Abstract
Pangenome references address biases of reference genomes by storing a representative set of diverse haplotypes and their alignment, usually as a graph. Alternate alleles determined by variant callers can be used to construct pangenome graphs, but advances in long-read sequencing are leading to widely available, high-quality phased assemblies. Constructing a pangenome graph directly from assemblies, as opposed to variant calls, leverages the graph’s ability to represent variation at different scales. Here we present the Minigraph-Cactus pangenome pipeline, which creates pangenomes directly from whole-genome alignments, and demonstrate its ability to scale to 90 human haplotypes from the Human Pangenome Reference Consortium. The method builds graphs containing all forms of genetic variation while allowing use of current mapping and genotyping tools. We measure the effect of the quality and completeness of reference genomes used for analysis within the pangenomes and show that using the CHM13 reference from the Telomere-to-Telomere Consortium improves the accuracy of our methods. We also demonstrate construction of a Drosophila melanogaster pangenome.
A pangenome refers to the set of genes present across a population or species. The patterns of presence and absence of genes from the pangenome in individual samples, typically prokaryotes, provide a rich context for understanding genes in populations1. Eukaryotic genomes can likewise be combined into pangenomes, which can be expressed in terms of variation throughout the entire genome rather than just genes. Eukaryotic pangenomics is growing in popularity, due in part to its potential to reduce reference bias in resequencing projects as compared to single-reference-based approaches2.
A pangenome can be represented as a set of variants against a single reference3, but technological advances in long-read sequencing are now making it possible to produce high-quality de novo genome assemblies of samples under study, allowing for variation to be studied within its full genomic context instead of a set of variants4. Two themes that have emerged from recent studies of using reference-based variant calls are that reliance on a single reference genome can be a source of bias, especially for short-read sequencing projects, and representation of structural variation can be a challenge. Pangenomes and the software toolkits that work with them aim to address these issues.
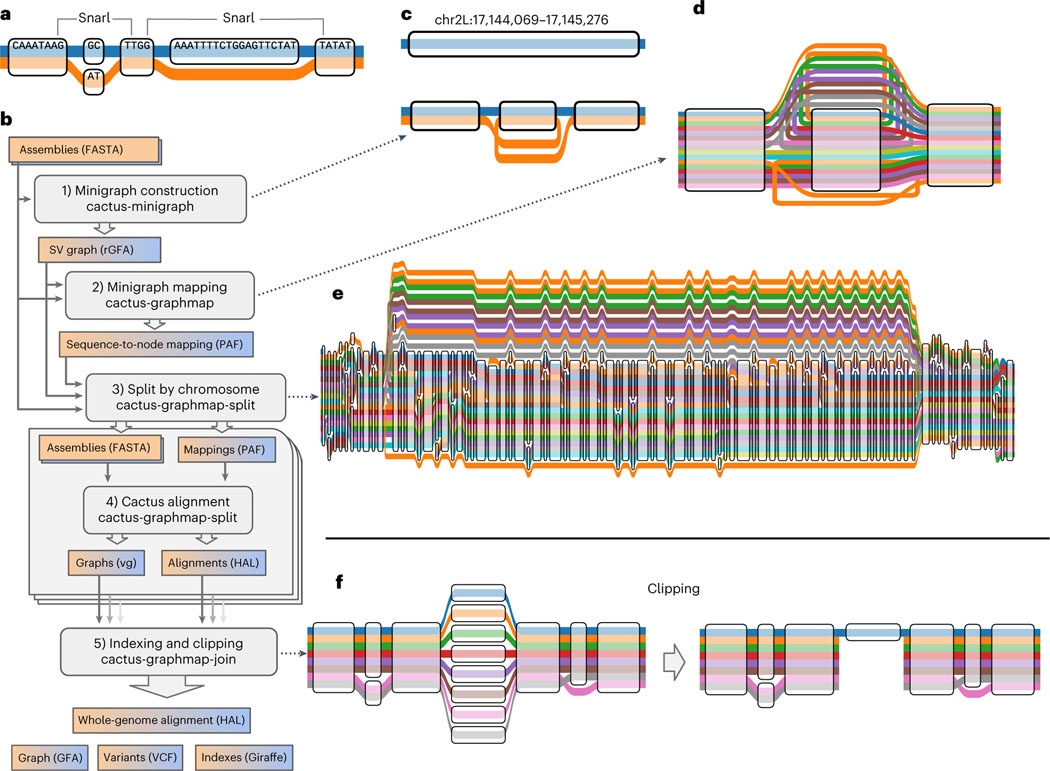
Sequence-resolved pangenomes are typically represented using graph models. There are two main classes of graph representation—sequence graphs and De Bruijn graphs—and several different methods have been published to generate each class. Different methods perform better for different applications, and there is no clear best practice. However, sequence graphs have generally proved more amenable for read mapping3,5,6. In a sequence graph, each node corresponds to a DNA sequence (Fig. 1a) or its reverse complement depending on the direction in which it is traversed. Sample haplotypes are stored as paths, and edges are bidirected to encode strandedness (that is, if an edge is incident to the forward or reverse complement sequence of a node). Sites of variation appear as bubbles, or snarls, which are defined by characteristic subgraphs7. Two snarls are indicated in the example graph in Fig. 1a, the left and right representing a 2-base substitution and 19-base deletion, respectively.
Fig. 1 |. Minigraph-Cactus pangenome construction.
a, ‘Tube Map’ view of a sequence graph shows two haplotypes as paths through the graph. The two snarls (variation sites defined by graph topology, also known as bubbles) are highlighted. b, The five steps and associated tools of the Minigraph-Cactus pipeline, which takes as input genome assemblies in FASTA format and outputs a pangenome graph, genome alignment, VCF and indexes required for mapping with vg Giraffe, illustrating the steps in the pipeline by example. c, SV graph construction using minigraph (as wrapped by Minigraph-Cactus) begins with a linear reference and adds SVs, in this case a single 1,204-bp inversion (at ch2L:17,144,069 in the D. melanogaster pangenome). d, The input haplotypes are mapped back to the graph with minigraph, in this example six of which contain the inversion allele from c. e, The minigraph mappings are combined into a base resolution graph using Cactus, augmenting the larger SVs with smaller variants—in this case, adding smaller variants within the inversion. f, An unaligned centromere is clipped out of a graph, leaving only the reference (blue) allele in that region. The other alleles are each broken into two separate subpaths but are otherwise unaffected outside the clipped region.
Phased variant call format (VCF) files can be thought of as sequence graphs. The variation graph (vg) toolkit makes this perspective explicit by supporting graph construction from VCF3. Using such graphs for mapping and variant calling reduces reference bias and improves accuracy over GRCh38 (refs. 3,6). These graphs can also be used to accurately genotype structural variants (SVs)5, but they are still limited to reference-based variant calls. For example, there is no satisfactory way in VCF 4.3 to directly represent variation nested within a large insertion. Now that they are becoming widely available8, high-quality assemblies can instead be used to directly construct a pangenome graph without the need to go through variant calls. This is equivalent to finding a whole-genome multiple alignment, which is known to be an extremely computationally challenging problem9. As such, multiple alignment algorithms must use heuristics for scaling with respect to both the number of input sequences and their combined length. Typically, the former is accomplished by decomposing the multiple alignment of N genomes into smaller subalignments that can be composed together and the latter by seed-and-extend heuristics10.
MultiZ11 was among the first methods able to align dozens of vertebrate genomes and is still used by the UCSC Genome Browser. It begins with a set of pairwise alignments of the input genomes to a given reference assembly and then uses progressive decomposition to merge the alignments according to their phylogenetic relationships. The pairwise alignments themselves are created with LASTZ, which uses a gapped seeding approach to find anchors, which are then chained and extended with dynamic programming12. Progressive Cactus is a more recent and scalable tool for large vertebrate-scale multiple alignments13. It also uses LASTZ, or the GPU-accelerated successor SegAlign14, to perform pairwise alignments. However, it does so by progressively reconstructing ancestral sequences using a phylogenetic guide tree. This eliminates the need for a global reference assembly, making Progressive Cactus reference independent. At each step, the LASTZ alignments are used as anchors to construct a cactus graph15, which, in turn, is used to filter and then refine the alignment.
Progressive Cactus was shown to be robust to small errors in the guide tree, but, like any progressive alignment approach, it still relies upon an accurate phylogenetic tree. Due to recombination, a single tree cannot reasonably represent the ancestry of any intraspecies genome set that one might want to use to construct a pangenome. Minigraph16 is a newer tool that uses an iterative sequence-to-graph mapping approach, similar to partial order alignment (POA)17, to construct a pangenome graph from a set of input genomes. It uses a generalization of minimap2ʼs minimizer-based seeding and chaining strategy18 and is similarly fast so long as the input genomes are relatively similar. Although minigraph can perform base-level alignment since version 0.17, it only includes SVs (≥50 base pairs (bp) by default) during graph construction. Excluding small variation prevents input genomes from being losslessly embedded as paths in the graph as well as the joint consideration of all types of variants with a single model.
