Summary
CircRNAs play multiple roles in a variety of cellular processes. We found that Circ-CDYL is highly enriched in early HCC plasma exosomes. Moreover, EpCAM+ HCC cells and exosomes had significant Circ-CDYL levels. We postulated that Circ-CDYL-enriched and EpCAM-positive exosomes would function as liver tumor-initiating exosomes (LTi-Exos). As predicted, intercellular transfer of LTi-Exos activates the HDGF-PI3K-AKT-mTOR and HIF1AN-NOTCH2 axes in recipient cells, promoting malignancy. Upstream, we found that the N6-methyladenosine (m6A) modification of Circ-CDYL exerted its action in HCC cells through a dual mechanism. First, it stimulated back-splicing processes via YTHDC1 to promote Circ-CDYL biogenesis. Second, it facilitates the active sorting of Circ-CDYL into exosomes via hnRNPA2/B1. Clinically, the combination of LTi-Exos and plasma alpha-fetoprotein (AFP) provides a promising early diagnostic biomarker for HCC with an AUC of 0.896. This study highlights the effect and mechanism by which m6A modification promotes hepatocarcinogenesis via modulation of the tumor microenvironment by LTi-Exos.
Subject areas: Biological sciences, Genetics, Molecular biology, Cancer
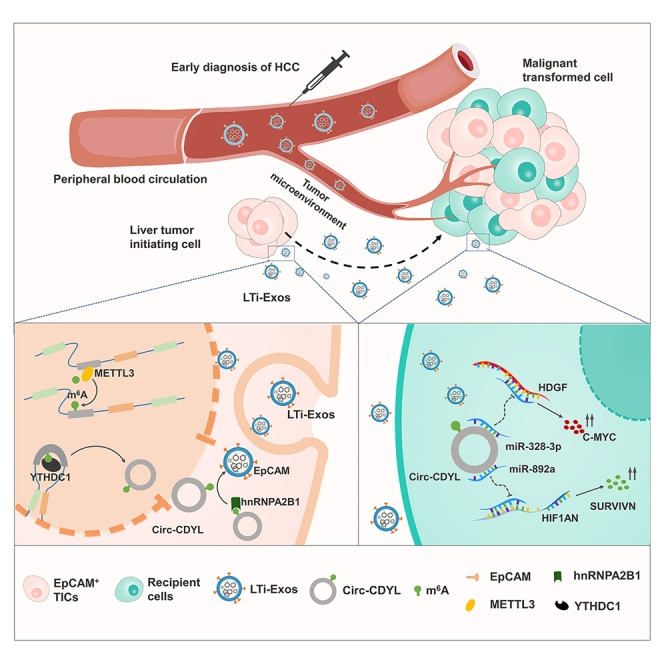
Graphical abstract
Highlights
-
•
Circ-CDYL is substantially enriched in the plasma exosomes of early HCC patients
-
•
Circ-CDYL-enriched and EpCAM-positive exosomes act as liver tumor-initiating exosomes
-
•
m6A modification promotes Circ-CDYL biogenesis by promoting back-splicing progress
-
•
m6A modification facilitates the sorting of Circ-CDYL into exosomes via hnRNPA2/B1
Biological sciences; Genetics; Molecular biology; Cancer
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related death worldwide in men, and its rising global prevalence highlights the need for effective strategies that can reduce the burden of tumors.1 Previous research has shown that neoplasm stage plays a crucial role in determining HCC prognosis: the 5-year survival rate for all-stage HCC patients is disappointingly low at 14% overall.2 However, the 5-year survival rate for patients with very early stage (Barcelona Clinic Liver Cancer stage 0, BCLC stage 0) and early stage (BCLC stage A) HCC approaches 90% and 50%–70% after active surgical treatment, respectively.3,4,5 A subgroup analysis revealed that the optimal median survival time for HCC patients with BCLC stage A is 53 months. The median survival time for patients with BCLC stages B/C/D is only 16, 7, and 3 months, respectively.6 Due to the significant difference in clinical outcome between patients with HCC at early and late stages, the importance of early screening and diagnosis for HCC high-risk populations has been emphasized. Unfortunately, although accumulating data encourage the early detection of HCC, the lack of specific symptoms in the early HCC,7 the suboptimal performance of ultrasound examination,8,9 and the insufficient biomarker AFP for HCC screening10,11 caused inevitable serious obstacles in the clinic. The inadequacies of currently available methods for HCC screening and early diagnosis necessitate the development of an innovative sufficient surveillance modality in clinical practice.
Exosomes are specialized, nanosized, membrane-bound endocytic vesicles released by various types of cells through inward budding of the limiting membrane of endosomes.12,13 Abundant cargoes of various DNA, mRNA, lncRNA, microRNA, and circRNA are encapsulated and stabilized in exosomes, which enables them to transmit intracellular cargoes and participate in intercellular communication when they fuse with target cells.14,15 Exosomes secreted by tumor cells are implicated in the malignant processes of cancer.15,16 The role of exosomes and their cargos in the diagnosis and progression of cancers such as pancreatic cancer,17 prostate cancer,18 and glioblastoma19 is anticipated to be crucial. However, the roles of liver cancer-derived exosomes in HCC malignancies, as well as the clinical significance of exosomal cargos in the diagnosis and treatment of HCC, are poorly understood.
The recently discovered noncoding RNA called circRNA, which has a structure that is covalently closed and does not have 3′ or 5′ free ends, is frequently expressed in a tissue- or developmental stage-specific manner.20,21 CircRNAs are unusually stable in biological processes due to their constitutive resistance to exonuclease activity.22 There is accumulating evidence that exosomes containing circRNA in body fluids have been identified as promising diagnostic cancer biomarkers due to their selective cargo wrapping and similarity to the cells from which they originated.12,13
One of the most common and abundant internal modifications in both coding and noncoding transcripts, such as circRNAs, is N6-methyladenosine (m6A) modification, which is defined as methylated adenosine at the N6 position.23 Multiple posttranscriptional processes involving circRNAs are affected by m6A modification, including back-splicing,24 export,25 stability,26 and translation.27 Importantly, dysregulation of m6A profiles has been implicated in the carcinogenesis and progression of HCC.28,29 Although the two-by-two interrelationship between m6A modification, circRNAs, and HCC has been repeatedly demonstrated, it is not well understood whether m6A modification can affect the initiation and progression of HCC by modifying circRNAs.
Our previous study clarified the intracellular function of Circ-CDYL, which is specifically elevated in the early HCC.30 We found that Circ-CDYL promotes HCC through regulating the expression of mRNAs encoding HDGF and HIF1AN by acting as the sponge of miR-892a and miR-328-3p, which enhances the stemness of HCC cells in sequence. However, the origin of aberrant expression of Circ-CDYL, and whether Circ-CDYL exerts intercellular function in early HCC remain unknown. This study demonstrates that plasma exosomes from patients with early HCC (BCLC stage 0 or A HCC) are enriched for Circ-CDYL, the circRNA that is most significantly increased at this stage. Intriguingly, more EpCAM-positive cells and exosomes were produced by HCC cells in which Circ-CDYL was overexpressed. Moreover, Circ-CDYL expression was higher in both EpCAM+ HCC cells and exosomes. Our working hypothesis was that this subpopulation of liver tumor-initiating exosomes (LTi-Exos) enriched in Circ-CDYL and EpCAM would initiate tumor growth in the liver via the microenvironment. Consequently, circulating LTi-Exos may serve as an ideal marker for the early diagnosis of HCC. Upstream, Circ-CDYL m6A RNA modification stimulated back-splicing via YTHDC1 to promote Circ-CDYL biogenesis in HCC cells and sorting Circ-CDYL into LTi-Exos via hnRNPA2/B1. STM2457 inhibition of the m6A writer METTL3 or Circ-CDYL methylation modification sites mutation inhibited the hepatocarcinogenic effect of LTi-Exos. Our data thus provide evidence that LTi-Exos may serve as a potential early screening biomarker and therapeutic target for HCC.
Results
Circ-CDYL is enriched in exosomes from early HCC patients and in EpCAM+ exosomes secreted from HCC cells
Targeting very early-stage HCC directly enables the precise identification of early diagnostic biomarkers and therapeutic targets. Our previous microarray assay analyzed the differentially expressed circRNAs across the genome in BCLC stage 0 HCC tissues and nontumorous adjacent tissues and determined that Circ-CDYL is the circRNA with the greatest increase. Annotation of the circRNA probe revealed that Circ-CDYL originated from the head-to-tail splicing of exon 2 of its parental gene, chromodomain Y-like (CDYL), a member of the Y chromosome gene family that is specific to primates.30
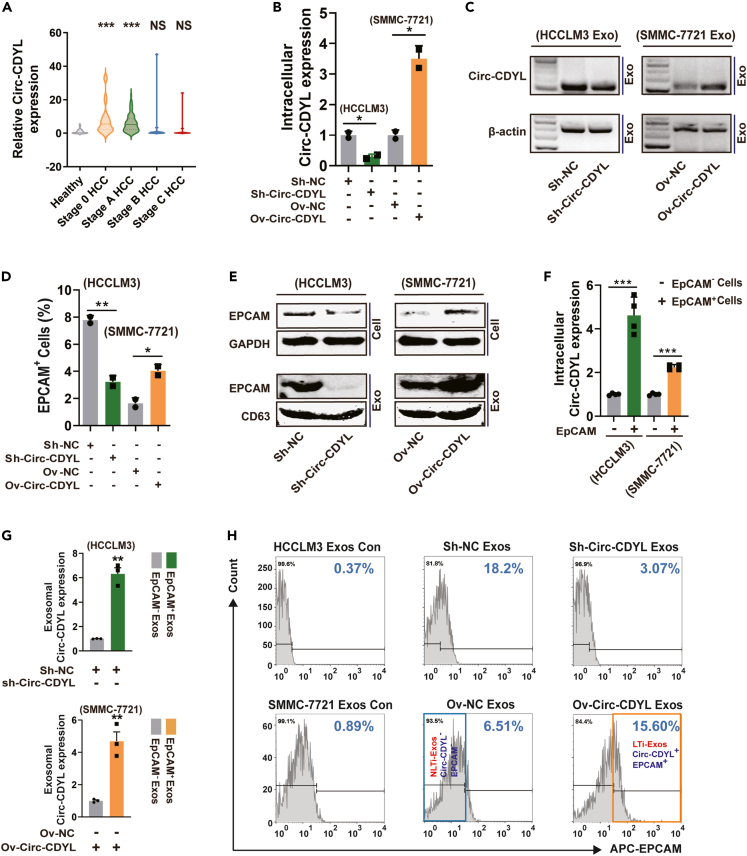
To learn more about the Circ-CDYL secretion pattern, we next examined exosomes derived from the plasma of HCC patients at various stages. Increased expression of Circ-CDYL was detected in exosomes isolated from BCLC stage 0 and A HCC patients compared with those isolated from healthy controls (Figure 1A). Contrary to expectations, patients with BCLC stages B and C HCC did not exhibit significantly elevated exosomal Circ-CDYL levels, which may have been due to the altered gene profile in each tumor stage.
Figure 1.
Circ-CDYL and EpCAM share close connections in HCC cells and exosomes
(A) The expression of Circ-CDYL was determined in exosomes isolated from the plasma of different stages of HCC patients (n = 24, 71, 21, and 20 for BCLC stage 0, A, B, C HCC, respectively) and 46 healthy donors.
(B) RNA expression level of Circ-CDYL determined by RT‒qPCR and depicted by relative quantity calculation using the 2-▵▵CT value. Stable Circ-CDYL knockdown HCCLM3 cells and Circ-CDYL-overexpressing SMMC-7721 cells were constructed by a lentivirus system.
(C) RNA expression of Circ-CDYL in exosomes isolated from the cell culture supernatant of Circ-CDYL knockdown and Circ-CDYL-overexpressing HCC cells, as determined using RT‒PCR followed by agarose gel electrophoresis.
(D) The percentage of EpCAM-positive (EpCAM+) cells determined by flow cytometry in Circ-CDYL-knockdown and Circ-CDYL-overexpressing HCC cells.
(E) Protein levels of EpCAM in the indicated cells and corresponding cell-derived exosomes.
(F) Circ-CDYL expression was determined in flow cytometry-sorted primary EpCAM+ and EpCAM− HCC cells using RT‒qPCR.
(G) Circ-CDYL expression was determined in flow cytometry-sorted primary EpCAM+ and EpCAM− exosomes purified from the cell culture supernatant of the indicated HCC cells.
(H) Flow cytometry analysis showed the percentage of EpCAM+ exosomes released from Circ-CDYL-ectopically expressed HCC cells. Exos, exosomes; Con, negative control using isotype antibody of anti-EpCAM; LTi, liver tumor-initiating; NLTi, nonliver tumor-initiating.
To explore the origin of plasma-derived exosomal Circ-CDYL, a stable Circ-CDYL-overexpressing cell line and a knockdown cell line were constructed with a lentivirus system using SMMC-7721 and HCCLM3 cells, respectively, based on the relative background expression of Circ-CDYL (Figure 1B). We then purified exosomes from the culture medium of the constructed HCC cell line using ultracentrifugation. Purified exosome samples exhibited similar sizes (diameters ranging from 20 to 200 nm) (Figure S1A) and typical cup-shaped morphology (Figure S1B). Furthermore, the identity of exosomes was confirmed by the positive exosomal protein markers TSG101, HSP70 and CD63 and the negative marker Calnexin (Figure S1C). Consistent with the expression pattern in parental cells (Figure 1B), the Circ-CDYL level was significantly higher in exosomes released from SMMC-7721 cells stably overexpressing Circ-CDYL (Figure 1C). Inhibition of Circ-CDYL expression in HCCLM3 cells led to the opposite effects (Figure 1C). These results indicated that the high exosomal Circ-CDYL originated from HCC cells with increased expression of Circ-CDYL.
