Abstract
INTRODUCTION:
Alzheimer’s disease (AD) is heterogeneous, both clinically and neuropathologically. We investigated whether polygenic risk scores (PRSs) integrated with transcriptome profiles from AD brains can explain AD clinical heterogeneity.
METHODS:
We conducted co-expression analysis and identified gene-sets (modules) which were preserved in three AD transcriptome datasets and associated with AD-related neuropathological traits including neuritic plaques (NPs) and neurofibrillary tangles (NFTs). We computed the module-based PRS (mbPRS) for each module and tested associations for mbPRSs with cognitive test scores, cognitively-defined AD subgroups, and brain imaging data.
RESULTS:
Of the modules significantly associated with NPs and/or NFTs, the mbPRSs from two modules (M6 and M9) showed distinct associations with language and visuospatial functioning, respectively. They matched clinical subtypes and brain atrophy at specific regions.
DISCUSSION:
Our findings demonstrate that polygenic profiling based on co-expressed gene-sets can explain heterogeneity in AD patients, enabling to genetically-informed patient stratification and precision medicine in AD.
Keywords: Alzheimer disease, co-expression network, cognitive performance, precision medicine, module-based polygenic risk score, patient stratification, genetic subtyping
1. BACKGROUND
Late onset Alzheimer’s disease (AD) is a complex disorder with clinical and neuropathological heterogeneity [1, 2]. Types of clinical heterogeneity include progression rate, predominant cognitive symptoms, and whether psychotic symptoms manifest [1]. AD neuropathology can also be varied with complications of other neuropathological traits beyond plaques and tangles [1, 2]. Clinical and neuropathological heterogeneity may have contributed to the repeated failure of AD clinical trials [3]. Classification of heterogeneous AD patients into biologically-relevant subgroups may improve our understanding of biological mechanisms underlying the variability of cognitive symptoms and trajectories of decline, as well as lead to development of subgroup-specific treatment options [4].
Different AD subtypes have been previously proposed based on neuropsychological and neuropathological characteristics [5–7], domain-specific cognitive functions, MRI brain imaging data [4], and metabolic profiling [8]. However, our understanding of molecular mechanisms underlying disease heterogeneity is still limited. Recent report illustrates that genetic variants with large effect sizes can distinguish six cognitively-defined subgroups of AD when compared with elderly controls [9]. A previous study showed that polygenic risk scores (PRSs) derived from clusters (i.e., gene-sets) in genome-wide association studies (GWASs) of type 2 diabetes (T2D)-related phenotypes have successfully classified T2D patients into different subtypes [10]. These studies demonstrate that PRSs from biologically connected gene-sets may explain disease heterogeneity and improve scientific understanding of biological mechanisms underlying disease subtypes. In addition, co-expression network analyses have shown to be useful for identifying biologically-connected and disease-relevant gene-sets using transcriptome data [11, 12]. Taken together, these findings led to the hypothesis that network analysis utilizing transcriptome data of AD brains could capture biologically relevant gene-sets responsible for distinct disease subtypes and PRSs derived from the gene-sets could explain clinical heterogeneity of AD.
In this study, we identified modules (sets of biologically relevant genes) by co-expression analysis and thereby generated module-based PRSs of AD patients. Then, using domain-specific cognitive functions, previously defined AD cognitive subgroups, and brain imaging data, we evaluated whether the module-based PRSs can explain cognitive impairment heterogeneity among the AD patients.
2. METHODS
2.1. Sources of RNA sequencing data in autopsied AD brains for network analysis
Co-expression analysis was performed using previously generated gene expression data from the dorsolateral prefrontal cortex (DLPFC) area of 65 autopsy-confirmed non-Hispanic white AD cases from the Framingham Heart Study and Boston University Alzheimer’s Disease Research Center (FHS/BUADRC) [13]. Details of procedures for quality control (QC) of RNA sequencing (RNA-Seq) data and neuropathological AD diagnosis are presented in Supplementary Information and previously reported elsewhere [13]. Additional RNA-Seq datasets for validation were obtained from the CommonMind portal (http://www.synapse.org) including post-QC normalized gene expression data (version #1) from the DLPFC area of 363 neuropathologically-confirmed AD cases in the Religious Orders Study and Rush Memory and Aging Project (ROSMAP) [14] and from temporal cortex area of 82 autopsy-confirmed AD cases in the Mayo Clinic Study of Aging (MAYO) [15].
2.2. Identifying Preserved and AD-associated Modules
Co-expression gene-sets (i.e., modules) were generated using the transcriptome data from the 65 AD brains in FHS/BUADRC using the Weighted Gene Co-expression Network Analysis (WGCNA) approach, which computes pairwise correlations for all gene pairs and clusters genes by the correlated expression levels [16]. Transcriptome data of AD-free controls were not included in our co-expression study because our interest is to identify gene-sets related to the disease heterogeneity, not the disease risk (e.g., cases versus controls). Details of co-expression module construction were presented in Supplementary Information and previously described [17]. Preservation of the discovery modules was evaluated in the two independent validation datasets, including ROSMAP and MAYO datasets, using z-summary statistics [16]. We considered a module to be preserved if z-summary scores greater than 5.0 in both validation datasets [16]. Among the preserved modules, we selected AD-associated modules by enrichment analyses using gene-sets for AD-related outcomes consisting of AD-related neuropathological traits [18] including neuritic plaques (NP) and neurofibrillary tangles (NFT) and AD-risk [19]. We used AD associated genes for enrichment analyses that contained at least one single nucleotide polymorphism (SNP) with P<10−3 located within +/− 20 kilobases from the gene for0 one of the AD phenotypes (NP, NFT, or AD-risk). Significant enrichment P-values<0.05 were applied using the Fisher’s exact test after false discovery rate (FDR) correction. Based on the result from the enrichment analysis for each module, we assigned the AD phenotypes (NP, NFT, or AD-risk) for which the module was most significantly enriched and used to calculate module based polygenic risk scores. The selected AD-associated modules were considered to generate module based polygenic risk scores.
We also examined expression coherence and cellular signatures of genes in each of the AD-associated modules using single cell RNA-seq data in five different cell types (astrocytes, microglia, oligodendrocytes, endothelia, and neurons) from the temporal lobe area (Gene Expression Omnibus ID: GSE67835) [20] and single nucleus RNA-seq data in seven cell types (astrocytes, microglia, oligodendrocytes, pericytes, endothelia, and excitatory/inhibitory neurons) from the prefrontal cortex in the ROSMAP [21]. Details of methods for deriving cell-type-specific gene-sets and their expression profiling are presented in Supplementary Information and reported elsewhere [13]. Enrichment of cell-type-specificity for each AD-associated module was tested using the Fisher’s exact test.