Here we present Minigraph-Cactus, a new pangenomics pipeline that combines minigraph’s fast assembly-to-graph mapping with a modified version of Cactus’s base aligner, alongside several key improvements in vg3,6, to produce base-level pangenome graphs at the scale of dozens to hundreds of vertebrate haplotypes. In addition to representing variation consistently at all resolutions, we show that these graphs can be used to improve upon the state of the art for short-read and long-read mapping, variant calling and SV genotyping.
Results
Minigraph-Cactus pangenome pipeline
The Minigraph-Cactus pangenome pipeline has been added to the Cactus software suite. Like Progressive Cactus13, it is implemented using Toil19, which allows it to be run either locally or via distributed computation on clusters, including those provisioned in the cloud. The pipeline consists of five steps, which can be run individually or together in a single workflow, as shown in Fig. 1b, which are used to generate a graph in both graphical fragment assembly (GFA) and VCF formats, as well as indexes required to map reads using vg Giraffe6.
Minigraph SV graph construction
The pipeline begins with the construction of an initial SV-only graph using minigraph as described in ref. 16. By default, only variants affecting 50 bp of sequence or more are included. This is an iterative procedure that closely resembles POA: a ‘reference’ assembly is chosen as an initial backbone and then augmented with variation from the remaining assemblies in turn. Figure 1c shows an example of an inversion being augmented into a reference chromosome. Minigraph does not collapse duplications. If two copies of a gene are present in the graph after adding i genomes, but there are three copies in the i + 1th genome, then an additional copy will be added to the graph. This is a key difference between minigraph and other approaches (including Progressive Cactus) that would tend to collapse all copies of the gene into a single sequence in the absence of outgroup information to determine the ancestral state. By keeping different gene copies separate, minigraph trades greater graph size for reduced path complexity (fewer cycles).
Minigraph contig mapping
Minigraph generalizes the minimizer-based seeding and chaining concepts from minimap2 (ref. 18) for use on sequence graphs. For this current work, we generalized it to produce base-level alignments between contigs and graphs (but not base-level graphs). In this step of the pipeline, each assembly, including the reference, is mapped back to the SV graph independently (Fig. 1d). The results are concatenated into a single graphical alignment format (GAF) file, which is then filtered to remove spurious alignments (see Methods for details). By re-aligning each assembly to the same graph in this step as opposed to re-using the iterative mappings created during construction, we mitigate an issue in the latter where orthologous sequences can be aligned to inconsistent locations when mapped to different versions of the graph.
Splitting by chromosome
Minigraph does not introduce interchromosomal events during graph construction, so every node in the SV graph is connected to exactly one chromosome (or contig) from the reference assembly. This information is used to split the mappings obtained in the previous step into chromosomes. If a contig maps to nodes from multiple chromosomes, it is assigned to the chromosome to which the most of its bases align. Thresholds (detailed in Methods) are used to filter out contigs that cannot be confidently assigned to any reference chromosome. Such contigs will be excluded from the constructed graph. Graph construction proceeds on each reference chromosome independently, which serves to increase parallelism and reduce peak memory usage (per job). These computational advantages are required to construct a 90-sample human pangenome graph on current hardware, but smaller datasets could be run all at once if desired, avoiding this step entirely.
Cactus base alignment
At its core, Cactus is a procedure for combining a set of pairwise alignments into a multiple alignment13,20. It begins by ‘pinching’ exactly matching aligned bases together in the pairwise alignments to form an initial sequence graph (Fig. 1a). This sequence graph is then transformed into a Cactus graph (Supplementary Fig. 1a–c), whose cycles represent the ‘chains’ of alignment within the sequence graph15, each chain being a maximal sequence of gapless alignments blocks (nodes in the sequence graph) unbroken by rearrangements (see Paten et al.7 for a formal definition). The topology of the Cactus graph is first used to remove candidate spurious or incomplete alignments corresponding to short alignment chains visited by large numbers of sequences. Interstitial unaligned sequences that share common anchors at their ends are then aligned together. This process as a whole remains unchanged at a conceptual level when using Cactus to construct pangenome alignments, but substantial changes to each step were required by the increase in the number of input genomes. Cactus does not typically align more than four genomes (two ingroups and two outgroups) at a time when computing progressive alignments, so scaling to 90 Human Pangenome Reference Consortium (HPRC) samples (and beyond) required the underlying graph structures to be rewritten to use less memory as well as completely replacing the algorithm for interstitial sequence alignment. In brief, the previous all-pairs approach, which scales quadratically with the number of genomes, was replaced with a POA approach that scales linearly (Methods).
Cactus natively outputs genome alignments in hierarchical alignment (HAL) format21. HAL files can be used to create assembly hubs on the UCSC Genome Browser, or to map annotations between genomes22, but they are not suitable for most pangenome graph applications, which expect GFA or vg. We, therefore, created a new tool, hal2vg, to convert HAL alignments into vg format (Methods). These graphs contain the underlying structural variation from the SV graph constructed by minigraph along with smaller variants, and the input haplotypes are represented as paths (Fig. 1e).
Indexing and clipping
The final step of the pipeline combines the chromosome-level results and performs some post-processing. This includes reassigning node IDs so that they are globally unique across different chromosome graphs and collapsing redundant sequence (nodes whose removal does not affect the set of possible haplotype sequences that can be represented by the graph) where possible using gaffix23 (Supplementary Fig. 1d). Nodes are also replaced with their reverse complement as necessary to ensure that reference paths only ever visit them in the forward orientation. The original SV graph produced by minigraph remains embedded in the results at this stage, with each minigraph node being represented by a separate embedded path.
Minigraph-Cactus (in common with all multi-sequence alignment tools that we know of24) cannot presently satisfactorily align highly repetitive sequences, such as satellite arrays, centromeres and telomeres, because they lack sufficiently unique subsequences for minigraph to use as alignment seeds. As such, these regions will remain largely unaligned throughout the pipeline and will make the graph difficult to index and map to by introducing vast amounts of redundant sequence. We recommend clipping them out for most applications and provide the option to do so by removing paths with >N bases that do not align to the underlying SV graph constructed with minigraph (Fig. 1f). In preliminary studies of mapping short reads and calling small variants (see below), we found that even more aggressively filtering the graph helps improve accuracy. For this reason, an optional allele frequency filter is included to remove nodes of the graph present in fewer than N haplotypes and can be used when making indexes for vg Giraffe.
In all, up to three graphs are produced while indexing:
Full graph: useful for storing complete sequences and performing liftover (translation between corresponding haplotypes); difficult to index and map to because of unaligned centromeres. These graphs are typically created only as intermediate results and are not directly used in any of the results in this report.
Default graph: clip out all stretches of sequences ≥10 kb that do not align to the minigraph. The intuition is that large SVs not in minigraph are under-alignments of sequence not presently alignable and not true variants. The 10-kb threshold is arbitrary but empirically was found to work well. This graph is ideal for studying variation and exporting to VCF and can be effectively indexed for read mapping. These graphs are used in all results unless otherwise explicitly stated.
Allele frequency-filtered graph: remove all nodes present in fewer than N haplotypes. This filter increases accuracy for short-read mapping and variant calling, as shown in Supplementary Figs. 7 and 8, respectively. These graphs are used for mapping with vg Giraffe.
Graph (2) is a subgraph of graph (1), and graph (3) is a subgraph of graph (2). They are node ID compatible, in that any node shared between two of the graphs will have the same sequence and ID. Unless otherwise stated, all results below about the graphs themselves are referring to the default graphs, whereas all results pertaining to short-read mapping and small-variant calling were performed on the allele frequency-filtered graphs.