Stem-like cells such as tumor-initiating cells (T-ICs) within tumors are a unique subpopulation of cells responsible for the initiation and progression of cancer. Liver T-ICs are characterized by several cell marker molecules, such as epithelial cell adhesion molecule (EpCAM), PROM1 (CD133) and CD24.31,32,33 We further examined whether Circ-CDYL, which is markedly highly expressed in very early-stage HCC, could affect the characteristics of liver T-ICs. HCC cells collected from nonattached spheroids, where T-ICs are enriched,18 showed higher expression of Circ-CDYL (Figure S2A). To further explore which subpopulation of T-ICs was associated with Circ-CDYL, we detected the proportion of T-ICs with liver stem-like cell markers in constructed HCC cell lines using flow cytometry assays. We observed that overexpression of Circ-CDYL especially elevated the proportion of EpCAM+ (Figure 1D) but not CD133+ and CD24+ liver T-ICs (Figures S2B and S2C). As a result, the proportion of EpCAM+ cells decreased when Circ-CDYL expression was inhibited in HCCLM3 cells (Figures 1D, S2B, and S2C). Consistently, ectopic overexpression of Circ-CDYL resulted in higher protein levels of EpCAM in HCC cells, while knockdown of Circ-CDYL showed the opposite effect. The exosomal level of EpCAM was synchronized with that of intracellular EpCAM (Figure 1E). This trend was similar to the expression pattern of exosomal Circ-CDYL synchronized with intracellular Circ-CDYL (Figures 1B and 1C).
To ascertain the relationship between Circ-CDYL and EpCAM, we further explored the correlation between the expression of these two molecules at both cellular and exosomal scales. Statistically, we found that the intracellular expression level of Circ-CDYL was higher in EpCAM+ HCC cells than in EpCAM− cells (Figure 1F). The exosomal level of Circ-CDYL in sorted EpCAM+ exosomes was higher than that in EpCAM− exosomes (Figure 1G). Circ-CDYL-overexpressing SMMC-7721 cells produced a significantly higher proportion of EpCAM+ exosomes (15.60% vs. 6.51%, Figure 1H). Conversely, interference with Circ-CDYL expression in HCCLM3 cells led to a significantly decreased proportion of EpCAM+ exosomes (18.20% vs. 3.07%, Figure 1H). In addition, when flow cytometry analysis was executed, we found that the proportion of EpCAM+ exosomes was markedly higher than the proportion of EpCAM+ parent cells. In SMMC-7721 cells and HCCLM3 cells, EpCAM+ cells accounted for a mean of 1.64% and 7.78%, respectively (Figure 1D). However, in SMMC-7721-secreted exosomes and HCCLM3-secreted exosomes, EpCAM+ exosomes accounted for 6.51% and 18.2%, respectively (Figure 1H). This result indicated that EpCAM has an enriching effect in the exosomes secreted by HCC cells.
Based on these results, it is reasonable to assume that Circ-CDYL and EpCAM are very closely related at both the HCC cell and exosome scales, and both may together constitute a specific HCC cell or exosome subtype. Because EpCAM+ liver cells are defined as liver tumor-initiating cells,32 we postulated that this subpopulation of Circ-CDYL-rich and EpCAM+ exosomes will work as liver tumor-initiating exosomes (LTi-Exos, orange frame, Figure 1H). Accordingly, another subpopulation of EpCAM− exosomes with low levels of Circ-CDYL could be regarded as non-liver tumor initiating exosomes (NLTi-Exos, Blue Frame, Figure 1H).
LTi-Exos promote early hepatocarcinogenesis through the tumor microenvironment
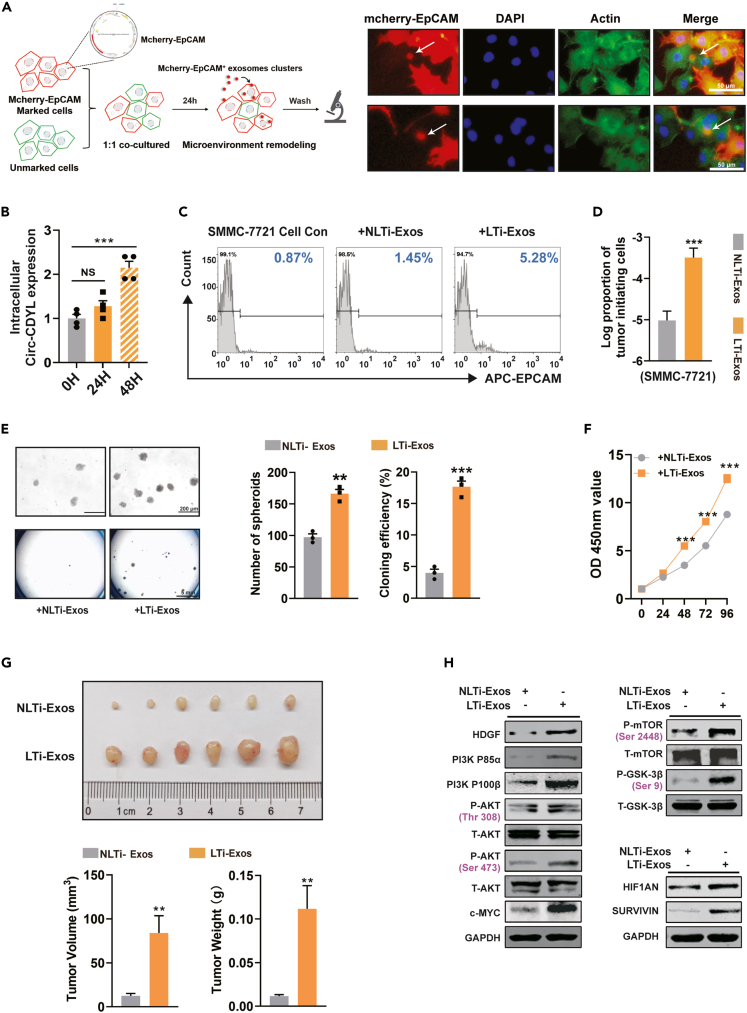
Exosomes perform their functions by delivering biomolecules, including lipids, proteins, and RNAs, to recipient cells.34 To investigate the effects and functions of LTi-Exos-mediated intercellular communication, we utilized PKH26 to label the sorted LTi-Exos (orange frame, Figure 1H) and then incubated them with SMMC-7721 recipient cells. The recipient cells, as anticipated, took up the PKH26-labeled LTi-Exos in the culture supernatant (Figure S3A). To further simulate the process of natural formation and intercellular communication of LTi-Exos, HCC cells stably expressing mCherry-EpCAM fusion protein (red) were cocultured with recipient cells (green). After the cell culture supernatant was removed, we observed that EpCAM-positive exosome clusters were absorbed by recipient cells (Figure 2A).
Figure 2.
LTi-Exos boosted hepatocarcinogenesis via the tumor microenvironment by performing functions similar to tumor-initiating cells
(A) Diagram of LTi-Exos uptake. SMMC-7721 cells stably expressing mCherry-EpCAM fusion protein (marked cells, red) were cocultured with negative control cells (unmarked cells) at a ratio of 1:1. After 24 h, the cell supernatant was removed, and all cells were dyed with actin-tracker probe (green) and DAPI (blue). White arrows indicate EpCAM+ exosome cluster uptake by HCC cells.
(B) The expression levels of Circ-CDYL in SMMC-7721 cells after incubation with LTi-Exos for 24 h and 48 h were determined.
(C) The percentage of EpCAM+ cells was determined by flow cytometry analysis 48 h after treatment with LTi-Exos or NLTi-Exos. Con, negative control using an isotype antibody against EpCAM.
(D) The proportion of T-ICs in SMMC-7721 cells was assessed by limiting dilution assay after incubation with LTi-Exos or NLTi-Exos for 48 h.
(E) Determination of spheroid formation in low-adhesion plates (upper). Colony formation was evaluated (below).
(F) SMMC-7721 cell proliferation was evaluated by CCK8 assay at different time points after incubation with LTi-Exos or NLTi-Exos.
(G) After incubation with LTi-Exos or NLTi-Exos for 48 h, 5×105 SMMC-7721 cells were subcutaneously injected into male nude mice (n = 6). The tumors were harvested, and their volume and weight were measured at 3 weeks after injection.
(H) Western blot showing the expression levels of HDGF, P85α/P110β regulatory subunit of PI3K, phosphorylation of AKT and mTOR, c-MYC, HIF1AN, and SURVIVIN.
Next, we functionally verified whether LTi-Exos can play a role in tumor initiation. After incubation, recipient HCC cells showed elevated expression of Circ-CDYL (Figure 2B), indicating that the oncogenic Circ-CDYL cargo was delivered into the recipient cell by exosomes. After 48 h of LTi-Exos treatment, more recipient HCC cells were transformed into EpCAM+ T-ICs than NLTi-Exos-treated cells (Figure 2C). In addition, the proportion of tumor-initiating cells estimated by limiting dilution assay increased (Figures 2D and S3B). LTi-Exos also significantly promoted spheroid formation (Figure 2E, upper), colony formation (Figure 2E, lower), and in vitro and in vivo cell proliferation (Figures 2F and 2G) of recipient cells.
Mechanistically, we verified whether LTi-Exos can cause the activation of Circ-CDYL-driven signal transduction pathways in recipient cells.30 We found that LTi-Exos elevated HDGF expression, activated the PI3K P85 and P110 subunits and enhanced AKT phosphorylation at the Thr308 and Ser473 sites in sequence, which stimulated the downstream targets mTORC1, GSK-3β and C-MYC more efficiently than NLTi-Exos (Figures 2H and S2D). Similarly, LTi-Exos increased the expression of HIF1AN and SURVIVIN more than NLTi-Exos (Figures 2H and S2D).
Overall, LTi-Exos performed functions similar to those of tumor-initiating cells to significantly promote early hepatocarcinogenesis through the tumor microenvironment.
m6A modification of Circ-CDYL regulates the stem-like properties of HCC cells
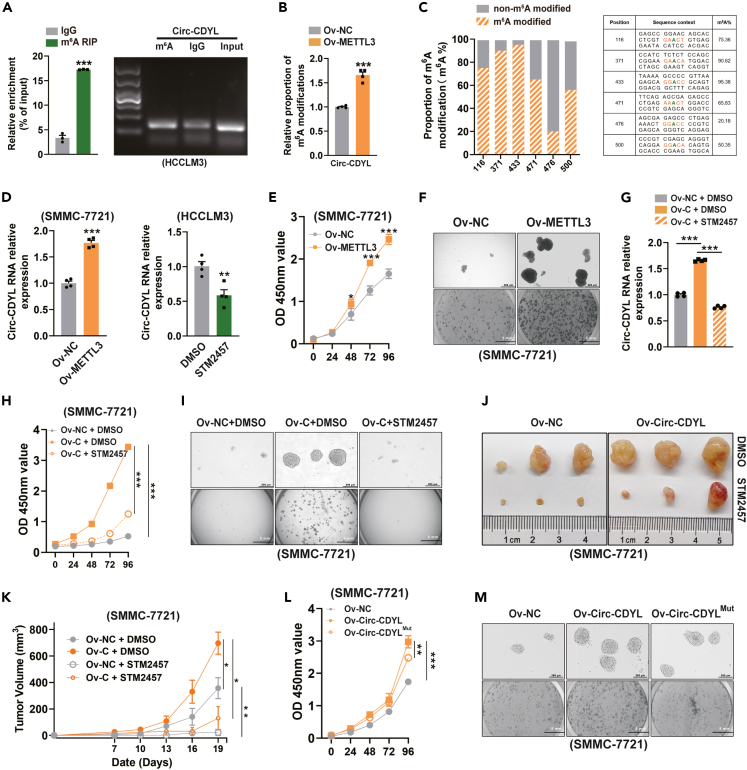
The most common RNA base modification, m6A modification, may be essential for circRNA biogenesis and function. The consensus motif “RRm6ACH” (R = G or A; H = A, C or U) is favored for the alterations.35 To explore whether the aberrantly expressed Circ-CDYL in early HCC is also subject to m6A modification, we first performed bioinformatic prediction. Several “very high confidence” potential m6A modification sites were found in Circ-CDYL using the SRAMP (http://www.cuilab.cn/sramp/) analysis tools. Using a systematic analysis, six sites with the most potential were chosen for further study (combined score >0.70). (Figures S4A and S4B). Furthermore, m6A RNA immunoprecipitation (MeRIP) assays validated the m6A enrichment in the Circ-CDYL sequence (Figure 3A).
Figure 3.
m6A modification of Circ-CDYL by METTL3 aided in HCC tumorigenesis
(A) MeRIP assay followed by RT‒qPCR showed an enrichment of Circ-CDYL using an anti-m6A antibody.
(B) The relative proportion of m6A-modified Circ-CDYL was determined using the MazF-qPCR assay in METTL3-overexpressing SMMC-7721 cells and negative control cells.
(C) Ligase-dependent absolute quantification of m6A modification at the predicted “very high confidence” m6A-modified sites in Circ-CDYL.
(D) Relative Circ-CDYL expression was determined in METTL3-overexpressing cells (left) and 48-h STM2457 (5 μM)/DMSO (1:1000 v/v)-treated cells (right) using RT‒qPCR.
(E) Cell proliferation was determined in the indicated HCC cells using a CCK-8 assay.
(F) Spheroid formation (upper) and colony formation (below) of the indicated HCC cells.
(G) Relative Circ-CDYL expression was determined in HCC cells after the indicated treatment.
(H and I) Cell proliferation (H), spheroid formation (I, upper) and colony formation (I, below) of HCC cells with the indicated treatment.
(J and K) Tumors were collected from day 19 post tumor implantation in nude mice that had been treated with either STM2457 (50 mg/kg) or vehicle (J). Growth curve of tumor were shown (K).
(L and M) Cell proliferation (L), spheroid formation (M, upper) and colony formation (M, below) of indicated HCC cell lines.
m6A modification is installed by a multicomponent methyltransferase complex (MTC), which is composed of core component methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) heterodimers.23 A previous study showed that METTL3 regulates the expression of RNA in a m6A-dependent manner.36 To explore the function of m6A modification in Circ-CDYL, a stable METTL3-overexpressing HCC cell line was constructed with a lentivirus system using SMMC-7721 cells (Ov-METTL3) (Figure S5A). Overexpression of METTL3 significantly increased the level of m6A modification in Circ-CDYL (Figure 3B). Ligase-dependent absolute quantification analysis (Figure S5B) was presented for quantifying the m6A ratio at the 6 predicted modified loci with “very high confidence” from extracted total RNA (Figure 3C). The methylation fractions of the endogenous specific sites were determined to range from 20.18% to 95.38% in the HCC cell line, among which over 90% of the Circ-CDYL transcript was m6A-modified at sites 371 and 433, and approximately 75% of Circ-CDYL was modified at site 116 in HCC cells (Figure 3C).
