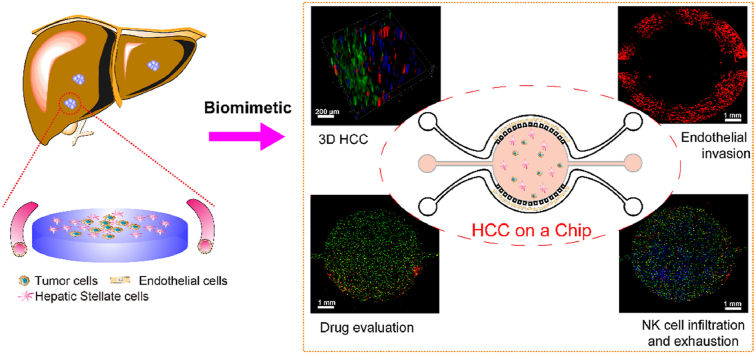
Abstract
Hepatic stellate cells (HSCs) represent a significant component of hepatocellular carcinoma (HCC) microenvironments which play a critical role in tumor progression and drug resistance. Tumor-on-a-chip technology has provided a powerful in vitro platform to investigate the crosstalk between activated HSCs and HCC cells by mimicking physiological architecture with precise spatiotemporal control. Here we developed a tri-cell culture microfluidic chip to evaluate the impact of HSCs on HCC progression. On-chip analysis revealed activated HSCs contributed to endothelial invasion, HCC drug resistance and natural killer (NK) cell exhaustion. Cytokine array and RNA sequencing analysis were combined to indicate the iron-binding protein LIPOCALIN-2 (LCN-2) as a key factor in remodeling tumor microenvironments in the HCC-on-a-chip. LCN-2 targeted therapy demonstrated robust anti-tumor effects both in vitro 3D biomimetic chip and in vivo mouse model, including angiogenesis inhibition, sorafenib sensitivity promotion and NK-cell cytotoxicity enhancement. Taken together, the microfluidic platform exhibited obvious advantages in mimicking functional characteristics of tumor microenvironments and developing targeted therapies.
KEY WORDS: HCC-on-a-chip model, Hepatic stellate cells, Tumor microenvironment, Endothelial invasion, Sorafenib resistance, NK cell Exhaustion, LIPOCALIN-2, Personalized anti-cancer therapies
Graphical abstract
A hepatocellular carcinoma (HCC)-on-a-chip was developed to mimic liver cancer and critical events, including tumor cell proliferation, endothelial cell invasion, tumor cell drug resistance, NK cell infiltration and exhaustion were assessed.
1. Introduction
As the primary malignancy of hepatocytes, hepatocellular carcinoma (HCC) is the most common form of primary liver cancer and remains a highly lethal disease1,2. Meanwhile, HCC drug resistance is an impenetrable gap that requires the discovery of potential mechanism and the development of corresponding therapeutic drugs. HCC progression and drug effectiveness is strongly influenced by the surrounding tumor microenvironment (TME), which comprises hepatic stellate cells (HSCs), tumor-associated macrophages (TAMs), stromal and endothelial cells, and the underlying extracellular matrix (ECM)3, 4, 5. Targeting TME of HCC patients is an effective strategy for tumor therapies, which is involved in controlling tumor metastasis and overcoming drug resistance6,7. Therefore, understanding the complexity of the TME in HCC is crucial to designing novel combinatorial therapy for therapeutic interventions.
As a significant component of hepatic TME, activated HSCs are thought to contribute to the occurrence and development of HCC by producing ECM proteins and inflammatory cytokines8,9. High expression of alpha-smooth muscle actin (α-SMA), an HSC activation marker, was reported to have important impacts on cancer progression by altering the composition of the ECM and triggering signaling cascades in hepatoma cells, which was associated with poor prognosis in HCC patients10,11. However, the detailed crosstalk between cancer cells and HSCs in tumor progression and mechanisms underlying tumor drug resistance remain elusive.
Possessing human-specific biophysical and biochemical factors, the TME are hard to recapitulate either by conventional in vitro planar cell models or in vivo animal models, which greatly hindered decoding the complexity and functional characteristics of in vivo TME12,13. By offering innovative biomimetic platforms, tumor-on-a-chip technology takes advantages in presenting integrated features to mimic TME including 3D scaffolding, multicellular culture, and a vasculature system to simulate dynamic flow in vivo14,15. Therefore, it is on great demand to utilize the specific microfluidic chip model to study the detailed crosstalk between cancer cells and HSCs in tumor progression by integrating tumor microvasculature and tumor-stromal microenvironment factors16, 17, 18, 19.
LIPOCALIN-2 (LCN-2), also known as neutrophil gelatinase-associated lipocalin (NGAL), is a small and secretory protein that is upregulated and involved in the development of serious types of cancer20. Recently, LCN-2 has been demonstrated to mediate key roles in drug resistance and immunomodulation of tumor cells via binding siderophore-complexed ferric iron with high affinity21,22, making it a potential therapeutic target for regulating the TME of liver cancer. In this study, we have proposed a novel strategy to recapitulate the liver cancer by tri-culturing HCC cells with HSCs and endothelial cells (or NK cells) forming a reconstituted liver cancer microenvironment and assess the function of HSCs on tumor progression and anti-tumor therapy. Using this HCC-on-a-chip model, we have investigated the interaction between cancer cells and HSCs, through which to regulate several key processes including endothelial cell invasion, drug resistance and immune exhaustion. Activated HSCs were proved to enhance endothelial cell invasion and contribute to HCC drug resistance in the 3D biomimetic HCC chip. Moreover, natural killer (NK) cell-mediated infiltration and cytotoxicity to tumor cells were inhibited, accompanied with elevated NK-cell exhaustion level. Additionally, we have identified a key factor LIPOCALIN-2 (LCN-2) in remodeling TME and affecting HCC progression through cytokine array and RNA sequencing analysis. Lastly, robust anti-tumor effects were determined by LCN-2 targeted therapy both in vitro 3D biomimetic chip and in vivo mouse model. LCN-2 ablation was shown to inhibit angiogenesis, promote sorafenib sensitivity and NK-cell cytotoxicity. Taken together, the microfluidic HCC-on-a-chip platform exhibited great potential on recapitulating and characterizing HCC TME, with promising applications in enabling personalized anti-cancer therapies.
2. Materials and methods
2.1. Chip fabrication and preparation
The fabrication protocols of the tri-cell culture microfluidic device were previously reported16. Briefly, the microfluidic device was fabricated according to standard photolithography and soft lithography procedures. The negative photoresist SU-8 (MicroChem, Westborough, MA, USA) pattern on the silicon wafer was fabricated with a photomask. The silicon wafer was then silanized with trimethylchlorosilane (Thermo Fisher Scientific, Waltham, MA, USA) to facilitate polydimethylsiloxane (PDMS) mold release. PDMS prepolymer (Dow Corning, Wiesbaden, Germany) was poured onto the silicon wafer and cured at 80 °C for 1 h. The devices were then cut out by a razor blade, the fluidic connection ports were punched, and bonding to a glass slide was done after oxygen plasma of both surfaces and conformal contact.
2.2. Cell culture in the HCC-on-a-chip model
HCC cell line HCCLM3 was acquired from CellCook BioTech (Guangzhou, China) and cultured in RPMI-DMEM (KGM12800N, KeyGEN BioTech, Nanjing, China) supplemented with 10% FBS (FSP500, Excell BioTech, Shanghai, China). LX2 cell line was acquired from Procell BioTech (Wuhan, China) and cultured in RPMI-DMEM supplemented with 10% FBS. HUVECs were acquired from Procell BioTech and cultured in the ECM (1001, Sciencell BioTech, Carlsbad, CA, USA). Patient-derived hepatocellular carcinoma cells (Liver-C-01-20170726) were obtained from the Shanghai Outdo BioTech (Shanghai, China) and cultured in DMEM supplemented with 10% FBS.
NK-92 cell line was acquired from Procell BioTech and cultured in the special medium (CM-0530, Procell BioTech) containing 12.5% FBS, 12.5% horse serum, 0.2 mmol/L inositol, 0.1 mmol/L β-mercaptoethanol, 0.02 mmol/L folic acid, 100 U/mL recombinant IL-2. Firstly, the microfluidic device was coated with poly-d-lysine (1 mg/mL), and then washed 4 times with deionized water; hydrophobicity was restored in dryer at 95 °C overnight. Next, the HCCLM3 cells alone or combined with LX2 cells were resuspended in Matrigel (356234, Corning, NY, USA) at a total density of 10 × 106/mL, injected in the central chamber of the microfluidic device using a syringe, and incubated for 30 min at 37 °C for cross-linking. After this, the lateral channels were coated using a 10% Matrigel solution and incubated for 30 min at 37 °C. HUVECs at the same density (10 × 106 cells/mL) were seeded on the lateral channels using a pipette and adhered for 6 h. Then, the cells were exposed to normal medium during the experiment. On Days 1 and 5, the cell culture was evaluated using Thunder wide field high resolution imaging system (Leica Biosystem, Wetzlar, Germany).
2.3. Live cell staining
HCCLM3, LX2, HUVECs and NK-92 cells were stained with fluorescent markers to monitor cell migration. More specifically, we labeled HCCLM3, LX2, HUVECs and NK-92 cells with CFDA-SE (green; 65-0850-84, Thermo Fisher Scientific), CM-Dil (red; C7001, Thermo Fisher Scientific), and CMF2HC (blue; C12881, Thermo Fisher Scientific), respectively. Briefly, the cells were counted and suspended in PBS at 1 million cells/mL. CellTracker labeling agent (3 μL) was added per milliliter of cell suspension. The cell suspension was placed in the incubator for 20 min and then centrifuged (400×g, 5 min, Scilogex llc, Rocky Hill, CT, USA) and washed twice with medium to remove the staining excess.
2.4. Cell isolation from the chip model
After each culture time, the Matrigel from the central chamber was retrieved by punching with a 4 mm sterile biopsy punch and placed in 24-well plates. The samples were washed with PBS solution, and the Matrigel was depolymerized by incubation with 1 mL of Cell Recovery Solution (354253, BD Biosciences, Franklin Lake, NJ, USA) for 30 min at 4 °C. After complete release from the Matrigel, the cells were transferred to an Eppendorf tube and centrifuged to a pellet by centrifugation at 300×g for 5 min. The cells were washed with cold PBS and centrifuged again. The different cells were isolate by flow cytometry (FACSAria™ Fusion, BD system). Different cells were transferred to medium for subculture experiments or were lysed for RNA-seq analysis.
2.5. RNA isolation and qPCR
Total RNA was harvested and isolated using TRIzol according to the instruction manual. Total RNA was reverse transcribed with an HiScript III 1st Strand cDNA Synthesis Kit (R312-01, Nanjing Vazyme Biotech, Nanjing, China). The resulting cDNA was used for real-time PCR using the ChamQ Universal SYBR qPCR Master Mix (Q711-02, Nanjing Vazyme Biotech). GAPDH was used as an internal control. Real-time PCR and data collection were performed on a CFX96 instrument (Bio-Rad, Hercules, CA, USA). Primers for qPCR are listed in Supporting Information Table S1.
