Abstract
HBV infection profoundly escalates hepatocellular carcinoma (HCC) susceptibility, responsible for a majority of HCC cases. HBV-driven immune-mediated hepatocyte impairment significantly fuels HCC progression. Regrettably, inconspicuous early HCC symptoms often culminate in belated diagnoses. Nevertheless, surgically treated early-stage HCC patients relish augmented five-year survival rates. In contrast, advanced HCC exhibits feeble responses to conventional interventions like radiotherapy, chemotherapy, and surgery, leading to diminished survival rates. This investigation endeavors to unearth diagnostic hallmark genes for HBV-HCC leveraging a bioinformatics framework, thus refining early HBV-HCC detection. Candidate genes were sieved via differential analysis and Weighted Gene Co-Expression Network Analysis (WGCNA). Employing three distinct machine learning algorithms unearthed three feature genes (HHIP, CXCL14, and CDHR2). Melding these genes yielded an innovative Artificial Neural Network (ANN) diagnostic blueprint, portending to alleviate patient encumbrance and elevate life quality. Immunoassay scrutiny unveiled accentuated immune damage in HBV-HCC patients relative to solitary HCC. Through consensus clustering, HBV-HCC was stratified into two subtypes (C1 and C2), the latter potentially indicating milder immune impairment. The diagnostic model grounded in these feature genes showcased robust and transferrable prognostic potentialities, introducing a novel outlook for early HBV-HCC diagnosis. This exhaustive immunological odyssey stands poised to expedite immunotherapeutic curatives' emergence for HBV-HCC.
Keywords: HBV-HCC, HBV, Tumor viruses, Tumor immunotherapy, Diagnostic model
1. Introduction
Primary liver cancer is among the most prevalent malignant tumors worldwide [[1], [2], [3]], and HCC constitutes 90% of primary liver cancers [4]. HCC ranks sixth in terms of incidence and third in terms of mortality among all types of malignancies worldwide [5], causing 6 million deaths annually worldwide [6]. However, most patients with HCC are diagnosed at an advanced stage due to the lack of early signs and symptoms [7]. Patients in the early stages of HCC have the option of surgical resection and have a 5-year survival rate of 30–70% [8]. In contrast, for patients with advanced HCC, surgical resection is not a viable option and 90% of them are left with the only options of radiotherapy or chemotherapy. Unfortunately, the 5-year survival rate for these patients is less than 10% [[9], [10], [11], [12], [13], [14]]. This results in many HCC patients not receiving effective treatment. Therefore, early prevention and diagnosis of HCC are crucial to enhance the prognosis of patients with HCC [15]. About 2 billion people worldwide are infected with HBV [16]. HBV infection is a major contributor to hepatocellular carcinoma, and more than 50% of HCC is caused by HBV infection [17,18]. Therefore, it is particularly important to prevent HBV patients from progressing to HBV-HCC. Currently, various serological tests are of clinical value for the early diagnosis of HBV-HCC, but the drawback of their low diagnostic sensitivity and specificity is also very obvious [19]. In the disease progression of HBV-HCC, HBV directly contributes to the development of HCC by increasing viral load and integrating DNA into the host genome [20]. However, HBV DNA integration is considered oncogenic in85% of tissue samples from patients with HBV-HCC, and the differences caused by this DNA integration can be detected at the transcriptional level [20,21]. Therefore, the search for new diagnostic feature genes at the genetic level is crucial to improve the early diagnosis and prognosis of HBV-HCC patients.
The synergy between high-throughput sequencing and machine learning not only helps to fully explore intricate biological processes, but also has the potential to reveal new insights not available through traditional methods [[22], [23], [24], [25]]. This integrated approach leverages the ability of high-throughput sequencing to generate large amounts of biological data and the ability of machine learning to decipher complex patterns, thus paving the way for transformative advances in biomedical research [[26], [27], [28]]. While some studies have focused on serological diagnostics of HBV-HCC, such as serum pentraxin 3 and Des-gamma-carboxyprothrombin, few have analyzed and searched for diagnostic feature genes associated with HBV-HCC at the genetic level [29,30]. Therefore, the primary objective of this study was to identify novel diagnostic feature genes to enhance the effectiveness of early detection in HBV-HCC patients, as well as to elucidate the potential molecular mechanisms underlying its development. Additionally, immune infiltration analysis was performed to understand the role played by the immune microenvironment in the progression of HBV to HBV-HCC. Consensus clustering of HBV-HCC was also performed, and immune infiltration analysis was conducted to investigate differences in the immune microenvironment among different subtypes of HBV-HCC.
The aim of this study was to perform differential analysis to identify differentially expressed genes (DEGs) between HBV and HBV-HCC. We employed a weighted gene co-expression network analysis (WGCNA) to screen for genes that are associated with HBV-HCC, and identified the intersection of these two sets of genes as the candidate gene pool. From this pool, we used three machine learning algorithms to obtain three HBV-HCC-related feature genes. To gain insight into the role played by these feature genes in the HBV-HCC process, we conducted correlation tests, GeneMANIA and single gene GSEA analyses. Subsequently, we constructed and validated a diagnostic model for HBV-HCC using the three feature genes. Furthermore, We conducted an analysis of immune infiltration to examine the variations in the immune microenvironment between HBV and HBV-HCC. Lastly, we utilized feature genes to classify HBV-HCC into subtypes and investigated dissimilarities in the immune microenvironment and fundamental molecular mechanisms between these subtypes. Our findings offer novel ideas and perspectives for the diagnosis and treatment of HBV-HCC.
2. Materials and methods
2.1. Collection and processing of raw data
The primary objective of this investigation was to explore the early detection of HBV-HCC and the contribution of the immune microenvironment in the disease progression. To achieve this, we retrieved two datasets, namely GSE121248 and GSE55092, from the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) for the analysis. The training set GSE121248 consisted of 37 paraneoplastic HBV samples and 70 HBV-HCC samples, while the validation set GSE55092 comprised of 91 paraneoplastic HBV samples and 49 HBV-HCC samples. We utilized GSE121248 for the preliminary investigation before identifying the feature genes and subsequently constructing the diagnostic model, which we validated using GSE55092. Additionally, we combined the two datasets, GSE121248 and GSE55092, to investigate the immune microenvironment profile between HBV and HBV-HCC. We also pooled HBV-HCC samples from both datasets to classify the subtypes of HBV-HCC and to explore the immune microenvironmental differences among the subtypes and the underlying molecular mechanisms.
2.2. Identification of differential genes
To discern the DEGs between HBV and HBV-HCC, we conducted differential gene expression analysis on the training set using the “limma” R package, applying filter parameters of |log2FC|>1 and FDR<0.05. The identified DEGs were presented in the form of heat maps and volcano maps created with the R packages “pheatmap” and “ggplot2″, respectively.
2.3. Gene set enrichment analysis (GSEA)
The R packages “clusterProfiler” and “enrich” were utilized for the GSEA enrichment analysis of genes. The top five functional pathways with the highest and lowest enrichment levels were selected for visualization. The biological significance of the genes in various functional pathways was elucidated by their respective enrichment levels.
2.4. Weighted gene co-expression network analysis (WGCNA)
To identify co-expressed genes associated with HBV-HCC, we utilized the WGCNA method. Firstly, the expression matrix of GSE121248 was used as input data, and co-expression networks were constructed using the R package “WGCNA”. Cluster trees and trait heat maps of the samples were created to detect any significant abnormal samples. To ensure a good average connectivity of the co-expression network, we set the scale-free topology index R^2 to 0.9. When the soft threshold β was set to 9, the network met the scale-free condition with good average connectivity. The genes were clustered hierarchically based on their similarity, and multiple modules were obtained by combining those with high gene similarity. A total of 18 modules were identified through the heat map of modules and traits. The gene co-expression modules with the strongest correlation with HBV-HCC were identified by plotting the module affiliation (MM) vs. gene significance (GS) scatter plot of the core co-expression module, revealing a high correlation with HBV-HCC.
2.5. Identification and enrichment analysis of candidate genes
To identify candidate genes associated with HBV-HCC, we obtained the intersection of DEGs and highly correlated modular genes with HBV-HCC using a Venn diagram. We then performed Disease Ontology (DO) enrichment analysis on the candidate genes using the R package “DOSE” to gain insight into their role in the HBV-HCC process by examining their level of enrichment in different diseases. Additionally, We utilized the R packages “clusterProfiler” and “enrich” to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to gain a deeper understanding of the molecular mechanisms involved in the progression of HBV to HBV-HCC. This was achieved by evaluating the enrichment levels of candidate genes in various biological processes and pathways.
2.6. Construction of protein-protein interactions (PPI) network
PPI prediction was performed to explore the interrelationships between candidate genes. The STRING database version 11.5 (https://cn.string-db.org) was used, and the minimum required interaction score was set to medium confidence (0.400). Disconnected nodes were hidden to obtain the final PPI network constructed by the candidate genes. This network was visualized using Cytoscape, An open-source software platform was used for the visualization of molecular interaction networks and integration with gene expression profiles and other state data. The PPI network provided insights into the potential functional relationships and pathways involved in the development of HBV-HCC, which could be further explored using functional enrichment analysis.
2.7. Three machine learning algorithms to screen for HBV-HCC feature genes
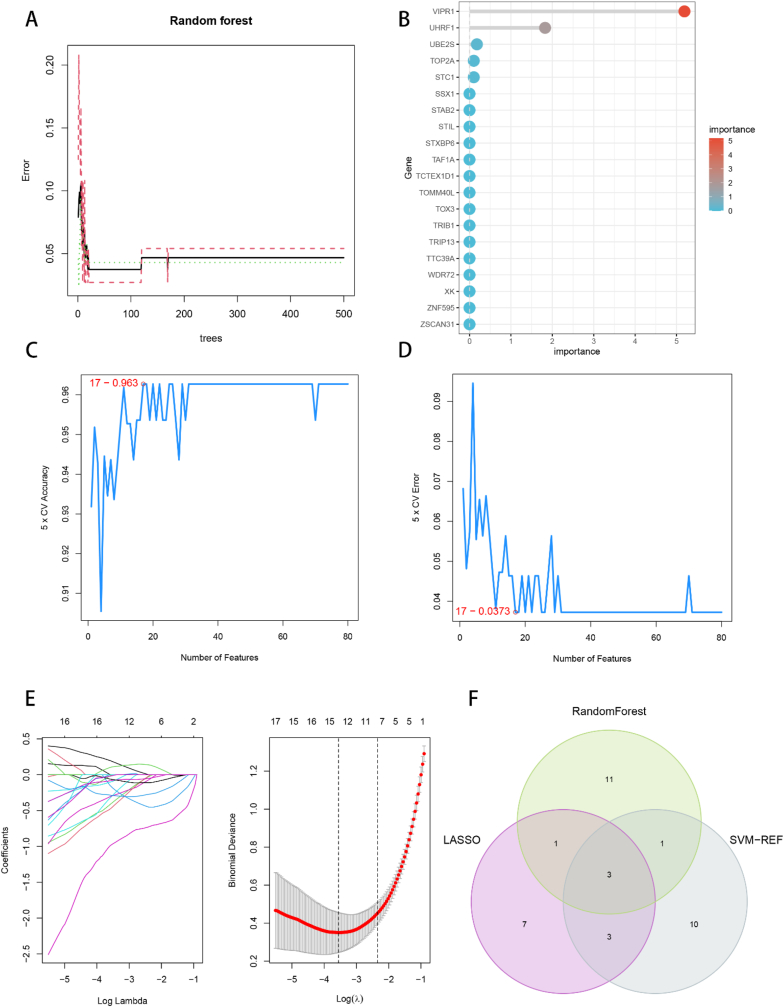
To further discern the feature genes linked to HBV-HCC, we employed three sophisticated machine learning techniques: random forest (RF), selected support vector machine recursive feature elimination (SVM-RFE), and least absolute shrinkage and selection operator (LASSO) logistic regression [[31], [32], [33]]. The RF algorithm was utilized to screen the feature genes, and the R package “randomForest” was leveraged to compute the number of decision trees and the error rate. The optimal number of decision trees was chosen when the error rate stabilized, and the importance of the candidate genes was ranked. We selected 16 genes with importance scores exceeding 1.5 for RF algorithm screening. The SVM-RFE algorithm screened the feature genes through the R packages “e1071″ and “caret” to train the candidate genes, and computed the highest correct rate and the lowest error rate of 17 for cross-validation. Consequently, the top 17 genes were regarded as the feature genes of SVM-RFE. The LASSO algorithm was employed to filter the feature genes. We utilized the R package “glmnet” to identify the optimal penalty parameter λ for 10-fold cross-validation, and obtained 14 feature genes. We obtained the three feature genes related to HBV-HCC by intersecting the three machine learning feature genes utilizing the Venn diagram.
