Abstract
In recent years, there has been a surge in the innovative modification and application of the viral vector-based gene therapy field. Significant and consistent improvements in the engineering, delivery, and safety of viral vectors have set the stage for their application as RNA interference (RNAi) delivery tools. Viral vector-based delivery of RNAi has made remarkable breakthroughs in the treatment of several debilitating diseases and disorders (e.g., neurological diseases); however, their novelty has yet to be fully applied and utilized for the treatment of cancer. This review highlights the most promising and emerging viral vector delivery tools for RNAi therapeutics while discussing the variables limiting their success and suitability for cancer therapy. Specifically, we outline different integrating and non-integrating viral platforms used for gene delivery, currently employed RNAi targets for anti-cancer effect, and various strategies used to optimize the safety and efficacy of these RNAi therapeutics. Most importantly, we provide great insight into what challenges exist in their application as cancer therapeutics and how these challenges can be effectively navigated to advance the field.
Keywords: RNA interference, RNAi therapeutics, viral vectors, cancer therapeutics, microRNA, silencing RNA, short hairpin RNA, DNA viruses, RNA viruses, oncolytic viruses, replicating viruses, non-replicating viruses, integrating viruses, adenovirus, AV, adeno-associated virus, AAV, herpes simplex virus, HSV, retroviruses, vesicular stomatitis virus, VSV
Graphical abstract
Ilkow and colleagues highlight that while recent advances in viral vector-based gene therapy show potential in cancer treatment, their full potential remains largely untapped. This review explores promising RNA-interference delivery methods, their efficacy, and their challenges in cancer therapy.
An introduction to RNA interference mechanisms and delivery methods
What is RNA interference?
RNA interference (RNAi) describes the mechanism of gene expression knockdown by disrupting cellular messenger RNA (mRNA) levels using short sequences of non-coding RNA. This phenomenon was first described by Andrew Fire and Craig C. Mello in 1998 when they described genetic interference via the injection of double-stranded RNA (dsRNA) in a C. elegans nematode model.1 In the following years the silencing mechanism was slowly elucidated, including the discovery of silencing intermediates such as small interfering RNA (siRNA) and identification of enzymes responsible for RNA cleavage and other regulatory pathways.2 In a physiological setting, endogenous RNAi effectors are used in the immune response, particularly in anti-viral defense to knock down essential viral proteins, thereby limiting virus propagation.3 When applied to the current research landscape, RNAi has since become the standard for transient gene knockdown studies.
The three most common types of RNAi species are siRNA, short hairpin RNA (shRNA), and microRNA (miRNA), each type differing in features including base-pair length, structure, and mechanism of gene regulation.2 When dealing with single gene knockdown, siRNA is typically used given that it is almost fully complementary to its target mRNA, thus conferring maximum specificity. These 21- to 23-nt RNA sequences, with a 2-nt overhang at the 3′ end, result either from the cellular processing of dsRNA by Dicer, a specialized ribonuclease III-like enzyme, or can directly be artificially synthesized.4 Similarly, shRNA is typically 50–70 bp, with dsRNA bridged by a single-strand loop with a 3′ overhang.5 This effector also inhibits protein translation through the same direct mRNA-degrading mechanism but is significantly more efficient.6,7 On the other hand, miRNA species exist endogenously; they are first processed within the nucleus and then exported to the cytoplasm where they are further processed by the Dicer complex. Unlike siRNA, mature miRNA effectors are capable of silencing multiple mRNA targets via partial complementation to the 3′ untranslated region (UTR) to repress its translation.8 Among these, there also exist other RNAi effectors such as piwi-interacting RNA and guide RNA for CRISPR-Cas9 applications.9,10
The development of RNAi therapeutics
Since its discovery, the therapeutic potential of RNAi has always been highly touted. Given that its target specificity is largely based upon genetic sequence, short sequences of non-coding RNA can be designed against virtually any cellular target, including targets without an available pharmacological inhibitor.11 As such, the concept of versatile, post-translational knockdown therapeutics was poised to revolutionize the entire field of gene therapy and become a powerful tool for targeting the “undruggable” targets. Despite challenges related to site-specific delivery, knockdown efficacy, and potential off-target toxicities, the United States Food and Drug Administration (FDA) approved the first RNAi-based therapeutic in August 2018. Patrisiran (Onpattro) uses lipid nanoparticles that deliver a small interfering RNA (siRNA) to knock down transthyretin (TTR) gene expression for the treatment of polyneuropathy in patients with hereditary TTR-mediated amyloidosis.12 The FDA has since approved two other RNAi-based therapeutics: givosiran (Givlaari) for acute hepatic polyuria in 2019 and lumasiran (Oxlumno) for primary hyperoxaluria in 2020. Moreover, currently seven other siRNA-based therapies are undergoing phase 3 clinical trial investigation.13 With respect to cancer therapy, there are many potential therapeutic roles for RNAi. In personalized medicine, genetic screening for overexpressed or overactive cancer driver mutations can identify effective knockdown targets tailored to each tumor. For example, a phase 1 trial of exosome-delivered siRNA targeting KrasG12D in KRAS-mutated pancreatic ductal adenocarcinoma is currently under way (NCT03608631).
Preferential delivery of RNAi by viral vectors
Although many therapeutic RNAi delivery strategies have been developed, one of the largest obstacles to their clinical application is the effective delivery of the RNAi effectors for potent gene knockdown. A vehicle, such as a viral vector or nanoparticles, is required, as naked RNA molecules are rapidly degraded by nucleases present in the extracellular milieu. Accordingly, a competent delivery strategy should encompass the following qualities: (1) the ability to safeguard the RNAi effector in extracellular space; (2) the ability to seamlessly penetrate the cellular membrane; and (3) the ability to release the RNAi effector into the cytoplasm when appropriate.14 The simplest delivery vehicle is liposome-mediated transfection or lipofection, which describes the packaging of the RNAi effector into a phospholipid bilayer complex that merges with the cellular membrane and releases the effector into the cytoplasm. Representing the simplest form of RNAi delivery, this strategy is very well established for routine use in experimental applications in laboratory settings. Despite this, its applications are limited by low target specificity and its reduced stability in vivo. Meanwhile, nanoparticle-based delivery technologies are being continuously investigated and improved by using newly developed nanomaterials with greater stability including nanotubes, quantum dots, and dextran cages.15
To further improve cell selectivity, nanoparticles can be conjugated to biomolecules such as peptides or antibodies for targeted delivery of RNAi to specific cell types or diseased cells. In a hallmark study by Song et al., protein-encased siRNA conjugated to HIV type 1 (HIV-1) envelope antigen-binding fragments (Fab) demonstrated target specificity to HIV envelope-expressing melanoma cells in vivo.16 Similar strategies, however, are notably reliant on non-specific electrostatic interactions between the RNAi carrier and the biomolecule, leaving them prone to aggregation events and subsequent unpredictable pharmacokinetics.17 The majority of these RNAi delivery methods may also be limited by off-site toxicity. For example, nanoparticles that fail to extravasate from the blood to the site of interest often end up accumulating in the liver. While recent technological advancements have shown glimpses of accomplishing selective delivery to mitigate toxicities, these strategies have complicated preparation procedures and are often expensive to manufacture.15,18
As a simple solution to alleviate these concerns of low stability and specificity, viral vectors represent an intriguing and naturally occurring option.19 First, viruses are stable in extracellular environments and already excel at efficiently delivering genetic material to cellular targets. Viral vectors such as retroviruses, lentiviruses, and adenoviruses have well-characterized modes of transmission and gene transfer mechanisms, unlike the aforementioned physical means of RNAi transfection.20 For example, adenoviruses have already been extensively explored in cancer gene therapy, delivering genes that trigger apoptosis (e.g., p53) or stimulating anti-tumor immune responses (e.g., interleukin-2 [IL-2]).21 Second, especially in the field of cancer therapy, several virus types have natural tumor tropisms.22,23 In the process of attaining neoplasticity, early on cancer cells often lose immunoregulatory mechanisms, thus becoming exquisitely susceptible to viral infection. This phenomenon is the basis of oncolytic virotherapy.24 Similar to nanoparticle-antibody conjugation, the tissue selectivity of viral vectors can also be further accomplished through “pseudotyping,” which is the incorporation of envelope material of other viral types to modulate its natural tissue tropism,25 or through “retargeting,” which involves the reprogramming of viral surface-exposed components with single-chain variable fragment antibodies (scFvs) or other cell-targeting moieties.26 Finally, with the recent advances in the field of synthetic biology, the production of genetically engineered viral vectors has become increasingly simple and cost-effective, making it an attractive therapeutic option with the capacity to deliver multiple RNAi species at once.27 In this review (summarized in Figure 1), we present viruses as optimal vectors for RNAi therapeutics and summarize the current strategies employed to target obstacles limiting their success in the clinic.
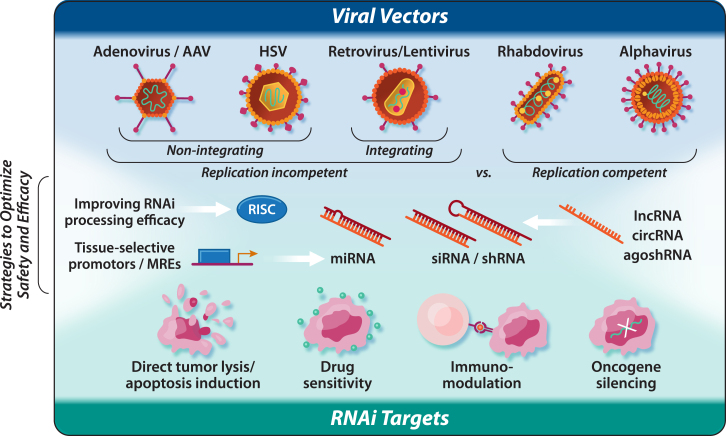
Figure 1.
Viruses as optimal vectors for RNA interference delivery
A graphical depiction of the major sections of the review. The selection of different viral vectors is first outlined, which can broadly be divided into replication-incompetent vs. replication-competent vectors. Classification of RNA interference targets with anti-cancer effects are then summarized. Finally, strategies to optimize the safety and efficacy of these RNA-interference-expressing viral vectors is explored.
The versatility of viral vectors for RNAi delivery
Considerations in using viruses as RNAi delivery systems
There has undoubtedly been a recent surge in the innovative modification and application of viral vector-based gene therapy.19 The last decade has seen significant and consistent improvements in the engineering, delivery, and safety of viral vectors as viable RNAi delivery tools in the clinic. Classically, the most common viral vector gene delivery candidates have proved to be the retroviruses (e.g., gammaretroviruses [γ-retroviruses] and lentiviruses), herpes simplex viruses (HSVs), adenoviruses (AdVs), and adeno-associated viruses (AAVs).28 These five main classes of viral vectors can be categorized into two groups according to whether their genomes integrate into host cellular chromatin (e.g., retroviruses and lentiviruses) or persist in the cell nucleus predominantly as extrachromosomal episomes (e.g., AAV, AdV, and HSV). More recently, self-replicating cytoplasm RNA viruses (e.g., vesicular stomatitis virus [VSV]) have also emerged as promising candidates for gene delivery, especially within the field of oncolytic virotherapy.29,30 Moreover, some of these classes have already been tested for the delivery of RNAi. Selection for their use in cancer therapy will depend on differences in key features, namely their efficacy, specificity, stability, and safety. In this section, we discuss features of the most common viral vector-based RNAi delivery agents, the integrating and non-integrating viral vectors, as well as introducing the emerging class of self-replicating cytoplasmic RNA viruses as it pertains to their application as RNAi delivery tools, both inside and outside the field of cancer therapeutics (Table 1).
Table 1.
