Abstract
Detailed knowledge of the genetic variations in diverse crop populations forms the basis for genetic crop improvement and gene functional studies. In the present study, we analyzed a large rice population with a total of 10 548 accessions to construct a rice super-population variation map (RSPVM), consisting of 54 378 986 single nucleotide polymorphisms, 11 119 947 insertion/deletion mutations and 184 736 presence/absence variations. Assessment of variation detection efficiency for different population sizes revealed a sharp increase of all types of variation as the population size increased and a gradual saturation of that after the population size reached 10 000. Variant frequency analysis indicated that ∼90% of the obtained variants were rare, and would therefore likely be difficult to detect in a relatively small population. Among the rare variants, only 2.7% were predicted to be deleterious. Population structure, genetic diversity and gene functional polymorphism of this large population were evaluated based on different subsets of RSPVM, demonstrating the great potential of RSPVM for use in downstream applications. Our study provides both a rich genetic basis for understanding natural rice variations and a powerful tool for exploiting great potential of rare variants in future rice research, including population genetics and functional genomics.
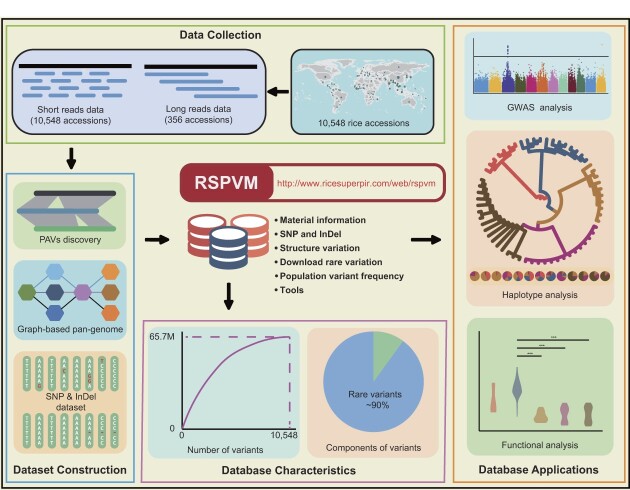
Graphical Abstract
Graphical Abstract.
Introduction
Rice is one of the most important food crops in the world, feeding more than half of the global population (1). Natural genomic variation is an important resource for genetic improvement and modern breeding methods to form new high-yield, high-quality rice varieties. Genomic variation has therefore long been a subject of intensive research. Common genomic variation types include single nucleotide polymorphisms (SNPs), insertion/deletion mutations (InDels) and large structural variations (SVs), all of which contribute extensively to gene functions and phenotypic traits in rice. For example, variations in the coding sequence of sd1 that alter the amino acid sequence are known to reduce rice plant height to varying degrees (2,3); a 1212-bp deletion can increase rice grain width and weight by regulating GW5 expression (4,5). Climatic and environmental changes throughout the world give great significance to explorations of natural variations in rice that could be exploited to enhance its ecological adaptability and improve quality and yield.
Use of larger-scale populations can allow a more comprehensive molecular characterization of genomic variations, especially rare variants, than smaller populations. For example, a super-large dataset of 64 000 human exomes demonstrated that human height and weight are largely affected by rare variants (6), which would likely be difficult to detect in a small population. Rare variants in rice have not yet been effectively used. It is therefore important to characterize genetic variations among a large-scale rice population including multiple subpopulations. Several previous rice variation datasets have been generated from thousands of accessions (e.g. 3010, 4726 or 5152 accessions) (7–9). However, those studies primarily focused on simple sequence variants such as SNPs (8); few similar studies have included large-scale SV detection. Although SVs are typically identified from long sequencing reads, current tools support SV identification from large-scale datasets composed of short sequencing reads, which are significantly less expensive to generate. At present, researchers have been generating and accumulating genomic sequencing data for nearly two decades, and these data could now be combined to form a broad rice genomic variation database with a super-large sample size.
In this study, we curated a dataset of both short and long genomic sequencing reads derived from a total of 10 548 cultivated and wild rice accessions; using these data, we constructed the first 10 000-level database of rice variation map (RSPVM) with a sample size of 10 548. The database contained a total of 54 378 986 SNPs, 11 119 947 InDels and 184 736 presence/absence variations (PAVs); 84% of the SNPs and 92% of the InDels both were rare variants, which would be difficult to detect in small-scale populations. Through evaluation of this database, we further demonstrated the great potential of this large variation dataset for studying population structure, genetic diversity, allele distribution and functional diversity in plants.
Materials and methods
Material collection and identification of variation dataset
We collected relevant resequencing data from public database, including NCBI, GSA and ENA (Supplementary Table S1). Quality control of short sequencing reads were conducted by using Trimmomatic (10) (v.0.39 parameter: MINLEN: 75 LEADING: 20 TRAILING: 20 SLIDINGWINDOW: 5:20; MINLEN = 40, while the read length is <75 bp). The reads were mapped to Nipponbare genome (MSU v.7.0) (11) with BWA software (12) (v.0.7.17-r1188) and then were used for SNP calling in Sentieon software (13) (v.sentieon-genomics-202112.02). Genetic variant annotation and functional effect prediction were conducted by using SnpEff (14) (v.4.3t). The long reads of Pacbio and Nanopore from 356 rice accessions were collected (Supplementary Table S2), mapped to the Nipponbare genome (MSU v.7.0) (11) with minimap2 (16) and NGMLR (17) and were further used for SV calling using Sniffles (17) (v.1.0.11, parameters: -l 50 -genotype) and cuteSV (18) (v.1.0.13, parameters: –max_cluster_bias_INS 100 –diff_ratio_merging_INS 0.3 –max_cluster_bias_DEL 200 –diff_ratio_merging_DEL 0.5 -l 50 -L 1000000 –genotype -S for PacBio reads and –max_cluster_bias_INS 100 –diff_ratio_merging_INS 0.3 –max_cluster_bias_DEL 100 –diff_ratio_merging_DEL 0.3 -l 50 -L 1000000 –genotype -S for Nanopore reads). Raw SV results from the two softwares were combined for each accession and further merged to call SVs for the entire population in SURVIVOR software (19) (v.1.0.7, parameters: 1000 1 1 -1 -1 50). The SVs with lengths from 50 bp to 1 Mb were filtered for constructing the graph-based pan-genome by using the vg software (20) (v.1.36.0). PAVs calling were conducted with short sequencing reads and the pan-genome by using vg giraffe (21) and SURVIVOR (v.1.0.7, parameters: 1000 1 1 -1 -1 50). PAVs with low-quality or unexpected length (>1 Mb or <50 bp) were removed. From 1000–10 000 samples, each increase of 1000 samples was a gradient with 50 replicates per gradient set to detect saturation of SNP, InDel and PAV.
