Abstract
Sugar–sugar glycosyltransferases play important roles in constructing complex and bioactive saponins. Here, we characterized a series of UDP-glycosyltransferases responsible for biosynthesizing the branched sugar chain of bioactive steroidal saponins from a widely known medicinal plant Paris polyphylla var. yunnanensis. Among them, a 2′-O-rhamnosyltransferase and three 6′-O-glucosyltrasferases catalyzed a cascade of glycosylation to produce steroidal diglycosides and triglycosides, respectively. These UDP-glycosyltransferases showed astonishing substrate promiscuity, resulting in the generation of a panel of 24 terpenoid glycosides including 15 previously undescribed compounds. A mutant library containing 44 variants was constructed based on the identification of critical residues by molecular docking simulations and protein model alignments, and a mutant UGT91AH1Y187A with increased catalytic efficiency was obtained. The steroidal saponins exhibited remarkable antifungal activity against four widespread strains of human pathogenic fungi attributed to ergosterol-dependent damage of fungal cell membranes, and 2′-O-rhamnosylation appeared to correlate with strong antifungal effects. The findings elucidated the biosynthetic machinery for their production of steroidal saponins and revealed their potential as new antifungal agents.
KEY WORDS: Paris polyphylla var. yunnanensis, Steroidal saponins, Glycosyltransferases, Sugar chain elongation, Antifungal activity
Graphical abstract
Six UDP-glycosyltransferases involved in the biosynthesis of steroidal saponins in Paris polyphylla var. yunnanensis were characterized. The potential of steroidal saponins as antifungal agents was disclosed.
1. Introduction
Saponins are highly diversified steroid and triterpenoid glycoside natural products possessing broad biological activities and important commercial applications in various fields such as pharmaceuticals, foods, and cosmetics1. One of the most important modifications of saponins is glycosylation, which is catalyzed almost exclusively by uridine diphosphate-dependent glycosyltransferases (UGTs) belonging to glycosyltransferase family 1 (GT1)2. Since glycosylation is an important contributor to the structural complexity and diversity, chemical stability, water solubility, and biological activity of saponins, the roles of UGTs in saponin formation have attracted considerable interest. Enzymes catalyzing the elongation of existing sugar chains form a distinct group within the UGT family that often leads to the unique characteristics of saponins, such as better biological activity and taste, as exemplified by ginsenosides and steviosides3,4. However, the identification of sugar–sugar glycosyltransferases that catalyze the addition of new sugar units to already-bound sugar residues remains challenging. UGTs are a highly divergent, polyphyletic and multigene family, and the saponin intermediates that accept additional sugars occur only in trace amounts in saponin-containing organisms. Phylogenetically, most of the characterized plant sugar–sugar glycosyltransferases belong to UGT73, UGT74, UGT76, or UGT94 subfamily. For instance, UGT74G1 and UGT76G1 have been identified as the catalysts for the sugar chain elongation of steviosides (diterpenoid glycosides) in Stevia rebaudiana5,6, UGTPg29, Pn3-31, and Pn3-32 for ginsenosides (triterpenoid glycosides) in Panax ginseng7,8, and GAME17, GAME18, and GAME2 for steroidal glycoalkaloids in potato9. However, considering the large numbers of steroidal saponins in plants, GTs catalyzing sugar–sugar linkages to form steroidal polyglycosides have been very much understudied.
The genus Paris belonging to the family Melanthiaceae includes famous traditional medicinal herbs worldwide10. In particular, Paris polyphylla Smith var. yunnanensis (Franch.) Hand.-Mazz. and Paris polyphylla Smith var. chinensis (Franch.) Hara. are raw materials of traditional Chinese medicine Rhizoma Paridis (“Chonglou” in Chinese) that have been documented in the Chinese Pharmacopoeia since 197711. Rhizoma Paridis has served as an important ingredient in more than 100 kinds of Chinese patented medicines and Chinese medicine prescriptions (https://db.yaozh.com/), including the well-known “Yunnan Baiyao” series and “Gongxuening” capsules that are used to stop wound bleeding and abnormal uterine bleeding, respectively. Because of increasing market demands, over-exploitation, and habitat destruction in the past few decades, wild populations of the genus Paris have become rare or even endangered. Steroidal saponins (named Paridis saponins, PSs hereafter) are the major bioactive components of Rhizoma Paridis, and more than 200 naturally occurring steroidal saponins (mainly spirostanol-type) have been isolated and identified from different Paris species12, 13, 14, 15. Among them, PSI, PSII, and PSVII are considered as important marker compounds to evaluate the quality of Rhizoma Paridis and their preparations16. In addition, PSs exhibit extensive pharmacological activities, including antitumor17, anti-angiogenesis18, hemostatic19, immuno-stimulating20, antifungal21, anthelmintic22, and antiviral23 properties, which have attracted considerable attention in drug discovery. However, the availability of PSs for both research and medicinal use remains a major limitation owing to diminishing natural populations, slow growth (7–8 years) of cultivated plants, and the tedious and costly processes needed to separate natural mixtures. Chemical synthesis of steroidal saponins has also been restricted by several challenges, including the structural complexity of the aglycones as well as the poor regio- and stereo-selectivity of glycosylation reactions24. By contrast, biotechnological production using synthetic biology and enzymatic conversion, as has been successfully used to produce ample amounts of artemisinic acid, ginsenosides and other natural products25,26, could be used to target specific PSs in an eco-friendly manner.
Investigation of the biosynthesis of PSs has begun despite the large genomes of Paris species (approximately 80 Gb) remaining unsequenced and the genetic manipulation of these plants not yet being established. For the two main aglycones of PSs, diosgenin and pennogenin (C17α-OH diosgenin), cytochrome P450 enzymes involved in their formation from cholesterol have recently been identified from P. polyphylla var. yunnanensis as well as Trigonella foenum-graecum and Dioscorea zingiberensis, two plants producing dioscin as a major saponin27, 28, 29, 30. However, there are still major gaps in understanding the subsequent glycosylation steps contributing to the major structure and bioactivity variations of this group of compounds. Approximately 90 spirostanol-type saponins derived from diosgenin and pennogenin, including mono-, di-, tri-, and tetra-glycosides, have been identified from the genus Paris. Their sugar chains are often composed of d-glucose, l-rhamnose, l-arabinose, d-xylose, d-apiose, d-galactose or l-fucose. It has been proved that the numbers and types of sugar moieties and their linkages have crucial impacts on the chemical properties, pharmacological activities and bioabsorbabilities of PSs31, 32, 33. Therefore, UGTs have attracted considerable interest in the construction of PSs, but the dedicated UGTs responsible for sugar–sugar glycosylation remain elusive and are the last remaining undescribed steps of the full PS biosynthetic pathway.
Here, we report the functional characterization of a series of six UDP-glycosyltransferases from P. polyphylla var. yunnanensis, including four novel sugar–sugar glucosyltransferases, which could catalyze a cascade of glycosylation to yield steroidal diglycosides and triglycosides. Moreover, the antifungal action of PSs against four widely spread strains of human pathogenic fungi, Trichophyton rubrum, Epidermophyton floccosum, Microsporum gypseum, and fluconazole-resistant Candida albicans are also described.
2. Materials and methods
2.1. Plant materials and transcriptome analysis
Paris polyphylla var. yunnanensis plants were collected from the wild (Locality: Luquan, Yunnan, China; N 24° 58′ 36.78″, E 102° 28′ 35.27″; Elevation, 1879 m. Habitat: an evergreen broadleaf forest dominated by Quercus glaucoides). The voucher specimen (Collection Number: JiYH2013104) was identified by Prof. Yunheng Ji and deposited at Kunming Institute of Botany, Chinese Academy of Sciences. The plants were repotted and grown in a greenhouse. RNA-seq was performed using total RNA extracted from the whole plants of P. polyphylla var. yunnanensis on Illumina sequencer in Solexa RNA (paired-end) mode. More detailed information on transcriptome analysis was described in Supporting Information Materials and methods.
2.2. Molecular cloning and heterologous expression
UGT genes of interest were amplified by PCR from the first strand cDNA synthesized by EasyScript® First-Strand cDNA Synthesis SuperMix (TransGen Biotech) using gene-specific primers listed in Supporting Information Table S1. The genes were then subcloned to pCold-TF vector by ClonExpress® Ultra One Step Cloning Kit (Vazyme Biotech) and transferred into the Escherichia coli strain Rosetta (DE3). After confirmation by colony-PCR and DNA sequencing, the positive transformants were induced to express target proteins under 0.3 mmol/L isopropyl-β-d-thiogalactopyranoside (IPTG) at 16 °C. Ni-NTA agarose columns (Qiagen) were used to purify recombinant proteins according to the manufacturer's instructions, and elution buffer (pH 8.0) containing 0.5 mol/L NaCl, 250 mmol/L imidazole and 5 mmol/L DTT was applied to purify target proteins. Both crude lysates and purified proteins were analyzed by SDS-PAGE. The purified proteins were concentrated in 30 kDa ultrafiltration centrifugal tubes (Millipore) and then treated with HRV 3C protease (Takara) at 18 °C for 2 h to remove the TF tags. The recombinant proteins with TF tag fusion or deletion were analyzed by SDS-PAGE. The protein concentrations were quantified using a BCA Protein Assay Kit (NCM Biotech). More detailed information was described in Supporting Information Materials and methods.
2.3. Enzyme assays in vitro
The recombinant UGT proteins with TF tag fusion or deletion were used to assay the enzyme activity in vitro, while the lysate harboring empty pCold-TF vector was used as a negative control. The reaction was performed in a final volume of 200 μL containing 50 mmol/L Tris-HCl (pH 8.0), 50 μmol/L sugar acceptor, 100 μmol/L UDP-glucose or UDP-rhamnose, and 100 μg recombinant proteins. The reaction mixture was incubated at 37 °C for 6 h. After concentration under reduced pressure using a concentrator (Eppendorf), the reaction products were dissolved with 100 μL methanol and centrifuged at 12,000 rpm for 10 min for the subsequent product analysis.
The supernatant (20 μL) was analyzed by HPLC with a diode array detector (HPLC‒DAD, Agilent 1260 system) equipped with an Agilent ZORBAX SB-C18 analytical column (5 μm, 4.6 mm × 250 mm) at a flow rate of 1 mL/min and column temperature at 35 °C. The enzymatic products of PpUGT 1 and 2 were separated with a linear gradient of 44% to 86% acetonitrile in water (0–18 min) and 86% to 100% acetonitrile in water (18.01 to 22 min) followed by washing with 100% acetonitrile for 12 min, and detected at 195 nm wavelength. The enzymatic products of PpUGT 3–6 were separated with a linear gradient of 5% to 100% acetonitrile in water for 30 min.
Meanwhile, all enzymatic products of PpUGTs were subjected to UPLC‒MS/MS analysis in the positive ion mode on an Agilent 1290 Infinity LC coupled with 6530 Q-TOF MS equipped with an electrospray ionization device. UPLC conditions and the injection volumes were consistent with those of HPLC‒DAD analysis with a mobile phase of 0.1% formic acid in water (A) and acetonitrile (B). The Q-TOF source parameters were set as follows: scan range, 200–1400 Da; dry gas flowrate, 8.0 L/min; gas temperature, 300 °C; nebulizer pressure, 35 psig; VCap, 3.5 kV. The software Qualitative Analysis B.06.00 was used for data analysis.