Available viral vectors for RNA interference delivery
| Genus | Main representative | Genome (sense) | Genome size (kb) | Immunogenicity | Duration of expression | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| Gamma (γ) retrovirus | murine leukemia virus (MLV) | ssRNA (+) | ∼7–12 | low | long term/permanent | persistent gene transfer in most tissues—broad cell tropism | integration might induce oncogenesis |
| RRV; confer a considerable degree of natural specificity for tumors (cancer therapy) | unable to transduce non-dividing cells (advantageous for cancer therapy) | ||||||
| RRV; non-cytolytic—allowing for persistent gene expression in transduced cells | site-specific delivery in vivo | ||||||
| Lentivirus | human immunodeficiency virus (HIV) 1 | ssRNA (+) | ∼10 | low | long term/permanent | persistent gene transfer in most tissues—broad cell tropism | integration might induce oncogenesis |
| capable of transducing dividing and non-dividing cells | site-specific delivery in vivo | ||||||
| extensive experience optimizing RNAi expression via ex vivo engineering of stem cells | |||||||
| Adenovirus | adenovirus (AdV) 5 | dsDNA | ∼26–45 | high | long term (cell type dependent) | efficient transduction of target cells at a low multiplicity of infection | immune response to viral proteins (CRAdV) |
| high probability of pre-existing immunity (AdV 1, 2, 5, 6) | |||||||
| selective and potent cancer-cell killing properties (oncolytic) | liver is often default destination | ||||||
| Adeno-associated virus | adeno-associated virus (AAV) serotype 2 | ssDNA | <5 | low | long term (cell type dependent) | low immunogenicity and no pathogenicity | low transduction efficiency |
| broad cell tropism | difficult to generate high titers | ||||||
| liver is often default destination | |||||||
| Herpes simplex virus | herpes simplex virus (HSV) type I | dsDNA | ∼150 | high | short to medium term | well suited as oncolytic vector and CNS applications (retrograde axonal transport) | risk of recombination with latently herpes simplex virus-infected cells |
| suitability as an RNAi delivery tool has been limited to in vitro investigations | |||||||
| Vesiculovirus | vesicular stomatitis virus (VSV) | ssRNA (−) | 11 | high | short term | cytoplasmic replication allowing high expression levels and potential non-canonical processing of RNAi | very sensitive to the anti-viral action of interferon |
| apoptosis induction, oncolytic in cancer gene therapy applications | neurotoxicity associated with viral glycoprotein | ||||||
| genetic structure allows for easy manipulation | |||||||
| high titer production | |||||||
| Flavivirus | West Nile virus (WNV), tick-borne encephalitis virus (TBEV) | ssRNA (+) | 11 | medium | short term | cytoplasmic replication allowing high expression levels and potential non-canonical processing of RNAi | toxicity (non-cytopathic vectors available) |
| infect neurons in primary and cell lines, could be good candidates for gene therapy in CNS | pre-existing immunity mainly in (sub)tropical countries | ||||||
| high titer production | |||||||
| Alphavirus | Sindbis virus (SINV), Semliki Forest virus (SFV), Venezuelan equine encephalitis virus (VEE) | ssRNA (+) | 11 | high | short term | cytoplasmic replication allowing high expression levels and potential non-canonical processing of RNAi | toxicity due to viral replication (non-cytopathic vectors overcome this limitation) |
| apoptosis induction, oncolytic in cancer gene therapy applications | |||||||
| high titer production |
Retroviruses as integrating viruses for the delivery of RNAi
Characterized by their unique ability to efficiently integrate their viral genome into host cells, retroviruses are a family of enveloped, positive-sense, single-stranded RNA (ssRNA) viruses defined by the enzymatic activities of reverse transcriptase and integrase.31 While several retroviruses have been investigated for various gene therapy applications, γ-retroviruses and lentiviruses are the most extensively studied and frequently modified for use as replication-incompetent vectors to deliver RNAi to mammalian cells.32,33 Their genome is largely non-overlapping and thus relatively amenable to manipulation, while the separation of cis (i.e., packaging signal) and trans (i.e., gag, pol, env) elements generates a simple recombinant retroviral system with up to 8 kb of transgene coding capacity suitable for the easy production of replication-defective recombinant RNAi retrovirus.31 Great advancements in the engineering of retroviral production systems, notably the advent of self-inactivating vectors, have greatly increased the safety profile of these vectors as RNAi delivery tools in vivo. Upon their activation, any subsequent spread is abolished and the induction of immune-related responses following transduction is minimized as no viral proteins are synthesized.34,35 These advancements ultimately result in relatively lower immunogenicity profiles compared to other viral platforms.36
Retroviruses replicate through a double-stranded DNA (dsDNA) intermediate and integrate their genomes stably into the host cells’ DNA, a unique feature that allows for long-term expression of RNAi molecules.31 While the γ-retroviruses (e.g., murine leukemia virus [MLV]) are only capable of integrating their viral genome into the host cells’ genomic DNA during the mitotic phase of the cell cycle, lentiviruses are capable of inducing stable and long-term gene silencing in both dividing and non-dividing cells.31 As such, lentiviral vectors serve as a more attractive option for the delivery of RNAi to the central nervous system (CNS), where they have been shown to efficiently transduce CNS neurons and mediate RNA silencing in the brain and spinal cord in vivo to successfully ameliorate several animal models of CNS diseases/disorders. The first studies to use lentiviral-mediated delivery of RNAi to pre-clinically treat CNS disorders employed an shRNA-based lentiviral approach to silence a disease-causing gene (SOD1) in mouse models of familial amyotrophic lateral sclerosis (ALS). Silencing the expression of SOD1 by lentiviral delivery of shSOD1 to familial ALS mice increased motor neuron survival, improved motor performance, and successfully delayed the onset and slowed down the progression of the disease.37 Indeed, retroviral delivery of RNAi has also shown remarkable pre-clinical success in the treatment of several neurodegenerative diseases and CNS disorders including, but not limited to, Huntington’s disease,38 Parkinson’s disease,39,40 Alzheimer’s disease,41,42 prion disease,43,44 and spinal cord injury.45
Lentiviruses offer the potential to transduce stem cells, making them particularly attractive tools for the delivery of RNAi to these non-proliferating or slowly proliferating (and often difficult to transduce) cell types. As such, lentivirus-based RNAi delivery has become a useful tool for the in vitro or ex vivo engineering of immune cells in the treatment of chronic viral infection. Hematopoietic stem cells (HSCs) can be engineered to resist viral infections through transduction with a lentivirus-encoding anti-viral RNAi effector.46,47 This approach was first pioneered by transplanting human CD34+ HSCs transduced with a lentivirus expressing an anti-HIV-1 shRNA (shRNA against rev) into thymus and liver grafts in humanized SCID (severe combined immunodeficiency disease) mice.48 These studies demonstrate that transduced T cells and macrophages isolated from mice were shown to resist HIV-1 challenge.48,49,50 Later this approach was adopted to transduce CD34+ HSCs with single or bispecific lentiviral constructs expressing shRNAs against the host cell factors, CCR-5 and/or CXCR-4, which subsequently gave rise to progeny macrophages resistant to HIV-1.51 In non-human primates, a stable reduction of CCR-5 in progeny T cells transduced with shCCR-5 ex vivo was observed, and shRNA transgene expression was sustained for over a year in vivo.52 RNAi has also been combined with other types of gene therapy approaches in a single lentiviral vector.47,53 For example, people living with HIV-1/AIDS suffering from malignant lymphomas may undergo autologous transplantation with peripheral blood-derived CD34+ hematopoietic progenitor cells transduced with lentivirus encoding the 3-RNA-based anti-HIV-1 moieties (Tat/Rev shRNA, TAR decoy, and CCR5 ribozyme) as treatment. Importantly, it is well documented that the vector persists in multiple cell lineages with prolonged siRNA expression, albeit at low levels, for up to 24 months.46 Using similar approaches, lentiviral RNAi delivery systems have been employed to express anti-viral RNAi mediators for the treatment of many chronic viral infections including, but not limited to, encephalitogenic flavivirus54,55 and Coxsackie B infections,56 as well as targeting the viral oncogenes E6 and E7 in human papillomavirus transformed carcinomas.57,58
Despite the recent successes discussed above, the in vivo application of retroviral-based RNAi delivery so far has been largely limited to local administration (i.e., treatment of neurological disease) or ex vivo approaches (i.e., HSC programming) owing to safety and efficacy limitations associated with systemic delivery.59,60 The irreversible and stable nature of retroviral integration means that targeting recombinant retroviral particles to desired cell types is essential for the safe and effective systemic administration of retroviral-RNAi therapies. Currently, retroviral cell targeting cannot be accomplished using retroviral vectors pseudotyped with the glycoprotein of VSV (VSV-G) given its broad tissue tropism through binding to the ubiquitously expressed cell-surface low-density lipoprotein receptor.61,62 Several diverse approaches have instead been used to alter retroviral tropism and/or develop highly targeted retroviral delivery systems, including the incorporation of heterologous attachment glycoproteins, single-chain and bispecific antibody adaptors, and genetic-based systems that alter glycoprotein tropism.63 While these changes improved target cell specificity, in many cases they were also accompanied by reduced transduction efficiency.64,65,66 Subsequently, the development of highly targeted retroviral delivery systems has remained one of the largest obstacles for the systemic delivery and clinical applicability of retroviral-based delivery of RNAi in the treatment of many human diseases including cancer.
In the context of treating human malignancies, lentiviruses do not have any natural tumor tropism; therefore, they require a targeted delivery strategy for the delivery of RNAi to tumor cells (e.g., protease-activated Env proteins).67,68,69 However, the γ-retroviruses possess a stringent requirement for cell division to achieve productive infection and preferentially replicate in cells with defective innate immunity, making them uniquely well suited for use in cancer therapy.70,71,72 By retaining all of the elements necessary for viral replication, retroviral replicating vectors (RRVs) based on γ-retroviruses (i.e., MLV) are capable of transmitting genes via exponential in situ amplification and are currently being pursued as therapeutic agents for cancer.73,74,75,76,77,78 While many other virus types being investigated for this purpose are inherently cytolytic, RRVs confer a considerable degree of natural specificity for tumors without the immediate induction of cytolysis which can contribute to longer-lasting therapeutic efficacy and be particularly advantageous for RNAi therapeutics.79 Furthermore, RRV’s non-lytic replication cycle does not trigger immediate anti-viral immune responses, allowing for sustained viral replication and therapeutic transgene expression into the tumor microenvironment.79 These factors alongside a growing safety and drug activity record in humans suggest that these delivery vectors could allow effective use of RNAi strategies in human cancers; however, this potential utility has yet to materialize.
Adenovirus, adeno-associated virus, and herpes simplex virus vectors as non-integrating vectors for the delivery of RNAi
While integrating viruses offer the potential for stable, long-term transgene expression through their capacity to integrate in the host cell genome, this unique feature can also pose great genotoxic risk and in some cases even induce oncogenesis.80,81 Non-integrating vectors specifically share a reduced risk of genotoxicity, offering a safer profile in vivo and in vitro. In contrast to retroviruses, their genomes exist and replicate efficiently as episomes during infection producing high yet transient expression of transgenes; however, expression can still be retained for long periods in post-mitotic tissues.80 AdV, AAV, and HSV vectors are three examples of non-integrating viruses that have been employed for RNAi delivery.82,83,84 All three of these vectors are capable of transducing or infecting dividing and non-dividing cells, thus offering excellent potential for RNAi delivery to cells in the CNS and other difficult-to-transduce cell types, such as stem cells.
Herpesviruses are an important family of dsDNA viruses known for their elaborate and large genome (152 kb), which encodes more than 80 gene products. Several genes involved in HSV replication, virulence, and immune evasion are non-essential for the viral life cycle in in vitro cell cultures. These genes can be deleted or modified, alone or in combination, to create attenuated and/or safer HSV mutants. In the context of cancer therapy, many of these mutants present with a reduced ability to replicate in normal quiescent cells but can grow efficiently in tumor or dividing cells, setting the stage as cancer therapeutic RNAi delivery vehicles.85,86 Recent efforts have also been made to further modify the envelope of the HSV-1 virion to target specific receptors that selectively increase infectivity of tumor cells bearing corresponding receptors.87,88 Although well exploited as an oncolytic virotherapy platform, HSV-1 has unfortunately been less explored as an RNAi delivery vehicle compared to the retroviruses AdV and AAV. However, HSV amplicon vectors expressing shRNA have been used recently to mediate post-transcriptional silencing of epidermal growth factor receptor (EGFR), which is frequently activated in human glioblastoma cells,84 and to inhibit the expression of BK polyomavirus (BKV) T antigen and tumorigenicity of BKV-transformed cells in vitro.89
Adenoviruses are non-enveloped viruses containing a dsDNA genome that provide efficient transduction of target cells at a low multiplicity of infection and have well-established methods for manipulation and propagation.90 Compared to retroviruses, these vectors have more established manufacturing capabilities.90 A recombinant AdV (rAdV) encoding shRNA (rAdV-shAbcc2) has been employed in vivo to target the murine ATP-binding cassette multidrug resistance protein 2 (Abcc2), a protein involved in the transport of bilirubin out of liver cells and into the bile. C57/BL6 mice injected with rAdV-shAbcc2 showed significant impairment of Abcc2 function for up to 3 weeks, as reflected by high levels of processed shRNA targeting Abcc2, specific reduction of Abcc2 mRNA, and increased serum bilirubin levels. These results were the first of several to indicate that AdV vectors can be used to express sufficient levels of shRNA capable of silencing target genes in the liver of mice.91
In the field of cancer gene therapy, AdV has gained considerable attention because of its selective and potent cancer-cell killing properties, amplified transgene expression, and additional therapeutic efficacy by shedding of virus progeny. The first strategy of oncolytic AdV armed with RNAi involved the use of conditionally replicating AdV (CRAdV) encoding shRNA against firefly luciferase. This proof-of-principle study demonstrated that siRNAs expressed from CRAdV could suppress the expression of firefly luciferase while the efficiency of silencing increased during viral replication.92 Zhang et al. later adopted the oncolytic AdV-RNAi platform to achieve siRNA-mediated gene silencing that led to tumor cell death.83 The authors engineered a novel oncolytic AdV carrying a mutant Kras siRNA transgene (AdV-siRNA Kras) which demonstrated an additive tumor growth-inhibitory response on human cancer cells through siRNA-mediated Kras knockdown and AdV-mediated cancer cell lysis. In a subcutaneous mouse xenograft model of H79 pancreatic cancer, daily intratumoral injections of AdV-siRNA Kras significantly reduced tumor growth (85.5% growth reduction) relative to parental AdV (47.8% growth reduction) or AdV expressing siRNA targeting GFP (44.1% growth reduction). Tumors were characterized by marked downregulation of Ras-signaling-related gene expression (AKT2, GSK3β, E2F2, and MAP4K5) and cell-cycle blockage reflecting potent siRNA Kras transgene activity.83 Since then, AdV-mediated delivery of RNAi effectors with anti-angiogenic (e.g., vascular endothelial growth factor [VEGF], IL-8) and anti-tumor properties (e.g., Ki67, MYCN) as well as the ability to sensitize cancer cells to chemotherapeutics (e.g., Survivin, Akt) have been tested the treatment of breast cancers,93 bladder cancer,94 neuroblastomas,95 prostate cancers,96 pancreatic cancers,83 lung cancers,97 colorectal cancers,97,98,99 and hepatocellular carcinomas100,101 in the pre-clinical setting.