2.6. RNA-seq analysis
Duplicate RNA samples of the isolated cells were subjected to mRNA sequencing at the LC-Bio Technology Co., Ltd., Hang zhou, China. After the mRNA profiles were generated, StringTie was used to estimate the expression levels of all transcripts by calculating Fragments Per Kilobase of exon model per Million mapped reads (FPKM). The differentially expressed mRNAs were detected with the threshold of |log2(fold change) |>1 and with q-value <0.05 by R (version 3.6.3, https://www.r-project.org/) LPEseq package. RNA sequencing data that support the findings of this study have been deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE208007.
2.7. Immunofluorescence
Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and washed again with PBS. Permeabilization of cells was done with 0.1% Triton X-100 (T8200, Solarbio Science & Technology Co., Ltd., Beijing, China) for 15 min at room temperature, followed by washing with PBS. Anti-alpha smooth muscle actin (α-SMA) (1:100, ab7817, Abcam, USA) was injected through the channels, let to incubate for 5 h at room temperature, washed with PBS, and then incubated with Fluorescein Isothiocyanate (FITC)-conjugated secondary antibodies (1:100; Abcam) for 3 h, washed with PBS, and then incubated with DAPI (1:100 in PBS; Beyotime Biotechnology, Shanghai, China) diluted in PBS for 10 min at room temperature. Images were captured by the Thunder wide field high resolution imaging system (Leica Biosystem).
2.8. Endothelial cell invasion
During cell culture, bright-field images were taken with an inverted microscope to analyze and quantify the extent of endothelial cells remaining on the lateral microchannels or responding to present medium in the Matrigel in the central chamber. Three images of each time point (Days 0, 2, 3, and 5) were analyzed. The length was measured using ImageJ, from the outer limit of the microchannels/hydrogel interface to the furthest point of the elongated cells. After cell culture, the cells in the chip were washed with PBS, and the cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and washed twice with PBS. Permeabilization of cells was done with 0.1% Triton X-100 for 15 min at room temperature, followed by washes with PBS. For F-Actin staining, the Phalloidin-iFluor 594 (ab176757, Abcam) was diluted in PBS with 1% BSA and was subsequently incubated for 1 h. The samples were washed with PBS and kept in PBS in the dark at 4 °C until image analysis. For Von Willebrand Factor (vWF) staining, primary antibody for vWF (ab287962, Abcam) diluted in PBS with 1% BSA was subsequently incubated overnight at 4 °C. Samples were washed with PBS and incubated with the corresponding secondary antibody Alexa Fluor 488 conjugated (1:500; Proteintech Group, Wuhan, China) for 3 h at room temperature. The samples were then washed with PBS, incubated with DAPI (Beyotime Biotechnology) for 10 min at room temperature, and lastly washed twice with PBS. The samples were kept in PBS in the dark at 4 °C until image analysis. Images were captured by the Thunder wide field high resolution imaging system (Leica Biosystem).
2.9. Drug diffusion in the chamber
To quantify the drug diffusion in the Matrigel, we used FITC-Dextran (MW = 10,000, F0918, Tokyo Chemical Industry, Japan) in PBS perfused through one side channel. The experiment was performed under Thunder wide field high resolution imaging system (Leica Biosystem) for time-lapse imaging of the gradient formation.
2.10. Cell viability
To measure cell viability in chip model, stock solutions of prodidium iodide (PI; 2 mg/mL) (P4864, Sigma–Aldrich, USA) were prepared following the supplier instructions. Working solutions were prepared by dissolving at 1:1000 and 1:500, respectively. After the drug treatment, the medium was removed and PI solution was added to incubate for 15 min and then washed with PBS. The SuperView 488 Caspase-3 Assay Kit (40273ES25, Yeasen Biotechnology, Shanghai, China) labeling the apoptotic cells in green also was used to detect the apoptosis in the chip. Afterward, cell viability was evaluated by fluorescent imaging using a Thunder wide field high resolution imaging system. In 2D assays, the cells were washed twice with PBS to remove floating dead cells. Next, viable tumor cells were stained with the CAM solution (C2012, Beyotime Biotechnology) (no PI was used). To measure the 50% growth inhibition (GI50) in plate, the HCCLM3 cells seeded in 96 well plates were treated with different concentrations of sorafenib for 48 h. Then cell viability was measured using the Cell Counting Kit-8 (GK10001, YIFEIXUE BIO TECH, Nanjing, China) assay.
2.11. Drug treatments
To measure the effects of different drugs on the cancer cell, 25 or 50 μmol/L of sorafenib (S7397, Selleck chemicals, Houston, TX, USA), 5-fluorouracil (5-FU) (HY-P90006, MedChemExpress, NJ, USA), oxaliplatin (S1224, Selleck chemicals) were added in one channel for 48 h. To measure how LCN-2 inhibition affected the antitumor therapy capacity in the chip model, the Matrigel was mixed with 100 ng/mL human LCN-2 antibody (MAB1757, R&D Bio-Techne, Los Angeles, CA, USA), and cell viability was assessed after treatment.
2.12. NK-92 cells invasion and cocultured with HCCLM3 in the HCC-on-a-chip model
After the HCCLM3 cells (labeled with CFDA-SE, green) cultured in the tumor-on-a-chip for 3 days, the NK-92 cells were loaded in one channel for another 48 h, then the PI was added in the same channel for 15 min and then washed with PBS. For NK-92 cells cocultured with/without HCCLM3 and LX2 cells in the HCC-on-a-chip model, NK-92 cells (labeled with CFDA-SE, green) alone or coculture with HCCLM3 cells and LX2 cells for 1 and 3 days, then the PI was added in both channels for 15 min and then washed with PBS. Images were captured by the Thunder wide field high resolution imaging system (Leica Biosystem).
2.13. NK-92 cells mitochondrial imaging
For mitochondrial imaging, fresh isolated NK cells were stained with MitoTracker Green (C1048, Beyotime Biotechnology) for 30 min, followed by Hoechst 33342 (C0031, Solarbio Science & Technology Co., Ltd.) for nuclear staining. Images were captured by the Thunder wide field high resolution imaging system (Leica Biosystem). The mean relative length of mitochondria in each group was obtained by analyzing the length of each mitochondria in a single cell and statistically analyzing 10 cells.
2.14. Flow cytometry analysis
For CD107a detection, the NK cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and washed twice with PBS. Then stained with PE/Dazzle™594 anti-human CD107a antibody (328645, Biolegend, San Diego, CA, USA) for 30 min in dark before flow cytometry (Gallios, Beckman, Bria, CA, USA) analysis. For PI and Annexin V staining, the cells were harvested and the pellets were washed twice with cold cell staining buffer (2% FBS in PBS). 1 × 106 cells were resuspended in 500 μL Annexin V Binding Buffer with 5 μL AnnexinV-FITC and 5 μL PI Viability Staining Solution (KGA108, KeyGEN BioTech). The cells were gently vortexed and then incubated at room temperature in the dark for 15 min before flow cytometry (Beckman) analysis.
2.15. Cytokine array and enzyme-linked immunosorbent assay (ELISA)
The conditioned medium from HCCLM3 cells alone, LX2 cells alone or co-culture in the chip were collected and centrifuged at 2500×g, 4 °C for 10 min (Scilogex llc, USA). The clear culture medium was collected for further use. The whole blood of the patients was collected from the vein and placed at room temperature for 30 min, followed by centrifugation at 1000×g for 15 min. The serum was collected for further use. Whole blood was collected from the hearts of mice immediately after euthanasia, and placed at room temperature for 30 min, followed by centrifugation at 1000×g for 15 min. The serum was collected for further use. The cytokine array assay (Human XL Cytokine Array Kit, ARY022B, R&D Systems) and ELISA kits (Human LCN-2 ELISA Kit, 70-EK1116-96, MultiSciences, Hangzhou, China) were performed according to the manufacturer's protocols.
2.16. Bioinformatics
TCGA (http://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/) datasets were utilized to analyze LCN-2 expression in the HCC and non-tumor tissues. The gene expression values were expressed as log2(FPKM +1), and a paired or unpaired t-test was performed to compare two groups. For correlation analyses of LCN-2 expression in liver cancer cell lines with the sensitivity to sorafenib, we used the Liver Cancer Model Repository (LIMORE) dataset (https://www.picb.ac.cn/limore/; dose responses are expressed as Emax values) consisting of 81 human liver cancer cell lines and the Liver Cancer Cell Line (LCCL) dataset (https://lccl.zucmanlab.com/hcc/home; dose responses are expressed as GI50 values) consisting of 28 human liver cancer cell lines. Linear relationships between gene expression and drug response were determined by the Pearson correlation analysis. Immune cell infiltration in HCC tissue was evaluated using the CIBERSORT algorithm. The LM22 gene file of CIBERSORT was used to define 22 immune cell subsets and analyse LCN-2 high expression HCC tissue and LCN-2 low expression HCC tissue data, which were obtained from the CIBERSORT web portal (http://CIBERSORT.stanford.edu/). This default signature matrix of 100 permutations was used in this algorithm. The visualisation of the results from CIBERSORT was carried out using the R packages, corplot, vioplot, ggplot2 and ggpubr.
2.17. Clinical sample preparation
Fresh hepatocellular carcinoma tissues were obtained from Affiliated Hospital of Nanjing University of Chinese Medicine (Nanjing, China). The tissues were fixed in 10% formalin for 7 days at room temperature. The samples were then dehydrated, paraffin-embedded and sectioned at 6 μm for staining. The serum of both the non-cancer donors and hepatocellular carcinoma were obtained from Affiliated Hospital of Nanjing University of Chinese Medicine. The collection and use of human samples were approved by the Ethics Committee of Affiliated Hospital of Nanjing University of Chinese Medicine (IRB number: 2021NL-099-01) following the Declaration of Helsinki Ethical Guidelines.
2.18. Immunohistochemistry (IHC) of hepatocellular carcinoma tissues
For tissue samples, the slides were deparaffinized in the xylene and degraded alcohols. After blocking with 20% horse serum, the slides were incubated with the LCN-2 primary antibody at 4 °C overnight and rinsed with PBS, followed by incubation with a biotinylated universal secondary antibody. After PBS washing, the slides were incubated with the avidinbiotin detection complex and then developed with 3,3′-diaminobenzidine (DAB) solution. Immunohistochemical staining was semiquantitatively analyzed using image J. The percentage of positive cells was scored on a scale of 1–6: 1 if 0%–10% of cells were positive, 2 if 11%–30% of cells were positive, 3 if 31%–50% of cells were positive, 4 if 51%–70% of cells were positive, 5 if 71%–90% of cells were positive, and 6 if 91%–100% of cells were positive.
2.19. shRNA transfection
shRNA targeting human LCN-2 was cloned into the pLV3ltr-Puro-U6 vector with restriction enzymes BamHI and EcoRI. shRNA transfection was performed using Lipofectamine™ 3000 transfection reagent (L3000015, Invitrogen, Thermo Fisher Scientific) following manufacturer's instruction.
