Abstract
Ubiquitin-specific proteases (USPs) are crucial for controlling cellular proteostasis and signaling pathways but how deubiquitination is selective remains poorly understood, in particular between paralogues. Here, we developed a fusion tag method by mining the Protein Data Bank and trapped USP11, a key regulator of DNA double-strand break repair, in complex with a novel engineered substrate mimetic. Together, this enabled structure determination of USP11 as a Michaelis-like complex that revealed key S1 and S1′ binding site interactions with a substrate. Combined mutational, enzymatic, and binding experiments identified Met77 in linear diubiquitin as a significant residue that leads to substrate discrimination. We identified an aspartate “gatekeeper” residue in the S1′ site of USP11 as a contributing feature for discriminating against linear diubiquitin. When mutated to a glycine, the corresponding residue in paralog USP15, USP11 acquired elevated activity toward linear diubiquitin in-gel shift assays, but not controls. The reverse mutation in USP15 confirmed that this position confers paralog-specific differences impacting diubiquitin cleavage rates. The results advance our understanding of the molecular basis for the higher selectivity of USP11 compared to USP15 and may aid targeted inhibitor development. Moreover, the reported carrier-based crystallization strategy may be applicable to other challenging targets.
Keywords: ubiquitin-specific protease, crystal structure, cysteine protease, deubiquitylation (deubiquitination), protease, selectivity, ubiquitin, deubiquitinase, fusion tag
Ubiquitin-specific proteases (USPs) are an integral part of the enzymatic network that regulates key cellular events by altering the ubiquitination status of a wide range of proteins (1). Substrates for deubiquitination have a “distal ubiquitin” moiety in common that is typically conjugated to a target’s lysine residue and binds to the protease extensive S1 binding pocket. This leaves the challenge of ubiquitin conjugated target discrimination to other regions of these proteases. A key region for target interaction is the S1′ site, which however appears shallow in the canonical USP fold (Fig. S1). How specific substrates are selected by each of the 56 USP enzymes encoded in the human genome remains an important question in the field and is a major factor for inhibitor development.
USP11 regulates multiple important cellular functions, including DNA double-strand break repair, cell cycle progression (2, 3, 4, 5), and signaling pathways such as transforming growth factor beta signaling (6, 7). Moreover, USP11 is involved in viral ribonucleic acid replication (8) and is dysregulated in different types of cancer including breast, ovarian, pancreatic cancer, melanoma, and myeloid leukemia. USP11 inhibition is a promising treatment strategy in synthetic lethality approaches (9, 10, 11, 12, 13, 14), yet at present structural information on USP11 as a platform for rational inhibitor design is lacking.
USP11 has two distantly related paralogs, USP4 and USP15, which display higher sequence identity with each other (56.9% over the entire sequence) than USP11 (41.4% shared sequence identity with both, USP4 and USP15). The paralogs share the same modular structure consisting of an N-terminal domain present in USPs and a ubiquitin-like domain (15, 16, 17), followed by the USP protease domain composed of subdomains D1 and D2 interspersed by an insertion (Fig. 1A). In contrast to USP11, catalytic domain structures of USP4-D1D2 (18) and USP15-D1D2 (19, 20) in the free form or bound to an inhibitor are available but the molecular basis of substrate interactions is currently unknown. While these paralogs function in related or common pathways, USP11, USP4, and USP15 show significant differences in regulation and substrate specificity (15, 18, 21). For example, ancillary domains do not significantly modulate the catalytic activity of USP11 or USP15 using ubiquitin-7-amino-4-methylcoumarin or diubiquitin substrates, but in USP4 they facilitate ubiquitin discharge (15, 18). USPs are generally promiscuous in cleaving ubiquitin chains (1, 22). However, for USP11, a preference for Lys63-, Lys6-, Lys33-, and Lys11-linked over Lys27-, Lys29-, Lys48-linked poly-ubiquitin chains has been observed in vitro consistent with its role in DNA damage repair. Linear ubiquitin chains are particularly poor substrates for USP11 (15, 22, 23). In contrast, USP15 readily accepts different types of ubiquitin chains (24) and a broader range of substrates (25). How this paralog-specific selectivity is achieved is currently unknown.
Figure 1.
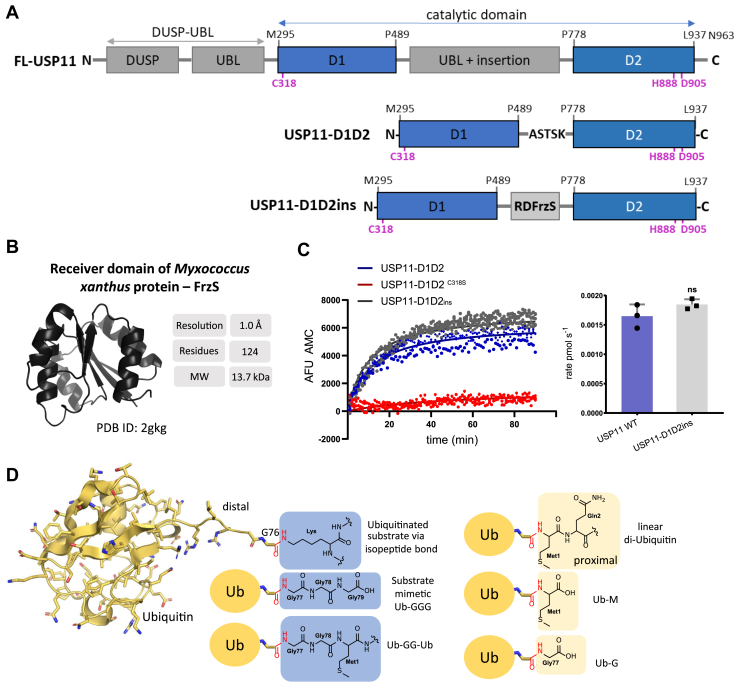
Design of USP11 catalytic domain and ubiquitin substrate constructs. A, top: schematic representation of the hUSP11 domain structure (UniProt P51784). The catalytic subdomains D1 and D2 are depicted in blue and the catalytic triad residues marked in magenta. Additional domains are shown in gray. Below: schematic representation of the USP11-D1D2 (residues Met295-Leu937) construct with the insertion replaced by an ASTSK linker or the RDFrzS loop insertion tag shown in light gray to engineer USP11-D1D2ins. B, receiver domain structure from the Myxococcus xanthus protein FrzS (26) used as an insertion tag. C, progress curves of USP11-D1D2 ubiquitin-7-amino-4-methylcoumarin cleavage in comparison with USP11-D1D2 C318S and USP11-D1D2ins constructs with a bar chart (unpaired two-tailed t test; n = 3 independent experiments; error = SD) highlighting that the insertion does not significantly affect the catalytic activity. D, ubiquitin C-terminal tail modified substrates as used in this study with respective tail modifications shown as chemical structures. USP, ubiquitin-specific protease.
Here, we present the first structure of the USP11 catalytic domain and discern differences with paralogs USP4 and USP15. The structure of USP11 trapped with an engineered USP substrate revealed critical binding interactions and led to the identification of an unexpected structural feature that contributes to USP11’s substrate selectivity. We also introduce the RDFrzS domain as a novel fusion tag that can be incorporated into loop regions to improve crystallization outcomes.
Results
A novel loop insertion tag strategy utilized for the crystallization of USP11
Previous strategies for replacing an insert region containing significant disorder to crystallize USP11 generated the USP11-D1D2 construct (Fig. 1A) (15, 19), but did not yield any diffraction quality crystals. Therefore, a novel insert–substitution strategy was developed. Using search parameters such as size, availability of a high-resolution structure (<1.6 Å), and expression yield, we mined the Protein Data Bank (PDB) and identified the N-terminal 13.7 kDa receiver domain (RD) of bacterial Myxococcus xanthus social motility protein FrzS (26) as a potential candidate tag for carrier-driven crystallization. The RDFrzS structure (PDB ID: 2gkg) was determined to an atomic resolution of 1 Å and adopts a globular fold consisting of a central β-sheet surrounded by five α-helices (Fig. 1B). Moreover, the RDFrzS protein can be highly concentrated and has been crystallized in multiple different space groups, indicating high solubility and crystallizability (26). We reasoned that due to the proximity of its N- and C-terminal Cα atoms (∼6.3 Å), RDFrzS can be inserted to replace predicted flexible loop regions or insertions in target proteins, such as USP11. Two surface entropy reducing (SER) modifications to the RDFrzS sequence were also incorporated. The final construct, schematically depicted in Figure 1A consisted of the USP11 catalytic core (residues Met295-Pro489 and Pro778-Leu937), with the two subdomains linked by the novel RDFrzS fusion tag, which substitutes the 289-aa long insertion between Pro489 and Pro778 (USP11-D1D2ins). Measurements of the catalytic activity of USP11-D1D2ins compared to the original catalytic core protein USP11-D1D2 (15) did not show any significant differences, confirming that the catalytic function of USP11-D1D2ins is equivalent to USP11-D1D2 (Fig. 1C).