We next investigated the effects of m6A modification on Circ-CDYL and found that the expression of Circ-CDYL was remarkably elevated by METTL3 (Figure 3D). In contrast, inhibiting METTL3 with STM2457, a selective and orally active METTL3 inhibitor, decreased the endogenous expression of Circ-CDYL in HCC cells (Figure 3D). These results indicated that the expression of Circ-CDYL could be positively regulated by METTL3 in a m6A-dependent manner.
As ectopic overexpression of Circ-CDYL enhanced the stem-like properties of HCC cells,30 we next investigated whether METTL3-mediated upregulation of Circ-CDYL has a similar effect. We found that overexpression of METTL3 significantly promoted malignant cell proliferation, spheroid growth and colony formation of HCC cells in vitro (Figures 3E–3F and S6A). Furthermore, the elevated level of Circ-CDYL generated by ectopic overexpression could also be reversed by STM2457 (Figure 3G). As anticipated, STM2457 significantly inhibited the increased malignant proliferation, self-renewal, and colony formation ability of HCC cells driven by ectopic overexpression of Circ-CDYL (Figures 3H, 3I, and S6B). In addition, subcutaneous injection of Circ-CDYL-overexpressing HCC cells into mice led to an increase in tumor formation and growth rate, whereas administration of STM2457 in vivo was able to inhibit this effect (Figures 3J and 3K).
By constructing either wild-type Circ-CDYL or mutated m6A-modified sites of Circ-CDYL stably overexpressing HCC cells, we were able to confirm the HCC-promoting function of m6A modification of Circ-CDYL. Notably, we found that overexpression of wild-type Circ-CDYL, but not the m6A modification sites mutant equivalents, could promote spheroid growth, colony formation and cell proliferation (Figures 3L, 3M, and S6C). These data showed that METTL3-mediated m6A modification of Circ-CDYL promoted HCC tumorigenesis.
m6A modification of Circ-CDYL regulates its biogenesis by promoting back-splicing
Several mechanisms, including back-splicing events24 and stabilization,26 are used by m6A modification to regulate transcript levels. We further studied the mechanism of m6A-dependent upregulation of Circ-CDYL. Actinomycin D pulse-chase experiments demonstrated that neither METTL3 overexpression nor inhibition had a notable effect on the half-life of Circ-CDYL (Figure S7A). We next examined whether the m6A modification regulated the back-splicing events of Circ-CDYL and found that METTL3 overexpression significantly increased Circ-CDYL expression, paralleled by a decrease in its precursor transcript (pre-CDYL) (Figure 4A, left set), while METTL3 inhibition led to the downregulation of Circ-CDYL expression and accumulation of pre-CDYL (Figure 4A, right set). These results suggested a role of METTL3 and m6A modification in the regulation of back-splicing events, which leads to the biogenesis of Circ-CDYL.
Figure 4.
Circ-CDYL back-splicing reaction regulated by METTL3 and YTHDC1-dependent m6A modification
(A–C) Relative RNA levels of the circular transcript (Circ-CDYL), linear transcript (Lin-CDYL), and precursor (Pre-CDYL) in METTL3-disturbed HCC cells (METTL3 overexpression or inhibited by STM2457, respectively) (A), YTHDC1 overexpression (Ov-YTHDC1) or interference cells (Si-YTHDC1) (B), and METTL3 overexpression only or combined overexpression of METTL3 and YTHDC1 cells (C). Values were normalized against β-actin and expressed as relative quantity with respect to negative control treatment set to a value of 1.
(D–G) Cell proliferation (D, E), spheroid formation (F, G, upper) and colony formation (F, G, below) of HCC cells with the indicated treatments.
(H) RIP assay using anti-YTHDC1 antibody, and IgG as a negative control. The enrichment of Circ-CDYL was determined by RT‒qPCR assay and agarose gel electrophoresis.
YTH domain containing 1 (YTHDC1) has been confirmed to regulate the back-splicing of circRNAs as an m6A reader.24,37 Thus, we then examined the involvement of YTHDC1 in the back-splicing of Circ-CDYL. YTHDC1 silencing (Figure S7B) in the HCCLM3 cell line resulted in a decrease in Circ-CDYL and was accompanied by an increase in pre-CDYL (Figure 4B, left set). Intriguingly, overexpression of YTHDC1 in the SMMC-7721 cell line (Figure S7C) decreased the pre-CDYL level but had a moderate effect on the level of Circ-CDYL (Figure 4B, right set). This phenomenon may be due to the low level of m6A modification in SMMC-7721 cells, resulting in the limited effect of YTHDC1 alone on Circ-CDYL expression. To confirm this hypothesis, we simultaneously increased the levels of METTL3 and YTHDC1 in SMMC-7721 cells and found a significant additive effect (Figure 4C).
Functionally, as expected, we found that simultaneous overexpression of METTL3 and YTHDC1 led to enhanced stem-like properties of HCC cells as elevated expression of Circ-CDYL did, while silencing of YTHDC1 led to the opposite effects (Figures 4D–4G). Moreover, RIP-qPCR with an anti-YTHDC1 antibody revealed that Circ-CDYL was enriched in the immunoprecipitated fraction (Figure 4H), indicating that YTHDC1 binds to regions of circRNA that undergo circularization, which consistent with the results of a previous study.24
Briefly, these results indicated a role for METTL3 and YTHDC1-dependent m6A modification in the regulation of the back-splicing reaction of Circ-CDYL, which increased the cellular expression level of Circ-CDYL by promoting its biogenesis and subsequently augmented the stem-like characteristics of HCC.
m6A modification of Circ-CDYL increased the exosome sorting of Circ-CDYL, thus enhancing the stemness of recipient cells in HCC
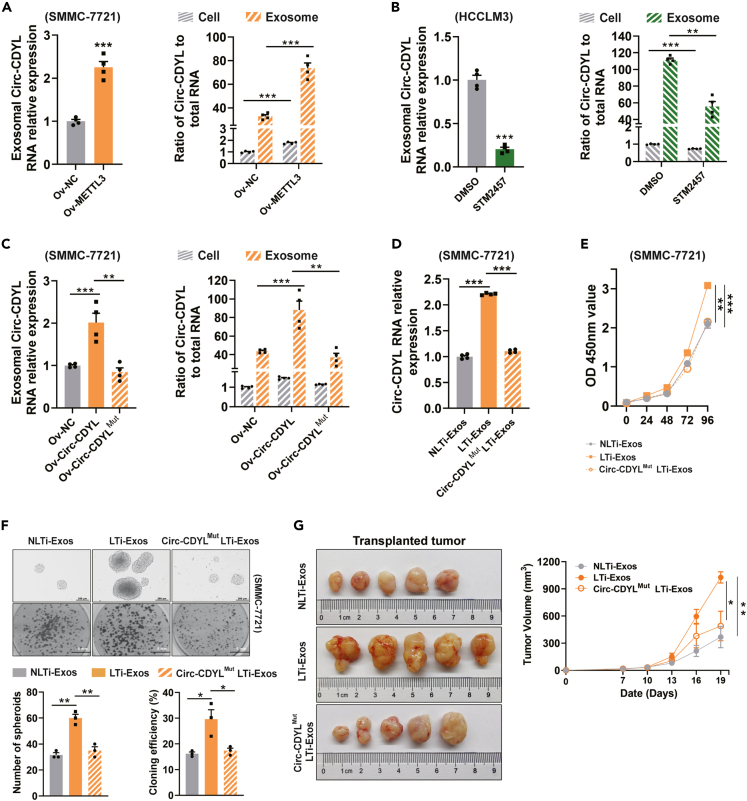
We next examined whether m6A modification affected exosomal Circ-CDYL. Consistent with the intracellular Circ-CDYL level, we also detected increased Circ-CDYL levels in exosomes derived from the METTL3-overexpressing cell line (Figure 5A, left) and decreased Circ-CDYL levels in METTL3-inhibited cells and corresponding exosomes (Figure 5B, left). Surprisingly, elevated levels of METTL3 resulted in a higher fold change in Circ-CDYL levels in exosomes than in cells (Figure 5A, right), while exosomal Circ-CDYL decreased more than cellular Circ-CDYL when METTL3 was specifically inhibited (Figure 5B, right), indicating that METTL3-dependent m6A modification promoted the active sorting of Circ-CDYL into exosomes. However, ectopic overexpression of m6A sites-mutated Circ-CDYL could not increase exosomal Circ-CDYL as much as the wild-type equivalent (Figure 5C), suggesting that the exosome sorting of Circ-CDYL, at least partly, is m6A modification dependent.
Figure 5.
LTi-Exos promote the stemness of recipient cells through m6A modification-dependent sorting of Circ-CDYL
(A–C) Exosomal Circ-CDYL levels were determined in METTL3-overexpressing HCC cells (Ov-METTL3) (A, left), STM2457-treated HCC cells (B, left), and HCC cells overexpressing wild-type Circ-CDYL and Circ-CDYL with m6A-modified sites mutations. The ratio of cellular and exosomal Circ-CDYL to total cellular and exosomal RNA of the indicated cells, respectively, was calculated using 2−ΔΔCT from RT‒qPCR (A–C, right). Ratios were normalized against β-actin and expressed as relative quantity with respect to negative control treatment set to a value of 1.
(D) The expression levels of Circ-CDYL were determined in SMMC-7721 cells after incubation with LTi-Exos derived from cells overexpressing wild-type Circ-CDYL or LTi-Exos derived from cells overexpressing m6A sites-mutated Circ-CDYL (Circ-CDYLMut LTi-Exos) for 48 h.
(E and F) Determination of cell proliferation by CCK-8 assay (E), spheroid formation in low-adhesion plates (F, upper), and colony formation (F, below).
(G) SMMC-7721 cells (5×105) were subcutaneously injected into male nude mice. 5 μg of the indicated exosomes derived from HCC cell supernatant were intratumorally injected every 3 days after tumor cell xenograft. Representative images of tumors harvested from mice in different treatment groups 19 days after injection (left) and the tumor growth curve (right) are shown.
Furthermore, LTi-Exos generated from cells with ectopic overexpression of m6A sites-mutated Circ-CDYL (Circ-CDYLMut LTi-Exos) failed to elevate the Circ-CDYL level in recipient cells as LTi-Exos did (Figure 5D) and thus were unable to promote malignant proliferation, self-renewal, and colony formation of recipient HCC cells (Figures 5E and 5F). Meanwhile, LTi-Exos promoted tumorigenesis of xenografts derived from HCC cell lines in vivo more effectively than NLTi-Exos and Circ-CDYLMut LTi-Exos (Figure 5G). These results indicated that LTi-Exos promote the stemness of recipient cells through m6A modification-dependent sorting of exosomal Circ-CDYL in HCC.
Activity of m6A-modified Circ-CDYL loading into exosomes is controlled by hnRNPA2/B1
Next, we investigated the molecular machinery that regulates the sorting of Circ-CDYL into exosomes. Heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2/B1) has been reported as an “m6A reader”38,39 and a ubiquitous protein that controls RNA trafficking to exosomes.40 Subsequently, we determined whether hnRNPA2/B1 controlled the loading of m6A-modified Circ-CDYL into exosomes. RNA antisense purification (RAP) assays showed that hnRNPA2/B1 in HCC cells was pulled down by a probe that specifically targeted Circ-CDYL. However, m6A sites-mutated Circ-CDYL could not enrich hnRNPA2/B1 effectively (Figure 6A). Moreover, specific binding of hnRNPA2/B1 to Circ-CDYL was validated by immunoprecipitation of hnRNPA2/B1 followed by RT‒qPCR of Circ-CDYL (Figure 6B). Circ-CDYL was amplified from hnRNPA2/B1 but not IgG immunoprecipitates (Figure 6B), indicating an interaction between Circ-CDYL and hnRNPA2/B1 protein.
Figure 6.
hnRNPA2/B1 controls the dynamic sorting of m6A-modified Circ-CDYL into exosomes
(A) RNA antisense purification (RAP) assays using a biotinylated antisense probe of Circ-CDYL or negative control probe were performed using Circ-CDYL-overexpressing HCC cells (SMMC-7721 Ov-Circ-CDYL) (left), and m6A sites-mutated Circ-CDYL-overexpressing HCC cells (SMMC-7721 Ov-Circ-CDYLMut) (right), followed by Western blot assays using an anti-hnRNPA2/B1 antibody.
(B) RIP assays using an anti-hnRNPA2/B1 antibody were performed. The immunoprecipitated Circ-CDYL was determined by RT‒qPCR, followed by agarose gel electrophoresis.
(C–E) Exosomal Circ-CDYL levels were determined in the HCCLM3 cell line (C, left), Circ-CDYL-overexpressing HCC cells (SMMC-7721 Ov-Circ-CDYL) (D, left), and m6A sites-mutated Circ-CDYL-overexpressing HCC cells (SMMC-7721 Ov-Circ-CDYLMut) (E, left) after hnRNPA2/B1 interference (Si-hnRNPA2/B1). The ratio of cellular and exosomal Circ-CDYL to total cellular and exosomal RNA of the indicated cells, respectively, was calculated using 2−ΔΔCT from RT‒qPCR (C–E, right). Ratios were normalized against β-actin and expressed as relative quantity with respect to negative control treatment set to a value of 1.
HnRNPA2/B1 silencing (Figure S7D) significantly decreased the levels of Circ-CDYL in exosomes derived from HCC cell lines with relatively high expression of Circ-CDYL (HCCLM3-and Circ-CDYL-overexpressing SMMC-7721 cells, respectively) (Figures 6C and 6D, left). Notably, the change in exosomal Circ-CDYL after hnRNPA2/B1 silencing was significant compared with that in the cellular Circ-CDYL level (Figures 6C and 6D, right), indicating that hnRNPA2/B1 could specifically stimulate the active exosome sorting of Circ-CDYL. However, when m6A modification sites on Circ-CDYL were mutated (Circ-CDYLMut), hnRNPA2/B1 had a less significant effect on the exosomal level of Circ-CDYL than the wild-type equivalent (Figure 6E). These results indicated that hnRNPA2/B1 regulates the active loading of m6A-modified Circ-CDYL into exosomes.
