Abstract
A new method for screening microbial colonies endowed with antiviral activity is described. It is based on close contact between microbial agar cultures and agar-covered virus-infected-cell monolayers and allows the screening of large numbers of colonies in just a few months.
In the last few years, more and more efforts have been made by pharmaceutical companies and university research centers to find safe and efficient drugs for the treatment of viral respiratory diseases, herpesvirus infections, and in particular retroviral infections (1, 8, 10). New types of antiviral compounds have been discovered, and novel, more potent, and selective derivatives of known compounds have been synthesized (9, 11, 14, 15).
The screening of natural products from microorganisms, plants, and algae, etc., still promises results, and some new active compounds have recently been described (3, 4, 12, 13).
One of the main problems in screening antiviral products from environment microorganisms is the difficulty of testing a high number of colonies in a short time, which requires that infected-cell cultures be challenged with microbial extracts.
With the aim of overcoming this problem, we devised a very simple method of screening microbial environmental colonies grown on suitable media directly on virus-infected-cell monolayers.
Vero, HEp2, MDCK, and Flow 2002 cells (all from ICN-Flow) were used for growing and plaquing cytopathic viruses and also for cytotoxic assays. Herpes simplex virus type 1 (HSV-1), HSV-2, vaccinia virus, poliovirus Sabin type 1, influenza A virus (strain WSN), and vesicular stomatitis virus were used for the screenings. All were purchased from the National Institutes of Health (Bethesda, Md.). The rich agar (AR) medium used for growing environmental microorganisms in the screening of antiviral activity had the following composition: 2 g of yeast extract, 10 g of Casamino Acids, 4 g of soluble starch, 11 g of purified agar (Oxoid), 6 g of NaCl, 0.4 g of KCl, 0.2 g of CaCl2 · 2H2O, and 0.1 g of MgSO4 · 7H2O in 1 liter of deionized water. This medium was capable of supporting the growth of most aerobic environmental microorganisms in 3 to 5 days at 25 to 28°C. The toxicity of AR medium on cell monolayers was minimal (5).
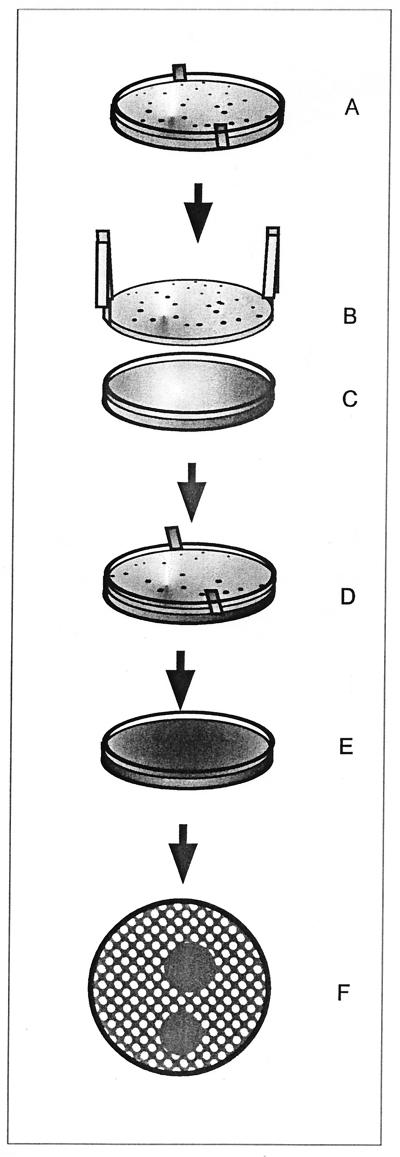
The various steps of the screening test for antiviral activity in environmental microorganisms are indicated in Fig. 1. Environmental samples of water and soil, animal feces, and other substances were diluted in normal saline and cultured in 9-cm-diameter petri dishes (with a thickness of about 6 mm) containing 20 ml of atoxic AR medium. The cultures were kept for 3 to 5 days at 25 to 28°C, until between 200 and 300 microbial colonies per plate were grown. In the bottom of the medium, a 9-cm-diameter dialysis membrane (molecular size exclusion, 10,000 Da; Medicell International Ltd., London, United Kingdom), which had been prepared previously with two margins jutting out at opposite sides, carefully washed, and sterilized by autoclaving at 121°C for 15 min, was positioned (Fig. 1A). The dialysis membrane allowed us not only to lift and move the agar culture but also to avoid aspecific toxicity due to bacterial hydrolytic enzymes. After colony growth, an entire culture was picked up (Fig. 1B) and put onto a cell monolayer in a 10-cm-diameter petri dish (Fig. 1C) which had previously been infected with a virus (HSV-1, HSV-2, poliovirus Sabin type 1, influenza A virus [WSN], vaccinia virus, or vesicular stomatitis virus) and covered with 20 ml of purified-agar (Oxoid)-solidified culture medium (Fig. 1D). After 20 to 30 min of contact, the microbial culture was removed (Fig. 1E) and saved and the cell monolayer was incubated for a further 2 to 3 days, after which the viral plaques were stained with neutral red and counted (Fig. 1F). Zones of inhibition of the viral plaques were scored, and the microbial colonies corresponding to the zones of inhibition were isolated in pure culture for further characterization (2).