USP11 trapping with an engineered substrate mimetic
To gain insights into substrate recognition, we engineered a novel substrate mimetic comprising ubiquitin extended by three C-terminal glycine residues. The design principle underlying the ubiquitin-triple-gly (Ub-GGG) substrate mimetic in comparison with other substrates is illustrated in Figure 1D. Ub-GGG represents a close substrate mimetic of ubiquitin conjugated to a substrate’s lysine via an isopeptide bond as this extension can adopt the extended shape of a lysine side chain and like an isopeptide bond harbors a CH2 group right next to the scissile bond. To trap a Michaelis complex intermediate for crystallization studies, we mutated the USP11 active site cysteine and confirmed that the mutant was inactive (Fig. 1C). USP11-D1D2ins C318S was coexpressed with Ub-GGG and complex formation was confirmed by gel filtration (data not shown). The complex was crystallized using sparse matrix crystallization screening, and the largest crystals were obtained in 100 mM Tris-Bicine pH 8.5, 30 mM sodium nitrate, 30 mM sodium phosphate dibasic, 30 mM ammonium sulphate, 11.25% (v/v) MPD; 11.25% (v/v) PEG 1000; 11.25% (w/v) PEG 3350 with 5 mM cadmium chloride after extensive optimizations. Crystals belonged to space group P 21 21 2 and diffracted to 2.44 Å resolution.
Structure of the USP11–Ub-GGG substrate complex
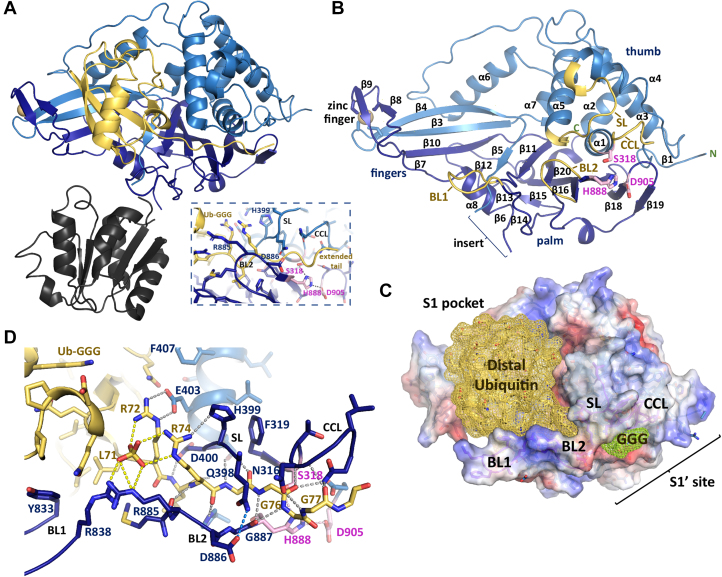
The structure of the USP11 catalytic core domain in complex with Ub-GGG was solved by molecular replacement. Data collection and refinement statistics are shown in Table 1. Both copies of USP11 in the asymmetric unit (ASU) superimpose well with differences seen in the position of the inserted crystallization tag RDFrzS (Fig. S2A). The RDFrzS tag is involved in several crystal contacts (Fig. S2B). USP11-D1D2 adopts the canonical USP protease fold with thumb, palm, and finger regions (Fig. 2A). The zinc finger (Cys468, Cys471, Cys802, Cys805) coordinates a cadmium ion, due to the crystallization additive CdCl2. The catalytic triad residues C318S, His888, and Asp905 are within hydrogen-bonding distance and adopt an active conformation (Fig. 2B). Electron density maps show both copies of the USP11 catalytic core interact with Ub-GGG in the S1 binding site with clear electron density for the entire extended glycine tail observed (Fig. S3). The buried surface area formed comprises ∼ 2286 Å2, including ∼46 H-bonding interactions and ten salt bridges according to PISA (27). The active site loop regions, namely catalytic cleft loop (CCL, T312-NLGNTS-F319), switching loop (SL, S391-QFLGYQQHDS-Q402), blocking loop 1 (BL1, S832-YTKFS-R838), and blocking loop 2 (BL2, G882-GMR-D886) engage in Ub-GGG substrate interactions and are well ordered (Fig. 2B).
Table.
Data collection and refinement statistics
| USP11-D1D2insC318S in complex with Ub-GGG | |
|---|---|
| Data collection | |
| Space group | P 21 21 2 |
| Cell dimensions | |
| a, b, c | 94.32 Å, 186.10 Å, 75.76 Å |
| Resolution (outer shell) | 48.0–2.44 Å (2.52–2.44) |
| Rmerge (outer shell)a | 0.40 (2.66) |
| Rpim (outer shell)b | 0.114 (0.782) |
| I/σI (outer shell) | 7.6 (1.45) |
| CC1/2 (outer shell)c | 0.988 (0.437) |
| Completeness (outer shell) | 99.8 (98.3) % |
| Redundancy (outer shell) | 13.4 (13.3) |
| Total Reflections (outer shell) | 675,489 (60,034) |
| Wilson B-factor | 33.7 Å2 |
| Refinement | |
| Resolution range | 48.0–2.44 Å |
| No. of unique reflections | 50,405 (4955) |
| Rwork/Rfreed | 0.173/0.230 |
| No. atoms | |
| Protein | 8850 |
| Other | 42 |
| Water | 505 |
| B-factors (Å2) | |
| Protein | 44.3 |
| Ligand | 49.4 |
| Water | 39.9 |
| Ramachandran | |
| Favoured regions (%) | 96.85 |
| Allowed regions (%) | 2.78 |
| Outliers (%) | 0.37 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0044 |
| Bond angles (°) | 0.81 |
Rmerge = Ʃh Ʃi | Ii(h) - Ī(h) |/Ʃh ƩiIi(h).
Rpim= Ʃh (1/N-1)1/2 Ʃi | Ii(h) - Ī(h) |/Ʃh ƩiIi(h),h is the given reflection, Ī(h) is the average intensity of each reflection, and i is the ith measurement of reflection h.
CC1/2 is the Pearson correlation coefficient between random half-datasets.
Rwork = Ʃh | Fobs(h) – Fcalc(h) |/ƩhFobs(h); Rfree corresponds to the Rwork based on 5% of the data excluded from refinement.
Figure 2.
USP11 crystal structure in complex with substrate Ub-GGG. A, crystal structure of USP11-D1D2ins with the USP11 catalytic core depicted in blue, Ub-GGG in yellow, and the RDFrzS insertion in dark gray. The yellow sphere represents the cadmium ion present in the structure. Inset shows a zoomed in view of the extended substrate tail region. B, cartoon representation of USP11-D1D2 catalytic core without Ub-GGG and the insertion shown, with the active site loops and secondary structure elements labeled; light blue D1, dark blue D2, catalytic triad residues pink in stick representation, and the BL1 (blocking loop 1), BL2 (blocking loop 2), SL (switching loop), and catalytic cleft loop (CCL) are labeled in dark yellow. C, USP11-D1D2 surface representation colored according to the electrostatic potential with Ub-GGG in yellow mesh representation. The extended glycine tail residues are colored in lime and the active site loop and S1, S1′ regions are indicated. D, close-up view of the Ub-GGG tail–binding channel showing key interactions. Likely H-bonding interactions are shown as black dashed lines and key residues are labeled. USP, ubiquitin-specific protease; Ub-GGG, ubiquitin-triple-gly.
The substrate-binding channel is in a closed conformation. Several noticeable features characterize the engagement of USP11 with the substrate’s C-terminal tail (Fig. 2, C and D). The Ub-GGG NH group of Gly77 that is equivalent to a lysine side chain’s NH group in an isopeptide bond forms a hydrogen-bonding interaction with USP11 Gly887 from the BL2 region. The Gly77 CH2 group (equivalent to a lysine’s side chain Cε) engages in van der Waals contacts with the catalytic histidine. The C-terminal Gly78 and Gly79 of the triple-gly extension, display higher B-factors and extend outward from the catalytic core in slightly different orientations in the two copies of the ASU (Fig. S2A). USP11 Asn313 in the CCL region interacts with the carbonyl group of the Ub-GGG Gly76 and USP11 Asn316 forms a hydrogen-bonding interaction with Ub-GGG Gly78. The USP11 SL loop Gln398 side chain closes over the substrate’s C-terminal tail by forming hydrogen-bonding interactions with the Asp886 side chain and Gly887 from the BL2 region (Fig. 2D). USP11 Phe831 is part of a hydrophobic region that accommodates Ub-GGG Val70, Leu71, and Leu73. Ub-GGG Arg72 forms an electrostatic interaction with USP11 Glu403 in helix α5. Density consistent with a phosphate ion (from the crystallization mother liquor) forms part of the interface between USP11 and the substrate’s C-terminal tail and the presence of 10 mM phosphate slightly increased the reaction rate (Fig. S4). In the USP11 BL1 region, only Tyr833 and Arg838 interact with the substrate through hydrophobic interactions with Ile36 and hydrogen-bonding interactions with Gln40, respectively. Together, the structure reveals key molecular features of the Michaelis complex intermediate by capturing the interactions of USP11 with a trapped, engineered USP substrate.
