10):1557–1562. doi: 10.1111/j.1349-7006.2007.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Y., et al. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J. Exp. Clin. Cancer Res. 2016;35(1):120. doi: 10.1186/s13046-016-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ren Y., et al. Gene silencing of claudin-6 enhances cell proliferation and migration accompanied with increased MMP-2 activity via p38 MAPK signaling pathway in human breast epithelium cell line HBL-100. Mol. Med. Rep. 2013;8(5):1505–1510. doi: 10.3892/mmr.2013.1675. [DOI] [PubMed] [Google Scholar]
- 154.Guo Y., et al. Apoptosis signal-regulating kinase 1 is associated with the effect of claudin-6 in breast cancer. Diagn. Pathol. 2012;7:111. doi: 10.1186/1746-1596-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guo Y., et al. CLDN6-induced apoptosis via regulating ASK1-p38/JNK signaling in breast cancer MCF-7 cells. Int. J. Oncol. 2016;48(6):2435–2444. doi: 10.3892/ijo.2016.3469. [DOI] [PubMed] [Google Scholar]
- 156.Song P., et al. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J. Exp. Clin. Cancer Res. 2019;38(1):354. doi: 10.1186/s13046-019-1359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang M., et al. CLDN6 promotes chemoresistance through GSTP1 in human breast cancer. J. Exp. Clin. Cancer Res. 2017;36(1):157. doi: 10.1186/s13046-017-0627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang M., et al. CLDN6 enhances chemoresistance to ADM via AF-6/ERKs pathway in TNBC cell line MDAMB231. Mol. Cell. Biochem. 2018;443(1–2):169–180. doi: 10.1007/s11010-017-3221-8. [DOI] [PubMed] [Google Scholar]
- 159.Martínez-Estrada O.M., et al. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem. J. 2006;394(Pt 2):449–457. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim T.H., et al. Down-regulation of claudin-2 in breast carcinomas is associated with advanced disease. Histopathology. 2008;53(1):48–55. doi: 10.1111/j.1365-2559.2008.03052.x. [DOI] [PubMed] [Google Scholar]
- 161.Tabariès S., et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene. 2011;30(11):1318–1328. doi: 10.1038/onc.2010.518. [DOI] [PubMed] [Google Scholar]
- 162.Kolokytha P., et al. Claudin-3 and claudin-4: distinct prognostic significance in triple-negative and luminal breast cancer. Appl. Immunohistochem. Mol. Morphol. 2014;22(2):125–131. doi: 10.1097/PAI.0b013e31828d9d62. [DOI] [PubMed] [Google Scholar]
- 163.Escudero-Esparza A., Jiang W.G., Martin T.A. Claudin-5 is involved in breast cancer cell motility through the N-WASP and ROCK signalling pathways. J. Exp. Clin. Cancer Res. 2012;31(1):43. doi: 10.1186/1756-9966-31-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang Y., et al. The expression and prognostic significance of claudin-8 and androgen receptor in breast cancer. OncoTargets Ther. 2020;13:3437–3448. doi: 10.2147/OTT.S242406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liao J., et al. CLDN10 single nucleotide polymorphism rs1325774 alters the risk of breast cancer in south Chinese women. Medicine (Baltim.) 2018;97(49) doi: 10.1097/MD.0000000000013187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shen Y., et al. MiR-205 suppressed the malignant behaviors of breast cancer cells by targeting CLDN11 via modulation of the epithelial-to-mesenchymal transition. Aging (Albany NY) 2021;13(9):13073–13086. doi: 10.18632/aging.202988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meng L., et al. Biomarker discovery to improve prediction of breast cancer survival: using gene expression profiling, meta-analysis, and tissue validation. OncoTargets Ther. 2016;9:6177–6185. doi: 10.2147/OTT.S113855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang Y., et al. Interleukin-18 enhances breast cancer cell migration via down-regulation of claudin-12 and induction of the p38 MAPK pathway. Biochem. Biophys. Res. Commun. 2015;459(3):379–386. doi: 10.1016/j.bbrc.2015.02.108. [DOI] [PubMed] [Google Scholar]
- 169.Iravani O., et al. Prognostic significance of Claudin 12 in estrogen receptor-negative breast cancer. J. Clin. Pathol. 2016;69(10):878–883. doi: 10.1136/jclinpath-2015-203265. [DOI] [PubMed] [Google Scholar]