2.4. Product purification and structure characterization
To isolate the glycosylated products of PpUGTs, 40 mL crude enzymes prepared from 500 mL E. coli cultures were used for the enzymatic reaction, and more than 3 mg of each sugar acceptor substrate (for detailed information see the Supporting Information Materials and methods) was used. The reaction mixture was incubated at 37 °C overnight and concentrated using a rotary evaporator. The residue was dissolved in methanol and chromatographed on a Sephadex LH-20 column (chloroform/methanol, 1:1) and then purified using semi-preparative HPLC (column: ZORBAX SB-C18, 5 μm, 9.4 mm × 250 mm) with different acetonitrile/H2O elution systems (55%, 40%, 42%, 65%, 80%, 65%, 40%, 55%, 44%, respectively) to yield compounds 5 (tR = 15.9 min, 2.3 mg), 14 (tR = 8.9 min, 4.5 mg), 21 (tR = 10.3 min, 1.0 mg), 41 (tR = 16.0 min, 0.8 mg), 43 (tR = 11.4 min, 1.3 mg), 44 (tR = 15.4 min, 4.0 mg), 45 (tR = 8.9 min, 1.0 mg), 46 (tR = 19.5 min, 0.8 mg), and 53 (tR = 11.5 min, 3.0 mg). Compounds 23a (tR = 9.5 min, 2.5 mg) and 26 (tR = 8.6 min, 3.5 mg) were obtained using linear gradient aqueous acetonitrile from 22% to 64% (0–14 min). NMR spectra were recorded on Bruker AV-600 or AV-800 spectrometer, with TMS as the internal standard. Other products including compounds 3, 13, gracillin, PSI, PSII, PSIII, PSVI and PSVII were identified through comparison with an authentic standard purchased from Chengdu HerbSubstance Co., Ltd. or Chengdu Must Bio-technology Co. Ltd.
2.5. Conversion rate analysis
The enzyme reaction system was the same as described above. The enzymatic products were analyzed using HPLC‒DAD with water (A) and acetonitrile (B) as mobile phase. For the products of 8‒10, 15–18, and 39–40, a gradient of 10%–95% B in A (0–28 min) followed by isocratic 95% of B (28.01–33 min) was used. For the products of 27‒32 and 38, a gradient of 34%–100% of B (0–22 min) followed by isocratic 100% of B (22.01–37 min) was used. And, for the products of 6 and 15 catalyzed by the PpUGT 4–6 mutants, a gradient of 22%–73% of B (0–17 min) was followed by isocratic 95% of B (17.01–22 min). The injection volume was 30 μL. Conversion rates in percentage (%) were calculated from peak areas of enzymatic products and substrates using HPLC‒DAD according to the literature34.
2.6. Kinetic studies
For determination of the kinetic parameters of PpUGTs, the in vitro enzyme assays were performed in a final volume of 100 μL containing 50 mmol/L Tris-HCl (pH 8.0), UDP-sugar donor (2 mmol/L UDP-glucose or 1.5 mmol/L UDP-rhamnose), varying concentrations of sugar acceptor (2.5, 5, 10, 20, 40, 60, 80, 100, 200, 400 μmol/L of diosgenin for PpUGT 1–2, trillin for PpUGT3, PSVI for PpUGT 4–6) and purified recombinant proteins (100 μg of PpUGT1, 100 μg of PpUGT2, 50 μg of PpUGT3, 50 μg of PpUGT4, 300 μg of PpUGT5 or 50 μg of PpUGT6). The mixtures were incubated at 37 °C for 30 min (for PpUGT 1, 2, 3, and 5) or 20 min (for PpUGT 4 and 6). The reactions were quenched with 100 μL methanol and then concentrated. The products were dissolved with 100 μL methanol and centrifuged, and 40 μL of supernatants catalyzed by PpUGT 4 and 5 were analyzed using HPLC‒DAD as described above, while 10 μL of supernatants catalyzed by PpUGT 1, 3 and 6 or 20 μL of supernatants catalyzed by PpUGT2 were analyzed using UPLC‒MS/MS. The mobile phase for UPLC‒MS/MS analysis was 0.1% formic acid in water (A) and acetonitrile (B). For the products of PpUGT 1–3, a linear gradient of 50% to 95% of B (0 to 15 min) was used, while for the products of PpUGT6, the mobile condition was the same as that of PpUGT 4 and 5. All experiments were performed in triplicate. Calibration curves of trillin, PSV, and PSVI were generated with six gradients of standard samples (3.125, 6.25, 12.5, 25, 50, and 100 μmol/L for trillin and PSV, while 5, 10, 20, 40, and 80 μmol/L for PSVI) using HPLC‒DAD (for PSVI) or UPLC‒MS/MS (for trillin and PSV) with the same method as described above. The linearity of the standard curve was generated by plotting the peak area versus concentration. The equations and correlation coefficients obtained from the linearity study were y = 2037.6χ + 27222.0 (R2 = 0.997) for trillin, y = 2583.4χ + 4436.9 (R2 = 0.996) for PSV, and y = 658.9χ + 10.5 (R2 = 0.999) for PSVI. The product yield of each enzyme reaction was then calculated according to the peak areas and calibration curves. The KM values were calculated by fitting the initial rate to the non-linear Michaelis‒Menten regression equation using GraphPad Prism 8.035.
2.7. Analysis of gene expression and steroidal saponin contents in plants
Leaves, stems, rhizomes, and fibrous roots of P. polyphylla var. yunnanensis were separately harvested and ground into powders using liquid nitrogen from mature plants growing in the greenhouse in April. Total RNA was extracted using the SteadyPure Plant RNA Extraction Kit (Accurate Biology). Real-time qPCR was performed using UltraSYBR mixture reagent (with ROX) (TRANSgen) on an Applied Biosystems 7500 instrument (Life Technologies) according to the product manual. The actin gene was used as an internal standard and relative expression levels were calculated using the ΔΔCT method. Real-time qPCR experiments were carried out with three independent biological replicates, each of which was performed in three technical replicates. Meanwhile, the powders of leaves, stems, rhizomes, and fibrous roots were extracted with methanol and analyzed using UPLC‒MS/MS. The equations and correlation coefficients obtained from the linearity study were y = 441.5χ ‒ 20.1 (R2 = 0.9936) for compound 6, y = 2894.8χ ‒ 999.4 (R2 = 0.9986) for compound 14, y = 254.9χ + 22.0 (R2 = 0.9999) for PSI, y = 422.8χ ‒ 19.7 (R2 = 1) for PSII, and y = 477.3χ + 323.8 (R2 = 0.9978) for PSVII. The contents of steroidal saponins were calculated according to the peak area and calibration curves. More detailed information for compound quantitation was described in Supporting Information Materials and methods.
2.8. Molecular docking, site-directed mutagenesis, and enzyme assay
The Uni-fold server was used to build 3D protein models of PpUGT 1 and 3–6 (https://hermite.dp.tech/home). The structure alignment was performed using PyMOL. Aglycones and sugar donors were docked into the binding pocket of PpUGT models using the AutoDockTools-1.5.6 software. The receptor–ligand interactions were predicted by Discovery Studio 4.0. All structures in the figures were generated using PyMOL. Mutants were constructed using overlap extension PCR and then subcloned into a pCold-TF vector. The primers are listed in Supporting Information Table S1. The recombinant proteins of these mutants were individually purified and quantified, and the in vitro enzyme assay was performed as described above.
2.9. Antifungal activity assay
T. rubrum ATCC4438, E. floccosum (CBS 566.94), and M. gypseum (CBS 118893) were bought from the Hospital for Skin Diseases, Institute of Dermatology Chinese of Medical Sciences, Peking Union Medical College. The C. albicans strain was donated by the Dermatological Department of the First Affiliated Hospital of Kunming Medical University. 199 μL microbe suspension and 1 μL test compound (dissolved in DMSO) with different concentrations were added into 96-well plates and incubated at 28 °C for 5 days for dermatophytes or at 37 °C for 24 h for fluconazole-resistant C. albicans. The absorbance value at 625 nm was recorded by a microplate reader (SpectraMax plus 384, MD, USA). Terbinafine hydrochloride or amphotericin B was used as the positive control, while DMSO and microbe suspension were used as negative and blank controls, respectively. Each experiment was carried out in triplicate. Statistical analysis was conducted using GraphPad Prism 8.0 software. More detailed information of antifungal assay was described in Supporting Information Materials and methods.
Fungal spore germination of T. rubrum was performed according to the literature with suitable modifications36. Different concentrations of compound 13 (2.5, 5, 10, and 20 μmol/L) and PSI (0.25, 0.5, 1, and 2 μmol/L) in DMSO were used, and samples containing the same concentration of DMSO were used as negative controls. The number of germinated and ungerminated spores was determined in a hemocytometer and the percentage of germinated spores was calculated. The experiment was performed in triplicate.
2.10. Sorbitol protection, cellular leakage, and ergosterol antagonist assays
Sorbitol protection and cellular substance leakage assays were performed according to the previous report36. MIC was measured according to CLSI guidelines (M38-A2) in the presence and absence of sorbitol (0.8 mol/L) and ergosterol (250 μg/mL) in RPMI-1640 culture medium (GIBCO BRL, Barcelona, Spain). The plates were incubated at 28 °C and MIC values were determined after 5 days. For the cellular leakage assay, the suspension of spores was prepared and adjusted to 5 × 106 spores/mL with cold MOPS buffer (Sangon, Shanghai, China). Compound 13 was added to the suspension to final concentrations of 20, 40, and 80 μmol/L, respectively, and PSI with concentrations of 5, 10, and 20 μmol/L was used. Samples containing the same concentration of DMSO were used as negative controls. 2% sodium dodecyl sulfate (SDS) was used as a reference compound since it could cause 100% cellular leakage. After treatment for 96 h at 28 °C, the release of intracellular nucleic acid was determined by measuring the absorbance of cell-free supernatant at 260 nm with a UV–Visible spectrophotometer. The rate of released intracellular material was calculated by comparing the OD260 values with that of the lysing agent. The experiment was performed in triplicate.
2.11. Endogenous ergosterol content assay
T. rubrum (1 × 106 spores/mL) was inoculated in the PDB medium and incubated with different concentrations of compound 13 (20 and 40 μmol/L) and PSI (0.625 and 1.25 μmol/L) at 28 °C for 5 days. Terbinafine hydrochloride (10 μmol/L) and DMSO were used as positive and negative controls, respectively. Endogenous sterols were extracted with n-hexane (3 × 3 mL) by vigorous vortex mixing, and 10 mg/mL of cholesterol was added as an internal standard. The organic phase was dried under nitrogen and derivatized with 0.4 mL N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA)/trimethylsilyl (TMS) (99:1) and 0.1 mL anhydrous pyridine for 1 h at 80 °C. Derivatized sterols were analyzed by gas chromatography–tandem mass spectrometry (GC/MS) on an Agilent Technologies 8890/7000D equipped with an Agilent 19091S-433 capillary column (HP-5MS 5% Phenyl Methyl Silox, 30 m × 250 μm × 0.25 μm). All assays were performed in triplicate, and ergosterol content was calculated according to the peak areas.