Structural differences between USP11 and the paralogs USP4 and USP15
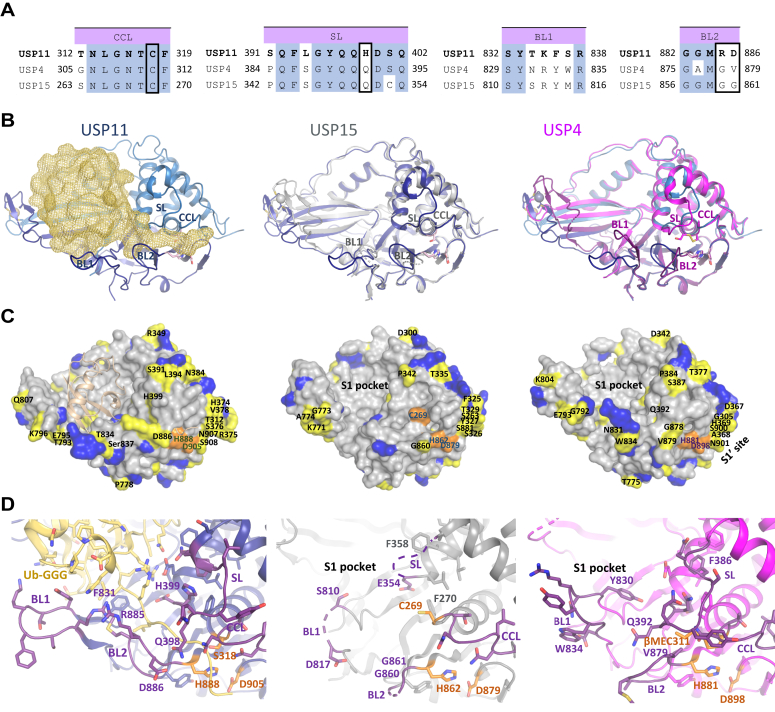
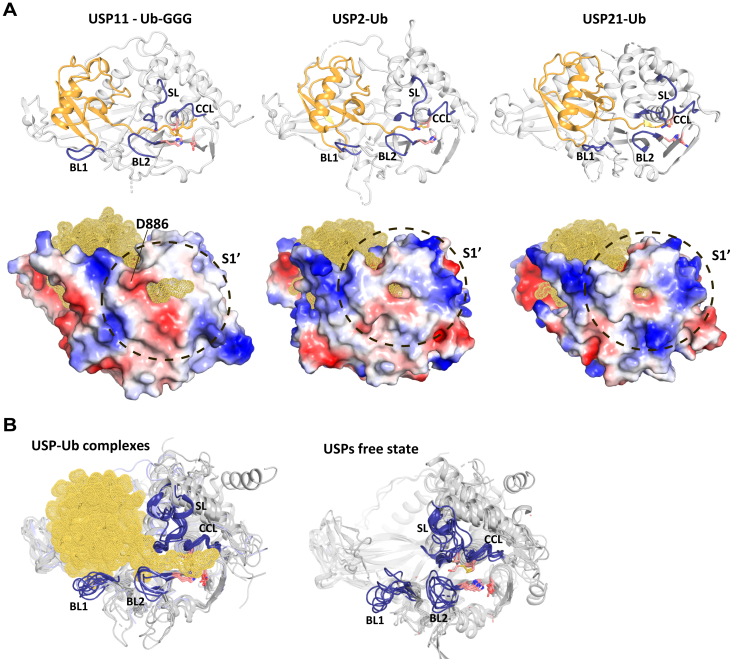
The USP11 catalytic core shares ∼66% sequence identity with the paralogs USP4 and USP15, respectively. Structures of USP15 in the free state or bound to either a small molecule or truncated and modified ubiquitin variant inhibitors (19, 20) and USP4 as a β-mercaptoethanol–bound adduct (18) have been solved. No USP4 or USP15 structure in complex with a reaction product or substrate has been reported to date. Hence, the following comparisons reflect differences between paralogs as well as the substrate-bound versus free state. The structure of USP11 in complex with the substrate Ub-GGG can be superimposed with USP4 (PDB ID: 2y6e) with a RMSD of 0.7 Å (over 1791 atoms out of 2735) and with USP15 in the free state (PDB ID: 6gha) with an RMSD of 0.7 Å (over 1719 atoms out of 2603), respectively. The active site CCL and SL loop regions share high-sequence conservation between the three paralogs, whereas the BL1 and BL2 loop regions are less well conserved (Figs. 3A and S5).
Figure 3.
Comparison of paralogues USP11, USP4, and USP15.A, sequence alignments of hUSP11 (UniProt P51784), hUSP4 (UniProt Q13107), and hUSP15 (Q9Y4E8-2) active site loop regions: catalytic cleft loop (CCL), switching loop (SL), blocking loop 1 (BL1), and blocking loop2 (BL2). Blue shading denotes identical residues between the paralogs and boxed residues highlight the basis for generated mutants in Figure 4. B, comparison of the USP11 catalytic core (blue) in complex with Ub-GGG (left, yellow mesh) superimposed onto USP15 (middle, gray, PDB: 6gha) and USP4 structures (right, magenta, PDB: 2y6e) in cartoon representation with active site loops labeled. C, comparison of USP11, USP15, and USP4 catalytic core structures in surface representation in the same order as in B highlighting sequence conservation: gray residues are identical between all paralogs, yellow residues are different in USP11 compared to USP4 and USP15, and blue residues have similar properties in USP11 compared to USP4 and USP15. The catalytic triad residues are shown in orange. D, active site region of USP11 (blue) in complex with Ub-GGG (yellow) shown in the same orientation as USP15 (gray, middle) and USP4 (magenta, right) highlighting the different conformations of the loop regions depicted in dark purple, catalytic triad residues in orange, and additional residues of interest are labeled. USP, ubiquitin-specific protease; Ub-GGG, ubiquitin-triple-gly.
Catalytic cleft, BL1 and 2, and SL regions
The CCL regions in the structures of USP11 (Thr312-Phe319) and USP4 (Gly305-Phe312) adopt largely similar conformations. The first turn of helix α1 harbors the catalytic cysteine that is in hydrogen-bonding distance with the other catalytic triad residues. In the USP15 free state, residues Asn267-Phe270 including the active site cysteine adopt a catalytically incompetent conformation (Fig. 3, B–D). In contrast, the catalytic triad histidine and aspartate closely superimpose in all available paralog structures (USP11 His888 and Asp446, USP4 His881 and Asp898, USP15 His862 and Asp879). The SL regions (USP11 Ser391-Gln402, USP4 Pro384-Gln395, USP15 Pro342-Gln353) adopt different conformations, likely attributable to the presence and absence of ubiquitin (Fig. 3D). USP11 harbours a QHD sequence in the SL loop (residues 398–400) that interacts with the substrate as opposed to the more typical “QQD” box in USP4 and USP15. The USP11 SL His399 side chain interacts with Ub-GGG Arg74. BL1 loop conformations also differ associated with the substrate-binding status (Fig. 3, B and D): USP11 Phe831 interacts with Ub-GGG Leu71 and Leu73. In the absence of a substrate’s distal ubiquitin moiety equivalent residues in USP4 (Phe828) and USP15 (Phe809) block the Leu73 binding pocket. The BL1 loop in the USP11 complex structure is rearranged to accommodate the substrate, whereby Tyr833 and Arg838 form most of the contacts with Ub-GGG (Ile36, Gln40, Leu71, Arg72). In the USP4 structure, Tyr830 blocks the substrate C-terminal tail binding channel by forming a hydrogen-bonding interaction with Asp393 in the SL. In USP15, large parts of the BL1 are flexible in the free state. The BL2 region is one of the least conserved active site loops (USP11 Tyr881-Gly887, USP4 Tyr874-Gly880, USP15 Tyr855-Gly861) and adopts different conformations in the structures. USP11 residue Arg885 (Fig. S4) is not conserved in either USP4 (Gly878) or USP15 (Gly859). In addition, USP11 Asp886 (USP4 Val879 and USP15 Gly860) contributes to BL2 differences by interacting with Gln398 in the SL loop (Fig. 3D).
USP11 S1 and S1′ pockets
In general, residues in the S1 distal ubiquitin binding pocket are relatively well conserved. The S1′ binding region that engages either a proximal Ub moiety in a polyubiquitin chain or different substrates linked to the C terminus of Ub displays considerable differences (Figs. 3C and S5).
At the rim of the S1 pocket, helix α4 is shifted outward to accommodate the Ub-GGG substrate in USP11 compared to the free paralog structures. USP11 Tyr424 and Glu426 between helices α5 and α6 at the top of the S1 pocket interact with Ub-GGG. At the bottom of the S1 binding pocket, USP11 Phe462 and Leu484 face Ub-GGG His68 and show flipped conformations compared to USP4 (Phe455, Leu477) and USP15 (Phe413, Leu435). Furthermore, in USP11 Cys482 and Tyr483 interact with the Ile44 patch of Ub-GGG. The finger regions in USP11 (Lys463-Thr477 and Thr789-Leu817) accommodate the distal ubiquitin core of the substrate. This “open hand” conformation sees shifts of approximately 9 Å compared to equivalent Cα positions in USP15 and USP4, which adopt a “closed hand” conformation in the absence of a substrate. The target substrate–specific S1′ site is characterized by low sequence conservation between the paralogs (Fig. 3C). Nevertheless, USP11 main chain residues His373-Ser376 superimpose well onto USP4 Arg366-His369 and USP15 Lys324-Tyr327. Furthermore, several other differences occur in the S1′ region as indicated in Figure 3.
Paralog-specific differences in product and substrate-binding parameters
To further investigate paralog-specific differences, we utilized site-directed mutagenesis coupled with isothermal titration calorimetry (ITC) and focused on differences in the catalytic site loop regions (Fig. 3A). Notably, in USP11 residues His399 (SL) as well as Arg885 and Asp886 (BL2) are not conserved in USP4 and USP15. To investigate these differences, we generated “chimeras” introducing USP15/USP4 or USP15-like mutations into USP11 (Figs. 3A and 4A).
Figure 4.