LTi-Exos hold promise for early HCC detection and surveillance
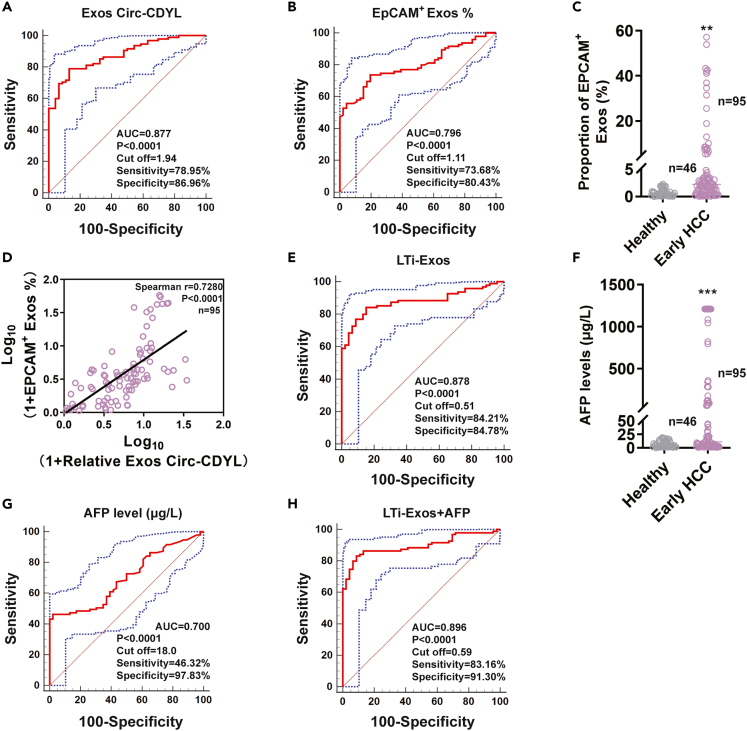
Given that Circ-CDYL plays important roles in the carcinogenesis of HCC and is specifically highly expressed in exosomes purified from the plasma of early HCC patients, we further elucidated whether Circ-CDYL could act as an early detection and surveillance biomarker for HCC in the clinic. When the diagnostic accuracy of exosomal Circ-CDYL was estimated with receiver operating characteristic (ROC) analysis in early HCC samples (Table S1), it presented an area under the curve (AUC) of 0.88 (95% CI: 0.81–0.93), with a sensitivity of 78.95% and specificity of 86.96% (Figure 7A and Table S2). Circ-CDYL expression was closely associated with EpCAM levels, as shown before (Figures 1D–1G). We next analyzed the relationship between Circ-CDYL and EpCAM in HCC clinical samples. As expected, early HCC patients had higher proportions of EpCAM+ exosomes in plasma than healthy controls, which could effectively distinguish between healthy and early HCC with a sensitivity of 73.68% and specificity of 80.43% (Figures 7B and 7C). The proportions of EpCAM+ exosomes were positively correlated with the exosomal level of Circ-CDYL (Figure 7D), indicating their potential synergistic effects and that these two biomarkers can be used together in clinical practice.
Figure 7.
The combination of LTi-Exos and current screening methods based on AFP shows promise for the early detection of HCC
(A) ROC curve analysis was performed for early HCC (n = 95, 24 BCLC stage 0 HCC plus 71 BCLC stage A HCC) and healthy donors (n = 46) by estimating plasma exosomal Circ-CDYL expression.
(B) ROC curve analysis was performed for early HCC (n = 95) and healthy people (n = 46) by estimating the proportion of EpCAM+ exosomes.
(C) The proportion of EpCAM+ exosomes was estimated in the plasma of early HCC patients and healthy controls using flow cytometry assays.
(D) Pearson correlation analysis between exosomal Circ-CDYL expression and the proportion of EpCAM+ exosomes in the plasma of early HCC (n = 95).
(E) ROC curve analysis was performed for early HCC (n = 95) and healthy donors (n = 46) by combined EpCAM+ exosomes with exosomal Circ-CDYL estimating (LTi-Exos).
(F) AFP levels were estimated in the plasma of early HCC patients and healthy controls.
(G and H) ROC curve analysis was performed for early HCC patients (n = 95) and healthy donors (n = 46) by estimating the AFP level (G) and AFP combined with LTi-Exos (H).
By integrated assessment of EpCAM+ exosomes with highly expressed exosomal Circ-CDYL, this LTi-Exos biomarker enhanced the AUC to 0.88 (95% CI: 0.81–0.93), with a sensitivity of 84.21% and a specificity of 84.78% when diagnosing early HCC (Figure 7E). Comprehensive comparison of diagnostic accuracy via Youden’s index (sensitivity+specificity-1) showed that the LTi-Exos biomarker, which combined the EpCAM+ exosome proportion with exosomal Circ-CDYL levels, showed higher diagnostic efficacy (LTi-Exos Youden’s index = 0.6899) than single factor uses (Exos Circ-CDYL Youden’s index = 0.6591; EpCAM+ Exos % Youden’s index = 0.5411) (Table S2).
α-fetoprotein (AFP), a traditional diagnostic biomarker of HCC, was also elevated in the plasma of early HCC patients in our study (Figure 7F). However, the AUC of the AFP biomarker was disappointingly low at 0.70 (95% CI: 0.62–0.77), with a sensitivity of 46.32% and specificity of 97.83% for the diagnosis of early HCC (Figure 7G). The Youden’s index of AFP was lower than that of the LTi-Exos (AFP Youden’s index = 0.4415; LTi-Exos Youden’s index = 0.6899) (Table S2). We then combined the newly identified LTi-Exos with traditional AFP to diagnose early HCC and found that the AUC increased to the optimal 0.90 (95% CI: 0.83–0.94), with a sensitivity of 83.16% and specificity of 91.30% (Figure 7H). Logistic regression analysis also indicated that LTi-Exos combined with AFP are both independent markers for the discrimination of early HCC patients, with odds ratios (ORs) of 382.00 (95% CI: 58.38–2499.40) and 494.27 (95% CI: 74.48–3280.05), respectively (Table S2).
Taken together, LTi-Exos combined with conventional screening tools using AFP may have great promise for the early diagnosis of HCC or for surveillance of the possibility of HCC occurrence in the healthy population.
Discussion
Despite the rapid development in cancer therapy, advanced HCC patients have limited therapeutic options in the clinic and thus have a poor prognosis. Fundamentally, insufficient sensitivity and specificity of the traditional surveillance test for HCC undermined the performance of early HCC diagnosis. It is noteworthy to elucidate the underlying mechanism of tumorigenesis and investigate early diagnostic and surveillance biomarkers for HCC.
By using a tissue microarray, the genome-wide differential expression of circRNAs in BCLC stage 0 HCC tissues and nontumorous adjacent tissues was analyzed. Among the identified circRNAs, Circ-CDYL was the most markedly upregulated circRNA in tumor tissues of very early stage HCC. The CDYL gene is located on the Y chromosome and encodes a protein that assists in gene transcription suppression and negatively regulates mammalian spermatogenesis and nervous system morphogenesis.41,42 Circ-CDYL is generated by the head-to-tail splicing junction of the second exon of the CDYL gene. The specific high expression level of Circ-CDYL in very early stage HCC indicated a potential oncogenic function of Circ-CDYL in liver neoplasm initiation.
Exosomes are a group of extracellular vesicles that carry and transfer bioactive molecules, including circRNAs, to mediate intercellular communication. A recent study showed that tumor-derived exosomes are enriched in the plasma of patients.43 CircRNAs were discovered to be transported by exosomes, which have been proposed as possible biomarkers for cancer.34,43 Therefore, we investigated the extracellular release pattern of Circ-CDYL by directly studying plasma-derived exosomes from patients with early HCC. Elevated levels of Circ-CDYL were detected in exosomes isolated from early HCC patients compared with those isolated from healthy donors. These findings point to the high expression of exosomal Circ-CDYL in early HCC as a potential tool for developing a combined screening or early diagnostic biomarker for high-risk populations.
EpCAM is a popular marker of liver tumor-initiating cells.32 An in vitro assay confirmed that the exosomal Circ-CDYL RNA level and EpCAM protein level were similar to those in their parental cells. Higher intracellular Circ-CDYL expression levels were detected in EpCAM+ cells and undifferentiated spheroids, indicating that it may originate from liver T-ICs. Intriguingly, the proportion of EpCAM+ exosomes was markedly higher than that of EpCAM+ HCC cells. Furthermore, the proportion of EpCAM+ exosomes increased when estimating the exosomes isolated from Circ-CDYL-overexpressing HCC cells, indicating the selective enrichment of Circ-CDYL and EpCAM cargoes sorted in producer cells before exosome release. In this study, we found that overexpression of Circ-CDYL increased the intracellular and exosomal EpCAM expression of HCC cell lines (Figure 1E). Preliminary study has been reported that EpCAM is a target of AKT/GSK-3β/β-catenin signaling pathway,44 which driven by Circ-CDYL as shown in this study (Figure 2H). Thus, we proposed that Circ-CDYL increased the EpCAM+ exosomes because exosomes derived from Circ-CDYL-overexpressed cells encapsulated more EpCAM.
Tumor-derived exosome cargoes can be transmitted into other cells, and recipient cells may subsequently take on oncogenic properties.45 Based on the evidence that Circ-CDYL-rich and EpCAM+ exosomes perform functions similar to those of liver tumor-initiating cells to promote the stem-like phenotypes of HCC, they have been termed liver tumor-initiating exosomes (LTi-Exos). We provided evidence for this concept by elevating the expression level of Circ-CDYL, increasing the proportion of EpCAM+ cells, and promoting self-renewal in recipient cells after incubation with LTi-Exos. Moreover, LTi-Exos significantly promoted the malignant growth of HCC cells both in vitro and in vivo. We demonstrated in our previous study that Circ-CDYL promoted HCC by elevating the expression of HDGF and HIF1AN, which led to upregulation of C-MYC and SURVIVIN via activation of the PI3K-AKT signaling pathway and derepression of SURVIVIN transcription via the NOTCH pathway.30 Mechanistically, LTi-Exos also activated the signal pathway that elevated intracellular expression of Circ-CDYL did.
m6A modification is deposited by the core heterodimeric complex composed of methyltransferase-like-3 (METTL3) and −14 (METTL14), with METTL3 being the sole catalytic subunit, and it can be reversed by the m6A demethylases FTO and ALKBH5.23 Emerging studies have demonstrated that m6A modifications are widespread in circRNAs, and the same m6A “writer, reader and eraser” machinery is involved as in mRNAs.23 MeRIP assays showed that Circ-CDYL was enriched in the m6A-modified fraction. Bioinformatics analysis using SRAMP showed several “very high confidence” predicted m6A sites on Circ-CDYL, among which the sites with combined score >0.7 were chosen, and were further confirmed by ligase-dependent absolute quantification analysis.
METTL3 was identified as a m6A methyltransferase approximately three decades ago, and it is widely expressed in human tissues and plays a role in various physiological and pathological processes.23 The elevated expression and oncogenic and cancer-promoting functions of METTL3 were reported in several types of cancer, including HCC.23,36 For instance, METTL3 was demonstrated to promote HCC progression by inhibiting SOCS2 expression via YTHDF246 and was also found to regulate the epithelial-mesenchymal transition (EMT) of HCC cells by promoting the translation of SNAIL via YTHDF1.36 In this study, we demonstrated that overexpression of METTL3 could increase the ratio of m6A-modified Circ-CDYL and elevate the expression of Circ-CDYL, while specifically inhibiting METTL3 enzyme activity using STM2457 led to the opposite effect. As expected, METTL3 enhanced the stem-like properties of the HCC cell line, and inhibiting METTL3 using STM2457 could restrain tumor growth of HCC both in vitro and in vivo.
m6A modification regulates various aspects of RNA at the posttranscriptional level by enlisting m6A-reader proteins.37 In this study, we determined that m6A modifications can regulate the action of Circ-CDYL via two distinct mechanisms: the first involves intracellular biogenesis and the preference for either linear or back splicing of pre-CDYL, and the second involves extracellular export via encapsulation into exosomes.
YTHDF1-3 and YTHDC2 are cytoplasmic m6A-reader proteins that regulate mRNA stability and translation, whereas YTHDC1 is located in the nucleus and has been demonstrated to regulate pre-mRNA splicing and export.37 In particular, YTHDC1 was shown to correlate with the deposition of m6A in specific exons on pre-mRNA.24,26 Circ-CDYL is generated from exon 2 of its parental gene, CDYL.30 The interaction between YTHDC1 and Circ-CDYL demonstrated by RNA Binding Protein Immunoprecipitation Assay (RIP) assay indicated that YTHDC1 may bind to exon 2 of CDYL. In agreement with previous evidence, we found that the alternative choice between linear and circular splicing of CDYL can be regulated by YTHDC1, which contributes to the increase in Circ-CDYL. Moreover, we found that YTHDC1 overexpression in SMMC-7721 cells was not able to significantly affect the back-splicing of Circ-CDYL alone, but when METTL3 and YTHDC1 were synergistically overexpressed, the back-splicing preference of Circ-CDYL was observed, emphasizing that the combined action of METTL3 and YTHDC1 is possibly required in the back-splicing reaction of Circ-CDYL.
Consistent with the cellular expression level of Circ-CDYL being positively related to its exosomal level, we found that the METTL3-dependent m6A modification of Circ-CDYL synergistically elevated its cellular expression and exosomal levels. The ratio of exosomal to cellular Circ-CDYL levels was greatly higher in the METTL3-overexpressing HCC cell line than in the negative control cells, indicating METTL3-dependent active regulation of the sorting of Circ-CDYL to exosomes. However, cells ectopically overexpressing Circ-CDYL with mutated m6A modification sites failed to produce exosomes with significantly higher levels of Circ-CDYL than their wild-type counterparts. LTi-Exos sorted from exosomes originating from cells ectopically overexpressing Circ-CDYL with mutated m6A modification sites (Circ-CDYLMut LTi-Exos) failed to enhance cell proliferation in vitro and in vivo and spheroid formation as well as LTi-Exos from the wild-type counterpart. Moreover, ectopic overexpression of wild-type Circ-CDYL enhanced the stemness of HCC cells, while m6A sites-mutated Circ-CDYL failed to do so. The possible mechanisms may be as follows: (1) The active sorting of Circ-CDYL depends on its m6A modification status. Exosomes from m6A sites-mutated cells incorporated less Circ-CDYL than their wild-type counterparts, which may in sequence lead to lower levels of liver cancer-promoting Circ-CDYL in receiver cells than their wild-type counterparts. (2) m6A modification of mature Circ-CDYL may be associated with cell stem-like phenotypes of liver cancer cells, so the Circ-CDYLMut LTi-Exos-treated recipient cells could not develop significantly enhanced stemness as the cells uptake LTi-Exos did. 3) Both the LTi-Exos and Circ-CDYLMut LTi-Exos released from the parental cells may regulate the stem-like properties of adjacent recipient cells, leading to integrated stemness alterations in HCC cells. In any case, HCC stem-like properties may be transferred via exosomes in the tumor microenvironment.