2.20. Immunoblotting analysis
Total cellular proteins were extracted using lysis buffer (R0030-100, Solarbio Science & Technology Co., Ltd.) supplemented with protease inhibitor cocktail (P0100, Solarbio Science & Technology Co., Ltd.). The extracted proteins were quantified using the BCA protein assay kit (23227, Thermo Fisher Scientific). The protein samples were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane (PVDF), blocked and tested with the indicated primary antibodies and corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies (YSFA01/YSFA02, YIFEIXUE Bio Tech). Enzymatic signals were visualized with ECL detection reagent (BL520A, Biosharp, Hefei, China). The following antibodies were used: LCN-2 and GAPDH (1:10,000, MB001, Bioworld Technology, Nanjing, China).
2.21. In vitro clonogenic assay
The HCCLM3 cells were seeded in 6-well or 12-well tissue culture plates at single-cell densities. After treatment with conditional medium and sorafenib (7.5 μmol/L) for 10 days, the cells were fixed and stained with crystal violet (0.05% w/v in formalin). The colony number was counted under an optical microscopy.
2.22. Tube formation assays
A 96-well plate was coated with 50 μL Matrigel (Corning) and incubated at 37 °C for 30 min. 2 × 104 HUVECs cells suspended in 50 μL conditional medium from different chip model was then seeded. After 8 h incubation at 37 °C, tube formation was examined under a microscope and complete tubular structures were counted. ‘Total Branching’ was analyzed using Image J.
2.23. Nude mice xenograft experiments
BALB/c-Nude mice (5-week-old, male) were purchased from GemPharmatech (Nanjing, China). Animal studies were approved by the Institutional and Local Committee on the Care and Use of Animals of Nanjing University of Chinese Medicine (approval number: 202202A035, Nanjing, China). All animals received humane care according to the National Institutes of Health (USA) Guidelines. We complied with the principles of the 3Rs and respected the highest ethical and animal welfare standards in carrying out the animal experiments. The mice were subcutaneously inoculated with HCCLM3 cells with or without LX2 cells (1:1) (total 1 × 106) suspended in 0.1 mL of Matrigel into the flanks of the mice. Tumor volumes were calculated using the formula: (length × width2)/2. After 7 days, when the tumors reached 50–100 mm3, the mice were randomly assigned to five treatment groups: (I) HCCLM3 (5% DMSO+45% PEG400 + 50% H2O, oral gavage); (II) sorafenib + HCCLM3 (sorafenib 30 mg/kg, diluted in 5% DMSO+45% PEG400 + 50% H2O, oral gavage); (III) HCCLM3+LX2 (5% DMSO+45% PEG400 + 50% H2O, oral gavage); (IV) sorafenib + HCCLM3+LX2 (sorafenib 30 mg/kg, diluted in 5% DMSO+45% PEG400 + 50% H2O, oral gavage); (V) LCN-2 antibody + sorafenib + HCCLM3+LX2 (sorafenib 30 mg/kg, diluted in 5% DMSO+45% PEG400 + 50% H2O, oral gavage + LCN-2 neutralizing antibody, intraperitoneal injection). The mice were euthanized after 3 weeks of drug treatment, and the tumors were weighing with analytical balance.
2.24. Statistical analysis
All data analyses were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA) or SPSS version 18 (IBM SPSS Statistics GradPack, Armonk, NY, USA). Data were presented as mean ± SEM. Two-tailed Student's t test was performed when only 2 groups were compared. One-way ANOVA followed by Dunnett's posttests were used to compare differences between multiple groups unless otherwise stated. ∗P < 0.05, ∗∗P < 0.01.
3. Results and discussion
3.1. HCC-on-a-chip
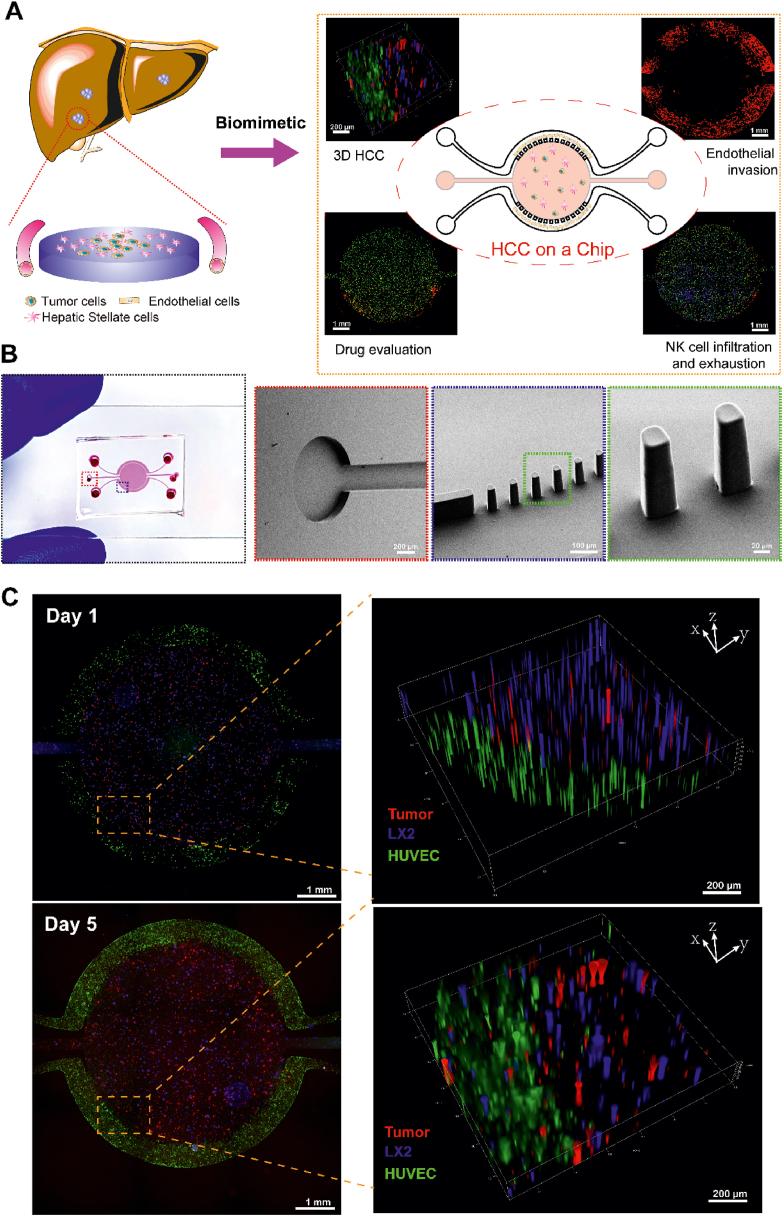
TME plays vital roles in the tumor malignant biological behaviors such as cell proliferation, angiogenesis, drug resistance and immune escape23, 24, 25. As one of hallmarks of tumor, aberrant angiogenesis is closely associated with tumor proliferation, drug resistance and metastasis26. Solid tumors can generate a harsh microenvironment including nutrient depletion, acidic pH and waste product accumulation, which play important roles in affecting immune responses27,28. Therefore, it is on great demand to develop a biomimetic platform to precisely mimic cellular heterogeneity, dynamicity, and intercellular cross-talk within human TME. Microfluidic tumor-on-a-chip technology has provided a unique tool to perform tumor pathology study and screen personalized anti-cancer drugs13. Here a three-channel microfluidic device was developed to culture three types of cells in a spatially organized manner to mimic HCC microenvironments and study the roles of diverse cells in remodeling the TME by altering the secretome, proteome and metabolome. By using this HCC-on-a-chip model, several key events were evaluated including endothelial cell invasion, drug resistance, immune cells infiltration and exhaustion (Fig. 1A).
Figure 1.
HCC-on-a-chip. (A) Schematic representation of the HCC-on-a-chip to mimic liver cancer. The HSCs are one of the key components in the HCC microenvironments and blood vessels are essential for nutrition supply and metabolites elimination. The chip was designed to contain a central chamber to mimic tumor microenvironments and side channels to mimic blood vessels for nutrients supply. Critical events including tumor cell proliferation, endothelial cell invasion, tumor cell drug resistance, NK cell infiltration and exhaustion were assessed in the HCC-on-a-chip model. (B) Structure of the HCC-on-a-chip. The whole chip was 16 mm × 7 mm (length × width) in dimension containing a round central chamber and two lateral channels. The central chamber was 5 mm in diameter and 120 μm in depth, with a separate inlet and outlet for hydrogel injection. The two lateral channels were not interconnected, allowing for the perfusion of different solutions. Scanning electron microscopy (SEM) of microbarriers and magnified structure of delineated area were shown. (C) The fluorescence microscopy images of HCCLM3 cancer cells (red), LX2 cells (blue) and HUVEC cells (green) were shown on Days 1 and 5 in the HCC-on-a-chip model. Scale bar: 1 mm. Representative areas were displayed with magnified three-dimensional pictures.
The whole chip is 16 mm × 7 mm (length × width) in dimension containing a central chamber which is 5 mm in diameter and 120 μm in depth. The central chamber is surrounded by two lateral channels with a width of 100 μm and trapezoidal barriers are designed between them with a gap from 50 to 60 μm (Fig. 1B, Supporting Information Figs. S1 and S2). The design of the trapezoidal structure and distance of the gap can trap the Matrigel inside the central chambers without leaking into the side channels during the Matrigel loading process. The HCC cells (i.e., HCCLM3) and HSCs (i.e., LX2) were mixed in Matrigel in a 1:1 ratio (total density 10 × 106 cells/mL) and injected into the central chamber to build the 3D scaffolding and multicellular culture system. After Matrigel polymerization, the side channels were lined with endothelial cells [i.e., human umbilical cord endothelial cells (HUVECs)] (total 10 × 106 cells/mL), and perfused with culture medium to generate a blood vessel surrogate that nourished the cells29. To evaluate the feasibility of this system for long-term biometric 3D cell culture, we labeled different cells with different live cell staining fluorescent dyes and examined cell viability during 5 days' co-culture course. As shown in Fig. 1C, the HUVECs grew with a microvasculature morphology after 5 days’ culture, which may be affected by the cytokines secreted by HSCs and HCCLM3 cells in the central chamber. Meanwhile, the HCCLM3 cells and LX2 cells in the central micro-chamber interacted with each other and grew well under favorable conditions. Under bright-field microscopy, we observed the LX2 cells had transformed into fibroblast-liked morphology and the HCCLM3 cells grew on top of the LX2 cells, which was difficult to be detected in a 2D plate culture system (Supporting Information Fig. S3). Taken together, we have successfully built a HCC-on-a-chip platform to mimic the HCC microenvironments, which could be used to evaluate various key tumor biology events, including angiogenesis, drug resistance, immune infiltration and escape processes.