- 170.Zhang D., et al. Twist1 accelerates tumour vasculogenic mimicry by inhibiting Claudin15 expression in triple-negative breast cancer. J. Cell Mol. Med. 2020;24(13):7163–7174. doi: 10.1111/jcmm.15167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 171.Martin T.A., et al. Claudin-16 reduces the aggressive behavior of human breast cancer cells. J. Cell. Biochem. 2008;105(1):41–52. doi: 10.1002/jcb.21797. [DOI] [PubMed] [Google Scholar]
- 172.Martin T.A., et al. Claudin-20 promotes an aggressive phenotype in human breast cancer cells. Tissue Barriers. 2013;1(3) doi: 10.4161/tisb.26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shimada H., et al. Downregulation of lipolysis-stimulated lipoprotein receptor promotes cell invasion via claudin-1-mediated matrix metalloproteinases in human endometrial cancer. Oncol. Lett. 2017;14(6):6776–6782. doi: 10.3892/ol.2017.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pan X.Y., et al. Expression of claudin-3 and claudin-4 in normal, hyperplastic, and malignant endometrial tissue. Int. J. Gynecol. Cancer. 2007;17(1):233–241. doi: 10.1111/j.1525-1438.2006.00748.x. [DOI] [PubMed] [Google Scholar]
- 175.Shimada H., et al. The roles of tricellular tight junction protein angulin-1/lipolysis-stimulated lipoprotein receptor (LSR) in endometriosis and endometrioid-endometrial carcinoma. Cancers. 2021;13(24) doi: 10.3390/cancers13246341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kojima M., et al. Prognostic significance of aberrant claudin-6 expression in endometrial cancer. Cancers. 2020;12(10) doi: 10.3390/cancers12102748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li X., et al. Downregulation of claudin-7 potentiates cellular proliferation and invasion in endometrial cancer. Oncol. Lett. 2013;6(1):101–105. doi: 10.3892/ol.2013.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang X., et al. Tight junction protein claudin-6 inhibits growth and induces the apoptosis of cervical carcinoma cells in vitro and in vivo. Med. Oncol. 2015;32(5):148. doi: 10.1007/s12032-015-0600-4. [DOI] [PubMed] [Google Scholar]
- 179.Zhang X., et al. Expression of apoptosis signal-regulating kinase 1 is associated with tight junction protein claudin-6 in cervical carcinoma. Int. J. Clin. Exp. Pathol. 2015;8(5):5535–5541. [PMC free article] [PubMed] [Google Scholar]
- 180.Rahman A., et al. Reduced claudin-12 expression predicts poor prognosis in cervical cancer. Int. J. Mol. Sci. 2021;22(7) doi: 10.3390/ijms22073774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Qin W., et al. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587(9):1434–1439. doi: 10.1016/j.febslet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 182.Rangel L.B., et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin. Cancer Res. 2003;9(7):2567–2575. [PubMed] [Google Scholar]
- 183.Agarwal R., D'Souza T., Morin P.J. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65(16):7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 184.Ogawa M., et al. Epidermal growth factor modulates claudins and tight junctional functions in ovarian cancer cell lines. Histochem. Cell Biol. 2012;138(2):323–338. doi: 10.1007/s00418-012-0956-x. [DOI] [PubMed] [Google Scholar]
- 185.Lin X., et al. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shang X., et al. Claudin-3 and claudin-4 regulate sensitivity to cisplatin by controlling expression of the copper and cisplatin influx transporter CTR1. Mol. Pharmacol. 2013;83(1):85–94. doi: 10.1124/mol.112.079798. [DOI] [PubMed] [Google Scholar]
- 187.Salomon D.S., et al. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. Hematol. 1995;19(3):183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 188.Santin A.D., et al. Treatment of chemotherapy-resistant human ovarian cancer xenografts in C.B-17/SCID mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer Res. 2005;65(10):4334–4342. doi: 10.1158/0008-5472.CAN-04-3472. [DOI] [PubMed] [Google Scholar]
- 189.Yoshida H., et al. Claudin-4: a potential therapeutic target in chemotherapy-resistant ovarian cancer. Anticancer Res. 2011;31(4):1271–1277. [PubMed] [Google Scholar]