2.3. Genotypic and phenotypic data in ADNI
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a longitudinal study assessing clinical, neuroimaging, genetic, and biomarker data from participants in various stages of cognitive impairment including cognitively normal (CN), mild cognitive impairment (MCI), and AD. Genetic and phenotypic data of ADNI participants were obtained from the LONI website (http://adni.loni.usc.edu). We used the ADNI genetic data for computing mbPRSs and phenotype data for evaluating the relationships between mbPRSs and cognitive impairment heterogeneity. Genome-wide genotype data from two different arrays (ADNI-1, n=679 and ADNI-GO/2, n=397) were imputed using the Haplotype Reference Consortium data. Details of quality control (QC), imputation, and population substructure procedures are described in the Supplementary Information. Characteristics of the sample after QC are presented in Table S1.
Since clinical spectrum of AD can be largely affected by impairment of specific cognitive functions [22], we hypothesized that deficits in particular cognitive domains explain at least in part disease heterogeneity (i.e., cognitive impairment heterogeneity). To explore this heterogeneity, we used domain-specific cognitive tests at the last exam from the ADNI dataset (Table S1): logical memory immediate (LIMMTOTAL) and delayed (LDELTOTAL) recall tests for memory; trail marking test A/B (TRAASCOR and TRABSCOR) for executive functioning; category fluency animal score (CATANIMSC) and Boston naming test total (BNTTOTAL) score for language; and clock test total score (COPYSCORE) for visuospatial functioning. Cognitive test scores were adjusted for age, sex, and education using linear regression, and the residuals derived from the regression models were ranked-transformed as previously described [17].
2.4. Computing and assessing polygenic risk scores for AD-associated modules in ADNI
We selected SNPs in each AD-associated module from the enrichment analysis for the assigned AD outcome and generated module based polygenic risk scores (mbPRSs) using effect estimates of the selected SNPs for NP, NFT, or AD-risk from the enrichment analysis. For comparison, we also generated PRSs for NP, NFT, and AD-risk in a conventional approach, which aggregates effect estimates of SNPs with P<0.001 across the genome, defined as genome-wide PRS (gwPRS). Details about computing these two types of PRSs (gwPRS and mbPRSs) are included in the Supplementary Information.
After generating those PRSs in ADNI, we evaluated correlations among the mbPRSs and gwPRSs. To assess relevance of those PRSs to disease stages/progression, we stratified ADNI sample by disease stages (CN, MCI, and AD) at the last exam and compared mean values of PRSs between different disease stages. We also tested associations between PRSs and conversion status for AD progression (e.g., CN to MCI or AD; MCI to AD) excluding AD at baseline using logistic regression models after adjusting for age, sex, the first four PCs and the array information.
Next, we conducted association tests with mbPRSs or gwPRSs for specific cognitive domains using rank-transformed cognitive test scores as quantitative outcomes in linear regression models after adjusting the first four PCs and genotype platform as covariates. We followed up the nominally significant modules (P<0.05) with domain-specific cognitive test scores as cognitive impairment heterogeneity (CIH) modules.
We also attempted to replicate the associations between mbPRSs of the selected CIH modules and domain-specific cognitive test scores among 134 AD cases in FHS (dbGaP Study Accession ID: phs000056.v5.p3). Details of sample characteristics, imputation, computation of mbPRSs, and association tests with cognitive test scores in Neuropsychological Test Battery in FHS is described in the Supplementary Information.
2.5. Validating CIH modules with cognitively-defined AD subtypes in ADNI
Previously, 672 AD cases in ADNI have been classified into cognitively-defined subtypes based on relative impairments at the time of AD diagnosis [9], consisting of 196 as AD-Memory, 16 as AD-Executive functioning, 52 as AD-Language, 91 as AD-Visuospatial functioning, and 317 other domains (Table S2). Details about these cognitively-defined subgroups are described in Supplementary Information and reported elsewhere [9]. We evaluated whether mbPRSs of CIH modules are linked into one of the four cognitively-defined subgroups (AD-Memory, AD-Executive, AD-Language, and AD-Visuospatial domains). Each subject was assigned into a membership of one subgroup coded as 1, and otherwise coded as 0 excluding subjects with overlapping memberships. We tested association between mbPRSs and a dichotomized membership of cognitively-defined subtypes in a logistic regression model adjusting for age, sex, the first four PCs, and genotype platform as covariates.
2.6. Brain imaging (MRI) data analysis with mbPRSs of the CIH modules in ADNI
To understand the relationships between our CIH modules and brain atrophy at specific locations, we tested the association between mbPRSs and surface-based cortical thickness of AD patients using general linear models after adjusting age, sex, magnetic field strength, and intracranial volume as covariates [23]. Detail information about brain imaging data processing for surface-based measure of cortical thickness in ADNI are described elsewhere [23].
2.7. Biological functions of genes in the CIH modules
Gene-ontology (GO) analyses were conducted to discern biological pathways of AD-associated genes in CIH modules using the Ingenuity Pathway Analysis software (QIAGEN, Redwood, CA). We also looked up associations between the CIH module-genes and AD-related neuropathological traits including Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) score and Braak stage, and quantitative measures of proteins including Aβ42, phosphorylated Tau at 181 (pTau181) and 231 (pTau231), postsynaptic density protein 95 (PSD95), C4a, C4b, and PPP2CA/B from prefrontal cortex area of autopsied brains (FHS/BUADRC) [13].
3. RESULTS
3.1. AD-associated modules in AD brains were preserved in independent studies
The overall study design including module selection process was provided in Figure S1. Eighty-three modules were identified in the discovery dataset (FHS/BUADRC), and 29 of these modules were preserved in the two validation datasets (Figure 1A). Fourteen of the 29 preserved modules (M1-M14) contained genes that were significantly enriched in at least one of the AD gene-sets (NP, NFT, or AD-risk) with FDR<0.05 (Figure 1B and Table 1). Interestingly, only four modules (M1–3 and M11) were nominally enriched in the AD-risk gene-set, and all 14 modules were at least three orders of magnitude more significantly enriched in either NP or NFT gene-sets (Table 1). These findings may imply that our modules derived from the transcriptome datasets of AD brains (without AD-free controls) would capture the gene-sets for underlying changes in AD pathology, rather than the overall disease risk. Therefore, we selected one outcome, either NP or NFT, but not AD-risk to compute fourteen mbPRSs (NP-linked modules: M2, M5, M6, M10, and M13; NFT-linked modules: M1, M3, M4, M7–9, M11, M12, and M14), according to its most significantly enriched gene-set and the GWAS summary data of the selected outcome (NP or NFT).
Figure 1.