3.2. HSCs activation and endothelial cell invasion
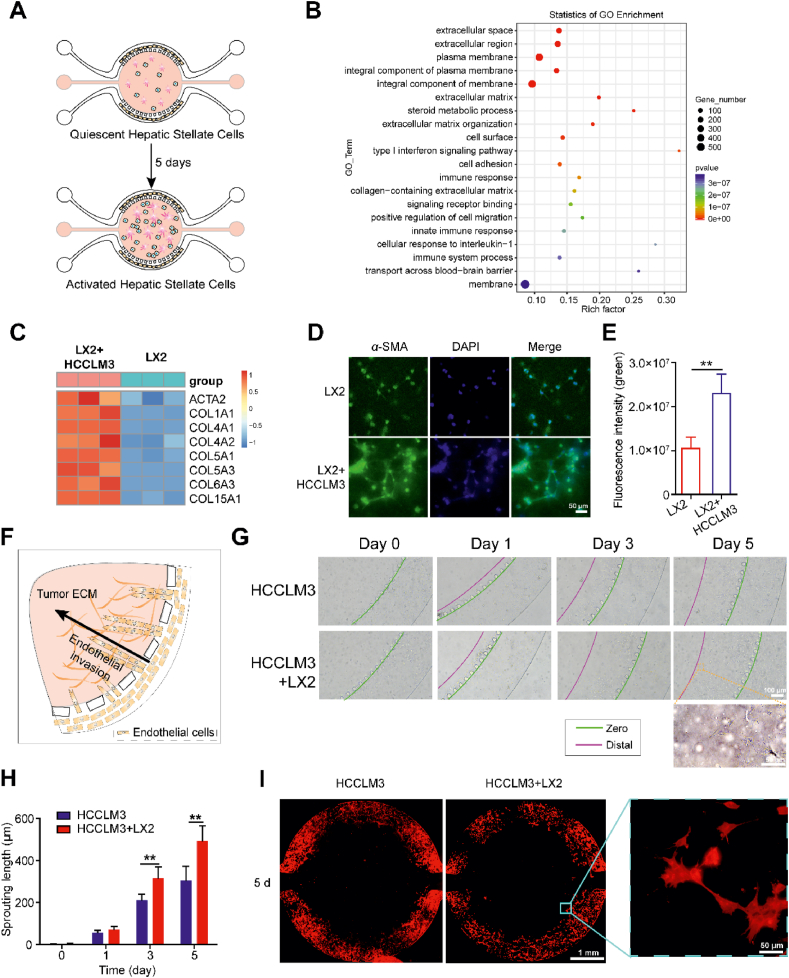
Numerous studies have shown that HCC is highly vascularized and endothelial cell invasion process can be induced by the interaction of tumor cell and HSCs30,31. Therefore, understanding the mechanisms of endothelial cell invasion and identifying the key factors involving in this process are critical to develop precise therapy strategies to cure HCC. We first tested whether HSCs were activated and played a key role in angiogenesis by the regulation of HCC microenvironments. The activation process of HSCs and the molecular changes were evaluated in the 3D HCC-on-a-chip settings (Fig. 2A). Non-labeled LX2 cells were cultured with tumor cells (labeled with CFDA-SE, green) in the collagen mixture with a 1:1 ratio (total density 10 × 106 cells/mL) for 5 days. After 5 days’ culture, we degraded the collagen hydrogel and isolated the total cells. Then, HCCLM3 cells, which were labeled in green fluorescence beforehand, were selectively removed to isolate pure LX2 cells using flow cytometry and cell sorting (Supporting Information Fig. S4). Total RNA from the isolated LX2 cell was extracted and sent for RNA-seq analysis to investigate the molecular changes of HSCs. In comparison to the LX2 cell culture alone group, the co-culture group in the HCC-on-a-chip model exhibited up-regulated genes and signaling pathways associated with several key processes, including cell adhesion and extracellular matrix regulation (Fig. 2B). Interestingly, we observed that multiple genes associated with HSCs activation (e.g., ACTA2, COL1A1, and COL1A4) were up-regulated in the isolated LX2 cells cocultured with HCCLM3 cells in the HCC-on-a-chip model (Fig. 2C). RT-qPCR results confirmed the key HSCs activation markers were highly expressed in the LX2 cells coculture with HCCLM3 cells in the chip model (Supporting Information Fig. S5). Moreover, the protein expression level of α-SMA (encoded by ACTA2 gene) was greatly increased after culture with HCCLM3 cells for 3 days in the HCC-on-a-chip model, which was accompanied by LX2 cells connected with each other in a mesenchymal-like phenotype (Fig. 2D and E). Taken together, these results indicate that LX2 cells were activated in co-cultured with HCCLM3 cells in the HCC-on-a-chip platform.
Figure 2.
Activated HSCs contributed to endothelial cell invasion. (A) Scheme of hepatic stellate cell activation in the HCC-on-a-chip. LX2 cells (no label) and tumor cells (labeled with CFDA-SE, green) were mixed with the Matrigel at a 1:1 ratio (total density 10 × 106 cells/mL) and injected in the HCC-on-a-chip for 5 days. After 5 days' culture, HCCLM3 cells were selectively removed which were labeled in green fluorescence forehand for the isolation of pure LX2 cells by flow cytometry cell sorting. (B) The RNA-seq was performed to analyze the total RNA from the isolated LX2 cells via chip (n = 3 per group). Gene Ontology enrichment analysis was applied to show the upregulated genes and enriched biological processes in LX2 cells from the cell co-culture group. (C) Heat map of multiple genes associated with HSCs activation were shown in the indicated groups (n = 3 per group). (D) Representative image showing α-SMA immunofluorescence staining of LX2 cells in the HCC-on-a-chip model (n = 3 per group). Scale bar: 50 μm. (E) Quantification of the α-SMA fluorescence intensity in indicated groups. (F) Schematic representation of HUVEC in the lateral channels invading towards the central chamber in response to the tumor cytokines microenvironments. (G) Representative bright-field images of endothelial cell sprout in the microfluidic device were shown on Days 1, 3 and 5. (H) Quantification of endothelial cell invasion length at determined time points in the indicated groups (n = 3 per group). (I) Representative images of endothelial cell sprout after F-actin staining. Scale bar: 1 mm. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01.
Based on the HCC-on-a-chip model, we set out to evaluate the endothelial cell invasion process induced by the interaction of activated HSCs and HCC cells. Firstly, we cultured HCCLM3 cells with/without LX2 cell in the HCC-on-a-chip model for 5 days, and collected the conditional medium. Then, the conditional medium was mixed with the Matrigel and injected into the central chamber to build the tumor cytokines microenvironments. After Matrigel polymerization, the HUVECs were lined in the lateral channels to mimic the endothelial cell invasion for 5 days. The endothelial cell invasion was oriented directly toward the central chamber which contained the cytokines from the HCC TME (Fig. 2F). After 5 days, the distance of endothelial cell invasion in the chip model reached nearly 550 μm and the endothelial cell invasion induced by the conditional medium from co-culture cells were farther than the conditional medium from HCCLM3 cells alone (Fig. 2G and H). Furthermore, we stained F-actin in HUVEC in the chip after 5 days, and the results showed the endothelial cell invasion was oriented uniformly toward the central chamber from both the channels in the whole chip, which was hard to be observed in a 2D plate. We also found the leading tip cells were replete with filopodia-like protrusions, morphologically recapitulating in vivo endothelial cell sprouting. With the tip cells continuing to invade and extend in the 3D matrix, they became longer and began to form interconnected branches, indicating the process occurring in the early stages of tumor development (Fig. 2I). VEGF inhibition has been proved an effective method to inhibit malignant progression of liver cancer. Bevacizumab, a VEGF inhibitor, was added to the central chamber to verify the reliability of the model. Results show that the endothelial cell invasion was restrained with the bevacizumab treated, confirming the effectiveness of the model for researching angiogenesis (Supporting Information Fig. S6).
In addition, we performed immunofluorescent staining of Von Willebrand Factor (vWF) in HUVEC cells to evaluate the endothelial properties, which was specifically expressed in endothelial cells and shown as a biological biomarker of endothelial function32. There was no obvious difference in the vWF expression between HUVEC cells invaded into the central chamber and cells stayed in the side channel, which indicated the endothelial properties of the HUVEC were not lost via culture in the chip (Supporting Information Fig. S7). Taken together, the HCC-on-a-chip model exhibited a robust platform to conduct studies in tumor microenvironment-induced angiogenesis and the activated HSCs were examined to facilitate endothelial cell invasion in the 3D biochip.
3.3. Assessment of drug resistance via chip
Drug resistance is a major reason for the failure of tumor targeted therapy33,34, but the efficient evaluation strategy is still lacking with the ability to assess pharmacological activity and decode drug resistance mechanism considering the complexity of tumor microenvironment35. Here we aimed to use the HCC-on-a-chip platform to investigate the killing effects of anti-tumor drugs and find a way to reverse tumor drug resistance. Firstly, to elucidate the molecule diffusion on the chip model, we used FITC-labeled Dextran to mimic drug penetration and verify the diffusion performance with the ability to form concentration gradients in TME containing different stroma cells (Supporting Information Fig. S8A). The HCCLM3 cells co-cultured with/without LX2 cells were mixed with the collagen mixture in the central chamber and endothelial cells were lined in the sided channels for 3 days. After 3 days, the medium was removed and FITC-labeled Dextran was introduced in one side of the chip for 6 h (Fig. S8B‒S8D). The diffusion process was profiled at different time points and the fluorescence intensity was quantified. FITC-labeled Dextran was first distributed in the side channel and then diffused into the Matrigel and reached the central microchamber after 6 h (Fig. S8D).