Detection of rare and deleterious variants
Allele frequencies for both SNPs and InDels were calculated with VCFtools (15) (v.0.1.16). These variants with MAF <0.01 were defined as rare variants. PCR experiments were conducted to verify seven and eight selected PAVs from short- and long-read datasets, respectively (Supplementary Figure S1, Supplementary Table S3). Primers were showed in supplementary information (Supplementary Table S3). The non-redundant (nr) protein sequence database was downloaded from NCBI (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/) and was used for annotating the functional effects of the genomic variations with SIFT4G (22). These variants with SIFT_SCORE <0.05 were annotated as deleterious variations.
Analysis of population structure
To construct a core variant dataset, the VCFtools (15) (v.0.1.16) was used to remove the variant site with a high missing rate (–max-missing 0.9, –maf 0.05) and the remained dataset was further filtered to remove the rice accessions with missing genotypes >20%. Genotype imputation of missing sites and phasing were performed using Beagle (23,24) (v.5.4). The results then were filtered based on linkage disequilibrium (–indep 50 5 2) in plink (25) (v.v1.90b6.26). Phylogenetic trees were constructed using FastTree (26) (v.2.1.11) with default parameters, and were visualized by iTOL (27) (v.6.3.1). Principal component analysis (PCA) was conducted with plink (25) (v.v1.90b6.26). Population structure of the rice accessions was estimated by using ADMIXTURE (28) (v.1.3.0).
Haplotype analysis of known functional genes
All mutant loci in the transcript region were used to analyze the haplotypes of functional genes. And the nonsynonymous mutant loci were used to estimate functional haplotypes (alleles) and construct the phylogenetic tree in FastTree (26) (v.2.1.11). The significance of differences between phenotypic traits of different haplotype groups was calculated by using a t-test.
Conduction of the Tools module in RSPVM
Four phenotypic datasets were collected from previous studies and denoted as 3kRice (7), BMCPB (29), SCLS-CN-Mix (30) and SCLS-NE-GJ (30), respectively. Those phenotypic data were combined with the genetic variant data in this study to conduct genome-wide association study (GWAS) analysis. The vcftools (15) (v.0.1.16) were used to filter the variants (–max-missing 0.9, –maf 0.05,–min-alleles 2 –max-alleles 2). The first five principal components and matrix of IBS kinship were calculated by using plink (25) (v.v1.90b6.26) (–pca 10) and EMMAX (31) (v.beta-07Mar2010) (emmax-kin -v -h -d 10), respectively, and further used as covariates for GWAS analysis. GWAS was performed using a mixed linear model in EMMAX (31) software (v.beta-07Mar2010). The threshold for GWAS was calculated using the Bonferroni test (0.05/SNPs). The SNPhub package (32) was used to construct the SNP and InDel, Variation map, Haplotype network, Sequence maker, Phylogenetic tree and Visualization of variant frequency sections in RSPVM. The geneHapR pacage (33) was used to construct the ANOVA (analysis of variance) of haplotypes section.
Results
Construction of a large genomic variation dataset from a 10 000-level population
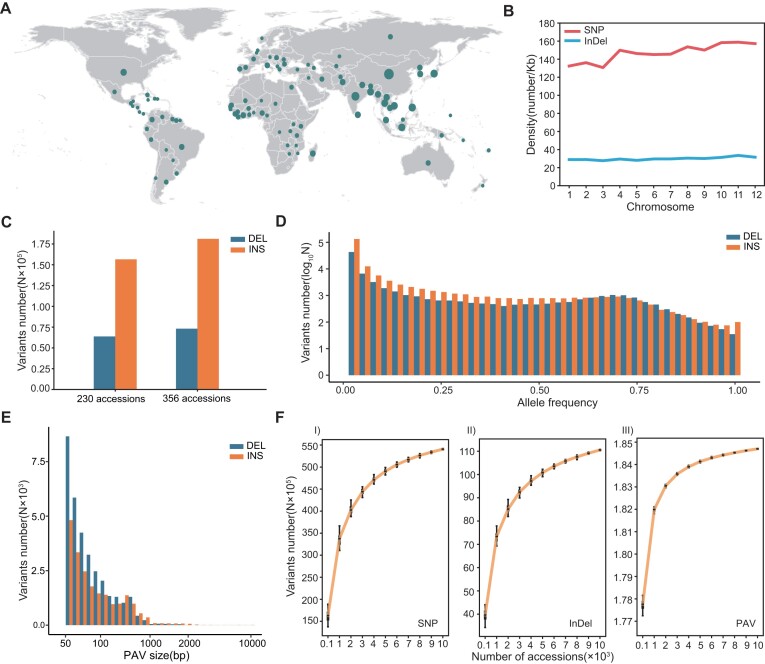
Resquencing data (7,30,34–50) were collected for a total of 10 548 accessions of Asian cultivated rice (Oryza sativa) and Asian wild rice (Oryza rufipogon) from 98 countries in four continents (Figure 1A). These data were used to generate a super-large rice genomic variation dataset. Using Nipponbare (MSU v.7.0) (11) as the reference genome, a total of 54 378 986 SNPs and 11 119 947 InDels were identified among all accessions, with average densities of 146 SNPs/Kb and 30 InDels/Kb. Chromosome 11 showed the highest variation density, with 159 SNPs/Kb and 34 InDels/Kb; chromosome 3 had the lowest variation density, with 131 SNPs/Kb and 28 InDels/Kb (Figure 1B). This indicated a potentially abnormal distribution of genetic diversity between chromosomes. To accurately identify PAVs from the resequencing dataset, a graph-based pan-genome was generated using long sequencing reads from 356 cultivated and wild rice accessions. This dataset constituted a 15.4% increase in the number of genomes compared to the 230 Asian rice accessions included in our