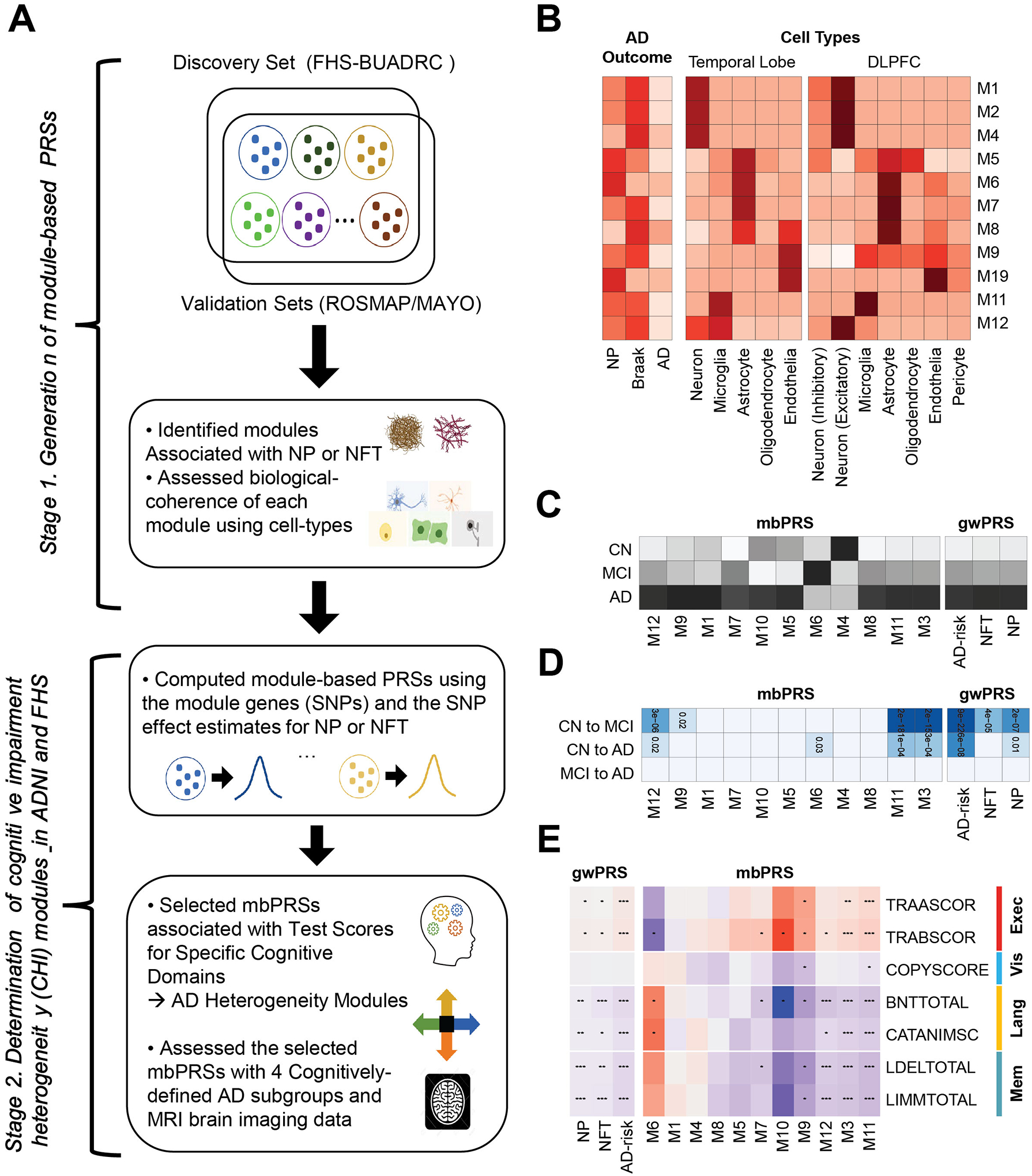
A. Schematic of our study design. We constructed co-expression modules (sets of genes), selected AD-associated modules, and generated module-based polygenic risk scores for explaining the AD heterogeneity, which were tested and evaluated with gene-sets for AD-related neuropathological traits (NP and NFT human brain cell-types, cognitive test scores, cognitively-defined AD subgroups, and brain MRI imaging data. B. Enrichment analysis. The strength of enrichment results with the eleven AD-associated modules for AD phenotypes (NP, NFT, and AD-risk; left) and cell type specific gene-sets in temporal lobe (middle) and dorsal lateral prefrontal cortex (DLPFC; right). The darker color indicates the more significant enrichment P-value. C. Heatmap of mean values for module-based PRSs (mbPRSs) and genome-wide PRSs (gwPRSs) across the disease stages including clinical normal (CN), mild cognitive impairment (MCI), and AD (the darker color indicates the larger mean value of the PRS). D. Associations between PRSs (mbPRSs and gwPRSs) and disease progression (CN to MCI, CN to AD, MCI to AD) (the darker color indicates the more significant P-value). E. Associations between PRSs (mbPRSs and gwPRSs) and seven test scores for four cognitive domains (executive functioning, visuospatial functioning, language, memory). Red and blue mean positive and negative effect directions, respectively. Dots in the cells indicate the strength of associations).
Table 1.
AD associated preserved networks in AD brains
| Module | Z-summary | Enrichment P-value | |||
|---|---|---|---|---|---|
| ROSMAP | MAYO | NP | NFT | AD-RISK | |
| M1 | 40.56 | 20.71 | 3.74X10−6 | 3.26X10−9 | 0.04 |
| M2 | 37.45 | 20.68 | 3.50X10−8 | 2.57X10−7 | 0.002 |
| M3 | 35.48 | 18.59 | 5.38X10−5 | 8.36X10−7 | 0.01 |
| M4 | 32.32 | 18 | 3.42X10−2 | 1.25X10−5 | 0.07 |
| M5 | 29.83 | 15.91 | 1.46X10−6 | 2.92X10−4 | 0.40 |
| M6 | 26.56 | 14.46 | 4.70X10−6 | 0.09 | 0.09 |
| M7 | 25.84 | 14.23 | 2.65X10−3 | 2.47X10−4 | 0.12 |
| M8 | 20.52 | 14.07 | 0.47 | 0.02 | 0.20 |
| M9 | 20.51 | 12.71 | 4.34X10−3 | 8.89X10−4 | 0.37 |
| M10 | 18.25 | 12.64 | 0.02 | 0.16 | 0.16 |
| M11 | 17.32 | 11.81 | 8.70X10−4 | 7.48X10−4 | 0.01 |
| M12 | 17.06 | 10.55 | 1.00 | 0.05 | 0.95 |
| M13 | 15.57 | 8.59 | 0.02 | 0.03 | 0.81 |
| M14 | 14.79 | 7.81 | 0.06 | 0.04 | 0.48 |
Module is a co-expressed gene network in the discovery from the Framingham Heart Study and Boston University Alzheimer’s Disease Center (FHS/BUADRC) study.
Z-summary is a network preservation score of a module from the discovery to at least one of two validation datasets, the Religious Orders Study and Rush Memory and Aging Project (ROSMAP) and the Mayo Clinic Study of Aging (MAYO).
Enrichment p-value for a module were computed using genes in the given module containing a SNP with P<10−3 from Beecham et al. for neuritic plaque [NP] and neurofibrillary tangles [NFT] [18] and from Kunkle et al. for AD-risk [19].