2.12. Ultrastructure analysis and RNA sequencing of T. rubrum
For scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analyses, hyphal samples exposed to PSI (0.625 μmol/L) or DMSO (control) were collected and then fixed in 2.5% glutaraldehyde at 4 °C overnight. SEM studies were performed using a scanning electron microscope (Hitachi SU-8010, Tokyo, Japan), while TEM studies were performed using a transmission electron microscope (Hitachi H-7650, Tokyo, Japan).
T. rubrum treated with 12.5 μL of PSI (20 mmol/L in DMSO) or 12.5 μL of DMSO was collected for RNA sequencing. Three biological replicates were performed for each treatment. Genes with a fold change ≥ 2.00 and FDR ≤ 0.05 were considered as differentially expressed genes (DEGs), which were analyzed using DESeq2 software37. To obtain further insight into metabolic pathways and functional classification of all annotated DEGs, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed.
2.13. Phylogenetic analysis
Protein sequences from different species were retrieved from the NCBI database (Supporting Information Table S2). A phylogenetic tree was built using MEGA-X using the Neighbor-Joining method. The amino acid sequences were aligned using Clustal W. Bootstrap values (based on 1000 replicates) were indicated at the tree nodes.
2.14. GenBank accession numbers
The sequences of PpUGT 1–6 have been deposited in the GenBank database under the following accession numbers: PpUGT1 (OP198194), PpUGT2 (OP198195), PpUGT3 (OP198196), PpUGT4 (OP198197), PpUGT5 (OP198198), and PpUGT6 (OP651003).
3. Results
3.1. Cloning and heterologous expression of candidate sugar‒sugar glycosyltransferases
To discover UGTs responsible for sugar chain elongation in the biosynthesis of PSs, transcriptome sequencing of one-year-old to five-year-old plants of P. polyphylla var. yunnanensis was individually performed using Solexa Paired-end sequencing technology (Supporting Information Tables S3 and S4) because the contents of PSs were reported to be closely related to plant age38. As a result, 793 unigenes that were predicted to encode glycosyltransferases in the transcriptome library were found, of which 104 belonged to glycosyltransferase family 1 (GT1). The expression levels of 41 putative UGT unigenes were considered substantial as they were higher than 5 reads per kilobase of exon model per million mapped reads (RPKM) in at least one sequenced sample (Supporting Information Fig. S1, Table S5). Because the marker compound PSVII was found to be mainly accumulated in the leaves (Fig. S1) and the RNA quality of the leaves was superior to that of the rhizomes in our experiments, the cDNA of P. polyphylla var. yunnanensis leaves was used to clone the target genes. As a result, 12 full-length cDNA sequences (PpUGT 1–12) were successfully amplified. A neighbor-joining phylogenetic tree of characterized plant sugar–sugar glycosyltransferases and PpUGT 1–12 was computed, including PpUGT73CR1 and Dz3GT1, two sterol glucosyltransferases identified from P. polyphylla and Dioscorea zingiberensis, respectively39,40 (Supporting Information Fig. S2). From the tree, PpUGT 3–8 and 11 were evolutionarily related to sugar–sugar rhamnosyl or glucosyl transferases, while PpUGT 1, 2, 9, 10 and 12 had close phylogenetic relationships with steroidal 3-O glucosyltransferases belonging to UGT80 and UGT73 subfamilies. PpUGT 1 and 2 were found to be mostly related to Dz3GT1, while PpUGT3 was close to DzGT1, a 2′-O-rhamnosyltransferase identified from D. zingiberensis41. Representatives of both groups were selected as candidates for further study and subcloned into the pCold-TF vector and heterologously expressed in the E. coli Rosetta (DE3) strain. SDS-PAGE analysis revealed that the recombinant proteins with expected molecular mass were presented in the soluble fraction after induction with IPTG at low temperature (Supporting Information Fig. S3). The recombinant proteins were then purified on Ni-NTA agarose columns for further functional characterization.
3.2. Functional characterization of two sterol 3-O-β-glucosyltransferases and a steroid glucoside 2′-O-rhamnosyltransferase
The majority of PSs, including the marker compounds PSI, PSII, and PSVII, possess an α-l-rhamnosyl-(1→2)-glucoside residue at the 3-position on the steroid nucleus. To start construction of this residue, we first characterized two sterol 3-O-β-glucosyltransferases PpUGT 1 and 2 (named UGT80A40 and UGT80A41, respectively) that both convert the steroid aglycones diosgenin (1) and pennogenin (2) into diosgenin 3-O-glucoside (3, trillin) and pennogenin 3-O-glucoside (4) respectively (Fig. 1A, Supporting Information Figs. S4 and S5). However, neither UGT80A40 nor UGT80A41 could catalyze subsequent glycosylations. UGT80A40 and UGT80A41 showed 99% identity with sterol 3-O-β-glucosyltransferases UGT80A33 and UGT80A34, respectively, identified from P. polyphylla var. yunnanensis recently40. However, the N-terminus of UGT80A40 was 34 amino acids shorter than sterol 3-O-β-glucosyltransferase UGT80A33. The apparent KM values of UGT80A40 and UGT80A41 towards diosgenin were 6.1 ± 0.7 and 5.9 ± 0.5 μmol/L respectively (Supporting Information Fig. S6, Table S6), lower than those of UGT80A33 and UGT80A34, indicating that they might be natural variants in different collections of P. polyphylla var. yunnanensis.
Figure 1.
Functional characterization of Paris polyphylla UDP-glycosyltransferases after heterologous expression. (A) Consecutive glycosylation reactions catalyzed by the six characterized UDP-glycosyltransferases in the biosynthesis of Paridis saponins. UGT80A40 and UGT80A41 glucosylate diosgenin (1) and pennogenin (2), and the resulting monoglycosides 3 and 4 are converted to rhamnosylated diglycosides 5 and 6 by UGT73CE1. These diglycosides are then converted to triglycosides 13 and 14via a second glucosylation catalyzed by UGT91AH 1–3. (B) HPLC‒DAD analysis of UGT73CE1 catalyzing the conversion of 3 and 4 to generate 5 and 6, respectively. (C) HPLC‒DAD analysis of UGT91AH 1–3 catalyzing the conversion of 5 and 6 into 13 and 14, respectively. The product peaks are highlighted in grey frames. For the control assays, an empty vector was expressed.
To identify rhamnosyltransferases for transferring rhamnose to the glucose moiety of diosgenin and pennogenin glucosides, in vitro enzymatic assays were performed using UDP-rhamnose as the sugar donor. The results showed that PpUGT3 (named UGT73CE1) could convert compounds 3 and 4 to PSV (5) and PSVI (6) respectively, whose structures were identified by comparison with commercially available standards (Fig. 1A‒B and Fig. S5). Furthermore, product 5 was purified from preparative-scale enzymatic reaction and identified using 1D- and 2D-NMR spectra that displayed the HMBC correlation between H-1′′ (δH 5.21) and C-2′ (δC 79.0) (Supporting Information Figs. S7–S10). Therefore, UGT73CE1 was characterized as a steroid glucoside 2′-O-rhamnosyltransferase, capable of transferring a rhamnosyl group to the C-2′ position of diosgenin and pennogenin 3-O-glucosides. UGT73CE1 contained a 1455 bp ORF encoding a protein of 484 amino acids. The apparent KM values of UGT73CE1 towards trillin was 7.1 ± 2.1 μmol/L (Fig. S6, Table S6).
To investigate the sugar acceptor selectivity of UGT73CE1, 16 natural products (compounds 7–11 and S1–S11) from our compound library, including steroid, triterpenoid, diterpenoid, monoterpenoid and flavonoid monoglucosides were selected to incubate with this enzyme and UDP-rhamnose (Fig. 2A and Supporting Information Fig. S11). However, only daucosterol (7), the glucosylated product of β-sitosterol catalyzed by UGT80A40 and 41, generated a product, which was identified as rhamnosylated daucosterol (12) based on its [M+H]+ and [M+Na]+ ions at m/z 723.4 and 745.1, respectively. The other substrates could not be glycosylated by UGT73CE1 from UPLC‒MS/MS analyses. Furthermore, UGT73CE1 could not accept UDP-glucose or UDP-arabinose as a sugar donor, when using compound 3 as a sugar acceptor (Supporting Information Fig. S12). These results indicated that UGT73CE1 could specifically elongate one rhamnose moiety at the C-2′ position of steroidal 3-O-glucosides.
Figure 2.
Substrate specificity of Paris polyphylla UDP-glycosyltransferases towards sugar acceptors. (A) Structures of sugar acceptors and their glycosylated products of UGT73CE1 and UGT91AH 1–3. The substrates and products of UGT73CE1 are in black, and UGT91AH 1–3 in red. (B) Structures of sugar acceptors and their glycosylated products of UGT80A40 and 41. The substrates and products of UGT80A40 and 41 are in blue, and UGT80A40 only are in purple. Red asterisks indicate previously undescribed compounds. (C) The conversion rate in percentage (%) of compounds 5–6, 8–10, and 15–18 catalyzed by UGT91AH 1–3. (D) The conversion rate in percentage (%) of substrates 27–32 and 38–40 catalyzed by UGT80A40 and 41.
3.3. Functional characterization of three steroid glucoside 6′-O-glucosyltransferases involved in the biosynthesis of steroidal triglycosides
To screen for glycosyltransferases responsible for the subsequent glycosylation steps, PSV (5) and PSVI (6), which were considered as putative biosynthetic precursors of the major PSs including I, II, III, VII and H, were tested as the substrates. When 5 or 6 was incubated with PpUGT 4–6 and UDP-glucose, the products (13 and 14, respectively) found in the reaction mixtures had an additional glucose residue (Fig. 1C and Fig. S5). Through comparison with authentic standards using HPLC‒DAD and UPLC‒MS/MS, compound 13 was recognized as diosgenin-3-O-rhamnosyl-(1→2)-[glucosyl-(1→6)]-glucoside. Product 14 was purified from the enzymatic reactions, and its structure was characterized as trikamsesuquiside B (pennogenin-3-O-rhamnosyl-(1→2)-[glucosyl-(1→6)]-glucoside) by interpreting its 1D- and 2D-NMR spectra that showed the HMBC correlation between H-1‴ (δH 4.39) and C-6′ (δC 69.6) (Supporting Information Figs. S13–S21). As expected, PpUGT 4–6 could not catalyze glucosylation of the steroid aglycones diosgenin (1) and pennogenin (2), indicating that they were sugar–sugar glycosyltransferases which function as steroid glucoside 6′-O-glucosyltransferases, distinct from the sugar-sterol glycosyltransferases UGT80A40 and 41. PpUGT 4–6 contain ORFs of 1398, 1404 and 1401 nucleotides encoding 465, 467, and 466 amino acids respectively, and their protein sequences shared 72%–90% identity. PpUGT 4–6 were named UGT91AH 1–3 according to the UGT Naming Committee. They are the first functionally characterized glucoside 6′-O-glucosyltransferases belonging to the UGT91 subfamily. Among themselves, UGT91AH 1–3 showed differences in enzyme properties. The apparent KM values of UGT91AH 1–3 toward 6 were 6.4 ± 1.3, 13.7 ± 0.7, and 12.4 ± 0.2 mmol/L, respectively (Fig. S6, Table S6).