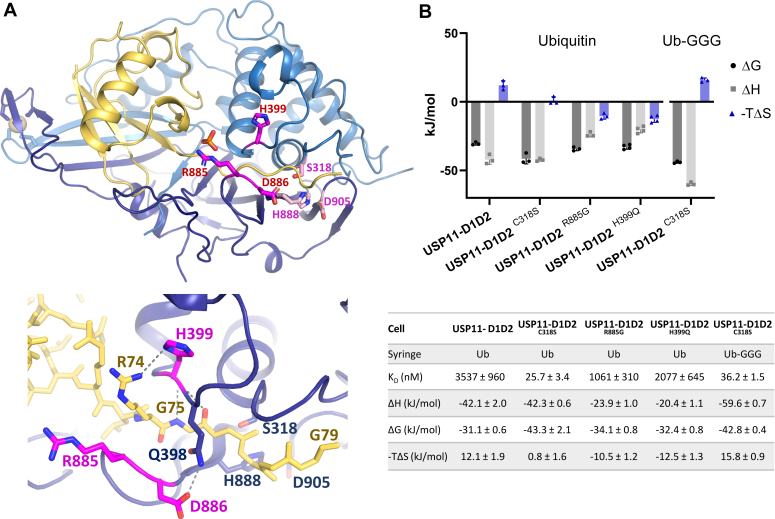
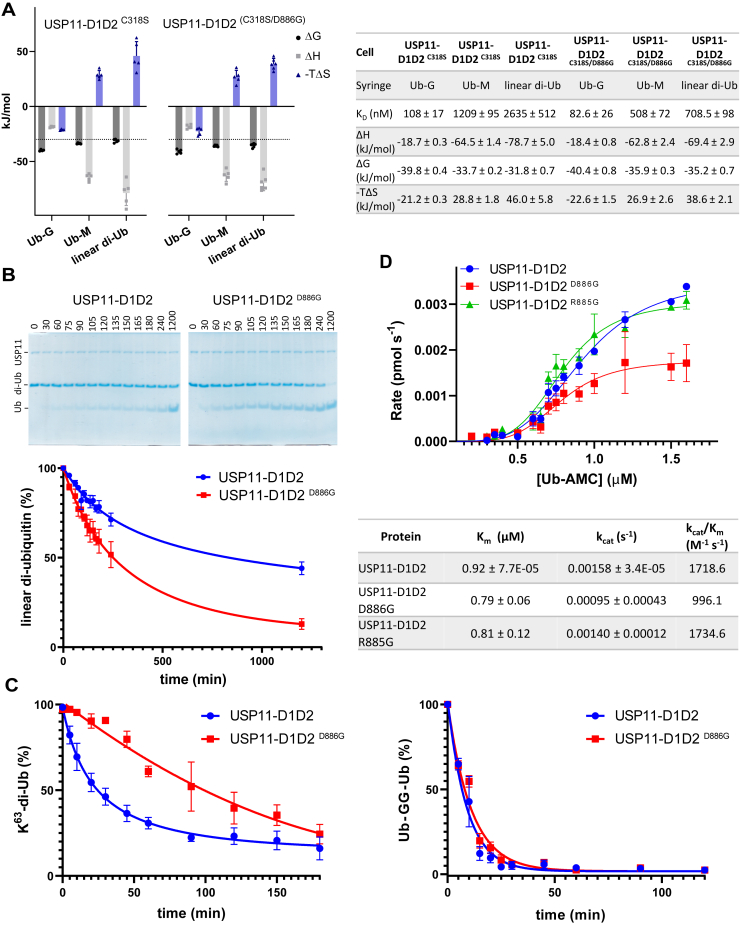
USP11 interactions with product ubiquitin and impact of mutations.A, structure of USP11 in complex with the Ub-GGG substrate with residues subjected to mutagenesis to probe paralog-specific differences shown in magenta stick representation and a zoom in view below highlighting putative H-bonding interactions. B, thermodynamic parameters from ITC binding assays for USP11-D1D2 or indicated USP11-D1D2 mutants with mono-ubiquitin (product) or Ub-GGG, in graphical (error = SD) and table (n = 3–4 independent experiments; error = SEM) format. USP, ubiquitin-specific protease; Ub-GGG, ubiquitin-triple-gly.
Binding of the product mono-ubiquitin to USP11-D1D2 revealed an exothermic binding behavior and a KD of ∼3.5 μM compared to 1.1 μM for USP11-D1D2 R885G and ∼2 μM for USP11-D1D2 H399Q, respectively, showing that USP11-D1D2 R885G displayed slightly higher affinity for mono-ubiquitin (Fig. 4B). The USP11 catalytic triad mutant C318S showed a higher affinity for ubiquitin than WT, an observation that has been reported for other USPs (19, 28) (Fig. 4B). USP11-D1D2 C318S interacted with high affinity with both mono-Ub and substrate Ub-GGG but a ∼50-fold reduction in affinity was observed for the linear di-Ub substrate (Figs. 4B and 5A and S6). We also investigated USP11-D1D2 C318S binding to Ub-M, a maximally truncated linear di-Ub substrate with only the first methionine of the proximal ubiquitin moiety present (Fig. 1D). Binding assays with Ub-M unexpectedly still displayed a KD that was significantly higher than Ub-GGG as well as Ub-G. Ub-G is equivalent to Ub-M, except for lacking the methionine side chain (Figs. 1D and 5A). This showed that the active site mutant USP11-D1D2 C318S binds considerably less tightly to linear di-Ub and Ub-M compared to Ub-GGG, Ub-G, or the reaction product mono-Ub. The data suggests that the presence of the start methionine of the proximal ubiquitin moiety in linear diubiquitin (the P1′ residue) is a key contributor to the significant reduction in the affinity observed for this substrate.
Figure 5.
Impact of paralog-specific differences in the BL2 region on USP11 substrate binding and cleavage.A, thermodynamic parameters from ITC binding assays for USP11-D1D2 C318S and USP11-D1D2 (C318S/D886G) interactions with Ub-G (minimal substrate), ubiquitin-methionine (Ub-M, truncated linear di-Ub substrate), and linear diubiquitin (poor substrate), in graphical (error = SD) and tabulated format (error = SEM; n ≥ 4). B, representative gels from gel shift assays comparing linear diubiquitin cleavage between the USP11 catalytic core and the USP11 D886G mutant with densitometric analysis shown below (error = SEM; n = 3). C, densitometric analyses of Lys63-linked di-Ub chain and Ub-GG-Ub chain cleavage assays (error = SEM; n = 3). D, kinetic data for USP11-D1D2 in comparison with USP11-D1D2 BL2 region D886G and R885G mutants (error = SD; n = 3). BL, blocking loop; ITC, isothermal titration calorimetry; USP, ubiquitin-specific protease.
An aspartate in the BL2 region modulates USP11’s substrate selectivity
USP11 is known to have a preference for Lys6- and Lys63-linked ubiquitin chains, whereas USP15 is more promiscuous. For example, linear diubiquitin is a very poor substrate for USP11 (15, 22, 23) and to a lesser degree USP4 (24, 29), but is readily cleaved by USP15.
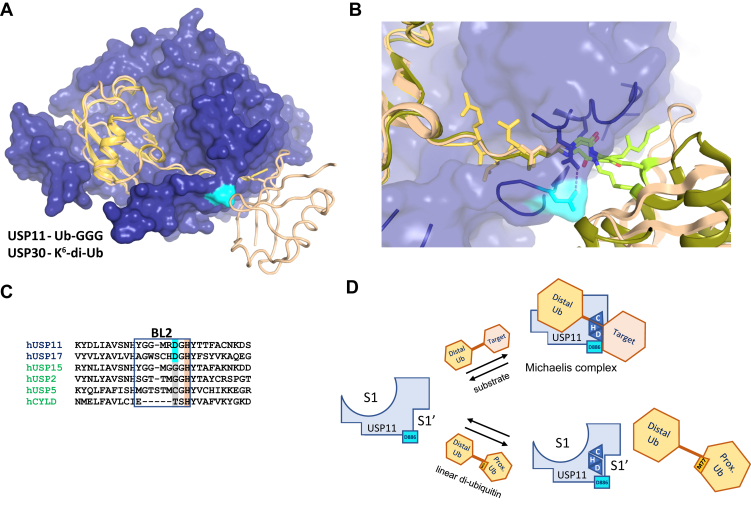
The BL2 region forms part of the S1′ binding site and is not well conserved between the paralogs. In the crystal structure, aspartate residue Asp886 is central to the USP11 S1′ region’s charge and shape (Figs. 4A, 6A, and 7A). The conformation of the Asp886 side chain is defined by forming hydrogen-bonding interactions with the side chain of Gln398 in the SL. Asp886 is thus involved in closing the substrate glycine tail binding channel by linking the SL and BL2 loops in the Michaelis complex and protrudes from the surface (Fig. 7A). We therefore hypothesized Asp886 may contribute to USP11’s ability to discriminate against linear diubiquitin. To test this, we generated variant USP11-D1D2 D886G, which is equivalent to residue Gly860 in USP15. USP11-D1D2 (C318S/D886G) bound linear diubiquitin and Ub-M with a higher affinity than when Asp886 was present confirming that Asp886 contributes to substrate interactions (Figs. 5A and S6). To assess the impact of this residue on the catalytic activity, we conducted enzymatic assays monitoring the gel-shift upon linear diubiquitin substrate cleavage for the WT versus the USP11-D1D2 D886G mutant. The data showed that the USP11-D1D2 D886G mutant was significantly more active against linear diubiquitin compared to the WT protein (Fig. 5B). Cleavage assays with Lys63-linked diubiquitin (Lys63-di-Ub) also displayed differences between WT and USP11-D1D2 D886G but with the opposite effect to linear di-Ub (Fig. 5C). In contrast, gel-shift assays with a linear di-Ub substrate engineered with an additional two glycine residues at the C terminus of the distal ubiquitin moiety (Ub-GG-Ub; Fig. 1D) resulted in rapid cleavage of the substrate irrespective of the D886G mutation. These results further indicate that the nature of the residue in the P1′ position of the substrate and its environment are important (Figs. 5C and S7). Enzymatic assays using minimal substrate ubiquitin-AMC displayed kinetic parameters consistent with slightly lower activity (Fig. 5D). This shows that USP11 with the D886G mutation despite displaying a slightly lower activity than WT using ubiquitin-7-amino-4-methylcoumarin as a substrate is significantly more active against linear di-Ub. The same is not observed for Lys63-di-Ub or Ub-GG-Ub. We next investigated the reciprocal G860D mutation in paralog USP15, which contains a glycine at the equivalent position to USP11 Asp886 (Figs. 3A and 6A). The results of the gel shift assay showed that the mutation has a considerable impact on USP15-D1D2’s rate of cleaving linear di-Ub (Figs. 6B and S7). Hence, this data reveals that Asp886 located in the BL2 loop of USP11 contributes to selective catalysis.