Fourteen modules were selected when a module in the FHS/BUADRC study was preserved with Z-summary>5 in both validation datasets, ROSMAP and MAYO, and significant at p-value<0.05 with at least one of the gene-sets for NP, NFT, or AD-risk.
All 14 AD-associated modules were significantly enriched in specific cell-types, where these results were consistent between temporal lobe and prefrontal cortex regions (Figure 1B and Table S3). M1 to M4 modules were predominantly enriched in excitatory neurons (best P with M2 from prefrontal cortex=4.4×10−188), M6 and M7 in astrocytes (best P with M7 from prefrontal cortex =1.3×10−97), M10 in endothelia (P from temporal lobe=9.9×1087), and M11 in microglia (P from temporal cortex=1.7×10−129). The other five modules (M5, M8, M9, M12 and M13) were significantly enriched in more than one cell type, while M5 and M8 (astrocytes), M9 (endothelia), M12 (neurons), and M13 (microglia) were significantly enriched in at least one cell type in both brain regions.
3.2. Module-based PRSs explained heterogeneity in cognitive functions among AD patients
Of 14 preserved and AD associated modules, we computed mbPRSs for nine NFT-linked modules (M1, M3, M4, M7–9, M11, M12, and M14) using NFT, while mbPRSs of the remaining five NP-linked modules (M2, M5, M6, M10, and M13) using NP. Three modules including M2, M13, and M14 were excluded due to their low standard error (<0.05) and/or their extremely skewed distributions for the following analyses (Table S4), leading to 11 modules for further evaluation.
In comparison, three genome-wide PRSs (gwPRSs) were significantly correlated with mbPRSs of the M3, M11, and M12 modules (correlation r2≥0.1), while the rest of eight mbPRSs were not correlated with those for gwPRSs (r2<0.01) (Figure S2). The mean values of all three gwPRSs were sequentially increased from CN to MCI and AD (Figure 1C). In contrast, the mean values of mbPRS were varied across the disease stages, which the mean values of modules (M3–6, and M10) were smaller in MCI or AD stages than in the CN stage (Figure 1C). For the disease progression, two gwPRSs (NFT and AD-risk) and three mbPRSs (M3, M11, and M12) were significantly associated with the progressions from CN to both MCI and AD (Figure 1D). None of PRSs were associated with the progression from MCI to AD. Interestingly, NFT-gwPRS and M9-mbPRS were associated with the progression from CN to MCI, while M6-mbPRS was associated with the progression from CN to AD. All three gwPRSs were significantly associated with most cognitive test scores, except for the visuospatial domain (COPYSCORE), with the consistent effect directions across cognitive tests. This indicates that the gwPRSs are not likely to differentiate cognitive impairment heterogeneity among AD cases (Figure 1E).
Five mbPRSs for M3, M6, M9, M11, and M12 were robustly associated (all tests in domain with P-value<0.05), while two mbPRSs for M7 and M10 were nominally associated with only one cognitive test in domain (Figure 1E and Table S5). Four mbPRSs for M1, M4, M5, and M8 showed no association (P-value>0.05) with any cognitive test scores (Figure 1E and Table S5). Of the 5 mbPRSs with robust associations for cognitive domains, mbPRSs for M3 and M11 were strongly associated with the 3 cognitive domains except for the visuospatial functioning, indicating that mbPRSs from M3 and M11 did not differentiate cognitive impairment heterogeneity. The M6-mbPRS was nominally associated with all two language-domain test scores (BNTTOTAL P-value=0.03 and CATANIMSC P-value=0.01). The M9-mbPRS was associated with all tests in memory and executive function domains (P<0.05), as well as with visuospatial functioning domain (COPYSCORE P-value=0.05). The M12-mbPRS was strongly associated with language (best P with BNTTOTAL=2.2×10−6) and memory (best P with LIMMTOTAL=4.4×10−6) domains (Table 2). Therefore, we prioritized M6, M9, and M12 as CIH modules and attempted to validate the associations between mbPRSs of the CIH modules (M6, M9, and M12) and the cognitive test scores among the AD cases in FHS (Table S6). We replicated nominally significant associations (P<0.05) between the M6-mbPRS and two language-domain cognitive test scores in AD cases from the FHS (BNT30 P-value=0.03 and BNT30cue P-value=0.03; Table S7). Although we did not find associations of the rest two modules (M9 and M12) with the cognitive test scores in FHS, three CIH modules (M6, M9, and M12) were further tested with cognitively-defined subgroups [9] and brain atrophy among AD patients.
Table 2.
Associations between cognitive test scores and three module-based PRSs
| Cognitive Domain | Cognitive Test | M6 | M9 | M12 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BETA | SE | P-value | BETA | SE | P-value | BETA | SE | P-value | ||
| Executive functioning | TRAASCOR | −0.16 | 0.12 | 0.17 | 0.18 | 0.06 | 1.08×10−3 | 0.02 | 0.02 | 0.15 |
| TRABSCOR | −0.26 | 0.12 | 0.03 | 0.18 | 0.06 | 1.58×10−3 | 0.03 | 0.02 | 0.05 | |
| Visuospatial functioning | COPYSCORE | 0.03 | 0.12 | 0.79 | −0.12 | 0.06 | 0.049 | −0.01 | 0.02 | 0.77 |
| Language | BNTTOTAL | 0.26 | 0.12 | 0.03 | −0.17 | 0.06 | 2.57×10−3 | −0.08 | 0.02 | 2.23×10−6 |
| CATAANIMSC | 0.29 | 0.12 | 0.01 | −0.11 | 0.06 | 0.05 | −0.04 | 0.02 | 0.01 | |
| Memory | LDELTOTAL | 0.22 | 0.12 | 0.06 | −0.18 | 0.06 | 1.14×10−3 | −0.07 | 0.02 | 4.55×10−5 |
| LIMMTOTAL | 0.18 | 0.12 | 0.12 | −0.15 | 0.06 | 7.18×10−3 | −0.08 | 0.02 | 4.42×10−6 | |
LIMMTOTAL and LDELTOTAL: logical memory immediate and delayed recall tests; TRAASCOR and TRABSCOR: trail marking test A/B. CATANIMSC: category fluency animal score. BNTTOTAL: Boston naming test total. COPYSCORE: clock test total score
Beta estimates (BETA), standard error (SE), and P-values were calculated with module-based PRSs of the M2, M6, and M9 modules for domain specific cognitive functions.
3.3. Module-based PRS associations with cognitively-defined AD subtypes and brain atrophy
Of the three CIH mbPRSs, the mbPRSs of M6 and M9 showed nominal associations at P<0.05 with odds ratio (OR)>1.0 with previously defined cognitive subtypes for AD-Language (OR=5.5; P=0.01) and AD-Visuospatial functioning (OR=1.9; P=0.04), respectively (Table 3). The M12-mbPRS was associated with none of the cognitive subtypes (Table 3). In contrast, the gwPRSs for NP and AD-risk failed to differentiate any of the subgroups, while only NFT-gwPRS was nominally associated with the AD-Memory subgroup (OR=1.01; P=0.04).