The expression profiles of these characterized UGT genes were analyzed by real-time qPCR in different tissues of P. polyphylla var. yunnanensis, and the contents of enzymatic products as well as the marker compounds PSI, PSII, and PSVII were quantified using UPLC‒MS/MS concomitantly (Fig. S1). The expression of UGT91AH 1 and 3 along with UGT73CE1 and UGT80A40 was found predominantly in rhizomes. UGT91AH2, on the other hand, showed the highest expression level in fibrous roots, followed by rhizomes, while UGT80A41 was expressed mainly in leaves. The triglycosidic product 14 was found to be accumulated mainly in fibrous roots and rhizomes, consistent with the transcript profiles of UGT91AH 1–3, UGT73CE1, and UGT80A40, which suggested that these genes were likely involved in the biosynthesis of compound 14. The diglycosidic product 6 was detected exclusively in the leaves. In addition, PSI and PSII were found to be accumulated mainly in rhizomes, while the content of PSVII was the highest in the leaf, followed by fibrous root and rhizome.
3.4. Exploring the sequence of glycosylation steps in the assembly of steroidal triglycosides
Considering that many of PSs are tri- or tetra-glycosides containing a common α-l-rhamnosyl-(1→2)-glucoside moiety, we are motivated to investigate glycosylation sequence through a comparison of the substrate preferences of PpUGTs. Steroidal saponins with a 4′-arabinose moiety instead of a 2′-rhamnosyl moiety as their second sugar residues were tested as substrates for UGT73CE1. Neither diosgenin-3-O-arabinosyl-(1→4)-glucoside (S1) nor pennogenin-3-O-arabinosyl-(1→4)-glucoside (S2), were accepted by UGT73CE1 (Supporting Information Fig. S22), suggesting that 2′-rhamnosylation could not follow 4′-arabinosylation. In addition, the steroidal monoglycoside 3 that lacks 2′-rhamnose was tested as a substrate for UGT91AH 1–3. As a result, UGT91AH 1–3 could glycosylate compound 3, but the product, diosgenin-3-O-glucosyl-(1→6)-glucoside (previously isolated from P. polyphylla var. yunnanensis42), was only detectable at trace levels using UPLC‒MS/MS. Therefore, UGT91AH 1–3 preferred the steroidal diglycosides 5 and 6 as substrates over the monoglycoside 3 (Supporting Information Table S7). This result implied that the 6′-glucosylation likely proceeded after 2′-rhamnosylation. This result gives further support to the role of 2′-rhamnosylation likely as a general second glycosylation step in PS formation. However, no enzymatic product could be detected when UGT91AH 1–3 were incubated with steroidal di/tri/tetra-glycosides S1, S2, and PSI, PSII, and PSIII, all of which are glycosylated at the C-4′ position.
3.5. Probing substrate specificity of the characterized glucosyltransferases
A total of 18 structurally diverse natural products were incubated with UDP-glucose to determine the substrate specificity of UGT91AH 1–3 (Fig. 2A and Fig. S11), including steroidal glycosides (7), triterpenoid glycosides (11, 18), diterpenoid glycosides (8–10, 15–17, S3), iridoid glycosides (S4 and S5), flavonoid glycosides (S6–S8) and phenolic glycosides (S9–S11). None of UGT91AH 1–3 could accept the steroidal monoglycoside 7. However, specific ion peaks with m/z values 162 higher than the substrate could be detected using UPLC‒MS/MS when UGT91AH 1–3 were individually incubated with the dammarane and cucurbitane triterpenoid glycosides 11 and 18 and the labdane and rearranged abietane diterpenoid glycosides 8–10 and 15–17 (Supporting Information Fig. S23). Thus, the products 19–26 were tentatively identified as the corresponding 6′-O-glucoside congeners including a rare ginsenoside 22. Among them, compounds 21, 23a and 26 were purified from the preparative-scale enzymatic reactions and confirmed as eremostachiin-6′-O-glucoside, phlomioside IV-6′-O-glucoside, and picfeltarraenin IB-6′-O-glucoside, respectively, by their 1D and 2D NMR spectra (Supporting Information Figs. S24–S44), all of which were undescribed compounds, while compounds 19, 20, 23b, 24 and 25 were putatively undescribed compounds. The results further confirmed UGT91AH 1–3 as 6′-O-glucosyltransferases. The glucosylation rate of 10 was higher than that of 15‒16 (Fig. 2C), indicating that UGT91AH 1–3 preferred diterpenoid monoglycosides over the diglycosides. Therefore, the conversion rate of glycosides catalyzed by UGT91AH 1–3 was likely related to the sugar numbers of glycosyl moiety. Intriguingly, two enzymatic products were generated when 15 was offered as a substrate, while only one product was detected when using 16 as a substrate. Compared to 16, substrate 15 has a glucosyl residue instead of rhamnose at the 2′-O position. Therefore, it was concluded that the UGT91AH 1–3 must have catalyzed the transfer of glucose on either of the two existing glucosyl moieties (C-6′-O or C-6″-O) of this diterpenoid glycoside. However, none of the tested phenolic glycosides and iridoid glycosides was accepted as a substrate by UGT91AH 1–3.
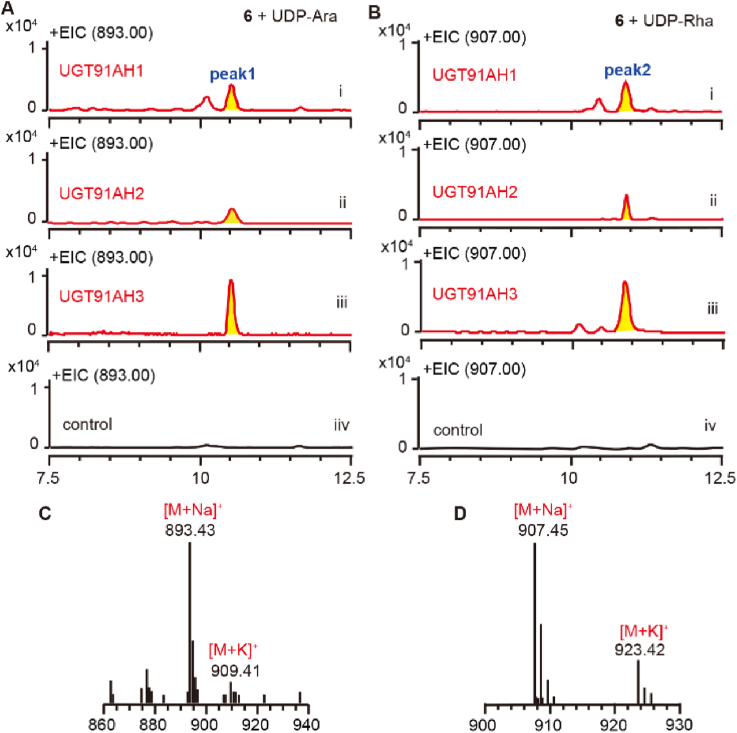
UDP-rhamnose and UDP-arabinose were used to test the sugar donor specificity of UGT91AH 1–3 using compound 6 as a sugar acceptor. Intriguingly, product peaks as yet unidentified were observed in UPLC‒MS/MS chromatograms when UGT91AH 1–3 were incubated with UDP-rhamnose or UDP-arabinose (Fig. 3). In addition, picfeltarraenin IB (18) was also used as an acceptor to determine the function of UGT91AH2 since its conversion rate catalyzed by UGT91AH2 was higher than compound 6. The results also indicated the glycosyltransferase activity of UGT91AH2 towards UDP-rhamnose and UDP-arabinose (Supporting Information Fig. S45). These findings demonstrated that UGT91AH 1–3 could accept UDP-rhamnose and UDP-arabinose as substrates, though the conversion rate for UDP-glucose was higher.
Figure 3.
Substrate specificity analysis of UGT91AH 1–3 towards sugar donors. (A–B) UPLC‒MS/MS analysis of the reaction mixtures containing recombinant UGT91AH 1–3 proteins using compound 6 and UDP-arabinose (UDP-Ara) or UDP-rhamnose (UDP-Rha) as substrates. Controls are the analysis of the reaction mixture harboring an empty vector. (C–D) Positive ESI-MS of the specific product peaks 1 and 2 in (A) and (B). Ion peaks at m/z 893 and m/z 907 represent an arabinose and a rhamnose addition to compound 6 (m/z 761), respectively.
For comparison, the substrate specificity of the two sterol 3-O-β-glucosyltransferases UGT80A40 and UGT80A41 was also investigated. Both UGT80A40 and 41 could accept a panel of steroids (27–32 for UGT80A40 and 41, while 33–35 only for UGT80A40) (Fig. 2B). None of these compounds had the 5,6-spiroketal moiety in the C-17 side chain as their native sterol substrates 1 and 2, so the substrate selectivity of UGT80A40 and 41 seems to be independent of this moiety. Curiously, no glycosylation products of cholesterol (S12), α-ecdysone (S13), and (+)-ecdysterone (S14) Fig. S46 in the presence of UGT80G40 and 41 were detected in our HPLC‒DAD and UPLC‒MS/MS analyses (Supporting Information Fig. S46). Two triterpenoids, a cycloartane 36 and a lupane 37, could be glycosylated to their corresponding mono-3-O-glucosides by both UGT80A40 and 41, while the ursane triterpenoid 38 could be glycosylated to its mono-3-O-glucoside by UGT80A40, although all of these reactions occurred at low efficiency (Supporting Information Table S8). In addition, the abietane compounds 39 and 40 could be converted into their mono-3-O-glucosides 52 and 53, respectively, by both UGT80A40 and UGT80A41 (Fig. 2B and Supporting Information Fig. S47). The enzymatic products 41, 43–46, and 53 were purified from preparative-scale enzymatic reactions, and their structures were elucidated by HR-ESI-MS and NMR spectral analysis. Compounds 43, 45, 46, and 53 were previously undescribed and their structures were further confirmed by 2D NMR (including 1H‒1H COSY, HSQC, HMBC, and ROESY) experiments. The HMBC spectra of 41, 43‒46, and 53 showed 1H‒13C long-range correlations of H-1′/C-1′ of the glucose with C-3/H-3 of the aglycones (Supporting Information Figs. S48–S76), providing solid evidence for the proposed structures. Compounds 42 and 47–52 were tentatively identified through UPLC‒MS/MS analysis based on the presence of characteristic [M+H]+ or [M+Na]+ ions (Fig. S47), and 51 and 52 were putatively previously undescribed compounds. Both UGT80A40 and 41 showed UDP-glucose specificity because no specific product was detected when using UDP-rhamnose or UDP-arabinose as a sugar donor and ergosterol peroxide (27) or topsentisterol D3 (29) as a sugar acceptor. Considering their ability to glycosylate a series of plant and fungal sterols including the ubiquitous β-sitosterol, as well as widely distributed plant cycloartane, lupane, and ursane triterpenoids and abietane diterpenoids, UGT80A40 and 41 can be considered very versatile plant sterol and triterpene glucosyltransferases, distinct from UGT91AH 1–3 that are sugar–sugar 6′-O-glucosyltransferases.