Human pangenome reference graphs
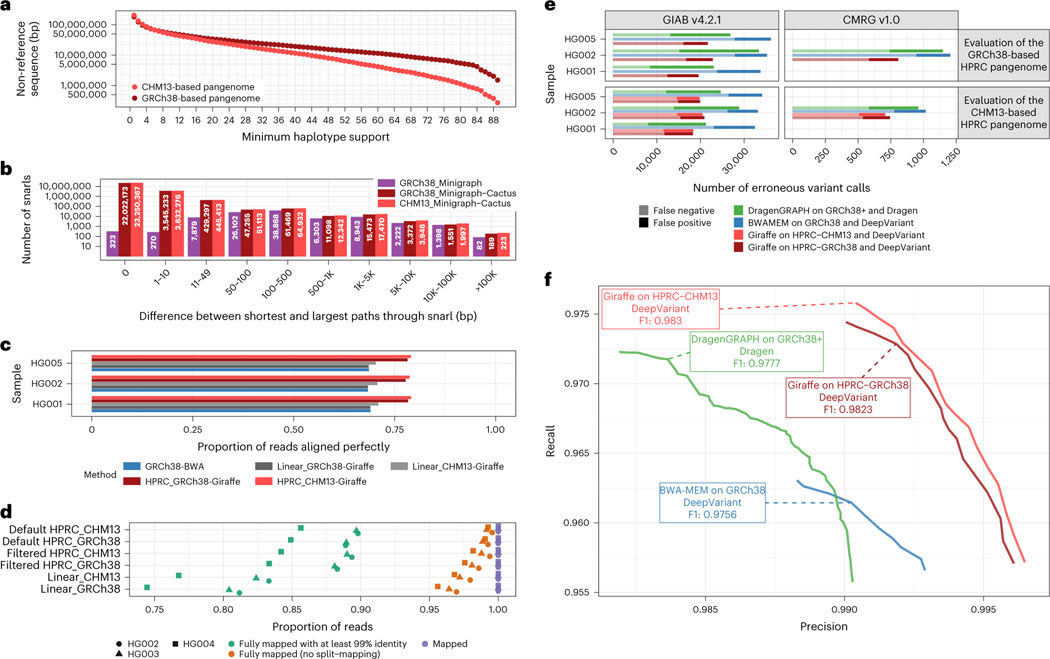
The Minigraph-Cactus pipeline was originally developed to construct a pangenome graph for the assemblies produced by the HPRC. In its first year, this consortium released 47 diploid assemblies25. For evaluation purposes, we held out three samples when generating the graph: HG002, HG005 and NA19240. The remaining 44 samples (88 haplotypes) and two reference genomes (GRCh38 and CHM13 (version 1.1))26 were used to construct the graph, with 90 haploid genomes total. Because the construction procedure is dependent on the reference chosen for the graph, we ran our pipeline twice independently on the same input assemblies, once using GRCh38 as the reference and once using CHM13. The CHM13-based graph includes more difficult and highly variant regions, such as in the acrocentric short arm of chr21, that are not represented in the GRCh38-based graph. This makes it slightly bigger than the GRCh38-based graph, both in terms of total sequence and in terms of nodes and edges (Supplementary Table 1). The final pangenomes have roughly 200× more nodes and edges than the SV graphs from minigraph, showing the amount of small variation required to embed the haplotype paths. Figure 2a shows the amount of non-reference sequence as a function of how many haploid genomes contain it (the same plot for total sequence can be found in Supplementary Fig. 2). The rise in the leftmost points (support = 1) is due to private sequence, only present in one sample, and may also contain alignment artifacts that often manifest as under-alignments affecting a single sample. The plot clearly shows that the CHM13-based graph has less non-reference sequence present across most samples, an apparent consequence of the improved completeness of CHM13 over GRCh38. The distribution of allele sizes within snarls (variant sites in the pangenome defined by graph topology; Fig. 2b) highlights the amount of small variation added relative to minigraph alone. The total time to create and index each HPRC pangenome graph was roughly 3 d (Supplementary Table 4). We compared the variants in the VCF representation of the graph to a benchmark set of variant calls produced from Hi-Fi reads mapped to GRCh38, for each sample in the graph. The average precision and recall across confident (~90% of autosomal genome) regions was 97.91% and 96.66% (see Liao et al.25 for the full evaluation).
Fig. 2 |. Evaluating GRCh38-based and T2T-CHM13-based human pangenomes.
a, The amount of non-reference sequence in the HPRC graphs by the minimum number of haplotypes it is contained in. b, Distribution of the size of the snarls (variation sites, also known as bubbles) for the GRCh38-based minigraph and GRCh38-based and CHM13-based Minigraph-Cactus pangenomes. Note that, in the case of overlapping variants, snarls can be much larger than any single event that they contain. c,e,f, ~30× Illumina short reads for three GIAB samples were mapped using three approaches: BWA-MEM on GRCh38 (blue), vg Giraffe on the linear pangenomes with GRCh 38 or CHM13 (gray) and vg Giraffe on the GRCh38-referenced or CHM13-referenced HPRC pangenome (red). c, Proportion of the reads aligning perfectly to the (pan-)genome for each sample (y axis). d, Number of Hi-Fi reads mapped to the linear, filtered and default (unfiltered by allele frequency) pangenomes. For each sample and pangenome, three points show the number of mapped reads (purple square), reads mapped without being split (orange triangle) and reads fully mapped with at least 99% identity. e,f, Short variants were called with DeepVariant after projecting the reads to GCRh38 from the GRCh38-based pangenome (dark red) or the CHM13-based pangenome (light red). The results when aligning reads with BWA-MEM (blue) or using the Dragen pipeline (green) are also shown. e, The number of erroneous calls (false positive in dark, false negative in pale) is shown on the x axis across samples from GIAB (y axis). Left: GIAB version 4.2.2 high-confidence calls. Right: CMRG version 1.0. When evaluating the CHM13-based pangenome (bottom panels), regions with false duplications or collapsed in GRCh38 were excluded. f, The graph shows the precision (x axis) and recall (y axis) for different approaches using the CMRG version 1.0 truth set for the HG002 sample (bottom-right panel in e). The curves are traced by increasing the minimum quality of the calls.
Mapping to the HPRC graphs
We benchmarked how well the pangenome graphs could be used as drop-in replacements for linear references in a state-of-the-art small-variant (<50-bp) discovery and genotyping pipeline. To do so, we used Illumina short reads (~30× coverage) from three Genome in a Bottle (GIAB) samples: HG001, HG002 and HG005. All mapping experiments were performed on filtered HPRC graphs with a minimum allele frequency of 10%, meaning that nodes supported by fewer than nine haplotypes were removed. This threshold was chosen to maximize variant calling sensitivity and mapping speed for the Giraffe/DeepVariant pipeline (Supplementary Figs. 9 and 10, respectively). We found that reads aligned with higher identity when mapped to the pangenomes using Giraffe, compared to the traditional approach of mapping reads with BWA-MEM on GRCh38. We also mapped reads to the linear references with Giraffe and achieved similar results to using BWA. On average, 78.1% and 78.9% of reads aligned perfectly for the GRCh38-based and CHM13-based pangenomes, respectively, compared to 68.7% when using BWA-MEM on GRCh38 (Fig. 2c). Similarly, reads mapped to the pangenomes had higher alignment scores (Supplementary Fig. 6). Mapping to the pangenomes resulted in a slight drop in mapping confidence, from about 94.9% to 94.1% of reads, with a mapping quality greater than 0 (Supplementary Fig. 7) in those samples. This is expected as the pangenome contains more sequence than GRCh38, including complex regions and large duplications that are more fully represented, which naturally and correctly reduces mapping confidence for some reads. The same trend is observed when the pangenome is not filtered by frequency (Supplementary Fig. 7). We also compared the alignment of long Hi-Fi reads, mapped with GraphAligner27. Mapping to the pangenomes results in more long reads mapped fully (that is, no split mapping) and with high identity (Fig. 2d).
Variant calling with the HPRC graphs
We used the short-read alignments to call variants with DeepVariant28. To prepare them for DeepVariant, the graph alignments were projected onto GRCh38 using the vg toolkit. Note that, even though the CHM13-based graph did not use GRCh38 as the initial reference, the graph does contain GRCh38. Thus, the CHM13-based graph can also be used in this pipeline.
Both pangenomes constructed with Minigraph-Cactus outperform current top-performing methods (Fig. 2e,f). We note that reads in regions that are falsely duplicated or collapsed in GRCh38 cannot be unambiguously projected from their corrected alleles in CHM13. For this reason, these regions were removed from the benchmark when evaluating the CHM13-based pangenome. Unsurprisingly, the CHM13-based pangenome offers the largest gains in variant calling in challenging regions like those assessed by the Challenging Medically Relevant Genes (CMRG) truth set (Fig. 2e)29. Figure 1f shows the precision and recall curves and the CHM13-based pangenome-based variant calls versus state-of-the-art methods based on linear references for the CMRG benchmark. The CHM13-based and GRCh38-based pangenomes have F1 scores of 0.9830 and 0.9823, respectively, compared to 0.9777 and 0.9756 of Dragen and BWA-MEM DeepVariant, respectively. This gain in F1, although modest, still corresponds to hundreds of variants in these regions (Fig. 2e). The frequency-filtered pangenomes performed better than using the default pangenomes (Supplementary Fig. 8). We also tested projecting and calling variants on CHM13. Although the benchmarking protocol is still preliminary for CHM13, we observed a clear improvement when using the pangenome compared to aligning the reads to CHM13 only (Supplementary Fig. 11). Some specific regions, including the major histocompatibility complex (MHC) region and segmental duplications, also have better variant calls on the CHM13-based graph (Supplementary Fig. 12).
SV genotyping with the HPRC graphs
PanGenie is a state-of-the art tool for genotyping human structural variation using short reads30. It uses a hidden Markov model (HMM) that combines information from known haplotypes in a pangenome (as represented by phased VCF) along with k-mers from short reads to infer genotypes and, as such, does not require any read mapping. Minigraph-Cactus can output phased VCF representations of pangenome graphs that can be used as input to PanGenie (see Methods for more details). We evaluated this process by genotyping a cohort of 368 samples from the 1000 Genomes Project31 (1KG) comprising 20 trios randomly selected from each of the five superpopulations, along with the samples present in the graphs. We repeated this process independently on three different graphs: the GRCh38-based and CHM13-based HPRC pangenomes as well the version 2.0 PanGenie lenient variant set produced by the Human Genome Structural Variation Consortium (HGSVC)32. This latter graph was made by constructing reference-based variant calls for each sample and then merging similar variants together into single consensus variants, exactly the process that our pipeline is designed to avoid. The number of variants in each graph is given in Supplementary Table 3.
