Abstract
Study Objectives.
We investigated whether genetic risk for insomnia and sleep duration abnormalities are associated with AUD and alcohol consumption. We also evaluated the causal relationships between sleep- and alcohol-related traits.
Methods.
Individual-level phenotype and genotype data from the Million Veteran Program were used. Polygenic risk scores (PRS) were computed using summary statistics from two recent discovery GWAS of insomnia (N=453,379 European-ancestry (EA) individuals) and sleep duration (N=446,118 EAs) and tested for association with lifetime AUD diagnosis (N=34,658 EA cases) and past-year Alcohol Use Disorders Identification Test-Consumption scale scores (AUDIT-C, N=200,680 EAs). Bi-directional two-sample Mendelian Randomization (MR) analyses assessed causal associations between the two sleep traits and the two alcohol-related traits.
Results.
The insomnia PRS was positively associated with AUD at 2/9 PRS thresholds, with p<0.01 being the most significant (OR=1.02, p=3.48x10−5). Conversely, insomnia PRS was negatively associated with AUDIT-C at 6/9 PRS thresholds (most significant threshold being p=0.001 (β=−0.02, p=5.6x10−8). Sleep duration PRS was positively associated with AUDIT-C at 2/9 PRS thresholds, with the most significant threshold being p = 1 x 10−6 (β=0.01, p=0.0009). MR analyses supported a significant positive causal effect of insomnia on AUD (14 SNPs; β=104.14; SE=16.19; p=2.22x10−5), although with significant heterogeneity. MR analyses also showed that shorter sleep duration had a causal effect on the risk of AUD (27 SNPs; β=−63.05; SE= 3.54; p=4.55x10−16), which was robust to sensitivity analyses.
Conclusion.
The genetic risk for insomnia shows pleiotropy with AUD, and sleep continuity abnormalities have a causal influence on the development of AUD.
Keywords: Sleep initiation and maintenance disorders, sleep duration, alcohol drinking, alcohol use disorder, mendelian randomization analysis
1. Introduction
1.1. Alcohol consumption.
Alcohol is a psychoactive substance that is consumed by nearly two-thirds of adults in the United States (US) (Boersma et al., 2020). Among these individuals, almost a third (29.9%) indulge in heavy drinking, defined as the consumption of ≥14 standard drinks a week or >4 drinks on any day in men and ≥ 7 drinks a week or >3 drinks on any day in women (Grant et al., 2017). Heavy drinking is a potentially modifiable risk factor for traumatic injuries and medical, psychiatric, and sleep-related disorders (Dawson et al., 2008; Gordon et al., 2008; N.I.A.A.A.). The most common sleep-related disorders that accompany heavy alcohol consumption and alcohol use disorder (AUD) are insomnia and short (insufficient) sleep duration.
1.2. Insomnia AUD, and alcohol consumption.
Insomnia is a disorder of sleep continuity that consists of difficulty falling asleep or staying asleep, or early morning awakening that leads to impaired functioning. Insomnia has a bi-directional causal relationship with alcohol consumption (Haario et al., 2013). Similarly, the presence of either insomnia or AUD increases the risk of the other disorder (Chakravorty et al., 2016b; Crum et al., 2004; Janson et al., 2001). Insomnia associated with heavy drinking may increase the risk of precipitating or aggravating psychiatric disorders (Chakravorty et al., 2013; Chaudhary et al., 2020), and suicidal behavior (Chakravorty et al., 2014; Kolla et al., 2018). Insomnia co-occurring with AUD is associated with an impaired quality of life, psychosocial problems, suicidal behavior and an increased risk of relapse to drinking during early recovery (Chakravorty et al., 2016a). A Mendelian Randomization (MR) study evaluating the bi-directional relationship between insomnia and AUD in a GWAS of 46,568 individuals showed a significant causal effect of insomnia on the development of AUD, but no causal effect of AUD on insomnia (Pasman et al., 2020). Further, the study did not show a causal effect of insomnia on alcohol consumption or vice-versa. Given the high prevalence of insomnia in individuals with AUD (Chakravorty et al., 2016a) and the sleep disruptive effects of alcohol consumption (Arnedt et al., 2011), these findings are clinically important, but they require replication. In addition, the independent association between abnormal sleep duration and pathological drinking in these individuals needs to be evaluated independently.
1.3. Sleep duration abnormalities AUD, and alcohol consumption.
Sleep duration, another aspect of sleep health, is the total duration of sleep obtained, either during nocturnal sleep or across a 24-hour period (Kline, 2013) with a range of 7–9 hours recommended to support optimal health in adults (Consensus Conference et al., 2015a). Abnormalities of sleep duration, especially short sleep duration (≤6 hours a night), are associated in cross-sectional studies with an increased risk of adverse health consequences such as mortality, suicide, physical injuries, cardio-metabolic and psychiatric comorbidities (Consensus Conference et al., 2015b), and heavy drinking and AUD (Chaput et al., 2012; John et al., 2005; Krueger and Friedman, 2009). As of now, it is unclear whether short sleep duration is a cause or a consequence of heavy drinking or AUD. Furthermore, are genetic risk variants associated with sleep duration abnormalities also associated with pathological alcohol consumption?
1.4. Polygenic risk scores of these sleep traits and alcohol-related traits.
Genetic factors influence variation in both alcohol- and sleep-related traits (Wray and Visscher, 2008). Recent estimates of the SNP-based heritability (i.e., that attributable to common genetic variation) among European-ancestry (EA) individuals is 7–11% for alcohol consumption based on the Alcohol Use Disorders Identification Test-Consumption (AUDIT-C) measure (Kranzler et al., 2019; Sanchez-Roige et al., 2019), 6–16% for AUD (Kranzler et al., 2019; Vrieze et al., 2013), 7–17% for insomnia (Jansen et al., 2019; Lane et al., 2019), and 10% for sleep duration (Dashti et al., 2019). A polygenic risk score (PRS), an individual’s estimated genetic predisposition for a given trait, is computed as the sum of the alleles associated with the trait, weighted by the risk allele effect sizes (Choi et al., 2020). PRS have been used to validate genetic links to disease, dissect pleiotropic associations across traits, and provide a quantitative measure of an aggregated genetic burden for illness in an individual, although no such studies involving PRS exist at this interface of sleep- and alcohol-related traits. Here, we evaluated the associations between insomnia or sleep duration-related PRS and two alcohol-related phenotypes (AUD and alcohol consumption) to investigate whether the genetic risk for sleep-related traits is associated with alcohol-related traits and whether the association is indicative of pleiotropy, although the PRS does not explain the nature of causal relationships between them.
1.5. Causal relationships between sleep- and alcohol-related traits.
The availability of large biobanks such as the United Kingdom Biobank (UKB) and the Million Veteran Program (MVP), which include a range of phenotypic data, has allowed researchers to conduct genome-wide association studies (GWAS) for traits like insomnia, sleep duration, AUD, and alcohol consumption (Dashti et al., 2019; Jansen et al., 2019; Lane et al., 2019). In addition to serving as discovery samples for the calculation of PRS, summary statistics from these GWASs can be used to conduct Mendelian Randomization (MR) studies to understand causal relationships between traits. MR studies can provide evidence supportive of causal relations because genetic variants are not substantially modifiable and are free from reverse confounding (Davies et al., 2018; Hemani et al., 2018). An MR study uses genetic variation identified in GWAS to create exposure and outcome instruments that determine the causal effect of the exposure (e.g., insomnia) on an outcome (e.g., AUD) (Davey Smith and Hemani, 2014; Hemani et al., 2018).
1.6. Gaps in the current literature.
The above literature shows that despite the heritability of these sleep- and alcohol-related traits, and the clinical association between these traits, it is unclear whether a genetic predisposition for insomnia or sleep duration abnormalities increases the risk for AUD or alcohol consumption. In a recent study, insomnia had a causal effect on the development of AUD, but there was no causal effect of AUD on insomnia. Also, neither insomnia nor the level of alcohol consumption had causal effects on one another (Pasman et al., 2020). Further, a growing body of cross-sectional studies has shown a negative relationship between sleep duration abnormalities and pathological drinking, but the causal pathways underlying these relationships are unclear.
In this study, we attempt to untangle these relationships and extend the literature at the interface of sleep- and alcohol-related traits. As a first step, we evaluated the association between PRS for two common sleep-related traits (insomnia and sleep duration) and two alcohol-related traits (AUD and alcohol consumption). As a next step, we repeated the MR analysis between insomnia and AUD using data from a GWAS of AUD in a sample of 202,004 individuals to evaluate the bidirectional relationship between these traits. We also re-assessed bidirectional relationships both between insomnia and alcohol consumption using data from a GWAS of alcohol consumption involving 200,680 individuals. Finally, we explored the bi-directional relationships between habitual reported sleep duration and both AUD and alcohol consumption. We evaluated AUD and AUDIT-C as separate alcohol-related traits as they have overlapping, but unique genetic architectures (Kranzler et al., 2019) and thus may have different associations with sleep-related phenotypes. We hypothesized that genetic risk for insomnia is positively associated with AUD and alcohol consumption, and despite a bidirectional causal effect between insomnia and AUD no such relationship exists for alcohol consumption. We also hypothesized that genetic risk for sleep duration is inversely associated with AUD and alcohol consumption and vice-versa. These analyses could improve our understanding of these relationships and help in the development of individualized care at this important clinical interface (Torkamani et al., 2018). If the PRS of insomnia or sleep duration is associated with AUD or drinking, then future PRS from GWAS with larger sample sizes could be used to identify people at risk for AUD or unhealthy drinking during the premorbid state before the onset of insomnia or alcohol use, similar to the recent studies in substance use disorders (Kranzler et al., 2023), and in cardiovascular disorders (O’Sullivan et al., 2022). If either insomnia or sleep duration abnormalities are a risk factor for the development of AUD or heavy drinking, then treating insomnia or improving sleep duration could decrease the risk of developing AUD or heavy drinking.