3.6. Exploring molecular mechanisms of glycosyltransferases and identification of residues crucial for catalytic activity
Since these characterized UDP-glycosyltransferases showed different sugar acceptor and donor selectivities and regional specificities, we were motivated to investigate their catalytic mechanisms. The 3D structures of UGT80A40 with glucosyl donor specificity, UGT73CE1 with rhamnosyl donor specificity, and UGT91AH 1–3 with sugar donor promiscuity, were modeled (https://hermite.dp.tech/home). All these PpUGTs were predicted to adopt the GT-B fold consisting of two separated but flexibly linked Rossmann-like domains and a catalytic cleft located between these two domains. Multiple sequence alignments showed a characteristic plant secondary product glycosyltransferase (PSPG) motif in the C-terminal Rossmann domain of these PpUGTs (Supporting Information Fig. S77). As for UGT73CE1 and UGT91AH 1–3, the residues W22, D/E43, and Q44 in the PSPG motif that have been reported to be involved in the formation of hydrogen bonds with the sugar moiety of the donor were conserved43. The last amino acid in the PSPG motif of UGT73CE1 was also found to be Q rather than H which has been reported to play a key role in sugar donor recognition in rhamnosyltransferases by interacting with the rhamnose ring44,45. However, UGT80A40 and 41 showed significant differences from UGT91AH 1–3 in their PSPG motifs by incorporation of 16 additional residues.
Protein‒ligand binding sites of these UDP-glycosyltransferases were predicted by docking the sugar acceptor and donor into the active pockets in the N-domain and C-domain (Supporting Information Fig. S78). The residue H21 of UGT73CE1 and H20 of UGT91AH 1–3 were predicted to form a hydrogen bond with the 2′-hydroxyl function of trillin (3) and the 6′-hydroxyl function of PSV (5), respectively (Fig. 4A and Fig. S78), which could deprotonate the hydroxyl function to activate them for nucleophile attack to the UDP-sugar donor. In addition, several residues of UGT73CE1 were predicted to interact with trillin (3) (Fig. S78, Table S9), which might be crucial for positioning the sugar acceptor in the active pocket. For UGT91AH 1–3, residues predicted to interact with the sugar acceptor involved several aromatic amino acids (Fig. S78, Table S9). Through comparison of the residues in the active pockets, it could be found that the numbers of aromatic residues interacting with the sugar acceptors in the glucosyltransferases UGT91AH 1–3 and UGT80A40 were greater than that in the rhamnosyltransferase UGT73CE1. Besides, the 42nd amino acid in the PSPG box was found to be different in UGT73CE1 and UGT91AH 1–3 (T386, Y376, F378, and Y377, respectively) by comparing those amino acids within 4 Å of the glycosyl moiety of acceptor substrates (Fig. 4A). Furthermore, the potential structural differences between rhamnosylation and glucosylation in UGT73CE1 and UGT91AH 1–3 were explored through comparison of the residues within 5 Å of the glycosyl moiety of sugar donor. Four residues in the rhamnose donor UGT73CE1, W293, A145, L146, and T386, were found to be different from the amino acids at the corresponding positions in the 6′-O-glucose donors UGT91AH 1–3 (Supporting Information Fig. S79, Table S9). Through protein model alignment, A145 and L146 in UGT73CE1, L142 and F143 in UGT91AH 1 and 3, and L145 and F146 in UGT91AH2 were predicted to stabilize the sugar donor, similar to P147 and I148 in the rhamnosyltransferase AtUGT89C1 (6IJA)46. To test the role of these residues, altogether 34 mutants (H20A, I124A, H125A, I151A, M154A, M203A, I206A, T386F, and W293E for UGT73CE1; H20A, Y121A, F143A, F151A, F204A, F207A, F207L, F207M, Y376A, Y376G, Y376L, Y376T, and E285W for UGT91AH1; H20A, F209L, F209M, F378G, F378L and F378T for UGT91AH2; H20A, F121A, Y377A, Y377G, Y377L, and Y377T for UGT91AH3) and two double mutants (A145G/L146P for UGT73CE1 and L142A/F143L for UGT91AH1) were constructed (Table S9), all of which produced soluble recombinant proteins (Supporting Information Figs. S80 and S81). Compound 3 and UDP-arabinose were used as substrates for the enzyme assay of UGT73CE1 mutants, while PSVI (6) and UDP-glucose were used as the substrates for the mutants of UGT91AH 1–3 because 6 has a similar chemical structure to 5 except for the hydroxy at C-17 but was glycosylated by UGT91AH 1–3 at a higher rate (Fig. 2C). All mutants resulted in a dramatic reduction or even complete loss in enzyme activity compared to the native enzymes (Fig. 4B‒D and Supporting Information Figs. S81–S83, Table S9), suggesting that these amino acids are crucial for catalytic activity. The sugar donor selectivity of the mutants W293E and A145G/L146P of UGT73CE1 and E285W and L142A/F143L of UGT91AH1 were assayed using UDP-glucose, UDP-arabinose or UDP-rhamnose as the sugar donor. UPLC‒MS/MS analysis indicated that the mutant UGT73CE1A145G/L146P accepted UDP-rhamnose specifically at lower efficiency than native UGT73CE1, but was unable to use UDP-glucose and UDP-arabinose (Fig. S82). The other three mutants had no detectable enzyme activity at all (Figs. S82 and S83). Accordingly, H21/H20 of UGT91AH 1–3 was required for specific binding of the sugar moiety of the acceptor substrate, with the rhamnosyltransferases and the glucosyltransferases utilizing similar active sites. Notably, H20 of UGT91AH 1 and 2 could also form a hydrogen bond with the 6′-hydroxyl function of compounds 8, 10, or 16 (Supporting Information Fig. S84), which could explain the substrate promiscuity of UGT91AH 1–3. The aromatic amino acids of UGT91AH 1–3 surrounding the sugar acceptors are important for acceptor recognition or stabilization, and the reason for more aromatic residues existing in the active pockets of glucosyltransferases compared to rhamnosyltransferases deserves further investigation.
Figure 4.
Analysis of amino acids surrounding the glycosyl moiety of sugar acceptor within 4 Å and the conversion rates catalyzed by UGT91AH 1–3 mutants. (A) Amino acids surrounding the glycosyl moiety of compound 3 within 4 Å of UGT73CE1 (a), and compound 5 within 4 Å of UGT91AH 1–3 (b–d). Hydrogen bonds are shown with green dashed lines, while π‒alkyl interaction and π stacks are displayed with violet dashed lines. (B–D) The conversion rate in percentage (%) of substrates 6 and 15 catalyzed by wild type UGT91AH 1–3 and their mutants when using UDP-glucose as the sugar donor. Substrate 15 was converted to two products, 23a and 23b.
Another feature conserved in the models of UGT91AH 1–3 was a hydrophobic interaction between the 5″-methyl group of sugar acceptor 5 and the aromatic rings of Y187, F183, and F180 in UGT91AH 1–3, respectively (Fig. 4A), which might contribute to β-(1→6)-glycosylation through stabilizing the 2′-rhamnosyl residue of the sugar acceptor. To test this proposition, these aromatic residues were individually mutated to more flexible amino acids, and eight mutants were created. The mutants UGT91AH1Y187A and UGT91AH1Y187M exhibited increased β-(1→6)-glucosylation activity toward 6, with conversion rates of 80% and 68%, respectively, while the conversion rate of native UGT91AH1 was around 60%. However, the other six mutants showed decreased catalytic activity (Fig. 4B‒D, Supporting Information Fig. S85 and Table S10). The results indicated that the molecular mechanisms of UGT91AH 1–3 for β-(1→6)-glucosylation were not the same, and increased catalytic activity of UGT91AH1Y187A and UGT91AH1Y187M might be due to the flexible amino acids leaving larger spaces for sugar acceptors.
The substrate recognition and regiospecificity of UGT91AH 1–3 were further investigated using compound 15 as a substrate because it could be converted into two enzymatic products, a 6′-O-glucosyl derivative 23a and a 6″-O-glucosyl derivative 23b. The H20A mutations of UGT91AH 1–3 drastically reduced or even abolished the production of 23a, but had less impact on 23b production (Fig. S85, Table S10). The results implied that PpUGT4-6 might employ different amino acids in controlling the production of 23a and 23b. The strong effect of the mutation at this site might have been expected given its predicted H bonding with the 6′-OH function, as mentioned above. The other mutants also showed a dramatic decrease in 6′-O-glycosylation activity with less change or a slight increase in 6″-O glycosylation activity (Fig. 4B‒D and Fig. S85, Table S10). The mutant UGT91AH1Y187A displayed a unique pattern with a 2.8-fold increase in the conversion rate of 23a formation, coupled with an obvious decrease in 23b formation, which may reflect its increased catalytic activity toward the substrate 6 compared to native UGT91AH1 (see previous paragraph).
3.7. Antifungal activity of Paridis saponins against three dermatophytes and fluconazole-resistant Candida albicans
Considering the topical usage of Rhizoma Paridis to treat abscesses and sores in traditional medicine16, we evaluated the inhibitory effects of the enzymatic steroidal glycosides and other representative PSs against four widespread strains of human pathogenic fungi, including three very common dermatophytes Trichophyton rubrum, Epidermophyton floccosum and Microsporum gypseum, and a clinical fluconazole-resistant C. albicans strain (Supporting Information Fig. S86). It was found that compounds S1, 13, gracillin, PSI, PSII, PSV (5), PSVI (6), PSVII, and PSH showed significant inhibitory activity against the three dermatophytes, with MIC50 values ranging from 0.07 to 24.08 μmol/L (Table 1). In particular, the triglycosidic product 13 showed obvious anti-dermatophyte activity, with MIC50 values much lower than its diglycosidic precursor 5. Meanwhile, PSI and PSII were found to show the strongest antifungal activity against T. rubrum, the most prevalent and primary causative pathogen of worldwide dermatophytosis47, with MIC50 values of 0.12 ± 0.02 and 0.37 ± 0.01 μmol/L, respectively, which were approximately 12- and 4-fold more potent than terbinafine hydrochloride (MIC50 = 1.54 ± 0.03 μmol/L). The antifungal activity of gracillin and PSVII against T. rubrum were comparable to that of terbinafine hydrochloride. Compound S2 exhibited moderate antifungal activity against E. floccosum and M. gypseum, but was not active against T. rubrum. However, no obvious antifungal activity was observed for compound 14 and PSIII against any of the three tested dermatophytes. Against the clinical fluconazole-resistant C. albicans strain, PSI, PSII, PSIII, and PSVII showed significant inhibitory activity, with MIC50 values ranging from 1.79 to 8.84 μmol/L (Table 1). Moreover, PSI, PSIII, and PSVII significantly prevented the biofilm formation and morphological transition from yeast to hyphae of C. albicans in a dose-dependent manner (Supporting Information Fig. S87).
Table 1.