previously reported rice pan-genome (49) (Figure 1C). A total of 315 655 SVs, including 254 051 PAVs, were detected in the 356 rice accessions; 94% of the PAVs had a relatively low frequency (<0.05) (Figure 1D). Using the pan-genome as a reference, we here performed PAV detection from short sequencing reads of a 10 000-level population for the first time. From the 10 548 accessions, we identified a total of 184 736 PAVs: 116 371 insertions and 68 365 deletions. The lengths of 58% of the PAVs ranged from 51 bp to 1 kb, and only 0.87% of the PAVs exceeded 10 kb (Figure 1E). These results indicated that shorter PAVs were more readily detected with this method. Saturation of the three types of variants was tested by randomly sampling subsets of the entire variation dataset 50 times. The numbers of SNPs, InDels and PAVs all initially showed rapid increases along with the sample size, then gradually stabilized until the sample size surpassed 10 000 (Figure 1F). The number of identified SNP increased by 61, 22 and 7% when 10 000 samples were used compared to 1000, 3000 and 6000 samples, respectively. Compared to SNPs and InDels, the number of PAVs identified reached saturation at a lower sample size (Supplementary Figure S2). These results strongly demonstrated the necessity and advantages of establishing a super-large-scale variation dataset.
Figure 1.
Identification and evaluation of the genomic variants. (A) Geographical distribution characteristics of the 10 548 accessions in this study (nmin = 1, nmax = 4620). (B) Distribution of SNP and InDel on different chromosomes. (C) Comparison of deletion (DEL) and insertion (INS) detected in 356 rice accessions in this study and a previously reported population with 230 rice accessions. This comparison presented the additional detected variations caused by a larger population. (D) Allele frequency distribution for PAV sites in 356 rice accessions. (E) Size distribution characteristics of PAV in 10 548 accessions. (F) Saturation curves of different variations in 10 548 accessions.
Rare and deleterious variants
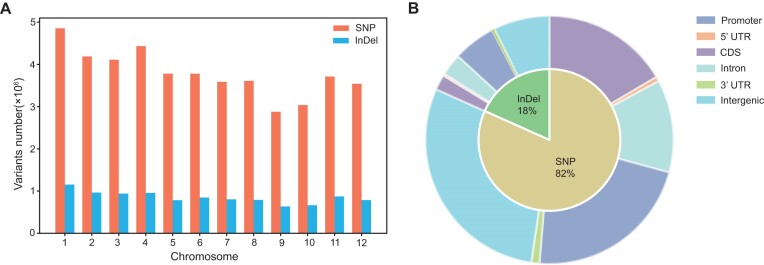
The allele frequencies of specific variants were investigated to understand the distribution patterns in a super-large population. Variations with a minor allele frequency (MAF) <0.01 were classified as rare variations, which are expected to be difficult to accurately detect in a small population. A total of 45 509 726 SNPs (84% of the total number identified) and 10 197 265 InDels (92%) were classified as rare variants, with per-chromosome averages of 3 792 477 SNPs (ranging from 2 878 047 to 4 855 139) and 849 772 InDels (ranging from 634 897 to 1 153 153) (Figure 2A). Of these, ∼19% of the rare variations were in a coding sequence, indicating that these rare variants may have genetic and phenotypic functional effects (Figure 2B). A total of 5 758 803 (12.65%) rare variants were non-synonymous mutations locating in 55 343 genes, in which 4011 genes are previously reported to be related to important traits such as plant growth and development, yield related traits and rice quality characteristics, biotic and abiotic stress response (51–54). This indicates that these rare variants could contribute largely to the genetic and phenotypic diversity in rice.
Figure 2.
Distribution characteristics of the rare variants. (A) Distribution of the rare SNP and InDel on different chromosomes. (B) Distribution characteristics of rare variants over different gene structures.
Deleterious variations during crop domestication have cumulative effects that are crucial for understanding potential crop improvement methods (55,56). Predictions of deleterious SNP sites were performed using SIFT4G (22), which revealed a total of 1 486 089 deleterious variant sites. These were unevenly distributed across genes; 3513 genes contained more than 100 deleterious SNPs each, whereas 32 881 genes had fewer than 10 deleterious SNPs each. This suggested gene-specific preferential accumulation of deleterious SNPs. We combined the deleterious variant data with the rare variant data and found that only 2.7% of the rare variants identified here were predicted to be deleterious. These results revealed the powerful advantages of using a super-large dataset for mining both rare and deleterious variants.