Oncolytic AdV vectors undoubtedly possess the capacity to deliver RNAi species to tumor cells for efficient gene knockdown; however, a major limitation to the clinical use of vectors is the host immune response.102,103,104 Neutralizing antibodies and pre-existing immunity represent two significant barriers to repeated vector administration of AdV-based delivery of RNAi.105 Low-level expression of viral vector genes in such settings almost always results in the generation of immune responses directed against AdV-transduced cells and ultimately in the loss of transgene expression. On the other hand, AAV is highly valued for its lack of pathogenicity in multiple vertebrate species, including human and non-human primates.106,107 Owing to their relatively low immunogenicity and their ability to mediate persistent gene expression, AAV vectors are the most actively investigated gene therapy vehicles, currently being tested in several human gene therapy trials.19,28
AAVs are a unique group of non-enveloped single-stranded DNA (ssDNA) viruses characterized by their reliance on helper viruses (i.e., adenoviruses) to support their propagation. In the most commonly used recombinant AAV (rAAV) systems, all AAV protein-coding sequences are removed to incorporate a payload that is flanked by AAV inverted terminal repeats.108 When designing vectors for gene replacement therapies, the relatively limited packaging capacity (∼4.7 kb) of AAV typically represents a disadvantage; however, this does not apply to RNAi-based applications. Separate or combined packaging constructs containing AAV rep and cap genes alongside adenoviral helper genes required for replication are provided in trans to produce replication-deficient AAV virions. While in quiescent cells, AAV’s stable transgene expression can be observed for multiple years; in rapidly dividing cells such as tumor cells, episomal AAV is gradually diluted and in some cases even lost over the repeated rounds of cell division. Of note, the AAV genome cannot replicate along with the host cell DNA, leading to loss of the transgene expression.80 Reports on the stability and duration of transgene expression are variable, and the exact molecular processes involved in establishing stable gene transduction remain under investigation. As such, it is very difficult to predict with certainty the duration of transgene expression from AAV, particularly in the case of cancers. Nonetheless, AAV is the first RNAi-based gene therapy viral delivery system to be used in humans in clinical settings (NCT01899092). TT-034, an AVV-based RNAi product for the treatment of hepatitis C virus (HCV) infection, is composed of an AAV8 vector carrying three different anti-HCV shRNAs that cleave the 5′ UTR and two coding NS5B regions in the HCV genome. The clinical trial data demonstrate that TT-034 is well tolerated, safe, and can effectively transduce hepatocytes and concurrently express three anti-HCV shRNAs in human subjects infected with HCV.82 As a result of the increasing competitive landscape in HCV treatment and the time required to get TT-034 to market, TT-034 has received limited partnering interest, preventing its clinical advancement. Despite this fact, TT-034 has provided a major proof-of-concept AAV-based RNAi delivery platform and has built a solid foundation for the use of viral vector-based RNAi delivery platforms, particularly AAV, in the treatment of human disease.
AAV has also shown pre-clinical promise for the delivery of RNAi therapeutics to treat cancers. For instance, silencing of the human telomerase reverse transcriptase using an AAV vector-based approach was shown to restore apoptosis in human oral squamous cells both in vitro and in vivo.109 Furthermore, expression of endogenous RNAi mediators such as miRNA-7 from AAV decreased tumor growth in human glioblastoma mouse xenograft models through downregulation of the growth-promoting EGFR pathway and upregulation of death receptor pathways.110 AAV-based delivery of RNAi has also been employed to target the expression of cancer-promoting miRNAs (e.g., miRNA-21). AAV can mediate stable expression of an shRNA targeting miRNA-21 and thus attenuate HT29 human colon carcinoma and PC3 human prostate tumor growth in mice.111
One of the main challenges with AAV-based cancer gene therapy is improving the AAV-specific transduction of cancer cells. Efficient targeting of cells and tissues beyond the liver remains a challenge for both AdV- and AAV-based RNAi delivery. Systemic administration of AdV and AAV vectors often results in liver retention, thus representing a key barrier when other organs are the intended targets.108,112 Fortunately, the engineering of novel AAV capsids has been a constant pursuit to improve and expand AAV biodistribution and transduction efficiency.108 To date, 12 different AAV serotypes have been translated into rAAV-based delivery systems. These vectors have variable tropism due to the differential binding of viral capsid proteins to specific cell-surface receptors. Many attempts have also been made to increase the target specificity of rAAV vectors using natural discovery,113 rational design (e.g., capsid/host cell biology),114,115 and directed evolution techniques (e.g., error-prone PCR),116,117 setting the stage for the future of highly targeted rAAV/AdV RNAi delivery systems.
Cytoplasmic RNA viruses for the delivery of RNAi
Although less explored, self-replicating cytoplasmic RNA viruses (cRNA) represent another valuable option for the delivery of RNAi therapeutics. Unlike retroviruses and lentiviruses, cRNA viruses have an ssRNA genome that replicates without reliance on any DNA intermediates. As such, viral genomic sequences do not integrate into the host cell genome, and their transcription and replication are restricted to the cytoplasm. Although many cRNA viruses have proved to have off-target toxicity, several cRNA viruses lack toxicity or can be genetically modified to be used as safe viral vectors for therapeutic delivery.30,118 Moreover, many cRNA viruses possess an inherent oncolytic capacity, making them an attractive tool for the delivery of RNAi to treat human malignancies.119,120 Compared to integrating RNA viruses, cRNA viruses ultimately pose less of an oncogenic risk due to the lack of viral sequence integration into the host genome. Because of their high replicative capacity, cRNA viruses such as alphaviruses, flaviviruses, and rhabdoviruses provide both efficient delivery and high-level expression of transgenes.30,118 These viral vectors are of ample use for delivering therapeutic payloads, including vaccine development and gene therapy-based immunotherapy.30
Historically, the restriction of cRNA virus transcription to the cytoplasm was initially theorized to prevent the adequate processing of certain RNAi intermediates by preventing access to canonical miRNA processing elements in the nucleus (e.g., Drosha/DGCR8). However, cytoplasmic RNA viruses can induce the accumulation of RNAi processing machinery (e.g., Drosha) in the cytoplasm.121 The accumulation of Drosha in the cytoplasm following infection with cRNA viruses expressing RNAi species allows Drosha to act on viral RNA in the cytoplasm to produce pre-miRNA that is subsequently processed into double-stranded RNAi effectors that can engage their target.29 This theory has been supported, for example, by the discovery of several cRNA virus-derived small RNAs and a functional mature miRNA-like structure (KUN-miR-1) expressed from the Kunjin strain of West Nile virus (WNVKUN).122 Another potential barrier for cRNA virus delivery of RNAi species is that the potential excision of RNAi precursors from the viral RNA genome can also destroy the viral genome and thus reduce the efficiency of viral replication. However, the presence of a functional and naturally occurring miRNA precursor element in the tick-borne encephalitis virus (TBEV, a cytoplasmic RNA virus) genome was shown to have no measurable negative impact on viral replication.29 To date, multiplexed high-throughput sequencing has revealed populations of small RNAs (10–60 nt long) produced in cells following infection with six different cytoplasmic RNA viruses. While the secondary structure of these RNAs differs from traditional miRNAs, populations of virally produced small RNAs that exist as duplexed siRNAs have been identified and strand-selective loading of viral siRNAs onto Argonaute complexes observed.123 Although the structure of these small RNAs suggests that miRNA processing may occur through alternative non-canonical pathways, these findings demonstrate that RNAi effectors can be expressed from cRNA viruses. By developing a better understanding of the structure and biogenesis of viral small RNAs, future work may exploit the intrinsic nature of these species for the development of targeted RNAi delivery by cRNA viruses.
Recent discoveries in the field of viral delivery have shown that both positive-sense and negative-sense cytoplasmic RNA viruses including TBEV, Sindbis virus, and VSV have been engineered to produce RNAi intermediates and subsequently induce post-transcriptional gene silencing of target genes.29,124,125 These replicating viral vectors represent a safe delivery method for RNAi and a potent strategy for the induction of transcriptional gene silencing, due to their capacity to express RNAi species without integrating viral sequences into the host genome.125,126 Similarly, we recently employed a Sindbis virus library to perform an in vitro screen based on virus-encoded artificial miRNAs (amiRs) targeting ∼16,000 mammalian genes to identify amiRs that can confer a replicative advantage to oncolytic virus (OV) platforms. Results revealed that amiRNA, termed amiR-4, targets ARID1A, a protein involved in chromatin remodeling and an important player in mediating resistance to OV replication. An OV backbone armed with amiR-4 enhanced OV replication and survival of tumor-bearing xenograft and immunocompetent murine models.127 While still at early stages of pre-clinical development, recent evidence highlights that replicating cRNA viruses can be employed as delivery vehicles for RNAi-based gene silencing for therapeutic interventions in various diseases, including the treatment of human cancers. More research is needed to unfold the full potential of cRNA viruses as safe delivery vehicles for RNAi therapeutics.
Applying RNAi to cancer therapy
Viral-delivered miRNA targets for cancer therapy
Given the powerful and versatile capabilities of miRNAs as biodrugs, many academic and pharmaceutical research groups are exploring the application of miRNA delivery via viral vectors for the treatment of cancer. The intended application of these different therapeutic miRNAs can be broadly classified into three modes of action: (1) induction of tumor lysis, (2) inhibition of tumor processes, and (3) sensitization to other therapy regimens. Delivery of RNAi effectors against these targets by non-replicating viral vectors has shown promise across different pre-clinical cancer models. While only a handful of examples will be covered here, a list of all recently tested viral vectors delivering RNAi payloads in vivo is included in Figure 2.
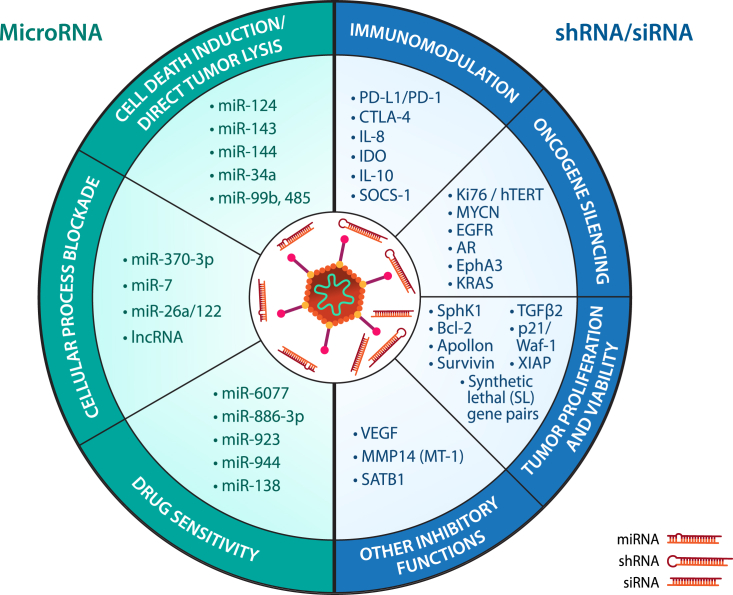
Figure 2.
The therapeutic potential of RNAi species and their link to cancer therapy
(Left) The delivery of microRNA by viral vectors can be divided into three major mechanisms of action: cell-death induction/direct tumor lysis, cellular process blockade, and drug sensitivity, each aiming to induce greater cell death and discourage neoplastic growth in the infected tumor cells. The endogenous and multi-targeting nature of microRNA gives delivery of this RNAi species tremendous versatility in cancer therapy. (Right) The delivery of shRNA/siRNA by viral vectors can be divided into four major mechanisms of action: immunomodulation, oncogene silencing, tumor proliferation and viability, and other inhibitory functions. If the correct gene is chosen, the strength of single gene knockdown by shRNA/siRNA can also confer profound anti-neoplastic activity in infected cancer cells. All targets listed have been experimentally demonstrated to confer therapeutic advantage in vivo over their respective unarmed virus controls.
A logical starting point for increasing cancer cell destruction would be to induce inherent cell-death programs such as apoptosis, necrosis, and pyroptosis. Control of these processes is a delicate balance between the promotion and inhibition of growth and relies on many host cell factors. miRNAs can influence these pathways at multiple steps, either by silencing anti-apoptotic factors or by promoting expression of pro-apoptotic effectors.128 For example, the Bcl-2 family is a commonly targeted family of proteins that operate as guardians to the apoptotic cascade. Delivery of miR-122, known to downregulate the expression of Bcl-2 members, by an AAV vector demonstrated markedly increased cell death in hepatocellular carcinoma cells and increased in vivo therapeutic efficacy in human liver mouse xenograft models.129 Similarly, miR-34a delivered by an adenovirus vector also blocked expression of Bcl-2 in liver cancer and multiple myeloma models. In addition, miR-34a-expressing viral vectors showed significant tumor regression in pre-clinical models using immunodeficient mice.130 Other miRNAs that target apoptotic effectors and can be expressed from viral vectors for cancer therapeutic purposes include miR-143, which targets Kras and miR-144, and thus negatively controls the expression of the TP53-inducible glycolysis apoptosis regulator factor.131,132
Delivery of miRNAs by AAV to modulate key components of uncontrolled cellular replication can also be utilized to limit cell growth. For example, in a study by Kota et al., the authors encoded miR-26a, a cell-cycle regulatory miRNA, into an AAV vector (AAV-miR-26a).133 Upon treatment with AAV-miR-26a, while most cells expressed miR-26a, liver tumor cells had markedly reduced levels of miR-26a and displayed reduced cell-cycle control. Indeed, using AAV-miR-26a as a replacement therapy approach, the authors found that the systemic administration of AAV-delivered miR-26a induced G1 cell-cycle arrest by inhibiting cyclins D2 and E2, which resulted in reduced tumor growth of a hepatocellular carcinoma mouse model. Other targets seek to replace downregulated tumor-suppressing miRNAs (e.g., miR-370-3p) responsible for controlling tumorigenesis and tumor migration.134
Finally, miRNA can be deployed to modulate processes that sensitize tumor cells to existing chemotherapy drugs or other therapeutic modalities. Expression of a subset of miRNAs (e.g., miR-886, -923, -944, -138) was found to correlate with both response to cisplatin and overall improved survival in bladder cancer.135 Applying a similar proof of concept, another group separately identified that miR-6077, through GLUT1 repression, lowered the half-maximal inhibitory concentration (IC50) required for anlotinib, a tyrosine kinase inhibitor, to achieve its anti-tumor effect in patient-derived cell lines of lung adenocarcinoma.136 Accordingly, this addition of novel miRNA-expressing viral delivery appears poised as an avenue to supplement existing current treatment regimens already used in the clinic.
Viral-delivered siRNA/shRNA targets for cancer therapy
Compared to miRNA, siRNA and shRNA differ by focusing on the knockdown of a single gene target as opposed to multiple targets. However, like many other single-target therapeutics, the application of siRNA and shRNA can have a profound therapeutic effect with a smaller side-effect profile if the appropriate target is selected. Here, we present four categories of explored RNAi targets: (1) immunosuppressive genes, (2) oncogenes, (3) genes promoting cell death, and (4) other cancer-promoting molecules. A comprehensive list of virally delivered siRNA or shRNA cancer therapeutics tested in pre-clinical studies can be found in Figure 2.
