Abstract
Twelve strains of Escherichia coli O157:H7 were isolated from 9 of 25 beef samples purchased from retail stores in Malaysia. These strains produced Shiga toxin 2 with or without Shiga toxin 1 and had the eae gene and a 60-MDa plasmid. The antibiograms and the profiles of the arbitrarily primed PCR of the strains were diverse, suggesting that the strains may have originated from diverse sources.
Escherichia coli O157:H7 can cause hemorrhagic colitis and other diseases through consumption of food and water and by human-to-human transmission (5). Infection by E. coli O157:H7 has become a very important food-borne disease in developed countries (4). One of the common modes of transmission appears to be the consumption of contaminated beef and related products (5). Isolation rates of E. coli O157:H7 from ground-beef samples in North America ranged from none to 3.7% (5). There have been reports on the isolation of this organism in other parts of the world (4). In Asia, isolation of E. coli of serogroup O157 has so far been reported in Japan, India, and China (5, 6, 15). The strains isolated in Asian countries other than Japan have not been well characterized. Other Asian countries do not seem to be exempt from the E. coli O157:H7 infection, although a study in Thailand failed to isolate E. coli of serogroup O157 from beef samples (14). We therefore investigated in this study whether E. coli O157:H7 is distributed in retailed beef in Malaysia.
Isolation of E. coli O157:H7.
Beef originally imported from India through an importer and sold as tenderloin was purchased from four retail stores in Malaysia at certain intervals. Twenty-five beef samples thus obtained were examined in this study. A 25-g portion of each beef sample was homogenized in a stomacher with 225 ml of medium (Difco Laboratories, Detroit, Mich.) for 2 min and then incubated statically at 37°C for 4 h. The culture was diluted in tryptone water (1% tryptone, 0.5% NaCl), inoculated onto MacConkey agar (Oxoid, Ltd., Basingstoke, England), and incubated overnight at 37°C. Twenty to 50 colonies per sample were selected and screened for lactose fermentation (blue-black colony with a greenish metallic sheen) on eosin methylene blue agar (Oxoid) and for sorbitol nonfermentation (colorless colony) on sorbitol MacConkey agar (Oxoid) and the same medium containing cefixime (50 μg/liter) and potassium tellurite (2.5 mg/liter) (2, 16). The isolates thus selected were subjected to the standard biochemical tests for identification of E. coli. The tests included conventional indole–methyl red–Voges-Proskauer–citrate and lysine decarboxylase tests, TSI reactions, and examination with an API20E test strip (Biomerieux Vitek, Inc., Hazelwood, Mo.). The identified strains were then screened for the absence of β-glucuronidase with sorbitol MacConkey agar with added 5-bromo-4-chloro-3-indoxyl-β-d-glucuronic acid cyclohexylammonium salt (0.1 mg/ml) and Fluorocult O157:H7 medium (Merck, Darmstadt, Germany). The β-glucuronidase-negative strains were further screened for the presence of the O157 antigen by the latex agglutination test with the E. coli O157 test kit (Oxoid) and the Serobact test kit E. coli O157 (Medvet Science Pty Ltd., Adelaide, Australia) according to the manufacturers’ specifications.
Sixty-three strains were obtained by the method described above. The O:H serotypes of these strains were determined by agglutination tests with antisera. The test strain was grown in tryptic soy broth (Difco) without shaking at 37°C for 5 h for O serotyping. The cells were collected by centrifugation and suspended in physiological saline. A part of this viable-cell suspension was heated at 121°C for 15 min to obtain the heat-killed suspension. The viable-cell and heat-killed-cell suspensions were tested by the agglutination test. The test strains were passed three to four times through heart infusion (Difco)-based semisolid medium containing 0.5% agar to enhance the motility before H serotyping. The strains were then grown in tryptic soy broth as described above, and the cells were fixed by adding formaline (1% final concentration). The cell suspension was prepared by centrifugation as described above, and the agglutination test was performed. Antisera contained in a commercially available O:H serotyping kit (Escherichia coli antisera SEIKEN; Denka Seiken Co., Ltd., Tokyo, Japan) was utilized for O:H serotyping. The antiserum specific to O157 antigen was used to confirm the O serotype. A set of antisera for H serotyping (against 22 different H antigens) was used to determine the H serotype. Fourteen of the 63 strains (listed in Table 1) agglutinated with the anti-O157 serum when the viable-cell suspensions were tested. However, 2 of the 14 strains, MA38 and MA39, gave negative results when the heat-killed suspensions were tested. These were judged to be non-O157 strains. All 14 strains were typed to H7. The 12 O157:H7 strains had been isolated from 9 of 25 beef samples. Three of the samples were found to contain two different O157:H7 strains in each of the samples: MA17 and MA21, MA29 and MA32, and MA51 and MA53. Presumptive E. coli counts of the nine O157:H7-positive samples ranged between 1.2 × 104 and 3.7 × 104 per gram of sample as determined by the method of Harley and Prescott (7).