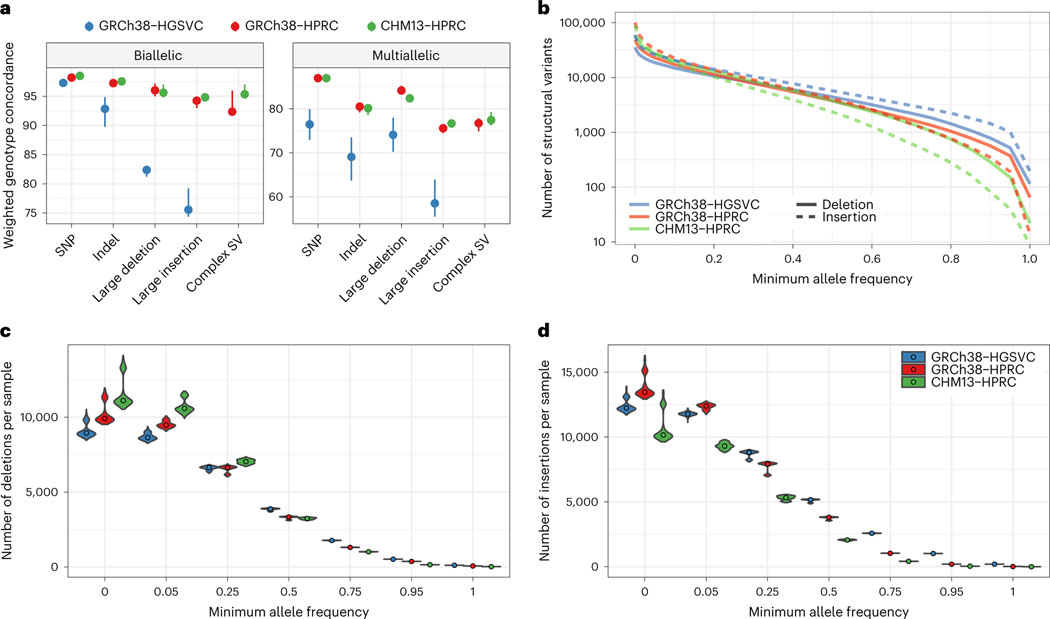
To measure PanGenie’s accuracy on each graph, we performed a leave-one-out experiment on five samples from the graphs. For each selected sample, its genotypes and private variants were removed from the VCF, which was then re-genotyped with PanGenie using short reads from that sample. These genotypes were then compared back to those from the original graph, effectively measuring how closely the haplotypes from short-read genotyping correspond to the original, assembly-based haplotypes. Due to their disjoint sample sets, different samples were used for the HPRC (HG00438, HG00733, HG02717, NA20129 and HG03453) and HGSVC (HG00731, HG00512, NA19238, NA19650 and HG02492). The results are shown in Fig. 3a, which shows the weighted genotype concordance30 across different types of variants, with the Minigraph-Cactus HPRC graphs showing much higher accuracy across all SV variant types than the HGSVC. This improvement can be attributed to the higher quality and number (44 versus 32) of the HPRC versus HGSVC assemblies as well as the more exact representation of variation, SVs in particular, in the multiple alignment-based Minigraph-Cactus graphs, which would explain the increased delta for SV insertions in particular. This more exact representation also explains why the HPRC graph-based genotypes have fewer very common SVs (allele frequency > 20%) (Fig. 3b), despite containing considerably more variants (Fig. 3c,d). As with the short-read variant calling results, the CHM13-based HPRC graph performs generally better than the GRCh38-based graph (Supplementary Fig. 13).
Fig. 3 |. Comparing pangenome SV genotyping.
a, Leave-one-out PanGenie validation measures the concordance of haplotypes as genotyped by short reads with the haplotypes created using genome assembly. The dots show the medians of five samples independently validated in this way. The error bars extend to the minimum and maxiumum values. Note that different samples were used for the HGSVC graph than for the HPRC graphs. b, log-scaled number of SVs given a minimum allele frequency in the PanGenie genotypes. c, The number of SV deletions genotyped per sample, stratified across six minimum allele frequency thresholds. The violin plots show the distribution across 368 samples, whereas the dots represent the median. d, The number of SV insertions genotyped per sample, stratified across six minimum allele frequency thresholds.
Drosophila melanogaster pangenome
We created a Drosophila melanogaster pangenome to demonstrate Minigraph-Cactus’s applicability to non-human organisms. We used 16 assemblies, including the reference, dm6 (ISO1), 14 geographically diverse strains described in ref. 33 and one additional strain, B7. Their sizes range from 132 Mb to 144 Mb. The allele frequency-filtered graph, used for all mapping experiments, was created by removing nodes appearing in fewer than two haplotypes, leading to a minimum allele frequency of ~12.5% (compared to 10% in the human graph), and was used only for mapping and genotyping, where private variation in the graph is less helpful. The amount of sequence removed by clipping and filtering is shown in Supplementary Fig. 17. The relatively small input meant that we could align it with Progressive Cactus using an all-versus-all (star phylogeny) rather than progressive alignment, and the results are included for comparison. In all, we produced five D. melanogaster graphs whose statistics are shown in Supplementary Table 2, a process that took roughly 5 h for the pangenomes (Supplementary Table 4) and 19 h for the progressive Cactus alignments (Supplementary Table 5). As in human, adding base-level variants to the SV graph increases its number of nodes and edges by roughly two orders of magnitude. The graph created from the Progressive Cactus alignment has roughly 45% more nodes and edges and over double the total node length (Supplementary Table 2). This is partially explained by the fact that it contains all the sequence filtered out during pangenome construction (Supplementary Fig. 17) along with interchromosomal alignments.
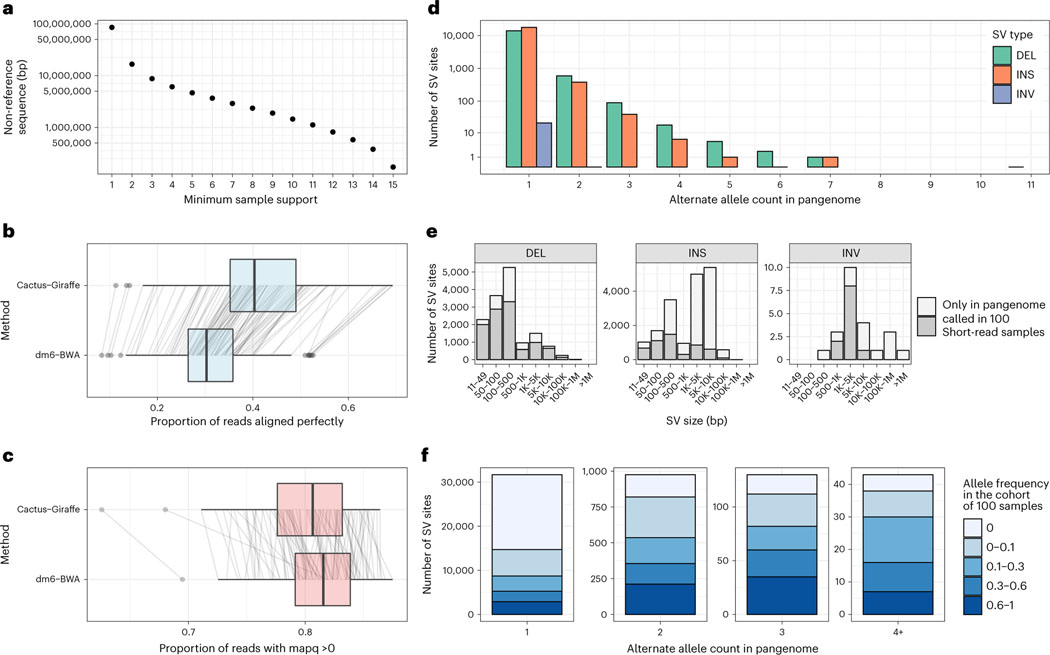
The ‘core’ genome size, which we define as the total length of all nodes present in all samples, of the Minigraph-Cactus pangenome is 110 Mb (Supplementary Fig. 13, first column), which is roughly half the total size of the graph. This reflects a high diversity among the samples: private transposable element (TE) insertions are known to be abundant in this species33. This diversity is also shown in Fig. 4a, which graphs the amount of non-reference sequence by the minimum number of samples it is present in, where the private TE insertions would account for much of the nearly 10× difference between the first and second columns. The trend for the number of non-reference nodes is less pronounced (Supplementary Fig. 15), which implies that the non-reference sequence is accounted for by larger insertion events and smaller variants tend to be more shared. We used the snarl subgraph decomposition7 to compute the variant sites within each graph—that is, subgraphs equivalent to individual single-nucleotide polymorphisms (SNPs), insertions and deletions (indels) and SVs. Supplementary Fig. 16 shows the pattern of nesting of the variant sites in the various graphs.