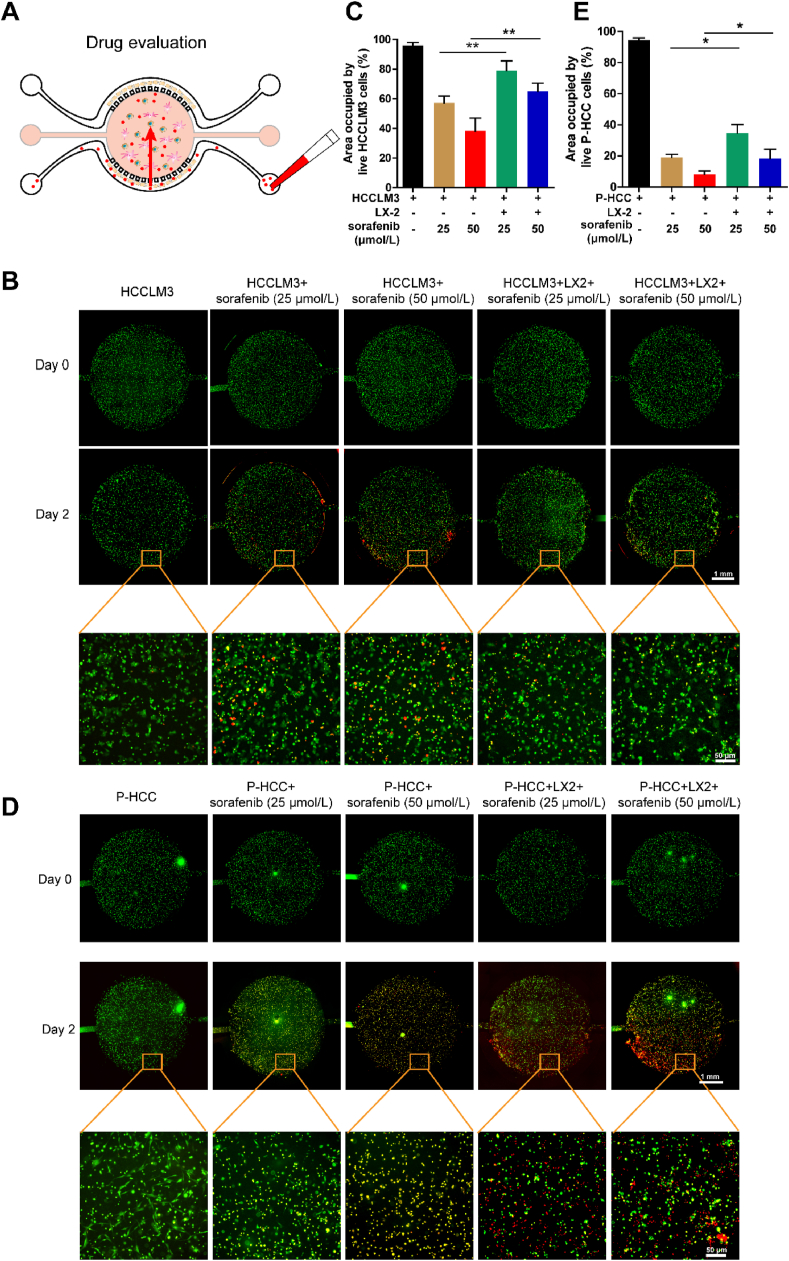
Then, we used the HCC-on-a-chip model to investigate the response of targeted drug (sorafenib)36 and chemotherapeutic drugs (5-FU and oxaliplatin) which were frequently used in clinic for HCC therapy (Fig. 3A). HCCLM3 cells (labeled with CFDA-SE, green) were cultured with/without LX2 cells in the HCC-on-a-chip model for 3 days, and then drugs were added to one side channel of the chip to investigate the tumor cell killing effect at different concentrations. After 48 h, the propidium iodide (PI) was added at the same channel to label the dead cells. The control group without drug treatment showed negligible levels of cell death rates, demonstrating the ability of the chip model on supporting a stable and viable culture environment for HCCLM3 cells (Fig. 3B). Under sorafenib treatment, the HCCLM3 cells exhibited a higher cell viability rate in culture with LX-2 cells compared with cells culture alone (Fig. 3C, Supporting Information Fig. S9A and S9B). The same phenomena were observed when HCCLM3 cells were treated with 5-FU, which also showed a higher cell viability rate when cultured with LX2 in the 3D matrix device (Supporting Information Fig. S10A and S10B). However, there was no significant difference when cells were treated with another chemotherapeutic drug oxaliplatin (Fig. S10C and S10D). These results indicate that the drug killing effect was weakened on HCCLM3 cells at the presence of LX2 cells in the HCC-on-a-chip model, which demonstrated an important role of LX2 cells in mediating drug resistance.
Figure 3.
Assessment of Sorafenib resistance via chip. (A) Schematic representation of drug killing effects on liver cancer cells in the HCC-on-a-chip model. The HCCLM3 cells (labeled in green) were cultured with or without LX2 in the HCC-on-a-chip model for 3 days, then specific drugs were added from one side channel of the chip to investigate the tumor cell killing effect at different concentrations. After 48 h, the PI was added at the same channel to label the dead cells. (B) Representative fluorescence microscopy images of live/dead HCCLM3 cells were shown in the chip after the treatment of sorafenib for 2 days. Scale bar: 1 mm. (C) Quantification of the area occupied by the live HCCLM3 cells (green) in the indicated groups from B (n = 5 per group). (D) Representative fluorescence microscopy images of live/dead patient-derived liver cancer cells (green) were shown in the chip after the treatment of sorafenib for 2 days. (E) Quantification of the area occupied by the patient-derived liver cancer cells (green) in indicated groups from D (n = 3 per group). Scale bar: 1 mm. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01.
Due to the heterogeneous and physiological characteristics, patient-derived cells can present emulated features and capture real situations of in vivo physiology. Thus, we also evaluated the potential of the platform for the concept of precision medicine by culturing patient-derived HCC cells in the device. The role of HSCs in response to the sorafenib and the interaction of HSCs with patient-derived HCC cells were assessed in the TME via chip (Supporting Information Fig. S11). The results reveal the viability of the patient-derived HCC cells in the model were decreased than the HCCLM3 cells under drug treatment at both tested concentrations (Fig. 3D and E), demonstrating the fragility of the patient-derived HCC cells. To expect, the viability of the patient-derived HCC cells was increased after sorafenib treatment in culture with LX2 cells in the model (Fig. 3D and E). Taken together, these results suggest the HCC-on-a-chip model was a powerful platform to evaluate the effects of anti-tumor drugs, through which HSCs were proved to affect tumor microenvironment and contribute to sorafenib resistance both in culture with HCC cell line and patient-derived cells.
3.4. On-chip evaluation of NK cell infiltration and exhaustion
In the past years, immunotherapy has demonstrated great success in fighting against hematological cancers (e.g., acute myeloid leukemia) such as chimeric antigen receptor (CAR) T/natural killer (NK) cells and immune checkpoint inhibitors (ICIs) based therapies37,38. However, due to the specific and complexed signatures of tumor microenvironments generated by the interaction of tumor cells and surrounding stroma cells, moderate success of immunotherapy has been achieved in solid tumors39,40. Thus, it is critical to develop effective immunotherapies by understanding the consequences of the environmental stress on immune cells which were generated by the interaction between tumor cells and stroma cells.
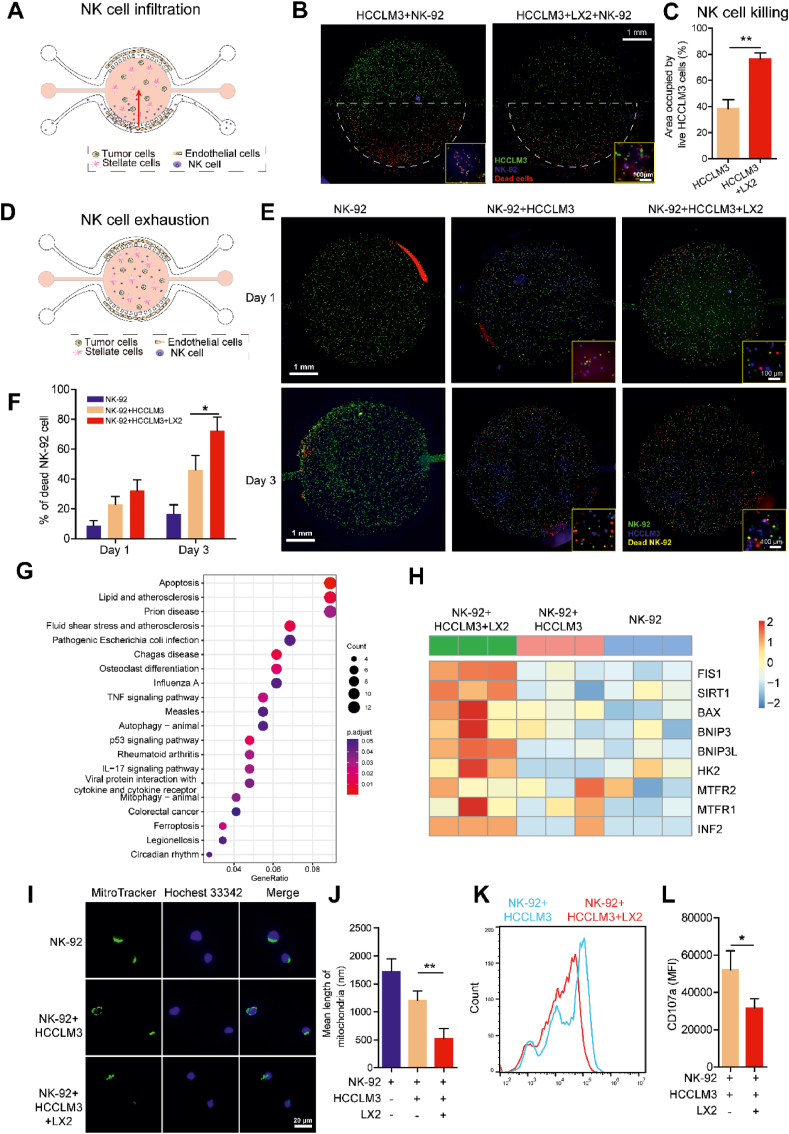
To address the important issue, we used the HCC-on-a-chip platform to evaluate NK cell infiltration and exhaustion for the initial test. Firstly, to evaluate the potential of the platform to study tumor immunotherapy, HCCLM3 cells were mixed with or without LX2 at a 1:1 ratio (total density 10 × 106 cells/mL) and loaded into the central chamber for 3 days to build up the co-culture chip model. Then the NK-92 cells were perfused through one side channel of the chip at a concentration of 5 × 106/mL to evaluate the migration ability to the central chamber and the tumor cell killing effect for 2 days (Fig. 4A). We found that the migration ability of NK-92 cells was decreased towards the tumor microenvironment in the central chamber at the present of LX2 cells (Supporting Information Movies S1 and S2), demonstrating NK-92 cells activity was hindered by the crosstalk between LX2 and HCCLM3 cells. We also tested HCCLM3 cell viability after NK-92 cells penetrated through the Matrigel for 2 days. The fluorescent staining showed the viability of HCCLM3 cells was increased in present with LX2 cells, which indicated LX2 cells played a role in the impairment of the capacity of NK-92 cells to destroy tumor cells in this model (Fig. 4B and C). Notably, though the NK cells could migrate to the central chamber, the NK cell density across the chamber was not homogeneous, which was observed to generate a gradient of NK cell density in the chip with most of the NK-92 cells concentrated in the proximal 2 mm area. To evaluate the NK cell exhaustion induced by the HCC microenvironment, we directly embedded NK-92, HCCLM3 and LX2 cells at a 1:1:1 ratio (total density 10 × 106 cells/mL) in the central chamber (Fig. 4D). We used an apoptosis fluorescent dye in combination with multiple live cell trackers to clearly detect cell–cell interactions and test NK-92 cell viability in the microchip for 1 and 3 days. The results showed the viability of NK-92 cells were decreased at the present of LX2 cells in the HCC-on-a-chip model for 1 and 3 days (Fig. 4E and F). Taken together, the infiltration ability of NK-92 cells was decreased by the interaction between HCCLM3 and LX2 cells, which may generate a harsh microenvironment to induce NK cell exhaustion.
Figure 4.