FIG. 1.
Schematic representation of our method of rapidly screening environmental microbial colonies for antiviral activity. The steps of the screening method are as follows: environmental microbes are cultured on AR agar plates (A); the microbial culture is picked up by using the edges of a dialysis membrane (B) and transferred onto an agar-covered virus-infected-cell monolayer (C); the colony-containing agar slant is placed onto the virus-infected agar-covered cells (D); the upper colony-containing agar culture is removed and saved, and the cell culture is incubated for 2 to 3 days (E); and cell monolayers are stained to show viral plaques and zones of plaque inhibition (F).
All the microbial colonies corresponding to the halo of viral plaque reduction were picked up and isolated in pure culture in a new plate of AR medium for characterization. A slight modification of the above-described method was used for this purpose. A virus-infected subconfluent cell monolayer was used, with about one-third of the number of cells used in the first screening, for the precise evaluation of cytotoxic activity. Onto these cultures we put an AR disk on which single colonies had been grown and then cut with the edge of a 10-mm-diameter glass tube. After 20 to 30 min of contact, the agar disks supporting the microbial colonies were removed and the cell cultures were incubated for 2 to 3 days until the viral plaques were evident and the cell monolayer became confluent.
Selectivity of the active microbial products appeared as (i) a lack of morphological alteration of cells, (ii) a growth rate of cells which was similar to that of control cells and reached confluence within 3 days, and (iii) a normal uptake of vital stain used to bring the viral plaques into evidence. Biologically active microorganisms underwent a process of extraction and purification of their antiviral substances (5, 6).
Table 1 reports the results achieved in 6 months of screening by this rapid method. About 105 microbial colonies were analyzed in all. This means that one technician was able to process between 20 and 30 plates per week for one or two viruses. Each microbial culture plate was generally used for only one cell-virus combination. Good results could be obtained with all the different viruses and cell lines indicated above, although more than 80% of assays were performed with both a single DNA virus (HSV-1) and a single RNA virus (poliovirus Sabin type 1).
TABLE 1.
Approximate numbers of microbial colonies screened in 6 months by a single operator and isolation of microorganisms with apparent antiviral activities
| Microorganisms | Total no. of colonies screened | No. of active colonies in preliminary screening | No. of colonies with apparently selective activity | No. of new compounds isolated and identifieda |
|---|---|---|---|---|
| All types | ∼96,000 | 585 | 26 | 5 |
| Gram positive | ∼62,000 | 220 | 9 | 4 |
| Gram negative | ∼30,000 | 325 | 6 | 1 |
| Fungi or actinomycetes | ∼4,000 | 40 | 11 |
Among the microbial colonies selected, five new chemical formulae were identified and their chemicals were purified from some bacilli and bacteria (5–7). The original new compound 5-(p-methoxy-phenyl)-3-acetate-1,2,4-pentatriole (Karalicin) was patented.
The novel method of screening microbial colonies for antiviral activities presented in this work shows interesting advantages: an ease of execution and the possibility of analyzing greater numbers of microbial colonies than the number described so far in any report. In fact, in our test, it is possible to challenge microbial cultures directly with agar-covered infected-cell monolayers and obtain results which can be read within 2 to 3 days. With this system it is possible for only one operator to screen thousands of microbial colonies in just a few months. Furthermore, this method allows the detection of antiviral activities in those colonies which rapidly lose their productivity after their first passage in test media. These colonies can be missed by traditional screening methods with subculture-broth extracts.