2. Materials and methods
2.1. Overview.
We used GWAS summary statistics from the UKB and individual-level data from MVP to construct polygenic risk scores for insomnia and sleep duration and test their association with alcohol-related phenotypes (Choi et al., 2020). We used MVP GWAS summary statistics for alcohol-related phenotypes and UKB summary statistics for insomnia and sleep duration to test their causal relations.
2.2. Standard protocols and informed consent:
We used summary level data from the UKB, a large biobank study investigating the contributions of genetic predisposition and environmental exposure to the development of disease. All UKB subjects provided written informed consent to participate and make their de-identified data publicly available (Sudlow et al., 2015). The MVP is an observational cohort study and biobank supported by the U.S. Department of Veterans Affairs (VA) (Gaziano et al., 2016). Phenotypic data were collected from MVP subjects using questionnaires, the VA electronic health record (EHR), and a blood sample for genetic analysis. The MVP provided access to patient-level AUDIT-C scores and AUD diagnoses, GWAS summary statistics, and approved the data analysis. The MVP study followed all relevant ethical regulations for research with human subjects and obtained informed consent from all participants.
2.3. Data source.
2.3.1. Insomnia GWAS:
We obtained summary statistics from the published GWAS of insomnia in EA participants in the UKB (N=453,379) (Lane et al., 2019). This study assessed insomnia symptoms using the question (data field 1200), “Do you have trouble falling asleep at night or do you wake up in the middle of the night?” The responses were primarily evaluated as a binary case-control variable, with cases reporting insomnia symptoms as “usually” or “sometimes” and controls endorsing “never/rarely.” This insomnia question has a sensitivity of 98% and a specificity of 96% in discriminating questionnaire-defined insomnia disorder cases from unaffected controls (Hammerschlag et al., 2017).
2.3.2. Sleep duration GWAS:
We obtained the GWAS summary statistics for sleep duration in EA participants from the UKB (N=446,118). The study assessed sleep duration using the question (data field 1160), “About how many hours sleep do you get in every 24 hours?” The response was recorded as a whole integer within a range of 1 to 23, but responses <3 and >18 hours were excluded from the GWAS analysis (Dashti et al., 2019). This self-reported sleep duration meaure is correlated with accelerometric recorded sleep duration (Jones et al., 2019).
2.3.3. Alcohol-related GWAS:
We used the GWAS summary statistics for AUDIT-C score and AUD in EA participants from the MVP (AUDIT-C [N= 200,680]; AUD [N = 202,004])(Kranzler et al., 2019).
2.3.4. Alcohol phenotypes in the Million Veteran Program (MVP):
We used the individual-level AUDIT-C scores (Bush et al., 1998) and AUD diagnostic codes from the VA EHR. The AUDIT-C consists of the first three items of the Alcohol Use Disorders Identification Test and measures typical quantity (question 1) and frequency (question 2) of drinking, and frequency of heavy drinking (question 3). The AUDIT-C is a mandatory annual assessment for all veterans seen in primary care within the VA health system. Alcohol-related disorders were identified using International Classification of Diseases (ICD), 9th revision (ICD-9) codes 303.X (dependence) and 305–305.03 (abuse) and ICD, 10th revision (ICD-10) codes F10.1 (abuse) and F10.2 (dependence). EA participants with at least one inpatient or two outpatient alcohol-related ICD-9/10 codes (N = 34,658) from 2000 to 2018 were assigned a diagnosis of AUD (Kranzler et al., 2019).
2.4. Genetic Correlations:
We used the cross-trait Linkage Disequilibrium (LD) score regression to calculate genome-wide genetic correlations (rg) between insomnia, sleep duration, and alcohol use phenotypes, using precomputed LD scores for European individuals from the 1000 Genomes Project (Bulik-Sullivan et al., 2015).
2.5. Polygenic risk scores (PRS):
We used the clumping and thresholding method (Choi et al., 2020) to create the PRS for insomnia and sleep duration in EA individuals (N=209,020). The PRS was calculated using the dosage information for each variant under an additive model as the sum of all alleles carried, weighted by the effect size of the allele in the GWAS. We performed p-value informed clumping using the 1000 Genomes EA participants as the LD background, with r2 = 0.1 and a distance threshold of 250 kb. The PRS was computed for nine p-value thresholds (p ≤ 0.000001, 0.00001, 0.0001, 0.001, 0.01, 0.05, 0.1, 0.5, and 1) and standardized the PRS with a mean = 0, and SD = 1. Linear regression (for AUDIT-C score as the outcome) and binary logistic regression (for AUD cases and controls) evaluated their association with PRS as the predictor after adjusting for covariates that included age, sex, and the first five principal components of their genetic ancestry (PCs) (Figure 1). A Bonferroni-corrected p-value (0.05/9=0.0056) was used to account for multiple testing.
Figure 1.
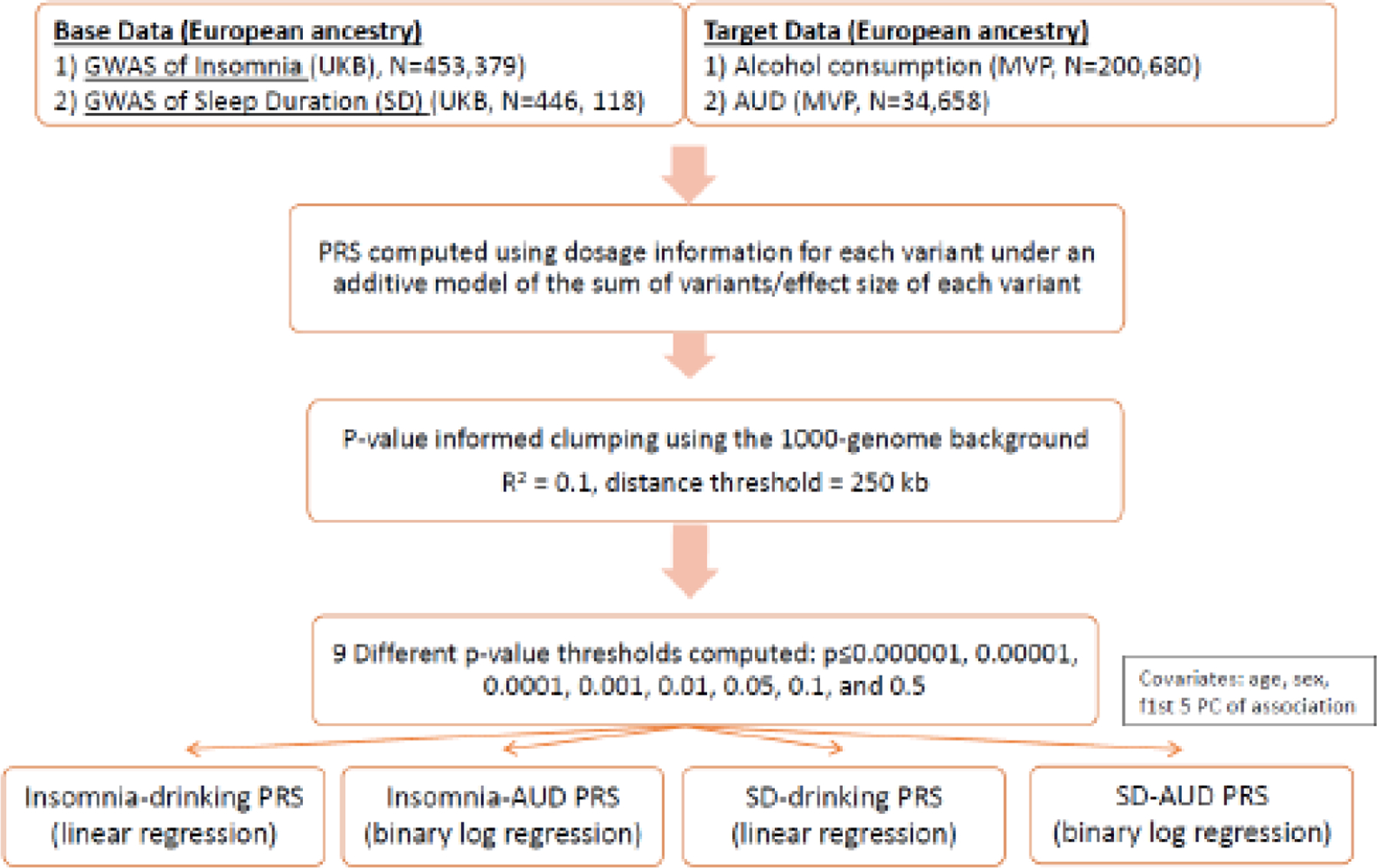
PRS methodology in the current study
Footnote: GWAS = genome-wide association study; UKB = United Kingdom Biobank; MVP = Million Veteran Program; PRS = Polygenic Risk Score; AUD = Alcohol Use Disorder; PC = Principal Components.