Fig. 4 |. A D. melanogaster pangenome.
a, Amount of non-reference sequence by minimum number of haplotypes it occurs in for the D. melanogaster pangenome. b,c, Reads mapped by two approaches (y axis): ‘Cactus-Giraffe’, where short reads are aligned to the pangenome using vg Giraffe, and ‘dm6-BWA’, where reads were mapped to dm6 using BWA-MEM. The box plots show the median (center line), upper and lower quartiles (box limits) up to 1.5× interquartile range (whiskers) and outliers (points). The lines connect the same sample between the two approaches. The x axis shows the proportion of reads that align perfectly (b) or the proportion of reads with a mapping quality (mapq) above 0 (c). d, Distribution of the alternate allele count across each SV site. The x axis represents the number of assemblies in the pangenome that support an SV. The y axis is log-scaled. e, The size distribution (x axis) of different SV types (panels). The SV sites are separated in two groups: SV sites that were called in at least one sample from the cohort of 100 samples with short reads (dark gray) and SV sites present only in the pangenome (light gray). f, Fraction of SVs of different frequency in the cohort of 100 samples (color) compared to their frequency in the pangenome (x axis). DEL, deletions; INS, insertions; INV, inversions.
Short-read mapping
The Drosophila melanogaster Genetic Reference Panel (DGRP) consists of 205 inbred genomes34, unrelated to the 16 strains used to construct the pangenome. We used short reads from this dataset to evaluate mapping performance for our pangenome graph. We selected 100 samples for our evaluation, filtering the dataset to include only samples with a single Sequence Read Archive (SRA) accession and Illumina sequencing with >15× coverage. We mapped these samples to the allele frequency-filtered pangenome graph with vg Giraffe in ‘fast’ mode and to dm6 using BWA-MEM. We counted the number of mapped reads, reads with perfect alignment and reads with a mapping quality above 0. We found that the number of reads aligning perfectly markedly increased (Fig. 4b), with, on average, 41.1% of the reads aligning perfectly to the pangenome compared to, on average, 31.0% when aligning reads with BWA on dm6. As in our results in human presented above, we observed a decrease in the number of reads mapped with a mapping quality above 0 when mapping to the pangenome (80.0% versus 81.1% on average; Fig. 4c).
Small variants
We projected pangenomic mappings to dm6 and used FreeBayes35 (in the absence of a high-quality DeepVariant model) to call variants on these mappings and those from BWA-MEM (Methods). We then compared the variant calls that were called by both approaches and those that were called by only one. Although variant sites called by both methods showed similar quality scores, there were more sites unique to our pangenomic approach compared to sites found only by mapping reads to the linear dm6 genome. This increase was observed across different quality thresholds (Supplementary Fig. 18a,c). Overall, that meant that slightly more variants are called when mapping short reads to the pangenome and projecting them to dm6. For example, on average, 740,696 small variants had a quality above 0.1 compared to 738,570 when reads were mapped to the dm6 with BWA-MEM (Supplementary Fig. 18b). For genotype quality above 10, 705,320 small variants were called versus 700,385 (Supplementary Fig. 18d). We also noticed a lower rate of heterozygous variants called when mapping the reads to the pangenome first (13.2% versus 18.1%, on average, per sample; Supplementary Fig. 19). Due to the high inbreeding of these samples, we expect only a small fraction of variants to truly be segregating34.
SVs
The variant sites in the pangenome (snarls) were decomposed into canonical SVs based on the assembly paths in the pangenome (Methods). In the pangenome, most of the SVs are rare and supported by one or two assemblies (Fig. 4d). Of note, the known In(3R)C inversion36 is present in the pangenome, along with 23 other smaller inversions. SVs were also genotyped from the short-read alignments to the pangenome using vg5 (Methods). Even though the genotyping used short reads and the pangenome was frequency filtered, 47.8% of the SVs in the pangenome were found when genotyping the 100 samples (on the filtered pangenome) with short-read data. Both the full set of SVs in the pangenome and the subset genotyped from the short-read data span the full-size spectrum of indels and a few inversions (Fig. 4e). As expected, SVs that were seen in multiple assemblies in the pangenome tended to have higher allele frequencies in the cohort of 100 samples (Fig. 4f). Both rare and more common SVs spanned the full spectrum of SV size and repeat profile, from the shorter simple repeats and satellite variation to the larger TE polymorphisms of LTR/Gypsy, LTR/Pao and LINE/I-Jockey elements, among others (Supplementary Fig. 20).
Discussion
The coordinate system provided by the human reference genome assembly has been vital to nearly all research in human genetics, but it can also be a source of bias. This bias can take the form of unmappable reads in the presence of diverse samples37 or, more subtly, variant calls being skewed toward the reference allele3,6. Pangenome graphs have been shown to be effective at reducing reference bias, but their construction has, until now, been limited by tradeoffs. Either the graphs needed to be constructed from variant calls against a reference3,6,32 and, therefore, unable to properly represent nested variation while still suffering from some reference bias, or they were limited to only SVs16 and unable to effectively be used for short-read mapping with current tools6. The method we present here overcomes these issues by constructing a pangenome graph directly from a multiple genome alignment that represents nearly all the variation within its inputs.
The challenges of effectively leveraging pangenome graphs for human data do not end at construction. Tooling for analysis, such as read mapping and genotyping, which, by definition, is more complex for graphs than single reference genomes, is essential. To this end, we have ensured that graphs produced with Minigraph-Cactus are compatible with most state-of-the art pangenome tools (re-engineering the tools as necessary) such as vg3,6, Giraffe3,5,6, PanGenie30 and GraphAligner27. These tools are all free and open source. Graphs constructed with Minigraph-Cactus are also freely available for download from the Cactus website and through the HPRC38.
To demonstrate the usefulness of these graphs and tools, we showed that Illumina and Hi-Fi reads can be mapped with higher identity and fewer split mappings, respectively, to the pangenome than the linear reference. In the former case, the mappings are used to also improve accuracy of short-read variant calling, and we are hopeful that similar gains will be made with long reads when pangenomics tools for variant calling with them are developed. The representation of SVs in our multiple alignment-based graphs also show considerable improvements in genotyping accuracy when compared to previous methods that rely on merging reference-based calls.
In the case of DeepVariant and PanGenie, the pangenome graph is used in the context of existing reference-based formats, such as BAM and VCF. This allows users to augment their existing workflows with pangenomes with minimal changes, which we think will be key to fostering more widespread adoption of pangenomics methods. Still, such projections back to a linear reference can be lossy, especially in complex regions. Although GAF is being increasingly adopted as the standard read mapping format for pangenomes, there is no corresponding graph-based alternative to VCF in use that we are aware of, and the necessity of always projecting variants back to VCF for analysis is a bottleneck to reaching the full potential of pangenome graphs. True graph-based genotyping formats and tools are needed.
Minigraph-Cactus requires at least one chromosome-level input assembly to be used as a reference backbone, and, in general, the quality and usefulness of the pangenome will increase with the quality and completeness of all the input assemblies. We do not think that this will be a bottleneck for most species going forward as it will soon be routine to produce large numbers of reference or even ‘telomere-to-telmore’ quality genomes for many species due to advances in sequencing technology and assembly tools. In the present work, we quantified the impact of reference genome assembly quality on our pangenomes and their applications. Even though both GRCh38 and CHM13 are included in all HPRC graphs that we constructed, the choice of which to use as a reference backbone influences the topology and completeness of the graph, and, in virtually all genome-wide measures of mapping, variant calling and genotyping performance, we found the CHM13-based graph to be superior. In the case of variant calling with Giraffe/DeepVariant, we showed that the CHM13-based graph was able to improve upon the state-of-the art accuracy of the GRCh38-based graph, even when making calls on GRCh38. We, therefore, think that our pangenomes could help some users, who would otherwise be reluctant to switch to reference assemblies, to still take advantage of them.
Building upon previous work in pangenomics, the HPRC has shown that high-quality genome assemblies can be leveraged to provide a better window into structural variation as well as to reduce bias incurred by relying on a single reference. The pangenome graph representation has been fundamental to this work, but graph construction remains an active research area. The key challenges stem not just from the computational difficulty of multiple genome alignment, particularly in complex regions, but also from fundamental questions about the tradeoffs between complexity and usability. In developing Minigraph-Cactus, we sought a method to construct graphs with as much variation as possible while still serving as useful inputs for current pangenome tools, such as vg and PanGenie.
Some of the compromises made to make our method practical represent exciting challenges for future work in both pangenome construction and applications. Pangenomes from Minigraph-Cactus cannot be used, for instance, to study centromeres. The omission of interchromosomal events will likewise preclude useful cancer pangenomes or studies into acrocentric chromosome evolution39. We are also interested in ways to remove the necessity of filtering the graph by allele frequency to get optimal mapping performance by using an online method at mapping time to identify a subgraph that most closely relates to the reads of a given sample. We are also working on improving Cactus’s native chaining logic to reduce the need for some of the heuristic filters that we presently rely on to filter out low-quality mappings. Progressive Cactus alignments can be combined and updated, and, as datasets become larger, this functionality is becoming more necessary for pangenome alignments. Comprehensive tooling to update pangenomes by adding, removing or updating assemblies is an area of future work.