Antifungal activity of Paridis saponins against Trichophyton rubrum, Epidermophyton floccosum, Microsporum gypseum and fluconazole-resistant Candida albicans.
| Compound | MIC50 (μmol/L) |
|||
|---|---|---|---|---|
| T. rubrum | E. floccosum | M. gypseum | C. albicans | |
| 1 | – | – | – | >100 |
| 3 | – | – | – | >100 |
| 5 (PSV) | 18.76 ± 0.37 | 12.40 ± 0.69 | 6.89 ± 0.34 | 38.33 ± 0.08 |
| 6 (PSVI) | 24.08 ± 0.22 | 18.61 ± 1.19 | 14.28 ± 0.10 | 37.91 ± 0.05 |
| S1 | 3.91 ± 0.18 | 1.62 ± 0.03 | 2.18 ± 0.06 | >100 |
| S2 | >100 | 45.86 ± 5.25 | 33.07 ± 1.40 | >100 |
| 13 | 5.46 ± 0.23 | 2.41 ± 0.01 | 2.93 ± 0.18 | 51.20 ± 0.11 |
| 14 | >100 | >100 | >100 | >100 |
| Gracillin | 1.38 ± 0.03 | 0.31 ± 0.01 | 0.65 ± 0.001 | >100 |
| PSIII | >100 | >100 | >100 | 3.67 ± 0.05 |
| PSI | 0.12 ± 0.02 | 0.07 ± 0.001 | 0.09 ± 0.004 | 1.79 ± 0.03 |
| PSH | 3.55 ± 0.05 | 1.55 ± 0.04 | 2.01 ± 0.06 | 13.00 ± 0.05 |
| PSII | 0.37 ± 0.01 | 0.21 ± 0.02 | 0.12 ± 0.002 | 8.84 ± 0.98 |
| PSVII | 1.36 ± 0.003 | 1.46 ± 0.04 | 1.61 ± 0.007 | 5.80 ± 0.03 |
| Terbinafine hydrochloride/Amphotericin B | 1.54 ± 0.03 | 0.005 ± 0.0001 | 0.001 ± 0.0002 | 0.10 ± 0.001 |
Dash indicates that the inhibitory effect was not tested. Terbinafine hydrochloride and amphotericin B were served as positive controls for evaluating inhibitory activities against dermatophytes and C. albicans, respectively. A clinical fluconazole-resistant strain of C. albicans was used. MIC50 values record as mean ± standard deviation (SD). The MIC50 values lower than or comparable to that of the positive control are highlighted in gray boxes.
A structure‒activity relationship (SAR) analysis suggested that multiple glycosylation contributed to strong antifungal activity, since most steroidal saponins bearing di-, tri-, and tetra-glycosides exhibited significant activity but the aglycone diosgenin (1) and diosgenin-3-O-glucoside (3) showed no obvious activity. The rhamnosyl group at the C-2′ position played an important role in the antifungal potency of this class of compounds, as the activity of PSI and PSH was significantly higher than that of compounds S1 and S2. An arabinose group at C-4′ of the PSs appeared to be more important than the glucosyl group for the antifungal activity, but a rhamnosyl group substituted at the same position resulted in remarkably decreased activity, based on a comparison of the antifungal activity of gracillin and PSI and PSIII, which share the same diosgenin aglycone and 3-glucosyl and 2′-rhamnosyl substituents, but have different glycosylations at C-4′. Further rhamnosyl modification of the 4′-rhamnosyl group significantly improved anti-dermatophyte activity, but decreased inhibitory activity against fluconazole-resistant C. albicans, according to a comparison of the antifungal activity of PSII and PSIII. Intriguingly, hydroxylation at the C-17 position of the steroidal aglycone (as in the pennogenin) remarkedly diminished antifungal activity against the three dermatophytes, as shown from a comparison of the antifungal effects of compounds S1 and S2, 5 and 6, 13 and 14, PSI and PSH, as well as PSII and PSVII. Strangely, C-17 hydroxylation had no obvious influence on the antifungal activity against fluconazole-resistant C. albicans, since PSV and PSVI, and likewise, PSII and PSVII, showed similar activity.
We also began to explore the mechanism of action of the enzymatic product 13 and the most active PSI against the dermatophyte T. rubrum. Firstly, we evaluated the inhibitory effect of 13 and PSI on the spore germination of T. rubrum, since spore germination is a critical process for pathogenesis and has been considered as a viable target for antifungal drug development48. The results showed that compound 13 and PSI significantly inhibited spore germination in a dose-dependent manner (Fig. 5A). The inhibitory rate reached 95.6% and 96.7% when the spore was exposed to 20 μmol/L of compound 13 and 2 μmol/L of PSI, respectively. However, no obvious inhibition of T. rubrum spore germination was observed in the presence of 0.25 μmol/L of PSI, a concentration approximately two-fold higher than the MIC50 value, suggesting that other cellular processes might be involved in addition to spore germination. In addition, we evaluated the action of compound 13 and PSI on the cell wall and membrane, which are the principal targets of many antifungal agents. Osmotic protection and cellular substance leakage assays were performed to determine the influence of 13 and PSI on the integrity of the cell wall and membrane of T. rubrum. The MIC values of compound 13 and PSI against T. rubrum (10 and 0.625 μmol/L, respectively) in the presence of osmotic protectant sorbitol were identical to those of the compounds used alone, suggesting that these compounds did not act on the cell wall. However, compound 13 (40 and 80 μmol/L) and PSI (5, 10, and 20 μmol/L) caused significant leakage of intracellular nucleic acids, and the leakage rates increased with the increase of compound concentrations (Fig. 5B). Accordingly, compound 13 and PSI were likely involved in the disruption of the integrity or permeability of fungal cell membrane.
Figure 5.
Antifungal activities of the enzymatic triglycosidic product 13 and PSI against Trichophyton rubrum. (A) Percentages of spore germination of T. rubrum treated by compound 13 and PSI. (B) The release rate of intracellular materials from T. rubrum treated by compound 13 and PSI. (C) Endogenous ergosterol contents in T. rubrum after exposure to compound 13, PSI, and terbinafine hydrochloride (TH). (D and E) Scanning electron microscopy of T. rubrum hyphae exposed to control (D) or PSI (E). (F and G) Transmission electron microscopy of T. rubrum hyphae exposed to control (F) or PSI (G). (H) Pathway of ergosterol biosynthesis in fungi. Genes in red color were upregulated by PSI treatment, and those in black color showed no changes in expression level. (I) Gene ontology (GO) enrichment analysis for differentially expressed genes (DEGs) of T. rubrum treated by PSI. Data are presented as means ± SEM (n = 3 per group). ∗P < 0.05, ∗∗P < 0.01 vs. control.
Considering that ergosterol is the main steroid component of fungal cell membranes responsible for maintaining membrane integrity and function, the influence of compound 13 and PSI on fungal ergosterol content was determined. However, GC–MS analysis indicated that no obvious change in the endogenous ergosterol content of T. rubrum was observed after treatment of the fungus with compound 13 (20 and 40 μmol/L) or PSI (0.625 and 1.25 μmol/L) (Fig. 5C). By contrast, exposure to terbinafine hydrochloride caused a significant decrease of endogenous ergosterol of T. rubrum (P < 0.01), which agreed with its inhibition effect on ergosterol biosynthesis reported previously49. However, the addition of exogenous ergosterol led to the compromised antifungal activity of compound 13 and PSI. In the presence of 250 μg/mL of exogenous ergosterol, the MIC values of compound 13 and PSI were 40 and 20 μmol/L, which were 4- and 32-fold of those of the compounds used alone (10 μmol/L for compound 13, and 0.625 μmol/L for PSI), respectively.
To further investigate the effect on the cell membrane, the ultrastructure of T. rubrum hyphae treated with PSI was examined by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). SEM analysis showed that the hyphae of control T. rubrum displayed a smooth, plump, and uniform surface (Fig. 5D). After exposure to 0.625 μmol/L of PSI, T. rubrum hyphae became sunken and twisted (Fig. 5E). Under TEM, the control T. rubrum hyphae exhibited an intact cell wall and cell membrane (Fig. 5F). However, PSI (0.625 μmol/L) treatment led to the disruption of the hyphal cell membrane, while the cell wall remained intact (Fig. 5G). The morphological changes of the cell membrane were consistent with the leakage of cellular nucleic acids, which further suggested that PS exposure caused damage to the membrane.
Further RNA-sequencing analysis of T. rubrum treated with PSI showed no obvious influence on the expression of most ergosterol biosynthetic genes, except for ERG1, ERG11, ERG6, and ERG4 which were up-regulated (Fig. 5H), indicating that PSs did not act as inhibitors of ergosterol biosynthesis. Gene ontology (GO) analysis of differentially expressed genes (DEGs) showed that the biological process with the most changes in gene expression after PSI treatment was transmembrane transport, which included 38 DEGs (Fig. 5I and Supporting Information Fig. S88). The cellular component with the most altered DEGs was the membrane, and the molecular function with the most alteration was transmembrane transporter activity. The top 10 DEGs included a P-type Na+/K+ transporter (downregulation) and an ABC-type transporter (upregulation). However, PSI exposure showed no obvious influence on the expression of genes participating in other processes associated with cell membrane assembly, such as glycerophospholipid metabolism, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, sphingolipid metabolism, and glycosphingolipid biosynthesis. In addition, the pathways of stress response, oxidative phosphorylation, glycolysis, and the citrate cycle were not significantly affected by PSs (Supporting Information Fig. S89). All these results demonstrated that PSI acted principally on fungal cell membranes, likely by targeting ergosterol to influence transmembrane transport.
4. Discussion
Glycosylation is a critical step in the biosynthesis of steroidal saponins, a structurally unique group of natural products that are the major active constituents of a well-known, valuable traditional Chinese medicine Rhizoma Paridis. Through transcriptome and phylogenetic analysis combined with biochemical approaches, we have successfully characterized six PpUGTs responsible for serial glycosylation to produce steroidal diglycosides and triglycosides, which provide a foundation for the production of steroidal saponins through plant or microbial metabolic engineering. UGT80A40 and UGT80A41 are sterol 3-O-β-glucosyltransferases converting diosgenin and pennogenin into their corresponding 3-O-glucosides. Next in succession is UGT73CE1, a steroid glucoside 2′-O-rhamnosyltransferase that converts the monoglycosides into diglycosidic PSV and PSVI. Finally, UGT91AH 1–3 are three steroid glucoside 6′-O-glucosyltransferases that catalyze further 6′-O-glucosylation of PSV and PSVI to generate the triglycosides 13 and 14, respectively. UGT91AH 1–3 are the first functionally characterized glucoside 6′-O-glucosyltransferases belonging to the UGT91 subfamily. The biochemical function of the UGT91 subfamily has been largely unknown, with only a few rhamnosyltransferases and 2′-O-glucosyltransferases identified50, 51, 52. Our findings reveal new regioselectivity and sugar donor use in the sugar–sugar glycosylations catalyzed by this subfamily. Among the enzymes characterized, UGT80A40 and 41 were variants of the recently published UGT80A33 and UGT80A34, respectively39, isolated from a different collection of P. polyphylla var. yunnanensis. The differences between their amino acid sequences and KM values indicate the diversity of saponin biosynthetic enzymes in this taxon, which might be one of the major causes for the unstable quality of Rhizoma Paridis.