TABLE 1.
Characteristics of the 12 strains of E. coli O157:H7 and two related strains isolated from beef in Malaysia
| Strain | Presence ofa:
|
Resistance tob:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | CR | ZO | CB | CX | TE | KF | NA | AM | GM | KM | CM | SM | AP | RA | NO | VA | |
| MA1 | − | + | + | + | − | + | − | − | + | − | − | − | − | + | + | − | − | − | + |
| MA6 | − | + | + | − | − | + | + | − | − | − | + | + | + | − | + | − | + | − | − |
| MA7 | − | + | + | − | − | − | + | − | − | + | + | − | − | − | − | − | + | − | + |
| MA17 | + | + | + | − | − | + | + | − | + | + | − | − | − | − | + | − | + | − | + |
| MA40 | + | + | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + |
| MA43 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + |
| MA51 | + | + | + | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | + |
| MA53 | + | + | + | + | − | + | − | − | + | + | + | + | − | − | − | − | + | + | + |
| MA59 | + | + | + | + | − | − | − | − | − | − | + | − | − | − | + | + | − | − | + |
| MA21 | + | + | + | − | − | − | + | − | + | − | + | + | − | + | − | − | + | − | + |
| MA29 | − | + | + | + | − | − | − | − | + | + | + | + | − | − | + | − | + | − | + |
| MA32 | − | + | + | − | − | − | + | − | + | − | − | + | − | − | − | + | + | + | + |
| MA38 | − | − | − | + | − | − | + | − | + | − | − | − | − | − | − | − | + | − | + |
| MA39 | − | − | − | + | − | − | + | − | − | − | − | − | − | − | − | − | + | − | + |
+, present; −, absent.
+, resistant; −, sensitive. CR, ceftriazone; ZO, ceftrizomine; CB, carbenicillin; CX, cefuroxime; TE, tetracycline; KF, cephalotin; NA, nalidixic acid; AM, amoxicillin; GM, gentamicin; KM, kanamycin; CM, chloramphenicol; SM, streptomycin; AP, ampicillin; RA, rifampin; NO, norfloxacin; VA, vancomycin.
Virulence-associated traits.
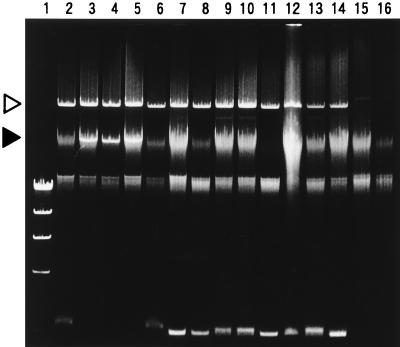
E. coli O157:H7 has important virulence attributes: production of Shiga toxin (Stx, synonymous with Vero toxin and Shiga-like toxin [3]) and attaching and effacing adherence (5). These characteristics of the test strains were examined as follows. To examine the ability to produce Shiga toxin, the test strain was grown in CAYE medium (2% Casamino Acids, 0.6% yeast extract, 0.25% NaCl, 0.87% K2HPO4, 0.005% MgSO4, 0.0005% FeCl3) with shaking (130 rpm) at 37°C for 15 h. The culture supernatant was then tested for the presence or absence of Stx1 and Stx2 by a reversed passive latex agglutination test with a commercially available kit (Verotox-F SEIKEN; Denka Seiken Co.). The presence or absence of the stx1 and stx2 genes in the test strain was tested by a PCR method. The test strain was grown on tryptic soy agar (Difco), and the growth was suspended in distilled water to achieve a slight turbidity (ca. 108 cells/ml). PCR was performed with a PCR amplification kit and