-
(1)
Targeting immunosuppressive molecules. The success of immune checkpoint inhibitors (ICIs) has placed this therapeutic modality at the forefront of modern cancer therapy development. By using monoclonal antibodies to inhibit immune checkpoint proteins, namely PD-1/PD-L1 and CTLA-4, cytotoxic (CD8+) T lymphocytes are re-engaged to recognize and kill cancer cells more efficiently.137 While many patients have successfully shown long-term remission, a fraction of patients remain resistant to ICIs.138 This resistance is mostly attributable to low tumor antigenicity, low infiltration of T cells in the tumor niche, or an overall poor immune response. The advantage of delivering ICI molecules using a viral vector is that viral backbones can intrinsically induce immune activation and upregulation of the antigen presentation machinery in the tumor microenvironment. Indeed, the combinational delivery of immune checkpoint blockade via oncolytic viral vectors has demonstrated efficacy in various mouse cancer models.139,140 Studies are already under way to incorporate into viral-based platforms RNAi effectors targeting immune checkpoint molecules. For example, the incorporation of RNAi against PD-L1 and CTLA-4 into a Newcastle disease oncolytic virus has demonstrated reduced tumor burden and improved overall survival in the poorly immunogenic B16-F10 syngeneic melanoma mouse model.141 Deploying siRNA/shRNA against multiple other immunotargets such as indoleamine 2,3-dioxygenase, IL-10, and suppressor of cytokine signaling 1 have also demonstrated increased activation of immune cells to stimulate a more robust anti-tumor response.142,143,144 Taken together, there is no shortage of immunosuppressive targets that can be downregulated by viral-mediated delivery of RNAi effectors to improve anti-cancer therapeutic efficacy.
-
(2)
Targeting oncogene addiction. Upon transformation of proto-oncogenes into oncogenes, tumors gain many of the characteristics essential for its pathogenesis, namely increased cell proliferation and survival. Naturally, it is hypothesized that the silencing of these genes would reverse this effect to limit tumor growth or increase cancer sensitivity to chemotherapy. As more than 700 oncogenes have been identified to date,145 there is a plethora of targets for viral RNAi delivery. For example, Li et al. engineered an AAV-expressing shRNA against MYCN, a known oncogene in 25% of neuroblastoma cases. By decreasing MYCN transcription factor levels and its downstream cell differentiation and proliferation programs, the virally delivered shRNA was shown to significantly reduce tumor burden through apoptosis induction in in vivo mouse xenograft neuroblastoma models.99,146 Additionally, knockdown of other well-established oncogenic markers by viral RNAi delivery have shown promise in pre-clinical studies, including EGFR in head and neck cancer147 and androgen receptor in prostate cancer.148
-
(3)
Cell-death induction. Similar to the miRNA targets mentioned above, siRNA/shRNA can be designed to target core cell components and thus either directly inhibit tumor proliferation or knock down anti-apoptotic effectors to induce cell death. Among other tested targets, adenovirus-delivered knockdown of Survivin, an established inhibitor of apoptosis, has shown efficacy in reducing tumor burden in murine models of colorectal and pancreatic cancers.149,150 Of similar interest, synthetic lethal gene pairs describe two unrelated mutations that do not impact the cell on their own but when present together lead to cell death.151 In this context, RNAi has typically been utilized preferentially as a screening tool to identify novel synthetic lethal gene pairs. For example, in human acute myeloid leukemia cells, the authors found that silencing Bcl-2 via lentiviral-delivered shRNA in cells featuring the isocitrate dehydrogenase-1 and -2 R132H mutation significantly decreased their viability.152 Exploration of delivery of these synthetic lethal pairs by viral vector represents yet another opportunity for investigation and development of virally expressed RNAi-based therapeutics.
-
(4)
Other RNAi therapeutic targets. In addition to targeting specifically cancer cells, virus-mediated RNAi delivery has been tested to modify the tumor microenvironment and discourage malignant growth. Multiple studies have previously demonstrated the use of viral vectors to target genes that inhibit angiogenesis. VEGF is a central signaling protein that initiates blood vessel formation and orchestrates tumor blood supply. By delivering a VEGF-targeting shRNA using AAV, the strategy demonstrated reduced tumor vascularization, blood vessel density, and blood vessel size. This potent anti-angiogenic effect led to improved overall survival of glioma-bearing mice.101
Utilizing RNAi to boost the therapeutic efficacy of oncolytic viruses
Most explored RNAi-delivering viral vectors employ viruses without replicating potential, meaning that any therapeutic activity can almost entirely be attributed to the cellular impact of the RNAi effector. While choosing viral vector delivery offers several advantages over other RNAi delivery platforms, herein lies an intriguing opportunity to synergize the effects of RNAi with the inherent killing ability of OVs given the multitude of platforms available, as outlined in “the versatility of viral vectors for RNAi delivery.” Indeed, several groups have already begun exploring this strategy. A study by Rovira-Rigau et al. screened an adenoviral library of 243 human miRNAs in human pancreatic cancers and identified that miR-99b and miR-485 repressed multiple target genes responsible for transcriptional regulation (e.g., ELF4, MDM2, and KLF8), allowing for the enhanced production of adenoviral proteins.153 Subsequently, when directly expressed by an oncolytic adenovirus, this viral enhancement was able to overwhelm tumor cells, leading to increased tumor cytotoxicity and an enhanced anti-tumor effect in various in vivo murine cancer models.
HSV-1 is one of the best characterized oncolytic viral platforms and has been modified to express RNAi effectors. An oncolytic HSV-1 has been engineered to target the apoptotic pathway and disrupt the cancer cell cycle by co-expressing siRNAs against Bcl-2 and Survivin. This recombinant HSV-1 vector showed decreased tumor volume growth in athymic nude mice bearing human breast adenocarcinomas.154 Through a similar mechanism of suppressing pro-survival proteins such as Bcl-2 and SIRT1, expression of miR-34a by a tumor-specific oncolytic vaccinia virus (VV-miR-34a) showed increased cytotoxicity in multiple myeloma cells. Although the VV-delivered expression of miR-34a did not show significant improvement in tumor regression alone, its co-administration with a vaccinia virus armed with another apoptosis inducer, SMAC, was able to achieve significantly improved survival of tumor-bearing mice.155
The field of oncolytic virotherapy continues to face obstacles in identifying an optimal combination of payloads to simultaneously increase viral spread and tumor cell killing, all while maintaining the initiation of a potent anti-tumor immune response for a durable cure. Some conventional payloads such as granulocyte-macrophage stimulating factor have been used with success to stimulate the immune system156,157; however, it is likely that more than one payload may be required to confer curative therapeutic effects. In addition to cytokines and cytotoxic proteins, RNAi effectors, like the examples outlined in this section, represent a novel class of payloads that can be explored for effective anti-cancer combinations.
Optimizing the viral-RNAi relationship for better safety and efficacy
Safety limitations to the viral-RNAi approach
As with any therapeutic strategy, concerns pertaining to safety and efficacy must be addressed before the therapy is able to move into clinical trials. The delivery of RNAi effectors using viral vectors alleviates many inherent concerns of using RNAi therapeutics including extracellular stability, tissue selectivity, and cellular uptake.158 Despite these advantages, several more breakthroughs are still required to increase the viability of viral-mediated RNAi delivery in cancer therapy. Starting with safety, excess accumulation leading to toxicity remains a concern following systemic delivery, especially in the liver. AAV-mediated in vivo delivery of high quantities of various shRNA were found to induce hepatotoxicity and, in some cases, death within 2 months of treatment.159 This occurs when the amount of exogenously delivered RNAi overwhelms the cell’s processing ability, resulting in cytotoxicity.160 Moreover, dose-dependent side effects can also arise at the administration site in response to the virus161; therefore, a delicate balance is required between reducing toxic side effects and maintaining good gene knockdown efficacy. As in the case of integrating viruses, the capacity for genome integration of lentivirus and retrovirus vectors into healthy host cells has been demonstrated to lead to side effects of leukoproliferation and malignancy.162,163 Finally, any replication-competent, unintended infection of healthy cells could cause necrosis of physiologically vital tissue (e.g., liver toxicity) and downstream adverse events.22 Fortunately, several innovative strategies exist to optimize the viral-RNAi relationship to improve RNAi processing and tissue selectivity and improve the safety profiles of these biotherapeutics.
Strategies to increase tissue selectivity
Strategies that maintain this “Goldilocks” level of RNAi expression over a prolonged period of time has been explored by rationally selecting the “type” of promoter that drives the specific RNAi effector expression. In viral vectors, RNAi effectors are typically expressed under RNA polymerase III promoters, such as H1 or U6 promoters, given their simple structure and well-understood features.164,165,166 In some contexts where expression is suboptimal, promoters with greater activity such as the cytomegalovirus (CMV) promoter can be considered.167,168 Additionally, these promoters can be swapped for tissue-selective promoters and thus increase on-target effects. For instance, the use of a liver-specific RNA polymerase II type promoter (ApoE/hAAT) for AAV-mediated delivery of shRNA in vivo showed decreased long-term hepatotoxicity and limited shRNA detection in other tissues (e.g., spleen, heart), even at very high doses, compared to the U6 promoter.169 Similar results were obtained from lentiviral-delivered RNAi using a neuron-specific polymerase II enolase promoter, which limited long-term brain tissue toxicity compared to a conventional CMV promoter in in vivo murine models.170 It is worthwhile discussing that this obstacle may potentially be completely bypassed by the selection of tumor-selective OVs with inherent tumor tropism, offering the benefit of less viral backbone modification.171 However, given that some oncolytic viruses rely on targets of apoptosis, rapid transcription, and rapid translation for selectivity, there still may be cell populations with rapid cell tumor where OV infection may be undesirable.172 As such, integration of tumor-specific promoters in these vectors may still be worth investigation as a redundant mechanism for safety.
Another potential strategy for selective targeting employs miRNA response elements (MREs), which are short target sequences typically found on mRNAs that are recognized by specific miRNA species. Superior complementarity pairing between MRE and mRNA leads to a greater likelihood of mRNA cleavage.173 Given that many miRNAs have their expression restricted to specific tissues or even particular cell types,174 MREs can be incorporated into the viral vector to reduce its expression in specific tissues or cells.175,176 For example, let-7 is a family of miRNA that operates as tumor suppressors and are subsequently downregulated in tumor cells. Indeed, in a study by Edge et al., infection of normal cells with an oncolytic VSV encoding let-7a MREs into the VSV-M gene showed repressed VSV infection in normal GM38 fibroblast cells but unaffected activity in lung A549 carcinoma cells, which express minimal let-7a levels. The let-7a engineered VSV platform did not cause weight loss in mice, and its anti-tumor activity was maintained at a comparable level to control VSV in a murine model of colon carcinoma.177 Modifications in MRE quantity, insertion location, or combinations of different MREs are continued avenues of investigation to ensure enhanced targeting efficacy of the viral vector.178
Strategies targeting the rate of RNAi processing
To successfully knock down a gene product via RNAi, intensive cooperation of many different cellular components is required. As a brief overview, the introduced RNAi effector is loaded into the RNA-induced silencing complex (RISC) and used as a guide to identify the target mRNA strand for silencing. Recognition of the respective complementary mRNA triggers endonucleolytic cleavage by the slicer Argonaute-2 (AGO2) to decrease availability of the mRNA transcript for subsequent protein translation.179 Given that the combined kinetics of RISC mRNA recognition and AGO2-mediated RNA degradation is finite, this represents the rate-limiting steps to viral-delivered RNAi efficacy when overwhelmed with exogenous RNAi effectors. In miRNA and shRNA processing, the Dicer enzyme (ribonuclease III) also comes into play to generate the RISC-compatible siRNA for downstream silencing and limits RNAi efficacy.179 Unengaged intracellular RNAi effectors then proceed to compromise cell viability through outcompeting physiological miRNA required for normal cell function for RISC processing or the accidental generation of off-target siRNA against vital cellular proteins.
Given these limitations in the physiological processing rate of RNAi effectors, strategies targeted at modifying these protein components can be considered to improve the safety and efficacy of delivered RNAi. The first option involves artificially increasing AGO2 expression to increase processing capacity. Co-expression of AGO2 along with RNAi effectors in the viral vector has been demonstrated to achieve greater knockdown efficacy without the same hepatotoxic effects. The knockdown was observed for 5 months after viral administration without any increase in circulating liver-damage markers.180
To counteract these excessive RNAi species, the cell uses two major transport karyopherins, exportin-5/XPO5 and exportin-1/CRM1, to reduce the levels of RNAi. In theory, by overexpressing these shuttle proteins, the capacity to nuclear export excess siRNA is increased to subsequently decrease any toxicity caused by oversaturation. XPO5 is of particular interest given its use in both shRNA and miRNA export mechanisms. Pioneering studies showed the promise of this approach by demonstrating improved shRNA silencing efficacy in cell lines stably overexpressing XPO5.181 However, while delivery of XPO5 overexpression along with the desired shRNA via AAV was found to double the duration of gene silencing in mouse models, its introduction also paradoxically increased mortality.182 The authors hypothesized that the increase in XPO5 precipitated the saturation of another downstream player, AGO2, which could lead to hepatotoxicity. Indeed, RNAi efficacy was found to be best when both XPO5 and AGO2 were co-overexpressed in the same viral vector.182 Thus, the combination of AGO2 and XPO5 remains a potential option to increase RNAi efficacy that warrants further investigation.
Prior to entering the RISC complex for AGO2-mediated splicing, dsRNA species, such as pre-miRNA, must undergo pre-processing by the Dicer complex to generate functional siRNAs. Within this processing step, two potential actionable approaches to increase RNAi efficacy could be implemented. The first is to bypass this rate-determining step altogether through “intelligent shRNA design.” A study by Liu et al. introduces the concept of “agoshRNA,” which describes the design of smaller shRNA with small loop sizes that can shunt its processing away from Dicer and become more reliant on the AGO2 endonuclease activity.183 Given its Dicer-independent miRNA processing, not only is RNAi processing efficacy expected to increase, but this approach could lead to a reduction in the levels of antisense RNA species available for off-target toxicity. While delivery of agoshRNA via viral vectors has not yet been explored, consideration of agoshRNA design over conventional shRNA could represent a simple strategy to improve the safety profile of viral vectors delivering RNAi.