Analysis of population structure
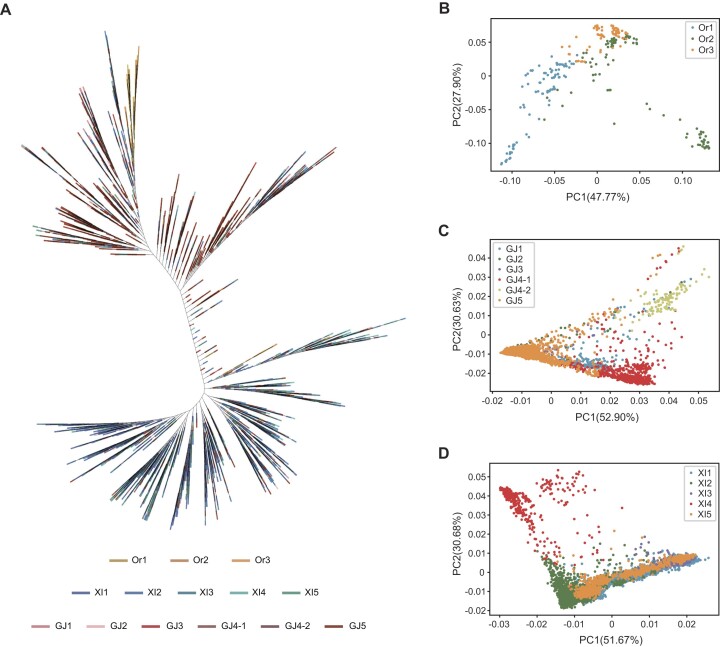
To further assess potential applications of the large variation map, we selected 9066 samples as a core collection from the entire population by filtering out samples containing only variants with high missing rate. For the population analysis, variations of the core collection were further filtered by adopting an LD-based SNP pruning procedure to produce a representative dataset consisting of 36 405 variants from the 9066 rice accessions. Based on this representative dataset, common wild rice and Asian cultivated rice accessions were classified as two distinct clusters. Common wild rice was divided into three subpopulations, Or1 (containing 246 samples), Or2 (99 samples) and Or3 (67 samples). Asian cultivated rice was divided into 10 subpopulations, namely the five indica subpopulations XI1 (1744), XI2 (1796), XI3 (282), XI4 (447) and XI5 (1026) and the six japonica subpopulations GJ1 (260), GJ2 (42), GJ3 (99), GJ4-1 (601), GJ4-2 (111) and GJ5 (2246). Most of the accessions in subpopulation XI4 were Aus rice and most of the GJ4-2 members were Basmati rice accessions. The neighbor-joining phylogenetic tree (Figure 3A) and PCA (Figure 3B–D) yielded consistent results; the subpopulations described before were clearly clustered into different clades of the phylogenetic tree and into distinct regions of the PCA map.
Figure 3.
Population analysis of the representative variant dataset containing 9066 rice accessions. (A) Phylogenetic tree of the 9066 wild and cultivated rice accessions based on the representative variant dataset. (B–D) Two-dimensional plotting of the first two principal components based on variations of wild rice (B), japonica rice (C) and indica rice (D).
Allelic genotypes and associated functional diversity
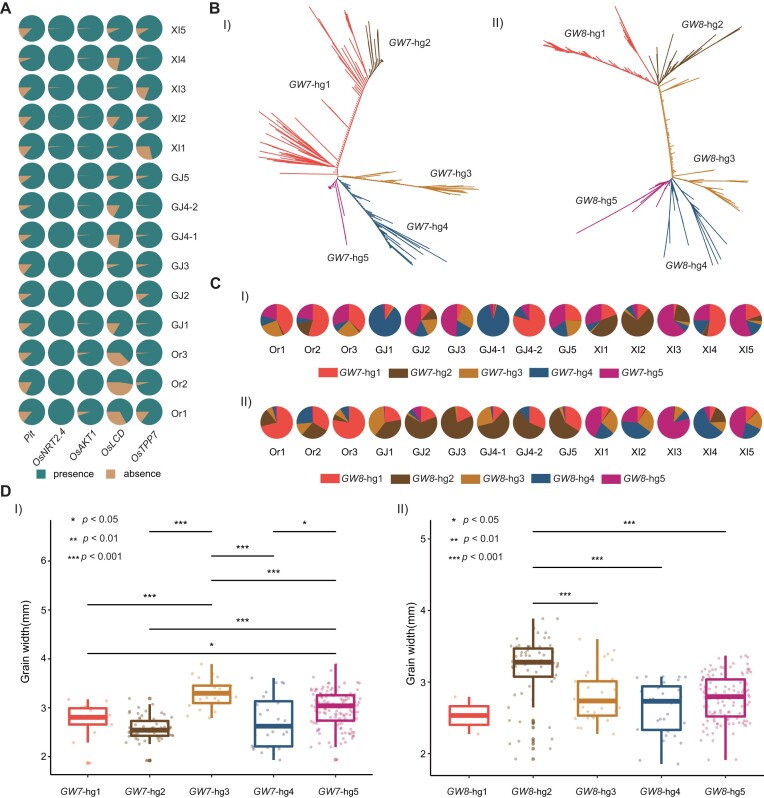
We further investigated allelic genotypes and associated functional variations in known genes caused by all variations in the core collections. A total of 8223 genes were included in the PAVs of the 9066 wild and cultivated rice accessions. Of these genes, five were selected and analyzed to determine the distinct PAV patterns between subpopulations (Figure 4A). The rice cadmium resistance gene OsLCD was present in a 34 708-bp deletion, which caused the widespread absence of this gene in numerous accessions in both the cultivated and wild rice subpopulations. However, it was retained in nearly all accessions in the XI1 and XI3 subpopulations. The rice high-affinity nitrate transporter protein gene OsNRT2.4 (57), which is an important gene related to nitrogen metabolism, was found to have a rare absence variation only in several accessions in the XI subpopulation. These PAVs of known functional genes provided a valuable basis for further use of diverse rice germplasm.
Figure 4.
Allele types of some important known genes across different subpopulations. (A) Presence and absence of five functional genes in different subpopulations due to large deletions. (B) Phylogenetic tree based on functional haplotype sequences of GW7 (I) and GW8 (II). (C) Population frequencies of GW7 (I) and GW8 (II) haplotype groups in different rice subpopulations. (D) The t-test analysis of the grain size between different haplotype groups based on 265 rice accessions.