2.6. Mendelian randomization analyses (MR):
We harmonized the GWAS summary statistics for insomnia and alcohol-related phenotypes before conducting genetic correlation analyses, bidirectional MR analyses, and sensitivity analyses. The primary MR method was random-effects inverse-variance weighted (IVW) regression for balanced horizontal pleiotropy and mode-based estimator for majority horizontal pleiotropy (Hemani et al., 2018; Yavorska and Burgess, 2017) with sleep (duration or insomnia) and alcohol-related variables (AUDIT-C score or AUD diagnosis) alternately used as exposure or outcome. We employed a stringent clumping threshold of r2 = 0.001. For dichotomous phenotypes (insomnia and AUD), a one-unit increase in the genetic instrument reflects a doubling in the odds of the exposure trait (Burgess and Labrecque, 2018). MR provides strong evidence for causality provided the following assumptions are met for the genetic instrument: (1) it is strongly associated with the exposure, (2) it is not associated with confounders, and (3) there is no horizontal pleiotropy, i.e., it only affects the outcome through its effect on the exposure (Davies et al., 2018). We performed sensitivity analyses using more robust genetic instruments for insomnia to address the first assumption. The use of randomly distributed alleles as the instrumental variables and the adjustment for population stratification in the GWAS satisfied the second assumption. We used multiple models robust to various forms of pleiotropy to evaluate the third assumption. The MR analyses included two studies of the causal effect of insomnia on alcohol consumption and AUD and two studies of the causal effect of alcohol consumption and AUD on insomnia (Figure 2). Thus, the four insomnia-related MR analyses yielded a Bonferroni-adjusted alpha threshold of 0.05/4 = 0.0125. Similarly, the four separate sleep duration-related MR analyses used a Bonferroni-adjusted alpha threshold of 0.05/4 = 0.0125. P-values lower than the corrected alpha thresholds represent significant evidence for causal effects, and p<0.05 provides nominal evidence for a causal effect. We used Cochrane’s Q-statistic to assess SNP effect heterogeneity (Bowden et al., 2018) and single-SNP analysis and the leave-one-out IVW analyses to evaluate the disproportionate effects of single SNPs in the models (Hemani et al., 2018). The F-scores for the sleep and alcohol-related instruments ranged from 40.06–90.71. Wherever horizontal pleiotropic effects were suspected based on the scatter plot results, median or mode-based estimators were applied, as applicable. The median-based approach provides an unbiased estimate in the presence of unbalanced horizontal pleiotropy, and the causal estimate from the mode-based estimator is unbiased if the SNPs contributing to the largest cluster are valid instruments. (Hemani et al., 2018) The data underlying this article are available in the article and in its online supplementary material.
Figure 2.
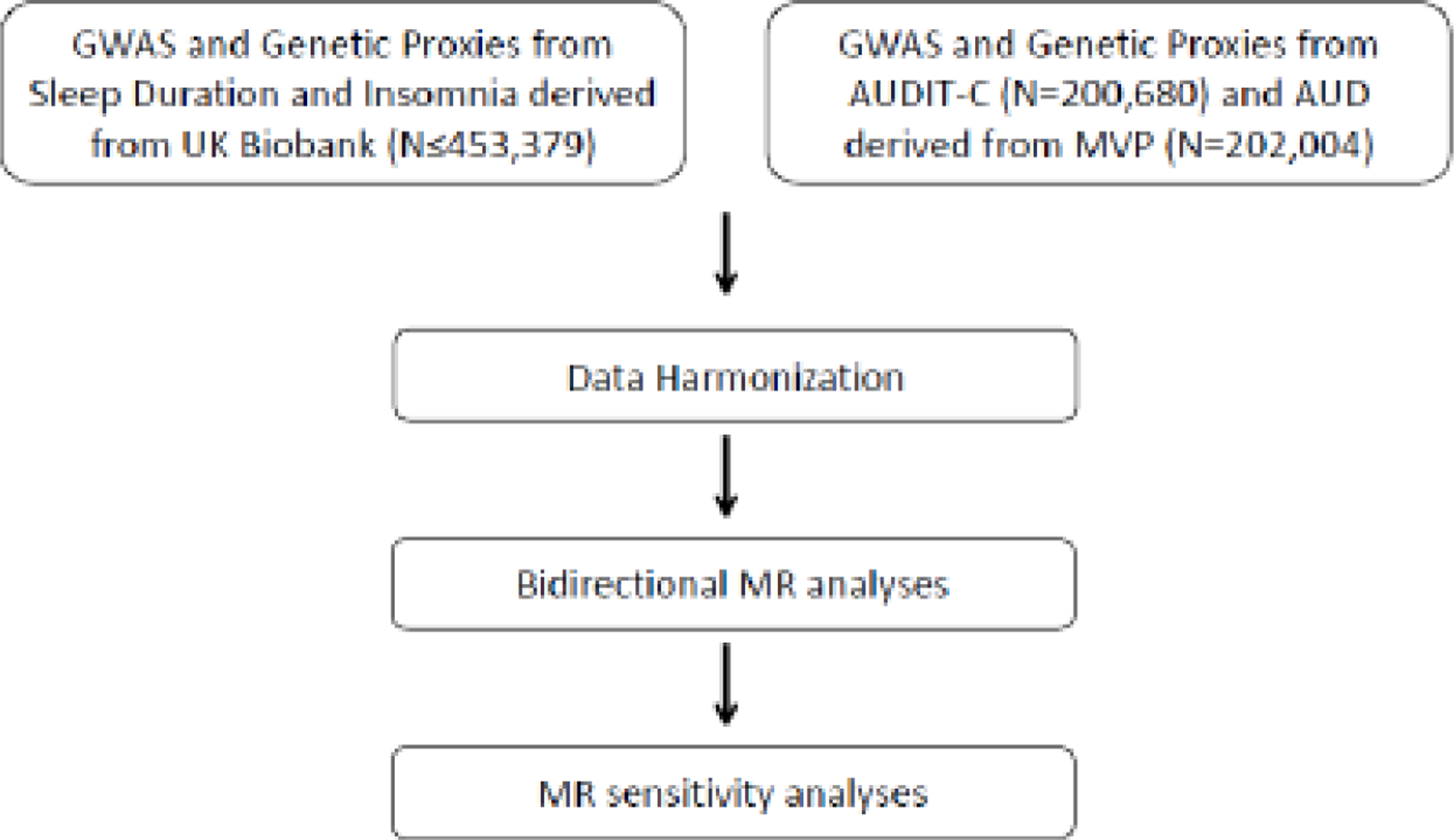
Mendelian Randomization analysis pipeline in the current study
Footnote: GWAS = gene-wide association study; AUDIT-C = Alcohol Use Disorder Identification Test – Consumption (a measure for drinking); AUD = Alcohol Use Disorder (“alcoholism”); MVP = Million Veteran Program; PC = Principal Components; MR = Mendelian Randomization study.
2.7. Statistical Software.
The genetic correlations were analyzed using LDSC v1.01,(Bulik-Sullivan et al., 2015) PRS analysis was conducted using PLINK version 2 (Purcell et al., 2007), and MR analysis was performed in R version 4.1.2(Team, 2018) using the TwoSampleMR package version 0.5.6 (Hemani et al., 2018).
We present this investigation using the STREGA-STROBE reporting recommendations.
3. Results
3.1. Genetic Correlations.
Insomnia was positively genetically correlated with AUD (rg = 0.17, p = 3.00 x 10−8), but negatively correlated with AUDIT-C (rg = −0.15, p = 1.27 x 10−5). Sleep duration was positively correlated with AUDIT-C (rg = 0.12, p = 1.59 x 10−4) but not AUD (rg = 0.05, p = 9.99 x 10−2; supplementary table 1.
3.2. Insomnia
3.2.1. Insomnia PRS is associated with alcohol consumption and AUD.
At two of the PRS thresholds there was a significant positive association of insomnia PRS with AUD, with the most significant being p<0.01 [Odds Ratio (OR) = 1.02, p = 3.48 x 10−5, Figure 3a and Supplementary Table 2]. In contrast, insomnia PRS was significantly associated with lower AUDIT-C scores for six of the nine thresholds, the most significant of which was p=0.001 (Beta = −0.02, p = 5.6 x 10−8; Figure 3b and Supplementary Table 2). A secondary analysis showed no relationship between the mean insomnia PRS score and the individual AUDIT-C scores from 0–12 (Supplementary Figure 1).