RNAi suppression strategies to increase efficacy
As miRNAs function as primary regulatory agents, it does not come as a surprise that the inverse, which is the suppression of RNAi effectors, can also be used to increase the anti-cancer therapeutic efficacy of viral vectors. The competitive endogenous RNA hypothesis suggests a potential regulatory network between mRNAs, miRNAs, and a set of long non-coding RNAs (lncRNAs) which contain miRNA binding sites and can sequester them.184 This hypothesis propelled efforts to use this concept to create more effective “miRNA sponge” strategies utilizing artificial lncRNAs. By delivering an artificial lncRNA designed to “sponge out” different known oncogenic miRNAs via oncolytic adenovirus, the resulting biotherapeutics were able to increase targeted endogenous mRNAs and significantly greater anti-tumor activity in in vivo models of hepatocellular carcinoma and diffuse large B cell lymphoma.185,186 Furthermore, viral delivery of the recently discovered circular lncRNAs, which feature greater stability due to resistance against endonucleolytic cleavage for more efficacious miRNA scavenging ability,187 represents yet another promising option for exploration.
Plants, fungi, and invertebrates naturally rely on RNAi to combat RNA and DNA virus infections.188,189 To counteract this RNAi-mediated anti-viral response, many viruses that infect these eukaryotic hosts have evolved virus-encoded suppressors of RNAi (VSRs).190 One such virus is the Nodamura virus (NoV), which primarily infects insects but is also highly virulent to certain mammals such as suckling mice and hamsters.191,192 NoV encodes a VSR known as B2, which binds dsRNA and inhibits processing by Dicer to prevent the production of anti-viral siRNAs.193,194 Similarly, influenza A virus encodes the NS1 protein,195,196 Ebola virus encodes VP35,197,198 HIV-1 virus encodes Tat,199,200 vaccinia virus encodes VP55,201 and encephalomyocarditis virus encodes 3A.202 Artificial incorporation of VSRs represents a related opportunity to increase production of their OV carriers, thereby increasing oncolytic efficacy. Indeed, several groups, including ours, have demonstrated that expression of VSRs such as B2 or VP55 in VSV203 or P19 (another plant virus RNAi inhibitor) in adenovirus204 increases OV production and tumor lytic efficacy. In the context of RNAi-expressing viral vectors, we need to acknowledge in future designs that RNAi can attack viral genomes and thus compromise the efficacy of these vectors as therapeutics and vectors to express payloads.
Conclusions
Viral vector delivery of RNAi effectors has been successfully used for other therapeutic applications as a powerful tool to knock down specific genes of interest; however, while exploration of its application for cancer therapy is under way, its clinical application remains limited. The intrigue in its continued exploration lies in the vast opportunity of combinations between different viral vector options, each with unique advantages and disadvantages, with different RNAi effectors. In this review, we outline each of the options for viral vectors and tested therapeutic targets to date. Moreover, we also offer innovative strategies that could potentially help overcome challenges faced by this therapeutic class such as modifying RNAi processing or exploitation of newer RNAi species (e.g., lncRNA, agoshRNA). Future directions for this field will continue to focus on identifying an optimal combination of virus and RNAi effectors that meet standards of therapeutic efficacy while retaining safety. Here we outline many options available, but a systematic approach to testing these combinations is reasonable for identifying candidates suitable for each clinical application. Nonetheless, from this multitude of avenues, we immediately foresee expanding RNAi delivery to replication-competent viral vectors with inherent tumor lytic abilities to have the potential for impressive synergy and, thus, potent therapeutic efficacy. The main limitations to this approach include the natural production of neutralizing antibodies against the virus, inhibiting both its oncolytic and knockdown capacity, as well as sufficient bioavailability of the virus at the tumor site.22,171 Strategies looking to overcome these obstacles in the form of optimal, context-dependent viral platform selection or combinational therapies with pharmacological compounds (“viral enhancers”) are under investigation.205,206 Nonetheless, given the multitude of options and strategies for researchers to bioengineer a breakthrough in viral vector delivery of RNAi for the treatment of cancer, it is not a matter of whether we will succeed, but when.
Acknowledgments
B.W. is supported by the Canadian Institutes of Health Research Doctoral Fellowship (CGS-D). R.B. is supported by the Ontario Graduate Scholarships. J.-S.D. is supported by the Terry Fox Research Institute (TFF 122868), the Canadian Institutes of Health Research (CIHR INI-147824 and grant #705952), and the Canadian Cancer Society supported by the Lotte & John Hecht Memorial Foundation (703014). C.S.I. is supported by grants from the Canadian Institutes of Health Research (#377104 and #183812), The Canadian Cancer Society (#840057), and the Canadian Cancer Society Innovation (#705973) and Impact (#IMP-14 and # 706162) grants as well as generous support from the Ontario Institute for Cancer Research, the Ottawa Regional Cancer Foundation, and the Ottawa Hospital Foundation.
Author contributions
All authors were involved in the writing and editing of the manuscript. B.W., R.B., R.R., and C.S.I. were involved in the initial conceptualization of this review. J.-S.D. and C.S.I. were involved in funding acquisition and study supervision.
Declaration of interests
The authors declare no competing interests.
References
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery M.K. In: RNA Interference, Editing, and Modification. Gott J.M., editor. Humana Press; 2004. RNA Interference; pp. 3–21. [DOI] [Google Scholar]
- 3.Stram Y., Kuzntzova L. Inhibition of Viruses by RNA Interference. Virus Genes. 2006;32:299–306. doi: 10.1007/s11262-005-6914-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 5.Taxman D.J., Moore C.B., Guthrie E.H., Huang M.T.-H. In: RNA Therapeutics Methods in Molecular Biology. Sioud M., editor. Humana Press; 2010. Short Hairpin RNA (shRNA): Design, Delivery, and Assessment of Gene Knockdown; pp. 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao D.D., Vorhies J.S., Senzer N., Nemunaitis J. siRNA vs. shRNA: Similarities and differences. Adv. Drug Deliv. Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 7.McAnuff M.A., Rettig G.R., Rice K.G. Potency of siRNA versus shRNA mediated knockdown in vivo. J. Pharm. Sci. 2007;96:2922–2930. doi: 10.1002/jps.20968. [DOI] [PubMed] [Google Scholar]
- 8.Lam J.K.W., Chow M.Y.T., Zhang Y., Leung S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes L.G.V., Guaman L.P., Vasconcellos S.A., Heinemann M.B., Picardeau M., Nascimento A.L.T.O. Gene silencing based on RNA-guided catalytically inactive Cas9 (dCas9): a new tool for genetic engineering in Leptospira. Sci. Rep. 2019;9:1839. doi: 10.1038/s41598-018-37949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouyang J.P.T., Folkmann A., Bernard L., Lee C.-Y., Seroussi U., Charlesworth A.G., Claycomb J.M., Seydoux G. P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev. Cel. 2019;50:716–728.e6. doi: 10.1016/j.devcel.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S.Y., Lopez-Berestein G., Calin G.A., Sood A.K. RNAi Therapies: Drugging the Undruggable. Sci. Transl. Med. 2014;6:240ps7. doi: 10.1126/scitranslmed.3008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoy S.M. Patisiran: First Global Approval. Drugs. 2018;78:1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M.M., Bahal R., Rasmussen T.P., Manautou J.E., Zhong X.B. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2021.114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmadzada T., Reid G., McKenzie D.R. Fundamentals of siRNA and miRNA therapeutics and a review of targeted nanoparticle delivery systems in breast cancer. Biophys. Rev. 2018;10:69–86. doi: 10.1007/s12551-017-0392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C.X., Parker A., Menocal E., Xiang S., Borodyansky L., Fruehauf J.H. Delivery of RNA Interference. Cell Cycle. 2006;5:2103–2109. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- 16.Song E., Zhu P., Lee S.-K., Chowdhury D., Kussman S., Dykxhoorn D.M., Feng Y., Palliser D., Weiner D.B., Shankar P., et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 17.Hamblett K.J., Senter P.D., Chace D.F., Sun M.M.C., Lenox J., Cerveny C.G., Kissler K.M., Bernhardt S.X., Kopcha A.K., Zabinski R.F., et al. Effects of Drug Loading on the Antitumor Activity of a Monoclonal Antibody Drug Conjugate. Clin. Cancer Res. 2004;10:7063–7070. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzer C., Dirin M., Winkler A.-M., Baumann V., Winkler J. Going beyond the liver: Progress and challenges of targeted delivery of siRNA therapeutics. J. Control Release. 2015;203:1–15. doi: 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021;6:53. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffer D.V., Koerber J.T., Lim K.i. Molecular Engineering of Viral Gene Delivery Vehicles. Annu. Rev. Biomed. Eng. 2008;10:169–194. doi: 10.1146/annurev.bioeng.10.061807.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relph K.L., Harrington K.J., Pandha H. Adenoviral Strategies for the Gene Therapy of Cancer. Semin. Oncol. 2005;32:573–582. doi: 10.1053/j.seminoncol.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Russell S.J., Peng K.-W., Bell J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M., Williamson C.T., Prudhomme J., Bebb D.G., Riabowol K., Lee P.W.K., Lees-Miller S.P., Mori Y., Rahman M.M., McFadden G., Johnston R.N. The viral tropism of two distinct oncolytic viruses, reovirus and myxoma virus, is modulated by cellular tumor suppressor gene status. Oncogene. 2010;29:3990–3996. doi: 10.1038/onc.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Critchley-Thorne R.J., Simons D.L., Yan N., Miyahira A.K., Dirbas F.M., Johnson D.L., Swetter S.M., Carlson R.W., Fisher G.A., Koong A., et al. Impaired interferon signaling is a common immune defect in human cancer. Proc. Natl. Acad. Sci. 2009;106:9010–9015. doi: 10.1073/pnas.0901329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacKenzie T.C., Kobinger G.P., Kootstra N.A., Radu A., Sena-Esteves M., Bouchard S., Wilson J.M., Verma I.M., Flake A.W. Efficient Transduction of Liver and Muscle after in Utero Injection of Lentiviral Vectors with Different Pseudotypes. Mol. Ther. 2002;6:349–358. doi: 10.1006/mthe.2002.0681. [DOI] [PubMed] [Google Scholar]
- 26.Poulin K.L., Lanthier R.M., Smith A.C., Christou C., Risco Quiroz M., Powell K.L., O’Meara R.W., Kothary R., Lorimer I.A., Parks R.J. Retargeting of Adenovirus Vectors through Genetic Fusion of a Single-Chain or Single-Domain Antibody to Capsid Protein IX. J. Virol. 2010;84:10074–10086. doi: 10.1128/JVI.02665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ungerechts G., Bossow S., Leuchs B., Holm P.S., Rommelaere J., Coffey M., Coffin R., Bell J., Nettelbeck D.M. Moving oncolytic viruses into the clinic: clinical-grade production, purification, and characterization of diverse oncolytic viruses. Mol. Ther. Methods Clin. Dev. 2016;3 doi: 10.1038/mtm.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Z., Anselmo A.C., Mitragotri S. Viral vector-based gene therapies in the clinic. Bioeng. Transl. Med. 2022;7:e10258. doi: 10.1002/btm2.10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouha H., Thurner C., Mandl C.W. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010;38:8328–8337. doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pijlman G.P., Suhrbier A., Khromykh A.A. Kunjin virus replicons: an RNA-based, non-cytopathic viral vector system for protein production, vaccine and gene therapy applications. Expert Opin. Biol. Ther. 2006;6:135–145. doi: 10.1517/14712598.6.2.135. [DOI] [PubMed] [Google Scholar]
- 31.Vargas J.E., Chicaybam L., Stein R.T., Tanuri A., Delgado-Cañedo A., Bonamino M.H. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives. J. Transl. Med. 2016;14:288. doi: 10.1186/s12967-016-1047-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham A.P., Andrews L.G., Tollefsbol T.O. In: Telomerase Inhibition Methods in Molecular Biology™. Andrews L.G., Tollefsbol T.O., editors. Humana Press; 2007. Retrovirus-Mediated RNA Interference; pp. 39–46. [DOI] [Google Scholar]
- 33.Brummelkamp T.R., Bernards R., Agami R. A System for Stable Expression of Short Interfering RNAs in Mammalian Cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 34.Yu S.S., Han E., Hong Y., Lee J.-T., Kim S., Kim S. Construction of a retroviral vector production system with the minimum possibility of a homologous recombination. Gene Ther. 2003;10:706–711. doi: 10.1038/sj.gt.3301892. [DOI] [PubMed] [Google Scholar]
- 35.Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-Inactivating Lentivirus Vector for Safe and Efficient In Vivo Gene Delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/JVI.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavazza A., Cocchiarella F., Bartholomae C., Schmidt M., Pincelli C., Larcher F., Mavilio F. Self-inactivating MLV vectors have a reduced genotoxic profile in human epidermal keratinocytes. Gene Ther. 2013;20:949–957. doi: 10.1038/gt.2013.18. [DOI] [PubMed] [Google Scholar]