- 190.Litkouhi B., et al. Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia. 2007;9(4):304–314. doi: 10.1593/neo.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gao P., et al. Association of CLDN6 and CLDN10 with immune microenvironment in ovarian cancer: a study of the claudin family. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.595436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lal-Nag M., et al. Claudin-6: a novel receptor for CPE-mediated cytotoxicity in ovarian cancer. Oncogenesis. 2012;1(11):e33. doi: 10.1038/oncsis.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim C.J., et al. High claudin-7 expression is associated with a poor response to platinum-based chemotherapy in epithelial ovarian carcinoma. Eur. J. Cancer. 2011;47(6):918–925. doi: 10.1016/j.ejca.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 194.Li Z., et al. Claudin 10 acts as a novel biomarker for the prognosis of patients with ovarian cancer. Oncol. Lett. 2020;20(1):373–381. doi: 10.3892/ol.2020.11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Szász A.M., et al. beta-catenin expression and claudin expression pattern as prognostic factors of prostatic cancer progression. BJU Int. 2010;105(5):716–722. doi: 10.1111/j.1464-410X.2009.08808.x. [DOI] [PubMed] [Google Scholar]
- 196.Ashikari D., et al. CLDN8, an androgen-regulated gene, promotes prostate cancer cell proliferation and migration. Cancer Sci. 2017;108(7):1386–1393. doi: 10.1111/cas.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fritzsche F.R., et al. Claudin-1 protein expression is a prognostic marker of patient survival in renal cell carcinomas. Clin. Cancer Res. 2008;14(21):7035–7042. doi: 10.1158/1078-0432.CCR-08-0855. [DOI] [PubMed] [Google Scholar]
- 198.Zhu Z., et al. Prognostic value and potential biological functions of CLDN8 in patients with clear cell renal cell carcinoma. OncoTargets Ther. 2020;13:9135–9145. doi: 10.2147/OTT.S266846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang W., et al. CLDN10 associated with immune infiltration is a novel prognostic biomarker for clear cell renal cell carcinoma. Epigenomics. 2021;13(1):31–45. doi: 10.2217/epi-2020-0256. [DOI] [PubMed] [Google Scholar]
- 200.Yang W., et al. Claudin-10 overexpression suppresses human clear cell renal cell carcinoma growth and metastasis by regulating ATP5O and causing mitochondrial dysfunction. Int. J. Biol. Sci. 2022;18(6):2329–2344. doi: 10.7150/ijbs.70105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Oku N., et al. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res. 2006;66(10):5251–5257. doi: 10.1158/0008-5472.CAN-05-4478. [DOI] [PubMed] [Google Scholar]
- 202.Sappayatosok K., Phattarataratip E. Overexpression of claudin-1 is associated with advanced clinical stage and invasive pathologic characteristics of oral squamous cell carcinoma. Head Neck Pathol. 2015;9(2):173–180. doi: 10.1007/s12105-014-0559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Melchers L.J., et al. Lack of claudin-7 is a strong predictor of regional recurrence in oral and oropharyngeal squamous cell carcinoma. Oral Oncol. 2013;49(10):998–1005. doi: 10.1016/j.oraloncology.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 204.Zhao X., et al. Expression profiles analysis identifies a novel three-mRNA signature to predict overall survival in oral squamous cell carcinoma. Am. J. Cancer Res. 2018;8(3):450–461. [PMC free article] [PubMed] [Google Scholar]
- 205.Xu Y.N., et al. Tight junction protein CLDN17 serves as a tumor suppressor to reduce the invasion and migration of oral cancer cells by inhibiting epithelial-mesenchymal transition. Arch. Oral Biol. 2022;133 doi: 10.1016/j.archoralbio.2021.105301. [DOI] [PubMed] [Google Scholar]
- 206.Li W.J., et al. Expression of claudin-1 and its relationship with lymphatic microvessel generation in hypopharyngeal squamous cell carcinoma. Genet. Mol. Res. 2015;14(4):11814–11826. doi: 10.4238/2015.October.2.15. [DOI] [PubMed] [Google Scholar]
- 207.Li W., et al. Prognostic significance of claudin-1 and cyclin B1 protein expression in patients with hypopharyngeal squamous cell carcinoma. Oncol. Lett. 2016;11(5):2995–3002. doi: 10.3892/ol.2016.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhou S., et al. Identification of claudin-1, -3, -7 and -8 as prognostic markers in human laryngeal carcinoma. Mol. Med. Rep. 2019;20(1):393–400. doi: 10.3892/mmr.2019.10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee K.W., et al. Twist1 causes the transcriptional repression of claudin-4 with prognostic significance in esophageal cancer. Biochem. Biophys. Res. Commun. 2012;423(3):454–460. doi: 10.1016/j.bbrc.2012.05.140. [DOI] [PubMed] [Google Scholar]
