replying to V. B. O’Donnell et al. Nature Communications 10.1038/s41467-023-41766-w (2023)
O’Donnell et al.1 assert that because they can achieve an area under the curve (AUC) of ≥2000 cps by integrating background noise, our criteria “lead to flawed … biomarker claims.” This assertion is based on the misapplication of the criteria that were described clearly, in our view, leading to erroneous results and therefore conclusions. In Gomez et al., the criterion described for peak identification and integration, and therefore calculation of the AUC, requires the presence of a distinct peak in the chromatogram as denoted by the following text in the methods: presence of a peak with a minimum area of 2000 counts. In the regions of chromatograms integrated by O’Donnell et al., there is no single discernible peak and therefore it would not meet the basic criterion for integration. To further clarify the application of the criteria employed in the analysis of data presented in Gomez et al.2, we provided an illustration that presents the decision pathway used for peak identification and integration (Supplementary Fig. 1). We believe that this aspect alone undermines O’Donnell et al.’s argument regarding the validity of our approach, since they do not demonstrate that blank samples yield a single discernible peak with an AUC ≥2000 cps. Nonetheless, and to further substantiate our argument, we also provide examples of chromatograms from our data analysis with a side-by-side comparison of AUC and signal-to-noise (s/n) ratios (see below and Supplementary Fig. 2). We present examples at the lower extreme of the identification spectrum, substantiating the argument that even the lower abundance peaks gave s/n ratios ≥5, with the signal obtained for most mediators identified being well above this threshold. In the Supplementary text we provide a discussion on the rationale behind the methodologies employed in Gomez et al.2.
O’Donnell et al. also argue that because we did not use s/n ratios as the cut-off parameter for determining the lower limits of quantitation (LLOQ) our analysis is flawed and that SPMs do not exist in biological systems. We respectfully disagree with this assertion and point of view. First, there is extensive documentation from many groups (including some of the co-authors of O’Donnell et al.3–10, also see the following recent reviews for a more comprehensive list of publications10,11) which identify and quantitate SPM in an array of biological systems. Second, while it is the case that the different entities mentioned by the authors have recommended the use of s/n ratios as an analytical criterion, this is not the only criterion they recommend.
Whilst we acknowledge that using s/n ratios is useful in determining the LLOQ/LLOD of LC-MS/MS assays, a review of the literature, including documents cited by O’Donnell et al., demonstrates that there are several approaches for the calculation of such parameters, and there are also different guidelines for cut-offs to be employed12,13. We note that independent of the approach used the methods need to be accurate and precise, aspects that are difficult to achieve with a pencil and ruler as employed by O’Donnell et al. in the reanalysis of our published dataset.
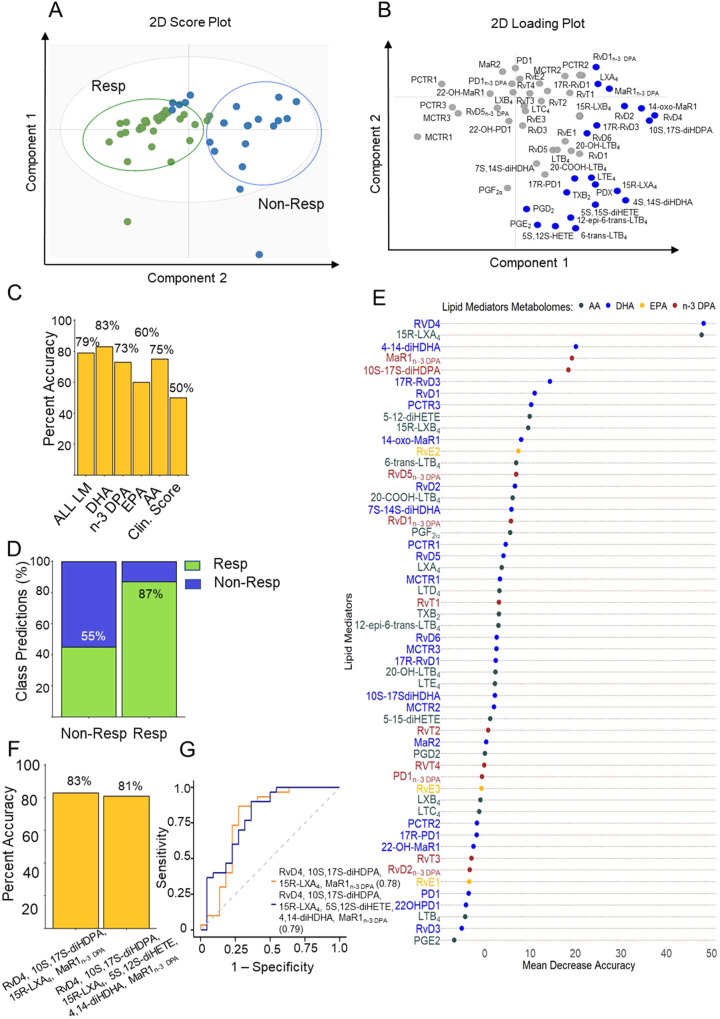
To demonstrate the robustness of the approach used in Gomez et al., we reanalyzed the underlying data from this publication using an orthogonal approach with the LLOQ set as a s/n ≥5. Due to (1) space limitations, (2) given that O’Donnell et al. claim that our approach does not support the utility of measuring SPMs as biomarkers and (3) since machine learning models are exquisitely sensitive to changes in the variables being used, in this response we show the results obtained from the reanalysis of data presented in Figure 1 of Gomez et al. Of note, both the accuracy and AUC values obtained using this orthogonal method gave essentially identical outcomes to those published in Gomez et al., supporting the robustness of the analysis performed in our publication (Fig. 1).