Figure 3:
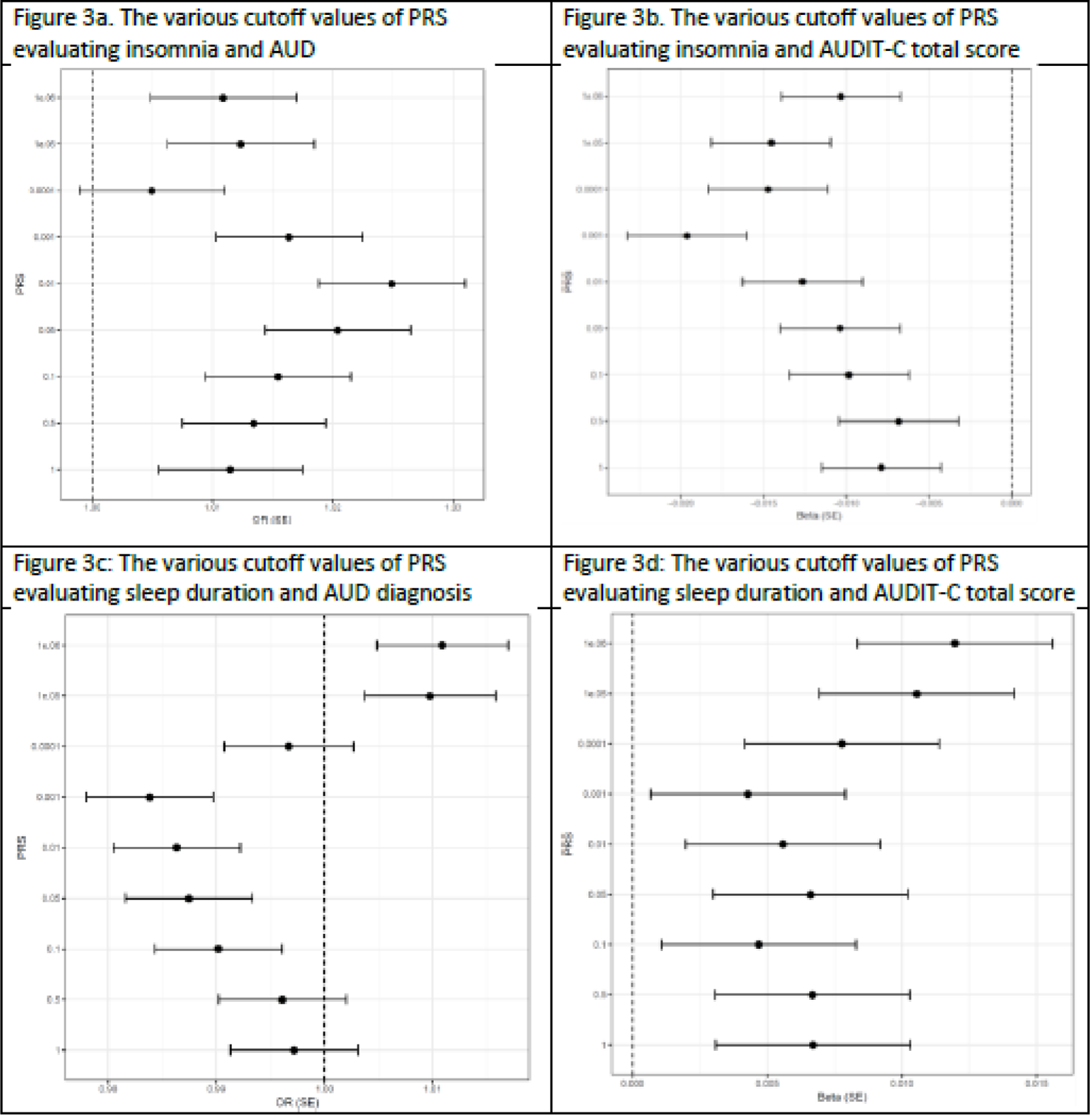
Polygenic Risk Score Analysis of sleep- and alcohol-related variables
Footnote: PRS = polygenic risk score; AUD = Alcohol Use Disorder; AUDIT-C = Alcohol Use Disorder Identification Test – Consumption; Beta = coefficient of regression; OR = odds ratio; SE = standard error.
3.2.2. MR analyses support the causal effects of insomnia on AUD.
The scatter plot showed heterogeneity and the resultant mode-based estimator showed a significant positive causal effect of insomnia on AUD (14 SNPs; beta = 104.14; SE = 16.19; p = 2.22 x 10−5, Figure 4, Supplementary Table 3, and Supplementary Figure 2). There was a significant weighted mode estimator in the sensitivity analyses for insomnia as an exposure (beta = −82.62, SE = 5.50, p = 1.38 x 10−9, Supplemental Figure 2). The leave-one-out analysis did not support excluding any SNPs from the analysis (Supplementary Table 3, Supplementary Figure 3). All 14 single-SNP analyses showed significant correlations, with p-values that ranged between 2.42 x 10−8 to 1.08 x 10−16. Of these, 7 SNPs were positively correlated, and 7 SNPs were negatively correlated. There was heterogeneity (Cochrane’s Q test p<0.0001) but no evidence of horizontal pleiotropy (p = 0.69).
Figure 4.
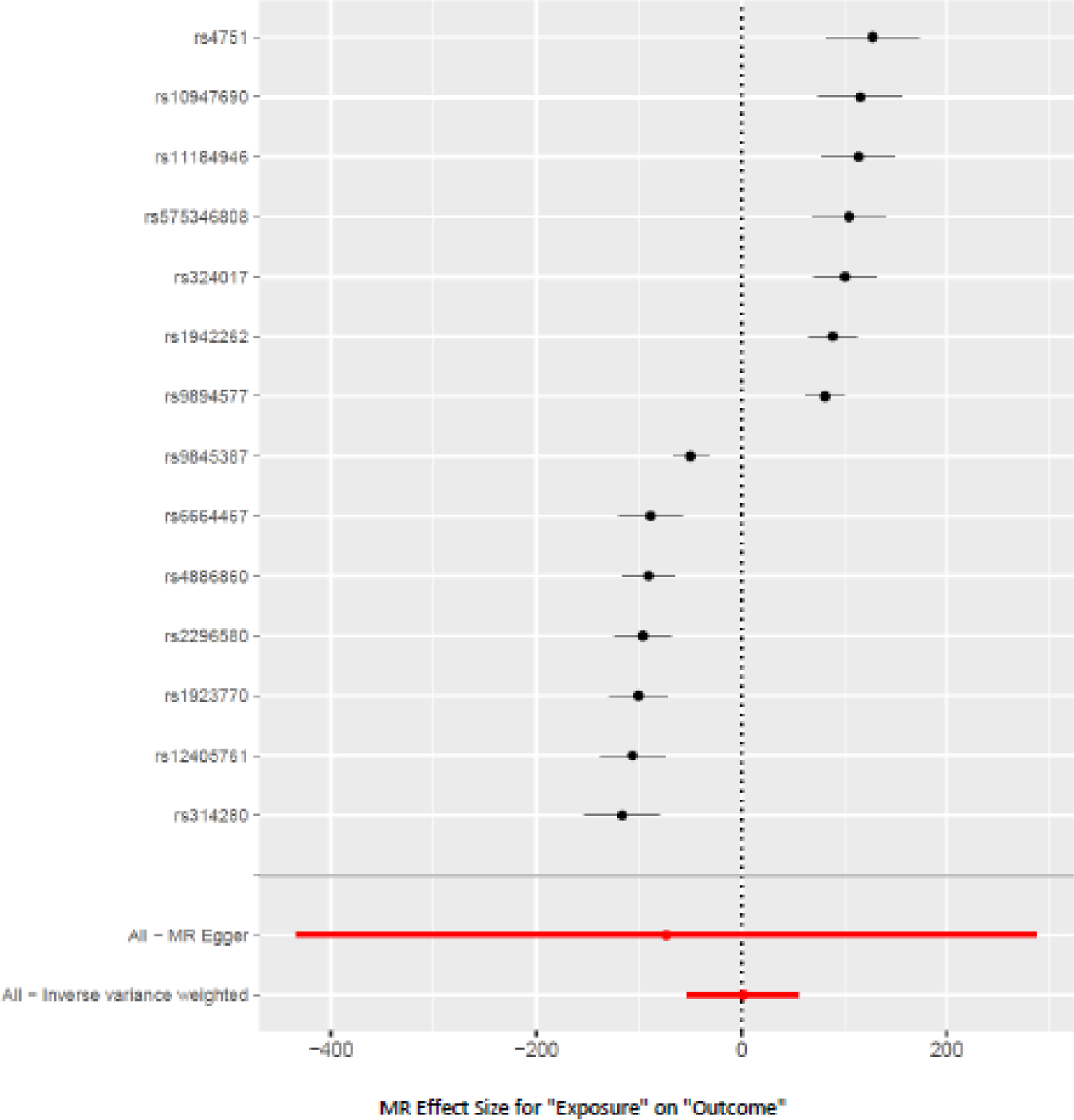
Forest Plot of the causal effect of Insomnia (exposure) on Alcohol Use Disorder (AUD, outcome)
3.2.3. Other findings.
The weighted median estimator showed a nominally positive causal effect of AUD on insomnia (10 SNPs; beta = 0.01; SE = 0.007; p = 0.01; Supplementary Table 4 and Supplementary Figures 4 and 5) although there was heterogeneity in the data (Cochrane’s Q test p = 6.24 x 10−8) but no horizontal pleiotropic effects. Sensitivity analyses for AUD showed a significant weighted mode estimator (beta = 0.02, SE = 0.008, p = 0.02).
The IVW estimator showed a nominally significant negative causal effect of insomnia on AUDIT-C score (16 SNPs; beta = −0.45; SE = 0.17; p = 0.008; Supplementary Figure 6). Sensitivity analysis using the Egger regression estimate, weighted median, simple mode, and weighted mode showed no association. Neither heterogeneity (p = 0.25) nor a horizontal pleiotropic effect (p < 0.80) was seen, Supplementary Table 5. There was no causal effect of AUDIT-C on insomnia using the IVW estimator (12 SNPs; p = 0.92; Supplementary Table 6).