Pangenomics has its origin in non-human species, and, as the assembly data become available, we will see pangenomes being produced for a wide array of organisms. Already there are data for a number of species, from tomato40 to cow41. In this work, we constructed a D. melanogaster pangenome as a proof of concept to show that our method can also be used on other non-human organisms. We hope that others will use the Minigraph-Cactus pipeline to produce useful graphs from sets of genome assemblies for their species of interest. Large-scale alignments are resource intensive, and the 90-human pangenomes required nearly 3 d to compute on a cluster. As such, we have made these alignments publicly available through the HPRC and will do the same for future releases.
Reference bias can also affect comparative genomics studies. For example, a genomic region can be of interest to a particular sample, but if that region happens to be missing from the reference genome due to intraspecies diversity or assembly errors, it would be absent from any alignments based solely on that reference. Therefore, we expect pangenome references to supplant single genome references for intraspecies population genomics studies; we also see this as the future in interspecies comparative genomics studies.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-023-01793-w.
Methods
HPRC graph construction
The HPRC version 1.0 graphs discussed here were created by an older version of the pipeline described above, with the main difference being that the satellite sequence was first removed from the input with dna-brnn42. This procedure is described in detail in ref. 25. The amount of sequence removed from the graph, and the reason it was removed, is shown in Supplementary Fig. 3. Roughly 200 Mb per assembly was excluded, most of which was flagged as centromeric (HSat2 or alpha satellite) by dna-brnn42. The ‘unassigned’, ‘minigraph-gap’ and ‘clipped’ categories denote the sequence that, respectively, did not map well enough to any one chromosome to be assigned to it; intervals >100 kb that did not map with minigraph; and intervals >10 kb that did not align with Cactus. Simply removing all sequence ≥10 kb that does not align with Cactus, as described in the methods above, amounts to nearly the same amount of sequence excluded (Supplementary Fig. 4). The 10-kb threshold was used for clipping because it was sufficient to remove all centromeres (as previously identified) with dna-brnn and also because it corresponds to the maximum length of an alignment that can be computed with abPOA. The ‘unassigned’ sequence is further characterized in Supplementary Fig. 5, showing it to be driven primarily by contigs featuring satellite sequence, segmental duplications43 and partial matches to acrocentric chromosomes. These are characteristics that were associated with potential assembly errors in the analysis of the same data in Liao et al.25. The exact commands to build HPRC graphs referred to in this figure are available at https://github.com/ComparativeGenomicsToolkit/cactus/blob/91bdd83728c8cdef8c34243f0a52b28d85711bcf/doc/pangenome.md#hprc-graph. They were run using the same Cactus commit: 91bdd83728c8cdef8c34243f0a52b28d85711bcf.
Filtering minigraph mappings and chromosome decomposition
Input contigs were labeled ‘unassigned’ above if they could not be confidently mapped to a single reference chromosome during the minigraph contig mapping phase of the pipeline. For a given contig, this determination was made by identifying the chromosome in the SV graph to which the highest fraction of its bases mapped with exact matches. If this highest fraction was at least three times higher than the second highest, and greater than or equal to a minimum threshold, the contig was assigned to that chromosome; otherwise, it was left unassigned (and omitted from the graph). The minimum threshold for chromosome assignment was 75% for contigs with length ≤100 kb, 50% for contigs with length in the range 100 kb–1 Mb and 25% with length >1 Mb. These values were chosen after empirical experimentation specifically to filter out spurious mappings as determined by VCF-based comparison with Hi-Fi-based DeepVariant calls25. Contigs filtered in this way are predominantly centromeric (and cannot be confidently mapped anywhere) or small fragments of acrocentric chromosome short arms (enriched for putative assembly misjoins25) or segmental duplications without enough flanking sequence to be correctly placed25. Such filtering is not needed on chromosome-level assemblies. It can also be easily relaxed if desired, for instance when working on small, very diverse assemblies.
Despite this filtering process, we found a small number of small contigs that, due to either misassembly or misalignment, confidently map across entire chromosome arms (one end of the contig maps near the centromere and the other near the telomere). The chromosome arm-spanning edges introduced by such mappings introduce topological complexities that can hinder downstream tools (for example, all variants on the spanned arm would be considered nested within a large deletion). To prevent this, any mapping that would introduce a deletion edge of 10 Mb or more (tunable by a parameter) relative to the reference path is removed. Finally, in rare cases, minigraph can map the same portion of a query contig to different target regions in the graph. When manually inspecting these cases, we found that they could lead to spurious variants in the graph when, as above, compared to variant calls directly from Hi-Fi-based DeepVariant calls (Liao et al.25). To mitigate these cases, we remove any aligned query interval (pairwise alignments are represented in terms of the query intervals, positions on the contig and target intervals, positions within the graph) that overlaps another by at least 25% of its length and whose mapping quality and/or block length is 5× lower than those of the other interval. We expect to obviate the need for both these filters with stricter chaining parameters within Cactus in the near future.
POA-based Cactus base aligner
We replaced the Base-level Alignment Refinement (BAR) algorithm that is used to create alignments between the interstitial sequences after the initial anchoring process20. In brief, the original algorithm has two stages. First, from the end of each alignment anchor (termed a block and defined by a gapless alignment of substrings of the input), it creates an MSA of the unaligned sequences incident with the anchor. Each such MSA has the property that the sequence alignment is pinned from the anchor point, but, because of rearrangement, the MSA is not necessarily global—that is, at the other end of the MSA from the starting anchor point, the different sequences may be non-homologous due to genome rearrangement. Second, the set of MSAs produced by the first step is refined by a greedy process that seeks to make the set of MSAs, which may overlap in terms of sequence positions, consistent, so resolving, at base-level resolution, the breakpoints of genome rearrangements. For details of this process, see the original paper20.
The replacement of the BAR algorithm achieved two things. First, we changed the process in the first step to create MSAs to use the abPOA MSA algorithm44. The previous algorithm was based upon the original Pecan MSA process and scaled quadratically with sequence number; in contrast, the new MSA process scales linearly and is overall faster even for small numbers of sequences. In this process, we updated abPOA to use the LASTZ default scoring parameters12, with the addition of a ‘long’ gap state not used by LASTZ but included within abPOA, whose value we chose to maximize performance on the Alignathon benchmark set45. Gap parameters were thus: short-gap-open, 400; short-gap-extend, 30; long-gap-open, 1,200; long-gap-extend, 1. Parameters for the long gap state were determined by empirical experimentation. Second, we fully reimplemented the second step of the BAR algorithm, both making it faster and removing various unnecessary bottlenecks that previously scaled superlinearly but that now all scale linearly with sequence number and length. Notably, this process did not materially affect the resulting alignments, as judged by extensive unit-level and system-level testing.
Conversion from multiple alignment to sequence graph
Cactus natively uses the HAL format21. We developed hal2vg, which converts HAL files to vg formats. It works for both Progressive Cactus and Minigraph-Cactus. It works in memory and, for large alignments, is reliant on having chromosomal decomposition of the HAL and simple topology to run efficiently. hal2vg begins by visiting the pairwise alignments in breadth-first order from the root of the underlying guide tree. Contiguous runs of exact matches in the pairwise are ‘pinched’ together to form nodes of a sequence graph using Cactus15, and the assemblies themselves are added as ‘threads’ to this graph. SNPs are stored in an auxiliary data structure and used to pinch together transitive exact matches as they arise. For example, if the pairwise alignments of a column (in the multiple alignment) are A > C and C > A, this structure will ensure that the two As are pinched together in the sequence graph (which, by definition, only represents exact matches within its nodes). Seqwish46 is a recent tool that also induces sequence graphs from sets of pairwise alignments but, because it does not transitively process SNPs in this way, will not work on tree-based sets of pairwise alignments as represented by HAL. Finally, once the sequence graph has been created in memory, it is serialized to disk, path by path, using libbdsg47, an application programming interface (API) for reading and writing sequence graphs in an efficient, vg-compatible binary format.
Conversion from sequence graph to VCF
By default, all graphs are output in GFA (version 1.1) as well as the vg-native indexes: xg, snarls and GBWT formats47,48. Because VCF remains more widely supported than these formats, we implemented a VCF exporter in vg (vg deconstruct) that is run as part of the Minigraph-Cactus pipeline. It outputs a site for each snarl in the graph. It uses the haplotype index (GBWT) to enumerate all haplotypes that traverse the site, which allows it to compute phased genotypes. For each allele, the corresponding path through the graph is stored in the AT (Allele Traversal) tag. Snarls can be nested, and this information is specified in the LV (Level) and PS (Parent Snarl) tags, which needs to be taken into account when interpreting the VCF. Any phasing information in the input assemblies is preserved in the VCF.