Fig. 1. Reanalysis of baseline plasma lipid mediator profiles supports their potential utility as biomarkers.
Plasma was collected from RA patients prior to the initiation of treatment with DMARDs and lipid mediator concentrations established using LC-MS/MS-based lipid mediator profiling (see Supplementary Methods for details). A, B OPLS-DA analysis of peripheral blood lipid mediator concentrations for DMARD responders (Resp) and DMARD non-Responders (Non-Resp). A Two-dimensional score plot with the gray circle representing the 95% confidence regions. B Two-dimensional loading plots. Lipid mediators with VIP score greater than 1 are highlighted in blue and upregulated in Non-Resp. Results are representative of n = 30 Resp and n = 22 Non-Resp. C Percentage accuracy score of prediction models based on the combination of all lipid mediators identified and quantified (AL LM) or individual fatty acid metabolomes as indicated. Clin. Score = clinical score (see methods from Gomez et al., for parameters included). D Classification predictions for each class (sensitivity and specificity) of the n-3 DPA model. Green indicates the samples that were predicted as Resp while blue indicates those patients predicted Non-Resp. Percentages indicate true positives (Resp class) and true negatives (Non-Resp class). E Relevance of lipid mediators in the prediction performance of the “ALL LM” model based on decreasing accuracy. F Percentage accuracy score of models using the indicated SPM. G Receiver operating characteristic (ROC) curves and AUC values for predictive models based on the indicated SPM. All the models were created using the random forest methodology (“randomForest” package from R).
The claim made by O’Donnell et al. that software developed by SCIEX yields inaccurate results would, in our view, require substantiation, especially since these authors appear to use SCIEX software for the calculation of s/n ratios in their publications6,14,15. We are also unclear of the scientific basis for the assertion that s/n should not be calculated after smoothing, especially because software from several vendors automatically performs this data-processing step. Furthermore, as noted elsewhere (SCIEX OS for Triple Quadrupole Systems Software User Guide [https://sciex.com/content/dam/SCIEX/pdf/customer-docs/user-guide/sciex-os-tnt-user-guide-en.pdf]) when applied appropriately smoothing increases the robustness of the s/n analysis by reducing the fluctuation in both the background signal and the signal for the peak of interest.
O’Donnell et al. also claim that they were able to obtain a spectrum that would match that of Maresin (MaR) 1 from a blank sample. We cannot know what contaminants there may have been in their instrumentation that may have contributed to their result. As can be observed from the data presented in Supplementary Fig. 3, the evaluation of blanks on our instrumentation did not yield MS/MS spectra that contained ions that could be linked with lipid mediator identification. Focusing on MaR1, when we extracted ions with an m/z of 359.4 corresponding to the MaR1 parent ion (and other dihydroxylated SPM derived from DHA) we did not observe any eluting at the retention time corresponding with that of MaR1 (and other dihydroxylated SPM). Furthermore, the evaluation of MS/MS spectra captured for molecules eluting before and after the retention time of MaR1 did not yield any of the ions reported by O’Donnell et al., suggesting that the spectra they reported arise from contaminants within their instrumentation. To further substantiate the utility of using MS/MS spectra for identification of SPM in our samples, we used the library match function in SCIEX OS. This analysis confirmed the presence of diagnostic MS/MS spectra in the plasma samples (Supplementary Fig. 4).
In summation, our view is that the critique made by O’Donnell and colleagues1 of our article is undermined by the misrepresentation of our criteria, potential contaminants in the system used, and by critical issues in the methodologies on which they are based. The reanalysis of the original data using orthogonal approaches and objective methodologies further substantiates both the presence of SPMs in human peripheral blood, in line with findings made by others3,5,6,16–18, and their utility as biomarkers. We welcome the opportunity to discuss results published in Gomez et al.2 and further demonstrate the strength of the identifications and conclusions.
Methods
Data acquisition, multivariate analysis and machine learning models were performed as detailed in Gomez et al.1. For the calculation of s/n ratios the AutoPeak and Noise filtering, and Relative Noise functions in SCIEX OS v2.1 were employed. See Supplementary Methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Prof Charles N. Serhan, Prof Mauro Perretti and Prof Roderick J. Flower for their insightful and invaluable feedback in the preparation of this manuscript.
Author contributions
E.A.G. and J.D. performed analyses and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
The data that support the figures and other findings within this paper are available from the corresponding authors upon request.