3.3. Sleep Duration
3.3.1. Sleep Duration PRS is not associated with AUD but is associated with alcohol consumption.
The PRS for sleep duration did not show an overall significant association with AUD (Bonferroni-adjusted p-value of 0.0056), though there was a non-significant trend for a negative association with AUD at a p-value cutoff of 0.001 (OR = 0.98, p = 0.006; Figure 3c and Supplementary Table 7). The PRS for sleep duration was positively associated with AUDIT-C at two significance thresholds, the most significant of which was p = 1 x 10−6 (Beta = 0.01, p = 0.0009; Figure 3d and Supplementary Table 7).
3.3.2. MR analyses support the causal effect of sleep duration on AUD but not the reverse.
The mode-based estimator using 27 SNPs showed a significant negative causal effect of sleep duration on AUD (beta = −63.05; SE = 3.54; p = 4.55 x 10−16; Supplementary Table 8). The causal effect of sleep duration on AUD was significant in sensitivity analyses with the weighted mode (beta = −51.66; SE = 2.68; p = 6.21 x 10−17) and weighted median estimator (beta = −21.78; SE = 3.80; p = 1.02 x 10−8) (Supplementary Table 8, Supplementary Figure 7). In single-SNP analyses all the SNPs had a significant effect, with p-values ranging from 1.41 x 10−8 to 1.55 x 10−13. None of the 27 SNPs in the leave-one-out sensitivity analysis resulted in a significantly unbalanced model (Supplementary Figure 8). The Cochrane’s Q test (p <0.0001) showed the presence of heterogeneity, with 14/27 SNPs showing an inverse association (Figure 5) but no horizontal pleiotropy was seen (p = 0.78). However, AUD was not causally associated with sleep duration using the IVW estimator (10 SNPs, p = 0.34 (Supplementary Table 9)).
Figure 5.
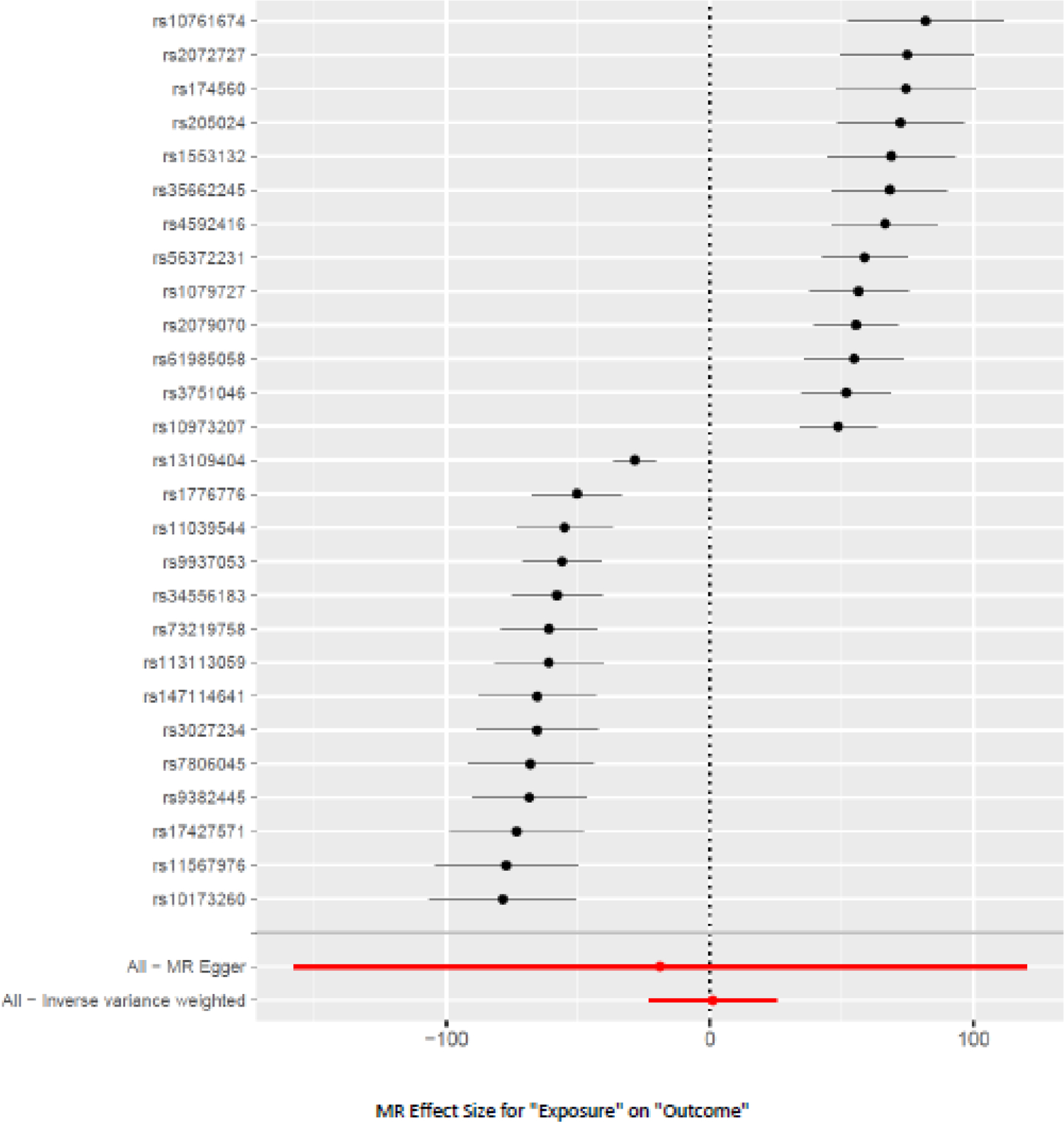
Forest Plot of the causal effect of sleep duration (exposure) on AUD (outcome)
3.3.3. Other findings.
AUDIT-C showed a nominally positive causal effect on sleep duration using the IVW estimator (12 SNPs, Beta = 0.10, SE = 0.04, p = 0.03 (Supplementary Table 10 and Supplementary Figures 9 and 10), although heterogeneity was seen (Cochrane’s Q-test p= 1.13 x 10−21), but no horizontal pleiotropic effect (p = 0.49). Sensitivity analyses showed no association and there was heterogeneity in the data. Sleep duration was not significantly causally associated with AUDIT-C using the mode-based estimator (Supplementary Table 11).
4. Discussion
Using data from four of the largest available GWASs of insomnia, sleep duration, AUD, and AUDIT-C scores in EA individuals, we evaluated whether genetic risk for sleep-related traits was associated with alcohol-related traits. We also investigated causal relationships between the sleep- and alcohol-related variables using bi-directional MR studies. We found that higher insomnia PRS was associated with greater risk of AUD and lower AUDIT-C score. MR studies showed a significant positive causal effect of insomnia on AUD, though the relationship was mitigated by heterogeneity. AUD also showed a small but statistically reliable positive causal effect on insomnia. The sleep duration PRS was positively associated with AUDIT-C. There was a nominally positive causal effect of AUDIT-C on sleep duration, but no causal effect of sleep duration on AUDIT-C. Finally, although shorter sleep duration had a positive causal effect on AUD, the reverse effect was not present.
Insomnia co-occurs with AUD and at a prevalence rate of 36–91% (Chakravorty et al., 2016a). Novel findings in this study help to elucidate this co-occurrence. First, we found a positive association between an insomnia PRS and AUD, suggesting genetic pleiotropy between the two disorders. This implies that there are common underlying biological pathways for insomnia and AUD, one such candidate for which is the orexinergic system that mediates wakefulness and drug use (Fragale et al., 2021). The insomnia PRS may also help in the future to characterize the course of individuals with insomnia that co-occurs with AUD based on their clinical presentation and long-term clinical outcomes for drinking or insomnia as shown recently for AUD (Kranzler et al., 2023), and for the response to treatment, as has been implemented in cardiovascular medicine (O’Sullivan et al., 2022).
Epidemiological studies have shown that insomnia is a risk factor for the development of AUD (Ford and Kamerow, 1989; Weissman et al., 1997). This association may result from a greater preference for alcohol in individuals with insomnia (Ancoli-Israel and Roth, 1999; Johnson et al., 1998; Kaneita et al., 2007; Roehrs et al., 1999), or the rapid development of tolerance to the hypnotic effect of alcohol (Roehrs and Roth, 2018). Our study also provided an important replication of Pasman and colleagues’ finding that insomnia has a causal effect on AUD. This finding implies that there is a need to treat persistent insomnia to avoid the development of AUD, i.e., before a patient resorts to frequent alcohol use to self-medicate the insomnia. Although insomnia had a causal relationship with AUD, the relationship may be confounded by other co-occurring conditions, such as psychiatric and metabolic disorders (Chaudhary et al., 2020; Jansen et al., 2019; Kolla et al., 2020). Heterogeneity in the MR analyses may be due to the use of a binary outcome variable, or variation in the diagnosis of AUD, as the diagnoses were derived from ICD-9 or ICD-10 clinical codes from the VA electronic health record rather than being obtained using a structured interview or another method aimed at ensuring diagnostic reliability. In contrast, a nominal causal effect was seen for AUD on insomnia, which is concordant with epidemiologic literature (Crum et al., 2004; Janson et al., 2001), but needs to be validated in future MR studies with larger samples.