2.8. GeneMANIA constructs feature gene interaction networks
To elucidate potential biological processes and pathways related to HBV-HCC, a co-expression network of feature genes and their corresponding similar genes was constructed using GeneMANIA (http://www.genemania.org/). The intrinsic association of genes was analyzed to gain insight into their potential biological roles.
2.9. Construction of a diagnostic model for HBV-HCC
An ANN model was developed as a diagnostic tool for HBV-HCC. To build the ANN model, “GeneScore” was used to score each sample of feature genes within the training set [34]. The scoring rule was based on the median value of the expression of all samples of each feature gene. For genes upregulated in the HBV-HCC group, a score of 1 was assigned above the median and 0 below, while genes downregulated in the HBV-HCC group had expression levels that scored 0 above the median and 1 below. The training set feature gene “GeneScore” scores were utilized as input data for the ANN model, which was constructed using the “neuralnet” package in R. The ANN model comprises of an input layer, a hidden layer, and an output layer. The input layer encompasses the feature gene “GeneScore”, the hidden layer represents the gene weight of the feature gene, and the output layer represents the sum of the product of the gene score and the gene weight (output layer calculation formula: Neural HBV-HCC = Σ(Gene Score*Gene Weight)). To assess the predictive performance of the ANN model, we generated a receiver operating characteristic (ROC) curve of the model utilizing the “PROC” package in R, which was measured by the area under the ROC curve (AUC) value. To validate the stability and generalizability of the ANN model built with the feature genes, we employed GSE55092 for validation. The validation set was scored by “GeneScore” in the same manner, and the ROC curve was plotted to obtain the AUC. The stability and generalizability of the ANN model could be determined by the size of the AUC.
2.10. Construction and evaluation of nomogram
In order to provide a more comprehensive evaluation of HBV-HCC from a clinical perspective, we developed a nomogram related to HBV-HCC that utilizes the R package “Rms” and feature genes. Each element of the nomogram is assigned a corresponding scoring scale, and the scores are summed to calculate the total score. Calibration curves and decision curve analysis (DCA) were utilized to visualize the results and evaluate the predictive performance of the nomogram.
2.11. Immune infiltration analysis
In order to investigate the immune microenvironment differences between HBV and HBV-HCC, we merged the expression matrices of both training and validation sets. The gene expression matrix was then utilized to estimate immune cell infiltration and immune function expression via the CIBERSORT algorithm. We quantified the proportion of the 22 immune cells infiltrating HBV and HBV-HCC samples using a stacked histogram generated with the R package “ggplot2”. We also generated a correlation heat map using the R package “corrplot” to examine the relationship between the 22 immune cells. To compare differences in immune cell infiltration and immune function expression across groups, we generated box plots to visualize these differences between HBV and HBV-HCC. Additionally, to investigate the association between immune cells and feature genes, we used “ggplot2″ to generate visualizations of the immune infiltration analysis. Finally, we performed immune infiltration analysis of HBV-HCC after subtyping, using the combined expression matrix of HBV-HCC samples from the training and validation sets as input data.
2.12. Consensus clustering of HBV-HCC
The expression matrices of HBV-HCC samples from the GSE121248 and GSE55092 datasets were integrated. The merged expression matrix was then used to extract the gene expression of HBV-HCC samples corresponding to the feature genes. Subsequently, the subtyping of HBV-HCC based on the expression of the feature genes was carried out using the “ConsensusClusterPlus” R package. To determine the optimal number of subtypes, the consensus matrix, consensus cumulative distribution function (CDF) and trace map were plotted, and the optimal number of clusters “k" was determined by assessing the cleanliness of the blank area between the blue modules and the relative change between the CDF trace map and the trace map. The scatter plots of samples between different subtypes were created using the “ggplot2″ R package to verify the effectiveness of the consensus clustering and to observe if the scatter plots were distinguishable between subtypes. The expression and expression differences of the feature genes among different subtypes were investigated using heat maps and box plots.
2.13. Gene set variation analysis (GSVA) of different subtypes of HBV-HCC
The analysis utilized the “c2. cp.kegg.symbols” gene set from the Molecular Feature Database (MSigDB) as a reference set. GSVA analysis was conducted on the various HBV-HCC subtypes to determine the absolute degree of their enrichment in different pathways. Pathway enrichment analysis was performed to shed light on the possible biological functions and pathways associated with the subtyping of HBV-HCC.
2.14. Statistical analysis
All statistical analyses in this study were executed through utilization of R software version 4.2.2. In this study, statistical significance was determined to exist when p-value was less than 0.05 and the false discovery rate (FDR) was less than 0.05. To compare the proportion of immune cell infiltration and levels of immune function expression between distinct groups, the application of the Wilcoxon rank sum test was utilized.
3. Results
3.1. Identification of differential genes between HBV and HBV-HCC
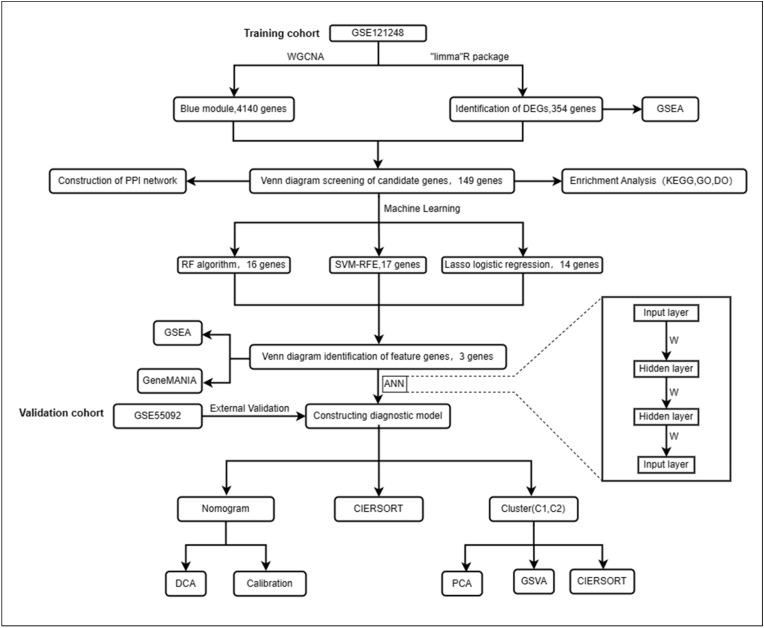
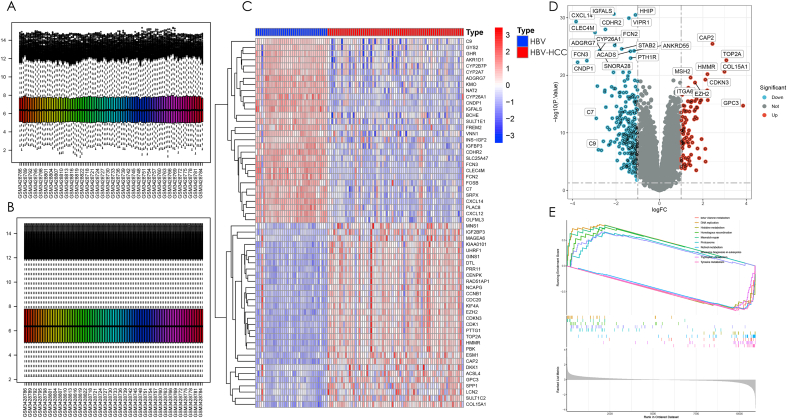
Fig. 1 depicts the study's workflow. The GSE121248 dataset was employed to identify DEGs between HBV and HBV-HCC. Initially, the expression matrix of the GSE121248 dataset was merged and batch-corrected to eliminate batch effects, and this process was visually demonstrated before and after correction (Fig. 2A and B). The batch-effect-corrected expression matrix was analyzed and visualized using heat and volcano plots (Fig. 2C and D), revealing a total of 344 genes with significant differences in expression between HBV patients and HBV-HCC patients (|log2FC|>1,FDR<0.05). Among these genes, 94 were up-regulated, and 250 were down-regulated. To determine the degree of enrichment of the screened DEGs in different functional pathways, GSEA for functional enrichment analysis was executed on the expression matrix following batch effect correction. The most and least significant enrichment pathways were reported as the top five pathways (Fig. 2E). The outcomes revealed that differential genes were enriched in pathways related to DNA replication, Homologous recombination, Mismatch repair, Proteasome, and Ribosome biogenesis in eukaryotes, all of which are related to genetic material replication and cell cycle regulation. The remaining five down-regulated pathways were all associated with the metabolism of substances linked to genetic material. Based on the enrichment outcomes, these differential genes may have a close relationship with the progression to HBV-HCC in HBV patients.
Fig. 1.
The workflow diagram of this study.
Fig. 2.
Identification and enrichment analysis of differential genes between HBV and HBV-HCC. (A) Raw data of the training set without batch correction. (B) Expression matrix of the training set after batch correction to remove batch effects. (C) Heat map of differential genes between HBV and HBV-HCC. The blue module represents HBV and the red module represents HBV-HCC, and the red color in the heat map represents up-regulated gene expression and blue color represents down-regulated gene expression. (D) Volcano plot of differential genes. Expression is up-regulated when |log2FC|>0,p < 0.05 and gene expression is down-regulated when |log2FC|<0,p < 0.05. Differential genes were screened by |log2FC|>1,p < 0.05, blue indicates down-regulation and red indicates up-regulation. (E) GSEA enrichment analysis of differential genes was performed, and the five pathways with the strongest enrichment significance were selected for display with the five weakest pathways.