On-chip analysis of NK cell infiltration and exhaustion. (A) Scheme of NK cell infiltration in the tumor-on-a-chip. The HCCLM3 cells (labeled in green) were mixed with LX2 cells (no label) at a 1:1 ratio (total density 10 × 106 cells/mL) and loaded into the central chamber for 3 days to build up the co-culture system. Then NK-92 cells (labeled in blue) were perfused through one side channel of the chip at the concentration of 5 × 106 cells/mL to evaluate the migration ability towards the central chamber and the tumor cell killing effects for 2 days. After 48 h, the PI staining was applied at the same channel to label the dead cells. (B) Fluorescence microscopy images showed the live and dead HCCLM3 cells after NK-92 cells were induced from the side channel of the chip. Scale bar: 1 mm. (C) Quantitative analysis of area occupied by live HCCLM3 cells (green) in the chip from B (n = 3 per group). (D) Scheme of NK cell exhaustion in the tumor-on-a-chip. The NK-92 cells (labeled in green) were mixed with HCCLM3 cells (labeled in blue) and LX2 (no label) at a 1:1:1 ratio (total density 10 × 106 cells/mL) and loaded into the central chamber to culture for 1 or 3 days. Then PI staining was applied to label the dead cells. (E) Fluorescence microscopy images showed the live and dead NK-92 cells in the chip. Scale bar: 1 mm. (F) Quantification of NK cell death in the indicated groups from E (n = 3 per group). (G) The NK-92 cells were mixed with HCCLM3 cells and LX2 cells at a 1:1:1 ratio and loaded into the central chamber for 3 days. After 3 days of culture, HCCLM3 cells and LX2 cells were selectively removed to isolate pure NK-92 cells using flow cytometry and cell sorting. Gene Ontology enrichment analysis was performed to show the upregulated genes and enriched biological processes in isolated NK-92 cells in the co-culture group via chip. (H) Heat map analysis of mitochondrial fission-related genes in isolated NK-92 cells from the indicated groups (n = 3 per group). (I) Fluorescence microscopy images of mitochondria (labeled with MitoTracker) in isolated NK-92 cells from chip in indicated groups. The nucleus was stained with Hoechst. Scale bar: 20 μm. (J) Quantification of the average lengths of the mitochondria in NK-92 cells of indicated groups from I (n = 10 per group). (K) Flow cytometry analysis of the CD107a expression in isolated NK-92 cells from indicated groups. (L) Quantification of the mean fluorescent intensity of CD107a expression level in isolated NK-92 cells of indicated groups from K (n = 3 per group). Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2023.04.010
The following are the supplementary data related to this article.
Next, we set out experiments to evaluate molecular changes in NK-92 cells in response to the tumor environment generated in the HCC-on-a-chip. After 3 days’ culture with the HCCLM3 and LX2 cells in the central chamber, we degraded the collagen hydrogel and carried out flow cytometry for the isolation of the unlabeled NK-92 cells from the cell suspension, which were sent for RNA-Seq. The GO analysis showed that multiple genes were up-regulated in NK-92 cells which were associated with signaling pathways including apoptosis, mitophage, autophage and ferroptosis in culture with HCCLM3 and LX2 cells (Fig. 4G). Interestingly, several key mitochondrial fission-related genes (e.g. FIS1, SIRT1)41,42 were noticed to be enriched and upregulated in NK cells (Fig. 4H). Previous studies have shown that mitochondrial fragmentation can limit NK cell-based tumor immunosurveillance which is induced by excess mitochondrial fission. To assess the potential effects on mitochondrial fragmentation of NK-92 cells induced by the interaction of HCCLM3 and HSCs, we analyzed mitochondrial morphology in isolated NK-92 cells from chip. We found that freshly isolated NK-92 cells had small fragmented mitochondria after culture with HCCLM3 and LX2 cells in the chip, indicating a high correlation of NK cell exhaustion with mitochondrial fragmentation (Fig. 4I and J). To confirm our results, we detected the expression of CD107a (also known as LAMP-1), which served as a substitute marker of cytotoxic activity expressed by NK cells on the cell surface upon degranulation43. The NK-92 cells showed decreased expression of CD107a after culture with HCCLM3 and LX2 cells in the chip (Fig. 4K and L), suggesting severe impaired function on NK cell activity. Taken together, these results show the activation and cytotoxic activity of NK cells were decreased after culture with HSCs and HCC cells, which was associated with the increased mitochondrial fragmentation in NK cells.
3.5. Identification of LCN-2 as a key factor in HCC development
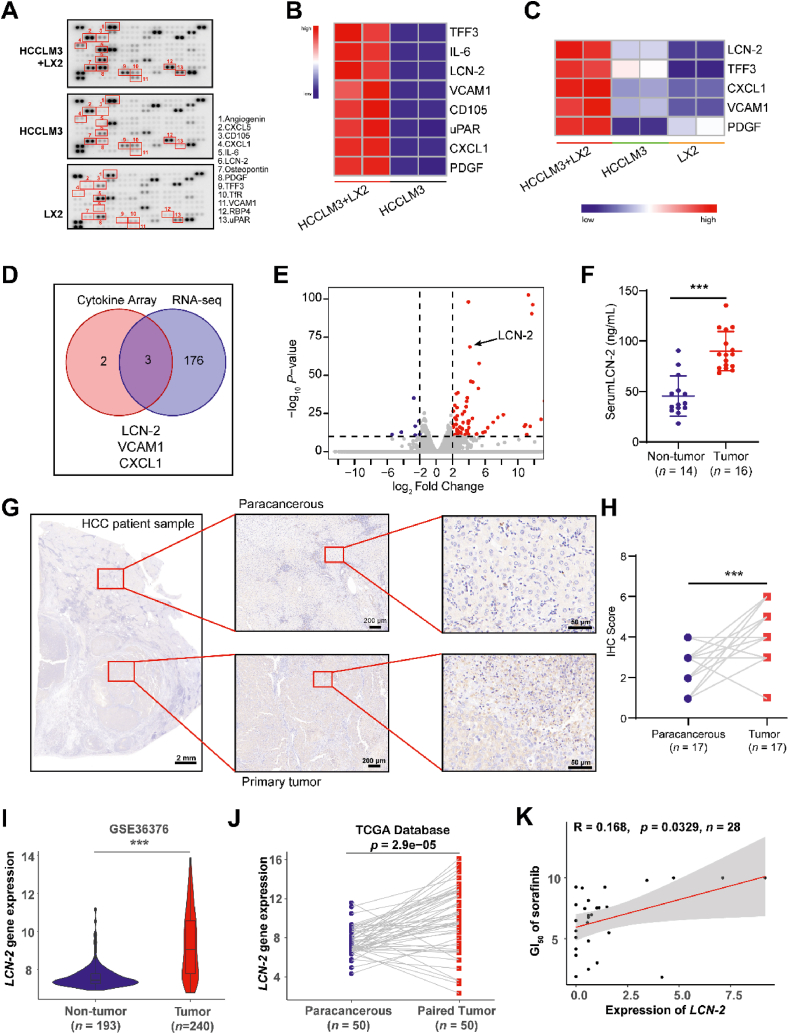
Based on the HCC-on-a-chip model, we found that HSCs were activated in the tumor microenvironment, which exhibited a critical role in enhancing endothelial evasion, drug resistance and NK cell exhaustion. Subsequently, we conducted experiments to investigate molecular changes and mechanism underlying the interaction between tumor cells and HSCs through secretomics and transcriptomics analysis in the biochip model. Firstly, HCCLM3 cells were cultured with LX2 cells in the HCC-on-a-chip model for 5 days and then the conditional medium was gathered from the chip for secretomics assessment. The results indicate that a total of 13 cytokines showed an obvious difference in the microenvironment generated by co-culture of the two types of cells (Fig. 5A and B, Supporting Information Fig. S12A). Interestingly, 5 candidate cytokines were shown to be induced by the interaction of LX2 and HCCLM3 cells, which showed a dramatic increase of secretion level in the microenvironment compared with each cell culture alone (Fig. 5C). To track the source and decipher the function of the increased cytokines, both LX2 and HCCLM3 cells were isolated from the cell mixture in the chip by flow cytometry sorting and sent for RNA-seq analysis. RNA sequencing results from LX2 cells showed upregulated genes after culture with HCCLM3 cells, but the top 5 induced candidate cytokines were not shown in the overlapped list (Fig. S12B and S12C). Bioinformatic analysis demonstrated upregulated genes and signaling pathways in the HCCLM3 cells after culture with LX2 in the chip model, which were mainly associated with ECM remodeling, cell adhesion, cell secretion, cell survival and cell stemness (Supporting Information Fig. S13). These molecular changes indicate the malignant enhancement of HCCLM3 cells after culture with LX2 cells. Notably, by combining the results of RNA-seq and secretomics analysis, three genes were examined with an overlap of increased expression in HCCLM3 cells induced by the interaction of activated LX2 cells (Fig. 5D). The Volcano plot analysis further confirmed the genes upregulated in HCCLM3 cells after culture with LX2 in the chip, by which the iron-binding protein LCN-2 exhibited as a top candidate with the highest change in expression (Fig. 5E).
Figure 5.
LCN-2 was identified as a key factor in HCC development. (A) The HCCLM3 cells and LX2 were cultured in the chip for 5 days and the conditional medium was gathered from the channel for cytokine array assessment. Each cell line cultured alone was set as control. Cytokines were indicated in red which showed a difference in secretion. (B) Heat map analysis showed the significant changed cytokines in the cell co-culture group compared with HCCLM3 cells culture alone. (C) Heat map analysis showed the significant changed cytokines in the cell co-culture group compared with each cell line culture alone. (D) The total RNA of isolated HCCLM3 cells from the HCC-on-a-chip was applied for RNA-seq assessment. Venn diagram indicated the overlapped genes with a dramatic increase by combining the results of RNA-seq in HCCLM3 cells and cytokines analysis in C. (E) The Volcano plot analysis of RNA-seq data in isolated HCCLM3 cells from chip. Three upregulated cytokines were marked which showed an obvious increase of secretion level in the cell co-culture system via chip. (F) LCN-2 expression levels in the serum of non-tumor (n = 14) and HCC patients (n = 16). (G) LCN-2 expression was detected by IHC in paracancerous and primary HCC tumor samples. Scale bar: 2 mm. (H) IHC score analysis of LCN-2 expression across paracancerous and primary tumor sites (n = 17 per group). (I) LCN-2 gene expression analysis in GSE36376 datasets from GEO database containing 193 non-tumor samples and 240 HCC tumor samples. (J) LCN-2 gene expression analysis in 50 paired normal and HCC samples from TCGA database. (K) Correlation between LCN-2 expression and sorafenib GI50, based on the liver cancer cell lines (n = 28) from CTRP. The linear relationship was determined by a two-tailed Pearson correlation analysis. ∗P < 0.05; ∗∗P < 0.01.