Another novel finding in this study was that sleep duration had a negative causal effect on AUD, i.e., individuals with insufficient sleep duration were at increased risk of developing AUD. These results are consistent with a prior epidemiologic study in which individuals with current AUD or those recovering from AUD were 1.3–1.9 times more likely to report short sleep duration (< 6 hours a night), than those without AUD (John et al., 2005). Screening for insufficient sleep duration during the premorbid or early during their drinking career may help to identify individuals at risk for developing AUD. However, we did not find that AUD had a causal effect on sleep duration abnormalities in, perhaps because individuals with AUD compensate for their sleep deficit by napping during the day (Currie et al., 2003).
The findings from PRS and MR analyses involving the AUDIT-C contradict the preponderance of existing epidemiological studies on sleep (Canham et al., 2015; Chaput et al., 2012; Haario et al., 2013; John et al., 2005; Krueger and Friedman, 2009), although they are in line with recent genetic studies showing that AUD and AUDIT-C have divergent effects, some of which may be due to confounding and non-linear relationships involving AUDIT-C (Kranzler et al., 2019). The PRS for insomnia was negatively associated with alcohol consumption, suggesting that the genetic risk for insomnia is associated with a propensity for lower alcohol consumption. Similarly, the genetic risk for sleep duration abnormalities was associated with an increased risk of alcohol consumption, implying that the genetic propensity for longer sleep duration is associated with greater alcohol consumption. Thus, underlying neurobiological pathways for sleep continuity disturbances could be protective against alcohol consumption, in contrast to the results of epidemiological studies (Canham et al., 2015; Haario et al., 2013). This finding warrants further study.
The MR studies of AUDIT-C showed that insomnia had a negligible negative causal effect on alcohol consumption, despite prior literature showing that individuals with insomnia prefer alcoholic beverages to non-alcoholic ones (Kaneita et al., 2007; Roehrs et al., 1999), that alcohol causes sleep fragmentation (Arnedt et al., 2011), and that there is a bidirectional relationship between alcohol consumption and insomnia (Haario et al., 2013). Larger study samples and additional genetic instruments may be needed to replicate the findings of epidemiological studies. The lack of a causal effect of drinking on insomnia replicates findings by Pasman and colleagues (Pasman et al., 2020). Additionally, alcohol consumption had a nominal causal effect on greater sleep duration, contrary to the decreased sleep duration seen in many prior studies (Chaput et al., 2012; Krueger and Friedman, 2009; Palmer et al., 1980). However, sleep duration did not have a causal effect on drinking. Because many of the findings involving alcohol consumption contradict the existing literature, heterogeneity in the AUDIT-C data could have contributed to the findings. These data were extracted from the clinical charts of Veterans screened annually for alcohol consumption. The clinical sample comprised of individuals with heavy drinking, moderate alcohol use, those who were abstinent after previously having AUD, and those who were lifetime abstainers. Veterans also have a higher prevalence of sleep disorders, such as insomnia, than the general population, which could lead to shorter sleep duration (Alexander et al., 2016), and insomnia can occur during all phases of the drinking career in individuals with AUD (Chakravorty et al., 2016a). This heterogeneity in drinking status could explain the lack of a dose-response relationship between insomnia PRS and AUDIT-C scale scores, as shown in Supplementary Figure 1. This assumption was also supported by the finding in the original GWAS in which AUD and AUDIT-C showed genetic correlations in opposite directions for many traits and disorders (Kranzler et al., 2019).
In contrast to prior findings of insomnia showing a clear causal effect on AUD (Pasman et al., 2020), we found the relationship to be heterogeneous. Our study used summary statistics from a larger sample of individuals (202,004 compared to 46,568 in the prior study (Pasman et al., 2020). Further, the AUD diagnoses in the MVP GWAS were clinically based and required the presence of either one instance of an inpatient diagnosis or two outpatient diagnoses made at different times. In contrast, the prior study (Pasman et al., 2020) used GWAS data from participants whose AUD diagnoses were made using clinician ratings or semi-structured interviews (Walters et al., 2018). Finally, the earlier study (Pasman et al., 2020) may have introduced confounding by using the Steiger filtering methodology where SNPs that explain more variance in the outcome than the exposure are excluded. In contrast, in the present analysis, SNPs that explained significantly more variance in the exposure (p<0.05) were retained in our MR methodology.
Our study has limitations. The currently available PRS are under-powered to serve as clinically useful predictors for these traits. We expect that future GWAS with larger sample sizes will lead to better predictive capability, especially in conjunction with other pertinent variables in a risk prediction model. Although we included sensitivity analyses to assess horizontal pleiotropy, we could not exclude this potential bias completely, using mode-based estimators as needed. Because we conducted our analyses using data from the UK Biobank and MVP the results are not representative of the general population because of selection biases in the samples as the US Veterans have a higher prevalence of insomnia and pathological drinking. Further, the use of AUDIT-C and ICD-9/-10 diagnosis of AUD from the patient charts and self-reported sleep data from the UKB may have introduced another potential bias in the investigation. Future studies should compute PRS using techniques such as Polygenic Prediction via Bayesian Regression and Continuous Shrinkage Priors (PRS-CS) – methods that are robust to varying genetic architectures and enable multivariate modeling of local linkage disequilibrium patterns. Furthermore, studies should investigate these relationships in ancestral groups other than European, evaluate accelerometric sleep duration, and use the effect estimates from the accelerometer based GWAS. Despite limitations, this is the first study to evaluate the association between PRS based on common sleep-related traits and alcohol-related traits. It is also the first to assess causal relationships between subjective sleep duration and alcohol-related traits. Our work here highlights the relationship between sleep and pathological drinking and suggests potential benefits to treating both disorders simultaneously in these individuals.
5. Conclusion
The genetic determinants of sleep traits and alcohol-related traits are partially overlapping. The genetic risk for insomnia has pleiotropic effects on alcohol consumption and AUD, and insomnia has a causal effect on AUD. Short sleep duration may causally influence AUD risk and interventions that increase sleep duration may be a promising therapeutic target for AUD.
Supplementary Material
Figure 6.
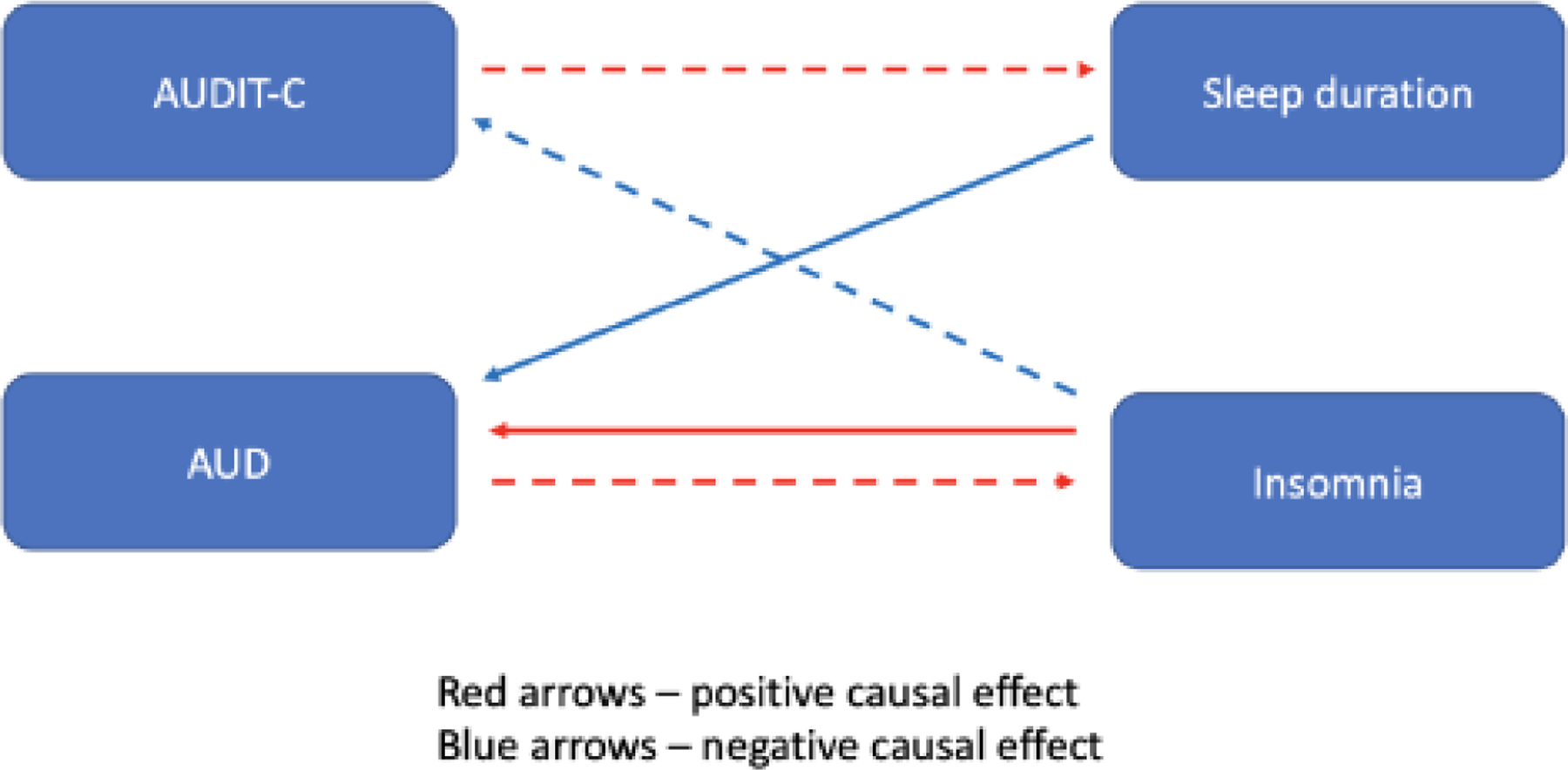
A conceptual diagram of the relationships between sleep (insomnia and sleep duration) and alcohol (AUD and AUDIT-C) variables
Footnote: AUD = Alcohol Use Disorder; AUDIT-C = Alcohol Use Disorder Identification Test – Consumption. Solid arrows depict statistically associations. Broken arrows depict nominally significant associations.