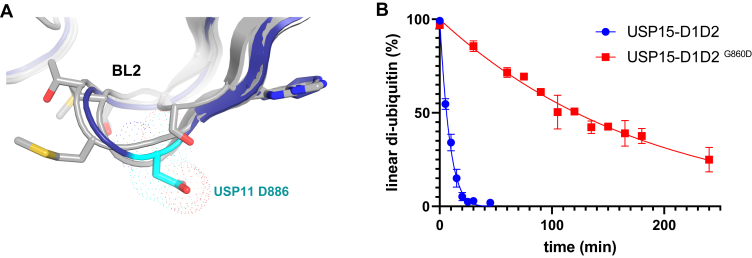
Figure 6.
Location of “gatekeeper residue” position in blocking loop 2 and impact of mutation on linear diubiquitin cleavage in USP15.A, closeup view of the USP11 Asp886 BL2 region (in blue) in a superposition with USPs that are known to readily cleave linear diubiquitin, namely CYLD (PDB ID: 2vhf, (62)), USP2 (PDB ID: 2hd5, (30)), and USP15 (PDB ID: 6gh9, (29)). B, densitometric analyses of linear di-Ub chain cleavage assays (error = SEM; n = 3) comparing USP15-D1D2 versus USP15-D1D2 G860D. USP, ubiquitin-specific protease.
Figure 7.
Impact of distal ubiquitin binding on USP structures.A, comparison of USP11 to the closest available USP–Ub complex structures of USP2 (PDB ID: 2ibi) and USP21 (PDB ID: 3i3t). Top: cartoon diagram of USP catalytic domains in gray with active site loops shown in blue and Ub-GGG or ubiquitin, respectively in light orange. Bottom: electrostatic surface representations of same enzyme as above displaying the S1′ site and active site cleft channel with Ub-GGG transiting in the complex structure of USP11 or the product ubiquitin seen through the channel in orange mesh representation for USP2 and USP21. B, left: superposition of catalytic core structures in the presence of a Ub species in the S1 pocket: USP2 (PDB ID: 2hd5, (30)), USP7 (PDB ID: 1nbf, (63)), USP11, USP14 (PDB ID: 2ayo, (64)), USP21 (PDB ID: 2y5b, (31)), USP30 (PDB ID: 5ohk, (32)), and USP46 (PDB ID: 5cvo, (65)). CCL, SL, BL1, and BL2 regions are highlighted in blue and only Ub-GGG is shown depicted in light orange. Right: superposition of USP catalytic core structures in the absence of a ubiquitin species in the S1-binding pocket (free states): USP4 (PDB ID: 2y6e, (18)), USP7 (PDB ID: 2f1z, (66)), USP8 (PDB ID: 2gfo, (67)), USP9X (PDB ID: 5wch, (68)), USP12 (PDB ID: 5k16, (37)), USP14 (PDB ID: 2ayn, (64)), and USP15 (PDB ID: 6gha, (19)). Active sites loops are depicted in blue and the catalytic triad residues are shown in light pink. USP, ubiquitin-specific protease; Ub-GGG, ubiquitin-triple-gly.
Structural comparisons of the USP11–Ub-GGG complex with other USPs
The phylogenetically closest USPs to USP11 for which structures are known apart from paralogs USP15 and USP4 are USP2, USP8, and USP21. No substrate trapped complexes are available for these USPs. Product bound structures of ubiquitin in the S1 site of USP2 (PDB ID: 2hd5; (30)) and USP21 (PDB ID: 2y5b; (31)) generally superimpose well with the USP11–substrate complex (43.0 % seq. id.; RMSD 0.97 Å over 308 aa and 41.1%; RMSD 1.2 Å over 302 aa with USP11, respectively). This includes the CCL, SL, and BL2 regions despite differences in the sequences. In contrast, the BL1 region adopts different conformations in the structures (Fig. 7, A and B) and USP11 residues Tyr833 and Arg838 that interact with the distal ubiquitin moiety are not conserved. Very few USP structures where substrates have been trapped are known to date and include USP30, USP22, and USP1–UAF1 complexes with Lys6-linked diubiquitin (32), mono-ubiquitinated H2B (33), and mono-ubiquitinated FANCI − FANCD2 (34), respectively. Amongst these, USP30 (selective for Lys6-linked poly-Ub, PDB ID: 5ohp) is the closest available USP catalytic core structure to USP11. Moreover, Lys6-linked Ub chains are good substrates for USP11 (15). A superposition of the USP30 structure in complex with Lys6-linked di-Ub and USP11 in complex with Ub-GGG shows good agreement (30.6% seq. id. and RMSD of 1.6 Å over 268 aa) despite differences in the sequences. The triple-gly tail in the Ub-GGG substrate aligns well with the Lys6-linked di-Ub lysine side chain and no major clashes of Lys6-linked di-Ub in this orientation with the USP11 catalytic core are apparent (Fig. 8A). Overall, these comparisons show that there are significant similarities in distal ubiquitin binding to the S1 site between closely related USPs and that the extended ubiquitin tail successfully mimics the lysine side chain in an isopeptide bond of a substrate. It also shows that the USP11 structure in complex with the substrate mimetic Ub-GGG captures the characteristics of a fully formed S1′ site.
Figure 8.
Ub-GGG as a substrate mimetic and the position of the “gatekeeper” residue.A, blue surface representation of USP11-D1D2 with Ub-GGG in yellow cartoon representation superimposed onto the USP30 (PDB ID: 5ohp, (32)) structure in complex with a Lys6-linked diubiquitin substrate (wheat color) highlighting that the triple glycine in Ub-GGG locates to the S1′ site. The location of BL2 USP11 Asp886 is highlighted in cyan and is with minor loop rearrangements compatible with linear Lys6-linked di-ub binding to USP11 in this orientation. B, USP11 transparent surface representation in complex with Ub-GGG (yellow) with the location of trapped substrates of Lys6-linked di-Ub (wheat color) as seen in complex with USP30 (PDB ID: 5ohp) and Ub-FANCD2 (olive color) as seen in complex with USP1 (PDB ID: 7ay1, (34)) superimposed. The position of the BL2 and CCL is shown and Asp886 highlighted in cyan. Scissile bond residues are highlighted in lime. C, sequence alignment of the BL2 region in USPs for which it is known that linear diubiquitin is either a very poor substrate (blue) or a good substrate (green) (29). The position of BL2 Asp886 is highlighted in cyan. D, schematic depiction of the location of residues Asp886 in the USP11 S1′ site as well as Met77 in linear diubiquitin that were found to influence USP11 cleavage rates for linear diubiquitin. BL, blocking loop; CCL, catalytic cleft loop; USP, ubiquitin-specific protease; Ub-GGG, ubiquitin-triple-gly.
Discussion
There are 56 USPs encoded in the human genome (35), but only for about a quarter structural information on the protease domain is available. Here, we added the structure of USP11 to the knowledge base. In contrast to the structures of the paralogs USP4 (18) and USP15 (19, 20), USP11 has been trapped with a novel engineered USP substrate developed in this study. The Michaelis complex structure of USP11 reveals the characteristics of the active conformation, while USP4 and USP15 structures (18, 19) adopt “closed hand” conformations that can only accommodate ubiquitin upon “opening up”. For USP15, a substrate-assisted mechanism with a reconfiguration of the catalytic triad has been proposed (19). Whether USP11 also displays an inactive catalytic triad configuration in the free state remains to be determined. Irrespectively, the comparisons suggest that substantial conformational changes are required to accommodate the substrate. These likely include rearrangement or flexible-to-order transitions of the catalytic site loops SL, BL1, and BL2. Moreover, they suggest that both the S1 and S1′ sites only fully form upon substrate binding by induced fit. Structures in different conformational states (free and S1 pocket occupied) are available for only a few USPs and include USP7 (36), USP12 (37, 38) and USP28 (39, 40). In general, the CCL, SL, BL1, and BL2 active site loop regions display significant conformational differences between USPs in the free states. When the S1 pocket is occupied by a distal ubiquitin moiety, the CCL and BL2 regions adopt very similar conformations, whereas more variability is still observed in the BL1 and SL regions (Fig. 7B). Sigmoidal kinetic behavior suggests that conformational changes are a key regulatory mechanism also for USP11. Very little is known about how USPs specifically recognize their respective substrates with a few exceptions (32, 33, 34, 41). Here, a novel substrate for deubiquitination was developed by constructing ubiquitin with three additional glycines (Ub-GGG) to mimic the lysine side chain. We show that Ub-GGG is a suitable substrate mimetic. Ub-GGG successfully mirrors the conformation of substrates harboring an isopeptide bond around the scissile bond (Fig. 8, A and B). This is consistent with the similarities of the extended nature of consecutive glycine residues and a lysine’s side chain.