EVT-1, EVT-2, EVS-1, and EVS-2 primers purchased from TaKaRa Biomedicals (Tokyo, Japan), and the amplicons of the specified sizes were detected by 1.5% agarose gel electrophoresis according to the manufacturer’s specifications. The presence or absence of the eae gene, which is necessary for attaching and effacing adherence (5), was examined by the DNA colony hybridization method with a polynucleotide probe. The 1-kb HindIII fragment isolated from pCDV443 was used as the probe. pCVD443 was constructed by the isolation of the 1-kb SalI-KpnI fragment of pCVD434 (8), the addition of the HindIII linkers to the isolated fragment by PCR, and the cloning of the fragment into the HindIII site of pUC19 in James B. Kaper’s laboratory (9). The probe DNA was labeled with [α-32P]dCTP, and hybridization was performed under high-stringency conditions as described previously (10). In addition, E. coli O157:H7 carries a plasmid of 60 MDa, a putative factor involved in adherence (5). We examined the presence or absence of the 60-MDa plasmid in the test strains. Plasmid DNA was extracted by the alkaline lysis method with a commercially available kit (FlexiPrep kit; Pharmacia Biotech Inc., Uppsala, Sweden) according to the manufacturer’s specifications, except that the DNA purification step with Sephaglas FP slurry was omitted. The DNA was resolved on a 0.7% agarose gel and visualized by staining with ethidium bromide and photographed with short-wave UV light. For detection of the eae gene and the 60-MDa plasmid, strain EDL933, an isolate from an outbreak in Michigan in 1982 (11, 13), was employed as the control strain. The results of the tests for the stx genes are presented in Table 1. The results of the detection of the Stxs and the respective genes for the 12 O157:H7 strains agreed (data not shown in Table 1). Seven of the 12 O157:H7 strains had the ability to produce both Stx1 and Stx2. The remaining five strains were capable of producing Stx2 only. The 12 strains had the eae gene (Table 1) and a 60-MDa plasmid (Fig. 1, lanes 3 to 14). Two non-O157 strains, MA38 and MA39, were unable to produce Stx and had neither the eae gene nor the 60-MDa plasmid.
FIG. 1.
Results of the plasmid assay. Lanes: 1, phage λ DNA digested with HindIII (molecular size markers); 2, EDL933 (control strain for 60-MDa plasmid); 3, MA1; 4, MA6; 5, MA7; 6, MA17; 7, MA40; 8, MA43; 9, MA51; 10, MA53; 11, MA59; 12, MA21; 13, MA29; 14, MA32; 15, MA38; and 16, MA39. The 60-MDa plasmid is indicated by the open triangle. The two smears at lower positions (above the largest [23 kb] λ HindIII marker) are residual chromosomal DNA. An apparently smaller plasmid in strains MA1, MA6, and MA7 is indicated by the solid triangle. These plasmids overlap the smear of chromosomal DNA (lanes 3 to 5).
Diversity of the strains.