SNPs were observed on 55 551 genes, and these SNPs generated a large number of haplotypes of functional genes. There was a total of 4522 haplotypes of GW7, a major quantitative trait locus controlling grain length and width in rice (58), in the high-quality dataset; the haplotype frequencies ranged from 1 to 700 accessions. To explore possible amino acid changes associated with these variants, nonsynonymous mutation analysis was conducted. This analysis yielded 2155 functional haplotypes that could be treated as potential alleles. The functional haplotypes were classified into five haplotype groups (GW7-hg1/2/3/4/5) based on neighbor-joining tree (Figure 4B), which showed distinct distributions between subpopulations (Figure 4C). For example, GW7-hg3 showed significantly higher grain width than other haplotype groups, and was mainly observed in members of the GJ2, GJ3 and GJ5 subpopulations. In contrast, GW7-hg2 was associated with the smallest grain width and was more concentrated in the XI1, XI2 and XI3 subpopulations (Figure 4D). In the entire variation dataset, there were 6923 haplotypes of GW8, an important gene that determines grain size, shape and quality (51,59). A total of 3730 functional haplotypes were identified for this gene, which were further classified into five haplotype groups (GW8-hg1/2/3/4/5) based on neighbor-joining tree (Figure 4B). These groups were unevenly distributed among 14 rice subpopulations (Figure 4C). Compared with other haplotype groups, GW8-hg2 was associated with higher grain width and was primarily distributed among members of XI4 and several GJ subpopulations. GW8-hg3, GW8-hg4 and GW8-hg5 were associated with relatively low grain width and were mainly observed in XI subpopulations (Figure 4D).
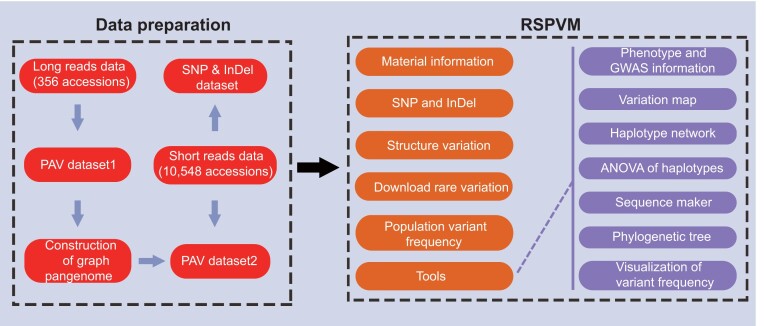
Configuration and usage of the variation database
Using the materials discussed before and the identified genomic variations, we constructed an online rice variation database, RSPVM (http://www.ricesuperpir.com/web/rspvm), which included all of the detected variations, the associated population frequencies, pan-genome sequences and metadata for all samples of the analyzed population (Figure 5). There were six sections of this database. The first was basic information for all samples used in this study, including the sample number, material name, classified population, accession number and data source. The second was a query and view service for SNPs and InDels (including rare variants) based on users’ specifications such as chromosome position, gene ID, range of MAF, etc. The third was a query and view service for SVs derived from long reads (356 accessions) and short reads (10 548 accessions). The fourth was the variation frequency for different rice populations. Two different search entries were provided to view and compare variant frequencies of different populations. The fifth was a download service for rare variations divided by chromosomes and variation types. The last contained a series of tools for analyzing variants, which could be summarized as follows: (i) phenotype and GWAS information. This dataset was generated by GWAS analysis based on phenotypic traits of 4790 accessions from previous studies (7,29,30) and the genetic variations in RSPVM. Phenotype values and frequencies, significant trait-associated loci for different traits could be obtained from this tool. (ii) Variation map, visualizing the variants according to the customized groups and accessions, chromosome regions or gene IDs, MAF, etc. (iii) Haplotype network, generating a haplotype network from variants in customized chromosome regions or genes. (iv) ANOVA of haplotypes, conducting an ANOVA analysis for a phenotypic trait between different haplotypes and visualizing them in heatmap and boxplot figures. (v) Sequence maker, generating sequences in FASTA format according to the customized accessions, and chromosome regions or gene IDs. (vi) Phylogenetic tree, generating a neighbor-joining (NJ) tree or multidimensional scaling (MDS) plot based on the user-specified variants. (vii) Visualization of variant frequency, visualizing the population frequency and functional annotation for the user-specified variants.
Figure 5.
Schematic roadmap of the RSPVM.
These applications will enable users to quickly obtain genomic variants for a gene of interest and to rapidly analyze frequency differences between subpopulations. Variations can also be analyzed in one target material compared to other accessions. These resources and functions are valuable tools for research involving population genetics, gene mining and rice breeding.
Discussion
Globally, there are abundant rice germplasm resources with very rich genetic and phenotypic diversity (60). As rice molecular genetics methods have been developed, screening of genomic variations at a population level has become essential for many research areas, e.g. population phylogenetics (61), genomics (62), pan-genomics (48–50), genetic diversity analysis, gene mining, allelic polymorphism analysis (63), and investigations into crop origins and domestication histories (45,64). However, the use of relatively small population sizes inestimably causes omission of rare mutations, resulting in the loss of a large amount of genetic information and biasing results.
To demonstrate potential applications of our dataset, we selected 52 genes with known functions (30) and analyzed the distributions and functions of their functional sites in our dataset. The proportions of functionally validated natural variants in each subpopulation were then analyzed (Supplementary Figure S3, Supplementary Table S4). Most of the results showed identical or similar distribution patterns compared to a previous report using a relatively small population (66 accessions) (44). However, in using a much larger population, we discovered many genes from different subpopulations that were previously reported (44) as absent in the corresponding subpopulations. For example, the nitrate-transporter gene NRT1.1B (65) was previously shown to be highly favorable alleles frequency only in indica rice and low favorable alleles frequency in japonica rice while in our study it also showed favorable alleles frequency in a few accessions of the japonica population. Similarly, different patterns were also found for sd1, SCM2 and other genes. These results systematically demonstrated the potential advantages of using 10 000-level data and extensive rare alleles for comprehensively understanding the functional variation of target genes.