The molecular basis of substrate selectivity is poorly understood for the majority of USPs. The S1′ site in the protease domain is assumed to play an important role in substrate recognition. Most USPs are considered promiscuous when it comes to differently linked poly-ubiquitin chains (1). However, amongst 52 active USPs, only USP2, USP5, CYLD, and USP15 display high activity toward linear di-Ub (29). Other USPs either show low activity or are largely inactive toward linear diubiquitin (29) (Fig. 8C). The difference in substrate selectivity between USP11 and the paralog USP15 despite significant sequence and structural similarities is especially intriguing. We noted that a position in the BL2 prior to the catalytic histidine may be of importance. The glycine prior to the catalytic histidine is almost absolutely conserved amongst USPs. This glycine is likely required for allowing the conformational changes to close the ubiquitin tail binding channel via an interaction with a glutamine from the SL region upon substrate binding. The position adjacent to this conserved glycine is more variable. USP11 has an atypical aspartate (Asp886) at this position in the BL2 (Figs. 6A and 8C). In the structure, the side chain of Asp886 is a distinctive feature of the S1′ site (Figs. 4A and 8B). Only USP17 isozymes, for which linear di-Ub is also a very poor substrate (42) have an Asp or Asn at this position. Comparison of USP11 and the paralog USP15 and their respective mutants showed that the presence or absence of an Asp in this “gatekeeper” position influences their ability to efficiently cleave linear diubiquitin. It is likely that charge-repulsion and/or steric reasons result in negative selection and the different reaction rates observed (Figs. 6A and 7A, schematic in Fig. 8D), although other factors such as different conformational dynamics and the rate of product release could also contribute. It is tempting to speculate that the nature of the residue in this position may also influence substrate selectivity in other USPs, but this remains to be determined. For example, we noted that USP2, another promiscuous USP similar to USP15, also has a glycine in this position (Figs. 7A and 8C).
The most defining characteristic of linear diubiquitin is that the C-terminal glycine of the distal ubiquitin is conjugated to the N-terminal methionine (the P1′ position of the substrate) of the proximal ubiquitin via a peptide bond, as opposed to conjugation to one of the seven internal lysine side chains via isopeptide bonds in different linkage-type poly-ubiquitin chains (Fig. 1D). The presence of the Met1 residue from the proximal ubiquitin moiety alone (as in linear diubiquitin) already causes a reduction in affinity for USP11, compared to Ub-G. This suggests that the Met1 residue of the proximal ubiquitin moiety is involved in causing linear diubiquitin to be a poor substrate for USP11, but other residues will also contribute. This is in line with structural models of USP11-D1D2 in complex with Ub-M from AlphaFold (43), which show Asp886 in close proximity to Met77 (equivalent to Met1 from the proximal ubiquitin moiety; Fig. S8).
For USP30, Ser477 in the S1′ site was identified as a substrate selectivity marker residue to explain Lys6-linkage preference (32). However, this position is occupied by an aspartate (USP11 Asp905) in USP11, USP4, and USP15, so does not account for paralogue specific differences in this subfamily of USPs. Individual USPs have evolved in different ways, and we are only just beginning to understand the molecular determinants for substrate selectivity amongst this family of cysteine proteases. For example, ancillary domains can also contribute to USP selectivity for certain substrates with different substrate-binding sites having been identified in USP11 (5, 44) and other USPs such as USP1 (45) and USP7 (46).
In this study a crystallizable stable USP11 catalytic domain construct was designed by use of a novel insertion tag. N-terminal fusion tags for carrier-driven crystallization have been used in isolated cases including maltose-binding protein (47), the macro domain (48) and thioredoxin (49). These tags can improve expression and solubility issues in addition to providing extra crystal contacts but suffer typically from increasing the flexibility, which can hinder crystallization. Loop insertion tags on the other hand do not suffer this disadvantage to the same extent. To our knowledge these have only been used in connection with solving the structures of membrane proteins, probably because the prediction of loop regions is more straight forward. A prominent example is T4 lysozyme, which was inserted to determine the structure of the human β2 adrenergic receptor (50) or the structure of the neurokinin 1 receptor where a disordered loop was replaced with a thermostable Pyrococcus abysii glycogen synthase domain (51). Here, we identified RDFrzS as a novel insertion tag by systematically mining the PDB database for suitable candidate proteins and proved its suitability as a crystallization tag for the case of the USP11 catalytic domain. Given that flexible large loops and insertions are common in many proteins and can be ever better predicted using ab initio folding and homology modeling methods (43, 52), this approach will be applicable to other targets that evaded crystallization to date.
Experimental procedures
Data base mining for the identification of the RDFrzS tag
The PDB was screened for candidate proteins or domains with properties that may aid solubility and crystallization efficiency and that are compatible with being inserted into loop regions. This was followed by visual inspection in PyMOL (PyMOL | pymol.org) and criteria included (i) size <400 amino acids, (ii) no disulphide bonds, (iii) no ligands, (iv) X-ray diffraction to < 1.6 Å resolution, (v) location of C and N termini no further than 10 Å apart, and (vi) globularity. Furthermore, factors such as concentration at which crystals were obtained, surface charge distribution, and B-factors were considered. The N-terminal RD of social motility protein FrzS from M. xanthus (RDFrzS; PDB ID: 2gkg (26)) was finally selected as a candidate insertion tag. The only other insertion tag tested was the structural 50S ribosomal protein L30e E90A variant from Thermococcus celer with PDB code 1w41 (53) (template plasmid kindly provided by Kaming Lee and Kam Bo Wong, The Chinese University of Hong Kong), but a USP11-L30e fusion construct did not yield any crystals.
Constructs, cloning, and mutagenesis
The constructs for USP11-D1D2, mono-Ub, and linear di-Ub have previously been described (15, 19). Throughout this manuscript USP11 residue numbering for isoform UniProt P51784 is used. The DNA sequence for RDFrzS (RD of M. xanthus social motility protein, residues 3–123) was ordered from GenScript with SER mutations K94A and K96S. The fusion construct of the USP11 catalytic core with the RDFrzS tag insertion (USP11-D1D2ins) was generated by restriction-free cloning in pET26b by replacing the ASTSK linker between the D1 and D2 domains (15) with the RDFrzS insertion tag sequence using the following primers fw: CCTTCTGCTACCTCAGTGTTCCACTGCCGGGTGCGAAAAAAATTCTGATTGTGGAAAG,rv: CTCAATGCACTCCTGCAGCCGCACCGGCGGAAAGCCAATCAGCGCG). An active site mutant and three loop mutants were generated using the Quick-Change mutagenesis protocol (C318S: Fw: CAATCTGGGCAACACGTCCTTCATGAACTCGGCCCTG, Rv: CAGGGCCGAGTTCATGAAGGACGTGTTGCCCAGATTG; H399Q: Fw: GGCTACCAGCAGCAAGACTCTCAGGAGCT, Rv: AGCTCCTGAGAGTCTTGCTGCTGGTAGCC; R885G: Fw: CCATTATGGGGGCATGGGTGATGGACACTACAC, Rv: GTGTAGTGTCCATCACCCATGCCCCCATAATGG) The active site mutant USP11-D1D2ins C318S was generated using the following primers: Fw: CAATCTGGGCAACACGTCCTTCATGAACTCGGCCCTG, Rv: CAGGGCCGAGTTCATGAAGGACGTGTTGCCCAGATTG). The active site mutant USP11-D1D2 D886G, as well as the double mutant USP11-D1D2 (C318S/D886G) were generated using site-directed mutagenesis using the Quick-Change mutagenesis protocol based on the USP11-D1D2 and USP11-D1D2 C318S templates (Fw: CATTATGGGGGCATGCGTGGTGGACACTACACAACATTTGC, Rv: GCAAATGTTGTGTAGTGTCCACCACGCATGCCCCCATAATG). The sequence for Ub-GGG (ubiquitin residues 1–76 extended by three glycines) was cloned into pCDFDuet-1 using NdeI and KpnI restriction endonucleases with primers Fw: GGGAATTCCATATGCAGATCTTCGTGAAGACTC and Rv: CGGGGTACCTCAACCACCACCCCCACCTCTGAGACGGAG. Ub-G was then generated by introducing a stop codon. Ub-GG-Ub was obtained as a synthetic gene (GenScript) and was cloned into pCDFDuet-1. USP15-D1D2 G860D was generated from a construct previously described (19).
Protein expression and purification
All proteins including USP11-D1D2, mutants C318S, R885G, D886G, (C318S/D886G), and H399Q, USP11-D1D2ins, USP11-D1D2ins C318S, USP15-D1D2, and mutants as well as ubiquitin constructs were expressed in Escherichia coli BL21(DE3)-Codon Plus cells using 2YT broth growth medium. The complex of USP11-D1D2ins C318S with substrate Ub-GGG was prepared from coexpression. All USP proteins and complexes were expressed and purified in the following way: After reaching an OD600 of ∼0.6, overexpression was induced with 0.5 mM IPTG and cultures grown for 16 h at 18 °C. For USP11 purifications, cells were harvested, resuspended in buffer A (50 mM Tris-Cl, pH 8, 300 mM NaCl, 20 mM imidazole, 10% glycerol), lysed by sonication, and clarified by centrifugation (24,000g, 1 h, 4 °C). The lysate was loaded onto a HiTrap chelating column (Cytiva) precharged with Ni2+ ions and protein eluted using an imidazole gradient (20–500 mM). The purest fractions were combined and concentrated, and proteins further purified by size-exclusion chromatography (SEC) on a Superdex 200 16/600 column (Cytiva) using SEC buffer: 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% (v/v) glycerol. Pure protein fractions were combined and concentrated.