Heterogeneous nuclear ribonucleoproteins (hnRNPs) control RNA splicing, export, modification, and localization at the transcriptional and posttranscriptional levels. In mammalian cells, hnRNP A2/B1 is a central part of the hnRNP complex. HnRNP A2/B1 controls how RNA is processed by binding to specific sequences. For example, it controls the sorting of miRNAs into exosomes by binding to specific motifs on RNA species.47 Moreover, hnRNPA2/B1 has been demonstrated to bind to m6A-modified sites in the transcriptome in vitro and in vivo and is regarded as an “m6A reader”.38 However, whether it can regulate the exosome sorting of m6A-modified circRNA is still not quite clear. Here, we found that m6A sites-mutated Circ-CDYL could not be effectively encapsulated into exosomes as wild-type Circ-CDYL, which further supports the m6A-dependent exosome sorting of Circ-CDYL. Furthermore, we found that less Circ-CDYL was packaged into exosomes in HCC cells after hnRNPA2/B1 interference, while hnRNPA2/B1 could not effectively regulate the exosome sorting of m6A sites-mutated Circ-CDYL as it did wild-type Circ-CDYL, indicating that hnRNPA2/B1 controls the exosome sorting of Circ-CDYL in a m6A modification-dependent mechanism. Recently, other research suggested that hnRNPA2/B1 regulates m6A-modified RNAs without binding to the m6A motif directly, which made it an “m6A switch” rather than an “m6A reader”.48 Uncovering the underlying mechanism of how hnRNPA2/B1 binds with and regulates m6A-bearing transcripts requires further study.
Exosomes have the appealing potential to be optimal biomarkers for screening diseases, including cancer, due to their reflection of both the genotype and phenotype of parental cells.43 Thus, we further investigated the possibility of early HCC diagnosis and surveillance utilizing these LTi-Exos. After analyzing the exosomes isolated from the plasma of BCLC stage 0 and stage A HCC patients, we proved the significantly higher proportion of EpCAM+ exosomes and elevated Circ-CDYL and AFP levels in early HCC patients. The LTi-Exos reached an AUC of 0.88, with a sensitivity of 84.21% and specificity of 84.78% when diagnosing early HCC. The diagnostic accuracy of LTi-Exos is better than that of traditional AFP, with an AUC of 0.70 and a sensitivity of 46.32%. When LTi-Exos were combined with AFP, BCLC stage 0/A HCC patient samples could be distinguished from healthy subjects with 83.16% sensitivity and 91.30% specificity, and the AUC was enhanced to optimal 0.90.
Collectively, we identified the oncogenic effects of METTL3-dependent m6A modification of Circ-CDYL in HCC. On the one hand, Circ-CDYL m6A modification elevates its cellular expression by promoting the back-splicing of Circ-CDYL via YTHDC1; on the other hand, Circ-CDYL m6A modification increases the exosomal level of Circ-CDYL by promoting the active sorting of Circ-CDYL into exosomes via hnRNPA2/B1, which increases the level of Circ-CDYL in recipient cells by regulating the tumor microenvironment. These two mechanisms synergistically contribute to cancer initiation and progression. Circ-CDYL-enriched and EpCAM-positive LTi-Exos could be a potential biomarker for early HCC detection. Combining these newly identified LTi-Exos with traditional AFP would be of great possibility for the early diagnosis of HCC by liquid biopsy. Furthermore, it may be used as a surveillance indicator for the healthy or high-risk population of liver cancer in the future.
Limitations of the study
There are also several limitations to our study, including the fact that we did not investigate how each m6A-modified site on Circ-CDYL contributes to its back-splicing and exosomes sorting. Future relevant research will be conducted.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-m6A antibody | Synaptic Systems | Cat#202003; RRID:AB_2279214 |
| biotin-labeled anti-CD63 mAb | Biolegend | Cat#353018; RRID:AB_2561676 |
| fluorescein-conjugated anti-EpCAM APC | Biolegend | Cat#369810; RRID:AB_2650907 |
| fluorescein-conjugated anti-CD133-APC | Biolegend | Cat#397906; RRID:AB_2876721 |
| fluorescein-conjugated anti-CD24-APC | Biolegend | Cat#382604; RRID:AB_2927986 |
| isotype control antibody | Biolegend | Cat#400222; RRID:AB_2891178 |
| rabbit polyclonal anti-CD63 | Proteintech | Cat#25682-1-AP; RRID:AB_2783831 |
| rabbit monoclonal anti-EpCAM | AbCAM | Cat#ab32392; RRID:AB_732181 |
| rabbit monoclonal anti-HDGF | AbCAM | Cat#ab131046; RRID:AB_11156674 |
| mouse mAb anti-PI3K (P85α) | Cell Signaling Technology | Cat#13666S; RRID:AB_2798288 |
| rabbit mAb anti-PI3K (P110β) | Cell Signaling Technology | Cat#3011S; RRID:AB_2165246 |
| rabbit monoclonal anti-p-Akt (Ser473) | Cell Signaling Technology | Cat#4060S; RRID:AB_2315049 |
| rabbit monoclonal anti-p-Akt (Thr308) | Cell Signaling Technology | Cat#13038S; RRID:AB_2629447 |
| rabbit monoclonal anti-AKT (pan) | Cell Signaling Technology | Cat#4691P; RRID:AB_915783 |
| rabbit monoclonal anti-C-MYC | Cell Signaling Technology | Cat#13987S; RRID:AB_2631168 |
| rabbit monoclonal anti-p-mTOR (Ser2448) | Cell Signaling Technology | Cat#5536P; RRID:AB_10691552 |
| rabbit monoclonal anti-mTOR | Cell Signaling Technology | Cat#2983P; RRID:AB_2105622 |
| rabbit polyclonal anti-p-GSK-3β (Ser9) | Abclonal | Cat#AP0358; RRID:AB_2771152 |
| rabbit polyclonal anti-GSK-3β | Abclonal | Cat#A0479; RRID:AB_2757212 |
| mouse polyclonal anti-HIF1AN | Santa Cruz | Cat#SC-271780; RRID:AB_10709435 |
| rabbit polyclonal anti-SURVIVIN | Proteintech | Cat#10508-1-AP; RRID:AB_2064048 |
| rabbit monoclonal anti-METTL3 | Cell Signaling Technology | Cat#86132; RRID:AB_2800072 |
| rabbit monoclonal anti-YTHDC1 | Cell Signaling Technology | Cat#77422; RRID:AB_2799899 |
| mouse monoclonal anti-hnRNPA2B1 | Cell Signaling Technology | Cat#9304; RRID:AB_10694208 |
| rabbit monoclonal anti-β-actin | Abclonal | Cat#AC026; RRID:AB_2768234 |
| mouse monoclonal anti-GAPDH | Santa Cruz | Cat#SC-47724; RRID:AB_627678 |
| mouse fluorescein-conjugated secondary antibody | Li-Cor | Cat#C926-32210; RRID:AB_621842 |
| rabbit fluorescein-conjugated secondary antibody | Li-Cor | Cat#926-32211; RRID:AB_621843 |
| Bacterial and virus strains | ||
| Circ-CDYL overexpression lentivirus | GeneChem | N/A |
| METTL3 overexpression lentivirus | Heyuan Biotechnology | N/A |
| YTHDC1 overexpression lentivirus | Heyuan Biotechnology | N/A |
| Biological samples | ||
| Human plasma using for exosomes analysis | Eastern Hepatobiliary Surgery Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol Reagent | Invitrogen | Cat#15596026 |
| STM2457 | MedChemExpress | Cat#HY-134836 |
| Actinomycin D | MedChemExpress | Cat#HY-17559 |
| DAPI | Beyotime | Cat# C1002 |
| Protease inhibitor cocktail and phosphatase inhibitor | Beyotime | Cat# P1046 |
| Western and IP buffer | Beyotime | Cat# P0013 |
| DMEM | BasalMedia | Cat#L110KJ |
| Fetal Bovine Serum | Biological Industries | Cat#04-001-1ACS |
| Critical commercial assays | ||
| RNA Antisense Purification Kit | BersinBio | Cat#Bes5103 |
| Magna RIP RNA-binding protein immunoprecipitation kit | Millipore | Cat#17-700 |
| Dynabeads kilobase BINDER kit | Invitrogen | Cat#60101 |
| Cell counting kit-8 (CCK-8) | Dojindo | Cat#Cat# CK04 |
| JetPRIME reagents | Poly plus | Cat#114-15 |
| exoEasy Maxi Kit | Qiagen | Cat#76064 |
| iScript™ cDNA Synthesis Kit | Bio-Rad | Cat#1708891 |
| PKH26 kit | Wayenbio | Cat#ESQ-R-001 |
| Actin-Tracker Green | Beyotime | Cat#C1033 |
| Experimental models: Cell lines | ||
| HCCLM3 | Cell Bank of Type Culture Collection of the Chinese Academy of Sciences | N/A |
| SMMC-7721 | Cell Bank of Type Culture Collection of the Chinese Academy of Sciences | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c-Nude | GemPharmatech Co. | D000521 |
| Oligonucleotides | ||
| Circ-CDYL RAP Probe sequence: Circ-CDYL1: TTCAACCTTTCCCGTTAACA-3′bio Circ-CDYL2: ACAATCCTTTCAACCTTTCC-3′bio |
RiboBio | N/A |
| NC siRNA: sense: UUCUCCGAACGUGUCACGUdTdT antisense: ACGUGACACGUUCGGAGAAdTdT |
Biotend Co. | N/A |
| YTHDC1 siRNA 1#: sense: ACGUCUAUCCACUUCAAGCCCdTdT antisense: GCUUGAAGUGGAUAGACGUGCdTdT |
Biotend Co. | N/A |
| YTHDC1 siRNA 2#: sense: AUACAGAUUGGAUUACGGCUGdTdT antisense: GCCGUAAUCCAAUCUGUAUUUdTdT |
Biotend Co. | N/A |
| HnRNPA2B1 siRNA 1#: sense: GGCUUUGUCUAGACAAGAAdTdT antisense: UUCUUGUCUAGACAAAGCCdTdT |
Biotend Co. | N/A |
| HnRNPA2B1 siRNA 2#: sense: GCUGCAAGACCUCAUUCAAdTdT antisense: UUGAAUGAGGUCUUGCAGCdTdT |
Biotend Co. | N/A |
| HnRNPA2B1 siRNA 2#: sense: GGACCAGGAAGUAACUUUAdTdT antisense: UAAAGUUACUUCCUGGUCCdTdT |
Biotend Co. | N/A |
| Recombinant DNA | ||
| Circ-CDYL overexpression, see STAR Methods | GeneChem | N/A |
| Circ-CDYLmut overexpression, see STAR Methods | GeneChem | N/A |
| EpCAM-mCherry fusion overexpression, see STAR Methods | GeneChem | N/A |
| METTL3 overexpression, see STAR Methods | Heyuan Biotechnology | N/A |
| YTHDC1 overexpression, see STAR Methods | Heyuan Biotechnology | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al.49 | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9 | Graphpad Software | https://www.graphpad.com |
| FlowJo | BD Biosciences Pharmingen | https://www.bdbiosciences.com |
| BioTek Gen5 system | BioTeck | N/A |
| LightCycler® 96 Real-Time PCR System | Roche | N/A |
| Other | ||
| Mastercycler | Eppendorf | Nexus GSX1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Hongyang Wang (hywangk@vip.sina.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
HCC patients and clinical samples
Early HCC patients’ plasma using for exosomes analysis in this study were obtained from patients who were diagnosed and received surgical resection at the Eastern Hepatobiliary Surgery Hospital, Shanghai, China. No patients received any preoperative anticancer treatment. All of these patients are Chinese (Asian), including 77 males and 18 females. The ages of these patients ranged from 30 to 70, with a mean age of 60. Detailed patient information is shown in Table S1.
HCC staging was determined by the Barcelona Clinic Liver Cancer staging system (BCLC). BCLC stage 0 HCC (very early stage HCC) was defined as a single lesion <2 cm without vascular involvement or metastasis. BCLC stage A HCC (early stage HCC) is defined by a single lesion between 2 and 5 cm or ≤3 lesions each <3 cm, without portal vein thrombosis or extrahepatic metastasis. Late-stage HCC was defined as the combination of intermediate (BCLC stage B HCC) or advanced HCC (BCLC stage C HCC). All research complied with the principles of the Declaration of Helsinki. Patient samples were obtained following informed consent according to an established protocol approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital.
Animals
The 4–6 weeks old male nude mice were housed in individual microisolator cages with free access to sterile water and irradiated normal food in a specific pathogen-free facility. Experiments were conducted by following the criteria outlined in the Guide for the Care and Use of Laboratory Animals, prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23; revised 1985). All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committee of the Laboratory Animal Research Center at the Eastern Hepato-biliary Surgery Hospital, Second Military Medical University.
Cell lines and cell culture
The HCC cell lines HCCLM3 and SMMC-7721 were obtained from Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai Institute of Cell Biology), and cultured at 37°C in an atmosphere containing 5% CO2 and routinely cultured in DMEM supplemented with 10% fetal bovine serum. Cells were passaged every 1-2 d to maintain logarithmic growth.
In vivo xenograft experiments
For the exosome in vitro treatment experiment, a total of 5×105 SMMC-7721 cells were injected subcutaneously into the flanks of male nude mice (4–6 weeks old, n = 6) after 48 hours of incubation with 1 μg of LTi-Exos or NLTi-Exos. The subcutaneous tumors were harvested 3 weeks after injection.
For the exosome in vivo treatment experiment, a total of 5×105 SMMC-7721 cells were injected subcutaneously into the flanks of male nude mice (4–6 weeks old, n = 5). Then, 5 μg of LTi-Exos or NLTi-Exos were injected intratumorally every 3 days after xenograft implantation. The subcutaneous tumors were harvested 19 days after injection. Tumor volume was calculated as follows: V (mm3) = width2 (mm2) × length (mm)/2.
Ethics approval and consent to participate
All research complied with the principles of the Declaration of Helsinki. Patient samples were obtained following informed consent according to an established protocol approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital. All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committee of the Laboratory Animal Research Center at the Eastern Hepato-biliary Surgery Hospital, Second Military Medical University.