To assess whether LCN-2 played an important role in HCC development, clinical samples were firstly used to examine the expression of LCN-2. Patient serum and HCC tumor tissues were collected and sent for detection respectively. The results demonstrated that LCN-2 was highly expressed in serum biopsy compared with samples from normal donors (Fig. 5F). Moreover, there was a significant increase in LCN-2 expression in patient tumor sites compared with paracancerous tissues (Fig. 5G and H). This observation was further validated in the GEO database (Fig. 5I) and paired HCC data based on the cancer genome atlas (TCGA) (Fig. 5J). To assess whether LCN-2 expression was associated with drug response, Cancer Therapeutics Response Portal (CTRP) was used which enabled analyzing the correlations between gene expression and the response to 481 compounds across different cancer cell lines44. According to the data analysis from liver cancer cell lines in CTRP, the expression level of LCN-2 was revealed to have a strong negative correlation with cell sensitivity to sorafenib treatment (Fig. 5K). Moreover, to assess the immune cell infiltration and activity in HCC tissue, the LM22 gene file of CIBERSORT45 was used to define and analyze 22 immune cell subsets from the database in LCN-2high and LCN-2low expression HCC tissues. The ability of NK cell infiltration was observed to decrease significantly in LCN-2high expression HCC tissues (Supporting Information Fig. S14). Taken together, LCN-2 was shown to act as a key factor in HCC development in the tumor-on-a chip model, which may contribute to sorafenib resistance and immune escape.
3.6. Targeting LCN-2 with robust anti-tumor effects
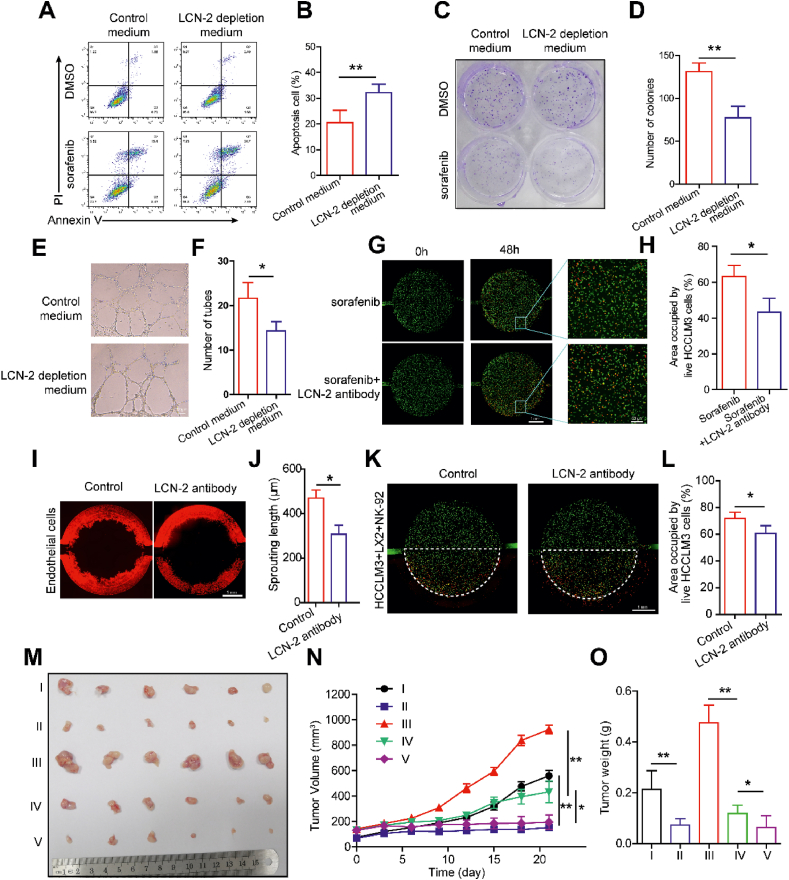
To further explore the role of LCN-2 in mediating endothelial invasion, drug resistance and NK exhaustion, we continued to investigate the therapeutic effects on inhibiting HCC development by targeting LCN-2. A shRNA plasmid was firstly used to knock down the expression of LCN-2 in HCCLM3 cells and the LCN-2 depleted cells were employed to conduct the loss-of-function experiments in the chip model. The results of qPCR and western blotting showed the successful knockdown of LCN-2 expression in HCCLM3 cells (Supporting Information Fig. S15A‒S15C). ELISA assay was also performed to detect the cell culture medium to confirm the depletion of secreted LCN-2 (Fig. S15D). Then, we embedded the LCN-2 depleted HCCLM3 cells cultured with LX2 in the HCC-on-a-chip for 5 days and gathered the culture medium from the both side channels. The conditional medium collected from the chip was used to treat HCCLM3 cells to test cell responses to sorafenib treatment. After the treatment of LCN-2 depleted medium, the apoptosis of HCCLM3 cells was increased under sorafenib treatment compared with control (Fig. 6A and B). The colony formation assay confirmed a similar trend in response to sorafenib treatment, whereby the viability of HCCLM3 cells was observed to decrease in LCN-2 depleted medium (Fig. 6C and D, Supporting Information Fig. S16A‒S16C). Moreover, the tubes of HUVEC in the plate were decreased with the treatment of LCN-2 depleted conditional medium (Fig. 6E and F). These results confirm the crucial role of LCN-2 in the tumor microenvironment, which impacts endothelial invasion and drug resistance.
Figure 6.
Targeting LCN-2 achieved robust anti-tumor effects. (A) The LCN-2 depleted HCCLM3 cells were cultured with LX2 in the chip for 5 days and the culture medium was gathered from side channels. Then the conditional medium was applied to wide-type HCCLM3 cells combined with sorafenib treatment for 48 h. Flow cytometry analysis was conducted to assess the apoptosis of HCCLM3 cells with indicated treatments. (B) Quantification of HCCLM3 apoptosis in indicated groups from A (n = 3 per group). (C) The clone formation of HCCLM3 cells with indicated treatments. (D) Quantification data from C. (E) The HUVECs were suspended in indicated conditional medium and seeded in the Matrigel for 8 h. Representative bright-field images of HUVEC tubes with indicated treatments. Scale bar: 100 μm. (F) Quantification of HUVEC tube formation from E (n = 3 per group). (G) The HCCLM3 cells (labeled with green) and LX2 cells (no label) were mixed with LCN-2 antibody in the Matrigel and cultured in the chip for 3 days. Then sorafenib was induced from side channel for treatment for 2 days. PI staining was applied to label the dead cells. Scale bar: 1 mm. (H) Quantification of the area occupied by the live HCCLM3 cells (green) from G (n = 3 per group). (I) Fluorescence microscopy images of endothelial cell invasion were shown in indicated groups via chip. Scale bar: 1 mm. (J) Quantification of endothelial cell invasion in indicated groups from I (n = 3 per group). (K) The HCCLM3 (labeled with green) and LX2 (no label) cells were mixed with LCN-2 antibody in the Matrigel and cultured in the chip for 3 days. Then NK-92 cells were induced from side channel for treatment for 2 days. PI staining was applied to label the dead cells. Fluorescence microscopy images showed the live and dead HCCLM3 cells in indicated groups. Scale bar: 1 mm. (L) Quantitative analysis of area occupied by live HCCLM3 cells (green) in the chip from K (n = 3 per group). (M) The xenografts were established by subcutaneous injection of HCCLM3 cells alone or the mixture of HCCLM3 and LX2 cells (at a ratio of 1:1) into the nude mice (n = 6 mice per group). When tumors reached 50–150 mm3 after 7 days, the mice were randomly divided to five groups and administrated with different treatments as below: I) HCCLM3; II) HCCLM3+sorafenib; III) HCCLM3+LX2; IV) HCCLM3+LX2+sorafenib; V) HCCLM3+LX2+sorafenib + LCN-2 antibody. Tumor pictures of indicated groups were shown at the end of the experiments. (N) The tumor growth curves of indicated groups were calculated in the BALB/c nude mice xenograft models. Tumor volumes were quantified. (O) The tumor weights of indicated groups were measured at the end of the experiments. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01.
To further confirm the anti-tumor therapeutic efficacy of targeting LCN-2, neutralizing antibody was used for test via HCC-on-a-chip assay. The LCN-2 antibody was mixed in the Matrigel before cells were loaded into the chamber to build the HCC-on-a-chip model. Live and dead staining results showed that the treatment of LCN-2 neutralizing antibody sensitized cancer cells to sorafenib treatment in the HCC-on-a-chip, indicating that targeting LCN-2 could improve the therapeutic efficacy of sorafenib treatment to HCC (Fig. 6G and H). In addition, the endothelial cell invasion was reduced effectively by the treatment of LCN-2 antibody in the HCC-on-a-chip (Fig. 6I and J). Moreover, in the presence of the LCN-2 neutralizing antibody treatment, NK-cell cytotoxicity to HCCLM3 cells was enhanced in the chip (Fig. 6K and L). Taken together, these results demonstrated targeting LCN-2 could effectively inhibit endothelial cell evasion and enhance the efficacy of anti-cancer therapies, including the sorafenib treatment and adoptive NK cell therapy.
Lastly, a mouse model was used to test the antitumor effects of LCN-2 targeted therapy. We implanted the cell mixture of HCCLM3 and LX-2 cells into nude mice to build the xenograft model and started the treatment when tumors reached 50–150 mm3. Xenografts bearing HCCLM3 cells alone were employed as control. The mice were randomly divided to five groups and administrated with different treatments as below: I) HCCLM3; II) HCCLM3+sorafenib; III) HCCLM3+LX2; IV) HCCLM3+LX2+sorafenib; V) HCCLM3+LX2+sorafenib + LCN-2 antibody. Sorafenib was given at 30 mg/kg body weight by oral gavage every other day until the endpoint of experiments. LCN-2 neutralizing antibody was administered at 75 μg per mouse by intraperitoneal injection twice a week until the endpoint of experiments (Supporting Information Fig. S17). The tumor in the HCCLM3+LX2 group grew much faster than that in the HCCLM3 group, indicating that the LX2 cells could support and promote tumor growth (Fig. 6M‒O). Moreover, the tumor in the HCCLM3+LX2+sorafenib group was bigger than that in the HCCLM3+sorafenib group, demonstrating that the LX2 cells decreased the cancer cell sensitivity to sorafenib treatment. Lastly, the combination of LCN-2 neutralizing antibody and sorafenib treatment achieved a much better anti-tumor effect than sorafenib treatment alone in the xenograft model bearing both HCCLM3 and LX2 cells. Collectively, these data suggested that targeting LCN-2 by neutralizing antibody could enhance cancer cell sensitivity to sorafenib treatment in the mouse model, which was consistent with the results observed in the HCC-on-a-chip model. To conclude, LCN-2 targeted therapy demonstrated robust anti-tumor effects both in vitro 3D biomimetic chip and in vivo mouse model validations, which exhibited obvious antitumor phenotypes such as angiogenesis inhibition, sorafenib sensitivity promotion and NK-cell cytotoxicity enhancement.