Crystallisation, data collection, and structure determination
Crystals of USP11-D1D2ins C318S in complex with Ub-GGG were grown using 8.6 mg/ml protein in SEC buffer at 20 °C using the sitting drop vapor diffusion method in the following condition: 100 mM Tris-Bicine pH 8.5, 30 mM sodium nitrate, 30 mM sodium phosphate dibasic, 30 mM ammonium sulphate, 11.25% (v/v) MPD; 11.25% (v/v) PEG 1000; 11.25% (w/v) PEG 3350 with 5 mM CdCl2. Needle-like crystals were cryoprotected with an additional 30% glycerol added to the mother liquor. Data from crystals containing the USP11-D1D2ins C318S in complex with Ub-GGG were collected at the Swiss Light Source beamline X06DA at a wavelength of 0.999995 Å and 100 K. Crystals belonged to space group P 21 21 2 with unit cell parameters of a = 94.32 Å, b = 186.10 Å, c = 75.76 Å. Data were processed using XDS (54) and AIMLESS (55), and the structure was solved by molecular replacement using coordinates of a truncated USP11 homology model generated by PHYRE2 (56), ubiquitin (PDB ID: 1ubi) and RDFrzS (PDB ID: 2gkg) as search models in PHASER (57). The crystal contained two USP11-D1D2ins C318S molecules in complex with Ub-GGG in the ASU. Both copies of the USP11 catalytic core superimpose well with an RMSD of 0.29 Å (over 2995 Cα atoms). Data collection statistics are shown in Table 1.
Model building, refinement, and validation
Model building was performed using COOT (cam.ac.uk) (58), while structure refinements were performed using REFMAC (59) and PHENIX (60). Model quality was assessed using MolProbity (61). For USP11-D1D2ins in chain A, seven N- and eight C-terminal residues and for USP11-D1D2ins in chain B eight N- and five C-terminal residues were not modeled due to flexibility. An overlay of the two copies in the ASU shows differences in the positioning of the RDFrzS tag but no significant differences between the two USP11 and Ub-GGG molecules. Additionally, all 79 residues of both Ub-GGG molecules were modeled. Density for glycerol, nitrate, phosphate, and cadmium ion ligands was observed in both copies of the complex. A disulphide bond links USP11 Cys428 residues from chain A and B, which is likely a crystallization artefact, as no dimerization was observed during purification. The absence of zinc was confirmed by a Zn K edge scan at DLS I24 compared to a scan for a crystal grown with ZnCl2 as an additive. In the final model 96.85% of residues are located in favored regions of the Ramachandran plot. Refinement statistics are shown in Table 1. Figures were generated using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Enzymatic assays and ITC
Kinetic parameters or time courses for USP11-D1D2 or mutants C318S, R885G, H399Q, D886G, or USP11-D1D2ins were determined using ubiquitin-AMC as fluorogenic substrate in buffer 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% (v/v) glycerol, 1 mM DTT. Deubiquitinase activity was measured in 384-well white plates in 30 μl reaction volume in triplicates using an EnVision 2104 multilabel plate reader at 25 °C (excitation: 355 nm, emission: 426/8 nm). Measurements were taken once per minute for 30 min. Data were fitted using nonlinear regression analysis in the GraphPad Prism software (Home - GraphPad) (allosteric sigmoidal model using the equation: Y = Vmax x Xh/(Khalfh + Xh)) to establish Khalf and kcat values. Gel-shift based linear diubiquitin cleavage assays were performed in 150 mM NaCl, 50 mM Tris–HCl, pH 7.5, 1 mM DTT, in triplicate at 25 °C. Reactions were started by adding linear di-Ub or Ub-GG-Ub to a final concentration of 5 μM to 400 nM of USP11-D1D2, USP11-D1D2 D886G, USP15-D1D2, or USP15-D1D2 G860D. Reactions were stopped by adding SDS-PAGE loading buffer and analyzed on 18% SDS-PAGE gels. Gels were stained with Colloidal Coomassie blue stain, scanned, and analyzed using ImageJ (https://imagej.nih.gov/ij/). Relative amounts of linear di-Ub, Lys63-di-Ub (UbiQ) or Ub-GG-Ub and mono-Ub for each time point were determined and plotted using GraphPad Prism. ITC data were measured using a PEAQ ITC instrument (Malvern). Different ubiquitin samples including mono-Ub, linear di-Ub, Ub-GGG, Ub-G, and Ub-M (200–500 μM) were titrated into USP11 protein samples (20–30 μM) in 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% (v/v) glycerol. Experimental settings were 25 °C, 180 s spacing of injections at a 750-rpm stirring speed. Analysis of the data was performed using the PEAQ ITC analysis software (MicroCal PEAQ-ITC Analysis Software v1.41 | Malvern Panalytical) (Malvern), fitting to a one-site binding model. Experiments were performed at least in triplicates.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Atomic coordinates and structure factors of the catalytic core of USP11 in complex with substrate Ub-GGG have been deposited in the Protein Data Bank (http://wwpdb.org/) with accession code 8OYP.
Supporting information
This article contains supporting information (43).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Crystallographic Data were obtained at Swiss Light Source (SLS, Paul Scherrer Institut, Villigen, Switzerland). We are grateful to colleagues at SLS for assistance with collecting data at beamline X06DA. We thank Hayley Gratton and Stephen Harper for preliminary work on expression constructs.
Author contributions
S. K. M., S. J. W., and S. G. C. methodology; S. K. M., M. P. M., S. B., M. H., I. D., and J. Z. investigation; S. K. M. and I. D. writing-original draft; S. K. M. formal analysis; S. K. M., M. P. M., J. Z., S. B., and I. D. visualization; M. P. M. validation; S. J. W., S. G. C., J. E., and I. D. writing-review and editing; J. E. and I. D. supervision; I. D. project administration.
Funding and additional information
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/H012656/1] (I. D.) Biotechnology and Biological Sciences Research Council (BBSRC) doctoral training studentship [grant number BB/M008770/1] (S. K. M., M. P. M., and S. J. W.).
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
Footnotes
Present address for Simon G. Caulton: Institute for Microbiology and Infection, School of Biosciences, University of Birmingham, Birmingham B15 2TT, UK.
Supporting information
References
- 1.Clague M.J., Urbe S., Komander D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019;20:338–352. doi: 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 2.Deng T., Yan G., Song X., Xie L., Zhou Y., Li J., et al. Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4678–4683. doi: 10.1073/pnas.1714938115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiltshire T.D., Lovejoy C.A., Wang T., Xia F., O'Connor M.J., Cortez D. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 2010;285:14565–14571. doi: 10.1074/jbc.M110.104745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurga M., Abugable A.A., Goldman A.S.H., El-Khamisy S.F. USP11 controls R-loops by regulating senataxin proteostasis. Nat. Commun. 2021;12:5156. doi: 10.1038/s41467-021-25459-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacko A.M., Nan L., Li S., Tan J., Zhao J., Kass D.J., et al. De-ubiquitinating enzyme, USP11, promotes transforming growth factor beta-1 signaling through stabilization of transforming growth factor beta receptor II. Cell Death Dis. 2016;7:e2474. doi: 10.1038/cddis.2016.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Salihi M.A., Herhaus L., Macartney T., Sapkota G.P. USP11 augments TGFbeta signalling by deubiquitylating ALK5. Open Biol. 2012;2 doi: 10.1098/rsob.120063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao T.L., Wu C.Y., Su W.C., Jeng K.S., Lai M.M. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 2010;29:3879–3890. doi: 10.1038/emboj.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhart R.A., Peng Y., Norris Z.A., Tholey R.M., Talbott V.A., Liang Q., et al. Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol. Cancer Res. 2013;11:901–911. doi: 10.1158/1541-7786.MCR-12-0699. [DOI] [PubMed] [Google Scholar]
- 10.Dwane L., O'Connor A.E., Das S., Moran B., Mulrane L., Pinto-Fernandez A., et al. A functional genomic screen identifies the deubiquitinase USP11 as a novel transcriptional regulator of ERalpha in breast cancer. Cancer Res. 2020;80:5076–5088. doi: 10.1158/0008-5472.CAN-20-0214. [DOI] [PubMed] [Google Scholar]
- 11.Park M.K., Yao Y., Xia W., Setijono S.R., Kim J.H., Vila I.K., et al. PTEN self-regulates through USP11 via the PI3K-FOXO pathway to stabilize tumor suppression. Nat. Commun. 2019;10:636. doi: 10.1038/s41467-019-08481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Wang J., Yan H., Zhang K., Liu Y. Upregulation of USP11 promotes epithelialtomesenchymal transition by deubiquitinating Snail in ovarian cancer. Oncol. Rep. 2019;41:1739–1748. doi: 10.3892/or.2018.6924. [DOI] [PubMed] [Google Scholar]
- 13.Kapadia B., Nanaji N.M., Bhalla K., Bhandary B., Lapidus R., Beheshti A., et al. Fatty Acid Synthase induced S6Kinase facilitates USP11-eIF4B complex formation for sustained oncogenic translation in DLBCL. Nat. Commun. 2018;9:829. doi: 10.1038/s41467-018-03028-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E.W., Seong D., Seo J., Jeong M., Lee H.K., Song J. USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ. 2015;22:1463–1476. doi: 10.1038/cdd.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper S., Gratton H.E., Cornaciu I., Oberer M., Scott D.J., Emsley J., et al. Structure and catalytic regulatory function of ubiquitin specific protease 11 N-terminal and ubiquitin-like domains. Biochemistry. 2014;53:2966–2978. doi: 10.1021/bi500116x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper S., Besong T.M., Emsley J., Scott D.J., Dreveny I. Structure of the USP15 N-terminal domains: a beta-hairpin mediates close association between the DUSP and UBL domains. Biochemistry. 2011;50:7995–8004. doi: 10.1021/bi200726e. [DOI] [PubMed] [Google Scholar]