Method details
RNA antisense purification (RAP) assay
For the RAP assay, an RNA Antisense Purification Kit (Bes5103, BersinBio, Guangzhou, China) was utilized. Biotinylated probes for Circ-CDYL and a negative control probe were produced by RiboBio Co. (Guangzhou, China).
RAP assays were performed according to the previous study.30 For each RAP experiment, lysate from 2∗108 cells, 40 pmol of probe, and 40 μL streptavidin-coated beads were used. HCC cell lines were crosslinked with 1 mg/mL 4′-aminomethyltrioxalen (AMT), a psoralen-derivative crosslinker used for fixing RNA‒RNA hybrids, for 15 minutes on ice and then lysed on ice. The homogenized lysate was hybridized with probes (mixture of probe Circ-CDYL1 and Circ-CDYL2) for 180 min. Following hybridization, the streptavidin-coated magnetic beads were resuspended in the lysate mixture and incubated at 37°C for 30 min with rotated mixing. Following incubation, RNA was eluted by resuspending the beads twice in 50 μL RAP Elution Buffer and incubating at 95°C for 2 min. Then, the eluate was collected. Finally, the eluate protein was collected, and Western blot assays were performed.
M6A-RNA binding protein immunoprecipitation (meRIP) assay
MeRIP was performed using a Magna RIP RNA-binding protein immunoprecipitation kit (17–700, Millipore) and a Dynabeads kilobase BINDER kit (60101, Invitrogen) according to the manufacturers’ instructions and previous study.25 In brief, Dynabeads were incubated with anti-m6A antibody (5 μg; Synaptic Systems, #202003) for 1 h at room temperature. Precleared cell lysates were incubated with antibody-conjugated beads for 1 h at 4°C. Beads were washed five times, and binding complexes were eluted and subjected to RT‒qPCR assay as described above.
Ligase-dependent absolute quantification of the m6A modification fraction at specific sites in RNAs
The ligase-dependent absolute quantification of m6A modification was performed by Aksomics (Shanghai, China). In brief, total RNA isolated from the HCCLM3 cell line was pretreated with RNase R. RNA spikes (100 pM) were added to 1 μg RNA and mixed with 20 nM probe L, 20 nM probe R, and 1×T3 ligation buffer (NEB). Then, the samples were incubated at 85°C for 3 min and cooled to 35°C for 10 min. The sample and the standard were subjected to real-time PCR. The RNA content corrected for m6A methylation of the target gene site for the sample= (Total cDNA concentration of target gene - cDNA concentration of target gene site not m6A methylated) ∗ RNA spike in RNA concentration/cDNA concentration concentration of RNA spike in.
MazF-dependent quantification of m6A modification in RNAs
The MazF-qPCR m6A modification assay was performed by Aksomics (Shanghai, China). In brief, total RNA isolated from the HCCLM3 cell line was pretreated with RNase R. Then, the RNA sample was divided into two equal parts, and MazF (20 U/μl) was added to one part for digestion. MazF-treated and untreated samples were simultaneously subjected to cDNA synthesis, followed by a qPCR assay. The relative m6A modification level of the samples was calculated by 2-▵▵CT.
CCK8, spheroid formation, and colony formation assays
CCK8: HCC cells were seeded in 96-well plates at a density of 2×103 cells per well. The viability of HCC cells was determined by Cell Counting Kit 8 (Dojindo, Japan) and measured at OD450 nm with the BioTek Gen5 system (BioTeck, USA) at 0, 24, 48, 72, 96 hours after seeding.
Spheroid formation: HCC cells were seeded in 6-well low-adhesion plates at a density of 3×103 cells per well. After 2 weeks, spheroids were observed using ImageJ software (NIH Image).
Colony formation: HCC cells were seeded in 6-well plates at a density of 3×103 cells per well. After 2 weeks, spheroids were observed using ImageJ software (NIH Image).
Limiting dilution: Cells were seeded into 96-well ultralow attachment microplates (Corning) at doses from 500 to 5 (8 wells per dose) and incubated under spheroid conditions for 2 weeks. Based on the frequency of wells without colonies, the proportion of T-ICs was determined using Poisson distribution statistics and L-Calc Version 1.1 software (Stem Cell Technologies, Inc., Vancouver, Canada).
Lentiviral infection
The lentivirus for overexpression of Circ-CDYL, Circ-CDYLmut and EpCAM-mCherry fusion was produced by GeneChem Company (Shanghai, China), and that for overexpression of METTL3 and YTHDC1 was produced by Heyuan Biotechnology (Shanghai, China). Lentiviral infection was carried out according to the manufacturers’ instructions. HCCLM3 cells and SMMC-7721 cells were infected with the concentrated virus at a multiplicity of infection (MOI) of 20 in the presence of 8 μg/mL polybrene (Sigma) for 10 hours. The expression of Circ-CDYL in the infected cells was validated by qRT‒PCR. Puromycin (1 ng/μl) was used to screen infected cells according to the manufacturers’ instructions.
For the Circ-CDYL and Circ-CDYLmut stable overexpression system, the “flank strategy” was used to form Circ-CDYL. Briefly, specific intron sequences at both ends of mature circRNAs were constructed, which could promote the formation of circles. The sequence (the capital letters) and the two ends of the loop arms (lowercase letters) needed to build the lentivirus are as follows.
Circ-CDYL
gccagaaatgtactgacgttcttaataattgataggtgcagacctttgcgttcttaattaaagagccactccgtggctgtaggcctctgcccacaagatacagagtcacatgctgcccccaggatgcaagctgatgcttttatagtaactgcagatgtaataacagattctaggtatttcttcagtaacgtatggaggcactgtgtcacataactctcgttcatgtgagtagcggaagatctctcttccatttacgcttcagatgttaaggtagtagaatttgtcacggtgataaaacgtgcttacaaggcagggttgtgtgaattgataaatttgatccaaaatgtattggtcatccatttcaatgattaacagcatctccattgaaagatgttagtgtaaaattaattaaaaccaagatttgaaaataacctgatcgctctagactgttttattacaatcaagtatgatgttctctgttgacctttttagttttaattgagtgcttctgaatgatacttgtgatacagatcatatgtttctttgatggcagtttcacaccacagaaggttctcagcgccttgtagctcattgctgtcaccactccaggctgttgcctgggtgcccgctgctggaaccctgggatgaccaggcacagctgcagatgcctcgagctgctccagccatgagtgtggaaaccaactcggtggtcaggcttcaacacagaaatagcagagagcccctttcagtcacaagaaacataagtcacctaatgcatatccacacgttctcagcataaaaatttagcatcttgtttgacttactttcagggaaattgtccacaacccaaatggcctattactattttattgaggttgcagaagagatcattttatcatctattgtggtttttgctatttttgatgtttcctaatgaaacttgcacttttccccccaaattttgatttttttaaaactatttttttccttttatgaacagGTTGAAAGGATTGTTGACAAAAGGAAAAATAAAAAAGGGAAGACAGAGTATTTGGTTCGGTGGAAAGGCTATGACAGCGAGGACGACACTTGGGAGCCGGAACAGCACCTCGTGAACTGTGAGGAATACATCCACGACTTCAACAGACGCCACACGGAGAAGCAGAAGGAGAGCACATTGACCAGAACAAACAGGACCTCTCCCAACAATGCTAGGAAACAAATCTCCAGATCCACCAACAGCAACTTTTCTAAGACCTCTCCTAAGGCACTCGTGATTGGGAAAGACCACGAATCCAAAAACAGCCAGCTGTTTGCTGCCAGCCAGAAGTTCAGGAAGAACACAGCTCCATCTCTCTCCAGCCGGAAGAACATGGACCTAGCGAAGTCAGGTATCAAGATCCTCGTGCCTAAAAGCCCCGTTAAGAGCAGGACCGCAGTGGACGGCTTTCAGAGCGAGAGCCCTGAGAAACTGGACCCCGTCGAGCAGGGTCAGGAGGACACAGTGGCACCCGAAGTGGCAGCGGAAAAGCCGGTCGGAGCTTTATTGGGCCCCGGTGCCGAGAGGGCCAGGATGGGGAGCAGGCCCAGGATACACCCACTAGTGCCTCAGGTGCCCGGCCCTGTGACTGCAGCCATGGCCACAGGCTTAGCTGTTAACGGGAAAGgtgagtgtctagggagctgctcaggctccgtggtgatcccagagggctcgggtccttctctagagatcaggccttgagctgggcagagtccggggcttctcacggcatctgctggagccttgcagagatttcgccttctcctttcttcgctggaagcctttgatatttctcttaattcggcaactagttttttctgtgtaattcgccatccacagactgtgctaccacactgacttgctttaaacagtatctccctctaactaggttattaggtgataagtttttacagcaagtttctgaagctgtctacctttcttcacctctgaagtgctgagtcttgtctcgtgttaagcgttttcttctaagtgtggccagcattgatacagaagtgcaggttcttattttgctcatctgtgtgaatacccaggaatgcctttcagtgggagcgttgctaacccagcagagcctttgggagccgggctcgtttctctgttgtgatcggcagtgcaggtgccattgcttcccccgtcagagcccagtctgcctggcaccggtggtgtcctctgttttactctcatccccagccccgctcctgggttc.
Circ-CDYLmut (bold letters refer to the mutant sites)
gccagaaatgtactgacgttcttaataattgataggtgcagacctttgcgttcttaattaaagagccactccgtggctgtaggcctctgcccacaagatacagagtcacatgctgcccccaggatgcaagctgatgcttttatagtaactgcagatgtaataacagattctaggtatttcttcagtaacgtatggaggcactgtgtcacataactctcgttcatgtgagtagcggaagatctctcttccatttacgcttcagatgttaaggtagtagaatttgtcacggtgataaaacgtgcttacaaggcagggttgtgtgaattgataaatttgatccaaaatgtattggtcatccatttcaatgattaacagcatctccattgaaagatgttagtgtaaaattaattaaaaccaagatttgaaaataacctgatcgctctagactgttttattacaatcaagtatgatgttctctgttgacctttttagttttaattgagtgcttctgaatgatacttgtgatacagatcatatgtttctttgatggcagtttcacaccacagaaggttctcagcgccttgtagctcattgctgtcaccactccaggctgttgcctgggtgcccgctgctggaaccctgggatgaccaggcacagctgcagatgcctcgagctgctccagccatgagtgtggaaaccaactcggtggtcaggcttcaacacagaaatagcagagagcccctttcagtcacaagaaacataagtcacctaatgcatatccacacgttctcagcataaaaatttagcatcttgtttgacttactttcagggaaattgtccacaacccaaatggcctattactattttattgaggttgcagaagagatcattttatcatctattgtggtttttgctatttttgatgtttcctaatgaaacttgcacttttccccccaaattttgatttttttaaaactatttttttccttttatgaacagGTTGAAAGGATTGTTGACAAAAGGAAAAATAAAAAAGGGAAGACAGAGTATTTGGTTCGGTGGAAAGGCTATGACAGCGAGGACGACACTTGGGAGCCGGAACAGCACCTCGTGATCTGTGAGGAATACATCCACGACTTCAACAGACGCCACACGGAGAAGCAGAAGGAGAGCACATTGACCAGAACAAACAGGACCTCTCCCAACAATGCTAGGAAACAAATCTCCAGATCCACCAACAGCAACTTTTCTAAGACCTCTCCTAAGGCACTCGTGATTGGGAAAGACCACGAATCCAAAAACAGCCAGCTGTTTGCTGCCAGCCAGAAGTTCAGGAAGAACACAGCTCCATCTCTCTCCAGCCGGAAGATCATGGACCTAGCGAAGTCAGGTATCAAGATCCTCGTGCCTAAAAGCCCCGTTAAGAGCAGGTCCGCAGTGGACGGCTTTCAGAGCGAGAGCCCTGAGAATCTGGTCCCCGTCGAGCAGGGTCAGGAGGTCACAGTGGCACCCGAAGTGGCAGCGGAAAAGCCGGTCGGAGCTTTATTGGGCCCCGGTGCCGAGAGGGCCAGGATGGGGAGCAGGCCCAGGATACACCCACTAGTGCCTCAGGTGCCCGGCCCTGTGTCTGCAGCCATGGCCACAGGCTTAGCTGTTAACGGGAAAGgtgagtgtctagggagctgctcaggctccgtggtgatcccagagggctcgggtccttctctagagatcaggccttgagctgggcagagtccggggcttctcacggcatctgctggagccttgcagagatttcgccttctcctttcttcgctggaagcctttgatatttctcttaattcggcaactagttttttctgtgtaattcgccatccacagactgtgctaccacactgacttgctttaaacagtatctccctctaactaggttattaggtgataagtttttacagcaagtttctgaagctgtctacctttcttcacctctgaagtgctgagtcttgtctcgtgttaagcgttttcttctaagtgtggccagcattgatacagaagtgcaggttcttattttgctcatctgtgtgaatacccaggaatgcctttcagtgggagcgttgctaacccagcagagcctttgggagccgggctcgtttctctgttgtgatcggcagtgcaggtgccattgcttcccccgtcagagcccagtctgcctggcaccggtggtgtcctctgttttactctcatccccagccccgctcctgggttc.
For the EpCAM-mCherry fusion, METTL3, and YTHDC1 stable overexpression system, the sequence of gene of interest to build the lentivirus was as follows.