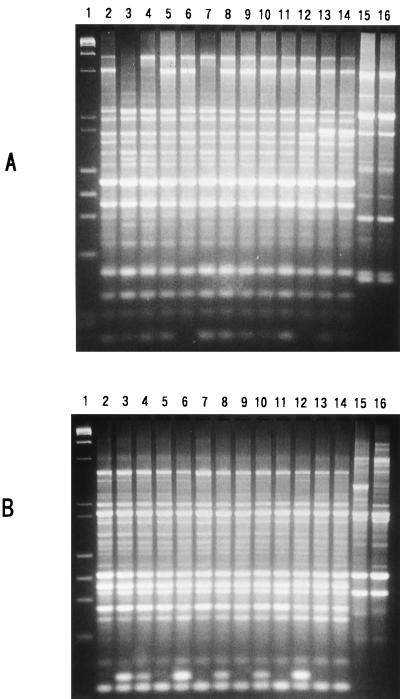
The 12 O157:H7 strains differed regarding some other characteristics. Of the five strains producing only Stx2, three strains had an additional plasmid apparently smaller than 60 MDa (Fig. 1, lanes 3 to 5). The antibiograms of the 12 strains were examined. Resistance or sensitivity of the test strain was examined on Mueller-Hinton agar (Oxoid) by the standard single-disk method (1) with each of the following antibiotic disks: ceftriazone (30 μg/ml, BBL Sensi-Disk; Becton Dickinson, Franklin Lakes, N.J.) ceftrizomine (30 μg/ml, BBL Sensi-Disk), carbenicillin (100 μg/ml, BBL Sensi-Disk), cefuroxime (30 μg/ml, BBL Sensi-Disk), tetracycline (30 μg/ml; Oxoid), cephalotin (30 μg/ml; Oxoid), nalidixic acid (30 μg/ml; Oxoid), amoxicillin (30 μg/ml, BBL Sensi-Disk), gentamicin (10 μg/ml, BBL Sensi-Disk), kanamycin (30 μg/ml; Oxoid), chloramphenicol (30 μg/ml; Oxoid), streptomycin (10 μg/ml; Oxoid), ampicillin (10 μg/ml; Oxoid), rifampin (15 μg/ml, BBL Sensi-Disk), norfloxacin (10 μg/ml, BBL Sensi-Disk), and vancomycin (30 μg/ml, BBL Sensi-Disk). The tests were done three times, and identical results were obtained for each strain and for all antibiotics. The results are summarized in Table 1. None of the strains showed identical antibiograms. The genetic difference among the strains was examined by an arbitrarily primed PCR (AP-PCR) method. Primer 1 (5′-d[GGTGCGGGAA]-3′), primer 2 (5′-d[GTTTCGCTCC]-3′), primer 3 (5′-d[GTAGACCCGT]-3′), primer 4 (5′-d[AAGAGCCCGT]-3′), primer 5 (5′-d[AACGCGCAAC]-3′), and primer 6 (5′-d[CCCGTCAGCA]-3′), included in RAPD analysis primer set (Pharmacia Biotech, Inc.), and the total DNAs extracted from the test strains were used for the AP-PCR assay as described previously (12). All 12 O157:H7 strains and the control strain, EDL933, had many amplicon bands in common, but strain-to-strain variation could be detected by the presence or absence of some other bands. Representative AP-PCR profiles are shown in Fig. 2. A detailed comparison of the AP-PCR profiles obtained with the six different primers indicated that the 12 O157:H7 strains could be classified into 11 different genetic types, as shown in Table 2 (genetic type numbers 2 to 12).
FIG. 2.
Representative results of the AP-PCR assay. The results obtained with primer 1 and primer 5 are shown in panels A and B, respectively. Lanes: 1, mixture of phage λ DNA digested with HindIII and phage φX174 DNA digested with HaeIII (molecular size markers); 2, EDL933 (a control strain); 3, MA1; 4, MA6; 5, MA7; 6, MA17; 7, MA40; 8, MA43; 9, MA51; 10, MA53; 11, MA59; 12, MA21; 13, MA29; 14, MA32; 15, MA38; and 16, MA39.
TABLE 2.
AP-PCR profile patterns and genetic types ofO157:H7 and related strains
| Strain | AP-PCR profile pattern observed with indicated primera
|
Genetic type no.b | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| EDL933 | A | A | A | A | A | A | 1 |
| MA1 | B | B | B | B | B | A | 2 |
| MA6 | C | B | C | B | B | A | 3 |
| MA7 | D | B | D | C | A | A | 4 |
| MA17 | E | A | E | A | B | A | 5 |
| MA40 | C | A | E | A | A | A | 6 |
| MA43 | D | A | E | A | B | A | 7 |
| MA51 | D | A | E | A | A | A | 8 |
| MA53 | D | A | E | A | B | A | 7 |
| MA59 | D | A | D | A | A | A | 9 |
| MA21 | E | A | E | A | B | A | 10 |
| MA29 | F | C | E | D | A | A | 11 |
| MA32 | F | C | E | A | A | A | 12 |
| MA38 | G | D | F | F | C | B | 13 |
| MA39 | G | D | G | F | D | B | 14 |
The designations of the AP-PCR profile patterns for each primer were arbitrarily assigned.