Competing interests
J.D. is an inventor on patents related to the composition of matter and/or use of pro-resolving mediators some of which are licensed by Brigham and Women’s Hospital or Queen Mary University of London for clinical development. E.A.G. is an inventor on patents related to the use of pro-resolving mediators as diagnostics licensed by Queen Mary University of London for clinical development.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-41767-9.
References
- 1.O’Donnell, V. et al. Failure to apply standard limit-of-detection or limit-of-quantitation criteria to specialized pro-resolving mediator analysis incorrectly characterizes their presence in biological samples. Nat. Commun.10.1038/s41467-023-41766-w (2023). [DOI] [PMC free article] [PubMed]
- 2.Gomez EA, et al. Blood pro-resolving mediators are linked with synovial pathology and are predictive of DMARD responsiveness in rheumatoid arthritis. Nat. Commun. 2020;11:5420. doi: 10.1038/s41467-020-19176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werner M, et al. Communication between human macrophages and epithelial cancer cell lines dictates lipid mediator biosynthesis. Cell Mol. Life Sci. 2020;77:4365–4378. doi: 10.1007/s00018-019-03413-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snodgrass RG, et al. Efferocytosis potentiates the expression of arachidonate 15-lipoxygenase (ALOX15) in alternatively activated human macrophages through LXR activation. Cell Death Differ. 2021;28:1301–1316. doi: 10.1038/s41418-020-00652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mas E, Croft KD, Zahra P, Barden A, Mori TA. Resolvins D1, D2, and other mediators of self-limited resolution of inflammation in human blood following n-3 fatty acid supplementation. Clin. Chem. 2012;58:1476–1484. doi: 10.1373/clinchem.2012.190199. [DOI] [PubMed] [Google Scholar]
- 6.Markworth JF, et al. Human inflammatory and resolving lipid mediator responses to resistance exercise and ibuprofen treatment. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;305:R1281–R1296. doi: 10.1152/ajpregu.00128.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mainka M, et al. On the biosynthesis of specialized pro-resolving mediators in human neutrophils and the influence of cell integrity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1867:159093. doi: 10.1016/j.bbalip.2021.159093. [DOI] [PubMed] [Google Scholar]
- 8.Liening S, Romp E, Werz O, Scriba GKE, Garscha U. Liquid chromatography-coupled mass spectrometry analysis of glutathione conjugates of oxygenated polyunsaturated fatty acids. Prostaglandins Other Lipid Mediat. 2019;144:106350. doi: 10.1016/j.prostaglandins.2019.106350. [DOI] [PubMed] [Google Scholar]
- 9.Ebert R, et al. Long-term stimulation of toll-like receptor-2 and -4 upregulates 5-LO and 15-LO-2 expression thereby inducing a lipid mediator shift in human monocyte-derived macrophages. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158702. doi: 10.1016/j.bbalip.2020.158702. [DOI] [PubMed] [Google Scholar]
- 10.Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serhan CN, Sulciner ML. Resolution medicine in cancer, infection, pain and inflammation: are we on track to address the next Pandemic? Cancer Metastasis Rev. 2023;42:13–17. doi: 10.1007/s10555-023-10091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serhan CN, Chiang N. Resolvins and cysteinyl-containing pro-resolving mediators activate resolution of infectious inflammation and tissue regeneration. Prostaglandins Other Lipid Mediat. 2023;166:106718. doi: 10.1016/j.prostaglandins.2023.106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheehan, T. L. & Yost, R. A. What’s the most meaningful standard for mass spectrometry: instrument detection limit or signal-to-noise ratio? Spectroscopy13, 16–22 (2017).
- 14.Evard H, Kruve A, Leito I. Tutorial on estimating the limit of detection using LC-MS analysis, part I: theoretical review. Anal. Chim. Acta. 2016;942:23–39. doi: 10.1016/j.aca.2016.08.043. [DOI] [PubMed] [Google Scholar]
- 15.Misheva M, et al. Oxylipin metabolism is controlled by mitochondrial beta-oxidation during bacterial inflammation. Nat. Commun. 2022;13:139. doi: 10.1038/s41467-021-27766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamon-Fava S, et al. Dose- and time-dependent increase in circulating anti-inflammatory and pro-resolving lipid mediators following eicosapentaenoic acid supplementation in patients with major depressive disorder and chronic inflammation. Prostaglandins Leukot. Ess. Fat. Acids. 2021;164:102219. doi: 10.1016/j.plefa.2020.102219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keeley EC, et al. Specialized proresolving mediators in symptomatic women with coronary microvascular dysfunction (from the Women’s Ischemia Trial to Reduce Events in Nonobstructive CAD [WARRIOR] Trial) Am. J. Cardiol. 2022;162:1–5. doi: 10.1016/j.amjcard.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uno H, et al. Immunonutrition suppresses acute inflammatory responses through modulation of resolvin E1 in patients undergoing major hepatobiliary resection. Surgery. 2016;160:228–236. doi: 10.1016/j.surg.2016.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the figures and other findings within this paper are available from the corresponding authors upon request.