We here used resequencing data from 10 548 rice accessions to build a comprehensive super-large variation database, RSPVM, containing more variations (e.g. 54 million SNPs) than a previously reported 3000-level database (29 million) (7), 4700-level database (14 million) (8) and 5000-level databse (18 million) (9). Providing abundance of rare variations, RSPVM is a powerful tool with great potential to enable and enhance many downstream studies. For example, a comprehensive understanding of genomic variations based on a 10 000-level population will yield better insights into genetic structure and diversity, more precise molecular fingerprints for germplasm identification, more functional variations and alleles of target genes for population genetics and functional genomics, and more informative loci and greater potential for whole-genome selection breeding compared to similar analyses using smaller populations. These potential applications reveal the broad prospective uses of our database. Some technical bottlenecks remain that prevent full use of the super-large variation dataset. For example, it remains a challenge to accurately estimate the contributions of rare variations in genome-wide association analyses, and many SVs (e.g. inversions) should be identified with long sequencing reads, which limits the possible number of input sequencing datasets.
Supplementary Material
Acknowledgements
Author contributions: L.S.: Conceptualization, Funding acquisition, Project administration, Supervision, Writing-review and editing. Q.Q.: Conceptualization, Funding acquisition, Writing-review and editing. T.W.: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing-original draft, Writing–review and editing. W.H.: Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing–original draft, Writing-review and editing. X.L.: Data curation, Formal Analysis, Methodology, Validation, Writing–original draft. C.Z.: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization. H.H.: Formal Analysis, Visualization. Q.Y.: Visualization, Validation, Formal Analysis. B.Z.: Visualization, Validation. H.Z.: Formal Analysis, Validation. Y.L.: Formal Analysis, Validation. H.W. Funding acquisition, Validation. Q.X.: Visualization. C.S.: Formal Analysis. X.L.: Formal Analysis. M.G.: Formal Analysis. X.W.: Formal Analysis. W.C.: Formal Analysis. Z.Z.: Formal Analysis. L.Y.: Formal Analysis. Y.L.: Formal Analysis. H.Q.: Formal Analysis. B.Z.: Visualization. X.Y.: Visualization. C.L.: Visualization. X.C.: Visualization. Y.C.: Visualization. Q.Z.: Visualization. X.D.: Visualization. L.G.: Conceptualization. Y.W.: Conceptualization. Y.Z.: Conceptualization. J.R.: Conceptualization.
Contributor Information
Tianyi Wang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475004, China; Shenzhen Research Institute of Henan university, Shenzhen 518000, China.
Wenchuang He, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xiaoxia Li, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Chao Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Huiying He, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Qiaoling Yuan, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Bin Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Hong Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Yue Leng, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Hua Wei, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Qiang Xu, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Chuanlin Shi, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xiangpei Liu, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Mingliang Guo, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xianmeng Wang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Wu Chen, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Zhipeng Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Longbo Yang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Yang Lv, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Hongge Qian, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Bintao Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xiaoman Yu, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Congcong Liu, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xinglan Cao, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Yan Cui, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Qianqian Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xiaofan Dai, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Longbiao Guo, State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 310006, China.
Yuexing Wang, State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 310006, China.
Yongfeng Zhou, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Jue Ruan, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Qian Qian, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 310006, China; Yazhouwan National Laboratory, No. 8 Huanjin Road, Yazhou District, Sanya City, Hainan Province 572024, China.
Lianguang Shang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; Yazhouwan National Laboratory, No. 8 Huanjin Road, Yazhou District, Sanya City, Hainan Province 572024, China.
Data availability
The long reads data and short reads data useful for this study were obtained from public databases (Supplementary Tables S1 and S2) and 126 genomic sequences were added to the blast panel of RiceSuperPIRdb (http://www.ricesuperpir.com/web/blast/blast1). All variation datasets used in this study could be found at RSPVM (available at http://www.ricesuperpir.com/web/rspvm).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
This work was supported by National Natural Science Foundation of China [32188102 to Q.Q., 32372148 to L.S.]; Guangdong Basic and Applied Basic Research Foundation [2023B1515020053 to L.S.]; Youth innovation of Chinese Academy of Agricultural Sciences [Y20230C36 to L.S.] and Shenzhen Science and Technology Program [20221008093339096 to H.W.]. Funding for open access charge: National Natural Science Foundation of China [32188102].
Conflict of interest statement. None declared.