Highlights.
Insomnia and sleep duration abnormalities are commonly associated with drinking and AUD
Polygenic risk score (PRS) analysis evaluates the genetic burden for a trait
Mendelian randomization studies investigate the causal relationship between traits
Insomnia PRS was associated with AUD at 2/9 thresholds and insomnia had a causal effect on AUD
Decreased sleep duration also had a causal effect on the risk for AUD
Acknowledgment:
This work was supported by the following grants: 1IK2CX000855 (S.C.); 1 I01 CX001957 (S.C.); I01 BX003341 (H.R.K); R01 HL143790 (P.R.G), R01 NR018836 (P.R.G.); 1K01 AA028292 (R.L.K.); and the Veterans Integrated Service Network 4 Mental Illness Research, Education, and Clinical Center. The content of this publication does not represent the views of the Department of Veterans Affairs, the US government, Perelman School of Medicine, or other participating institutions.
The content of this publication does not represent the views of the Department of Veterans Affairs, the US government, Perelman School of Medicine, or other participating institutions
Role of Funding Sources:
This work was supported by the following grants: 1IK2CX000855 (S.C.); 1 I01 CX001957 (S.C.); I01 BX003341 (H.R.K); R01 HL143790 (P.R.G), R01 NR018836 (P.R.G.); 1K01 AA028292 (R.L.K.); and the Veterans Integrated Service Network 4 Mental Illness Research, Education, and Clinical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: Dr. Chakravorty has received research support from AstraZeneca Pharmaceuticals and Teva Pharma. Dr. Gehrman has been a consultant for Eight Sleep, Fisher Wallace Laboratories, Eisai, Inc., and Idorsia Pharmaceuticals, Inc. He has obtained research support from Merck & Co. Dr. Kranzler is a member of advisory boards for Dicerna Pharmaceuticals, Sophrosyne Pharmaceuticals, Clearmind Medicine, and Enthion Pharmaceuticals; a consultant to Sobrera Pharmaceuticals; the recipient of research funding and medication supplies for an investigator-initiated study from Alkermes; a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by Alkermes, Dicerna, Ethypharm, Lundbeck, Mitsubishi, and Otsuka; and a holder of U.S. patent 10,900,082 titled: “Genotype-guided dosing of opioid agonists,” issued 26 January 2021. The other authors report no actual or potential conflicts of interest.
Declaration of Competing Interest
No conflict declared.
Conflict of Interest:
None of the authors report any conflict of interest with this investigation and manuscript.
References
- Alexander M, Ray MA, Hebert JR, Youngstedt SD, Zhang H, Steck SE, Bogan RK, Burch JB, 2016. The National Veteran Sleep Disorder Study: Descriptive Epidemiology and Secular Trends, 2000–2010. Sleep 39, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancoli-Israel S, Roth T, 1999. Characteristics of insomnia in the United States: results of the 1991 National Sleep Foundation Survey. I. Sleep 22 Suppl 2, S347–353. [PubMed] [Google Scholar]
- Arnedt JT, Rohsenow DJ, Almeida AB, Hunt SK, Gokhale M, Gottlieb DJ, Howland J, 2011. Sleep following alcohol intoxication in healthy, young adults: effects of sex and family history of alcoholism. Alcoholism, clinical and experimental research 35, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma P, Villarroel MA, Vahratian A, 2020. Heavy Drinking Among U.S. Adults, 2018 https://www.cdc.gov/nchs/products/databriefs/db374.htm.accessed on April 26, 2021. [PubMed]
- Bowden J, Hemani G, Davey Smith G, 2018. Invited Commentary: Detecting Individual and Global Horizontal Pleiotropy in Mendelian Randomization-A Job for the Humble Heterogeneity Statistic? Am J Epidemiol 187, 2681–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen C, Psychiatric Genomics C, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control, C., Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM, 2015. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Labrecque JA, 2018. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol 33, 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Kivlahan DR, McDonell MB, Fihn SD, Bradley KA, 1998. The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med 158, 1789–1795. [DOI] [PubMed] [Google Scholar]
- Canham SL, Kaufmann CN, Mauro PM, Mojtabai R, Spira AP, 2015. Binge drinking and insomnia in middle-aged and older adults: the Health and Retirement Study. Int J Geriatr Psychiatry 30, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S, Chaudhary NS, Brower KJ, 2016a. Alcohol Dependence and Its Relationship With Insomnia and Other Sleep Disorders. Alcohol Clin Exp Res 40, 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S, Chaudhary NS, Brower KJ, 2016b. Alcohol Dependence and Its Relationship With Insomnia and Other Sleep Disorders. Alcohol Clin Exp Res [DOI] [PMC free article] [PubMed]
- Chakravorty S, Grandner MA, Kranzler HR, Mavandadi S, Kling MA, Perlis ML, Oslin DW, 2013. Insomnia in alcohol dependence: predictors of symptoms in a sample of veterans referred from primary care. The American journal on addictions / American Academy of Psychiatrists in Alcoholism and Addictions 22, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S, Grandner MA, Mavandadi S, Perlis ML, Sturgis EB, Oslin DW, 2014. Suicidal ideation in veterans misusing alcohol: relationships with insomnia symptoms and sleep duration. Addictive behaviors 39, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput JP, McNeil J, Despres JP, Bouchard C, Tremblay A, 2012. Short sleep duration is associated with greater alcohol consumption in adults. Appetite 59, 650–655. [DOI] [PubMed] [Google Scholar]
- Chaudhary NS, Wong MM, Kolla BP, Kampman KM, Chakravorty S, 2020. The relationship between insomnia and the intensity of drinking in treatment-seeking individuals with alcohol dependence. Drug Alcohol Depend 215, 108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Mak TS, O’Reilly PF, 2020. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 15, 2759–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consensus Conference P, Watson NF, Badr MS, Belenky G, Bliwise DL, Buxton OM, Buysse D, Dinges DF, Gangwisch J, Grandner MA, Kushida C, Malhotra RK, Martin JL, Patel SR, Quan SF, Tasali E, 2015a. Joint Consensus Statement of the American Academy of Sleep Medicine and Sleep Research Society on the Recommended Amount of Sleep for a Healthy Adult: Methodology and Discussion. J Clin Sleep Med 11, 931–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consensus Conference P, Watson NF, Badr MS, Belenky G, Bliwise DL, Buxton OM, Buysse D, Dinges DF, Gangwisch J, Grandner MA, Kushida C, Malhotra RK, Martin JL, Patel SR, Quan SF, Tasali E, Non-Participating O, Twery M, Croft JB, Maher E, American Academy of Sleep Medicine, S., Barrett JA, Thomas SM, Heald JL, 2015b. Recommended Amount of Sleep for a Healthy Adult: A Joint Consensus Statement of the American Academy of Sleep Medicine and Sleep Research Society. J Clin Sleep Med 11, 591–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum RM, Ford DE, Storr CL, Chan YF, 2004. Association of sleep disturbance with chronicity and remission of alcohol dependence: data from a population-based prospective study. Alcoholism, clinical and experimental research 28, 1533–1540. [DOI] [PubMed] [Google Scholar]
- Currie SR, Clark S, Rimac S, Malhotra S, 2003. Comprehensive assessment of insomnia in recovering alcoholics using daily sleep diaries and ambulatory monitoring. Alcoholism, clinical and experimental research 27, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Dashti HS, Jones SE, Wood AR, Lane JM, van Hees VT, Wang H, Rhodes JA, Song Y, Patel K, Anderson SG, Beaumont RN, Bechtold DA, Bowden J, Cade BE, Garaulet M, Kyle SD, Little MA, Loudon AS, Luik AI, Scheer F, Spiegelhalder K, Tyrrell J, Gottlieb DJ, Tiemeier H, Ray DW, Purcell SM, Frayling TM, Redline S, Lawlor DA, Rutter MK, Weedon MN, Saxena R, 2019. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun 10, 1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Hemani G, 2014. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23, R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NM, Holmes MV, Davey Smith G, 2018. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson DA, Li TK, Grant BF, 2008. A prospective study of risk drinking: at risk for what? Drug Alcohol Depend 95, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford DE, Kamerow DB, 1989. Epidemiologic study of sleep disturbances and psychiatric disorders. An opportunity for prevention? JAMA 262, 1479–1484. [DOI] [PubMed] [Google Scholar]
- Fragale JE, James MH, Avila JA, Spaeth AM, Aurora RN, Langleben D, Aston-Jones G, 2021. The Insomnia-Addiction Positive Feedback Loop: Role of the Orexin System. Front Neurol Neurosci 45, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, Guarino P, Aslan M, Anderson D, LaFleur R, Hammond T, Schaa K, Moser J, Huang G, Muralidhar S, Przygodzki R, O’Leary TJ, 2016. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–223. [DOI] [PubMed] [Google Scholar]