- 210.Sung C.O., Han S.Y., Kim S.H. Low expression of claudin-4 is associated with poor prognosis in esophageal squamous cell carcinoma. Ann. Surg Oncol. 2011;18(1):273–281. doi: 10.1245/s10434-010-1289-4. [DOI] [PubMed] [Google Scholar]
- 211.Shi M., et al. Low expression of claudin-4: an indicator of recurrence in esophageal squamous cell carcinoma after Ivor Lewis esophagectomy? Med. Oncol. 2014;31(5):951. doi: 10.1007/s12032-014-0951-2. [DOI] [PubMed] [Google Scholar]
- 212.Tsunoda S., et al. Methylation of CLDN6, FBN2, RBP1, RBP4, TFPI2, and TMEFF2 in esophageal squamous cell carcinoma. Oncol. Rep. 2009;21(4):1067–1073. doi: 10.3892/or_00000325. [DOI] [PubMed] [Google Scholar]
- 213.Lioni M., et al. Dysregulation of claudin-7 leads to loss of E-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am. J. Pathol. 2007;170(2):709–721. doi: 10.2353/ajpath.2007.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Leotlela P.D., et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26(26):3846–3856. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- 215.Perez E.C., et al. The axis IL-10/claudin-10 is implicated in the modulation of aggressiveness of melanoma cells by B-1 lymphocytes. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gao L., et al. Promoter CpG island hypermethylation in dysplastic nevus and melanoma: CLDN11 as an epigenetic biomarker for malignancy. J. Invest. Dermatol. 2014;134(12):2957–2966. doi: 10.1038/jid.2014.270. [DOI] [PubMed] [Google Scholar]
- 217.Németh J., et al. High expression of claudin-1 protein in papillary thyroid tumor and its regional lymph node metastasis. Pathol. Oncol. Res. 2010;16(1):19–27. doi: 10.1007/s12253-009-9182-9. [DOI] [PubMed] [Google Scholar]
- 218.Aldred M.A., et al. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J. Clin. Oncol. 2004;22(17):3531–3539. doi: 10.1200/JCO.2004.08.127. [DOI] [PubMed] [Google Scholar]
- 219.Zhou Y., et al. CLDN10 is associated with papillary thyroid cancer progression. J. Cancer. 2018;9(24):4712–4717. doi: 10.7150/jca.28636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li H.P., et al. Inactivation of the tight junction gene CLDN11 by aberrant hypermethylation modulates tubulins polymerization and promotes cell migration in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2018;37(1):102. doi: 10.1186/s13046-018-0754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Liu B., et al. MiR-361-5p inhibits cell proliferation and induces cell apoptosis in retinoblastoma by negatively regulating CLDN8. Childs Nerv Syst. 2019;35(8):1303–1311. doi: 10.1007/s00381-019-04199-9. [DOI] [PubMed] [Google Scholar]
- 222.Hong L., et al. Overexpression of the cell adhesion molecule claudin-9 is associated with invasion in pituitary oncocytomas. Hum. Pathol. 2014;45(12):2423–2429. doi: 10.1016/j.humpath.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 223.Katsushima K., et al. Contribution of microRNA-1275 to Claudin11 protein suppression via a polycomb-mediated silencing mechanism in human glioma stem-like cells. J. Biol. Chem. 2012;287(33):27396–27406. doi: 10.1074/jbc.M112.359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang X., et al. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int. 2018;18:160. doi: 10.1186/s12935-018-0662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xu J., et al. Claudin 8 contributes to malignant proliferation in human osteosarcoma U2OS cells. Cancer Biother. Radiopharm. 2015;30(9):400–404. doi: 10.1089/cbr.2015.1815. [DOI] [PubMed] [Google Scholar]
- 226.Tian X., et al. The cytoplasmic expression of CLDN12 predicts an unfavorable prognosis and promotes proliferation and migration of osteosarcoma. Cancer Manag. Res. 2019;11:9339–9351. doi: 10.2147/CMAR.S229441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Watanabe M., et al. CLDN15 is a novel diagnostic marker for malignant pleural mesothelioma. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-91464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.