EpCAM-mCherry fusion
AGGTGTCCACTCCCAGGTCCAACTGCACCTCGGTTCTAAGCTTCTGCAGGTCGACTCTAGAGGATCCCGCCACCATGGCGCCCCCGCAGGTCCTCGCGTTCGGGCTTCTGCTTGCCGCGGCGACGGCGACTTTTGCCGCAGCTCAGGAAGAATGTGTCTGTGAAAACTACAAGCTGGCCGTAAACTGCTTTGTGAATAATAATCGTCAATGCCAGTGTACTTCAGTTGGTGCACAAAATACTGTCATTTGCTCAAAGCTGGCTGCCAAATGTTTGGTGATGAAGGCAGAAATGAATGGCTCAAAACTTGGGAGAAGAGCAAAACCTGAAGGGGCCCTCCAGAACAATGATGGGCTTTATGATCCTGACTGCGATGAGAGCGGGCTCTTTAAGGCCAAGCAGTGCAACGGCACCTCCATGTGCTGGTGTGTGAACACTGCTGGGGTCAGAAGAACAGACAAGGACACTGAAATAACCTGCTCTGAGCGAGTGAGAACCTACTGGATCATCATTGAACTAAAACACAAAGCAAGAGAAAAACCTTATGATAGTAAAAGTTTGCGGACTGCACTTCAGAAGGAGATCACAACGCGTTATCAACTGGATCCAAAATTTATCACGAGTATTTTGTATGAGAATAATGTTATCACTATTGATCTGGTTCAAAATTCTTCTCAAAAAACTCAGAATGATGTGGACATAGCTGATGTGGCTTATTATTTTGAAAAAGATGTTAAAGGTGAATCCTTGTTTCATTCTAAGAAAATGGACCTGACAGTAAATGGGGAACAACTGGATCTGGATCCTGGTCAAACTTTAATTTATTATGTTGATGAAAAAGCACCTGAATTCTCAATGCAGGGTCTAAAAGCTGGTGTTATTGCTGTTATTGTGGTTGTGGTGATAGCAGTTGTTGCTGGAATTGTTGTGCTGGTTATTTCCAGAAAGAAGAGAATGGCAAAGTATGAGAAGGCTGAGATAAAGGAGATGGGTGAGATGCATAGGGAACTCAATGCAGGAGGTGGAGGATCAATGGTGAGCAAGGGCGAGGAGGATAACATGGCCATCATCAAGGAGTTCATGCGCTTCAAGGTGCACATGGAGGGCTCCGTGAACGGCCACGAGTTCGAGATCGAGGGCGAGGGCGAGGGCCGCCCCTACGAGGGCACCCAGACCGCCAAGCTGAAGGTGACCAAGGGTGGCCCCCTGCCCTTCGCCTGGGACATCCTGTCCCCTCAGTTCATGTACGGCTCCAAGGCCTACGTGAAGCACCCCGCCGACATCCCCGACTACTTGAAGCTGTCCTTCCCCGAGGGCTTCAAGTGGGAGCGCGTGATGAACTTCGAGGACGGCGGCGTGGTGACCGTGACCCAGGACTCCTCCCTGCAGGACGGCGAGTTCATCTACAAGGTGAAGCTGCGCGGCACCAACTTCCCCTCCGACGGCCCCGTAATGCAGAAGAAGACCATGGGCTGGGAGGCCTCCTCCGAGCGGATGTACCCCGAGGACGGCGCCCTGAAGGGCGAGATCAAGCAGAGGCTGAAGCTGAAGGACGGCGGCCACTACGACGCTGAGGTCAAGACCACCTACAAGGCCAAGAAGCCCGTGCAGCTGCCCGGCGCCTACAACGTCAACATCAAGTTGGACATCACCTCCCACAACGAGGACTACACCATCGTGGAACAGTACGAACGCGCCGAGGGCCGCCACTCCACCGGCGGCATGGACGAGCTGTACAAGTAACTCGAGTCCATCGATACTAGTCTGTGGAATGTGTGTCAGTTAGGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAAAGCTCCCGGGAGCTTGTATATCCATTTTCGGATCTGATCGCCACCATGACCGAGTACAAGCCCACGGTGCGC.
METTL3
ATGTCGGACACGTGGAGCTCTATCCAGGCCCACAAGAAGCAGCTGGACTCTCTGCGGGAGAGGCTGCAGCGGAGGCGGAAGCAGGACTCGGGGCACTTGGATCTACGGAATCCAGAGGCAGCATTGTCTCCAACCTTCCGTAGTGACAGCCCAGTGCCTACTGCACCCACCTCTGGTGGCCCTAAGCCCAGCACAGCTTCAGCAGTTCCTGAATTAGCTACAGATCCTGAGTTAGAGAAGAAGTTGCTACACCACCTCTCTGATCTGGCCTTAACATTGCCCACTGATGCTGTGTCCATCTGTCTTGCCATCTCCACGCCAGATGCTCCTGCCACTCAAGATGGGGTAGAAAGCCTCCTGCAGAAGTTTGCAGCTCAGGAGTTGATTGAGGTAAAGCGAGGTCTCCTACAAGATGATGCACATCCTACTCTTGTAACCTATGCTGACCATTCCAAGCTCTCTGCCATGATGGGTGCTGTGGCAGAAAAGAAGGGCCCTGGGGAGGTAGCAGGGACTGTCACAGGGCAGAAGCGGCGTGCAGAACAGGACTCGACTACAGTAGCTGCCTTTGCCAGTTCGTTAGTCTCTGGTCTGAACTCTTCAGCATCGGAACCAGCAAAGGAGCCAGCCAAGAAATCAAGGAAACATGCTGCCTCAGATGTTGATCTGGAGATAGAGAGCCTTCTGAACCAACAGTCCACTAAGGAACAACAGAGCAAGAAGGTCAGTCAGGAGATCCTAGAGCTATTAAATACTACAACAGCCAAGGAACAATCCATTGTTGAAAAATTTCGCTCTCGAGGTCGGGCCCAAGTGCAAGAATTCTGTGACTATGGAACCAAGGAGGAGTGCATGAAAGCCAGTGATGCTGATCGACCCTGTCGCAAGCTGCACTTCAGACGAATTATCAATAAACACACTGATGAGTCTTTAGGTGACTGCTCTTTCCTTAATACATGTTTCCACATGGATACCTGCAAGTATGTTCACTATGAAATTGATGCTTGCATGGATTCTGAGGCCCCTGGCAGCAAAGACCACACGCCAAGCCAGGAGCTTGCTCTTACACAGAGTGTCGGAGGTGATTCCAGTGCAGACCGACTCTTCCCACCTCAGTGGATCTGTTGTGATATCCGCTACCTGGACGTCAGTATCTTGGGCAAGTTTGCAGTTGTGATGGCTGACCCACCCTGGGATATTCACATGGAACTGCCCTATGGGACCCTGACAGATGATGAGATGCGCAGGCTCAACATACCCGTACTACAGGATGATGGCTTTCTCTTCCTCTGGGTCACAGGCAGGGCCATGGAGTTGGGGAGAGAATGTCTAAACCTCTGGGGGTATGAACGGGTAGATGAAATTATTTGGGTGAAGACAAATCAACTGCAACGCATCATTCGGACAGGCCGTACAGGTCACTGGTTGAACCATGGGAAGGAACACTGCTTGGTTGGTGTCAAAGGAAATCCCCAAGGCTTCAACCAGGGTCTGGATTGTGATGTGATCGTAGCTGAGGTTCGTTCCACCAGTCATAAACCAGATGAAATCTATGGCATGATTGAAAGACTATCTCCTGGCACTCGCAAGATTGAGTTATTTGGACGACCACACAATGTGCAACCCAACTGGATCACCCTTGGAAACCAACTGGATGGGATCCACCTACTAGACCCAGATGTGGTTGCACGGTTCAAGCAAAGGTACCCAGATGGTATCATCTCTAAACCTAAGAATTTATAG.
YTHDC1
ATGGCGGCTGACAGTCGGGAGGAGAAAGATGGAGAACTTAATGTTCTGGATGATATTTTAACTGAAGTACCAGAACAAGATGATGAACTGTATAATCCAGAGAGTGAACAAGATAAAAATGAGAAAAAGGGATCAAAAAGAAAAAGTGATCGAATGGAATCTACTGATACCAAACGACAAAAGCCTTCTGTCCATTCTAGACAACTGGTTTCTAAGCCACTGAGCTCATCTGTTAGCAATAACAAAAGAATAGTTAGTACAAAAGGAAAGTCAGCCACAGAGTATAAAAATGAGGAATATCAAAGATCTGAAAGAAACAAGCGTCTAGATGCTGATCGGAAAATTCGTCTATCAAGTAGTGCCTCCAGAGAACCTTATAAGAATCAACCTGAAAAAACCTGTGTCCGGAAAAGGGATCCTGAAAGGAGGGCCAAATCTCCTACGCCAGATGGTTCTGAGAGAATTGGGCTTGAAGTGGATAGACGTGCAAGCAGATCCAGCCAGTCTTCTAAGGAAGAAGTGAACTCTGAAGAATATGGCTCTGACCATGAGACTGGCAGCAGTGGTTCTTCTGATGAGCAAGGGAACAACACTGAGAATGAGGAGGAAGGAGTGGAAGAAGATGTGGAGGAAGATGAAGAAGTAGAAGAAGATGCAGAAGAAGATGAAGAGGTGGATGAAGATGGAGAGGAGGAGGAGGAAGAGGAGGAGGAGGAAGAGGAGGAGGAGGAGGAGGAAGAAGAAGAATATGAACAGGATGAGAGAGACCAGAAAGAGGAGGGAAATGATTATGACACTCGAAGTGAGGCCAGTGACTCTGGTTCTGAATCTGTTTCCTTCACAGATGGGTCTGTCAGATCTGGTTCAGGCACAGATGGATCAGATGAGAAAAAGAAGGAAAGGAAGAGAGCTAGAGGCATATCTCCAATTGTTTTTGATAGAAGTGGAAGCTCTGCATCAGAGTCATATGCAGGTTCAGAAAAGAAGCATGAGAAATTATCATCTTCCGTTCGTGCTGTCCGAAAAGATCAAACCAGTAAACTCAAATATGTGCTTCAAGATGCAAGATTTTTCCTCATAAAGAGTAACAACCATGAGAATGTGTCTCTTGCCAAAGCGAAGGGTGTATGGTCCACGCTCCCTGTAAATGAGAAGAAATTAAATCTTGCATTTAGATCTGCAAGGAGTGTTATCTTAATATTTTCTGTCAGAGAGAGTGGAAAATTTCAAGGGTTTGCAAGACTTTCTTCAGAATCACATCACGGAGGATCTCCTATACACTGGGTGCTTCCAGCAGGAATGAGTGCTAAAATGCTGGGAGGTGTCTTTAAAATTGACTGGATTTGCAGGCGTGAATTACCCTTCACTAAGTCGGCTCATCTCACCAATCCTTGGAATGAACATAAACCAGTAAAGATCGGACGTGATGGACAGGAAATTGAACTTGAATGTGGAACCCAGCTTTGTCTTCTGTTTCCCCCCGATGAAAGTATTGACTTGTATCAGGTCATTCATAAAATGCGTCACAAGAGAAGAATGCATTCTCAGCCCCGATCACGAGGACGTCCATCCCGTCGAGAACCAGTCCGGGATGTGGGAAGGCGTCGACCAGAAGATTATGATATTCATAACAGCAGAAAGAAACCAAGGATTGACTATCCCCCTGAGTTTCACCAGAGACCAGGGTATTTAAAGGATCCACGATACCAGGAAGTGGACAGACGATTTTCAGGAGTTCGCCGAGATGTGTTTTTAAATGGGTCCTACAATGATTATGTGAGGGAATTTCATAACATGGGACCACCACCACCTTGGCAAGGAATGCCCCCTTACCCAGGAATGGAACAACCTCCACACCATCCTTACTATCAGCACCATGCTCCACCTCCTCAAGCTCATCCCCCTTACTCAGGACATCATCCAGTACCACATGAAGCAAGATACAGAGATAAACGAGTACATGATTATGATATGAGGGTGGATGATTTCCTTCGTCGCACACAAGCTGTTGTCAGTGGCCGGAGAAGTAGACCCCGTGAAAGAGACCGGGAACGAGAGCGAGACCGCCCTAGAGATAACAGACGAGACAGAGAGCGAGATAGAGGACGTGATAGAGAAAGAGAAAGAGAGCGATTATGTGATCGAGACAGAGACCGAGGGGAGAGAGGTCGATATAGAAGA.
siRNA transfection
The siRNAs for YTHDC1 and hnRNPA2B1 were purchased from Biotend Co. (Shanghai, China). Transfection was performed with JetPRIME reagents (Poly plus, Illkirch, France) according to the manufacturers’ instructions. Briefly, 2∗105 cells were seeded into 6-well plates 24 hours before transfection. For each well, 10 μl (20 μM storage concentration) siRNA was added to the JetPRIME buffer. Mix by pipetting up and down. Then, JetPRIME was added to the siRNA duplexes and homogenized by vortexing immediately for 1 second. Incubate for 10 minutes at room temperature to allow transfection complexes to form between duplexes and JetPRIME. Then, the transfection mix was added to 2 ml of cell culture medium to a final concentration of 100 nM siRNAs. Finally, homogenize by gently swirling the plate.
Exosomes purification
The exosomes were purified using ultracentrifuge according to previous study.50
For cell-derived exosomes purification, cell lines were cultured in DMEM containing 10% exosomes-depleted FBS. After 48 h of culture, the cell culture medium was collected, and centrifuge through the following step: (1) Centrifuge 10 min at 300 × g, 4°C. Discard the pellet. (2) Centrifuge 20 min at 2,000 × g, 4°C. Discard the pellet. (3) Centrifuge 30 min at 12,000 × g, 4°C. Discard the pellet. Filter the suspension through a 0.22-μm filter. (4) Centrifuge 90 min at 120,000 × g, 4°C. Discard the supernatant. (5) Resuspend the pellet with 1 mL PBS. Centrifuge 70 min at 120,000 × g, 4°C. Discard the supernatant and collect the exosomes pellet.
For plasma-derived exosomes purification, 3 mL of venous whole blood samples of patients were collected using an anticoagulation tube (containing EDTA) (GD050EK2, Gongdong). The blood sample were centrifuged 10 min at 1,500 × g, 4°C, to collect 1 mL of plasma, and centrifuge through the following step: (1) Dilute plasma with triple volume of PBS, centrifuge 30 min at 2,000 × g, 4°C. Discard the pellet. (2) Centrifuge 45 min at 12,000 × g, 4°C. Discard the pellet. Filter the suspension through a 0.22-μm filter. (3) Centrifuge 90 min at 120,000 × g, 4°C. Discard the supernatant. (4) Resuspend the pellet with 1 mL PBS. Centrifuge 70 min at 120,000 × g, 4°C. Discard the supernatant and collect the exosomes pellet.
Exosomes identification
The identification of exosomes was performed by Echobiotech (Beijing, China). Exosomes were isolated from the cell culture supernatant of HCCLM3 and SMMC-7721 cells using ultracentrifugation. Exosomes were examined and photographed by electron microscopy using negative staining. Exosomes markers were detected using Western blot assays. The size of the exosomes was tested with a ZETASIZER Nano series-Nano-ZS (Malvern, England).