The sequence of glycosylation steps in the assembly of PSs can now be proposed according to the products and substrate preferences of the characterized UDP-glycosyltransferases. After initial 3-O-glucosylation of the steroidal aglycones diosgenin and pennogenin, the 2′-rhamnosylation catalyzed by UGT73CE1 is suggested to be the second glycosylation step in the formation of the major PSs bearing an α-l-rhamnosyl-(1→2)-glucoside moiety because of the specific recognition of steroidal monoglycosides rather than diglycosides by UGT73CE1. Of note, most of PSs derived from diosgenin and pennogenin possess an α-l-rhamnosyl-(1→2)-glucoside residue12. Therefore, the glycosylation assembly process of PSs is distinct from that of steroidal glycoalkaloids in potatoes, in which 2′-rhamnosylation is considered a terminal step53. The 6′-glucosylation should proceed after 2′-rhamnosylation since UGT91AH 1–3 preferred steroidal diglycosides rather than monoglycosides. However, glycosylation at C-6′ and C-4′ cannot occur simultaneously or in succession in this class of natural products due to steric hindrance, which is the reason why previously identified PSs harbor only 6′-glycosyl or 4′-glycosyl moieties but not both12. Intriguingly, the products of UGT91AH 1–3 identified in our assays, 13 and 14, are not yet isolated from the genus Paris, although they have already been isolated from the closely related Lilium candidum54 and Trillium kamtschaticum55, also in the order Liliales. Considering the close phylogenetic relationship of these plants and that the affinity constants of UGT91AH 1–3 are comparable to those of other plant UGTs, compounds 13 and 14 should be minor rare steroidal glycosides in P. polyphylla, as compound 14 was indeed detected in all tissues especially rhizomes and fibrous roots, while compound 13 might only be produced at specific growth stages or under specific environmental conditions.
The rhamnosyltransferase UGT73CE1 displayed relatively low sugar acceptor selectivity with the capacity to accept 3-O-glucosyl steroids regardless of variation in the C-17 side-chain, but could not utilize steroidal diglycosides as well as triterpenoid, diterpenoid, monoterpenoid and flavonoid monoglucosides. However, the regional and sugar donor specificities of UGT73CE1 were stringent, and this enzyme catalyzed only β-(1→2)-rhamnosylation. By contrast, the other characterized glucosyltransferases showed high promiscuity in their sugar acceptor. UGT91AH 1–3 could accept steroidal monoglycosides as well as diglycosides, although not steroidal tri- or tetra-glycosides. The catalytic activity of UGT91AH 1–3 could be enhanced by the presence of a 2′-rhamnosyl moiety in steroidal diglycosides, but the existence of a 4′-glycosyl moiety hindered β-(1→6)-glucosylation probably due to the steric effect. These enzymes also glycosylated triterpenoid and diterpenoid glycosides but preferred monoglycosides over diglycosides. This was distinct from their preference for steroidal glycosides (diglycosides preferred), probably due to space constraints of the active pocket for the accommodation of sugar acceptors. Moreover, UGT91AH 1–3 could utilize UDP-rhamnose and UDP-arabinose besides UDP-glucose. The high substrate promiscuity of UGT91AH 1–3 provides enzyme templates to create engineered biocatalysts for β-(1→6)-glycosylation of valuable terpenoid saponins in the future. This is also true for UGT80A40 and 41, which could glycosylate a series of plant and fungal sterols, triterpenoids and diterpenoids. Ultimately, this study generated a panel of 24 terpenoid saponins, including 15 previously undescribed compounds based on the astonishing substrate promiscuity of these characterized UDP-glycosyltransferases.
Despite their general promiscuity, these UDP-glycosyltransferases show distinct patterns of selectivity for sugar acceptor and sugar donor allowing further study of the molecular mechanisms of plant UGTs. Both the rhamnosyltransferase UGT73CE1 and the glucosyltransferases UGT91AH 1–3 utilize His as a general acid to activate the nucleophilic hydroxyl group of sugar acceptors. Molecular docking and site-directed mutagenesis suggested that amino acids, especially aromatic residues interacting with the sugar acceptors via hydrogen bonds or π stacks, are crucial for substrate recognition or stabilization. The mutant UGT91AH1Y187A with increased catalytic efficiency was constructed, providing an effective strategy for enzyme engineering of UGTs. Intriguingly, PpUGT 4–6 could add two glucose to diterpenoid glycosides containing two glucosyl moieties both having a free 6-hydroxyl group, but the amnio acids controlling the catalysis of these two glucosyl additions were different. Moreover, the residues A145/L142, L146/F143, and W239/E285 in UGT73CE1/UGT91AH1 appeared to interact with the sugar acceptors and were therefore suggested to be crucial for sugar donor recognition. However, both the double and single mutants showed a drastic decrease or even loss of catalytic activity, highlighting that sugar donor-specific recognition is determined by protein structure rather than just conserved residues56.
Furthermore, we found for the first time that most PSs exhibited potent antifungal activity against three dermatophytes and fluconazole-resistant C. albicans, which are intractable clinical troubles with increased morbidity globally57,58. In particular, the triglycosidic product 13 showed obvious anti-dermatophyte activity. Meanwhile, PSI and PSII were found to be approximately 12- and 4-fold more potent than the antifungal drug terbinafine hydrochloride against T. rubrum. The results corroborate the traditional usage of Rhizoma Paridis in the treatment of infections. The sugar composition, aglycone type and connection site of the glycosyl chain greatly modulated the antifungal activity of PSs. In particular, the rhamnosyl group at the C-2′ position played an important role in potent antifungal activity.
The osmotic protection and nucleic acid leakage assays suggested that the antifungal activity of PSs was related to the disruption of the fungal cell membrane rather than the cell wall. The primary site of action of PSs on cell membranes was further confirmed by TEM and SEM studies. It has been proved that the most commonly used antifungal drugs, including azoles and polyenes, act primarily on cell membranes by either inhibiting ergosterol biosynthesis or binding membrane-embedded ergosterol59,60. However, the inhibition of ergosterol biosynthesis could be excluded in this study since PS exposure did not influence the endogenous content of ergosterol, although the expression of ergosterol biosynthetic genes of T. rubrum was upregulated. The antifungal activity of PSs against the fluconazole-resistant C. albicans strain further demonstrated that the targets of PSs and azole antifungal agents are different. However, the antifungal activity of PSs is still ergosterol dependent, according to the decreased antifungal effect of PSs in the presence of exogenous ergosterol. The up-regulated expression of ergosterol biosynthetic genes could be a compensatory response to cell membrane damage or ergosterol sequestration by PSs. Structurally, PSs contain a hydrophobic aglycone and a hydrophilic sugar chain that render them amphoteric, which is also characteristic of the polyene antibiotics amphotericin B, nystatin and natamycin. Whether PSs function through binding ergosterol and punching holes in cell membranes like amphotericin B remains to be investigated. Alternatively, PSs could be incorporated into fungal membranes directly by mimicking ergosterol, but the inactivity of aglycones contradicts this scenario. The fact that PS treatment enriched the GO biological process of transmembrane transport further supports the ability of these compounds to disturb cell membrane integrity and permeability. Therefore, PSs could serve as a promising candidate for the development of alternative therapeutic strategies against T. rubrum. This topic clearly warrants further in-depth investigation to learn more about the mode of action of these components of a renowned Chinese traditional medicine.
5. Conclusions
In conclusion, we have functionally characterized six UDP-glycosyltransferases involved in the serial glycosylation of steroidal aglycones to generate the triglycosides from a famous medicinal plant P. polyphylla var. yunnanensis. We revealed the catalytic specificity and astonishing substrate promiscuity of these UDP-glycosyltransferases and demonstrated the catalytic residues and mechanisms crucial for enzyme activity by molecular docking and site-directed mutagenesis experiments. We found for the first time that most Paridis saponins including the triglycosidic product 13 exhibited potent antifungal activity against three widespread human dermatophyte pathogens and a fluconazole-resistant Candida strain from the clinic. This class of natural products damaged the fungal cell membrane via an ergosterol-dependent mechanism. Take together, our work suggests that steroidal saponins in P. polyphylla var. yunnanensis are a promising new class of potential antifungal agents in the treatment of dermatophyte and fluconazole-resistant Candida infections, and the six characterized PpUGTs provided valuable catalytic tools for manipulating saponin production using synthetic biology or enzymatic approaches for further drug development.
Acknowledgments
The authors gratefully acknowledge Prof. Haiyang Liu (Kunming Institute of Botany, Chinese Academy of Sciences) for providing Paridis saponins for the structural elucidation of enzymatic products. This work was supported financially by the National Natural Science Foundation of China (Nos. 82222072 and U1902214), Yunnan Key Research and Development Program (No. 2019ZF011-2, China), and the Research Project of Sichuan Province (2022JDJQ0055, China).
Author contributions
Yan Liu and Shenghong Li designed experiments and analyzed the data. Yuegui Chen generated the majority of the presented data. Yunheng Ji performed plant RNAseq experiment. Yuegui Chen, Qin Yan and Xue Bai performed antifungal experiments. Desen Li, Rongfang Mu, Kai Guo, Minjie Yang and Yang Tao help with enzyme assay or NMR analysis. Yuegui Chen, Yan Liu, Jonathan Gershenzon and Shenghong Li wrote the manuscript.
Conflicts of interest
There are no conflicts to declare.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.05.033.
Contributor Information
Yan Liu, Email: liuyan@mail.kib.ac.cn.