Acknowledgments
This work was supported by a grant from the CNR of Italy, PF41 no. 95.00889, to R.P.
REFERENCES
- 1.Averett D R. Anti HIV compound assessment by two novel high capacity assays. J Virol Methods. 1989;23:263–276. doi: 10.1016/0166-0934(89)90159-6. [DOI] [PubMed] [Google Scholar]
- 2.Baron E J, Weissfeld A S, Fuselier P A, Brenner D J. Classification and identification of bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 249–264. [Google Scholar]
- 3.Giromes R, Jofre J T, Bosch A. Isolation of marine bacteria with antiviral properties. Can J Microbiol. 1989;35:1015–1021. doi: 10.1139/m89-169. [DOI] [PubMed] [Google Scholar]
- 4.Kamei Y, Yoshimizu M, Ezura Y, Kimura T. Screening of bacteria with antiviral activity from fresh water salmonid hatcheries. Microbiol Immunol. 1988;32:67–73. doi: 10.1111/j.1348-0421.1988.tb01366.x. [DOI] [PubMed] [Google Scholar]
- 5.Lampis G, Deidda D, Maullu C, Madeddu M A, Pompei R, Delle Monache F, Satta G. Sattabacins and sattazolins: new biologically active compounds with antiviral properties extracted from a Bacillus sp. J Antibiot. 1995;48:967–972. doi: 10.7164/antibiotics.48.967. [DOI] [PubMed] [Google Scholar]
- 6.Lampis G, Deidda D, Maullu C, Madeddu M A, Pompei R, Delle Monache F, Satta G. Karalicin, a new biologically active compound from Pseudomonas fluorescens/putida. I. Production, isolation, physico-chemical properties and structure elucidation. J Antibiot. 1996;49:260–262. doi: 10.7164/antibiotics.49.260. [DOI] [PubMed] [Google Scholar]
- 7.Lampis G, Deidda D, Maullu C, Madeddu M A, Pompei R, Delle Monache F, Satta G. Karalicin, a new biologically active compound from Pseudomonas fluorescens/putida. II. Biological properties. J Antibiot. 1996;49:263–266. doi: 10.7164/antibiotics.49.263. [DOI] [PubMed] [Google Scholar]
- 8.Netzer W J. Emerging tools for discovering drugs. Biotechnology. 1990;8:618–622. doi: 10.1038/nbt0790-618. [DOI] [PubMed] [Google Scholar]
- 9.Omura S. Philosophy of new drug discovery. Microbiol Rev. 1986;50:259–279. doi: 10.1128/mr.50.3.259-279.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prusoff W H, Limand T S, Zucker M. Potential of targets for antiviral chemotherapy. Antivir Res. 1986;6:311–328. doi: 10.1016/0166-3542(86)90014-8. [DOI] [PubMed] [Google Scholar]
- 11.Schwöbel W, Streissle G. Attempts to standardize the screening for antiviral drugs by in vitro tests. Chemotherapy. 1979;25:268–278. doi: 10.1159/000237850. [DOI] [PubMed] [Google Scholar]
- 12.Silver L, Bostiam K. Screening of natural products for antimicrobial agents. Eur J Clin Microbiol Infect Dis. 1990;9:455–461. doi: 10.1007/BF01964283. [DOI] [PubMed] [Google Scholar]
- 13.Take Y, Kubo T, Takemori E, Inouye Y, Nakamura S. Biological properties of streptonigrin derivatives. III. In vitro and in vivo antiviral and antitumor activities. J Antibiot. 1989;42:968–976. doi: 10.7164/antibiotics.42.968. [DOI] [PubMed] [Google Scholar]
- 14.Talenti A, Smith T H. Screening with a shell vial assay for antiviral activity against cytomegalovirus. Diagn Microbiol Infect Dis. 1989;12:5–8. doi: 10.1016/0732-8893(89)90036-9. [DOI] [PubMed] [Google Scholar]
- 15.Whitley R J, Alford C A. Developmental aspects of selected antiviral chemotherapeutic agents. Annu Rev Microbiol. 1978;32:285–300. doi: 10.1146/annurev.mi.32.100178.001441. [DOI] [PubMed] [Google Scholar]