Flow cytometry analysis of exosomes
Flow cytometry was used to analyze the exosomes phenotype derived from cultured HCC cell or HCC patient plasma. Exosomes purified from cell supernatant or HCC patient plasma were coincubated with biotin-labeled anti-CD63 mAb (353018, Biolegend, San Diego, CA). Next, streptavidin beads were added and incubated for 2 h at RT. The bead/anti-CD63Ab/exosomes complexes were then coincubated with the fluorescein-conjugated monoclonal antibodies anti-EpCAM APC (369810, Biolegend, San Diego, CA) or with the isotype control (400222, Biolegend, San Diego, CA) at 4°C in the dark overnight. The samples were analyzed using a Beckman MoFlo XDP™ flow cytometer (Beckman Coulter, USA). The analysis of the raw exosomes flow cytometry data was performed according to a previous study.51 The positive gate was set at the point where <2% of the isotype control was positive. The results are presented as relative fluorescence values (%).
For LTi-Exos and NLTi-Exos subpopulation sorting, the final volume of samples was increased to 1 ml using 0.5% BSA in PBS before sorting. The samples were analyzed using a Beckman MoFlo XDP™ flow cytometer (Beckman Coulter) and sorted using a Beckman MoFlo XDP™ flow cytometer smart sampler (Beckman Coulter).
Exosomal RNA isolation and PCR analysis
Exosomal total RNA was isolated from cell- or plasma-derived exosomes using the exoEasy Maxi Kit (76064, Qiagen) following the manufacturer’s instructions. The total amount of RNA for each reverse transcription reaction was 100 ng, and the complementary DNA template was prepared using an iScript™ cDNA Synthesis Kit (1708891, Bio-Rad) according to the manufacturer’s protocol. The complementary DNA was amplified in a reaction mix of SYBR Green (Takara) using a LightCycler® 96 Real-Time PCR System (Roche) according to the manufacturer’s instructions.
Exosomes labeling
Purified exosomes from HCC cell line supernatants were labeled using the red fluorescent lipid dye PKH26 kit (Wayenbio, ESQ-R-001) following the manufacturer’s instruction, and then incubated with SMMC-7721 cells at 37°C for 12 hours. The cocultured cells were then fixed and labeled using Actin-Tracker Green (Beyotime, C1033) following the manufacturer’s instruction.
STM2457 treatment
For in vitro treatment, HCC cell lines were treated with STM2457 at a final concentration of 5 μM or DMSO (1:1000 v/v) for 48 h. For in vivo treatment, xenografted nude mice were given intraperitoneal injections of 50 mg/kg STM2457 or vehicle every day.
Flow cytometry analysis of cells
Cells were collected and incubated for 30 min with 1% bovine serum albumin (BSA, Gibco) in PBS to block nonspecific antigens. Conjugated antibody (EpCAM-APC: 369810; CD133-APC, 397906; CD24-APC, 382604, Biolegend) incubation for cells was performed at 4°C for 10 minutes. The samples were analyzed using a Beckman MoFlo XDP™ flow cytometer (Beckman Coulter).
Western blot
Cell lysates were prepared with lysis buffer (Beyotime Biotechnology, China) and centrifuged at 12,000 × g for 20 min to collect the supernatant. Protein concentrations were determined using the bicinchoninic acid (BCA) assay. Immunoblotting was performed using specific primary antibodies as shown in key resources table. Immunocomplexes were incubated with the fluorescein-conjugated secondary antibody and then detected using an Odyssey fluorescence scanner (Li-Cor, Lincoln, NE).
Actinomycin D treatment
HCC cells were plated into six-well plates. After 4 h, cells were treated with actinomycin D (5 μg/mL) or DMSO (1:1000 v/v), and collected after 0, 1, 4, 8, and 12 hours of treatment.
RNA isolation and PCR analysis
Total RNA was isolated from cells using TRIzol Reagent (Invitrogen, USA) following the manufacturer’s instructions. The complementary DNA template was prepared using oligo (dT) random primers and Moloney murine leukemia virus reverse transcriptase (Promega, USA) according to the manufacturer’s protocol. After reverse transcription, the complementary DNA template was quantitated using real-time PCR technology. The primers used in this study were designed using the Primer-BLAST tool provided by www.ncbi.nlm.nih.gov. cDNA was amplified in a reaction mix of SYBR Green (Takara, Japan) with a LightCycler 96 Real-Time PCR System (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The mRNA levels were normalized against β-actin in cell and tissue lysates. Real-time PCR experiments for each gene were performed on 3 separate occasions.
Quantification and statistical analysis
All statistical analyses were performed with SPSS 18.0 software. Qualitative variables were analyzed by the chi-square test or Fisher’s exact test. For continuous variables that obey the normal distribution, Student’s t test was used to compare the differences. Otherwise, variables with an abnormal distribution were compared using a nonparametric test. Differences between groups were compared using analysis of variance (ANOVA) when applicable or a nonparametric test. Correlation analysis was performed using the Pearson correlation coefficient method. Unless otherwise specified, the results are presented as the means ± standard deviations (SD). All statistical tests were 2 sided, and p < 0.05 was considered statistically significant. In each graph, otherwise specifically mentioned, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS stands for “not significant”.
Acknowledgments
We gratefully acknowledge the support from the State Key Project on Infectious Diseases of China (2018ZX10723204-002-002), National Natural Science Foundation of China (82172896, 91859205, 81988101, 81830054, 81902940, 81902942), Natural Science Foundation of Shanghai (19ZR1400300), Shanghai Top Young Talents Program, Foundation of Shanghai Shenkang Hospital Development Center (SHDC2020CR2011A, SHDC12016127), Shanghai Key Laboratory of Hepato-biliary Tumor Biology, The Key Laboratory of Signaling Regulation and Targeting Therapy of Liver Cancer (SMMU), Ministry of Education, Shanghai, China. We would like to thank Yanting Yu, Qinyun Huang, Shanhua Tang, Linna Guo, Dan Cao for technical assistance.
Author contributions
H.Y.W. contributed to study concept design and supervised all works; L.L. and Y.X.T. contributed to the analysis and interpretation of data and drafting of the paper. Y.P.W. and J.B.F. contributed to the acquisition of data, H.Z. and Y.L. contributed to the acquisition of clinical samples. X.W.T., S.W.L., M.Y., F.Y.L., G.K.Z., H.H.Q., K.C.Z., P.H.Y., X.W.Y., Q.Y., S.N.G., and B.H.Z. contributed to technical and material support. All authors have read and approved the article.
Declaration of interests
The authors declare that they have no competing interests.
Published: October 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108022.
Contributor Information
Yexiong Tan, Email: yxtan1214@163.com.
Liang Li, Email: liliangyuanquan@163.com.
Hongyang Wang, Email: hywangk@vip.sina.com.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA. Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Li L., Chen J., Chen X., Tang J., Guo H., Wang X., Qian J., Luo G., He F., Lu X., et al. Serum miRNAs as predictive and preventive biomarker for pre-clinical hepatocellular carcinoma. Cancer Lett. 2016;373:234–240. doi: 10.1016/j.canlet.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forner A., Reig M.E., de Lope C.R., Bruix J. Current strategy for staging and treatment: the BCLC update and future prospects. Semin. Liver Dis. 2010;30:61–74. doi: 10.1055/s-0030-1247133. [DOI] [PubMed] [Google Scholar]
- 4.Canelo R., Hakim N.S., Ringe B. Experience with hystidine tryptophan ketoglutarate versus University Wisconsin preservation solutions in transplantation. Int. Surg. 2003;88:145–151. [PubMed] [Google Scholar]
- 5.Llovet J.M., Fuster J., Bruix J. Intention-to-treat analysis of surgical treatment for early hepatocellular carcinoma: resection versus transplantation. Hepatology. 1999;30:1434–1440. doi: 10.1002/hep.510300629. [DOI] [PubMed] [Google Scholar]
- 6.Cillo U., Vitale A., Grigoletto F., Farinati F., Brolese A., Zanus G., Neri D., Boccagni P., Srsen N., D'Amico F., et al. Prospective validation of the Barcelona Clinic Liver Cancer staging system. J. Hepatol. 2006;44:723–731. doi: 10.1016/j.jhep.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Forner A. Hepatocellular carcinoma surveillance with miRNAs. Lancet Oncol. 2015;16:743–745. doi: 10.1016/S1470-2045(15)00014-5. [DOI] [PubMed] [Google Scholar]
- 8.Poon D., Anderson B.O., Chen L.T., Tanaka K., Lau W.Y., Van Cutsem E., Singh H., Chow W.C., Ooi L.L., Chow P., et al. Management of hepatocellular carcinoma in Asia: consensus statement from the Asian Oncology Summit 2009. Lancet Oncol. 2009;10:1111–1118. doi: 10.1016/S1470-2045(09)70241-4. [DOI] [PubMed] [Google Scholar]
- 9.Gebo K.A., Chander G., Jenckes M.W., Ghanem K.G., Herlong H.F., Torbenson M.S., El-Kamary S.S., Bass E.B. Screening tests for hepatocellular carcinoma in patients with chronic hepatitis C: a systematic review. Hepatology. 2002;36(5 Suppl 1):S84–S92. doi: 10.1053/jhep.2002.36817. [DOI] [PubMed] [Google Scholar]
- 10.Zinkin N.T., Grall F., Bhaskar K., Otu H.H., Spentzos D., Kalmowitz B., Wells M., Guerrero M., Asara J.M., Libermann T.A., Afdhal N.H. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin. Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 11.Marrero J.A., Feng Z., Wang Y., Nguyen M.H., Befeler A.S., Roberts L.R., Reddy K.R., Harnois D., Llovet J.M., Normolle D., et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110–118. doi: 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L., et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S., Li Y., Chen B., Zhao J., Yu S., Tang Y., Zheng Q., Li Y., Wang P., He X., Huang S. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang P., Song F., Yang X., Yan X., Huang X., Qiu Z., Wen Z., Liang C., Xin X., Lei Z., et al. Exosomal MicroRNA Signature Acts as an Efficient Biomarker for Non-Invasive Diagnosis of Gallbladder Carcinoma. iScience. 2022;25 doi: 10.1016/j.isci.2022.104816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J., Wang X., Zhai S., Shi M., Peng C., Deng X., Fu D., Wang J., Shen B. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J. Hematol. Oncol. 2022;15:128. doi: 10.1186/s13045-022-01348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Movahedpour A., Khatami S.H., Karami N., Vakili O., Naeli P., Jamali Z., Shabaninejad Z., Tazik K., Behrouj H., Ghasemi H. Exosomal noncoding RNAs in prostate cancer. Clin. Chim. Acta. 2022;537:127–132. doi: 10.1016/j.cca.2022.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Rackles E., Lopez P.H., Falcon-Perez J.M. Extracellular vesicles as source for the identification of minimally invasive molecular signatures in glioblastoma. Semin. Cancer Biol. 2022;87:148–159. doi: 10.1016/j.semcancer.2022.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 21.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H., Weng H., Chen J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Timoteo G., Dattilo D., Centrón-Broco A., Colantoni A., Guarnacci M., Rossi F., Incarnato D., Oliviero S., Fatica A., Morlando M., Bozzoni I. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107641. [DOI] [PubMed] [Google Scholar]
- 25.Chen R.X., Chen X., Xia L.P., Zhang J.X., Pan Z.Z., Ma X.D., Han K., Chen J.W., Judde J.G., Deas O., et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo X., Chen Z., Gao W., Zhang Y., Wang J., Wang J., Cao M., Cai J., Wu J., Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng H., Chen B., Wei W., Guo S., Han H., Yang C., Ma J., Wang L., Peng S., Kuang M., Lin S. N6-methyladenosine (m6A) in 18S rRNA promotes fatty acid metabolism and oncogenic transformation. Nat. Metab. 2022;4:1041–1054. doi: 10.1038/s42255-022-00622-9. [DOI] [PubMed] [Google Scholar]
- 30.Wei Y., Chen X., Liang C., Ling Y., Yang X., Ye X., Zhang H., Yang P., Cui X., Ren Y., et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology. 2020;71:130–147. doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 31.Ma S., Chan K.W., Hu L., Lee T.K.W., Wo J.Y.H., Ng I.O.L., Zheng B.J., Guan X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita T., Ji J., Budhu A., Forgues M., Yang W., Wang H.Y., Jia H., Ye Q., Qin L.X., Wauthier E., et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee T.K.W., Castilho A., Cheung V.C.H., Tang K.H., Ma S., Ng I.O.L. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Li C., Ni Y.Q., Xu H., Xiang Q.Y., Zhao Y., Zhan J.K., He J.Y., Li S., Liu Y.S. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 2021;6:383. doi: 10.1038/s41392-021-00779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Csepany T., Lin A., Baldick C.J., Jr., Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 1990;265:20117–20122. [PubMed] [Google Scholar]
- 36.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S., et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 38.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang F., Tang X., Tang C., Hua Z., Ke M., Wang C., Zhao J., Gao S., Jurczyszyn A., Janz S., et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J. Hematol. Oncol. 2021;14:54. doi: 10.1186/s13045-021-01066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Z., Zhao R., Li B., Qi Y., Qiu W., Guo Q., Zhang S., Zhao S., Xu H., Li M., et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer. 2022;21:16. doi: 10.1186/s12943-021-01485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulligan P., Westbrook T.F., Ottinger M., Pavlova N., Chang B., Macia E., Shi Y.J., Barretina J., Liu J., Howley P.M., et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol. Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi C., Liu S., Qin R., Zhang Y., Wang G., Shang Y., Wang Y., Liang J. Coordinated regulation of dendrite arborization by epigenetic factors CDYL and EZH2. J. Neurosci. 2014;34:4494–4508. doi: 10.1523/JNEUROSCI.3647-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P.P., Wang P.Q., Qiao C.P., Zhang Q., Zhang J.P., Chen F., Zhang X., Xie W.F., Yuan Z.L., Li Z.S., Chen Y.X. Differentiation therapy of hepatocellular carcinoma by inhibiting the activity of AKT/GSK-3β/β-catenin axis and TGF-β induced EMT with sophocarpine. Cancer Lett. 2016;376:95–103. doi: 10.1016/j.canlet.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F., et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Chen M., Wei L., Law C.T., Tsang F.H.C., Shen J., Cheng C.L.H., Tsang L.H., Ho D.W.H., Chiu D.K.C., Lee J.M.F., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 47.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu B., Su S., Patil D.P., Liu H., Gan J., Jaffrey S.R., Ma J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3: Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 51.Theodoraki M.N., Yerneni S.S., Hoffmann T.K., Gooding W.E., Whiteside T.L. Clinical significance of PD-L1+ exosomes in plasma of head and neck cancer patients. Clin. Cancer Res. 2018;24:896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.