Shenghong Li, Email: shli@mail.kib.ac.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Osbourn A., Goss R.J., Field R.A. The saponins: polar isoprenoids with important and diverse biological activities. Nat Prod Rep. 2011;28:1261–1268. doi: 10.1039/c1np00015b. [DOI] [PubMed] [Google Scholar]
- 2.Yonekura-Sakakibara K., Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- 3.Yang T., Zhang J., Ke D., Yang W., Tang M., Jiang J., et al. Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations. Nat Commun. 2019;10:3214. doi: 10.1038/s41467-019-11154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei W., Wang P., Wei Y., Liu Q., Yang C., Zhao G., et al. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol Plant. 2015;8:1412–1424. doi: 10.1016/j.molp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Li S., Xiong Z., Wang Y. Pathway mining-based integration of critical enzyme parts for de novo biosynthesis of steviolglycosides sweetener in Escherichia coli. Cell Res. 2016;26:258–261. doi: 10.1038/cr.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z., Li J., Sun Y., Zhang P., Wang Y. Structural insights into the catalytic mechanism of a plant diterpene glycosyltransferase SrUGT76G1. Plant Commun. 2020;1 doi: 10.1016/j.xplc.2019.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Wang J., Shi Y., Li R., Fan F., Huang Y., et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Itkin M., Heinig U., Tzfadia O., Bhide A.J., Shinde B., Cardenas P.D., et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- 10.Li H., Su B., Zhang Z.Y., Yang Y.M. An assessment on the rarely medical Paris plants in China with exploring the future development of its plantation. J West China For Sci. 2015;44:1–7. [Google Scholar]
- 11.Cunningham A.B., Brinckmann J.A., Bi Y.F., Pei S.J., Schippmann U., Luo P. Paris in the spring: a review of the trade, conservation and opportunities in the shift from wild harvest to cultivation of Paris polyphylla (Trilliaceae) J Ethnopharmacol. 2018;222:208–216. doi: 10.1016/j.jep.2018.04.048. [DOI] [PubMed] [Google Scholar]
- 12.Wei J.C., Gao W.Y., Yan X.D., Wang Y., Jing S.S., Xiao P.G. Chemical constituents of plants from the genus Paris. Chem Biodiver. 2014;11:1277–1297. doi: 10.1002/cbdv.201300083. [DOI] [PubMed] [Google Scholar]
- 13.Guan X., Li R.S., Duan B.Z., Wang Y., Fan M., Wang S., et al. Advances in research on chemical constituents and pharmacological effects of Paris genus and prediction and analysis of quality markers. Chin Tradit Herb Drugs. 2019;50:4838–4852. [Google Scholar]
- 14.Wang Y., Fan Q., Xiang J., Huang H., Chen S., Liu B., et al. Structural characterization and discrimination of Paris polyphylla var. yunnanensis by a molecular networking strategy coupled with ultra-high-performance liquid chromatography with quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2020;34 doi: 10.1002/rcm.8760. [DOI] [PubMed] [Google Scholar]
- 15.Qin X.J., Zhang L.J., Zhang Y., Ni W., Yang X.Z., Yu Q., et al. Polyphyllosides A‒F, six new spirostanol saponins from the stems and leaves of Paris polyphylla var. chinensis. Bioorg Chem. 2020;99 doi: 10.1016/j.bioorg.2020.103788. [DOI] [PubMed] [Google Scholar]
- 16.Chinese Pharmacopoeia Commission . China Medical Science Press; Beijing: 2020. Chinese Pharmacopoeia; pp. 217–272. [Google Scholar]
- 17.Chen Y., Zhu J., Zhang W. Antitumor effect of traditional Chinese herbal medicines against lung cancer. Anti Cancer Drugs. 2014;25:983–991. doi: 10.1097/CAD.0000000000000127. [DOI] [PubMed] [Google Scholar]
- 18.Xiao X., Yang M., Xiao J.G., Zou J., Huang Q., Yang K.X., et al. Paris saponin II suppresses the growth of human ovarian cancer xenografts via modulating VEGF-mediated angiogenesis and tumor cell migration. Cancer Chemother Pharmacol. 2014;73:807–818. doi: 10.1007/s00280-014-2408-x. [DOI] [PubMed] [Google Scholar]
- 19.Guo L., Su J., Deng B.W., Yu Z.Y., Kang L.P., Zhao Z.H., et al. Active pharmaceutical ingredients and mechanisms underlying phasic myometrial contractions stimulated with the saponin extract from Paris polyphylla Sm. var. yunnanensis used for abnormal uterine bleeding. Hum Reprod. 2008;23:964–971. doi: 10.1093/humrep/den001. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X.F., Yan C., Huang J.J., Zhang Y.Z., Nie Z., Wang L.F., et al. Immuno-stimulating properties of diosgenyl saponins isolated from Paris polyphylla. Bioorg Med Chem Lett. 2007;17:2408–2413. doi: 10.1016/j.bmcl.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 21.Deng D., Lauren D.R., Cooney J.M., Jensen D.J., Wurms K.V., Upritchard J.E., et al. Antifungal saponins from Paris polyphylla Smith. Planta Med. 2008;74:1397–1402. doi: 10.1055/s-2008-1081345. [DOI] [PubMed] [Google Scholar]
- 22.Wang G.X., Han J., Zhao L.W., Jiang D.X., Liu Y.T., Liu X.L. Anthelmintic activity of steroidal saponins from Paris polyphylla. Phytomedicine. 2010;17:1102–1105. doi: 10.1016/j.phymed.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Pu X., Jing R., Ma X., Lu L., Li H. Polyphylla saponin I has antiviral activity against influenza A virus. Int J Clin Exp Med. 2015;8 [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Zhang X., Yu B. O-Glycosylation methods in the total synthesis of complex natural glycosides. Nat Prod Rep. 2015;32:1331–1355. doi: 10.1039/c5np00033e. [DOI] [PubMed] [Google Scholar]
- 25.Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 26.Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christ B., Xu C., Xu M., Li F.S., Wada N., Mitchell A.J., et al. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat Commun. 2019;10:3206. doi: 10.1038/s41467-019-11286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y., Gao L., Zhang X., Gao W. A cytochrome P450 monooxygenase responsible for the C-22 hydroxylation step in the Paris polyphylla steroidal saponin biosynthesis pathway. Phytochemistry. 2018;156:116–123. doi: 10.1016/j.phytochem.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J., Chen J., Liu X., Li X., Zhang W., Dai Z., et al. The origin and evolution of the diosgenin biosynthetic pathway in yam. Plant Commun. 2021;2 doi: 10.1016/j.xplc.2020.100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C., Yang Y., Tian J., Wu Y., An F., Li C., et al. 22R- but not 22S-hydroxycholesterol is recruited for diosgenin biosynthesis. Plant J. 2022;109:940–951. doi: 10.1111/tpj.15604. [DOI] [PubMed] [Google Scholar]
- 31.Seki H., Tamura K., Muranaka T. P450s and UGTs: key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol. 2015;56:1463–1471. doi: 10.1093/pcp/pcv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowles D., Isayenkova J., Lim E.K., Poppenberger B. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Frydman A., Liberman R., Huhman D.V., Carmeli-Weissberg M., Sapir-Mir M., Ophir R., et al. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication. Plant J. 2013;73:166–178. doi: 10.1111/tpj.12030. [DOI] [PubMed] [Google Scholar]
- 34.He J.B., Zhao P., Hu Z.M., Liu S., Kuang Y., Zhang M., et al. Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angew Chem Int Ed. 2019;58:11513–11520. doi: 10.1002/anie.201905505. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y., Wu Y., Zhuang Y., Liu T. Discovery of glycosyltransferases involved in the biosynthesis of ligupurpuroside B. Org Lett. 2021;23:7851–7854. doi: 10.1021/acs.orglett.1c02873. [DOI] [PubMed] [Google Scholar]
- 36.Tian J., Ban X., Zeng H., He J., Huang B., Wang Y. Chemical composition and antifungal activity of essential oil from Cicuta virosa L. var. latisecta Celak. Int J Food Microbiol. 2011;145:464–470. doi: 10.1016/j.ijfoodmicro.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y.Z., Li P. Effect of cultivation years on saponins in Paris Polyphylla var. yunnanensis using ultra-high liquid chromatography–tandem mass spectrometry and Fourier transform infrared spectroscopy. Plant Growth Regul. 2017;84:373–381. [Google Scholar]
- 39.Song W., Zhang C., Wu J., Qi J., Hua X., Kang L., et al. Characterization of three Paris polyphylla glycosyltransferases from different UGT families for steroid functionalization. ACS Synth Biol. 2022;11:1669–1680. doi: 10.1021/acssynbio.2c00103. [DOI] [PubMed] [Google Scholar]
- 40.Li J., Liang Q., Li C., Liu M., Zhang Y. Comparative transcriptome analysis identifies putative genes involved in dioscin biosynthesis in Dioscorea zingiberensis. Molecules. 2018;23:454. doi: 10.3390/molecules23020454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Mosongo I., Li H., Wu Y., Li C., Yang S., et al. Identification and characterization of a trillin rhamnosyltransferase from Dioscorea zingiberensis. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.713036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X., Wang L., Wang G.C., Wang H., Dai Y., Ye W.C., et al. New steroidal saponins and sterol glycosides from Paris polyphylla var. yunnanensis. Planta Med. 2012;78:1667–1675. doi: 10.1055/s-0032-1315239. [DOI] [PubMed] [Google Scholar]
- 43.Wang X. Structure, function, and engineering of enzymes in isoflavonoid biosynthesis. Funct Integr Genom. 2011;11:13–22. doi: 10.1007/s10142-010-0197-9. [DOI] [PubMed] [Google Scholar]
- 44.Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69:1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- 45.Ono E., Homma Y., Horikawa M., Kunikane-Doi S., Imai H., Takahashi S., et al. Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera) Plant Cell. 2010;22:2856–2871. doi: 10.1105/tpc.110.074625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zong G., Fei S., Liu X., Li J., Gao Y., Yang X., et al. Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism. Plant J. 2019;99:257–269. doi: 10.1111/tpj.14321. [DOI] [PubMed] [Google Scholar]
- 47.Ameen M. Epidemiology of superficial fungal infections. Clin Dermatol. 2010;28:197–201. doi: 10.1016/j.clindermatol.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Ortiz S.C., Huang M., Hull C.M. Spore germination as a target for antifungal therapeutics. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.00994-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balfour J.A., Faulds D., Terbinafine A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in superficial mycoses. Drugs. 1992;43:259–284. doi: 10.2165/00003495-199243020-00010. [DOI] [PubMed] [Google Scholar]
- 50.Casas M.I., Falcone-Ferreyra M.L., Jiang N., Mejia-Guerra M.K., Rodriguez E., Wilson T., et al. Identification and characterization of maize salmon silks genes involved in insecticidal maysin biosynthesis. Plant Cell. 2016;28:1297–1309. doi: 10.1105/tpc.16.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibuya M., Nishimura K., Yasuyama N., Ebizuka Y. Identification and characterization of glycosyltransferases involved in the biosynthesis of soyasaponin I in Glycine max. FEBS Lett. 2010;584:2258–2264. doi: 10.1016/j.febslet.2010.03.037. [DOI] [PubMed] [Google Scholar]
- 52.Li J., Halitschke R., Li D., Paetz C., Su H., Heiling S., et al. Controlled hydroxylations of diterpenoids allow for plant chemical defense without autotoxicity. Science. 2021;371:255–260. doi: 10.1126/science.abe4713. [DOI] [PubMed] [Google Scholar]
- 53.Mccue K.F., Allen P.V., Shepherd L., Blake A., Maccree M.M., Rockhold D.R., et al. Potato glycosterol rhamnosyltransferase, the terminal step in triose side-chain biosynthesis. Phytochemistry. 2007;68:327–334. doi: 10.1016/j.phytochem.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 54.Mimaki Y., Satou T., Kuroda M., Sashida Y., Hatakeyama Y. Steroidal saponins from the bulbs of Lilium candidum. Phytochemistry. 1999;51:567–573. doi: 10.1016/s0031-9422(99)00022-9. [DOI] [PubMed] [Google Scholar]
- 55.Ono M., Takamura C., Sugita F., Masuoka C., Yoshimitsu H., Ikeda T., et al. Two new steroid glycosides and a new sesquiterpenoid glycoside from the underground parts of Trillium kamtschaticum. Chem Pharm Bull. 2007;55:551–556. doi: 10.1248/cpb.55.551. [DOI] [PubMed] [Google Scholar]
- 56.Jiang N., Dillon F.M., Silva A., Gomez-Cano L., Grotewold E. Rhamnose in plants‒from biosynthesis to diverse functions. Plant Sci. 2021;302 doi: 10.1016/j.plantsci.2020.110687. [DOI] [PubMed] [Google Scholar]
- 57.Brown G.D., Denning D.W., Levitz S.M. Tackling human fungal infections. Science. 2012;336:647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 58.Tan J., Jiang S., Tan L., Shi H., Yang L., Sun Y., et al. Antifungal activity of minocycline and azoles against fluconazole-resistant Candida species. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.649026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scorzoni L., de Paula E., Silva A.C.A., Marcos C.M., Assato P.A., de Melo W.C.M.A., de Oliveira H.C., et al. Antifungal therapy: new advances in the understanding and treatment of mycosis. Front Microbiol. 2017;8:36. doi: 10.3389/fmicb.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci U S A. 2012;109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.