4. Conclusions
The TME contains a hard extracellular matrix and multiple stromal cells, which increases the difficulty of anti-tumor drug development. In the last decade, many researchers have focused on understanding the modulation of the TME to reveal the mechanisms of HCC. However, a fundamental gap in knowledge is the crosstalk between stromal cells and HCC cells. Typically, the transwell and conditioned medium have been used as common indirect modes to reveal the mechanisms of crosstalk, which lose the ECM and direct connection with different cells, limiting the development of new drugs. Recently, microfluidic chips have great advantages in the high-throughput screening and efficacy evaluation of new drugs by using their specific pore design and modularity. Especially, the organs-on-chips platforms can quickly, personalize, and simulate the physiological and pathological structure of the body in vitro, providing a new strategy for modeling the crosstalk of different cells. Currently, biomimetic reconstructions of the liver, kidney, lung, gut, and tumor have been successfully performed in microfluidic chips46,47, and these organ chips have been applied to disease models to explain the pathogenesis of diseases. Based on these advantages, organs-on-chips may provide a more powerful and human-resembling approach for drug screening compared with conventional animal-based assessments.
In summary, we have developed a powerful in vitro 3D multi-cell culture platform to evaluate the impact of HSCs in remodeling the TME on HCC progression. By utilizing this 3D biomimetic chip, we found HSCs played a critical role in several processes including endothelial cell invasion, drug resistance and immune cell exhaustion. In detail, activated HSCs could enhance endothelial cell invasion and contribute to HCC drug resistance. Moreover, NK cell-mediated cytotoxicity was inhibited and immune exhaustion level was elevated by the interaction of HSCs and tumor cells. Taken together, the observed on-chip phenotypes revealed the importance of microenvironments on tumor development, which became an attractive target to develop therapies by reprogramming TME.
By combining cytokines array and RNA sequencing analysis, the iron-binding protein LCN-2 was discovered as a key factor which may function in remodeling TME. Activated HSCs could increase the expression of LCN-2 in HCC cells and the secreted LCN-2 was observed to associate with HCC progression and sorafenib resistance. Importantly, LCN-2 was demonstrated to be a key regulator of HCC development by validating its expression in clinical samples, such as patient serum and HCC tumor tissues. LCN-2 targeted therapy demonstrated robust anti-tumor effects including angiogenesis inhibition, sorafenib sensitivity promotion and NK-cell cytotoxicity enhancement. To conclude, the multi-cell microfluidic HCC-on-a-chip platform exhibited great advantages in mimic functional characteristics of the in vivo TME, which could be an effective strategy for drug screening and mechanism research.
Acknowledgments
This study was funded by National Natural Science Foundation of China (Nos. 31901010 and 81803916, China); Natural Science Foundation of Jiangsu Province (grant No. BK20180128, China); The Priority Academic Program Development of Jiangsu Higher Education Institutions (Integration of Chinese and Western Medicine, China); Jiangsu Specially Appointed Professorship; Jiangsu Key Discipline Construction Fund of the 14th Five-Year Plan (Biology, China); The Graduate Research & Practice Innovation Program of Jiangsu Province (KYCX21_1743, KYCX22_1992, China).
Author contributions
Xin Han conceived the idea and designed the experiments. Peiliang Shen performed the majority of experimental work and data analysis. Yuanyuan Jia, Weijia Zhou, Weiwei Zheng assisted chip development and performed cell analysis. Yueyao Wu, Suchen Qu, Shiyu Du and Jia Sun assisted animal experiments. Huilian Shi and Siliang Wang collected the clinic sample for analysis. Peiliang Shen and Xin Han wrote the manuscript with feedback from all the authors. Siliang Wang and Huilian Shi supervised the project. All authors have approved the final version of the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.04.010.
Contributor Information
Siliang Wang, Email: wsl_dth@126.com.
Xin Han, Email: xhan0220@njucm.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Vogel A., Meyer T., Sapisochin G., Salem R., Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345–1362. doi: 10.1016/S0140-6736(22)01200-4. [DOI] [PubMed] [Google Scholar]
- 2.Lee T.K., Guan X.Y., Ma S. Cancer stem cells in hepatocellular carcinoma—from origin to clinical implications. Nat Rev Gastroenterol Hepatol. 2022;19:26–44. doi: 10.1038/s41575-021-00508-3. [DOI] [PubMed] [Google Scholar]
- 3.Li X.Y., Shen Y., Zhang L., Guo X., Wu J. Understanding initiation and progression of hepatocellular carcinoma through single cell sequencing. Biochim Biophys Acta Rev Cancer. 2022;1877 doi: 10.1016/j.bbcan.2022.188720. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y., et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. doi: 10.1186/s13046-018-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma A., et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–394 e21. doi: 10.1016/j.cell.2020.08.040. [DOI] [PubMed] [Google Scholar]
- 6.Tian X.P., et al. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics. 2019;9:1965–1979. doi: 10.7150/thno.30958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei C.Y., et al. PKCalpha/ZFP64/CSF1 axis resets the tumor microenvironment and fuels anti-PD1 resistance in hepatocellular carcinoma. J Hepatol. 2022;77:163–176. doi: 10.1016/j.jhep.2022.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Affo S., Yu L.X., Schwabe R.F. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiori M.E., Di Franco S., Villanova L., Bianca P., Stassi G., De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol Cancer. 2019;18:70. doi: 10.1186/s12943-019-0994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makino Y., et al. CTGF mediates tumor-stroma interactions between hepatoma cells and hepatic stellate cells to accelerate HCC progression. Cancer Res. 2018;78:4902–4914. doi: 10.1158/0008-5472.CAN-17-3844. [DOI] [PubMed] [Google Scholar]
- 11.Myojin Y., et al. Hepatic stellate cells in hepatocellular carcinoma promote tumor growth via growth differentiation factor 15 production. Gastroenterology. 2021;160:1741–1754 e16. doi: 10.1053/j.gastro.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Yao Y., et al. Multicompartmental scaffolds for coordinated periodontal tissue engineering. J Dent Res. 2022;101:1457–1466. doi: 10.1177/00220345221099823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu X., et al. Tumor-on-a-chip: from bioinspired design to biomedical application. Microsyst Nanoeng. 2021;7:50. doi: 10.1038/s41378-021-00277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aung A., Kumar V., Theprungsirikul J., Davey S.K., Varghese S. An engineered tumor-on-a-chip device with breast cancer-immune cell interactions for assessing T-cell recruitment. Cancer Res. 2020;80:263–275. doi: 10.1158/0008-5472.CAN-19-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho M.R., et al. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang G., et al. Unidirectional intercellular communication on a microfluidic chip. Biosens Bioelectron. 2021;175 doi: 10.1016/j.bios.2020.112833. [DOI] [PubMed] [Google Scholar]
- 18.Cui X., et al. Dissecting the immunosuppressive tumor microenvironments in glioblastoma-on-a-chip for optimized PD-1 immunotherapy. Elife. 2020;9 doi: 10.7554/eLife.52253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H., et al. Recapitulated crosstalk between cerebral metastatic lung cancer cells and brain perivascular tumor microenvironment in a microfluidic co-culture chip. Adv Sci. 2022;9 doi: 10.1002/advs.202201785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaberi S.A., et al. Lipocalin-2: structure, function, distribution and role in metabolic disorders. Biomed Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.112002. [DOI] [PubMed] [Google Scholar]
- 21.Rehwald C., et al. The iron load of lipocalin-2 (LCN-2) defines its pro-tumour function in clear-cell renal cell carcinoma. Br J Cancer. 2020;122:421–433. doi: 10.1038/s41416-019-0655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi Y., et al. Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis. Science. 2020;369:276–282. doi: 10.1126/science.aaz2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson N.M., Simon M.C. The tumor microenvironment. Curr Biol. 2020;30:R921–R925. doi: 10.1016/j.cub.2020.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bejarano L., Jordao M.J.C., Joyce J.A. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11:933–959. doi: 10.1158/2159-8290.CD-20-1808. [DOI] [PubMed] [Google Scholar]
- 25.Lei X., et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Ackerman D., Simon M.C. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayuso J.M., et al. Microfluidic tumor-on-a-chip model to evaluate the role of tumor environmental stress on NK cell exhaustion. Sci Adv. 2021;7 doi: 10.1126/sciadv.abc2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon J.S., et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A. 2015;112:214–219. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morse M.A., et al. The role of angiogenesis in hepatocellular carcinoma. Clin Cancer Res. 2019;25:912–920. doi: 10.1158/1078-0432.CCR-18-1254. [DOI] [PubMed] [Google Scholar]
- 31.Zhu B., et al. Activated hepatic stellate cells promote angiogenesis via interleukin-8 in hepatocellular carcinoma. J Transl Med. 2015;13:365. doi: 10.1186/s12967-015-0730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randi A.M., Smith K.E., Castaman G. von Willebrand factor regulation of blood vessel formation. Blood. 2018;132:132–140. doi: 10.1182/blood-2018-01-769018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasan N., Baselga J., Hyman D.M. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khalaf K., Hana D., Chou J.T., Singh C., Mackiewicz A., Kaczmarek M. Aspects of the tumor microenvironment involved in immune resistance and drug resistance. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.656364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash J., Pinzani M. Fibroblasts and extracellular matrix: targeting and therapeutic tools in fibrosis and cancer. Adv Drug Deliv Rev. 2017;121:1–2. doi: 10.1016/j.addr.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Huang A., Yang X.R., Chung W.Y., Dennison A.R., Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Targeted Ther. 2020;5:146. doi: 10.1038/s41392-020-00264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., et al. Long-term activity of tandem CD19/CD20 CAR therapy in refractory/relapsed B-cell lymphoma: a single-arm, phase 1-2 trial. Leukemia. 2022;36:189–196. doi: 10.1038/s41375-021-01345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski T.J., Biederstadt A., Rezvani K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat Rev Cancer. 2022;22:557–575. doi: 10.1038/s41568-022-00491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grosser R., Cherkassky L., Chintala N., Adusumilli P.S. Combination immunotherapy with CAR T cells and checkpoint blockade for the treatment of solid tumors. Cancer Cell. 2019;36:471–482. doi: 10.1016/j.ccell.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardiana S., Solomon B.J., Darcy P.K., Beavis P.A. Supercharging adoptive T cell therapy to overcome solid tumor-induced immunosuppression. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaw2293. [DOI] [PubMed] [Google Scholar]
- 41.Zheng X., et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat Immunol. 2019;20:1656–1667. doi: 10.1038/s41590-019-0511-1. [DOI] [PubMed] [Google Scholar]
- 42.Poznanski S.M., et al. Metabolic flexibility determines human NK cell functional fate in the tumor microenvironment. Cell Metabol. 2021;33:1205–12020 e5. doi: 10.1016/j.cmet.2021.03.023. [DOI] [PubMed] [Google Scholar]
- 43.Ng S.S., et al. The NK cell granule protein NKG7 regulates cytotoxic granule exocytosis and inflammation. Nat Immunol. 2020;21:1205–1218. doi: 10.1038/s41590-020-0758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rees M.G., et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman A.M., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dsouza V.L., Kuthethur R., Kabekkodu S.P., Chakrabarty S. Organ-on-chip platforms to study tumor evolution and chemosensitivity. Biochim Biophys Acta Rev Cancer. 2022;1877 doi: 10.1016/j.bbcan.2022.188717. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., Sun L., Zhang H., Shang L., Zhao Y. Microfluidics for drug development: from synthesis to evaluation. Chem Rev. 2021;121:7468–7529. doi: 10.1021/acs.chemrev.0c01289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.