References
- 1. Sasaki T., Burr B.. International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000; 3:138–141. [DOI] [PubMed] [Google Scholar]
- 2. Monna L., Kitazawa N., Yoshino R., Suzuki J., Masuda H., Maehara Y., Tanji M., Sato M., Nasu S., Minobe Y.. Positional cloning of rice semidwarfing gene, sd-1: rice “Green Revolution Gene” encodes a mutant enzyme Involved in gibberellin synthesis. DNA Res. 2002; 9:11–17. [DOI] [PubMed] [Google Scholar]
- 3. Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S.et al.. A mutant gibberellin-synthesis gene in rice. Nature. 2002; 416:701–702. [DOI] [PubMed] [Google Scholar]
- 4. Weng J., Gu S., Wan X., Gao H., Guo T., Su N., Lei C., Zhang X., Cheng Z., Guo X.et al.. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008; 18:1199–1209. [DOI] [PubMed] [Google Scholar]
- 5. Liu J., Chen J., Zheng X., Wu F., Lin Q., Heng Y., Tian P., Cheng Z., Yu X., Zhou K.et al.. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants. 2017; 3:17043. [DOI] [PubMed] [Google Scholar]
- 6. Akbari P., Gilani A., Sosina O., Kosmicki J.A., Khrimian L., Fang Y.Y., Persaud T., Garcia V., Sun D., Li A.et al.. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021; 373:eabf8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., Li M., Zheng T., Fuentes R.R., Zhang F.et al.. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018; 557:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao H., Li J., Yang L., Qin G., Xia C., Xu X., Su Y., Liu Y., Ming L., Chen L.L.et al.. An inferred functional impact map of genetic variants in rice. Mol. Plant. 2021; 14:1584–1599. [DOI] [PubMed] [Google Scholar]
- 9. Yan J., Zou D., Li C., Zhang Z., Song S., Wang X.. SR4R: an integrative SNP resource for genomic breeding and population research in rice. Genom. Proteom. Bioinformatics. 2020; 18:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawahara Y., Bastide M.d.l., Hamilton J.P., Kanamori H., McCombie W.R., Ouyang S., Schwartz D.C., Tanaka T., Wu J., Zhou S.et al.. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013; 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kendig K.I., Baheti S., Bockol M.A., Drucker T.M., Hart S.N., Heldenbrand J.R., Hernaez M., Hudson M.E., Kalmbach M.T., Klee E.W.et al.. Sentieon DNASeq variant calling workflow demonstrates strong computational performance and accuracy. Front. Genet. 2019; 10:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2014; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T.et al.. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C.. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018; 15:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang T., Liu Y., Jiang Y., Li J., Gao Y., Cui Z., Liu Y., Liu B., Wang Y.. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020; 21:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeffares D.C., Jolly C., Hoti M., Speed D., Shaw L., Rallis C., Balloux F., Dessimoz C., Bahler J., Sedlazeck F.J.. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017; 8:14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hickey G., Heller D., Monlong J., Sibbesen J.A., Siren J., Eizenga J., Dawson E.T., Garrison E., Novak A.M., Paten B.. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol. 2020; 21:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siren J., Monlong J., Chang X., Novak A.M., Eizenga J.M., Markello C., Sibbesen J.A., Hickey G., Chang P.-C., Carroll A.et al.. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science. 2021; 374:1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C.. SIFT missense predictions for genomes. Nat. Protoc. 2016; 11:1–9. [DOI] [PubMed] [Google Scholar]
- 23. Browning B.L., Tian X., Zhou Y., Browning S.R.. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021; 108:1880–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Browning B.L., Zhou Y., Browning S.R.. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 2018; 103:338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J.et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price M.N., Dehal P.S., Arkin A.P.. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009; 26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Letunic I., Bork P.. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021; 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alexander D.H., Novembre J., Lange K.. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009; 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhong H., Liu S., Sun T., Kong W., Deng X., Peng Z., Li Y.. Multi-locus genome-wide association studies for five yield-related traits in rice. BMC Plant Biol. 2021; 21:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., Chen Z., Zhang G., Lu H., Qin P., Qi M., Yu Y., Jiao B., Zhao X., Gao Q.et al.. Analysis of genetic architecture and favorable allele usage of agronomic traits in a large collection of Chinese rice accessions. Science China-Life Sci. 2020; 63:1688–1702. [DOI] [PubMed] [Google Scholar]
- 31. Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-y., Freimer N.B., Sabatti C., Eskin E.. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010; 42:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo W., Sun Q., Yao Y., Peng H., Xin M., Hu Z., Ni Z., Li X., Wang Z., Wang W.. SnpHub: an easy-to-set-up web server framework for exploring large-scale genomic variation data in the post-genomic era with applications in wheat. GigaScience. 2020; 9:giaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang R., Jia G., Diao X.. geneHapR: an R package for gene haplotypic statistics and visualization. BMC Bioinf. 2023; 24:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen W., Gao Y., Xie W., Gong L., Lu K., Wang W., Li Y., Liu X., Zhang H., Dong H.et al.. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014; 46:714–721. [DOI] [PubMed] [Google Scholar]
- 35. Qiu J., Zhou Y., Mao L., Ye C., Wang W., Zhang J., Yu Y., Fu F., Wang Y., Qian F.et al.. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Commun. 2017; 8:15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xia H., Luo Z., Xiong J., Ma X., Lou Q., Wei H., Qiu J., Yang H., Liu G., Fan L.et al.. Bi-directional selection in upland rice leads to its adaptive differentiation from lowland rice in drought resistance and productivity. Mol. Plant. 2019; 12:170–184. [DOI] [PubMed] [Google Scholar]
- 37. Gutaker R.M., Groen S.C., Bellis E.S., Choi J.Y., Pires I.S., Bocinsky R.K., Slayton E.R., Wilkins O., Castillo C.C., Negrao S.et al.. Genomic history and ecology of the geographic spread of rice. Nat. Plants. 2020; 6:492–502. [DOI] [PubMed] [Google Scholar]