- Gordon AJ, Fiellin DA, Friedmann PD, Gourevitch MN, Kraemer KL, Arnsten JH, Saitz R, Society of General Internal Medicine’s Substance Abuse Interest, G., 2008. Update in addiction medicine for the primary care clinician. J Gen Intern Med 23, 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Chou SP, Saha TD, Pickering RP, Kerridge BT, Ruan WJ, Huang B, Jung J, Zhang H, Fan A, Hasin DS, 2017. Prevalence of 12-Month Alcohol Use, High-Risk Drinking, and DSM-IV Alcohol Use Disorder in the United States, 2001–2002 to 2012–2013: Results From the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry 74, 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haario P, Rahkonen O, Laaksonen M, Lahelma E, Lallukka T, 2013. Bidirectional associations between insomnia symptoms and unhealthy behaviours. Journal of sleep research 22, 89–95. [DOI] [PubMed] [Google Scholar]
- Hammerschlag AR, Stringer S, de Leeuw CA, Sniekers S, Taskesen E, Watanabe K, Blanken TF, Dekker K, Te Lindert BHW, Wassing R, Jonsdottir I, Thorleifsson G, Stefansson H, Gislason T, Berger K, Schormair B, Wellmann J, Winkelmann J, Stefansson K, Oexle K, Van Someren EJW, Posthuma D, 2017. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet 49, 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC, 2018. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen PR, Watanabe K, Stringer S, Skene N, Bryois J, Hammerschlag AR, de Leeuw CA, Benjamins JS, Munoz-Manchado AB, Nagel M, Savage JE, Tiemeier H, White T, andMe Research T, Tung JY, Hinds DA, Vacic V, Wang X, Sullivan PF, van der Sluis S, Polderman TJC, Smit AB, Hjerling-Leffler J, Van Someren EJW, Posthuma D, 2019. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet 51, 394–403. [DOI] [PubMed] [Google Scholar]
- Janson C, Lindberg E, Gislason T, Elmasry A, Boman G, 2001. Insomnia in men-a 10-year prospective population based study. Sleep 24, 425–430. [DOI] [PubMed] [Google Scholar]
- John U, Meyer C, Rumpf HJ, Hapke U, 2005. Relationships of psychiatric disorders with sleep duration in an adult general population sample. J Psychiatr Res 39, 577–583. [DOI] [PubMed] [Google Scholar]
- Johnson EO, Roehrs T, Roth T, Breslau N, 1998. Epidemiology of alcohol and medication as aids to sleep in early adulthood. Sleep: Journal of Sleep Research & Sleep Medicine 21, 178–186. [DOI] [PubMed] [Google Scholar]
- Jones SE, van Hees VT, Mazzotti DR, Marques-Vidal P, Sabia S, van der Spek A, Dashti HS, Engmann J, Kocevska D, Tyrrell J, Beaumont RN, Hillsdon M, Ruth KS, Tuke MA, Yaghootkar H, Sharp SA, Ji Y, Harrison JW, Freathy RM, Murray A, Luik AI, Amin N, Lane JM, Saxena R, Rutter MK, Tiemeier H, Kutalik Z, Kumari M, Frayling TM, Weedon MN, Gehrman PR, Wood AR, 2019. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat Commun 10, 1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneita Y, Uchiyama M, Takemura S, Yokoyama E, Miyake T, Harano S, Asai T, Tsutsui T, Kaneko A, Nakamura H, Ohida T, 2007. Use of alcohol and hypnotic medication as aids to sleep among the Japanese general population. Sleep Medicine 8, 723–732. [DOI] [PubMed] [Google Scholar]
- Kline C, 2013. Sleep Duration. In: Gellman MD, Turner JR (Eds.), Encyclopedia of Behavioral Medicine Springer, New York, NY. pp. 1808–1810. [Google Scholar]
- Kolla BP, Foroughi M, Saeidifard F, Chakravorty S, Wang Z, Mansukhani MP, 2018. The impact of alcohol on breathing parameters during sleep: A systematic review and meta-analysis. Sleep Med Rev 42, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla BP, Mansukhani MP, Biernacka J, Chakravorty S, Karpyak VM, 2020. Sleep disturbances in early alcohol recovery: Prevalence and associations with clinical characteristics and severity of alcohol consumption. Drug Alcohol Depend 206, 107655. [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Feinn R, Xu H, Ho BL, Saini D, Nicastro OR, Jacoby A, Toikumo S, Gelernter J, Hartwell EE, Kember RL, 2023. Does polygenic risk for substance-related traits predict ages of onset and progression of symptoms? Addiction [DOI] [PMC free article] [PubMed]
- Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, Tsao PS, Klarin D, Baras A, Reid J, Overton J, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, Gelernter J, 2019. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 10, 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger PM, Friedman EM, 2009. Sleep duration in the United States: a cross-sectional population-based study. Am J Epidemiol 169, 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JM, Jones SE, Dashti HS, Wood AR, Aragam KG, van Hees VT, Strand LB, Winsvold BS, Wang H, Bowden J, Song Y, Patel K, Anderson SG, Beaumont RN, Bechtold DA, Cade BE, Haas M, Kathiresan S, Little MA, Luik AI, Loudon AS, Purcell S, Richmond RC, Scheer F, Schormair B, Tyrrell J, Winkelman JW, Winkelmann J, Sleep HAI, Hveem K, Zhao C, Nielsen JB, Willer CJ, Redline S, Spiegelhalder K, Kyle SD, Ray DW, Zwart JA, Brumpton B, Frayling TM, Lawlor DA, Rutter MK, Weedon MN, Saxena R, 2019. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet 51, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N.I.A.A.A., Rethinking Drinking https://www.rethinkingdrinking.niaaa.nih.gov/How-much-is-too-much/Whats-the-harm/What-Are-The-Risks.aspx.accessed on April 24, 2021.
- O’Sullivan JW, Raghavan S, Marquez-Luna C, Luzum JA, Damrauer SM, Ashley EA, O’Donnell CJ, Willer CJ, Natarajan P, American Heart Association Council on, G., Precision M, Council on Clinical, C., Council on Arteriosclerosis, T., Vascular, B., Council on Cardiovascular, R., Intervention, Council on, L., Cardiometabolic, H., Council on Peripheral Vascular, D., 2022. Polygenic Risk Scores for Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation 146, e93–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman JA, Smit DJA, Kingma L, Vink JM, Treur JL, Verweij KJH, 2020. Causal relationships between substance use and insomnia. Drug Alcohol Depend 214, 108151. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrs T, Papineau K, Rosenthal L, Roth T, 1999. Ethanol as a hypnotic in insomniacs: self administration and effects on sleep and mood. Neuropsychopharmacology 20, 279–286. [DOI] [PubMed] [Google Scholar]
- Roehrs T, Roth T, 2018. Insomnia as a path to alcoholism: tolerance development and dose escalation. Sleep 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23 andMe Research Team, t.S.U.D.W.G.o.t.P.G.C., Adams MJ, Howard DM, Edenberg HJ, Davies G, Crist RC, Deary IJ, McIntosh AM, Clarke TK, 2019. Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry 176, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R, 2015. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.C., 2018. R: A language and environment for statistical computing R Foundation for Statistical Computing., Vienna, Austria. [Google Scholar]
- Torkamani A, Wineinger NE, Topol EJ, 2018. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19, 581–590. [DOI] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG, 2013. Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet 43, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu SA, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen LS, Clarke TK, Chou YL, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, Foo JC, Fox L, Frank J, Giegling I, Gordon S, Hack LM, Hartmann AM, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Jan Hottenga J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Lai D, Ligthart L, Loukola A, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Meyers JL, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang JC, Webb BT, Wedow R, Wetherill L, Wills AG, andMe Research T, Boardman JD, Chen D, Choi DS, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye MA, Gabel W, Hayward C, Ising M, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Mannisto S, Muller-Myhsok B, Murray AD, Nurnberger JI, Palotie A, Preuss U, Raikkonen K, Reynolds MD, Ridinger M, Scherbaum N, Schuckit MA, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma DI, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AM, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer CJ, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak VM, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes HH, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nothen MM, Palmer AA, Pedersen NL, Penninx B, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen PH, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A, 2018. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 21, 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman MM, Greenwald S, Nino-Murcia G, Dement WC, 1997. The morbidity of insomnia uncomplicated by psychiatric disorders. Gen Hosp Psychiatry 19, 245–250. [DOI] [PubMed] [Google Scholar]
- Wray N, Visscher P, 2008. Estimating trait heritability. . Nature Education 1, 29. [Google Scholar]
- Yavorska OO, Burgess S, 2017. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 46, 1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