Table 3.
Membership assignment of three module-based PRSs in previously defined cognitive subtypes of AD in ADNI
| Source of PRS | Executive Functioning ST | Language ST | Memory ST | Visuospatial Functioning ST | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Genome-wide PRS (gwPRS) | ||||||||
| NP | 0.99 (0.93–1.0) | 0.69 | 0.99 (0.95–1.0) | 0.43 | 1.00 (0.99–1.00) | 0.57 | 1.00 (0.97–1.00) | 0.75 |
| NFT | 1.00 (0.91–1.10) | 0.98 | 0.99 (0.93–1.01) | 0.74 | 1.01 (1.00–1.11) | 0.04 | 0.96 (0.91–1.01) | 0.09 |
| AD-Risk | 1.01 (0.93–1.10) | 0.65 | 0.97 (0.91–1.00) | 0.24 | 1.00 (1.00–1.10) | 0.08 | 0.96 (0.91–1.00) | 0.07 |
| Module-based PRS (mbPRSs) | ||||||||
| M6 | 0.37 (0.04–3.60) | 0.39 | 5.50 (1.50–20.0) | 0.009 | 0.97 (0.44–2.20) | 0.95 | 0.88 (0.28–2.80) | 0.83 |
| M9 | 0.95 (0.34–2.60) | 0.92 | 0.81 (0.42–1.50) | 0.51 | 1.20 (0.84–1.80) | 0.29 | 1.91 (1.11–3.31) | 0.04 |
| M12 | 1.10 (0.78–1.50) | 0.66 | 0.92 (0.75–1.10) | 0.40 | 1.00 (0.92–1.20) | 0.56 | 1.00 (0.87–1.20) | 0.76 |
OR: odds ratio; CI: 95% confidence interval; P: P value. NP: neuritic plaques; NFT: neurofibrillary tangles. Cognitively-defined AD subtypes (ST) in ADNI have been previously defined [9]. Bold values with significance at P<0.01 and OR>1.0 indicate these modules are likely members of the corresponding cognitively-defined subtypes.
The mbPRSs of M6 and M9 were significantly associated with cortical thickness at specific brain locations (M6: bilateral frontal, parietal, and temporal lobes; M9: bilateral frontal lobes; Figure 2A). Particularly, the brain atrophy for the M6-mbPRS was localized at the Wernicke area where lesions have been associated with severe impairments of word comprehension [24].
Figure 2.
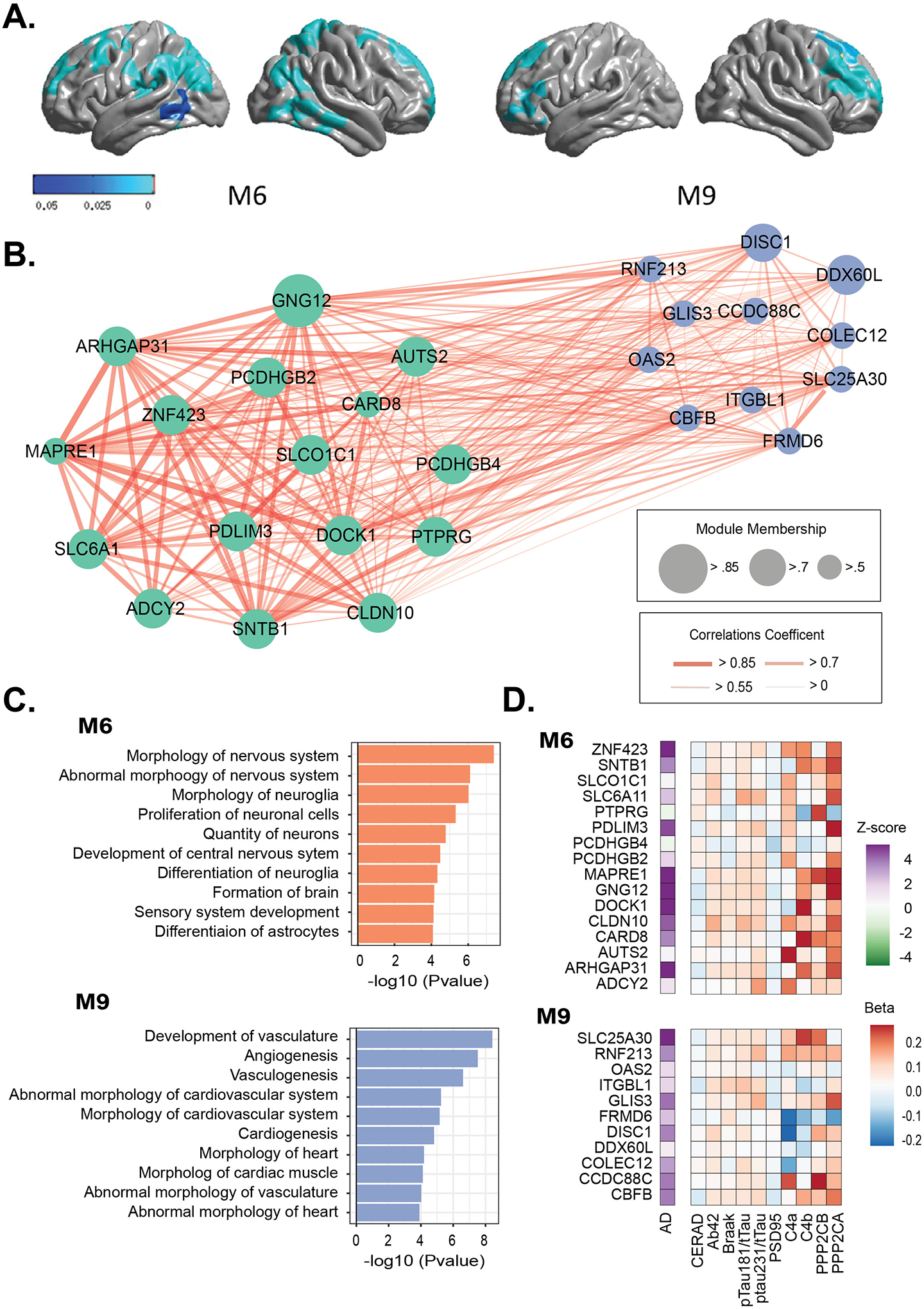
M6 and M9 were selected as cognitive impairment heterogeneity (CIH) modules. A. Association between cerebral cortical thickness and module-based polygenic risk scores for M6 and M9. P-value map with threshold at p<0.05 indicated that the darker blue color showed more significant P-value. B. Co-expression network of genes in M6 and M9. C. Pathways enriched for M6 and M9. D. Associations of the expression levels of genes in M6 and M9 with AD and AD-related neuropathological traits (previously published [13]).