- 38. Lv Q., Li W., Sun Z., Ouyang N., Jing X., He Q., Wu J., Zheng J., Zheng J., Tang S.et al.. Resequencing of 1,143 indica rice accessions reveals important genetic variations and different heterosis patterns. Nat. Commun. 2020; 11:4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mao D., Xin Y., Tan Y., Hu X., Bai J., Liu Z.-y., Yu Y., Li L., Peng C., Fan T.et al.. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl Acad. Sci. USA. 2019; 116:3494–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao N., Pan C., Li Y., Wu Y., Cai Y., Lu Y., Wang R., Yu L., Shi W., Kang H.et al.. Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 2021; 22:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng X., Pang H., Wang J., Yao X., Song Y., Li F., Lou D., Ge J., Zhao Z., Qiao W.et al.. Genomic signatures of domestication and adaptation during geographical expansions of rice cultivation. Plant Biotechnol. J. 2022; 20:16–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yano K., Yamamoto E., Aya K., Takeuchi H., Lo P.-c., Hu L., Yamasaki M., Yoshida S., Kitano H., Hirano K.et al.. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016; 48:927. [DOI] [PubMed] [Google Scholar]
- 43. Wang X., Wang W., Tai S., Li M., Gao Q., Hu Z., Hu W., Wu Z., Zhu X., Xie J.et al.. Selective and comparative genome architecture of Asian cultivated rice (Oryza sativa L.) attributed to domestication and modern breeding. J. Adv. Res. 2022; 42:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., Li C., Zhu C., Lu T., Zhang Z.et al.. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010; 42:961–967. [DOI] [PubMed] [Google Scholar]
- 45. Huang X., Kurata N., Wei X., Wang Z.-X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W.et al.. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012; 490:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Verma R.K., Chetia S.K., Sharma V., Baishya S., Sharma H., Modi M.K.. GWAS to spot candidate genes associated with grain quality traits in diverse rice accessions of North East India. Mol. Biol. Rep. 2022; 49:5365–5377. [DOI] [PubMed] [Google Scholar]
- 47. Higgins J., Santos B., Khanh T.D., Trung K.H., Duong T.D., Doai N.T.P., Hall A., Dyer S., Ham L.H., Caccamo M.et al.. Genomic regions and candidate genes selected during the breeding of rice in Vietnam. Evolut. Appl. 2022; 15:1141–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qin P., Lu H., Du H., Wang H., Chen W., Chen Z., He Q., Ou S., Zhang H., Li X.et al.. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell. 2021; 184:3542. [DOI] [PubMed] [Google Scholar]
- 49. Shang L., Li X., He H., Yuan Q., Song Y., Wei Z., Lin H., Hu M., Zhao F., Zhang C.et al.. A super pan-genomic landscape of rice. Cell Res. 2022; 32:878–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang F., Xue H., Dong X., Li M., Zheng X., Li Z., Xu J., Wang W., Wei C.. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes. Genome Res. 2022; 32:853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., Zeng R., Zhu H., Dong G., Qian Q.et al.. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012; 44:950–954. [DOI] [PubMed] [Google Scholar]
- 52. Kuroha T., Nagai K., Gamuyao R., Wang D.R., Furuta T., Nakamori M., Kitaoka T., Adachi K., Minami A., Mori Y.et al.. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science. 2018; 361:181–186. [DOI] [PubMed] [Google Scholar]
- 53. Wang J., Zhou L., Shi H., Chern M., Yu H., Yi H., He M., Yin J., Zhu X., Li Y.et al.. A single transcription factor promotes both yield and immunity in rice. Science. 2018; 361:1026–1028. [DOI] [PubMed] [Google Scholar]
- 54. Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., Nakashima A., Takahashi H., Yoshida H., Wong H.L.et al.. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe. 2010; 7:362–375. [DOI] [PubMed] [Google Scholar]
- 55. Morrell P.L., Buckler E.S., Ross-Ibarra J.. Crop genomics: advances and applications. Nat. Rev. Genet. 2011; 13:85–96. [DOI] [PubMed] [Google Scholar]
- 56. Liu Q., Zhou Y., Morrell P.L., Gaut B.S.. Deleterious variants in Asian rice and the potential cost of domestication. Mol. Biol. Evol. 2017; 34:908–924. [DOI] [PubMed] [Google Scholar]
- 57. Li K., Zhang S., Tang S., Zhang J., Dong H., Yang S., Qu H., Xuan W., Gu M., Xu G.. The rice transcription factor Nhd1 regulates root growth and nitrogen uptake by activating nitrogen transporters. Plant Physiol. 2022; 189:1608–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., Fang Y., Zeng L., Xu E., Xu J.et al.. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015; 47:944–948. [DOI] [PubMed] [Google Scholar]
- 59. Wang S., Li S., Liu Q., Wu K., Zhang J., Wang S., Wang Y., Chen X., Zhang Y., Gao C.et al.. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015; 47:949–954. [DOI] [PubMed] [Google Scholar]
- 60. Chen R., Deng Y., Ding Y., Guo J., Qiu J., Wang B., Wang C., Xie Y., Zhang Z., Chen J.et al.. Rice functional genomics: decades' efforts and roads ahead. Sci. China Life Sci. 2022; 65:33–92. [DOI] [PubMed] [Google Scholar]
- 61. Qiu J., Jia L., Wu D., Weng X., Chen L., Sun J., Chen M., Mao L., Jiang B., Ye C.et al.. Diverse genetic mechanisms underlie worldwide convergent rice feralization. Genome Biol. 2020; 21:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shang L., He W., Wang T., Yang Y., Xu Q., Zhao X., Yang L., Zhang H., Li X., Lv Y.et al.. A complete assembly of the rice Nipponbare reference genome. Mol. Plant. 2023; 16:1232–1236. [DOI] [PubMed] [Google Scholar]
- 63. Wang Q., Tang J., Han B., Huang X.. Advances in genome-wide association studies of complex traits in rice. Theor. Appl. Genet. 2020; 133:1415–1425. [DOI] [PubMed] [Google Scholar]
- 64. Xie W., Wang G., Yuan M., Yao W., Lyu K., Zhao H., Yang M., Li P., Zhang X., Yuan J.et al.. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. USA. 2015; 112:E5411–E5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hu B., Jiang Z., Wang W., Qiu Y., Zhang Z., Liu Y., Li A., Gao X., Liu L., Qian Y.et al.. Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants. 2019; 5:401–413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The long reads data and short reads data useful for this study were obtained from public databases (Supplementary Tables S1 and S2) and 126 genomic sequences were added to the blast panel of RiceSuperPIRdb (http://www.ricesuperpir.com/web/blast/blast1). All variation datasets used in this study could be found at RSPVM (available at http://www.ricesuperpir.com/web/rspvm).