3.2. WGCNA construction of a gene co-expression network between HBV and HBV-HCC
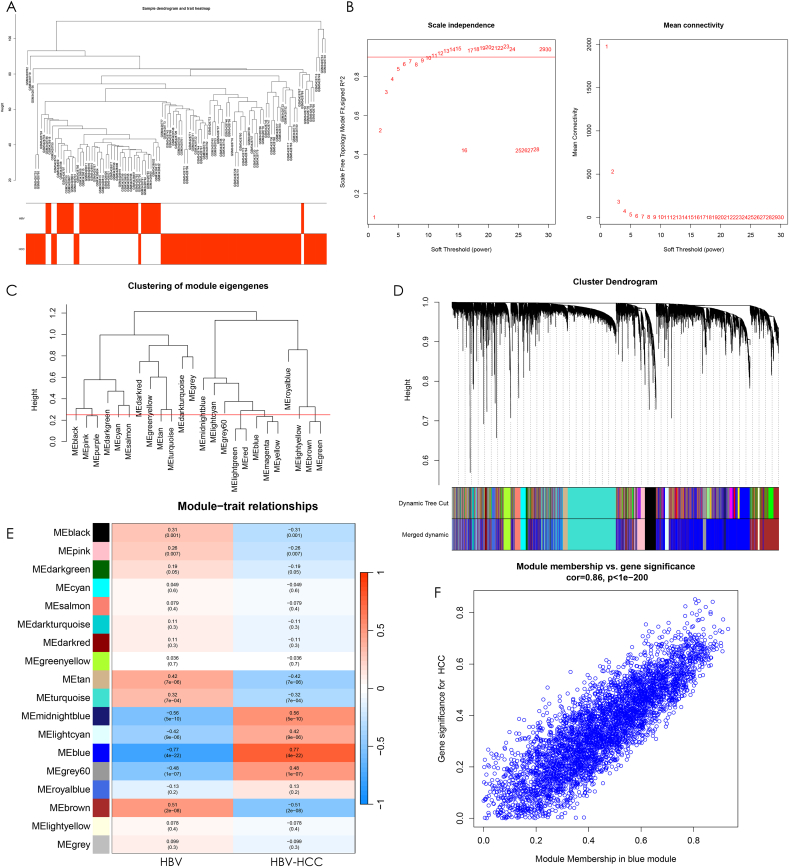
In order to identify co-expressed genes that are associated with HBV-HCC, a Weighted Gene Co-expression Network Analysis (WGCNA) was performed on the dataset. Firstly, a sample clustering tree with trait heat map display was conducted for HBV and HBV-HCC (Fig. 3A). Outliers were detected and removed. To ensure that the co-expression network had good average connectivity, a scale-free topology fit index R^2 > 0.9 was set, and the network displayed good average connectivity with a soft threshold β > 9 (Fig. 3B). A clustering height of 0.25 was set, and strongly associated modules were merged to produce a module clustering tree for the genes. The results showed that there were 18 modules with clustering height greater than 0.25 after clustering, which were used for the next step of the study (Fig. 3C). The 24 original modules were visually compared before and after merging of strongly associated modules, along with the sample clustering tree, which showed the 18 merged modules (Fig. 3D). To identify the gene co-expression modules that were strongly associated with HBV-HCC, a module-disease correlation heat map was plotted (Fig. 3E). The outcomes demonstrated that the blue module, composed of 4140 genes, exhibited the most robust correlation among the 18 modules with HBV-HCC (r = 0.77, p = 4e-22). In the MM versus GS scatter plot of the blue module for HBV-HCC, it was observed that the blue module displayed a high correlation of 0.86 and statistical significance (p-value < 1e-200) with HBV-HCC genes (Fig. 3F). All genes in this module will be used in the next step of the study.
Fig. 3.
Construction of gene co-expression network. (A) Cluster tree with heat map display for all samples in the training set, and the cluster tree samples corresponding to the red heat map belong to this clinical trait. (B) The left panel is set when the scale-free topological fit index R^2 = 0.9, and the best soft threshold β = 9 is chosen to obtain the best average connectivity of the co-expression network on the right panel. (C) The clustering height was set to 0.25 and the modules were clipped. The modules with high gene similarity were merged together to get 18 modules. (D) Sample clustering tree with the modules before and after merging, Dynamic Tree Cut for the original modules and Merged dynamic for the result after merging the strongly associated modules. (E) Heat map of correlations between modules and clinical traits. Red indicates positive correlation, blue indicates negative correlation, and the darker the color, the stronger the correlation. The numbers in parentheses are the p-values of correlations between modules and traits to test whether they are statistically significant with each other. The numbers above the brackets indicate the magnitude of correlation between modules and traits. (F) Scatter plot between blue module affiliation and HBV-HCC gene significance with a correlation between each other of cor = 0.86,p < 1e-200.
3.3. Identification and functional enrichment analysis of candidate genes and PPI network construction
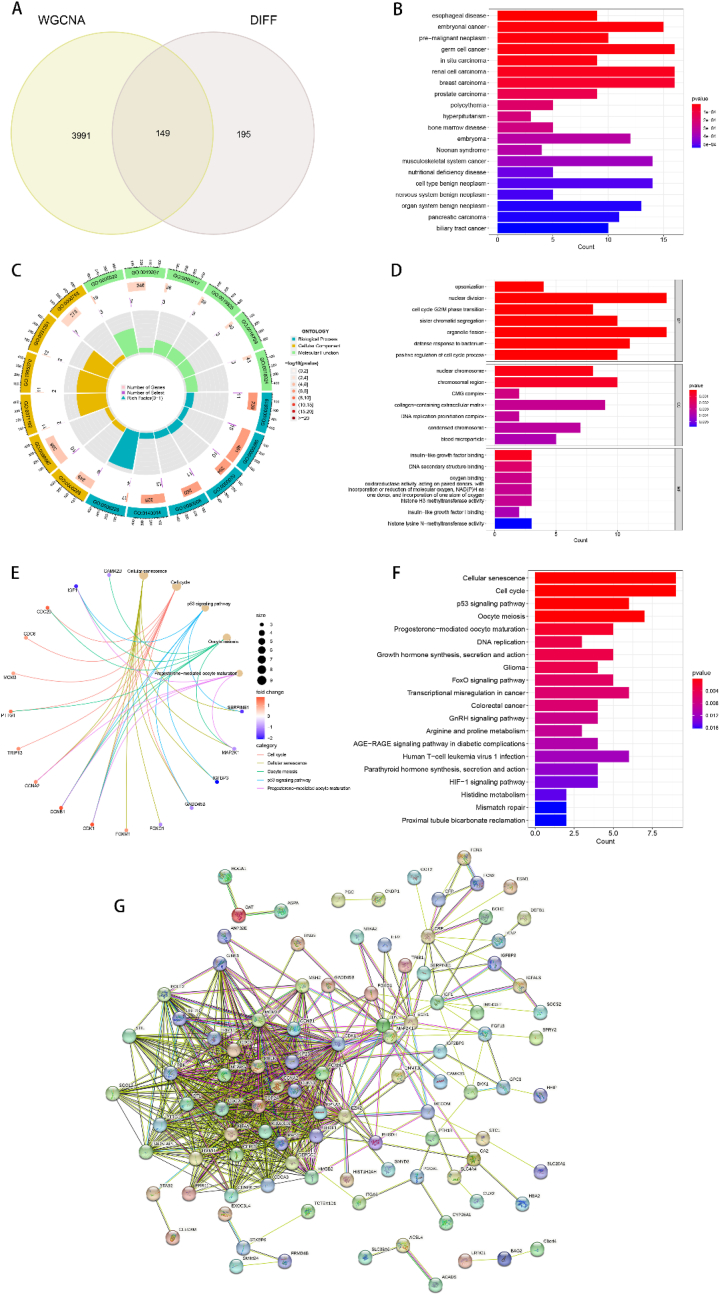
To further discern the feature genes distinguishing HBV from HBV-HCC, we utilized a Venn diagram to determine the intersections of DEGs with significantly correlated co-expression module genes. Subsequently, 149 genes were identified as candidate genes for further investigation (Fig. 4A). To elucidate the role of candidate genes in the progression from HBV to HBV-HCC, we conducted three enrichment analyses (DO, GO, KEGG) on the screened candidate genes. To comprehend the involvement of candidate genes in various diseases, we conducted DO enrichment analysis (Fig. 4B), revealing that candidate genes were predominantly enriched in a variety of cancers, implying their role in cancer development and formation. To obtain a more comprehensive comprehension of the biological mechanisms and signaling pathways associated with potential genes, we executed GO and KEGG enrichment analyses. GO enrichment analysis demonstrated that candidate genes were mainly involved in biological processes such as chromosome replication, segregation, and cell cycle regulation. Regarding molecular functions, candidate genes were mainly associated with functions related to chromosome and DNA replication, which are imperative in the formation of genetic material. As for cellular components, candidate genes were predominantly involved in the formation of cellular components regulating cell formation, metabolism, growth, and differentiation (Fig. 4C and D). The KEGG enrichment analysis results indicated that the candidate genes were primarily linked with the process of cellular senescence, cell cycle regulatory signaling pathways, and various cancer-related regulatory pathways (Fig. 4E and F). Additionally, we performed PPI network construction for the candidate genes, utilizing a medium confidence level of 0.4 as the minimum interaction score. Disconnected nodes were concealed to analyze the PPI, and the PPI network was found to contain 101 nodes and 624 edges (Fig. 4G).
Fig. 4.
Identification and analysis of candidate genes. (A) Wayne diagram of 149 intersecting genes obtained from DEGs and co-expressed genes with the strongest association with HBV-HCC as candidate genes. (B) Histogram of DO enrichment analysis of candidate genes. (C) Histogram of GO enrichment analysis of candidate genes, the outermost circle is the GO sequence number, the second circle is the number of genes enriched by that GO, the third circle is the number of select, and the last circle is the corresponding GO enrichment factor.(D) Histogram of GO enrichment analysis of candidate genes. (E) KEGG network diagram of the correlation between some candidate genes and biological processes. (F) Histogram of KEGG enrichment analysis of candidate genes. (G) PPI network of candidate genes, the circles indicate protein nodes, and the lines between each other indicate the existence of interrelationship between two proteins, the PPI network has 101 nodes and 624 edges.
3.4. Machine learning algorithms to screen for feature genes
To further refine the list of candidate genes and identify the feature genes specific to HBV-HCC, our study employed three machine learning algorithms. The RF algorithm utilized the number of decision trees in combination with the error rate, which stabilized around 200 trees (Fig. 5A). The candidate genes were then ranked by importance and displayed in a graph (Fig. 5B), with 16 genes having relative importance greater than 1.5 selected as feature genes. The SVM-RFE algorithm calculated and ranked the average risk of candidate genes, achieving the highest accuracy rate (Fig. 5C) and lowest error rate (Fig. 5D) when selecting the top 17 genes. Therefore, 17 feature genes were screened by the SVM-RFE algorithm. Finally, using LASSO logistic regression, 14 genes were selected as feature genes from the statistically significant variables (Fig. 5E). The intersection of the three machine learning algorithms was obtained using a Venn diagram, resulting in the identification of HHIP, CXCL14, and CDHR2 as the three feature genes specific to HBV-HCC (Fig. 5F).
Fig. 5.
Machine learning screening of feature genes. (A) Combination of number of decision trees and error rate of random forest algorithm. (B) Random forest calculates the top 20 genes for gene importance for ranking. (C, D) support vector machine recursive feature elimination (SVM-RFE) algorithm to screen biologic feature genes, and the point with the lowest accuracy and error rate is used as the number of feature genes screened by SVM-RFE. (E) LASSO algorithm for feature gene screening. (F) Wayne diagram to obtain the feature genes screened by three machine learning algorithms as the feature genes for HBV-HCC.