- 37.Raoul C., Abbas-Terki T., Bensadoun J.-C., Guillot S., Haase G., Szulc J., Henderson C.E., Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 38.Drouet V., Ruiz M., Zala D., Feyeux M., Auregan G., Cambon K., Troquier L., Carpentier J., Aubert S., Merienne N., et al. Allele-Specific Silencing of Mutant Huntingtin in Rodent Brain and Human Stem Cells. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0099341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sapru M.K., Yates J.W., Hogan S., Jiang L., Halter J., Bohn M.C. Silencing of human α-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp. Neurol. 2006;198:382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 40.Horvath L., van Marion I., Taï K., Nielsen T.T., Lundberg C. Knockdown of GAD67 protein levels normalizes neuronal activity in a rat model of Parkinson’s disease. J. Gene Med. 2011;13:188–197. doi: 10.1002/jgm.1555. [DOI] [PubMed] [Google Scholar]
- 41.Singer O., Marr R.A., Rockenstein E., Crews L., Coufal N.G., Gage F.H., Verma I.M., Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 42.Piedrahita D., Hernández I., López-Tobón A., Fedorov D., Obara B., Manjunath B.S., Boudreau R.L., Davidson B., LaFerla F., Gallego-Gómez J.C., et al. Silencing of CDK5 Reduces Neurofibrillary Tangles in Transgenic Alzheimer’s Mice. J. Neurosci. 2010;30:13966–13976. doi: 10.1523/JNEUROSCI.3637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeifer A., Eigenbrod S., Al-Khadra S., Hofmann A., Mitteregger G., Moser M., Bertsch U., Kretzschmar H. Lentivector-mediated RNAi efficiently suppresses prion protein and prolongs survival of scrapie-infected mice. J. Clin. Invest. 2006;116:3204–3210. doi: 10.1172/JCI29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White M.D., Farmer M., Mirabile I., Brandner S., Collinge J., Mallucci G.R. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc. Natl. Acad. Sci. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou H.-X., Li X.-Y., Li F.-Y., Liu C., Liang Z.-P., Liu S., Zhang B., Wang T.-Y., Chu T.-C., Lu L., et al. Targeting RPTPσ with lentiviral shRNA promotes neurites outgrowth of cortical neurons and improves functional recovery in a rat spinal cord contusion model. Brain Res. 2014;1586:46–63. doi: 10.1016/j.brainres.2014.08.048. [DOI] [PubMed] [Google Scholar]
- 46.DiGiusto D.L., Krishnan A., Li L., Li H., Li S., Rao A., Mi S., Yam P., Stinson S., Kalos M., et al. RNA-Based Gene Therapy for HIV with Lentiviral Vector–Modified CD34 + Cells in Patients Undergoing Transplantation for AIDS-Related Lymphoma. Sci. Transl. Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson J., Li M.-J., Palmer B., Remling L., Li S., Yam P., Yee J.-K., Rossi J., Zaia J., Akkina R. Safety and Efficacy of a Lentiviral Vector Containing Three Anti-HIV Genes—CCR5 Ribozyme, Tat-rev siRNA, and TAR Decoy—in SCID-hu Mouse–Derived T Cells. Mol. Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 48.Li M., Rossi J.J. RNA Silencing. Humana Press; 2005. Lentiviral Vector Delivery of siRNA and shRNA Encoding Genes into Cultured and Primary Hematopoietic Cells; pp. 261–272. [DOI] [PubMed] [Google Scholar]
- 49.Banerjea A., Li M.-J., Bauer G., Remling L., Lee N.-S., Rossi J., Akkina R. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 2003;8:62–71. doi: 10.1016/S1525-0016(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 50.Li M.-J., Kim J., Li S., Zaia J., Yee J.-K., Anderson J., Akkina R., Rossi J.J. Long-Term Inhibition of HIV-1 Infection in Primary Hematopoietic Cells by Lentiviral Vector Delivery of a Triple Combination of Anti-HIV shRNA, Anti-CCR5 Ribozyme, and a Nucleolar-Localizing TAR Decoy. Mol. Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 51.Anderson J., Akkina R. CXCR4 and CCR5 shRNA transgenic CD34+ cell derived macrophages are functionally normal and resist HIV-1 infection. Retrovirology. 2005;2:53. doi: 10.1186/1742-4690-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An D.S., Donahue R.E., Kamata M., Poon B., Metzger M., Mao S.-H., Bonifacino A., Krouse A.E., Darlix J.-L., Baltimore D., et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc. Natl. Acad. Sci. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barclay S.L., Yang Y., Zhang S., Fong R., Barraza A., Nolta J.A., Torbett B.E., Abedi M., Bauer G., Anderson J.S. Safety and Efficacy of a tCD25 Preselective Combination Anti-HIV Lentiviral Vector in Human Hematopoietic Stem and Progenitor Cells. Stem Cells. 2015;33:870–879. doi: 10.1002/stem.1919. [DOI] [PubMed] [Google Scholar]
- 54.Kumar P., Lee S.K., Shankar P., Manjunath N. A Single siRNA Suppresses Fatal Encephalitis Induced by Two Different Flaviviruses. Plos Med. 2006;3:e96. doi: 10.1371/journal.pmed.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar P., Wu H., McBride J.L., Jung K.-E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 56.Kim Y.-J., Ahn J., Jeung S.-Y., Kim D.-S., Na H.-N., Cho Y.-J., Yun S.-H., Jee Y., Jeon E.-S., Lee H., Nam J.H. Recombinant lentivirus-delivered short hairpin RNAs targeted to conserved coxsackievirus sequences protect against viral myocarditis and improve survival rate in an animal model. Virus Genes. 2008;36:141–146. doi: 10.1007/s11262-007-0192-y. [DOI] [PubMed] [Google Scholar]
- 57.Gu W., Putral L., Hengst K., Minto K., Saunders N.A., Leggatt G., McMillan N.A.J. Inhibition of cervical cancer cell growth in vitro and in vivo with lentiviral-vector delivered short hairpin RNA targeting human papillomavirus E6 and E7 oncogenes. Cancer Gene Ther. 2006;13:1023–1032. doi: 10.1038/sj.cgt.7700971. [DOI] [PubMed] [Google Scholar]
- 58.Zhou J., Li B., Peng C., Wang F., Fu Z., Zhou C., Hong D., Ye F., Lü W., Xie X. Inhibition of cervical cancer cell growth in vitro and in vivo by lentiviral-vector mediated shRNA targeting the common promoter of HPV16 E6 and E7 oncogenes. Antivir. Res. 2013;98:305–313. doi: 10.1016/j.antiviral.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 59.Dittgen T., Nimmerjahn A., Komai S., Licznerski P., Waters J., Margrie T.W., Helmchen F., Denk W., Brecht M., Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc. Natl. Acad. Sci. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris K.V., Rossi J.J. Lentiviral-mediated delivery of siRNAs for antiviral therapy. Gene Ther. 2006;13:553–558. doi: 10.1038/sj.gt.3302688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burns J.C., Friedmann T., Driever W., Burrascano M., Yee J.K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cronin J., Zhang X.-Y., Reiser J. Altering the Tropism of Lentiviral Vectors through Pseudotyping. Curr. Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei Y., Joo K.-I., Zarzar J., Wong C., Wang P. Targeting lentiviral vector to specific cell types through surface displayed single chain antibody and fusogenic molecule. Virol. J. 2010;7:35. doi: 10.1186/1743-422X-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang A., Chu T.H., Nocken F., Cichutek K., Dornburg R. Cell-Type-Specific Gene Transfer into Human Cells with Retroviral Vectors That Display Single-Chain Antibodies. J. Virol. 1998;72:10148–10156. doi: 10.1128/JVI.72.12.10148-10156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang A., Dornburg R., Dornburg R. In vivo cell type-specific gene delivery with retroviral vectors that display single chain antibodies. Gene Ther. 1999;6:1982–1987. doi: 10.1038/sj.gt.3301043. [DOI] [PubMed] [Google Scholar]
- 67.Bupp K., Roth M.J. Altering Retroviral Tropism Using a Random-Display Envelope Library. Mol. Ther. 2002;5:329–335. doi: 10.1006/mthe.2002.0546. [DOI] [PubMed] [Google Scholar]
- 68.Szécsi J., Drury R., Josserand V., Grange M.-P., Boson B., Hartl I., Schneider R., Buchholz C.J., Coll J.-L., Russell S.J., et al. Targeted retroviral vectors displaying a cleavage site-engineered hemagglutinin (HA) through HA–protease interactions. Mol. Ther. 2006;14:735–744. doi: 10.1016/j.ymthe.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 69.Duerner L.J., Schwantes A., Schneider I.C., Cichutek K., Buchholz C.J. Cell entry targeting restricts biodistribution of replication-competent retroviruses to tumour tissue. Gene Ther. 2008;15:1500–1510. doi: 10.1038/gt.2008.92. [DOI] [PubMed] [Google Scholar]
- 70.Seamon J.A., Jones K.S., Miller C., Roth M.J. Inserting a Nuclear Targeting Signal into a Replication-Competent Moloney Murine Leukemia Virus Affects Viral Export and Is Not Sufficient for Cell Cycle-Independent Infection. J. Virol. 2002;76:8475–8484. doi: 10.1128/JVI.76.16.8475-8484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roe T., Reynolds T.C., Yu G., Brown P.O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis P.F., Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawasaki Y., Tamamoto A., Takagi-Kimura M., Maeyama Y., Yamaoka N., Terada N., Okamura H., Kasahara N., Kubo S. Replication-competent retrovirus vector-mediated prodrug activator gene therapy in experimental models of human malignant mesothelioma. Cancer Gene Ther. 2011;18:571–578. doi: 10.1038/cgt.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Logg C.R., Tai C.-K., Logg A., Anderson W.F., Kasahara N. A Uniquely Stable Replication-Competent Retrovirus Vector Achieves Efficient Gene Delivery in Vitro and in Solid Tumors. Hum. Gene Ther. 2001;12:921–932. doi: 10.1089/104303401750195881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hiraoka K., Kimura T., Logg C.R., Tai C.-K., Haga K., Lawson G.W., Kasahara N. Therapeutic Efficacy of Replication-Competent Retrovirus Vector–Mediated Suicide Gene Therapy in a Multifocal Colorectal Cancer Metastasis Model. Cancer Res. 2007;67:5345–5353. doi: 10.1158/0008-5472.CAN-06-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu Y.C., Chen Y.J., Yu Y.R., Lai Y.H., Cheng J.C., Li Y.F., Shen C.H., Tai C.K. Replicating retroviral vectors for oncolytic virotherapy of experimental hepatocellular carcinoma. Oncol. Rep. 2012;28:21–26. doi: 10.3892/or.2012.1789. [DOI] [PubMed] [Google Scholar]
- 77.Tai C.-K., Wang W.J., Chen T.C., Kasahara N. Single-Shot, Multicycle Suicide Gene Therapy by Replication-Competent Retrovirus Vectors Achieves Long-Term Survival Benefit in Experimental Glioma. Mol. Ther. 2005;12:842–851. doi: 10.1016/j.ymthe.2005.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang W.J., Tai C.-K., Kasahara N., Chen T.C. Highly Efficient and Tumor-Restricted Gene Transfer to Malignant Gliomas by Replication-Competent Retroviral Vectors. Hum. Gene Ther. 2003;14:117–127. doi: 10.1089/104303403321070810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Logg C.R., Robbins J.M., Jolly D.J., Gruber H.E., Kasahara N. Methods in Enzymology. Elsevier; 2012. Retroviral Replicating Vectors in Cancer; pp. 199–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Athanasopoulos T., Munye M.M., Yáñez-Muñoz R.J. Nonintegrating Gene Therapy Vectors. Hematol. Oncol. Clin. North Am. 2017;31:753–770. doi: 10.1016/j.hoc.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Uren A.G., Kool J., Berns A., van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 82.Patel K., Kilfoil G., Wyles D.L., Naggie S., Lawitz E., Bradley S., Lindell P., Suhy D. Phase I/IIa Study of TT-034, a DNA-Directed RNA Interference (ddRNAi) Agent Delivered as a Single Administration for the Treatment of Subjects with Chronic Hepatitis C Virus (HCV) Mol. Ther. 2016;24:S102. doi: 10.1016/S1525-0016(16)33067-2. [DOI] [Google Scholar]
- 83.Zhang Y.-A., Nemunaitis J., Samuel S.K., Chen P., Shen Y., Tong A.W. Antitumor Activity of an Oncolytic Adenovirus-Delivered Oncogene Small Interfering RNA. Cancer Res. 2006;66:9736–9743. doi: 10.1158/0008-5472.CAN-06-1617. [DOI] [PubMed] [Google Scholar]
- 84.Saydam O., Glauser D.L., Heid I., Turkeri G., Hilbe M., Jacobs A.H., Ackermann M., Fraefel C. Herpes Simplex Virus 1 Amplicon Vector-Mediated siRNA Targeting Epidermal Growth Factor Receptor Inhibits Growth of Human Glioma Cells in Vivo. Mol. Ther. 2005;12:803–812. doi: 10.1016/j.ymthe.2005.07.534. [DOI] [PubMed] [Google Scholar]
- 85.Hu J.C.C., Coffin R.S. International Review of Neurobiology. Elsevier; 2003. Gene Therapy with Virus Vectors for specific Disease of the Nervous System; pp. 165–184. [DOI] [Google Scholar]
- 86.Post D.E., Fulci G., Chiocca E.A., Van Meir E.G. Replicative Oncolytic Herpes Simplex Viruses in Combination Cancer Therapies. Curr. Gene Ther. 2004;4:41–51. doi: 10.2174/1566523044577988. [DOI] [PubMed] [Google Scholar]
- 87.Grandi P., Spear M., Breakefield X.O., Wang S. Targeting HSV amplicon vectors. Methods. 2004;33:179–186. doi: 10.1016/j.ymeth.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 88.Grandi P., Wang S., Schuback D., Krasnykh V., Spear M., Curiel D.T., Manservigi R., Breakefield X.O. HSV-1 Virions Engineered for Specific Binding to Cell Surface Receptors. Mol. Ther. 2004;9:419–427. doi: 10.1016/j.ymthe.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 89.Sabbioni S., Callegari E., Manservigi M., Argnani R., Corallini A., Negrini M., Manservigi R. Use of herpes simplex virus type 1-based amplicon vector for delivery of small interfering RNA. Gene Ther. 2007;14:459–464. doi: 10.1038/sj.gt.3302878. [DOI] [PubMed] [Google Scholar]
- 90.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X., et al. Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narvaiza I., Aparicio O., Vera M., Razquin N., Bortolanza S., Prieto J., Fortes P. Effect of Adenovirus-Mediated RNA Interference on Endogenous MicroRNAs in a Mouse Model of Multidrug Resistance Protein 2 Gene Silencing. J. Virol. 2006;80:12236–12247. doi: 10.1128/JVI.01205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carette J.E., Overmeer R.M., Schagen F.H.E., Alemany R., Barski O.A., Gerritsen W.R., van Beusechem V.W. Conditionally Replicating Adenoviruses Expressing Short Hairpin RNAs Silence the Expression of a Target Gene in Cancer Cells. Cancer Res. 2004;64:2663–2667. doi: 10.1158/0008-5472.CAN-03-3530. [DOI] [PubMed] [Google Scholar]