HPRC graph mapping and variant calling
We used 30× Illumina NovaSeq PCR-free short-read data HG001, HG002 and HG005, available at gs://deepvariant/benchmarking/fastq/wgs_pcr_free/30x/. The reads were mapped to the pangenome using vg Giraffe (version 1.37.0). The same reads were mapped to GRCh38 with decoy sequences but no ALTs using BWA-MEM (version 0.7.17). To provide additional baselines, reads were also mapped with vg Giraffe to linear pangenomes—that is, pangenomes containing only the reference genome (GRCh38 or CHM13). The number of reads mapped with different mapping quality (or aligning perfectly) were extracted from the graph alignment file (GAF/GAM files) produced by vg Giraffe and from the BAM files produced by BWA-MEM.
Variants were called using the approach described in ref. 25. In brief, the graph alignments were projected to the chromosomal paths (chr 1–22, X, Y) of GRCh38 using vg surject. Once sorted with SAMtools (version 1.3.1), the reads were realigned using bamleftalign (FreeBayes version 1.2.0)35 and ABRA (version 2.23)49. DeepVariant (version 1.3)28 then called small variants using models trained for the HPRC pangenome25. We used the same approach when calling small variants using the CHM13-based pangenome and when projecting to CHM13 chromosomal paths.
Evaluation of small-variant calls.
Calls on GRCh38 were evaluated as in ref. 25—that is, using the GIAB benchmark and confident regions for each of the three samples50. For HG002, the CMRG truth set version 1.0 (ref. 29) was also used to evaluate small-variant calls in those challenging regions. The evaluation was performed by hap.py51 version 0.3.12 via the jmcdani20/hap.py:v0.3.12 docker image, except for Supplementary Fig. 9 which reports accuracy measures from rtg vcf eval v3.91 (ref. 52).
When evaluating calls made against the GRCh38 chromosomal paths using the CHM13-based pangenome, we excluded regions annotated as false duplications and collapsed in GRCh38. These regions do not have a well-defined truth label in the context of CHM13. We used the ‘GRCh38_collapsed_duplication_FP_regions’, ‘GRCh38_false_duplications_correct_copy’, ‘GRCh38_false_duplications_incorrect_copy’ and ‘GRCh38_population_CNV_FP_regions’ region sets available at https://github.com/genome-in-a-bottle/genome-stratifications.
To evaluate the calls made on CHM13 version 1.1, we used two approaches. First, the calls from CHM13 version 1.1 were lifted to GRCh38 and evaluated using the GRCh38 truth sets described above (GIAB version 4.2.1 and CMRG version 1.0). For this evaluation, we also lifted these GRCh38-based truth sets to CHM13 version 1.1 to identify which variants of the truth set are not visible on CHM13 because they are homozygous for the CHM13 reference allele. Indeed, being homozygous for the reference allele, those calls will not be present in the VCF because there are no alternate alleles to find. These variants were excluded from the truth set during evaluation. The second approach was to evaluate the calls in CHM13 version 1.1 directly. To be able to use the CMRG version 1.0 truth set provided by GIAB, we lifted the variants and confident regions from CHM13 version 1.0 to CHM13 version 1.1. The CMRG version 1.0 truth set focuses on challenging regions but still provides variant calls across the whole genome. Hence, we used those variants to evaluate the performance genome wide, although restricting to a set of confident regions constructed by intersecting the confident regions for HG002 from GIAB version 4.2.1 (lifted from GRCh38 to CHM13 version 1.1) and the alignment regions produced by dipcall in the making of the CMRG version 1.0 truth set (https://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/release/AshkenazimTrio/HG002_NA24385_son/CMRG_v1.00/CHM13v1.0/SupplementaryFiles/HG002v11-align2-CHM13v1.0/HG002v11-align2-CHM13v1.0.dip.bed). Finally, we used the preliminary HG002 truth set from GIAB on CHM13 version 2.0, which is equivalent to CHM13 version 1.1 with the added chromosome Y from HG002. The calls in this set were based on aligning a high-confidence assembly using dipcall53 (labeled in figure as ‘dipcall CHM13 v2.0’). Here again, we intersected the confident regions with the GIAB version 4.2.1 confident regions lifted from GRCh38 to CHM13.
In all experiments described above, the variants (VCF files) were lifted over using Picard (version 2.27.4)54 LiftoverVcf and the RECOVER_SWAPPED_REF_ALT option. Regions (BED files) were lifted with liftOver55.
Finally, we compared in greater detail the calling performance using the GRCh38-based and CHM13-based pangenomes by stratifying the evaluation across genomic region sets provided by GIAB (https://github.com/genome-in-a-bottle/genome-stratifications). These regions included, for example, different types of challenging regions, such as segmental duplications, simple repeats and TEs.
Alignment of long reads.
Hi-Fi reads from HG002, HG003 and HG004 were downloaded from the GIAB FTP site: ftp-trace.ncbi.nlm.nih.gov.:/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/PacBio_CCS_15kb_20kb_chemistry2/reads/m64011_190830_220126. fastq.gz /giab/ftp/data/AshkenazimTrio/HG003_NA24149_father/PacBio_CCS_15kb_20kb_chemistry2/reads/PBmixSequel729_1_A01_PBTH_30hours_19kbV2PD_70pM_HumanHG003.fastq.gz/giab/ftp/data/AshkenazimTrio/HG004_NA24143_mother/PacBio_CCS_15kb_20kb_chemistry2/uBAMs/m64017_191115_211223.hifi_reads.bam.
The reads were then aligned to the pangenomes (after being converted to FASTQ with SAMtools FASTQ in the case of HG004) using GraphAligner (version 1.0.13) with ‘-x vg’ with .gam output. We parsed the vg graph alignment map (GAM) output to extract the first record as primary alignment. By overlapping the other alignment records with the primary alignment, we identified reads with split mapping—that is, with part of the read mapped to a different location from the primary alignment. The alignment identity is reported by GraphAligner and was also extracted from the GAM.
SV genotyping with PanGenie
Variants corresponding to nested sites in the HPGRC graph-derived VCFs were decomposed as described in ref. 25 before running PanGenie version 2.1.0 with its default parameters. The HGSVC version 4.0 ‘lenient set’32 was also included but did not require decomposition. These three VCFs, annotated with all computed genotypes, are available for download at https://zenodo.org/record/7669083. The genotyped samples were chosen by randomly selecting 100 trios from the 1KG data, 20 from each superpopulation. Samples present in HPRC and HGSVC were also included, for a total of 368. High-coverage short reads from the 1KG (ref. 31) were used for genotyping. The leave-one-out experiments were performed as described in ref. 25, and, like in that work, variants were ‘collapsed’ using truvari collapse -r 500 -p 0.95-P 0.95 -s 50 -S 100000 from Truvari56 version 3.5.0 when comparing counts of genotyped variants (Fig. 3b–d). This is because near-identical insertions in the graph become completely separate variants in the VCF when, for the purposes of this comparison, we want to treat them the same. SV deletions (insertions) were sites with reference alleles of length ≥50 (1) and alternative alleles of length 1 (≥50). Sites that did not meet these criteria but had a reference or alternative allele of length ≥50 were classified as ‘SV Other’.
D. melanogaster graph construction
The D. melanogaster pangenome was created using Minigraph-Cactus using the procedure described in the ‘Minigraph-Cactus pangenome pipeline’ subsection. Progressive Cactus was run on the same input (which implies a star phylogeny) and was exported to vg with hal2vg.
D. melanogaster variant decomposition
The variant sites in the pangenome (snarls, also known as bubbles) were decomposed into canonical SVs using a script developed for the HPRC analysis25. In brief, each allele in the deconstructed VCF specifies the corresponding path in the pangenome. The script follows these paths and, comparing them with the dm6 reference path, enumerates each canonical variant (SNPs, indels and SVs). The frequency of each variant in the pangenome corresponds to the number of assemblies that traverse their paths.
D. melanogaster graph mapping and variant calling
The DGPR samples used are listed in Supplementary Table 6. Short reads were obtained using fasterq-dump –split 3 on the accessions in the last column of this table. Each read pair was mapped to the allele frequency-filtered graph with vg giraffe and to dm6 with BWA-MEM.
vg call was used to genotype variants in the pangenome. For each sample, these variant calls were decomposed into canonical SVs using the same approach described above on the HPRC deconstructed VCF. The SV calls were then compared to the SVs in the pangenome using the sveval package5, which matches SVs based on their types, sizes and location. Because SVs are genotyped using the same pangenome, they are expected to be relatively similar, and we can use standard ‘collapse’ criteria to cluster them in SV sites. Two SVs were matched if their regions had a reciprocal overlap of at least 90% for deletions and inversions; they were located at fewer than 100 bp from each other; and their inserted sequences were at least 90% similar for insertions. The same approach was used to cluster the SV alleles into the SV sites reported in the text and figures. The SV alleles were annotated with RepeatMasker (version 4.0.9)57. We assigned a repeat class to an SV if more than 80% of the allelic sequence was annotated as such. The 80% threshold was chosen by inspecting the distribution and observing a negligible number of events below this value.
We used vg surject to produce BAM files referenced on dm6 from the mappings to the pangenome and FreeBayes version 1.3.6 (ref. 35) (in the absence of a high-quality DeepVariant model) to call variants on these mappings and those from BWA-MEM. Single-sample VCFs were merged with bcftools merge.