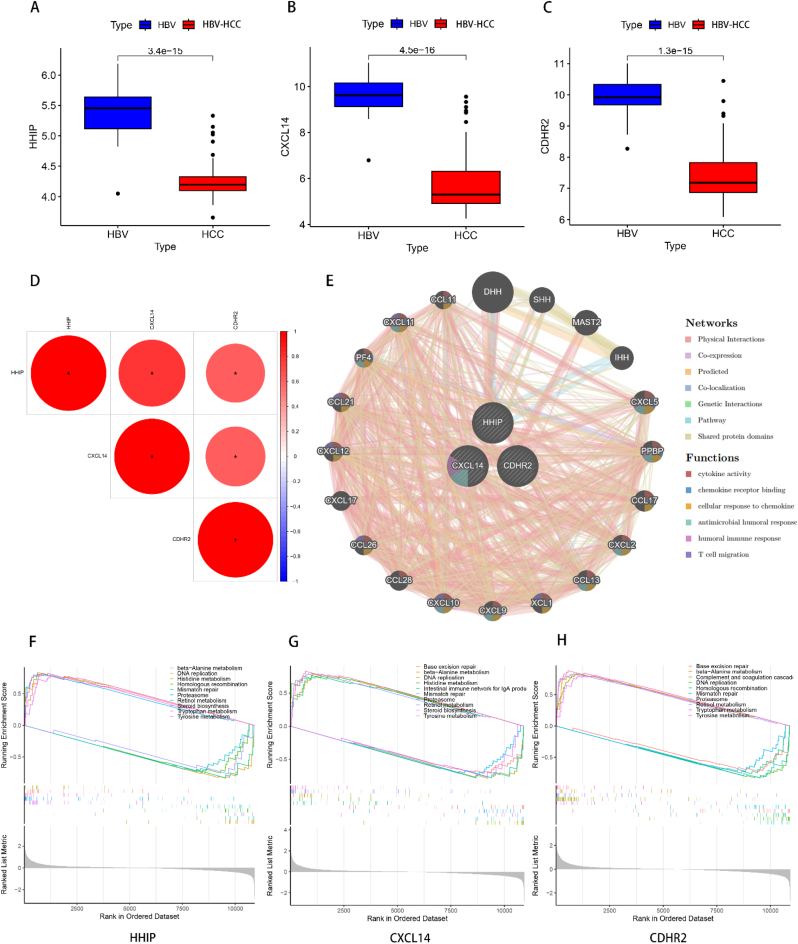
3.5. Feature gene expression differences, correlation and enrichment analysis
To further comprehend the variations in the expression of the three examined feature genes between HBV and HBV-HCC, we illustrated the expression of the feature genes among the distinct groups. The outcomes demonstrated that the expression of the 3 feature genes, namely HHIP, CXCL14, and CDHR2, was reduced in the HBV-HCC group as opposed to the HBV group, and the differences were all statistically significant (Fig. 6A–C). In order to understand the correlation among the feature genes, we executed a gene correlation test. The results displayed positive and statistically significant correlations among all three feature genes (Fig. 6D), indicating a significant functional resemblance among the three genes. The PPI network of the feature genes was established utilizing GeneMANIA to predict the genes with analogous functions to the feature genes. The findings revealed that the 3 feature genes were centrally located, while 20 analogous genes were peripherally located (Fig. 6E). Their functions were primarily enriched in cytokine activity, chemokine receptor binding, cellular response to chemokine, antimicrobial humoral response, humoral immune response, and T cell migration, implying that the feature genes and their analogs are implicated in microbial clearance and the corresponding immune response. We performed a single-gene GSEA enrichment analysis to investigate the role of each feature gene in disease progression. The results illustrated that all three feature genes were upregulated in various metabolic pathways, including beta-Alanine metabolism, Retinol metabolism, and Tyrosine metabolism, all of which are related to liver metabolism. In contrast, all three feature genes were enriched for downregulation in DNA replication, Homologous recombination, Mismatch repair, and Proteasome, which are all pathways associated with replication of cytogenetic material or protein regulation of the cell cycle (Fig. 6F–H). Nevertheless, upon analyzing the differences in gene expression between HBV and HBV-HCC, the expression of these feature genes was lower in the HBV-HCC than in the HBV. Therefore, the expression of these genes was anticipated to have a role in HBV-HCC, despite the results of the GSEA enrichment analysis.
Fig. 6.
Feature gene correlation analysis. (A–C) Box line plots of differential expression of HHIP, CXCL14 and CDHR2 in HBV versus HBV-HCC. The range of sizes of the box line plots indicates the range of their gene expression, while the black line in the plot indicates the mean of their expression. The values between the two box line plots are p-values, and the differences are statistically significant when p < 0.05. (D) Heat map of correlation between the three feature genes. Red represents positive correlation with each other, blue represents negative correlation with each other, and larger circles with redder colors represent greater correlation. *p < 0.05.(E) GeneMANIA network diagram of the three feature genes, the 20 genes in the outer circle are the similar genes of the feature genes, and the interconnected lines indicate the correlation between them, and the different color modules in the genes represent the different functional pathways enriched. (E–F) Single gene GSEA analysis of HHIP, CXCL14 and CDHR2, respectively, and the top five pathways with the highest and lowest enrichment significance were selected for display.
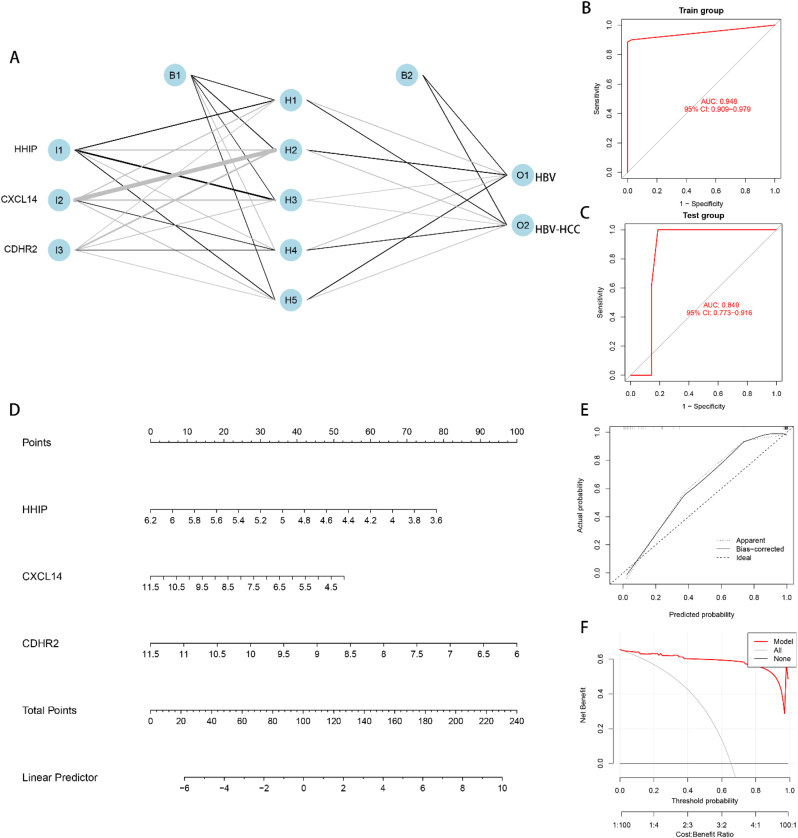
3.6. Construction and validation of artificial neural network (ANN) models, construction of nomogram models
The ANN diagnostic model was constructed utilizing the GSE121248 dataset combined with the screened 3 feature genes. The model comprised an input layer, a hidden layer, and an output layer (Fig. 7A). To validate the constructed model, GSE55092 dataset was utilized. The ROC curve analysis indicated that the constructed model had an AUC of 0.948 (95% CI = 0.909–0.979) (Fig. 7B). These results demonstrated that the ANN model had good predictive performance using HHIP, CXCL14, and CDHR2 as feature genes. To further validate the model, the dataset GSE55092 was used, and the ROC curve analysis was performed, demonstrating an AUC of 0.849 (95% CI = 0.773–0.916) (Fig. 7C). This validated the stability and generalizability of the model. To assess the risk of HBV conversion to HBV-HCC in a clinical setting, a nomogram model was constructed using the 3 feature genes (Fig. 7D). A scoring scale was assigned to each element in the nomogram, and the scores were added to predict the risk of morbidity based on the expression of each sample. The predictive performance of the nomogram was assessed using calibration curves, indicating that the nomogram exhibited favorable predictive capability for HBV-HCC (Fig. 7E). The decision curve analysis (DCA) showed that the nomogram model had the best prediction for the conversion of HBV to HBV-HCC at a threshold value of 0–1 (Fig. 7F).
Fig. 7.
Construction and validation of ANN model and nomogram model. (A) ANN model constructed using feature genes, containing input layer, hidden layer and output layer. (B) Training set ROC curve with AUC = 0.948, 95% CI = 0.909–0.979, used to illustrate whether the model has good prediction performance. (C) Validation set ROC curve, AUC = 0.849, 95% CI = 0.773–0.916, used to demonstrate whether the stability and generalization of the model are good. (D) Nomogram plot of the characteristic gene construction, each element followed by a scoring scale. The scores of each element are summed to obtain a total score to predict the risk of disease. (E) Calibration curve for the evaluation of nomogram prediction performance. The higher the overlap between the solid and dashed lines and the closer the diagonal line, the better the performance. (F) Decision curve analysis (DCA), which compares the clinical benefit between the nomogram model and other diagnostic indicators. the higher the AUC, the higher the clinical benefit in the range of possible thresholds from 0 to 1.
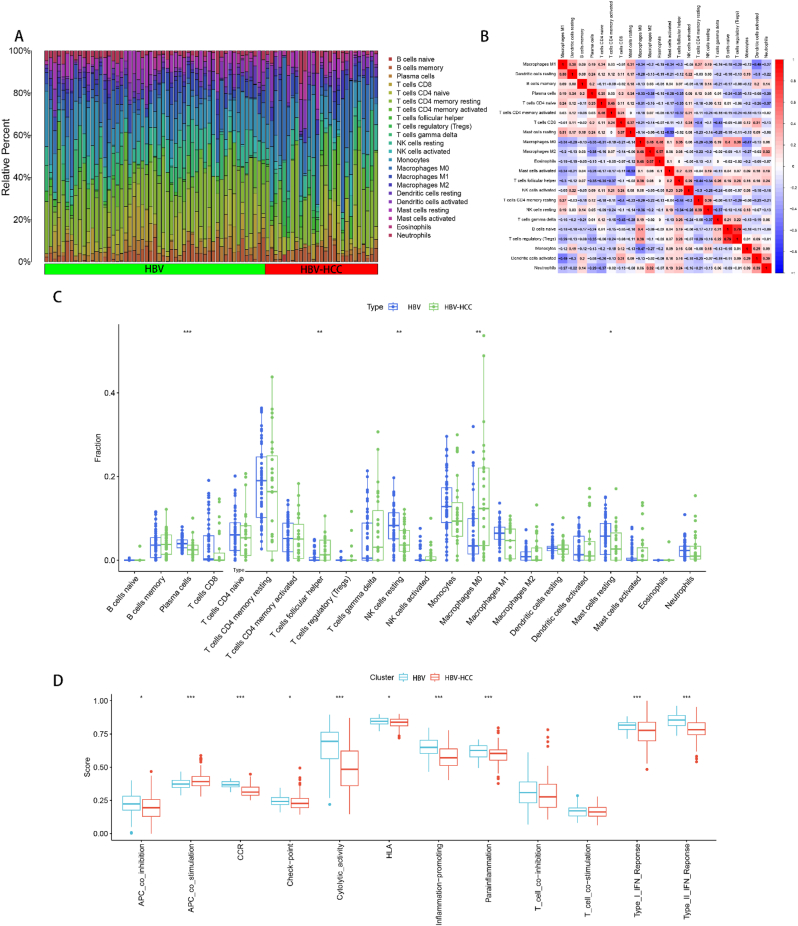
3.7. Analysis of immune cell and immune function infiltration between HBV and HBV-HCC
We combined the training and validation sets to acquire the merged expression matrix, and employed the CIBERSORT algorithm to scrutinize immune infiltration between HBV and HBV-HCC. Stacked histograms were used to visualize the percentage of 22 immune cells in each sample (Fig. 8A). By generating a heat map of infiltration correlation between 22 immune cells, we identified a significant positive correlation between B cells naive and T cells regulatory (Tregs), Macrophages M2 and Eosinophils. Additionally, we discovered a significant negative correlation between Mast cells resting and Mast cells activated, Macrophages M1 and Dendritic cells activated (Fig. 8B). Differential expression of 22 immune cell infiltrates was presented in box plots. T cells follicular helper and Macrophages M0 expression were found to be higher in the HBV-HCC group than in the HBV group, while Plasma cells, NK cells resting and Mast cells resting were higher in the HBV group than in the HBV-HCC group (Fig. 8C). We performed 12 immune function infiltration analyses to further investigate the immune infiltration between HBV and HBV-HCC. The results indicated that the expression level of APC_co_stimulation was greater in the HBV-HCC group compared to the HBV group. On the other hand, APC_co_inhibition, CCR, Check-point, Cytolytic_activity, HLA, Inflammation-promoting, and Parainflammation, Type_I_IFN_Reponse and Type_II_IFN_Reponse were expressed at higher levels in the HBV group than in the HBV-HCC group (Fig. 8D). All discrepancies were statistically significant with a p-value of less than 0.05.