3.4. Functional profiling of M6 and M9
Among the genes in M6 and M9, we focused on the GWAS genes (i.e., seed genes) containing a SNP with P<0.001 for NP or NFT (# of GWAS genes, M6=16 and M9=11; Figure 2B and Table S8). The seed genes in M6 were significantly enriched in pathways (Figure 2C and Table S9) including morphology of nervous system (P=4.0×10−8), abnormal morphology of nervous system (P=7.8×10−7) and differentiation of astrocytes (P=8.4×10−5), while the M9 seed genes were enriched in pathways including vascular system including development of vasculature (P=3.8×10−9), angiogenesis (P=3.0×10−8), and vasculogenesis (P=2.5×10−7) (Figure 2C and Table S9).
According to our previous report [13], the majority of the seed genes in M6 (best: DOCK1, P=3.0×10−7) and M9 (best: SLC25A30, P=6.3×10−6) were up-regulated in AD compared with control brains (Table S10). In addition, we observed significant associations between expression levels of the seed genes in M6 and M9 and AD-related protein levels (Figure 2D and Table S11). The seed genes in M6 were significantly associated (P<0.05) with CERAD scores, Braak stages, Aβ42, pTau181/tTau ratio, pTau231/tTau ratio, C4a, C4b, and PSD95 (Table S11), which the most significant association was observed with expression of ADCY2 with pTau181/tTau ratio (P-value=1.1×10−3). The expressions of the seed genes in M9 were nominally associated with Braak stages, Aβ42, pTau231/tTau ratio, and C4a levels with the best P-value between expression of DISC1 and C4a (P-value=1.0×10−3).
4. DISCUSSION
4.1. Key findings
The goal of this study was to identify gene-sets responsible for the biological mechanisms underlying AD heterogeneity. We generated modules (gene-sets) that were commonly observed in multiple transcriptome datasets of AD brains. We closely evaluated biological coherence and disease relevance of networks of genes (modules) using profiling of human brain cell types and genetics of AD neuropathology. Then, we selected the CIH modules (M6, M9, and M12), which are likely to explain the disease heterogeneity in cognitive impairment of the AD patients, by testing with domain-specific cognitive test scores in ADNI (clinic-based study) and in FHS (population-based study). We identified and validated two CIH modules (M6 and M9) that showed significant associations for language and visuospatial domains with matching cognitive AD subtypes (AD-Language and AD-Visuospatial), respectively. These results were further linked to atrophy in specific brain areas (M6: Wernicke’s area in temporoparietal cortex; M9: frontal cortex) which were previously reported to underpin for language comprehension [24, 25] and visuospatial deficit [26]. This study demonstrated the novel concept that can be generalizable and applicable to diverse populations, although not all the modules are available in all populations. The process and approach used in this study indicate that polygenic risk profiling in co-regulated and biologically connected genes provide one of unique and distinct frameworks to explain AD heterogeneity.
4.2. Advantage of mbPRSs for AD subgrouping
The three gwPRSs for NP, NFT, and AD-risk showed high correlations with each other and largely similar patterns from associations with disease conversion and cognitive test scores. In contrast, the mbPRSs showed almost no correlations with each other and were associated with the performance of specific cognitive domains. These findings after comparing our novel mbPRSs with conventional gwPRSs demonstrate that mbPRSs would be more useful for explaining the phenotypic heterogeneity in AD patients, while gwPRSs (i.e., traditional PRS) would be more relevant to predict the overall disease risk. Our mbPRSs successfully distinguished differences in clinical (cognitive domains) and structural brain imaging patterns, indicating representation of different disease mechanisms and thereby would be effective tools for dissecting the disease heterogeneity. The gwPRSs for NP and AD-risk failed to recognize the AD subgroups. Only the gwPRS for NFT discerned the most typical cognitive subgroup, AD-Memory domain. In contrast, newly identified mbPRSs for the M6 an M9 modules recognized different types of AD subgroups. This indicates that the conventional gwPRS approach is less likely to recognize differences between AD subtypes. Further, these results support our hypothesis that subgrouping genetic markers from gene-sets responsible for a distinct disease mechanism leading to an AD subtype is important for precision medicine and genome-guided clinical trials.
There have been huge efforts to improve predicting and distinguishing disease subtypes using polygenic profiling for early detection of subjects at risk [27–29]. Polygenic risk scores can be also useful to predict expected development of a disease or treatment responses in particular patient subgroups [30]. Our module-based polygenic profiling has innovative features compared to those previously conducted co-expression studies [11, 12] and conventional PRS approaches for AD [29–32]. First, our co-expression modules were developed from only AD brains excluding CN and MCI brains, while previous co-expression studies used transcriptome data of AD cases together with controls [11, 12]. Biological processes underlying disease heterogeneity in AD brains may be different from CN or MCI brains [33, 34]. Inclusion of non-AD transcriptome data would well differentiate gene-sets relevant to the disease risk but not explain disease heterogeneity. Second, previous polygenic profiling studies have generated PRSs by aggregating genetic estimates of genome-wide or most significant SNPs, which may have improved prediction rates [30] but cannot explain specific biological functions. In contrast, our mbPRSs are derived from biologically coherent gene-sets, which enable us to interpret biological functions of the modules and thereby provide insights on functional/mechanistic pathways for the AD subtypes. A previous study demonstrated genomic annotations at the single tissue level can improve our understanding on the etiology of complex human diseases [35]. A recent simulation study with failed AD trials confirms that the main failure reason is because variability between individuals’ in trials masks efficacy [3]. Therefore, our mbPRSs relevant to cell/tissue-level transcriptome profiles, brain imaging data, and cognitively-defined subgroups can be utilized for studying disease subtypes, prognosis, and response of treatment.
4.3. Role of omics and genetic profiling in AD subgrouping
Profiling using omics data including transcriptome data at tissue- or cell-level helped identify clinically and neuropathologically heterogeneous modules but also understand the biological functions of the modules. For example, the identified M6 module-genes were enriched for astrocytes, neuritic plaque scores, and language domain of cognitive function. This confirms the previous report that astrocytes are involved in amyloid clearance [36] and damaged astrocytes impact language domain among AD patients [37]. Our discovery showed that M9 module-genes are linked to endothelial cells, Braak stages, and visuospatial functioning in this study. Increased vascular inflammation in endothelial cells has been observed among AD patients with poor short-term visuospatial functioning [38].
Genes in the M6 and M9 modules have been previously reported for association with neurodegenerative diseases. Most of genes in the two modules have biological functions relevant to the nervous system or have been previously reported in genetic or experimental studies for neurodegenerative diseases. For example, SLC6A11 in M6 has been targeted for drug development of different neurodegenerative diseases including epilepsy [39]. GLIS3 in M9 has been associated with T2D [40, 41] and a longer life expectancy [42]. SNPs from GLIS3 in M9 showed genome-wide significant associations from GWASs for amyloid-β and phosphorylated tau proteins in cerebrospinal fluid (CSF) [43].