- 93.Doloff J.C., Waxman D.J., Jounaidi Y. Human Telomerase Reverse Transcriptase Promoter-Driven Oncolytic Adenovirus with E1B-19 kDa and E1B-55 kDa Gene Deletions. Hum. Gene Ther. 2008;19:1383–1400. doi: 10.1089/hum.2008.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J., Ramesh N., Chen Y., Li Y., Dilley J., Working P., Yu D.-C. Identification of human uroplakin II promoter and its use in the construction of CG8840, a urothelium-specific adenovirus variant that eliminates established bladder tumors in combination with docetaxel. Cancer Res. 2002;62:3743–3750. [PubMed] [Google Scholar]
- 95.Tanimoto T., Tazawa H., Ieda T., Nouso H., Tani M., Oyama T., Urata Y., Kagawa S., Noda T., Fujiwara T. Elimination of MYCN-Amplified Neuroblastoma Cells by Telomerase-Targeted Oncolytic Virus via MYCN Suppression. Mol. Ther. Oncolytics. 2020;18:14–23. doi: 10.1016/j.omto.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mao L.j., Zhang J., Liu N., Fan L., Yang D.r., Xue B.x., Shan Y.x., Zheng J.n. Oncolytic virus carrying shRNA targeting SATB1 inhibits prostate cancer growth and metastasis. Tumor Biol. 2015;36:9073–9081. doi: 10.1007/s13277-015-3658-x. [DOI] [PubMed] [Google Scholar]
- 97.Yoo J.Y., Kim J.-H., Kim J., Huang J.-H., Zhang S.N., Kang Y.-A., Kim H., Yun C.-O. Short hairpin RNA-expressing oncolytic adenovirus-mediated inhibition of IL-8: effects on antiangiogenesis and tumor growth inhibition. Gene Ther. 2008;15:635–651. doi: 10.1038/gt.2008.3. [DOI] [PubMed] [Google Scholar]
- 98.Luo Q., Basnet S., Dai Z., Li S., Zhang Z., Ge H. A novel E1B55kDa-deleted oncolytic adenovirus carrying microRNA-143 exerts specific antitumor efficacy on colorectal cancer cells. Am. J. Transl. Res. 2016;8:3822–3830. [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y., Zhang B., Zhang H., Zhu X., Feng D., Zhang D., Zhuo B., Li L., Zheng J. Oncolytic adenovirus armed with shRNA targeting MYCN gene inhibits neuroblastoma cell proliferation and in vivo xenograft tumor growth. J. Cancer Res. Clin. Oncol. 2013;139:933–941. doi: 10.1007/s00432-013-1406-4. [DOI] [PubMed] [Google Scholar]
- 100.Chen J., Yang L., Chen H., Yuan T., Liu M., Chen P. Recombinant adenovirus encoding FAT10 small interfering RNA inhibits HCC growth in vitro and in vivo. Exp. Mol. Pathol. 2014;96:207–211. doi: 10.1016/j.yexmp.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 101.Yoo J.Y., Kim J.-H., Kwon Y.-G., Kim E.-C., Kim N.K., Choi H.J., Yun C.-O. VEGF-specific Short Hairpin RNA–expressing Oncolytic Adenovirus Elicits Potent Inhibition of Angiogenesis and Tumor Growth. Mol. Ther. 2007;15:295–302. doi: 10.1038/sj.mt.6300023. [DOI] [PubMed] [Google Scholar]
- 102.Machitani M., Yamaguchi T., Shimizu K., Sakurai F., Katayama K., Kawabata K., Mizuguchi H. Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses. Pharmaceutics. 2011;3:338–353. doi: 10.3390/pharmaceutics3030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sakurai H., Kawabata K., Sakurai F., Nakagawa S., Mizuguchi H. Innate immune response induced by gene delivery vectors. Int. J. Pharm. 2008;354:9–15. doi: 10.1016/j.ijpharm.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 104.Zhu J., Huang X., Yang Y. Innate Immune Response to Adenoviral Vectors Is Mediated by both Toll-Like Receptor-Dependent and -Independent Pathways. J. Virol. 2007;81:3170–3180. doi: 10.1128/JVI.02192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mast T.C., Kierstead L., Gupta S.B., Nikas A.A., Kallas E.G., Novitsky V., Mbewe B., Pitisuttithum P., Schechter M., Vardas E., et al. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: Correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine. 2010;28:950–957. doi: 10.1016/j.vaccine.2009.10.145. [DOI] [PubMed] [Google Scholar]
- 106.Somanathan S., Breous E., Bell P., Wilson J.M. AAV Vectors Avoid Inflammatory Signals Necessary to Render Transduced Hepatocyte Targets for Destructive T Cells. Mol. Ther. 2010;18:977–982. doi: 10.1038/mt.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zaiss A.-K., Liu Q., Bowen G.P., Wong N.C.W., Bartlett J.S., Muruve D.A. Differential Activation of Innate Immune Responses by Adenovirus and Adeno-Associated Virus Vectors. J. Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao X., Zhang C., Le Z., Zeng S., Pan C., Shi J., Wang J., Zhao X. Telomerase reverse transcriptase interference synergistically promotes tumor necrosis factor-related apoptosis-inducing ligand-induced oral squamous cell carcinoma apoptosis and suppresses proliferation in vitro and in vivo. Int. J. Mol. Med. 2018;42:1283–1294. doi: 10.3892/ijmm.2018.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bhere D., Tamura K., Wakimoto H., Choi S.H., Purow B., Debatisse J., Shah K. microRNA-7 upregulates death receptor 5 and primes resistant brain tumors to caspase-mediated apoptosis. Neuro. Oncol. 2018;20:215–224. doi: 10.1093/neuonc/nox138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bhere D., Arghiani N., Lechtich E.R., Yao Y., Alsaab S., Bei F., Matin M.M., Shah K. Simultaneous downregulation of miR-21 and upregulation of miR-7 has anti-tumor efficacy. Sci. Rep. 2020;10:1779. doi: 10.1038/s41598-020-58072-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kattenhorn L.M., Tipper C.H., Stoica L., Geraghty D.S., Wright T.L., Clark K.R., Wadsworth S.C. Adeno-Associated Virus Gene Therapy for Liver Disease. Hum. Gene Ther. 2016;27:947–961. doi: 10.1089/hum.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu G., Luo L., Tai P.W., Qin W., Xiao Y., Wang C., Su Q., Ma H., He R., Wei Y., Gao G. High-Throughput Sequencing of AAV Proviral Libraries from the Human Population Reveals Novel Variants with Unprecedented Intra-and Inter-Tissue Diversity. Mol. Ther. 2016;24:S4. doi: 10.1016/S1525-0016(16)32816-7. [DOI] [Google Scholar]
- 114.Wang D., Li S., Gessler D.J., Xie J., Zhong L., Li J., Tran K., Van Vliet K., Ren L., Su Q., et al. A Rationally Engineered Capsid Variant of AAV9 for Systemic CNS-Directed and Peripheral Tissue-Detargeted Gene Delivery in Neonates. Mol. Ther. Methods Clin. Dev. 2018;9:234–246. doi: 10.1016/j.omtm.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tse L.V., Klinc K.A., Madigan V.J., Castellanos Rivera R.M., Wells L.F., Havlik L.P., Smith J.K., Agbandje-McKenna M., Asokan A. Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. 2017;114:E4812–E4821. doi: 10.1073/pnas.1704766114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wooley D.P., Sharma P., Weinstein J.R., Kotha Lakshmi Narayan P., Schaffer D.V., Excoffon K.J.D.A. A directed evolution approach to select for novel Adeno-associated virus capsids on an HIV-1 producer T cell line. J. Virol. Methods. 2017;250:47–54. doi: 10.1016/j.jviromet.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.-L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Usme-Ciro J.A., Campillo-Pedroza N., Almazán F., Gallego-Gomez J.C. Cytoplasmic RNA viruses as potential vehicles for the delivery of therapeutic small RNAs. Virol. J. 2013;10:185. doi: 10.1186/1743-422X-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hastie E., Grdzelishvili V.Z. Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J. Gen. Virol. 2012;93:2529–2545. doi: 10.1099/vir.0.046672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lundstrom K. Oncolytic Alphaviruses in Cancer Immunotherapy. Vaccines. 2017;5:9. doi: 10.3390/vaccines5020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shapiro J.S., Schmid S., Aguado L.C., Sabin L.R., Yasunaga A., Shim J.V., Sachs D., Cherry S., tenOever B.R. Drosha as an interferon-independent antiviral factor. Proc. Natl. Acad. Sci. 2014;111:7108–7113. doi: 10.1073/pnas.1319635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hussain M., Torres S., Schnettler E., Funk A., Grundhoff A., Pijlman G.P., Khromykh A.A., Asgari S. West Nile virus encodes a microRNA-like small RNA in the 3’ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012;40:2210–2223. doi: 10.1093/nar/gkr848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parameswaran P., Sklan E., Wilkins C., Burgon T., Samuel M.A., Lu R., Ansel K.M., Heissmeyer V., Einav S., Jackson W., et al. Six RNA Viruses and Forty-One Hosts: Viral Small RNAs and Modulation of Small RNA Repertoires in Vertebrate and Invertebrate Systems. Plos Pathog. 2010;6 doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shapiro J.S., Varble A., Pham A.M., tenOever B.R. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Langlois R.A., Shapiro J.S., Pham A.M., tenOever B.R. In Vivo Delivery of Cytoplasmic RNA Virus-derived miRNAs. Mol. Ther. 2012;20:367–375. doi: 10.1038/mt.2011.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.tenOever B.R. RNA viruses and the host microRNA machinery. Nat. Rev. Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- 127.Wedge M.-E., Jennings V.A., Crupi M.J.F., Poutou J., Jamieson T., Pelin A., Pugliese G., de Souza C.T., Petryk J., Laight B.J., et al. Virally programmed extracellular vesicles sensitize cancer cells to oncolytic virus and small molecule therapy. Nat. Commun. 2022;13:1898. doi: 10.1038/s41467-022-29526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Danial N.N., Korsmeyer S.J. Cell Death. Cell. 2004;116:205–219. doi: 10.1016/S0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 129.Yin L., Keeler G.D., Zhang Y., Hoffman B.E., Ling C., Qing K., Srivastava A. AAV3-miRNA vectors for growth suppression of human hepatocellular carcinoma cells in vitro and human liver tumors in a murine xenograft model in vivo. Gene Ther. 2021;28:422–434. doi: 10.1038/s41434-020-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lou W., Chen Q., Ma L., Liu J., Yang Z., Shen J., Cui Y., Bian X.W., Qian C. Oncolytic adenovirus co-expressing miRNA-34a and IL-24 induces superior antitumor activity in experimental tumor model. J. Mol. Med. 2013;91:715–725. doi: 10.1007/s00109-012-0985-x. [DOI] [PubMed] [Google Scholar]
- 131.Luo Q., Song H., Deng X., Li J., Jian W., Zhao J., Zheng X., Basnet S., Ge H., Daniel T., et al. A Triple-Regulated Oncolytic Adenovirus Carrying MicroRNA-143 Exhibits Potent Antitumor Efficacy in Colorectal Cancer. Mol. Ther. Oncolytics. 2020;16:219–229. doi: 10.1016/j.omto.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mu Y., Wang Q., Tan L., Lin L., Zhang B. microRNA-144 inhibits cell proliferation and invasion by directly targeting TIGAR in esophageal carcinoma. Oncol. Lett. 2020;19:3079–3088. doi: 10.3892/ol.2020.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.-W., Chang T.-C., Vivekanandan P., Torbenson M., Clark K.R., et al. Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lin L., Wang D., Qu S., Zhao H., Lin Y. miR-370-3p Alleviates Ulcerative Colitis-Related Colorectal Cancer in Mice Through Inhibiting the Inflammatory Response and Epithelial-Mesenchymal Transition. Drug Des. Devel. Ther. 2020;14:1127–1141. doi: 10.2147/DDDT.S238124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nordentoft I., Birkenkamp-Demtroder K., Agerbæk M., Theodorescu D., Ostenfeld M.S., Hartmann A., Borre M., Ørntoft T.F., Dyrskjøt L. miRNAs associated with chemo-sensitivity in cell lines and in advanced bladder cancer. BMC Med. Genomics. 2012;5:40. doi: 10.1186/1755-8794-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma D.B., Qin M.M., Shi L., Ding X.M. MicroRNA-6077 enhances the sensitivity of patients-derived lung adenocarcinoma cells to anlotinib by repressing the activation of glucose transporter 1 pathway. Cell. Signal. 2019;64 doi: 10.1016/j.cellsig.2019.109391. [DOI] [PubMed] [Google Scholar]
- 137.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jenkins R.W., Barbie D.A., Flaherty K.T. Mechanisms of resistance to immune checkpoint inhibitors. Br. J. Cancer. 2018;118:9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang G., Kang X., Chen K.S., Jehng T., Jones L., Chen J., Huang X.F., Chen S.-Y. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat. Commun. 2020;11:1395. doi: 10.1038/s41467-020-15229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hamilton J.R., Vijayakumar G., Palese P. A Recombinant Antibody-Expressing Influenza Virus Delays Tumor Growth in a Mouse Model. Cell Rep. 2018;22:1–7. doi: 10.1016/j.celrep.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 141.Vijayakumar G., McCroskery S., Palese P. Engineering Newcastle Disease Virus as an Oncolytic Vector for Intratumoral Delivery of Immune Checkpoint Inhibitors and Immunocytokines. J. Virol. 2020;94 doi: 10.1128/JVI.01677-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zheng X., Koropatnick J., Li M., Zhang X., Ling F., Ren X., Hao X., Sun H., Vladau C., Franek J.A., et al. Reinstalling Antitumor Immunity by Inhibiting Tumor-Derived Immunosuppressive Molecule IDO through RNA Interference. J. Immunol. 2006;177:5639–5646. doi: 10.4049/jimmunol.177.8.5639. [DOI] [PubMed] [Google Scholar]