Fig. 8.
Immune infiltration analysis between HBV and HBV-HCC. (A) Stacked histogram of the infiltration ratio of 22 immune cells between HBV and HBV-HCC in each sample. (B) Heat map of correlation between 22 immune cells, red indicates positive correlation, blue indicates negative correlation, and the values in the heat map are the magnitude of correlation between the corresponding immune cells. (C) Box plot of the difference in infiltration of 22 immune cells between HBV and HBV-HCC. (D) Box-line plot of the difference in expression of 12 immune functions between HBV and HBV-HCC. *p < 0.05, **p < 0.01, ***p < 0.001.
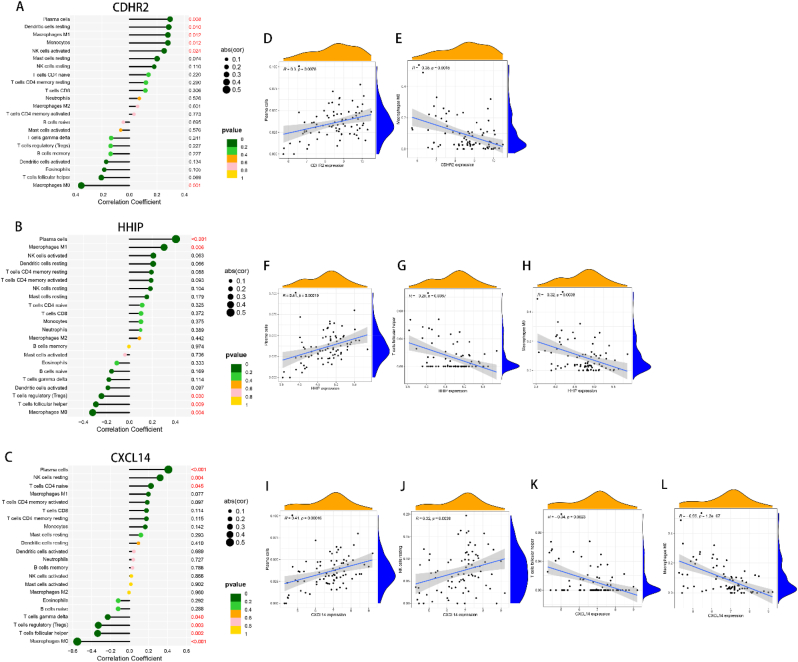
3.8. Correlation analysis of immune cell infiltration of feature genes
To investigate the correlation between the feature genes and immune cell infiltration, we presented heat maps depicting the correlations of CDHR2, HHIP, and CXCL14 with 22 immune cell types based on size and statistical significance. CDHR2 was positively correlated with Plasma cells, Dendritic cells resting, Macrophages M1 Monocytes, and NK cells activated, while being negatively correlated with NK cells activated (Fig. 9A). HHIP was positively correlated with Plasma cells and Macrophages M1, while being negatively correlated with T cells regulatory (Tregs), T cells follicular helper, and Macrophages M0 (Fig. 9B). CXCL14 was positively correlated with Plasma cells, NK cells resting, and T cells CD4 naïve, while being negatively correlated with T cells gamma delta, T cells regulatory (Tregs), T cells follicular helper, and Macrophages M0 (Fig. 9C). T cells follicular helper and Macrophages M0 expression levels were higher in the HBV-HCC group than in the HBV group, while Plasma cells, NK cells resting, and Mast cells resting were higher in the HBV group than in the HBV-HCC group (Fig. 8C). Scatter plots were used to visualize the immune infiltration of characteristic genes in the four immune cells that differed between HBV and HBV-HCC and were statistically significant. CDHR2 correlated with Plasma cells r = 0.3, p = 0.0078 (Fig. 9D), and negatively correlated with Macrophages M0 r = −0.35, p = 0.0015 (Fig. 9E). HHIP correlated with Plasma cells r = 0.41, p = 0.00019 (Fig. 9F), while being negatively correlated with T cells follicular helper correlation r = −0.29, p = 0.0087 (Fig. 9G), and Macrophages M0 correlation r = −0.32, p = 0.0038 (Fig. 9H). CXCL14 correlated with Plasma cells r = 0.41, p = 0.00016 (Fig. 9F), NK cells resting correlation r = 0.32, p = 0.0036 (Fig. 9F), and negatively correlated with T cells follicular helper correlation r = −0.34, p = 0.0023 (Fig. 9F) and Macrophages M0 correlation r = −0.55, p = 1.2e-07 (Fig. 9F).
Fig. 9.
Feature gene immune cell infiltration analysis. (A–C) Heat map of CDHR2, HHIP and CXCL14 with the degree of infiltration of 22 immune cells, respectively, with p < 0.05 being statistically significant. (D–L) Scatter plot visualization of immune cells with differential and statistically significant expression of feature genes between HBV and HBV-HCC.
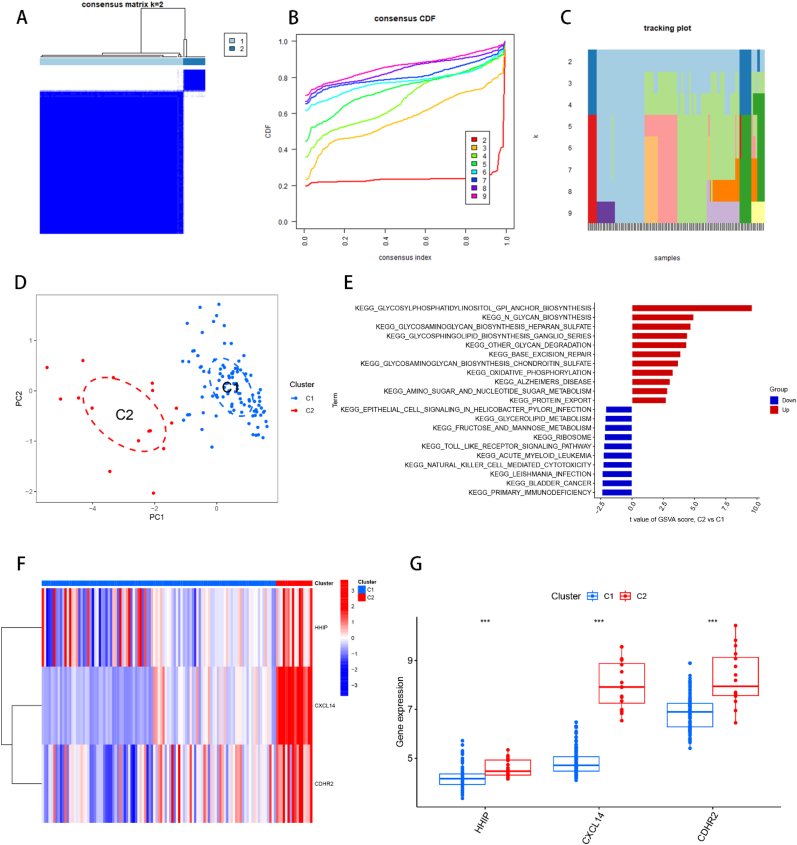
3.9. Consensus clustering of the identification of two subtypes of HBV-HCC
To classify the subtypes of HBV-HCC, we obtained an expression matrix by combining the HBV-HCC samples from both the training and validation sets. Subtyping was performed uitilizing the “ConsensusClusterPlus” R package based on the gene expression profiles of three feature genes: HHIP, CXCL14, and CDHR2. Through analysis of the consensus matrix plot, the consensus distribution function (CDF) plot, and the relative modification of the area under the CDF curve, we determined that the optimal subtyping number was k = 2, and the HBV-HCC samples were classified into C1 and C2 subtypes (Fig. 10A and B). Tracing plots from k = 2 to k = 9 were also generated to confirm the successful subtyping at k = 2 (Fig. 10C). PCA plots were plotted for both subtypes, which revealed that the two subtypes could be distinguished by clustering among the scatter plots (Fig. 10D). The Glycosylphosphatidylinositol (GPI)-anchor biosynthesis pathway played an important role in the subtyping of HBV-HCC, as demonstrated by the GSVA analysis of the two subtypes (Fig. 10E). The findings demonstrated that the expression levels of HHIP, CXCL14, and CDHR2 were greater in the C2 subtype as compared to the C1 subtype (Fig. 10F and G).
Fig. 10.
Consensus clustering of HBV-HCC samples. (A) Consensus matrix plot, the cleaner the blank area between the blue modules indicates the more successful analysis. (B) Cumulative distribution function (CDF) plot of consensus clustering, showing the relative change of consensus index from k = 2 to k = 9 with the change of CDF value, and the k value of the curve with the most stable change is the optimal fractal number. (C) Trace plot of k = 2 to k = 9. (D) PCA plot of HBV-HCC samples. The scatter plot allows visualization of the characteristic genes that classify HBV-HCC into two subtypes, C1 and C2. (E) GSVA analysis between C1 and C2, red represents the up-regulated functional pathway and blue represents the down-regulated functional pathway. (F) Heat map between C1 and C2 and feature genes, red indicates up-regulated gene expression and blue indicates down-regulated gene expression. (G) Box line plot of the difference in expression of feature genes between C1 and C2 of two subtypes of HBV-HCC. *p < 0.05, **p < 0.01, ***p < 0.001.