4.4. Limitations
Our study has several limitations. First, the sample size of discovery AD brains was modest. Therefore, we did not have statistical power for explaining the subtle phenotypic variations among AD patients, which might lead to detect modules associated with a few specific cognitive domains. In addition, our current study exclusively relied on cognitive test scores for prioritizing CIH modules, which may not be useful for detecting unknown or brain imaging-based subtypes of the disease. Second, our findings in ADNI may not represent AD heterogeneity in other populations. However, since one of modules was replicated in an independent study (FHS), there are shared mechanisms across diverse populations. Third, since we focused on AD patients, our sample size of AD subgroups remained underpower, so we could not apply multiple testing correction in current study. This limitation was mitigated by replicating one of the mbPRSs in FHS. Fourth, we limited our mbPRSs calculating using GWAS summary statistics for AD-risk and neuropathological outcomes regardless of available GWAS studies for CSF biomarker [44] or brain imaging data [1]. We decided to focus on neuropathological outcomes instead of biomarkers, since our goal is to explain AD heterogeneity by linking clinical subtypes to neuropathological outcomes. Fifth, we did not observe significant associations between PRSs and uncommon subgroups (e.g., AD-Executive). This may be because most of previously defined cognitive subtypes in ADNI were predominantly classified as subgroups of memory (31.8%), and executive functioning subgroup (2%) was relatively limited, especially in small datasets. In addition, datasets with GWAS for enough AD patients with carefully classified clinical phenotypes and clinically and/or pathologically defined subtypes were extremely limited. Finally, we recognize that the GWAS summary statistics for AD neuropathological traits (NP and NFT) in this study were generated based on genotype imputation using a previous reference panel (1000 genome) [18], which may affect quality and accuracy of our gene-sets and PRSs. However, we used common SNPs (MAF>5%) for constructing gene-sets and PRSs, and the imputation qualities of common SNPs are still relatively acceptable even in the previous reference panel [45]. Therefore, potential problems caused by low imputation quality would be largely limited in our study.
Future work in other independent GWAS sample with cognitively-defined subgroups (or relevant subgroups based on cognitive tests) will be required to validate our module-based subgrouping of AD patients. Furthermore, linking genetics of various AD-related phenotypes including endophenotypes would enhance to dissect further the disease heterogeneity [10]. Other AD-related GWAS summary data including cerebral amyloid angiopathy, hypertension, cholesterol, and insulin resistance can be added for extending AD phenotype gene-sets, which will lead us to detect novel gene-sets and to recognize other subgroups beyond AD-Language/Visuospatial domains.
4.5. Conclusion
In conclusion, PRSs developed using biologically coherent gene-sets and disease-related phenotypes can successfully differentiate cognitively-defined subgroups and brain region specific atrophy, which likely represent specific mechanistic pathways responsible for the corresponding disease subtypes. Classification of patients using genetic information will allow patient subgrouping and target prioritization for the subgroups, which may eventually lead to precision medicine in AD. However, AD heterogeneity explained by the specific polygenic risk profiles in this study does not mean that mbPRSs can predict subjects in different disease stages of AD or at risk to AD progression in future, since our mbPRSs can only differentiate AD patients into different cognitive subgroups. By comparing high- and low-risk groups of each mbPRSs using cognitively normal and MCI subjects, this aspect may be tested in future. Our study warrants further validations in large datasets.
Supplementary Material
Highlights.
Co-expression gene-network analysis in Alzheimer’s disease (AD) brains identified gene-sets (modules) associated with AD heterogeneity.
AD associated modules were selected when genes in each module were enriched for neuritic plaques and neurofibrillary tangles.
Polygenic risk scores from two selected modules were linked to the matching cognitively defined AD subgroups (language and visuospatial subgroups).
Polygenic risk scores from the two modules were associated with cognitive performance in language and visuospatial domains and the associations were confirmed in regional specific brain atrophy data.
AD associated genes in these modules were enriched in nervous and vascular systems.
RESEARCH IN CONTEXT.
Systematic review: We reviewed the literature using traditional (eg., PubMed) as well as preprinted (e.g., medRxiv) sources on studies about Alzheimer’s disease heterogeneity using genetic information.
Interpretation: Our coexpression network analysis among only AD brains without controls identified gene-sets (modules) which are likely to be responsible for AD heterogeneity. The polygenic risk scores derived from the modules associated with cognitive performance for certain domains (language and visuospatial functioning) were also associated with cognitively-defined AD subgroups for the matching domains and cortical thickness at the specific brain regions. These findings imply that genetics can be a useful source for dissecting the disease heterogeneity along with other resources including domain-specific cognitive measures, brain imaging scan, and neuropathological traits.
Future directions: Follow-up analysis will repeat the analysis in independent samples to validate our approach and findings.
Funding Sources
This study was supported by the National Institute on Aging (NIA) grants, U01-AG068057, U19-AG068753, P30-AG072978, and RF1-AG057519. GWAS summary statistics used in this study were distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24-AG041689-01). Collection of study data provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago was supported through funding by NIA grants P30-AG10161, R01-AG15819, R01-AG17917, R01-AG30146, R01-AG36836, U01-AG32984, U01-AG46152, and U01-AG61358, and funding from the Illinois Department of Public Health and the Translational Genomics Research Institute. Study data were also provided by Dr. Nilüfer Ertekin-Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center, and the Mayo Clinic Brain Bank. Collection of these data was supported through funding by NIH grants P50-AG016574, R01-AG032990, U01-AG046139, R01-AG018023, U01-AG006576, U01-AG006786, R01-AG025711, R01-AG017216, R01-AG003949, and R01-NS080820, and by funding from the CurePSP Foundation and the Mayo Foundation. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Conflicts of Interest
All authors report no conflicts of interest.
Consent statement
The study protocol, design, and performance of the current study were approved by the Boston University Institutional Review Board.
Data Availability Statement
Genotypes and clinical/neuropathological phenotype data are accessible by directly applying to the LONI portal for the ADNI at http://adni.loni.usc.edu. Summary statistics for GWAS studies can be accessed by applying directly to the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS), a NIA/NIH-sanctioned qualified-access data repository, under accession NG00075. Data supporting the findings of this study are available from the NIAGADS website (https://www.niagads.org/).