Genetic types were arbitrarily assigned on the basis of the combinations of different AP-PCR profile patterns.
In summary, E. coli O157:H7 strains possessing important virulence traits were shown to be distributed at a considerable frequency in the beef retailed in Malaysia. The differences in the antibiogram, plasmid profile, and AP-PCR profile among the strains suggest that the strains may have originated from diverse sources. This organism is significant to public health and needs to be surveyed in more detail in Malaysia and the neighboring areas.
Acknowledgments
We are grateful to James B. Kaper, Center for Vaccine Development, University of Maryland School of Medicine, for supplying the plasmid pCVD443 and strain EDL933.
This research was supported in part by a fund for researchers abroad from the Ministry of Education, Science, Sports and Culture, Japan, and by funds from the Academic Forum for Northeast Asia and Japan and from the Malaysian government through IRPA.
REFERENCES
- 1.Bauer A W, Kirby W M M, Sherris J C. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1996;45:493–496. [PubMed] [Google Scholar]
- 2.Bennett A R, MacPhee S, Betts R P. Evaluation of methods for the isolation and detection of Escherichia coli O157 in minced beef. Lett Appl Microbiol. 1995;20:375–379. doi: 10.1111/j.1472-765x.1995.tb01325.x. [DOI] [PubMed] [Google Scholar]
- 3.Calderwood S B, Acheson D W K, Keusch G T, Barrett T J, Griffin P M, Strockbine N A, Swaminathan B, Kaper J B, Levine M M, Kaplan B S, Karch H, O’Brien A D, Obrig T G, Takeda Y, Tarr P I, Wachsmuth I K. Proposed new nomenclature for SLT (VT) family. ASM News. 1996;62:118–119. [Google Scholar]
- 4.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 5.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 739–761. [Google Scholar]
- 6.Gupta S, Soni N K, Kaur P, Sood D K. Verocytopathic activity of Escherichia coli O157 & other ‘O’ serogroups isolated from patients of diarrhoea. Indian J Med Res. 1992;95:71–76. [PubMed] [Google Scholar]
- 7.Harley J P, Prescott L M. Bacterial count of a food product. In: Harley J P, Prescott L M, editors. Laboratory exercises in microbiology. 2nd ed. Iowa: WBC Publishers; 1993. pp. 174–175. [Google Scholar]
- 8.Jerse A E, Yu J, Tall B, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper, J. B. Personal communication.
- 10.Nishibuchi M, Ishibashi M, Takeda Y, Kaper J B. Detection of the thermostable direct hemolysin gene and related DNA sequences in Vibrio parahaemolyticus and other Vibrio species by the DNA colony hybridization test. Infect Immun. 1985;49:481–486. doi: 10.1128/iai.49.3.481-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien A D, Lively M E, Chen S, Rothman W, Formal S B. Escherichia coli O157:H7 strains associated with hemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (Shiga)-like cytotoxin. Lancet. 1983;i:702. doi: 10.1016/s0140-6736(83)91987-6. [DOI] [PubMed] [Google Scholar]
- 12.Okuda J, Ishibashi M, Abbott S L, Janda J M, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in the urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35:1965–1971. doi: 10.1128/jcm.35.8.1965-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 14.Suthienkul O, Brown J E, Seriwatana J, Tienthongdee S, Sastravaha S, Echeverria P. Shiga-like toxin-producing Escherichia coli in retail meats and cattle in Thailand. Appl Environ Microbiol. 1990;56:1135–1139. doi: 10.1128/aem.56.4.1135-1139.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada S, Matsushita S, Kai A, Sasaki M, Tsuji A, Kanemitsu T, Yamashita N, Anzai E, Kudoh Y. Detection of verocytotoxin from stool and serological testing of patients with diarrhea caused by Escherichia coli O157:H7. Microbiol Immunol. 1993;37:111–118. doi: 10.1111/j.1348-0421.1993.tb03187.x. [DOI] [PubMed] [Google Scholar]
- 16.Zadik, P. M., P. A. Chapman, and C. A. Siddons. 1993. Use of tellurite for the selection of verocytotoxigenic Escherichia coli O157. [DOI] [PubMed]