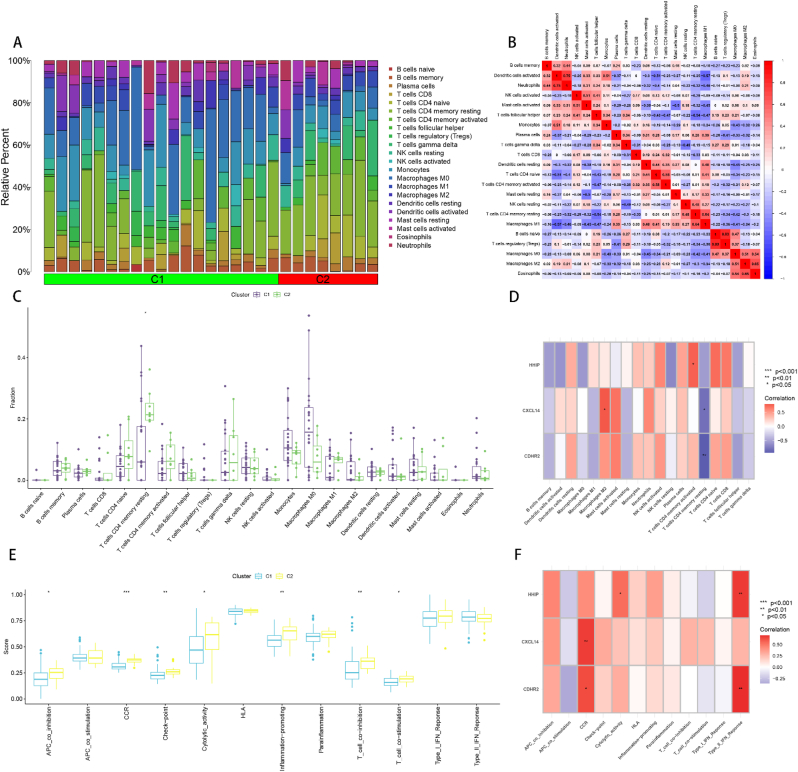
3.10. Immune infiltration analysis of two subtypes C1 and C2
In order to further investigate the role of the immune microenvironment in the differentiation of HBV-HCC subtypes, we conducted an analysis of immune infiltration in HBV-HCC samples categorized into two subtypes, C1 and C2. The proportion of 22 immune cells infiltrating in both C1 and C2 subtypes was determined using the CIBERSORT algorithm, and presented using stacked histograms (Fig. 11A). We then analyzed the correlation between the 22 immune cells using a correlation heat map, which revealed that B cells naive and T cells regulatory (Tregs), Dendritic cells activated and Neutrophils, Macrophages M2 and Eosinophils, T cells CD4 memory resting and Macrophages M1, T cells CD4 naive and T cells CD4 memory activated, NK cells activated and Mast cells activated, Macrophages M0 and Macrophages M2 were significantly and positively correlated. Additionally, we found a significant negative correlation between Macrophages M1 and Dendritic cells activated, T cells CD4 memory resting and T cells follicular helper (Fig. 11B). We performed a differential expression analysis of 22 immune cell infiltrates for both C1 and C2 subtypes, and observed that only the C2 subtype had a higher expression of T cell CD4 memory resting infiltration compared to the C1 subtype (Fig. 11C). Furthermore, we examined the expression of three feature genes in C1 and C2 subtypes. Our results indicated that, among the statistically significant results, HHIP was low in T cells CD4 memory activated, CXCL14 was low in Macrophages M2 and high in T cells CD4 memory resting, and CDHR was high in T cells CD4 memory resting (Fig. 11D). We also analyzed 12 immune function infiltrations between C1 and C2 subtypes, and identified that among the statistically significant results, APC_co_inhibition, CCR, Check-point, Cytolytic_activity, Inflammation-promoting, T_cell co inhibition and T_cell co-stimulation were higher in the C2 subtype compared to the C1 subtype (Fig. 11E). Finally, among the 12 immune functions among the 3 feature genes, we found that HHIP was highly expressed in Cytolytic_activity and Type_II_IFN_Response, CXCL14 was highly expressed in CCR, and CDHR2 was highly expressed in CCR and Type_II_IFN_Response (Fig. 11F).
Fig. 11.
Immune infiltration analysis between the two subtypes of HBV-HCC. (A) Stacked histogram of the infiltration ratio of 22 immune cells in each sample of the two subtypes C1 and C2. (B) Heat map of correlation between 22 immune cells, red indicates positive correlation, blue indicates negative correlation, and the values in the heat map are the magnitude of correlation between the corresponding immune cells. (C) Box plot of the difference in infiltration between C1 and C2 for the 22 immune cells. (D) Heat map of the correlation between characteristic genes and 22 immune cell infiltrates in HBV-HCC samples, red represents positive correlation and blue represents negative correlation. (E) Box-line plot of the difference in expression of 12 immune functions between C1 and C2. (F) Heat map of the correlation between feature genes and 12 immune cell infiltrates in HBV-HCC samples, red represents positive correlation and blue represents negative correlation. *p < 0.05, **p < 0.01, ***p < 0.001.
4. Discussion
Roughly 2 billion individuals across the world have contracted HBV, with 257 million individuals experiencing chronic hepatitis B infection [35]. HBV infection is a significant contributor to the development of HCC, and HBV-induced HCC represents a significant threat to global public health [36,37]. Due to the absence of overt early symptoms and specific biomarkers, a majority of patients with HCC receive a late-stage diagnosis, leading to a bleak prognosis [38]. As a result, the identification of validated diagnostic feature genes is imperative in enhancing the prognosis of patients with HBV-HCC. Furthermore, the chronic inflammatory response due to CHB infection constitutes a major risk factor for HCC [39], and this response can contribute to the progression and development of HCC by disrupting the immune microenvironment of the liver [40]. In this study, we endeavored to employ bioinformatics to screen for the diagnostic feature genes of HBV-HCC and investigate the role played by immune infiltration in the progression from HBV to HBV-HCC. To further explore the role of the immune microenvironment in the development of HBV-HCC, we utilized feature genes to subtype HBV-HCC by performing immune infiltration analysis of different HBV-HCC subtypes.
Our investigation ultimately identified three distinctive genes, HHIP, CXCL14, and CDHR2. HHIP encodes for hedgehog-interacting protein, which is generally down-regulated in various types of epithelial tumors [41]. It has been observed that the expression of HHIP is suppressed by promoter hypermethylation in HCC [42,43]. The up-regulation of miR-25-3p expression has been linked to the exosomes secreted by CHB patients, which suppresses HHIP expression by shifting miR-25-3p and promotes HCC development [44,45]. CXCL14, a member of the CXC chemokine family [46], acts as a tumor suppressor in specific cancer types and primes B cells, NK cells, dendritic cells, and monocytes to fight cancer [[47], [48], [49]]. CXCL14 expression is generally lower in HCC than in normal tissues, and overexpression can inhibit the migration, proliferation, invasion, and angiogenesis of HCC cells [50]. Moreover, CXCL14 expression is inhibited and reduced in advanced HBV-HCC tissues [47]. CDHR2, also known as procalcitonin 24 (PCDH24), belongs to the human non-classical calmodulin family [51]. CDHR2 is considered a potential tumor suppressor in hepatocellular carcinoma due to its role in epithelial cell contact inhibition and intercellular adhesion [52,53]. The expression of CDHR2 in HCC tissue samples was significantly lower than that in its paracancerous tissue samples [54]. The expression of these three feature genes was analyzed differently between HBV and HBV-HCC. The results showed that the expression of all three feature genes was lower in HBV-HCC than in HBV, and the correlation results showed a positive correlation between all three. GeneMANIA analysis of the feature genes showed that the feature genes were involved in microbial clearance and corresponding humoral immune processes with their analogs. However, since the expression of the feature gene was down-regulated in HBV-HCC, the microbial clearance and immune processes were correspondingly suppressed, leading to the development of HCC. Single-gene GSEA analysis of the feature genes showed that all three feature genes were enriched and up-regulated in various metabolic pathways such as beta-Alanine metabolism, Retinol metabolism, and Tyrosine metabolism, all of which are associated with liver metabolism [[55], [56], [57]]. In contrast, all three feature genes are enriched for the down-regulated processes of DNA replication, Homologous recombination, Mismatch repair, and Proteasome, pathways that are associated with the replication of cytogenetic material or protein regulation of the cell cycle [58]. Thus, the feature genes affect the course of HBV-HCC by regulating the replication of genetic material and the cell cycle.
The immune microenvironment of patients with HBV infection and HBV-HCC has been compared, patients with HBV-HCC exhibit a greater expression of M0 macrophages and T cells follicular helper compared to HBV patients, indicating a state of immune tolerance [59,60]. The immune tolerant microenvironment may promote immune response damage in both HBV infection and HCC, leading to metastasis and development of HCC [61]. The expression of plasma cells, NK cells resting and mast cells resting is higher in HBV patients, suggesting that chronic liver injury due to immune attack is less severe compared to HBV-HCC patients [62]. Results of immune cell infiltration analysis of the feature gene are consistent with the results of infiltration between HBV and HBV-HCC. Furthermore, the analysis of immune function between HBV and HBV-HCC showed that the APC_co_stimulation function is stronger in the HBV-HCC group, which may stimulate cellular immune processes and lead to more severe immunogenic damage to its cells [63,64]. In contrast, the HBV group showed upregulated expression of inflammatory response-related functions such as CCR, Inflammation-promoting, and Parainflammation, suggesting a gradual development of inflammatory response [65]. The Type_I_IFN_Reponse and Type_II_IFN_Reponse functions are central to the regulation of antiviral response and counteracting viral infection [66], which may have a suppressive effect on the progression of inflammatory response. Therefore, the combination of inflammatory and antiviral responses may lead to chronic inflammation in the HBV group. Chronic inflammatory response caused by HBV chronic infection is a significant risk factor for HCC [39], and it may promote the development and progression of HCC by disrupting the immune microenvironment of the liver [40]. Thus, the immune microenvironment of HBV contributes to the progression of HBV-HCC.
To gain further understanding of the differences between patients with HBV-HCC and provide potential directions for clinical management, our study employed three feature genes to classify HBV-HCC into two subtypes. Using HHIP, CXCL14, and CDHR2 as markers, we classified HBV-HCC into two subtypes, C1 and C2. Our analysis revealed that the C2 subtype showed higher expression levels of all three genes compared to the C1 subtype. Furthermore, gene set variation analysis (GSVA) showed that the C1 and C2 subtypes were significantly enriched in the glycosylphosphatidylinositol anchor (GPI-anchor) pathway. In contrast, Glypican-3 (GPC3), a member of the glypican family that attaches to the cell surface via the GPI-anchor, is expressed in approximately 70% of HCC cases [[67], [68], [69]]. Therefore, differential expression of the GPI-anchor pathway may be an important mechanism for subtype delineation in HBV-HCC.
Immune infiltration analysis was conducted to compare the immune microenvironments between C1 and C2 subtypes. Among the statistically significant findings, only the infiltration levels of resting memory CD4 T cells were higher in the C2 subtype than in the C1 subtype, indicating that cellular immunity in the C2 subtype was relatively quiescent compared to that in the C1 subtype. While the expression of feature genes was higher in the C2 subtype than in the C1 subtype, the correlation between feature genes and 22 immune cells resulted in lower expression of activated memory CD4 T cells and M2 macrophages and higher expression of resting memory CD4 T cells and CDHR in the C2 subtype. Thus, immune infiltration analysis of the feature genes showed that the C2 subtype had high levels of resting memory CD4 T cells and low levels of activated memory CD4 T cells and M2 macrophages. These results are consistent with the successful subtyping of the feature genes and indicate that the immune microenvironment differs between subtypes. Notably, there are differences in the immune microenvironment between C1 and C2 subtypes. For instance, in patients with HBV-HCC, T cells not only clear HBV but also cause damage to infected hepatocytes through their own specific immune response [70,71]. Therefore, patients with HBV-HCC of the C2 subtype may experience less damage than those of the C1 subtype.
Twelve immune function infiltrations were analyzed between C1 and C2 subtypes, and among the statistically significant results, APC co-inhibition, CCR, Check-point, Cytolytic activity, Inflammation-promoting, T cell co-inhibition, and T cell co-stimulation were higher in the C2 subtype than in the C1 subtype. Among them, the difference of T cell co-inhibition was greater than that of T cell co-stimulation, while the remaining immune functions were all associated with specific immunosuppression and inflammatory responses [65,72,73]. These 12 immune functions were also among the 3 feature genes, with HHIP highly expressed in cytolytic activity and type II IFN response, CXCL14 highly expressed in CCR, and CDHR2 highly expressed in CCR and type II IFN response. However, although the expression of characteristic genes was higher in the C2 subtype than in the C1 subtype, the C2 subtype was found to be highly expressed in these immune functions, which are associated with the development of inflammatory and antiviral responses [65,66].
Hence, the assessment from both viewpoints supports the notion that HBV-HCC patients belonging to the C2 subtype are likely to experience lower levels of impairment compared to those in the C1 subtype. Consequently, the immunological analyses conducted for both C1 and C2 subtypes reinforce the finding that patients with C2 subtype may experience less impairment than those with C1 subtype.
Despite using various bioinformatics and statistical methods to identify diagnostic feature genes associated with HBV-HCC and revealing the role of the immune microenvironment in the disease process, our study has some limitations that must be acknowledged. Firstly, our study is retrospective and requires further validation by future prospective studies. Secondly, the sample size used in our analysis was limited by the data obtained from the GEO database. Lastly, some potential mechanisms associated with HBV-HCC that were identified in our study may not have been fully elucidated, as existing research suggests.