- 17.Elliott P.R., Liu H., Pastok M.W., Grossmann G.J., Rigden D.J., Clague M.J., et al. Structural variability of the ubiquitin specific protease DUSP-UBL double domains. FEBS Lett. 2011;585:3385–3390. doi: 10.1016/j.febslet.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 18.Clerici M., Luna-Vargas M.P., Faesen A.C., Sixma T.K. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 2014;5:5399. doi: 10.1038/ncomms6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward S.J., Gratton H.E., Indrayudha P., Michavila C., Mukhopadhyay R., Maurer S.K., et al. The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. J. Biol. Chem. 2018;293:17362–17374. doi: 10.1074/jbc.RA118.003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teyra J., Singer A.U., Schmitges F.W., Jaynes P., Kit Leng Lui S., Polyak M.J., et al. Structural and functional characterization of ubiquitin variant inhibitors of USP15. Structure. 2019;27:590–605.e595. doi: 10.1016/j.str.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Vlasschaert C., Xia X., Coulombe J., Gray D.A. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol. Biol. 2015;15:230. doi: 10.1186/s12862-015-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faesen A.C., Luna-Vargas M.P., Geurink P.P., Clerici M., Merkx R., van Dijk W.J., et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Bailon M.P., Choi S.Y., Dufficy E.R., Sharma K., McNee G.S., Gunnell E., et al. Arginine methylation and ubiquitylation crosstalk controls DNA end-resection and homologous recombination repair. Nat. Commun. 2021;12:6313. doi: 10.1038/s41467-021-26413-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritorto M.S., Ewan R., Perez-Oliva A.B., Knebel A., Buhrlage S.J., Wightman M., et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat. Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossio V., Paulo J.A., Chick J., Brasher B., Gygi S.P., King R.W. Proteomics of broad deubiquitylase inhibition unmasks redundant enzyme function to reveal substrates and assess enzyme specificity. Cell Chem. Biol. 2021;28:487–502.e485. doi: 10.1016/j.chembiol.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser J.S., Merlie J.P., Jr., Echols N., Weisfield S.R., Mignot T., Wemmer D.E., et al. An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS. Mol. Microbiol. 2007;65:319–332. doi: 10.1111/j.1365-2958.2007.05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Morrow M.E., Morgan M.T., Clerici M., Growkova K., Yan M., Komander D., et al. Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins. EMBO Rep. 2018;19:e45680. doi: 10.15252/embr.201745680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi H., Yamanaka S., Kuwada S., Higaki K., Kido K., Sato Y., et al. A human DUB protein array for clarification of linkage specificity of polyubiquitin chain and application to evaluation of its inhibitors. Biomedicines. 2020;8:152. doi: 10.3390/biomedicines8060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renatus M., Parrado S.G., D'Arcy A., Eidhoff U., Gerhartz B., Hassiepen U., et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Y., Akutsu M., Reyes-Turcu F., Enchev R.I., Wilkinson K.D., Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gersch M., Gladkova C., Schubert A.F., Michel M.A., Maslen S., Komander D. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 2017;24:920–930. doi: 10.1038/nsmb.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan M.T., Haj-Yahya M., Ringel A.E., Bandi P., Brik A., Wolberger C. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rennie M.L., Arkinson C., Chaugule V.K., Toth R., Walden H. Structural basis of FANCD2 deubiquitination by USP1-UAF1. Nat. Struct. Mol. Biol. 2021;28:356–364. doi: 10.1038/s41594-021-00576-8. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Reverter D. Molecular mechanisms of DUBs regulation in signaling and Disease. Int. J. Mol. Sci. 2021;22:986. doi: 10.3390/ijms22030986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozen A., Rouge L., Bashore C., Hearn B.R., Skelton N.J., Dueber E.C. Selectively modulating conformational states of USP7 catalytic domain for activation. Structure. 2018;26:72–84.e77. doi: 10.1016/j.str.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Li H., Lim K.S., Kim H., Hinds T.R., Jo U., Mao H., et al. Allosteric activation of ubiquitin-specific proteases by beta-propeller proteins UAF1 and WDR20. Mol. Cell. 2016;63:249–260. doi: 10.1016/j.molcel.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dharadhar S., Clerici M., van Dijk W.J., Fish A., Sixma T.K. A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme. J. Struct. Biol. 2016;196:437–447. doi: 10.1016/j.jsb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauer F., Klemm T., Kollampally R.B., Tessmer I., Nair R.K., Popov N., et al. Differential oligomerization of the deubiquitinases USP25 and USP28 regulates their activities. Mol. Cell. 2019;74:421–435.e410. doi: 10.1016/j.molcel.2019.02.029. [DOI] [PubMed] [Google Scholar]
- 40.Gersch M., Wagstaff J.L., Toms A.V., Graves B., Freund S.M.V., Komander D. Distinct USP25 and USP28 oligomerization states regulate deubiquitinating activity. Mol. Cell. 2019;74:436–451.e437. doi: 10.1016/j.molcel.2019.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim R.Q., Geurink P.P., Mulder M.P.C., Fish A., Ekkebus R., El Oualid F., et al. Kinetic analysis of multistep USP7 mechanism shows critical role for target protein in activity. Nat. Commun. 2019;10:231. doi: 10.1038/s41467-018-08231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hjortland N.M., Mesecar A.D. Steady-state kinetic studies reveal that the anti-cancer target Ubiquitin-Specific Protease 17 (USP17) is a highly efficient deubiquitinating enzyme. Arch. Biochem. Biophys. 2016;612:35–45. doi: 10.1016/j.abb.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiliotopoulos A., Blokpoel Ferreras L., Densham R.M., Caulton S.G., Maddison B.C., Morris J.R., et al. Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11. J. Biol. Chem. 2019;294:424–436. doi: 10.1074/jbc.RA118.004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkinson C., Chaugule V.K., Toth R., Walden H. Specificity for deubiquitination of monoubiquitinated FANCD2 is driven by the N-terminus of USP1. Life Sci. Alliance. 2018;1 doi: 10.26508/lsa.201800162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rouge L., Bainbridge T.W., Kwok M., Tong R., Di Lello P., Wertz I.E., et al. Molecular understanding of USP7 substrate recognition and C-terminal activation. Structure. 2016;24:1335–1345. doi: 10.1016/j.str.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Waugh D.S. Crystal structures of MBP fusion proteins. Protein Sci. 2016;25:559–571. doi: 10.1002/pro.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild R., Hothorn M. The macro domain as fusion tag for carrier-driven crystallization. Protein Sci. 2017;26:365–374. doi: 10.1002/pro.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corsini L., Hothorn M., Scheffzek K., Sattler M., Stier G. Thioredoxin as a fusion tag for carrier-driven crystallization. Protein Sci. 2008;17:2070–2079. doi: 10.1110/ps.037564.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbaum D.M., Rasmussen S.G., Kobilka B.K. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoppe J., Ehrenmann J., Klenk C., Rucktooa P., Schutz M., Dore A.S., et al. Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat. Commun. 2019;10:17. doi: 10.1038/s41467-018-07939-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee C.F., Allen M.D., Bycroft M., Wong K.B. Electrostatic interactions contribute to reduced heat capacity change of unfolding in a thermophilic ribosomal protein l30e. J. Mol. Biol. 2005;348:419–431. doi: 10.1016/j.jmb.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCoy A.J. Acknowledging errors: advanced molecular replacement with phaser. Methods Mol. Biol. 2017;1607:421–453. doi: 10.1007/978-1-4939-7000-1_18. [DOI] [PubMed] [Google Scholar]
- 58.Casanal A., Lohkamp B., Emsley P. Current developments in coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2020;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winn M.D., Murshudov G.N., Papiz M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 60.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komander D., Lord C.J., Scheel H., Swift S., Hofmann K., Ashworth A., et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 63.Hu M., Li P., Li M., Li W., Yao T., Wu J.W., et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 64.Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin J., Schoeffler A.J., Wickliffe K., Newton K., Starovasnik M.A., Dueber E.C., et al. Structural insights into WD-Repeat 48 activation of ubiquitin-specific protease 46. Structure. 2015;23:2043–2054. doi: 10.1016/j.str.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 66.Hu M., Gu L., Li M., Jeffrey P.D., Gu W., Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avvakumov G.V., Walker J.R., Xue S., Finerty P.J., Jr., Mackenzie F., Newman E.M., et al. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J. Biol. Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 68.Paudel P., Zhang Q., Leung C., Greenberg H.C., Guo Y., Chern Y.H., et al. Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7288–7297. doi: 10.1073/pnas.1815027116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Atomic coordinates and structure factors of the catalytic core of USP11 in complex with substrate Ub-GGG have been deposited in the Protein Data Bank (http://wwpdb.org/) with accession code 8OYP.