To compare the variant calls by both approaches, we used bcftools58 (version 1.10.2) to normalize the VCFs (bcftools norm) and compare them (bcftools isec) to mark variant sites where both approaches call a variant and sites where only one approach does. We compared the number of calls in each category, across samples, and for different minimum variant quality thresholds (quality (QUAL) field or genotype quality (GQ) field).
Reporting Summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank A. D. Long for many suggestions and insights regarding the D. melanogaster data and the whole vg team for their work to create and maintain vg, upon which much of this work depends. B.P., A.N., J.M.E. and J.M. were partly supported by National Institutes of Health (NIH) grants R01HG010485, U24HG010262, U24HG011853, OT3HL142481, U01HG010961 (with H.L.) and OT2OD033761. H.L. was partly supported by NIH grant R01HG010040 and T.M. by U01HG010973. Computational infrastructure and support for running PanGenie were provided by the Centre for Information and Media Technology at Heinrich Heine University Düsseldorf.
Footnotes
Code availability
All source code for the Minigraph-Cactus pangenome pipeline, as well as release binaries, Docker images and user manuals, can be found at https://github.com/ComparativeGenomicsToolkit/cactus.
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-023-01793-w.
Peer review information Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
All data, software versions and commands are available at https://github.com/ComparativeGenomicsToolkit/cactus/tree/master/doc/mc-paper. HPRC graphs can be downloaded from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=pangenomes/freeze/freeze1/minigraph-cactus/. Consult the Data Portal for explanations of the different files: https://github.com/human-pangenomics/hpp_pangenome_resources/. Variant calls can be downloaded from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=publications/mc_2022/hprc-human/. SV genotyping results are available at https://doi.org/10.5281/zenodo.7669083. D. melanogaster graphs can be downloaded from https://s3-us-west2.amazonaws.com/human-pangenomics/index.html?prefix=publications/mc_2022/mc_pangenomes/16-fruitfly-mc-2022-05-26/. Consult the Data Portal for explanations of the different files: https://github.com/ComparativeGenomicsToolkit/cactus/tree/master/doc/mc-pangenomes. D. melanogaster mapping and calling results can be downloaded from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=publications/mc_2022/fruitfly/.
References
- 1.Eizenga JM et al. Pangenome graphs. Annu. Rev. Genomics Hum. Genet. 21, 139–162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miga KH & Wang T. The need for a human pangenome reference sequence. Annu. Rev. Genomics Hum. Genet. 22, 81–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrison E. et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat. Biotechnol. 36, 875–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abel HJ et al. Mapping and characterization of structural variation in 17,795 human genomes. Nature 583, 83–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickey G. et al. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol. 21, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirén J. et al. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science 374, abg8871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paten B. et al. Superbubbles, ultrabubbles, and cacti. J. Comput. Biol. 25, 649–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rautiainen M. et al. Telomere-to-telomere assembly of diploid chromosomes with Verkko. Nat. Biotechnol. 10.1038/s41587-022-01435-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Just W. Computational complexity of multiple sequence alignment with SP-score. J. Comput. Biol. 8, 615–623 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Kille B, Balaji A, Sedlazeck FJ, Nute M. & Treangen TJ Multiple genome alignment in the telomere-to-telomere assembly era. Genome Biol. 23, 182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchette M. et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris RS Improved Pairwise Alignment of Genomic DNA. PhD thesis, Pennsylvania State Univ. (2007). [Google Scholar]
- 13.Armstrong J. et al. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587, 246–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goenka SD, Turakhia Y, Paten B. & Horowitz M. SegAlign: a scalable GPU-based whole genome aligner. In SC20: International Conference for High Performance Computing, Networking, Storage and Analysis. 10.1109/sc41405.2020.00043 (IEEE, 2020). [DOI] [Google Scholar]
- 15.Paten B. et al. Cactus graphs for genome comparisons. J. Comput. Biol. 18, 461–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Feng X. & Chu C. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 21, 265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C, Grasso C. & Sharlow MF Multiple sequence alignment using partial order graphs. Bioinformatics 18, 452–464 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivian J. et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314–316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paten B. et al. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 21, 1512–1528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickey G, Paten B, Earl D, Zerbino D. & Haussler D. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29, 1341–1342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiddes IT et al. Comparative Annotation Toolkit (CAT)-simultaneous clade and personal genome annotation. Genome Res. 28, 1029–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doerr D. GFAffix. https://github.com/marschall-lab/GFAffix(2022). [Google Scholar]
- 24.Bzikadze AV & Pevzner PA TandemAligner: a new parameter-free framework for fast sequence alignment. Preprint at bioRxiv 10.1101/2022.09.15.507041 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Liao W-W et al. A draft human pangenome reference. Nature 10.1038/s41586-023-05896-x (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nurk S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rautiainen M. & Marschall T. GraphAligner: rapid and versatile sequence-to-graph alignment. Genome Biol. 21, 253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poplin R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Wagner J. et al. Curated variation benchmarks for challenging medically relevant autosomal genes. Nat. Biotechnol. 40, 672–680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebler J. et al. Pangenome-based genome inference allows efficient and accurate genotyping across a wide spectrum of variant classes. Nat. Genet. 54, 518–525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortiumet al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert P. et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372, eabf7117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty M, Emerson JJ, Macdonald SJ & Long AD Structural variants exhibit widespread allelic heterogeneity and shape variation in complex traits. Nat. Commun. 10, 4872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W. et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24, 1193–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrison E. & Marth G. Haplotype-based variant detection from short-read sequencing. Preprint at arXiv 10.48550/arXiv.1207.3907 (2012). [DOI] [Google Scholar]
- 36.Miller DE et al. Identification and characterization of breakpoints and mutations on Drosophila melanogaster balancer chromosomes. G3 (Bethesda) 10, 4271–4285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman RM et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 51, 30–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Human Pangenome Reference Consortium. HPRC Pangenome Resources. https://github.com/human-pangenomics/hpp_pangenome_resources (2022).
- 39.Guarracino A. et al. Recombination between heterologous human acrocentric chromosomes. Nature 10.1038/s41586-023-05976-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y. et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 606, 527–534 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonard AS et al. Structural variant-based pangenome construction has low sensitivity to variability of haplotype-resolved bovine assemblies. Nat. Commun. 13, 3012 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. Identifying centromeric satellites with dna-brnn. Bioinformatics 35, 4408–4410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Numanagic I. et al. Fast characterization of segmental duplications in genome assemblies. Bioinformatics 34, i706–i714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y. et al. abPOA: an SIMD-based C library for fast partial order alignment using adaptive band. Bioinformatics 37, 2209–2211 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Earl D. et al. Alignathon: a competitive assessment of whole-genome alignment methods. Genome Res. 24, 2077–2089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrison E. & Guarracino A. Unbiased pangenome graphs. Bioinformatics 39, btac743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eizenga JM et al. Efficient dynamic variation graphs. Bioinformatics 36, 5139–5144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sirén J, Garrison E, Novak AM, Paten B. & Durbin R. Haplotype-aware graph indexes. Bioinformatics 36, 400–407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mose LE, Wilkerson MD, Hayes DN, Perou CM & Parker JS ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics 30, 2813–2815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zook JM et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data 3, 160025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krusche P. et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol. 37, 555–560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cleary JG et al. Comparing variant call files for performance benchmarking of next-generation sequencing variant calling pipelines. Preprint at bioRxiv 10.1101/023754 (2015). [DOI] [Google Scholar]
- 53.Li H. et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat. Methods 15, 595–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.broadinstitute/picard. https://github.com/broadinstitute/picard
- 55.Kuhn RM, Haussler D. & Kent WJ The UCSC Genome Browser and associated tools. Brief. Bioinform. 14, 144–161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.English AC, Menon VK, Gibbs RA, Metcalf GA & Sedlazeck FJ Truvari: refined structural variant comparison preserves allelic diversity. Genome Biol. 23, 271 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smit AFA, Hubley R. & Green P. RepeatMasker Open-4.0. http://www.repeatmasker.org (2013–2015).
- 58.Danecek P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, software versions and commands are available at https://github.com/ComparativeGenomicsToolkit/cactus/tree/master/doc/mc-paper. HPRC graphs can be downloaded from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=pangenomes/freeze/freeze1/minigraph-cactus/. Consult the Data Portal for explanations of the different files: https://github.com/human-pangenomics/hpp_pangenome_resources/. Variant calls can be downloaded from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=publications/mc_2022/hprc-human/. SV genotyping results are available at https://doi.org/10.5281/zenodo.7669083. D. melanogaster graphs can be downloaded from https://s3-us-west2.amazonaws.com/human-pangenomics/index.html?prefix=publications/mc_2022/mc_pangenomes/16-fruitfly-mc-2022-05-26/. Consult the Data Portal for explanations of the different files: https://github.com/ComparativeGenomicsToolkit/cactus/tree/master/doc/mc-pangenomes. D. melanogaster mapping and calling results can be downloaded from https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=publications/mc_2022/fruitfly/.