5. Conclusion
Our study utilized a variety of bioinformatics analysis methods to identify and characterize three diagnostic feature genes, HHIP, CXCL14, and CDHR2, that are associated with HBV-HCC. The ANN and nomogram models we constructed based on these genes demonstrated strong predictive capability. Additionally, our analysis of the enrichment of the feature genes yielded valuable insights into the molecular mechanisms underlying the progression of HBV-HCC. By conducting immune infiltration analysis, we delved into the role played by the immune microenvironment in the progression of HBV-HCC. Furthermore, we successfully classified the subtypes of HBV-HCC based on the expression of the feature genes. Furthermore, our study provides evidence of potential differences in severity of impairment between patients with different subtypes of HBV-HCC based on immunological analyses. Our findings have important implications for the development of improved diagnostic and clinical treatments for HBV-HCC, with a focus on molecular mechanisms and the immune microenvironment.
Authors' contributions
SZ, CJ, HC and GT conceived the study. SZ and CJ drafted the manuscript. CJ, LJ, HQC, JH, ZX, JZ and LT performed the literature search and collected the data. SZ, LJ, HQC, JH, ZX and HC analyzed and visualized the data. HC, GY and GT helped with the final revision of this manuscript. All authors reviewed and approved the final manuscript.
Author statement
The datasets that were analyzed in this study can be found in the GEO (https://www.ncbi.nlm.nih.gov/geo/). However, if needed, the corresponding author of this study can provide access to the datasets used and/or analyzed in this research upon a reasonable request. All raw data is available at this website: https://www.jianguoyun.com/p/DRVHjCEQro7JCxiUy4EFIAA.
Funding
This study was supported by grants from the Luzhou Science and Technology Department Applied Basic Research Program (No: 2022-JYJ-145), the Sichuan Province Science and Technology Department of foreign (border) high-end talent introduction project (No: 2023JDGD0037), and Sichuan Provincial Medical Association (No: Q22027).
Declaration of competing interest
The authors confirm that the study was conducted with utmost transparency and without any commercial or financial relationships that could be perceived as a potential conflict of interest.
Contributor Information
Hao Chi, Email: chihao7511@gmail.com.
Guanhu Yang, Email: guanhuyang@gmail.com.
Gang Tian, Email: tiangang@swmu.edu.cn.
Data availability
I have shared the link to my data/code at the Attach File step.All raw data is available at this website: https://www.jianguoyun.com/p/DRVHjCEQro7JCxiUy4EFIAA
References
- 1.Li H., Wu Z., Chen J., Su K., Guo L., Xu K., Gu T., Jiang Y., Wang P., Zeng H., Chi H., He K., Han Y. Clinical and experimental medicine; 2022. External Radiotherapy Combined with Sorafenib Has Better Efficacy in Unresectable Hepatocellular Carcinoma: a Systematic Review and Meta-Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su K., Liu Y., Wang P., He K., Wang F., Chi H., Rao M., Li X., Wen L., Song Y., Zhang J., Gu T., Xu K., Li Q., Chen J., Wu Z., Li H., Huang W., Chen L., Tong J., Li H., Feng X., Chen S., Yang B., Jin H., Yang Y., Liu H., Yang C., Wu M., Xiong F., Peng K., Zhu L., Xu Y., Tang X., Tan Z., Luo X., Zheng H., Zhang Y., Guo L., Han Y. Heat-shock protein 90α is a potential prognostic and predictive biomarker in hepatocellular carcinoma: a large-scale and multicenter study. Hepatology international. 2022;16:1208–1219. doi: 10.1007/s12072-022-10391-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou Y., Guo C.G., Zhang M.M. Inhibition of human hepatocellular carcinoma tumor angiogenesis by siRNA silencing of VEGF via hepatic artery perfusion. Eur. Rev. Med. Pharmacol. Sci. 2015;19:4751–4761. [PubMed] [Google Scholar]
- 4.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 5.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 6.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 7.Truty M.J., Vauthey J.N. Surgical resection of high-risk hepatocellular carcinoma: patient selection, preoperative considerations, and operative technique. Ann. Surg Oncol. 2010;17:1219–1225. doi: 10.1245/s10434-010-0976-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau W.Y., Lai E.C., Lau S.H. The current role of neoadjuvant/adjuvant/chemoprevention therapy in partial hepatectomy for hepatocellular carcinoma: a systematic review. Hepatobiliary Pancreat. Dis. Int. : HBPD INT. 2009;8:124–133. [PubMed] [Google Scholar]
- 9.Zhang S., Yue M., Shu R., Cheng H., Hu P. Recent advances in the management of hepatocellular carcinoma. Journal of B.U.ON. : official journal of the Balkan Union of Oncology. 2016;21:307–311. [PubMed] [Google Scholar]
- 10.Ling C.Q., Liu Q., Li D.T., Yue X.Q., Hou F.G., Zhu D.Z., Yu C.Q., Chen Z., Zhai X.F., Yu Y. [Study of a qualitative diagnostic criterion for basic syndromes of traditional Chinese medicine in patients with primary liver cancer] Zhong xi yi jie he xue bao = Journal of Chinese integrative medicine. 2005;3:95–98. doi: 10.3736/jcim20050204. [DOI] [PubMed] [Google Scholar]
- 11.Shen K., Xi Z., Xie J., Wang H., Xie C., Lee C.S., Fahey P., Dong Q., Xu H. Guttiferone K suppresses cell motility and metastasis of hepatocellular carcinoma by restoring aberrantly reduced profilin 1. Oncotarget. 2016;7:56650–56663. doi: 10.18632/oncotarget.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei L., Owen D., Rosen B., Guo X., Cuneo K., Lawrence T.S., Ten Haken R., El Naqa I. A deep survival interpretable radiomics model of hepatocellular carcinoma patients. Phys. Med. : PM : an international journal devoted to the applications of physics to medicine and biology : official journal of the Italian Association of Biomedical Physics (AIFB) 2021;82:295–305. doi: 10.1016/j.ejmp.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei L., Simeth J., Aryal M.P., Matuszak M., Haken R.K.T., Cuneo K., Lawrence T.S., Cao Y. The effect of stereotactic body radiation therapy for hepatocellular cancer on regional hepatic liver function. Int. J. Radiat. Oncol. Biol. Phys. 2023;115:794–802. doi: 10.1016/j.ijrobp.2022.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su K., Guo L., Ma W., Wang J., Xie Y., Rao M., Zhang J., Li X., Wen L., Li B., Yang X., Song Y., Huang W., Chi H., Gu T., Xu K., Liu Y., Chen J., Wu Z., Jiang Y., Li H., Zeng H., Wang P., Feng X., Chen S., Yang B., Jin H., He K., Han Y. PD-1 inhibitors plus anti-angiogenic therapy with or without intensity-modulated radiotherapy for advanced hepatocellular carcinoma: a propensity score matching study. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.972503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Guo L., Su K., Li C., Jiang Y., Wang P., Chen J., Wu Z., Xu K., Gu T., Zeng H., He K., Chi H., Zhao W., Han L., Han Y. Construction and validation of tace therapeutic efficacy by alr score and nomogram: a large, multicenter study. J. Hepatocell. Carcinoma. 2023;10:1009–1017. doi: 10.2147/JHC.S414926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benarji K.A., Anitha A., Suresh B., Aparna V., Praveena A., Penumatsa L.A. Knowledge and attitude of dental students toward hepatitis B virus and its vaccination - a cross-sectional study. J. Oral Maxillofac. Pathol. : JOMFP. 2021;25:553. doi: 10.4103/jomfp.jomfp_387_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih C., Yang C.C., Choijilsuren G., Chang C.H., Liou A.T. Hepatitis B virus. Trends Microbiol. 2018;26:386–387. doi: 10.1016/j.tim.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A., Allen C., Al-Raddadi R., Alvis-Guzman N., Amoako Y., Artaman A., Ayele T.A., Barac A., Bensenor I., Berhane A., Bhutta Z., Castillo-Rivas J., Chitheer A., Choi J.Y., Cowie B., Dandona L., Dandona R., Dey S., Dicker D., Phuc H., Ekwueme D.U., Zaki M.S., Fischer F., Fürst T., Hancock J., Hay S.I., Hotez P., Jee S.H., Kasaeian A., Khader Y., Khang Y.H., Kumar A., Kutz M., Larson H., Lopez A., Lunevicius R., Malekzadeh R., McAlinden C., Meier T., Mendoza W., Mokdad A., Moradi-Lakeh M., Nagel G., Nguyen Q., Nguyen G., Ogbo F., Patton G., Pereira D.M., Pourmalek F., Qorbani M., Radfar A., Roshandel G., Salomon J.A., Sanabria J., Sartorius B., Satpathy M., Sawhney M., Sepanlou S., Shackelford K., Shore H., Sun J., Mengistu D.T., Topór-Mądry R., Tran B., Ukwaja K.N., Vlassov V., Vollset S.E., Vos T., Wakayo T., Weiderpass E., Werdecker A., Yonemoto N., Younis M., Yu C., Zaidi Z., Zhu L., Murray C.J.L., Naghavi M., Fitzmaurice C. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song T., Wang L., Su B., Zeng W., Jiang T., Zhang T., Sun G., Wu H. Diagnostic value of alpha-fetoprotein, Lens culinaris agglutinin-reactive alpha-fetoprotein, and des-gamma-carboxyprothrombin in hepatitis B virus-related hepatocellular carcinoma. J. Int. Med. Res. 2020;48 doi: 10.1177/0300060519889270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L., Ye S., Zhao X., Ji L., Zhang Y., Zhou P., Sun J., Guan Y., Han Y., Ni C., Hu X., Liu W., Wang H., Zhou B., Huang J. Molecular characterization of HBV DNA integration in patients with hepatitis and hepatocellular carcinoma. J. Cancer. 2018;9:3225–3235. doi: 10.7150/jca.26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S.Y., Zhang A., Lian J., Wang J., Chang T.T., Lin Y.J., Song W., Su Y.H. Recurrent HBV integration targets as potential drivers in hepatocellular carcinoma. Cells. 2021;10 doi: 10.3390/cells10061294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi H., Gao X., Xia Z., Yu W., Yin X., Pan Y., Peng G., Mao X., Teichmann A.T., Zhang J., Tran L.J., Jiang T., Liu Y., Yang G., Wang Q. FAM family gene prediction model reveals heterogeneity, stemness and immune microenvironment of UCEC. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1200335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao S., Zhang X., Gao F., Chi H., Zhang J., Xia Z., Cheng C., Liu J. Identification of copper metabolism-related subtypes and establishment of the prognostic model in ovarian cancer. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1145797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren Q., Zhang P., Lin H., Feng Y., Chi H., Zhang X., Xia Z., Cai H., Yu Y. A novel signature predicts prognosis and immunotherapy in lung adenocarcinoma based on cancer-associated fibroblasts. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1201573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng G., Chi H., Gao X., Zhang J., Song G., Xie X., Su K., Song B., Yang J., Gu T., Li Y., Xu K., Li H., Liu Y., Tian G. Identification and validation of neurotrophic factor-related genes signature in HNSCC to predict survival and immune landscapes. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi H., Yang J., Peng G., Zhang J., Song G., Xie X., Xia Z., Liu J., Tian G. Circadian rhythm-related genes index: a predictor for HNSCC prognosis, immunotherapy efficacy, and chemosensitivity. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1091218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S., Zhang L., Ji W., Shi Y., Lai G., Chi H., Huang W., Cheng C. Machine learning-based characterization of cuprotosis-related biomarkers and immune infiltration in Parkinson's disease. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1010361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S., Chi H., Ji W., He Q., Lai G., Peng G., Zhao X., Cheng C. A bioinformatics-based analysis of an anoikis-related gene signature predicts the prognosis of patients with low-grade gliomas. Brain Sci. 2022;12 doi: 10.3390/brainsci12101349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng H., Fan X., Wang X., Zeng L., Zhang K., Zhang X., Li N., Han Q., Lv Y., Liu Z. Serum pentraxin 3 as a biomarker of hepatocellular carcinoma in chronic hepatitis B virus infection. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-77332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z., Wu M., Lin D., Li N. Des-gamma-carboxyprothrombin is a favorable biomarker for the early diagnosis of alfa-fetoprotein-negative hepatitis B virus-related hepatocellular carcinoma. J. Int. Med. Res. 2020;48 doi: 10.1177/0300060520902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng H.H., Wei L., Cui S., Luo Y., Ten Haken R.K., El Naqa I. Machine learning and imaging informatics in oncology. Oncology. 2020;98:344–362. doi: 10.1159/000493575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y., Tseng H.H., Cui S., Wei L., Ten Haken R.K., El Naqa I. Balancing accuracy and interpretability of machine learning approaches for radiation treatment outcomes modeling. BJR open. 2019;1 doi: 10.1259/bjro.20190021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi H., Peng G., Yang J., Zhang J., Song G., Xie X., Strohmer D.F., Lai G., Zhao S., Wang R., Yang F., Tian G. Machine learning to construct sphingolipid metabolism genes signature to characterize the immune landscape and prognosis of patients with uveal melanoma. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.1056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi H., Chen H., Wang R., Zhang J., Jiang L., Zhang S., Jiang C., Huang J., Quan X., Liu Y., Zhang Q., Yang G. Proposing new early detection indicators for pancreatic cancer: combining machine learning and neural networks for serum miRNA-based diagnostic model. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1244578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho J.K., Jeevan-Raj B., Netter H.J. Hepatitis B virus (HBV) subviral particles as protective vaccines and vaccine platforms. Viruses. 2020;12 doi: 10.3390/v12020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang L.S.Y., Covert E., Wilson E., Kottilil S. Chronic hepatitis B infection: a review. JAMA. 2018;319:1802–1813. doi: 10.1001/jama.2018.3795. [DOI] [PubMed] [Google Scholar]
- 37.Su K., Shen Q., Tong J., Gu T., Xu K., Li H., Chi H., Liu Y., Li X., Wen L., Song Y., Guo Q., Chen J., Wu Z., Jiang Y., He K., Guo L., Han Y. Construction and validation of a nomogram for HBV-related hepatocellular carcinoma: a large, multicenter study. Ann. Hepatol. 2023;28 doi: 10.1016/j.aohep.2023.101109. [DOI] [PubMed] [Google Scholar]
- 38.Lu X., Li Y., Li Y., Zhang X., Shi J., Feng H., Gao Y., Yu Z. Advances of multi-omics applications in hepatic precancerous lesions and hepatocellular carcinoma: the role of extracellular vesicles. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1114594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu C., Zhou W., Wang Y., Qiao L. Hepatitis B virus-induced hepatocellular carcinoma. Cancer letters. 2014;345:216–222. doi: 10.1016/j.canlet.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 40.Ruff S.M., Shannon A.H., Beane J.D., Pawlik T.M. Highlighting novel targets in immunotherapy for liver cancer. Expet Rev. Gastroenterol. Hepatol. 2022;16:1029–1041. doi: 10.1080/17474124.2022.2150841. [DOI] [PubMed] [Google Scholar]
- 41.Katoh Y., Katoh M. Comparative genomics on HHIP family orthologs. Int. J. Mol. Med. 2006;17:391–395. [PubMed] [Google Scholar]
- 42.Tada M., Kanai F., Tanaka Y., Tateishi K., Ohta M., Asaoka Y., Seto M., Muroyama R., Fukai K., Imazeki F., Kawabe T., Yokosuka O., Omata M. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2008;14:3768–3776. doi: 10.1158/1078-0432.CCR-07-1181. [DOI] [PubMed] [Google Scholar]
- 43.Eichenmüller M., Gruner I., Hagl B., Häberle B., Müller-Höcker J., von Schweinitz D., Kappler R. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49:482–490. doi: 10.1002/hep.22649. [DOI] [PubMed] [Google Scholar]
- 44.Ouyang Y., Tang Y., Fu L., Peng S., Wu W., Tan D., Fu X. Exosomes secreted by chronic hepatitis B patients with PNALT and liver inflammation grade ≥ A2 promoted the progression of liver cancer by transferring miR-25-3p to inhibit the co-expression of TCF21 and HHIP. Cell Prolif. 2020;53 doi: 10.1111/cpr.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., Chi H., Zhao X., Pan R., Wei Y., Han Y. Role of exosomes in immune microenvironment of hepatocellular carcinoma. Journal of oncology. 2022;2022 doi: 10.1155/2022/2521025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hara T., Nakayama Y. CXCL14 and insulin action. Vitam. Horm. 2009;80:107–123. doi: 10.1016/S0083-6729(08)00605-5. [DOI] [PubMed] [Google Scholar]
- 47.Lin Y., Chen B.M., Yu X.L., Yi H.C., Niu J.J., Li S.L. Suppressed expression of CXCL14 in hepatocellular carcinoma tissues and its reduction in the advanced stage of chronic HBV infection. Cancer Manag. Res. 2019;11:10435–10443. doi: 10.2147/CMAR.S220528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjöberg E., Augsten M., Bergh J., Jirström K., Östman A. Expression of the chemokine CXCL14 in the tumour stroma is an independent marker of survival in breast cancer. Br. J. Cancer. 2016;114:1117–1124. doi: 10.1038/bjc.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong J., Chi H., Yang G., Zhao S., Zhang J., Tran L.J., Xia Z., Yang F., Tian G. Revolutionizing anti-tumor therapy: unleashing the potential of B cell-derived exosomes. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1188760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y., Chang Q., Wu X., Yu Y., Zhang H. Effect of chemokine CXCL14 on in vitro angiogenesis of human hepatocellular carcinoma cells. Arch. Physiol. Biochem. 2022;128:1316–1322. doi: 10.1080/13813455.2020.1769677. [DOI] [PubMed] [Google Scholar]
- 51.Ose R., Oharaa O., Nagase T. Galectin-1 and galectin-3 mediate protocadherin-24-dependent membrane localization of β-catenin in colon cancer cell line HCT116. Curr. Chem. Genom. 2012;6:18–26. doi: 10.2174/1875397301206010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okazaki N., Takahashi N., Kojima S., Masuho Y., Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis. 2002;23:1139–1148. doi: 10.1093/carcin/23.7.1139. [DOI] [PubMed] [Google Scholar]
- 53.Ose R., Yanagawa T., Ikeda S., Ohara O., Koga H. PCDH24-induced contact inhibition involves downregulation of beta-catenin signaling. Mol. Oncol. 2009;3:54–66. doi: 10.1016/j.molonc.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia Z., Huang M., Zhu Q., Li Y., Ma Q., Wang Y., Chen X., Li J., Qiu L., Zhang J., Zheng J., Lu B. Cadherin related family member 2 acts as A tumor suppressor by inactivating AKT in human hepatocellular carcinoma. J. Cancer. 2019;10:864–873. doi: 10.7150/jca.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahabiyeh L.A., Malkawi A.K., Wang X., Colak D., Mujamammi A.H., Sabi E.M., Li L., Dasouki M., Abdel Rahman A.M. Dexamethasone-induced perturbations in tissue metabolomics revealed by chemical isotope labeling LC-MS analysis. Metabolites. 2020;10 doi: 10.3390/metabo10020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leo M.A., Lieber C.S. New pathway for retinol metabolism in liver microsomes. J. Biol. Chem. 1985;260:5228–5231. [PubMed] [Google Scholar]
- 57.Leonhardi G. [Tyrosine metabolism and liver function] Klin. Wochenschr. 1952;30:168–173. doi: 10.1007/BF01478254. [DOI] [PubMed] [Google Scholar]
- 58.Vandewynckel Y.P., Coucke C., Laukens D., Devisscher L., Paridaens A., Bogaerts E., Vandierendonck A., Raevens S., Verhelst X., Van Steenkiste C., Libbrecht L., Geerts A., Van Vlierberghe H. Next-generation proteasome inhibitor oprozomib synergizes with modulators of the unfolded protein response to suppress hepatocellular carcinoma. Oncotarget. 2016;7:34988–35000. doi: 10.18632/oncotarget.9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D., Jin S., Chen P., Zhang Y., Li Y., Zhong C., Fan X., Lin H. Comprehensive analysis of cuproptosis-related lncRNAs for prognostic significance and immune microenvironment characterization in hepatocellular carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.991604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin W., Yang Q., Chi H., Wei K., Zhang P., Zhao G., Chen S., Xia Z., Li X. Ensemble deep learning enhanced with self-attention for predicting immunotherapeutic responses to cancers. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1025330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W., Han J., Wu H. Regulatory T-cells promote hepatitis B virus infection and hepatocellular carcinoma progression. Chronic diseases and translational medicine. 2016;2:67–80. doi: 10.1016/j.cdtm.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y., Tian Z. HBV-induced immune imbalance in the development of HCC. Front. Immunol. 2019;10:2048. doi: 10.3389/fimmu.2019.02048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuwabara T., Matsui Y., Ishikawa F., Kondo M. Regulation of T-cell signaling by post-translational modifications in autoimmune disease. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19030819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y., Wei K., Chi H., Xia Z., Li X. IL-7: a promising adjuvant ensuring effective T cell responses and memory in combination with cancer vaccines? Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1022808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teng P., Xu X., Ni C., Yan H., Sun Q., Zhang E., Ni Y. Identification of key genes in calcific aortic valve disease by integrated bioinformatics analysis. Medicine. 2020;99 doi: 10.1097/MD.0000000000021286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee A.J., Ashkar A.A. The dual nature of type I and type II interferons. Front. Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirakawa H., Kuronuma T., Nishimura Y., Hasebe T., Nakano M., Gotohda N., Takahashi S., Nakagohri T., Konishi M., Kobayashi N., Kinoshita T., Nakatsura T. Glypican-3 is a useful diagnostic marker for a component of hepatocellular carcinoma in human liver cancer. Int. J. Oncol. 2009;34:649–656. doi: 10.3892/ijo_00000190. [DOI] [PubMed] [Google Scholar]
- 68.Wang H.L., Anatelli F., Zhai Q.J., Adley B., Chuang S.T., Yang X.J. Glypican-3 as a useful diagnostic marker that distinguishes hepatocellular carcinoma from benign hepatocellular mass lesions. Arch. Pathol. Lab Med. 2008;132:1723–1728. doi: 10.5858/132.11.1723. [DOI] [PubMed] [Google Scholar]
- 69.Shirakawa H., Suzuki H., Shimomura M., Kojima M., Gotohda N., Takahashi S., Nakagohri T., Konishi M., Kobayashi N., Kinoshita T., Nakatsura T. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2009;100:1403–1407. doi: 10.1111/j.1349-7006.2009.01206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guidotti L.G., Chisari F.V. Immunobiology and pathogenesis of viral hepatitis. Annual review of pathology. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 71.Chi H., Zhao S., Yang J., Gao X., Peng G., Zhang J., Xie X., Song G., Xu K., Xia Z., Chen S., Zhao J. T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1137025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertram E.M., Dawicki W., Watts T.H. Role of T cell costimulation in anti-viral immunity. Semin. Immunol. 2004;16:185–196. doi: 10.1016/j.smim.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 73.Mott K.R., Allen S.J., Zandian M., Akbari O., Hamrah P., Maazi H., Wechsler S.L., Sharpe A.H., Freeman G.J., Ghiasi H. Inclusion of CD80 in HSV targets the recombinant virus to PD-L1 on DCs and allows productive infection and robust immune responses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
I have shared the link to my data/code at the Attach File step.All raw data is available at this website: https://www.jianguoyun.com/p/DRVHjCEQro7JCxiUy4EFIAA