- 143.Kim J.H., Kang T.H., Noh K.H., Bae H.C., Ahn Y.-H., Lee Y.-H., Choi E.Y., Chun K.-H., Lee S.-J., Kim T.W. Blocking the immunosuppressive axis with small interfering RNA targeting interleukin (IL)-10 receptor enhances dendritic cell-based vaccine potency: IL-10 receptor siRNA enhances DC-based vaccine potency. Clin. Exp. Immunol. 2011;165:180–189. doi: 10.1111/j.1365-2249.2011.04410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Subramanya S., Armant M., Salkowitz J.R., Nyakeriga A.M., Haridas V., Hasan M., Bansal A., Goepfert P.A., Wynn K.K., Ladell K., et al. Enhanced Induction of HIV-specific Cytotoxic T Lymphocytes by Dendritic Cell-targeted Delivery of SOCS-1 siRNA. Mol. Ther. 2010;18:2028–2037. doi: 10.1038/mt.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018;18:696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li Y., Zhang H., Zhu X., Feng D., Zhang D., Zhuo B., Zheng J. Oncolytic adenovirus-mediated short hairpin RNA targeting MYCN gene induces apoptosis by upregulating RKIP in neuroblastoma. Tumor Biol. 2015;36:6037–6043. doi: 10.1007/s13277-015-3280-y. [DOI] [PubMed] [Google Scholar]
- 147.Uehara N., Otsuki N., Kubo M., Kitamoto J., Kojima Y., Teshima M., Shinomiya H., Shirakawa T., Nibu K.I. Oncolytic effect of Midkine promoter–based conditionally replicating adenoviruses expressing EGFR siRNA in head and neck squamous cancer cell line T891. Cancer Rep. 2020;3:e1231. doi: 10.1002/cnr2.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sun A., Tang J., Terranova P.F., Zhang X., Thrasher J.B., Li B. Adeno-associated virus–delivered short hairpin-structured RNA for androgen receptor gene silencing induces tumor eradication of prostate cancer xenografts in nude mice: A preclinical study. Int. J. Cancer. 2010;126:764–774. doi: 10.1002/ijc.24778. [DOI] [PubMed] [Google Scholar]
- 149.Shen W., Wang C.-Y., Wang X.-H., Fu Z.-X. Oncolytic adenovirus mediated Survivin knockdown by RNA interference suppresses human colorectal carcinoma growth in vitro and in vivo. J. Exp. Clin. Cancer Res. 2009;28:81. doi: 10.1186/1756-9966-28-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Han Z., Lee S., Je S., Eom C.-Y., Choi H.J., Song J.J., Kim J.-H. Survivin silencing and TRAIL expression using oncolytic adenovirus increase anti-tumorigenic activity in gemcitabine-resistant pancreatic cancer cells. Apoptosis. 2016;21:351–364. doi: 10.1007/s10495-015-1208-z. [DOI] [PubMed] [Google Scholar]
- 151.Ye H., Zhang X., Chen Y., Liu Q., Wei J. Ranking novel cancer driving synthetic lethal gene pairs using TCGA data. Oncotarget. 2016;7:55352–55367. doi: 10.18632/oncotarget.10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chan S.M., Thomas D., Corces-Zimmerman M.R., Xavy S., Rastogi S., Hong W.-J., Zhao F., Medeiros B.C., Tyvoll D.A., Majeti R. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 2015;21:178–184. doi: 10.1038/nm.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rovira-Rigau M., Raimondi G., Marín M.Á., Gironella M., Alemany R., Fillat C. Bioselection Reveals miR-99b and miR-485 as Enhancers of Adenoviral Oncolysis in Pancreatic Cancer. Mol. Ther. 2019;27:230–243. doi: 10.1016/j.ymthe.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen X., Zhou Y., Wang J., Wang J., Yang J., Zhai Y., Li B. Dual silencing of Bcl-2 and Survivin by HSV-1 vector shows better antitumor efficacy in higher PKR phosphorylation tumor cells in vitro and in vivo. Cancer Gene Ther. 2015;22:380–386. doi: 10.1038/cgt.2015.30. [DOI] [PubMed] [Google Scholar]
- 155.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pol J., Kroemer G., Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2015.1115641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rehman H., Silk A.W., Kane M.P., Kaufman H.L. Into the clinic: Talimogene laherparepvec (T-VEC), a first-in-class intratumoral oncolytic viral therapy. J. Immunother. Cancer. 2016;4:53. doi: 10.1186/s40425-016-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tomar R.S., Matta H., Chaudhary P.M. Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene. 2003;22:5712–5715. doi: 10.1038/sj.onc.1206733. [DOI] [PubMed] [Google Scholar]
- 159.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 160.Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence. 2011;2:8. doi: 10.1186/1758-907X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schirrmacher V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review) Int. J. Oncol. 2019;13 doi: 10.3892/ijo.2018.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cornetta K., Lin T.-Y., Pellin D., Kohn D.B. Meeting FDA Guidance recommendations for replication-competent virus and insertional oncogenesis testing. Mol. Ther. Methods Clin. Dev. 2023;28:28–39. doi: 10.1016/j.omtm.2022.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Connolly J.B. Lentiviruses in gene therapy clinical research. Gene Ther. 2002;9:1730–1734. doi: 10.1038/sj.gt.3301893. [DOI] [PubMed] [Google Scholar]
- 164.Nie L., Das Thakur M., Wang Y., Su Q., Zhao Y., Feng Y. Regulation of U6 Promoter Activity by Transcriptional Interference in Viral Vector-Based RNAi. Genomics Proteomics Bioinformatics. 2010;8:170–179. doi: 10.1016/S1672-0229(10)60019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mäkinen P.I., Koponen J.K., Kärkkäinen A.M., Malm T.M., Pulkkinen K.H., Koistinaho J., Turunen M.P., Ylä-Herttuala S. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J. Gene Med. 2006;8:433–441. doi: 10.1002/jgm.860. [DOI] [PubMed] [Google Scholar]
- 166.Goguen R.P., Del Corpo O., Malard C.M.G., Daher A., Alpuche-Lazcano S.P., Chen M.J., Scarborough R.J., Gatignol A. Efficacy, accumulation, and transcriptional profile of anti-HIV shRNAs expressed from human U6, 7SK, and H1 promoters. Mol. Ther. Nucleic Acids. 2021;23:1020–1034. doi: 10.1016/j.omtn.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xia X.G., Zhou H., Ding H., Affar E.B., Shi Y., Xu Z. An enhanced U6 promoter for synthesis of short hairpin RNA. Nucleic Acids Res. 2003;31:100e–1100e. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ong S.T., Li F., Du J., Tan Y.W., Wang S. Hybrid Cytomegalovirus Enhancer–H1 Promoter-Based Plasmid and Baculovirus Vectors Mediate Effective RNA Interference. Hum. Gene Ther. 2005;16:1404–1412. doi: 10.1089/hum.2005.16.1404. [DOI] [PubMed] [Google Scholar]
- 169.Giering J.C., Grimm D., Storm T.A., Kay M.A. Expression of shRNA From a Tissue-specific pol II Promoter Is an Effective and Safe RNAi Therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 170.Nielsen T.T., Marion I.v., Hasholt L., Lundberg C. Neuron-specific RNA interference using lentiviral vectors. J. Gene Med. 2009;11:559–569. doi: 10.1002/jgm.1333. [DOI] [PubMed] [Google Scholar]
- 171.Hu P.-Y., Fan X.-M., Zhang Y.-N., Wang S.-B., Wan W.-J., Pan H.-Y., Mou X.-Z. The limiting factors of oncolytic virus immunotherapy and the approaches to overcome them. Appl. Microbiol. Biotechnol. 2021;105:5257. doi: 10.1007/s00253-021-11335-6. [DOI] [PubMed] [Google Scholar]
- 172.Singh P.K., Doley J., Kumar G.R., Sahoo A.P., Tiwari A.K. Oncolytic viruses & their specific targeting to tumour cells. Indian J. Med. Res. 2012;136:571–584. [PMC free article] [PubMed] [Google Scholar]
- 173.Jo M.H., Shin S., Jung S.-R., Kim E., Song J.-J., Hohng S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cel. 2015;59:117–124. doi: 10.1016/j.molcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- 174.Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stähler C., Meese E., Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kelly E.J., Hadac E.M., Greiner S., Russell S.J. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 176.Ylösmäki E., Hakkarainen T., Hemminki A., Visakorpi T., Andino R., Saksela K. Generation of a Conditionally Replicating Adenovirus Based on Targeted Destruction of E1A mRNA by a Cell Type-Specific MicroRNA. J. Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Edge R.E., Falls T.J., Brown C.W., Lichty B.D., Atkins H., Bell J.C. A let-7 MicroRNA-sensitive Vesicular Stomatitis Virus Demonstrates Tumor-specific Replication. Mol. Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- 178.Leber M.F., Baertsch M.-A., Anker S.C., Henkel L., Singh H.M., Bossow S., Engeland C.E., Barkley R., Hoyler B., Albert J., et al. Enhanced Control of Oncolytic Measles Virus Using MicroRNA Target Sites. Mol. Ther. Oncolytics. 2018;9:30–40. doi: 10.1016/j.omto.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Paroo Z., Liu Q., Wang X. Biochemical mechanisms of the RNA-induced silencing complex. Cell Res. 2007;17:187–194. doi: 10.1038/sj.cr.7310148. [DOI] [PubMed] [Google Scholar]
- 180.Börner K., Niopek D., Cotugno G., Kaldenbach M., Pankert T., Willemsen J., Zhang X., Schürmann N., Mockenhaupt S., Serva A., et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 2013;41:e199. doi: 10.1093/nar/gkt836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yi R., Doehle B.P., Qin Y., Macara I.G., Cullen B.R. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Grimm D., Wang L., Lee J.S., Schürmann N., Gu S., Börner K., Storm T.A., Kay M.A. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu Y.P., Schopman N.C.T., Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013;41:3723–3733. doi: 10.1093/nar/gkt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 185.Li X., Su Y., Sun B., Ji W., Peng Z., Xu Y., Wu M., Su C. An Artificially Designed Interfering lncRNA Expressed by Oncolytic Adenovirus Competitively Consumes OncomiRs to Exert Antitumor Efficacy in Hepatocellular Carcinoma. Mol. Cancer Ther. 2016;15:1436–1451. doi: 10.1158/1535-7163.MCT-16-0096. [DOI] [PubMed] [Google Scholar]
- 186.Su Y., Sun B., Lin X., Zhao X., Ji W., He M., Qian H., Song X., Yang J., Wang J., Chen J. Therapeutic strategy with artificially-designed i-lncRNA targeting multiple oncogenic microRNAs exhibits effective antitumor activity in diffuse large B-cell lymphoma. Oncotarget. 2016;7:49143–49155. doi: 10.18632/oncotarget.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rossbach O. Artificial Circular RNA Sponges Targeting MicroRNAs as a Novel Tool in Molecular Biology. Mol. Ther. Nucleic Acids. 2019;17:452–454. doi: 10.1016/j.omtn.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yan N., Chen Z.J. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jiang L., Wei C., Li Y. Viral suppression of RNA silencing. Sci. China. Life Sci. 2012;55:109–118. doi: 10.1007/s11427-012-4279-x. [DOI] [PubMed] [Google Scholar]
- 190.Bailey L., Newman J.F., Porterfield J.S. The Multiplication of Nodamura Virus in Insect and Mammalian Cell Cultures. J. Gen. Virol. 1975;26:15–20. doi: 10.1099/0022-1317-26-1-15. [DOI] [PubMed] [Google Scholar]
- 191.Bailey L., Scott H.A. The Pathogenicity of Nodamura Virus for Insects. Nature. 1973;241:545. doi: 10.1038/241545a0. [DOI] [PubMed] [Google Scholar]
- 192.Chao J.A., Lee J.H., Chapados B.R., Debler E.W., Schneemann A., Williamson J.R. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 193.Körber S., Shaik Syed Ali P., Chen J.C.-H. Structure of the RNA-Binding Domain of Nodamura Virus Protein B2, a Suppressor of RNA Interference. Biochemistry. 2009;48:2307–2309. doi: 10.1021/bi900126s. [DOI] [PubMed] [Google Scholar]
- 194.Maillard P.V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S.W., Voinnet O. Antiviral RNA Interference in Mammalian Cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lu Y., Wambach M., Katze M.G., Krug R.M. Binding of the Influenza Virus NS1 Protein to Double-Stranded RNA Inhibits the Activation of the Protein Kinase That Phosphorylates the eIF-2 Translation Initiation Factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 196.Bucher E., Hemmes H., de Haan P., Goldbach R., Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- 197.Cárdenas W.B., Loo Y.-M., Gale M., Hartman A.L., Kimberlin C.R., Martínez-Sobrido L., Saphire E.O., Basler C.F. Ebola Virus VP35 Protein Binds Double-Stranded RNA and Inhibits Alpha/Beta Interferon Production Induced by RIG-I Signaling. J. Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Haasnoot J., de Vries W., Geutjes E.-J., Prins M., de Haan P., Berkhout B. The Ebola Virus VP35 Protein Is a Suppressor of RNA Silencing. Plos Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bennasser Y., Jeang K.-T. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bennasser Y., Le S.-Y., Benkirane M., Jeang K.-T. Evidence that HIV-1 Encodes an siRNA and a Suppressor of RNA Silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 201.Backes S., Shapiro J.S., Sabin L.R., Pham A.M., Reyes I., Moss B., Cherry S., tenOever B.R. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li Y., Basavappa M., Lu J., Dong S., Cronkite D.A., Prior J.T., Reinecker H.-C., Hertzog P., Han Y., Li W.-X., et al. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2016;2 doi: 10.1038/nmicrobiol.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bastin D., Aitken A.S., Pelin A., Pikor L.A., Crupi M.J.F., Huh M.S., Bourgeois-Daigneault M.-C., Bell J.C., Ilkow C.S. Enhanced susceptibility of cancer cells to oncolytic rhabdo-virotherapy by expression of Nodamura virus protein B2 as a suppressor of RNA interference. J. Immunother. Cancer. 2018;6:62. doi: 10.1186/s40425-018-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Doerner J., Sallard E., Zhang W., Solanki M., Liu J., Ehrke-Schulz E., Zirngibl H., Lieber A., Ehrhardt A. Novel group C oncolytic adenoviruses carrying a microRNA inhibitor demonstrate enhanced oncolytic activity in vitro and in vivo. Mol. Cancer Ther. 2022;21:460–470. doi: 10.1158/1535-7163.MCT-21-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Forbes N.E., Krishnan R., Diallo J.-S. Pharmacological Modulation of Anti-Tumor Immunity Induced by Oncolytic Viruses. Front. Oncol. 2014;4:191. doi: 10.3389/fonc.2014.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Le Boeuf F., Diallo J.-S., McCart J.A., Thorne S., Falls T., Stanford M., Kanji F., Auer R., Brown C.W., Lichty B.D., et al. Synergistic Interaction Between Oncolytic Viruses Augments Tumor Killing. Mol. Ther. 2010;18:888–895. doi: 10.1038/mt.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]