References
- [1].Dong A, Toledo JB, Honnorat N, Doshi J, Varol E, Sotiras A, et al. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain. 2017;140:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qiu Y, Jacobs DM, Messer K, Salmon DP, Feldman HH. Cognitive heterogeneity in probable Alzheimer disease: Clinical and neuropathologic features. Neurology. 2019;93:e778–e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Anderson RM, Hadjichrysanthou C, Evans S, Wong MM. Why do so many clinical trials of therapies for Alzheimer’s disease fail? Lancet. 2017;390:2327–9. [DOI] [PubMed] [Google Scholar]
- [4].Ten Kate M, Dicks E, Visser PJ, van der Flier WM, Teunissen CE, Barkhof F, et al. Atrophy subtypes in prodromal Alzheimer’s disease are associated with cognitive decline. Brain. 2018;141:3443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 2011;10:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Crane PK, Trittschuh E, Mukherjee S, Saykin AJ, Sanders RE, Larson EB, et al. Incidence of cognitively defined late-onset Alzheimer’s dementia subgroups from a prospective cohort study. Alzheimers Dement. 2017;13:1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Groot C, Grothe MJ, Mukherjee S, Jelistratova I, Jansen I, van Loenhoud AC, et al. Differential patterns of gray matter volumes and associated gene expression profiles in cognitively-defined Alzheimer’s disease subgroups. Neuroimage Clin. 2021;30:102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mendez MF. Early-onset Alzheimer Disease and Its Variants. Continuum (Minneap Minn). 2019;25:34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mukherjee S, Mez J, Trittschuh EH, Saykin AJ, Gibbons LE, Fardo DW, et al. Genetic data and cognitively defined late-onset Alzheimer’s disease subgroups. Mol Psychiatry. 2020;25:2942–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Udler MS, Kim J, von Grotthuss M, Bonas-Guarch S, Cole JB, Chiou J, et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: A soft clustering analysis. PLoS Med. 2018;15:e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci. 2018;21:811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Panitch R, Hu J, Chung J, Zhu C, Meng G, Xia W, et al. Integrative brain transcriptome analysis links complement component 4 and HSPA2 to the APOE ε2 protective effect in Alzheimer disease. Mol Psychiatry. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D, et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci Data. 2018;5:180142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS, et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data. 2016;3:160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chung J, Wang X, Maruyama T, Ma Y, Zhang X, Mez J, et al. Genome-wide association study of Alzheimer’s disease endophenotypes at prediagnosis stages. Alzheimers Dement. 2018;14:623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10:e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Scheltens NM, Galindo-Garre F, Pijnenburg YA, van der Vlies AE, Smits LL, Koene T, et al. The identification of cognitive subtypes in Alzheimer’s disease dementia using latent class analysis. J Neurol Neurosurg Psychiatry. 2016;87:235–43. [DOI] [PubMed] [Google Scholar]
- [23].Nho K, Risacher SL, Crane PK, DeCarli C, Glymour MM, Habeck C, et al. Voxel and surface-based topography of memory and executive deficits in mild cognitive impairment and Alzheimer’s disease. Brain Imaging Behav. 2012;6:551–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mesulam MM, Thompson CK, Weintraub S, Rogalski EJ. The Wernicke conundrum and the anatomy of language comprehension in primary progressive aphasia. Brain. 2015;138:2423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Binder JR, Frost JA, Hammeke TA, Cox RW, Rao SM, Prieto T. Human brain language areas identified by functional magnetic resonance imaging. J Neurosci. 1997;17:353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nobre AC, Sebestyen GN, Gitelman DR, Mesulam MM, Frackowiak RS, Frith CD. Functional localization of the system for visuospatial attention using positron emission tomography. Brain. 1997;120 (Pt 3):515–33. [DOI] [PubMed] [Google Scholar]
- [27].Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol. 2017;82:311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017;14:e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chaudhury S, Brookes KJ, Patel T, Fallows A, Guetta-Baranes T, Turton JC, et al. Correction: Alzheimer’s disease polygenic risk score as a predictor of conversion from mild-cognitive impairment. Transl Psychiatry. 2019;9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wand H, Lambert SA, Tamburro C, Iacocca MA, O’Sullivan JW, Sillari C, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature. 2021;591:211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang T, Han Z, Yang Y, Tian R, Zhou W, Ren P, et al. Polygenic Risk Score for Alzheimer’s Disease Is Associated With Ch4 Volume in Normal Subjects. Front Genet. 2019;10:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Leonenko G, Sims R, Shoai M, Frizzati A, Bossu P, Spalletta G, et al. Polygenic risk and hazard scores for Alzheimer’s disease prediction. Ann Clin Transl Neurol. 2019;6:456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sanchez-Valle J, Tejero H, Fernandez JM, Juan D, Urda-Garcia B, Capella-Gutierrez S, et al. Interpreting molecular similarity between patients as a determinant of disease comorbidity relationships. Nat Commun. 2020;11:2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dahl A, Zaitlen N. Genetic Influences on Disease Subtypes. Annu Rev Genomics Hum Genet. 2020;21:413–35. [DOI] [PubMed] [Google Scholar]
- [35].Lu Q, Powles RL, Abdallah S, Ou D, Wang Q, Hu Y, et al. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 2017;13:e1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Thal DR. The role of astrocytes in amyloid beta-protein toxicity and clearance. Exp Neurol. 2012;236:1–5. [DOI] [PubMed] [Google Scholar]
- [37].Resende EPF, Nolan AL, Petersen C, Ehrenberg AJ, Spina S, Allen IE, et al. Language and spatial dysfunction in Alzheimer disease with white matter thorn-shaped astrocytes. Neurology. 2020;94:e1353–e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu X, Ma Y, Ouyang R, Zeng Z, Zhan Z, Lu H, et al. The relationship between inflammation and neurocognitive dysfunction in obstructive sleep apnea syndrome. J Neuroinflammation. 2020;17:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ricciarelli R, Argellati F, Pronzato MA, Domenicotti C. Vitamin E and neurodegenerative diseases. Mol Aspects Med. 2007;28:591–606. [DOI] [PubMed] [Google Scholar]
- [40].Dimitri P, Warner JT, Minton JA, Patch AM, Ellard S, Hattersley AT, et al. Novel GLIS3 mutations demonstrate an extended multisystem phenotype. Eur J Endocrinol. 2011;164:437–43. [DOI] [PubMed] [Google Scholar]
- [41].Barker A, Sharp SJ, Timpson NJ, Bouatia-Naji N, Warrington NM, Kanoni S, et al. Association of genetic Loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes. 2011;60:1805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dimitri P, Habeb AM, Gurbuz F, Millward A, Wallis S, Moussa K, et al. Expanding the Clinical Spectrum Associated With GLIS3 Mutations. J Clin Endocrinol Metab. 2015;100:E1362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K, et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017;133:839–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tijms BM, Gobom J, Reus L, Jansen I, Hong S, Dobricic V, et al. Pathophysiological subtypes of Alzheimer’s disease based on cerebrospinal fluid proteomics. Brain. 2020;143:3776–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes and clinical/neuropathological phenotype data are accessible by directly applying to the LONI portal for the ADNI at http://adni.loni.usc.edu. Summary statistics for GWAS studies can be accessed by applying directly to the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS), a NIA/NIH-sanctioned qualified-access data repository, under accession NG00075. Data supporting the findings of this study are available from the NIAGADS website (https://www.niagads.org/).


